Abstract
Nitric oxide synthases are the major sources of nitric oxide, a critical signaling molecule involved in a wide range of cellular and physiological processes. These enzymes comprise a family of genes that are highly conserved across all eukaryotes. The three family members found in mammals are important for inter- and intra-cellular signaling in tissues that include the nervous system, the vasculature, the gut, skeletal muscle, and the immune system, among others. We summarize major advances in the understanding of biochemical and tissue-specific roles of nitric oxide synthases, with a focus on how these mechanisms enable tissue adaptation and health or dysfunction and disease. We highlight the unique mechanisms and processes of neuronal nitric oxide synthase, or NOS1. This was the first of these enzymes discovered in mammals, and yet much remains to be understood about this highly conserved and complex gene. We provide examples of two areas that will likely be of increasing importance in nitric oxide biology. These include the mechanisms by which these critical enzymes promote adaptation or disease by 1) coordinating communication by diverse cell types within a tissue and 2) directing cellular differentiation/activation decisions processes.
Keywords: Nitic oxide synthase, Adaptation, Intracellular signaling, Intercellular signaling
1. Introduction
Nitric oxide (NO) is a short-lived, gaseous, free radical that was long thought of as an air pollutant [[1], [2], [3]]. In the latter part of the 20th century, many studies revealed important biological roles for NO, which is produced by a small number of physiological processes. These include pathways that were once thought to be inconsequential, such as nitrite reduction, that are emerging as important facets of nitroso group metabolism [[4], [5], [6]]. The best studied and potentially most relevant biological source of NO is the nitric oxide synthase (NOS) family. These enzymes oxidize l-arginine to produce NO. Biochemical, genetic, and phylogenetic analyses demonstrate the critical roles of NOS enzymes in a wide range of physiological contexts [4,7,8]. There are three highly conserved mammalian NOS genes: NOS1 or neuronal NOS, NOS2 or inducible NOS, and NOS3 or endothelial NOS.
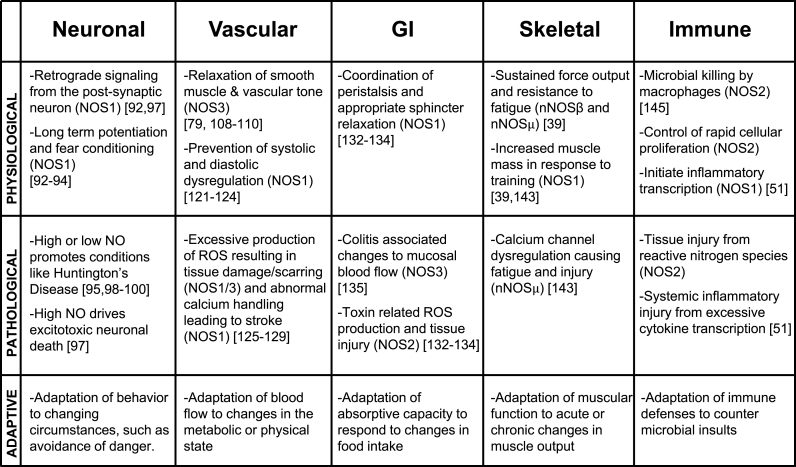
Early work on NO and NOS activity was recognized with the 1998 Nobel Prize for Physiology and Medicine, awarded to Drs. Furchgott, Ignarro, and Murad [2]. Much of this early research focused on defining the role of NO in intercellular communications such as the regulation of vascular tone (mediated by NOS3). This physiologically vital process results in relaxation of vascular smooth muscle cells through activation of soluble guanylyl cyclase and Protein Kinase G [[9], [10], [11], [12]]. Additionally, NO has been implicated in retrograde neuronal communications and regulation of synaptic plasticity (mediated by NOS1) [[13], [14], [15]], as well as in targeting pathogens with deadly NO-derived oxidants, such as peroxynitrite (mediated by NOS2) [16,17]. This review focuses on how NO signaling in these and other contexts can promote adaptive strategies in biochemical signaling, cellular behavior, and tissue function. We discuss how appropriate adaptation of these systems can improve fitness and how maladaptive changes can cause disease (see Fig. 3).
Fig. 3.
Diverse physiologic contexts and roles for Nitric Oxide Synthases in mammals. NO plays important roles in a wide range of tissues, where it exerts important regulation on physiological processes that are critical for tissue function. When NO signaling is dysregulated, these central tissue processes can become pathophysiological, causing acute or chronic disease. These vignettes illustrate a more general role for NO in adaptation to changes in the organism's need for each of the tissue-specific functions.
2. Basic biology of nitric oxide synthases
2.1. Regulation of NOS catalysis
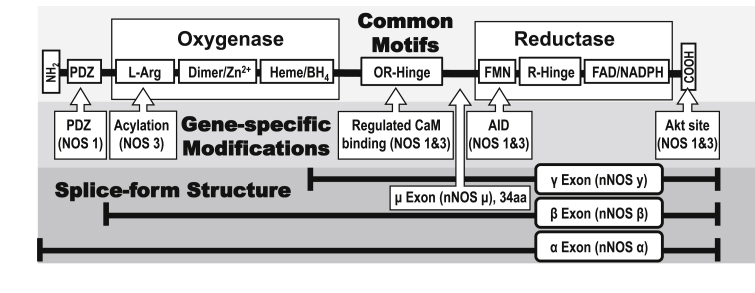
The enzymatic mechanism of NOS family members is highly conserved. All members catalyze the production of NO and l-citrulline from l-arginine and oxygen using two catalytic protein domains to promote two distinct biochemical activities. Each domain comprises a series of motifs that enable a complex series of highly regulated chemical reactions, while an additional two domains regulate this enzymatic activity (see Fig. 1). These four domains are as follows. First, the N-terminal oxygenase domain, where NO is produced, contains dimerization sites and binding sites for heme, tetrahydrobiopterin (BH4), and l-arginine. Second, the C-terminal reductase domain has binding sites for NADPH, which serves as the electron source, and the two electron carriers, flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN). A flexible hinge (R-Hinge) between the two flavin sites that regulates enzymatic activity is a site of diversity between NOS family members. The third and fourth regulatory domains also exhibit diversity among isoforms. The third regulatory domain being the oxygenase-reductase hinge (OR-Hinge), which controls the flow of electrons from the reductase to the oxygenase domain. This transfer is enabled by structural changes resulting from the binding of the calcium-dependent adaptor calmodulin (CaM). The fourth regulatory domain is the PSD/Disc-Large/ZO-1 (PDZ) domain, which changes the subcellular localization of the enzyme via protein-protein interactions, and therefore availability of substrates and access to NO targets (see Fig. 2).
Fig. 1.
Structural Diversity among NOS Enzyme Isoforms. The well-conserved protein domain structure of the NOS enzymes is depicted, both common and isoform-specific motifs are shown. Larger boxes indicate the oxygenase and reductase domains that perform the important enzymatic functions, while smaller boxes indicate the simplified positions of a variety of various functional motifs, although in some cases these are distributed in a more complex manner on the actual protein structure. Both the flexible hinge domains between the flavin binding sites in the reductase domain (R-Hinge) and the hinge between the oxidase and reductase domains (OR-Hinge) regulate the flow of electrons between domains, and thus enzymatic activity. Important differences between a number of NOS isoforms are indicated by arrow boxes. Isoform specific localization is directed by multiple motifs, including acylation (palmitoylation, myristoylation) and protein:protein interactions via the PDZ domain. The autoinhibitory (AID) and Akt phosphorylation sites confer regulatory constraint by cellular signaling pathways on NOS1 and NOS3 enzymatic activity, their absence contributes to the constitutive activity of NOS2. Several identified splice variants of NOS1 are indicated. nNOSα contains all the functional domains, while nNOSμ contains an additional 34 amino acid insertion between the flavin binding domain and the regulatory OR-Hinge. Conversely, nNOSβ lacks the PDZ domain and nNOSγ appears to lack enzymatic activity.
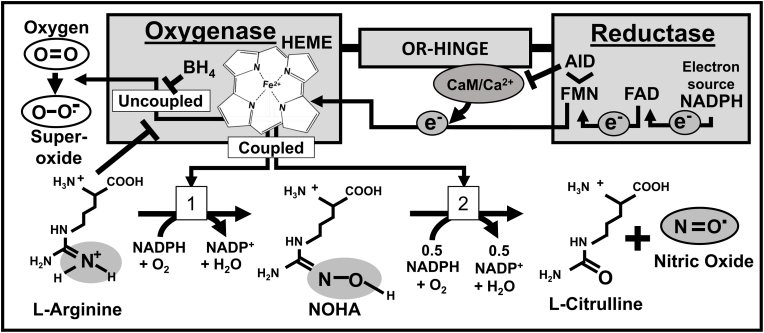
Fig. 2.
Conserved Catalytic Mechanism of NOS enzymes. The catalytic mechanism of NOS enzymes is shown superimposed on the protein modular domains where the activity occurs. The role of prosthetic groups in promoting electron flow is emphasized. Electrons are derived from NADPH, then transferred to FAD, and then FMN, before being transferred ultimately to the oxidase domain of the other protein chain in the NOS homodimer (shown here happening within a monomer for simplicity). Calcium promotes the binding of calmodulin (CaM) to the OR-Hinge, inducing a structural rearrangement that allows electron transfer. The autoinhibitory (AID) loop lies within the FMN binding domain of NOS1 and NOS3, it can dislodge CaM in the absence of Ca2+ and terminate enzymatic activity. When the reaction is “coupled”, the Heme/BH4 complex transfers electrons to molecular oxygen and promotes two distinct oxygenation reactions that convert 1)l-Arginine to N-hydroxyarginine (NOHA), and finally to 2)l-citrulline and NO. Loss of substrates or BH4 can cause “uncoupling”, or release of superoxide.
The biochemical reaction is initiated in the reductase domain, where NADPH is converted to NADP+. This liberates electrons to reduce FAD, which subsequently passes electrons to FMN. Although NOS monomers each contain oxygenase and reductase domains, they do not transfer electrons within the same protein chain. As a dimer, however, this transfer occurs efficiently when the FMN group of one monomer transfers electrons to the heme group in the oxygenase domain of the other monomer. This transfer is a rate-limiting step in the reaction, and a target of intensive regulation of the enzymatic activity of NOS family members. Binding by CaM to the linker domain promotes significant changes to the accessibility of the FMN domain, and this structural change permits electron transfer to the heme group. This was a novel role for CaM when it was discovered, and the precise molecular details of this mechanism remain an active area of study [18,19]. The reductase domain of NOS is similar to the NADPH-dependent diflavin reductase enzyme that provides electrons to the domains of cytochrome P450 enzymes (R-Hinge, see Fig. 1). [[20], [21], [22]]. This hinge domain demonstrates conserved differences between NOS family members. These variations control both the magnitude of NO production and the degree of uncoupling (or formation of superoxide, see Fig. 2) [[22], [23], [24]]. NO formation occurs in the oxygenase domain, where the heme-BH4 groups take electrons and transfer them to molecular oxygen, which then oxidizes l-arginine to N-hydroxyarginine. This intermediate is subsequently oxygenated again, converting it to l-citrulline in a process that liberates NO and completes the reaction (Fig. 2) [25,26].
This mechanism for NO production is highly sensitive to the availability of the reaction substrates, resulting in well-established regulation of the reaction by l-arginine levels [27]. Consumption of l-arginine through other metabolic pathways underlies several important control mechanisms for NO production. This substrate competition is well-documented, particularly in the immune response. For example, bacterial pathogens such as Helicobacter pylori possess gene products that starve macrophages of arginine and thereby block NOS-mediated antimicrobial activity [27,28]. A similar mechanism reinforces innate immune inflammatory behavior, or polarization, by repressing macrophage NO production through arginase-mediated arginine depletion [29,30]. Competition for substrates may also play a role in coupling NO production to metabolism via NADPH levels and to oxygen availability [28,31]. Post-translational modifications of NOS enzymes or their interactions with other proteins also modulate enzymatic activity and thereby couple NO production to major cellular signaling pathways [[32], [33], [34], [35]]. It is interesting to consider that alternative NO pathways, such as nitrite reduction to NO or transfer of NO groups between proteins, would be insensitive to these reagent limitations. This would allow certain types nitrogen radical signaling to occur in biochemical niches that did not favor NOS activity, and hence extend the signaling complexity of NO-dependent signal transduction.
2.2. Nitric oxide at the subcellular level
The regulatory roles of NO are complex, in part because NO concentrations can vary greatly in magnitude. Physiologically, concentrations of NO can vary from the low nanomolar up to the low micromolar. Lower concentrations of NO may not be sufficient to activate all signaling pathways, but instead will preferentially stimulate pathways triggered by highly reactive substrates for NO, such as heme-containing proteins [36]. This difference in biochemical reactivity enables multiple channels to transmit different information, effectively increasing the bandwidth of nitrogen radical signaling. NO signal diversity is further enriched by spatial variations in concentration. Gradients of NO were thought to be effectively flat across many cell diameters, principally because NO diffuses readily through barriers such as the cell membrane [36,37]. Although this may be true at higher concentrations, the cytoplasm of many cells contains molecules that rapidly consume NO, such as heme or superoxide from mitochondria. These factors can significantly limit the diffusion of NO and thus raise the possibility of physiologically important gradients in NO at the subcellular level [36].
A few lines of evidence support this concept. Some NOS enzymes occur as isoforms with distinct subcellular localizations [25,35,[38], [39], [40], [41], [42], [43]]. If subcellular gradients of NO are not possible, then NOS activity moving from one organelle to another should not have much effect on cellular signaling pathways. Muscle cells contain a splice variant of NOS1 that localizes close to the membrane to promote vasodilation and muscle performance. Despite the presence of other splice forms elsewhere in the cell, the loss of membrane associated NOS1 leads to ischemia and inflammation [38,39]. Even the high output enzyme, NOS2, is reported to localize to the apical surface of epithelial cells through interactions via its C-terminus [43]. Subcellular location may impose other types of regulation, such as substrate availability. Nonetheless, there is a complex landscape of subcellular peaks and valleys for the concentrations or activity of other physiologically important redox-active species, with important effects on NO signaling. Some of these molecules (e.g., superoxide) can react directly with NO, and others interact indirectly through intermediates (e.g., glutathione, thioredoxin, superoxide dismutase) [36,[44], [45], [46], [47], [48], [49], [50]]. Heterogeneous subcellular concentrations of NO are thought to transmit information about the functional state of the cell, resulting in important cellular changes. In the following sections, we discuss some of these processes, such as the plasticity of individual synapses in neurons, compartmentalized calcium signaling in muscle cells, and the transcriptional machinery in the nuclei of macrophages that controls inflammation [38,39,51]. Because of these diverse roles, NO can also integrate signals originating from diverse processes, making it an important target for therapeutic intervention.
2.3. Conservation and diversity in the NOS family
NOS genes have been identified in a wide variety of organisms, and the protein domains encoding the enzymatic machinery are highly conserved. Some bacterially expressed synthases share the same architecture as the vertebrate oxidase domain, indicating that its catalytic mechanism could accept electrons from alternate sources. Despite this, NOS proteins from plants and algae share the same essential domain architecture as the mammalian NOS genes, including both the oxygenase and diflavin reductase domains [[52], [53], [54]]. Mammals encode three separate NOS genes, and while they share the same basic architecture, they serve unique and non-redundant functions [54]. In fact, these three enzymes occur in all tetrapod genomes analyzed to date, suggesting that this arrangement was fixed around the time that the lineage that gave rise to land vertebrates split from teleost fishes [[53], [54], [55]]. NOS1, NOS2, and NOS3 (also known as nNOS, iNOS, and eNOS, respectively) exhibit mutual sequence divergence but high individual sequence conservation, despite ~400 million years of evolution [53,54,56].
All vertebrates have highly conserved NOS1 and NOS2 genes, and NOS1 is itself similar to the NOS genes in our closest invertebrate relatives. The tunicate Ciona intestinalis is an invertebrate chordate that shares much of the sequence and even much of the intron-exon boundaries of vertebrate NOS1. We review the role of NOS1 splice variants in one specialized signaling context in our discussion of skeletal muscle. These phylogenetic observations suggest that this splice form specialization has ancient origins. The inducible NOS2 gene likely arose through gene duplication and became fixed very early in vertebrate evolution. The loss of several autoinhibitory features enables high NO output, which is frequently associated with pathogen killing by mammalian innate immunity, although the enzyme itself arose at about the same time as the emergence of the acquired immune system [53]. It is perhaps unsurprising then that NOS2 also plays important roles in B and T lymphocytes [57]. The auto-inhibitory domain lost from NOS2 is important in suppressing enzymatic activity of NOS1 and NOS3 under basal conditions. This is accomplished by expelling calcium-free CaM from the enzyme, which blocks electron transfer to the oxygenase domain [58]. Although the endothelial-associated NOS3 gene is not found in fish genomes, it is highly conserved in all tetrapods. Thus, some have theorized that NOS3 played some role in adapting to breathing air, but this role remains unclear [53,54].
NOS3 contains several unique features, compared with NOS1, including the loss of the PDZ domain, the inclusion of acylation sites in the oxygenase domain that mediate membrane localization, and modification to the hinge domain within the reductase (Fig. 1). Chimeric protein studies demonstrate that replacing the reductase-hinge (R-Hinge) domain from NOS3 with that from NOS1 is sufficient to increase the NO output and coupling efficiency of the enzyme [22,23]. These conserved changes in NOS family members could thus permit different ratios of oxygen and nitrogen radicals. Nonetheless, the strong conservation of individual NOS genes suggests that these enzymes serve important, non-redundant roles [53,54].
Of the three genes, NOS1 was first to be identified. The original name, neuronal NOS (nNOS), reflects the cell type that was initially studied, though many cell types express NOS1. It is unique among the three genes because it encodes a PDZ protein:protein interaction domain on the amino terminus. This domain is important for subcellular localization, though it is not expressed in all splice variants of NOS1 [53,59]. NOS2 was originally named inducible NOS (iNOS) for its potent, stimulus-dependent transcription in immune cells, such as macrophages. NOS3, or endothelial NOS (eNOS), is an important regulator of the vasculature [16,[60], [61], [62]]. NOS1 and NOS3 are constitutively expressed, and they must be activated by cellular signaling pathways. These pathways include CaM-binding to the hinge domain in response to calcium influx, and phosphorylation by the phosphatidylinositol 3-Kinase (PI3K)/Akt signaling axis. This phosphorylation does not require calcium but instead appears to facilitate the interaction of CaM with the enzymes, thereby enhancing their catalytic activity [63,64]. The upstream regulators that promote calcium flux and PI3K activity vary widely across cell types but are frequently linked to critical functions of the cell.
NOS activity thus can couple NO production to signaling pathways that are specific to cell type. Unlike the signal-regulated family members, NOS2 is constitutively due to sequence changes that permit unregulated binding to CaM. This dysregulated binding enables high-output NO production at basal cytosolic calcium levels [25]. This can deplete reaction substrates, such as l-arginine, which can impose constraints on other l-arginine-dependent processes, for example. Accumulating NO rapidly reacts with any superoxide present to form the highly toxic radical peroxynitrite (ONOO−). Although critically important for killing invading pathogens, peroxynitrite can cause significant damage to the host tissue. Thus, the gene is tightly regulated at the level of mRNA transcription [16,59,61,62,65]. Recognition of microbial products, such as lipopolysaccharide or certain inflammatory cytokines, can promote transcription of NOS2, which involves complex transcriptional regulation that results from macrophage inflammatory polarization [[66], [67], [68]]. Interestingly, the NO from NOS2 has been shown to modify and inhibit the pro-inflammatory signaling pathways that promote its own transcription, which is probably one of the best-described examples of NO-mediated feedback signaling [51,69,70]. Genes in the NOS family can regulate themselves and each other using similarly complex feedback signaling. The high degree of evolutionary conservation suggest that these systems were fixed early in vertebrate evolution.
2.4. Signaling pathways activated by nitric oxide
NO transmits information by chemically modifying a diverse set of important cellular molecules. In fact, the addition of the NO group to protein underpins a signaling system that has important parallels with phosphorylation in terms of modifying protein function and propagating information [71]. NO modification of organic or inorganic moieties occurs through nitrosation or nitrosylation, respectively. Although frequently used interchangeably, these terms refer to distinct reactions. Nitrosation occurs indirectly via a nitrosonium ion, which reacts with the nucleophilic center of moieties, such as the thiol groups in cysteine residues. Nitrosation refers to both physiological signaling processes and injurious processes caused by excessive NO, but it does not apply to oxidative damage due to other molecules, such as NO2− or peroxynitrite (ONOO−) [71,72]. The more rapid nitrosylation reaction also involves the addition of the NO group, but to inorganic moieties, such as the iron in heme. Nitrosylation reactions are favored over nitrosation, and hence can occur at lower concentrations of NO. The NO moiety itself forms an excellent leaving group and can transfer to cysteines on a second protein, for example.
The NO group on the donor is thought to function like a nitrosonium ion in the reaction, which should therefore be referred to as transnitrosation [72]. This transfer reaction requires protein–protein interaction between the donor and recipient and depends on the redox state of both proteins. The reactivity of the cysteine thiol is determined by factors such as pH and protein structure, suggesting that transnitrosation reactions could be controlled by posttranslational modifications or even allosteric changes [[71], [72], [73], [74]]. More recent studies demonstrate that cascades of these nitroso transfer reactions can serve important signaling roles in cells [[71], [72], [73],75].
A canonical signaling role for NO modifications is the activation of soluble guanylate cyclase (sGC), which generates cyclic guanosine monophosphate (cGMP). The sensitivity of the heme group in sGC to nitrosylation by NO enables this reaction to occur at low concentrations of NO. Work with cGMP biosensors demonstrate responses to NO concentrations in the low picomolar range [76]. Further, these downstream signaling pathways can increase activity with increasing NO concentrations [[75], [76], [77], [78], [79]]. The best characterized role of cGMP is the activation of Protein Kinase G, but this is only one facet of its participation in far more complex cyclic purine signaling with cyclic adenosine monophosphate (cAMP). These second messengers direct activity of their respective ABC kinases (Protein Kinase G and Protein Kinase A), multiple ion channels, and even the phosphodiesterases which can feedback regulate the concentrations of the cyclic purines by degrading them [80].
Spatiotemporal regulation of cAMP-Protein Kinase A is highly refined, with the A Kinase Anchoring Proteins (AKAPs) shaping kinase activity at the subcellular level [81,82]. It is possible that NOS enzymes participate in similarly complex subcellular signaling cascades with cGMP, Protein Kinase G, and other adaptors. Regulation of ion channels by cGMP is critical for the well-studied role of NO in controlling smooth muscle, but cGMP-independent regulation of channels by NO also is important [80]. NOS1-derived NO nitrosates the ryanodine receptor in skeletal muscle, for example, which alleviates calcium-mediated feedback inhibition of these important channels. Loss of this circuit blocks adaptive changes of muscle to persistent loading, resulting in atrophy [39].
NOS enzymes participate in a wide array of these feedback signals, with important consequences for adaptive changes by cells and tissue [83]. NO from NOS2 inhibits NFκB signaling, and this feedback inhibits the transcription of NOS2 and other inflammatory mediators. The loss of NOS2 leads to exaggerated inflammatory responses and increased mortality in models of sepsis [51,69,84,85]. NO displays positive feedback, as well, sometimes even within the same regulatory pathway, using different concentrations of NO or different enzymes. Unlike NO from NOS2, we showed that NO from NOS1 increases proinflammatory transcription by nitrosating and inactivating SOCS1, an inhibitor of NFκB, leading to enhanced NOS2 expression and NO production [51,83]. In a model of airway sensitization, NOS3 nuclear localization regulates NOS2 activity [86,87].
NO regulates NOS activity in more complicated ways. NO changes how some cells flux calcium, which itself modifies NOS activity [41]. NO and superoxide react to form peroxynitrite, which readily oxidizes tetrahydrobiopterin. Loss of this prosthetic group inhibits NO production, allowing oxygen and nitrogen radical formation to act as potent feedback inhibition on NOS-mediated NO production [88]. NO also exerts profound effects on cellular metabolism by changing mitochondria. Cytochrome C oxidase, complex IV of the electron transport chain, accounts for ~90% of cellular oxygen consumption [89]. Nitrosylation of this molecule blocks its binding to oxygen and induces so-called metabolic hypoxia, rendering the cell incapable of consuming oxygen for oxidative phosphorylation [90]. This process is reversible, although peroxynitrite can target the same pathway irreversibly, a process that is involved in regulating cell survival [91,92]. A combination of these mechanisms participates in important adaptive signaling pathways involving intra- and inter-cellular signaling in a wide range of tissues and physiological systems.
3. Physiological contexts for NOS enzymes
3.1. Nervous system
Originally studied in neurons, the NOS enzymes are well characterized in these cells. In addition to anterograde synaptic signaling (from axon to dendrite), like most neurotransmitters, NO can mediate retrograde signals from the postsynaptic membrane back to the originating neuron (see Fig. 3). This modifies synaptic signaling and is thought to serve as an important feedback mechanism underpinning cellular processes, such as long-term potentiation, and neurological phenomena, such as fear conditioning [[93], [94], [95]]. The NO-mediated cGMP/Protein Kinase G pathway can modify other important signaling pathways, such as the MAP kinases, in this case leading to enhanced ERK-mediated transcription [94,96]. Multiple NOS isoforms, in this case NOS1 and NOS3, are involved in these complex, organ-wide responses [93].
Another recurring theme in the neuronal system is the role of NO in both physiologic and injurious processes, such as neurotoxicity [97]. NOS1 can be found post-synaptically in neurons, where it can be stimulated by calcium after stimulation by neurotransmitters like glutamate during axonal signaling. Excitotoxicity in these neurons leads to excessive production of NO, which can contribute to neuronal death [98]. Generally, low doses of NO produced from NOS1 are protective, whereas high doses contribute to disease [96,99,100]. Yet, even this is not always the case, as NO appears to serve both protective and toxic roles at both high and low levels, depending on the progression of neurological diseases, such as Huntington's and Parkinson's Diseases [101,102]. The mechanisms of NO-mediated toxicity can be linked to increased apoptosis after treatment with industrial pollutants like 2,3,7,8-Tetrachlorodibenzo-p-dioxin due to destabilization of the mitochondrial membrane and increased cytochrome P450 release [103].
Small molecule inhibitors that target NOS1 show some promise in ameliorating this mitochondrial injury [104]. This is good news, because additional mechanisms of NOS1-mediated mitochondrial injury may underlie other important neurological conditions. Nitrosation of important regulators (e.g., cdk5, Drp1, Parkin) exaggerate mitochondrial fission and decrease mitochondrial network size and cellular capacity to generate energy. This process is implicated both in ischemic injury and stroke, as well as neurodegenerative diseases such as amyotrophic lateral sclerosis, Alzheimer's, and Parkinson's [98,[105], [106], [107]]. Although specific mechanisms remain unclear, NOS1 has also been implicated in depression, where cerebrospinal fluid demonstrates decreased quantities of NO [99,108]. Neuronal NO plays important roles in both the central and peripheral nervous systems. Similar to the vascular system, neuronal NO permits nerves to communicate with and regulate other tissues, as discussed in the section covering the gastrointestinal tract.
3.2. Vascular system
Perhaps the best-known role of NO is in vasodilation, during which NO from endothelial cell expressed NOS3 relaxes neighboring smooth muscle cells and therefore vascular tone [80,109]. This mechanism was therapeutically targeted with nitroglycerin to treat chest pain long before it was understood [110,111]. The NO produced by the endothelial cells lining the blood vessel signals the smooth muscles to relax and vasodilate. This signaling partially explains low vascular resistance at rest [112,113]. NOS3−/− mice demonstrate poor vasodilation and develop hypertension, whereas their smooth muscle relaxation can be restored with chemical donors of NO even in the absence of the endothelium [[114], [115], [116], [117]]. This intercellular signal works through nitrosylation of the heme group of soluble guanylate cyclase in smooth muscle, leading to the synthesis of cGMP [77,78]. The potent second messenger promotes relaxation and vasodilation through multiple mechanisms, among which are behavioral changes of ion channels [80].
Feedback inhibition also is important in vascular tone regulation and occurs through multiple mechanisms. These include inhibition by direct nitrosation of NOS3 itself, movement of the enzyme through nitrosation of its interacting partners, and even a novel role for alpha hemoglobin in controlling NO diffusion in arterial endothelial cells [[118], [119], [120], [121]]. NOS3 is not the only family member involved in these tissues, however. NOS1 has a protective role in the heart by preventing systolic and diastolic dysregulation, oxidative stress, inflammatory tissue remodeling, and arrhythmia [[122], [123], [124], [125]]. NOS1 activity in non-endothelial cells also mediates vasodilation in response to indications of increased metabolic load, such as calcium signaling in muscle cells. This mechanism permits physiological adaptation by enhancing blood flow where it is needed, and it underpins pathological remodeling in response to high level NOS2 activity [[126], [127], [128], [129]].
Calcium-regulated NOS enzymes also have been implicated in cardiovascular disease. NOS1 has been implicated in stroke pathology resulting from abnormal calcium handling and oxidative stress [130]. An important pathological mechanism of NOS in the vasculature involves so-called uncoupling of NOS3, wherein the enzyme uncouples its oxidase and reductase activities normally via dissociation into monomers that generate superoxide radicals rather than NO. This promotes hypertension and vascular remodeling in part by disrupting angiotensin signaling [131,132]. As discussed later in this article, the vascular NO response can be used by tissues such as skeletal muscle to regulate their perfusion in response to activity.
3.3. Gastrointestinal tract
NOS enzymes play a critical role in coordinating the action of different cells and tissues throughout the body. The gastrointestinal (GI) tract is an excellent example. Approximately 50% of nerves in the GI tract contain NOS1, and NO is a vital inhibitory neurotransmitter in this organ. Thus, NOS1−/− mice demonstrate faster relaxation of the lower esophageal sphincter compared with wild types, and higher rates of gastroparesis, and partial paralysis of the stomach. Diseases with impaired release or decreased quantities of NO (e.g., achalasia, Hirschsprung's, hypertrophic pyloric stenosis) cause difficulty in relaxing portions of the gut [[133], [134], [135]]. Additionally, NOS1 in neurons of the colonic mesenteric plexus, produces NO in response to requirements for absorption, secretion, or general secretory functions [133]. NOS3 regulates mucosal blood flow [136].
NO is a critical regulator of key intestinal processes, and its dysregulation can result in disease. The physiological role and structure of the gut can make it accessible to microbial and environmental insults, including pathogens, alcohol, or pollutants in food. This can change the metabolic activity or expression of NOS enzymes, such as alcohol-induced overproduction of NOS2, which causes excess reactive oxygen species in the intestines [133,134,136]. This accessibility also makes the gut a good target for therapeutic interventions. Indeed, some current treatment strategies for GI disorders target NOS enzymes or NO directly. As in many tissues, multiple cell types (neurons, vascular, muscle, immune cells, etc.) are integrated in the digestive tract. NOS enzymes play specific roles in all of these systems, both under physiologically normal conditions as well as in disease [[80], [81], [82]]. NO serves a critical role in communicating and coordinating the action of diverse cell types to contribute to the physiological function of the organ.
3.4. Skeletal muscle
The story of NOS in skeletal muscle involves an intriguing role for splice variants in the adaptation of muscle to training. Both the NOS1 and NOS3 genes can produce alternative splice forms, and these variant protein structures can transmit distinct signals [137,138]. This is best characterized for the NOS1 gene, which sits on human chromosome 12, consists of 29 exons, and spans more than 240 kilobases. This highly conserved gene yields five different splice variants whose nomenclature relies on its alternate gene name, nNOS: nNOSα, nNOSβ, nNOSγ, nNOSμ, and nNOS2 [[139], [140], [141]]. The canonical translation start and stop sites are on exon 2 and exon 29, respectively. However, splice variant structure is determined by several factors, including alternative promoters, alternative transcription and translation start sites, exon deletions and insertions, and alternative polyadenylation signals [122,139]. Thus, nNOSα is almost full length, and nNOSμ even includes 34 additional amino acid insertions. In contrast, nNOSβ and nNOSγ lack the PDZ domain, and nNOSγ and nNOS2 demonstrate changes that modulate their catalytic efficiency [142,143].
The signaling that activates these different isoforms is thought to be similar, still requiring calcium and activity of the PI3K/Akt axis [63]. However, isoform localization can differ at the subcellular level, and these differences can change their enzymatic output or the effects of the NO produced. nNOSμ and nNOSβ are both present in skeletal muscle, and their localization differences are thought to be important for maintaining tissue integrity and functionality. Calcium signaing is integral for skeletal muscle contraction, and NOS1 enzymatic activity is triggered by these calcium transients. nNOSμ is thought to act as a sensor of muscle activity, conferring the ability to resist fatigue and adapt to persistent activation, as occurs in endurance training [39]. nNOSμ physically associates with the dystrophin glycoprotein complex (DGC), which tethers it to the sarcolemma. When the DGC fails to assemble, this membrane association is lost, as occurs in Duchenne muscular dystrophy and a murine dystrophy model. This localization is critical for the sensor function of nNOSμ, as NO from this site mediates vasodilation of nearby blood vessels and promotes blood flow and muscle performance. Blockade of this DGC-associated NOS activity leads to ischemia and inflammation, presumably resulting from poor perfusion of the metabolically active muscle tissue [39,144].
In contrast, nNOSβ is localized near the sarcolemmal Golgi, where it participates in sustaining muscle force output, possibly by regulating mitochondrial function. Both splice forms appear to play roles in the resistance of muscles to fatigue. It appears that their distinct subcellular localization allows these enzymes to perform both redundant and non-redundant roles that are essential for muscle tissue performance and adaptation. In fact, endurance training results in 60% higher NOS1 expression, whereas inactivity leads to loss of NOS1 and muscle atrophy. The loss of nNOSμ from the sarcolemma in the case of muscular dystrophy does not remove it from the cytosol, where it associates with ryanodine receptors on the sarcoplasmic reticulum. This calcium channel association enhances calcium leakage through direct nitrosation, which is thought to help shape and regulate the coupling of cellular excitation and muscular contraction [144,145]. However, dysregulation of this circuit caused by disease or overexertion leads to hypernitrosation of the channel and exaggerated calcium leakage, which causes weakness, fatigue, and ultimately damages the tissue [39]. Similarly, patients lacking nNOSμ altogether, such as those with Duchenne or Becker muscular dystrophies, have very low tolerances for exercise [39,141]. The intricate interplay of different NOS isoforms in distinct subcellular localizations maintains muscle integrity and promotes adaptation to new functional demands, and critically this work provides a model for developing a mechanistic understanding of these enzymes in other tissues.
3.5. Immune system
NO plays several roles in the immune response, but the best appreciated is certainly NOS2-dependent pathogen killing. Sustained, high-output NO enables macrophages to kill or control a wide array of pathogens [146]. Despite some debate about the precise role of NOS2 for human microbicidal activity, it is clear that this enzyme plays important roles in several immune cell types [147]. The most studied is the role in macrophages; however, NOS2 also regulates T cells, B cells, and myeloid derived suppressor cells [[146], [147], [148], [149]]. Yet, three generally accepted models for NOS2 in immunity involve producing sufficient NO for immune effector functions, such as peroxynitrite-mediated microbicidal activity, depletion of reaction substrates to suppress l-arginine-dependent cellular proliferation or signaling, and changing cellular signaling in an autocrine and paracrine fashion. Dysregulated innate immune responses occur when NOS2 is deleted, suggesting that these regulatory roles are critical for normal immune function [86,150,151].
In addition to the central role of NOS2, the other NOS enzymes have important roles in the immune system [57]. NOS1 has been implicated in a handful of reports. Given the neuronal expression of this isoform, it is not surprising that it was found to be involved in viral encephalitis. Its functions outside of the CNS, such as in dendritic cell activation and allergen-induced asthma, also have been reported [[152], [153], [154], [155], [156]]. In our own studies, we observe that NOS1 is essential for macrophage inflammatory transcription and host-tissue injury in an animal model of sepsis [51]. NOS1 is activated during stimulation of Toll-like Receptor 4, the resulting NO leads to nitrosation of Suppressor of Cytokine Signaling 1 (SOCS1) on two key cysteine residues resulting in its proteasomal degradation. In the absence of NOS1, SOCS1 protein is preserved and, instead, mediates proteasomal degradation of the p65 subunit of NFκB, leaving only p50 NFκB. This preponderance of transcriptionally active p50 leads to attenuation of pro-inflammatory cytokines, explaining the protection from tissue injury observed in models of systemic inflammatory response and sepsis [51].
NO-based feedforward signals also occur in this system. Loss of NOS1 leads to a blockage in the transcription of NOS2. Intriguingly, NOS1 is localized to the nucleus of macrophages, and SOCS1 also must be in the nucleus to degrade p65 [51,157]. Nevertheless, exogenously applied NO donors are sufficient to rescue SOCS1 degradation in the absence of NOS1, suggesting that paracrine NO signaling may play a role in regulating this inflammatory circuit. This work suggests the important role of NO in orchestrating immunity, including the earliest interactions of the host with a pathogen.
4. Conclusion
The highly conserved NOS family is central to the physiological regulation of cells and tissues in every animal in which it has been studied. The biochemistry of NO explains some of these observations. NOS enzymes demonstrate exquisite regulation in response to substrate availability, transcription, translation, and cellular and tissue signaling. As a highly diffusible and diatomic gas, NO is an ideal molecule for rapidly transmitting information. Yet, its highly reactive biochemistry enables cells to control its spread by capturing it with redox-active molecules, such as glutathione or heme. This transforms the highly mobile messenger into an NO group, whose movement is tightly regulated by nitroso transferase reactions between an array of cellular molecules.
Despite this mechanistic diversity, NO permits very different cell types to communicate and coordinates complex tissue functions, such as the vasodilatory effect or gastrointestinal peristalsis. This integrative quality also allows NO to coordinate how tissue responds to changing circumstances and enables adaptive changes, such as the muscle response to endurance training or the immune response to detection of a pathogenic insult. Dysregulation of NO signaling, however, can cause pathological processes in all of these tissues. These findings make NO an attractive target for therapeutic interventions, and some targeted treatments show promise. An improved understanding of the mechanisms of NO regulation and new approaches to targeting it with specificity will be key to realizing this potential.
Declaration of competing interest
The authors declare no conflict of interest regarding comments in this work.
Acknowledgements
The authors are grateful for support from the Dr. Nancy Laning Sobczak Fund for Breast Cancer (to BNG), the MCW Cancer Center through the Research and Education Program Fund, a component of the Advancing a Healthier Wisconsin endowment (to BNG and MGB), a grant from the MCW Research Affairs Committee (to BNG), RO1CA216882 (to MGB), RO1AI131267 (to MGB), RO1ES028149 (to MGB), DOD 71138LS (to MGB), the Wisconsin Breast Cancer Showhouse (to MGB and BNG). The authors thank Jennifer Keane for editing and critical reading of the manuscript.
Abbreviations
- AID
Autoinhibitory Domain
- AKAPs
A Kinase Anchoring Proteins
- AKT
Protein Kinase B
- BH4
Tetrahydrobiopterin
- Ca2+
Calcium
- CaM
Calmodulin
- Cdk5
Cyclin-dependent kinase 5
- cGMP
Cyclic Guanosine Monophosphate
- DGC
Dystrophin Glycoprotein Complex
- Drp1
Dynamin-1-like protein
- e−
Electron
- ERK
Extracellular signal-regulated kinases
- FAD
Flavin adenine dinucleotide
- FMN
Flavin mononucleotide
- GI
Gastro-intestinal
- L-Arg
Arginine
- LPS
Lipopolysaccharide
- MAPK
Mitogen-activated Protein Kinase
- NADP+
Nicotinamide adenine dinucleotide phosphate
- NADPH
Reduced nicotinamide adenine dinucleotide phosphate
- NFkB
nuclear factor kappa-light-chain-enhancer of activated B cells
- nNOS α, β, γ, μ, 2
Splice-variants of NOS1
- NO
Nitric Oxide
- NOHA
N-hydroxy-l-arginine
- NOS1
Nitric Oxide Synthase 1 (nNOS)
- NOS2
Nitric Oxide Synthase 2 (iNOS)
- NOS3
Nitric Oxide Synthase 3 (eNOS)
- O2
Molecular Oxygen
- ONOO
Peroxynitrite
- OR-Hinge
Flexible Hinge between the Oxygenase and Reductase domains
- PDZ
PSD/Disc-Large/ZO-1 Domain
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- R-Hinge
Flexible Hinge within the Reductase Domain
- ROS
Reactive Oxygen Species
- sGC
Soluble Guanylate Cyclase
- SOCS1
Suppressor of Cytokine Signaling 1
- TLR
Toll-Like Receptor
- Zn2+
Zinc
References
- 1.Brennan P.A., Moncada S. From pollutant gas to biological messenger: the diverse actions of nitric oxide in cancer. Ann. R. Coll. Surg. Engl. 2002;84:75–78. [PMC free article] [PubMed] [Google Scholar]
- 2.Ruth SoRelle. Nobel prize awarded to scientists for nitric oxide discoveries. Circulation. 1998;98:2365–2366. doi: 10.1161/01.CIR.98.22.2365. [DOI] [PubMed] [Google Scholar]
- 3.Giacoia Nitric oxide: an environmental pollutant as a therapeutic agent. J. Oklahoma State Med. Assoc. 1995;88(1):17–23. 1995. [PubMed] [Google Scholar]
- 4.Zweier H.L., Samouilov A., Liu X. Mechanisms of nitrite reduction to nitric oxide in the heart and vessel wall. Nitric Oxide. 2010;22(2):83–90. doi: 10.1016/j.niox.2009.12.004. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weitzberg M.H., Lundberg J.O. Nitrate-nitrite-nitric oxide PathwayImplications for anesthesiology and intensive care. Anesthesiology. 2010;113:1460–1475. doi: 10.1097/ALN.0b013e3181fcf3cc. [DOI] [PubMed] [Google Scholar]
- 6.Lundberg E.W., Gladwin M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008;7(2):156–167. doi: 10.1038/nrd2466. 2008. [DOI] [PubMed] [Google Scholar]
- 7.Jensen The role of nitrite in nitric oxide homeostasis: a comparative perspective. Biochimica et Biophysica Acta (BBA, Bioenergetics. 2009;1787:841–848. doi: 10.1016/j.bbabio.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Tiso A.N.S. Nitrate reduction to nitrite, nitric oxide and ammonia by gut bacteria under physiological conditions. PloS One. 2015;10 doi: 10.1371/journal.pone.0119712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold C.K.M., Katsuki S., Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. Unit. States Am. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph Loscalzo. The identification of nitric oxide as endothelium-derived relaxing factor. Circ. Res. 2013;113:100–103. doi: 10.1161/CIRCRESAHA.113.301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furchgott Endothelium-derived relaxing factor: discovery, early studies, and identification as nitric oxide. Biosci. Rep. 1999;19:235–251. doi: 10.1023/A:1020537506008. [DOI] [PubMed] [Google Scholar]
- 12.Rubanyi J.C.R., Vanhoutte P.M. Flow-induced release of endothelium-derived relaxing factor. Am. J. Physiol. 1986;2 doi: 10.1152/ajpheart.1986.250.6.H1145. [DOI] [PubMed] [Google Scholar]
- 13.Posada P.G.H.C. Role of nitric oxide in a fast retrograde signal during development. Dev. Brain Res. 1999;114:37–42. doi: 10.1016/S0165-3806(99)00016-4. [DOI] [PubMed] [Google Scholar]
- 14.Dawson V.L.D., Snyder S.H. A novel neuronal messenger molecule in brain: the free radical, nitric oxide. Ann. Neurol. 1992;32:297–311. doi: 10.1002/ana.410320302. [DOI] [PubMed] [Google Scholar]
- 15.Hardingham J.D., Fox K. The role of nitric oxide in pre-synaptic plasticity and homeostasis. Front. Cell. Neurosci. 2013;7 doi: 10.3389/fncel.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheschowitsch J.A.M., Sordi R., Barja-Fidalgo C., Assreuy J. Rapid NOS-1-derived nitric oxide and peroxynitrite formation act as signaling agents for inducible NOS-2 expression in vascular smooth muscle cells. Pharmacol. Res. 2015;100:73–84. doi: 10.1016/j.phrs.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Prolo M.N.Á., Ríos N., Peluffo G., Radi R., Romero N. Nitric oxide diffusion to red blood cells limits extracellular, but not intraphagosomal, peroxynitrite formation by macrophages. Free Radic. Biol. Med. 2015;87:346–355. doi: 10.1016/j.freeradbiomed.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Yokom Y.M., Lau M., Su M., Glukhova A., Osawa Y. Architecture of the nitric-oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J. Biol. Chem. 2014;289(24):16855–16865. doi: 10.1074/jbc.M114.564005. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abu-Soud D.J.S. Nitric oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc. Natl. Acad. Sci. U. S. A. 1993;90(22):10769–10772. doi: 10.1073/pnas.90.22.10769. 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyanagi C.X., Kim J.-J.P. NADPH–cytochrome P450 oxidoreductase: prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 2012;528:72–89. doi: 10.1016/j.abb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorren B.M. Nitric-oxide synthase: a cytochrome P450 family foster child. Biochimica et Biophysica Acta (BBA, General Subjects. 2007;1770:432–445. doi: 10.1016/j.bbagen.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Haque K.P., Tejero J., Aulak K.S., Fadlalla M.A., Mustovich A.T., Stuehr D.J. A connecting hinge represses the activity of endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104:9254–9259. doi: 10.1073/pnas.0700332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haque M.A.F., Aulak K.S., Ghosh A., Durra D., Stuehr D.J. Control of electron transfer and catalysis in neuronal nitric-oxide synthase (nNOS) by a hinge connecting its FMN and FAD-NADPH domains. J. Biol. Chem. 2012;287:30105–30116. doi: 10.1074/jbc.M112.339697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasquez-Vivar J., Shi Z., Jeong J.-W., Luo K., Sharma A., Thirugnanam K., Tan S. Neuronal vulnerability to fetal hypoxia-reoxygenation injury and motor deficit development relies on regional brain tetrahydrobiopterin levels. Redox Biol. 2019;29 doi: 10.1016/j.redox.2019.101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forstermann W.C.S. Nitric oxide synthases: regulation and function. Eur. Heart J. 2012;33(7):829–837. doi: 10.1093/eurheartj/ehr304. (2011) 37 –37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith E.S.U., Kulp D.W., Schief W.R., Marletta M.A. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:3577–3586. doi: 10.1073/pnas.1313331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaturvedi M.A., Lewis N.D., Algood H.M., Cover T.L., Kim P.Y. L-arginine availability regulates inducible nitric oxide synthase-dependent host defense against Helicobacter pylori. Infect. Immun. 2007;75(9):4305–4315. doi: 10.1128/IAI.00578-07. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig S.K.C., Daff S. Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J. Biol. Chem. 2002;277(37):33987–33994. doi: 10.1074/jbc.M203118200. 2002. [DOI] [PubMed] [Google Scholar]
- 29.Wijnands T.M.R.C., Hommen M.P.J., Meesters D.M., Poeze M. Arginine and citrulline and the immune response in sepsis. Nutrients. 2015;7:1426–1463. doi: 10.3390/nu7031426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rath I.M., Kropf P., Closs E.I., Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abu-Soud D.L.R., Stuehr D.J. Nitric oxide binding to the heme of neuronal nitric-oxide synthase links its activity to changes in oxygen tension. J. Biol. Chem. 1996;271:32515–32518. doi: 10.1074/jbc.271.51.32515. [DOI] [PubMed] [Google Scholar]
- 32.Corso-Diaz T.L. Krukoff, nNOS alpha and nNOS beta localization to aggresome-like inclusions is dependent on HSP90 activity. J. Neurochem. 2010;114(3):864–872. doi: 10.1111/j.1471-4159.2010.06813.x. 2010. [DOI] [PubMed] [Google Scholar]
- 33.Billecke D.I.D., Morishima Y., Murphy P.J., Dunbar A.Y., Pratt W.B. The role of hsp90 in heme-dependent activation of apo-neuronal nitric-oxide synthase. J. Biol. Chem. 2004;279(29):30252–30258. doi: 10.1074/jbc.M403864200. 2004. [DOI] [PubMed] [Google Scholar]
- 34.Oess A.I., Fulton D., Govers R., Muller-Esterl W. Subcellular targeting and trafficking of nitric oxide synthases. Biochem. J. 2006;396(3):401–409. doi: 10.1042/BJ20060321. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costas-Insua C., Merino-Gracia J., Aicart-Ramos C., Rodríguez-Crespo I. Chapter five - subcellular targeting of nitric oxide synthases mediated by their N-terminal motifs. In: Donev R., editor. Advances in Protein Chemistry and Structural Biology. Academic Press; 2018. pp. 165–195. [DOI] [PubMed] [Google Scholar]
- 36.Thomas J.L.H., Ridnour L.A., Cheng R.Y., Kesarwala A.H., Switzer C.H. Signaling and stress: the redox landscape in NOS2 biology. Free Radic. Biol. Med. 2015;87:204–225. doi: 10.1016/j.freeradbiomed.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancaster J.R. A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 38.Percival K.N.A., Huang P., Adams M.E., Froehner S.C. Golgi and sarcolemmal neuronal NOS differentially regulate contraction-induced fatigue and vasoconstriction in exercising mouse skeletal muscle. J. Clin. Invest. 2010;120(3):816–826. doi: 10.1172/JCI40736. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Percival nNOS regulation of skeletal muscle fatigue and exercise performance. Biophys Rev. 2011;3(4):209–217. doi: 10.1007/s12551-011-0060-9. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramadoss M.B.P., Magness R.R. Endothelial caveolar subcellular domain regulation of endothelial nitric oxide synthase. Clin. Exp. Pharmacol. Physiol. 2013;40(11):753–764. doi: 10.1111/1440-1681.12136. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou D.Y.Z. Neuronal nitric oxide synthase: structure, subcellular localization, regulation, and clinical implications. Nitric Oxide. 2009;20(4):223–230. doi: 10.1016/j.niox.2009.03.001. 2009. [DOI] [PubMed] [Google Scholar]
- 42.Maron T.M. Subcellular localization of oxidants and redox modulation of endothelial nitric oxide synthase. Circ. J. 2012;76:2497–2512. doi: 10.1253/circj.CJ-12-1207. [DOI] [PubMed] [Google Scholar]
- 43.Glynne P.A., Darling K.E.A., Picot J., Evans T.J. Epithelial inducible nitric-oxide synthase is an apical EBP50-binding protein that directs vectorial nitric oxide output. J. Biol. Chem. 2002;277:33132–33138. doi: 10.1074/jbc.M205764200. [DOI] [PubMed] [Google Scholar]
- 44.Beckman W.H.K. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am. J. Physiol. Cell Physiol. 1996;271:1424–1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 45.McBride V.B., Brown G.C. Superoxide dismutase and hydrogen peroxide cause rapid nitric oxide breakdown, peroxynitrite production and subsequent cell death. Biochimica et Biophysica Acta (BBA, Molecular Basis of Disease. 1999;1454:275–288. doi: 10.1016/S0925-4439(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 46.Ferret E.S., Negre O., Wollman E.E., Fradelizi D. Protective effect of thioredoxin upon NO-mediated cell injury in THP1 monocytic human cells. Biochem. J. 2000;346:759–765. doi: 10.1042/bj3460759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilgers V.K.-S., Subramani J., Chen L.C., Cuello L.G., Rusch N.J. Thioredoxin reverses age-related hypertension by chronically improving vascular redox and restoring eNOS function. Sci. Transl. Med. 2017;9:376. doi: 10.1126/scitranslmed.aaf6094. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue E.F.S., Nishikawa M., Park A.M., Kira Y., Imada I. Cross talk of nitric oxide, oxygen radicals, and superoxide dismutase regulates the energy metabolism and cell death and determines the fates of aerobic life. Antioxid Redox Signal. 2003;5(4):475–484. doi: 10.1089/152308603768295221. 2003. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Zulueta L.M.E., Mukhina G., Lebovitz R.M., Zwacka R.M., Engelhardt J.F., Oberley L.W., Dawson V.L., Dawson T.M. Manganese superoxide dismutase protects nNOS neurons from NMDA and nitric oxide-mediated neurotoxicity. J. Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drobyshevsky A., Luo K., Derrick M., Yu L., Du H., Prasad P.V., Vasquez-Vivar J., Batinic-Haberle I., Tan S. Motor deficits are triggered by reperfusion-reoxygenation injury as diagnosed by MRI and by a mechanism involving oxidants. J. Neurosci. 2012;32:5500–5509. doi: 10.1523/JNEUROSCI.5986-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baig S.V.Z., Mao M., Abreu A.L., Bakhshi F.R., Hart P.C. NOS1-derived nitric oxide promotes NF-kappaB transcriptional activity through inhibition of suppressor of cytokine signaling-1. J. Exp. Med. 2015;212(10):1725–1738. doi: 10.1084/jem.20140654. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeandroz D.W., Stuehr D.J., Lamattina L., Melkonian M., Tian Z. Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal. 2016;9(417):re2. doi: 10.1126/scisignal.aad4403. 2016. [DOI] [PubMed] [Google Scholar]
- 53.Aniello Andreakis S.D., Albalat R., Patti F.P., Garcia-Fernandez J., Procaccini G. Evolution of the nitric oxide synthase family in metazoans. Mol. Biol. Evol. 2011;28(1):163–179. doi: 10.1093/molbev/msq179. 2010. [DOI] [PubMed] [Google Scholar]
- 54.González-Domenech R.M.-C. Molecular evolution of nitric oxide synthases in metazoans. Comp, Biochem. Physiol. Part D Genomics Proteomics. 2010;5:295–301. doi: 10.1016/j.cbd.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Cioni E.A., Toni M. Nitric oxide and the neuroendocrine control of the osmotic stress response in teleosts. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20030489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamas P.A.M., Li G.K., Tempst P., Michel T. Endothelial nitric oxide synthase: molecular cloning and characterization of a distinct constitutive enzyme isoform. Proc. Natl. Acad. Sci. Unit. States Am. 1992;89:6348–6352. doi: 10.1073/pnas.89.14.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bogdan Regulation of lymphocytes by nitric oxide. Methods Mol. Biol. 2010;677:375–393. doi: 10.1007/978-1-60761-869-0_24. [DOI] [PubMed] [Google Scholar]
- 58.Daff I.S., Shimizu T. The 42-amino acid insert in the FMN domain of neuronal nitric-oxide synthase exerts control over Ca(2+)/calmodulin-dependent electron transfer. J. Biol. Chem. 1999;274:30589–30595. doi: 10.1074/jbc.274.43.30589. [DOI] [PubMed] [Google Scholar]
- 59.Mungrue D.S.B., Stewart D.J., Husain M. From molecules to mammals: what's NOS got to do with it? Acta Physiol. Scand. 2003;179 doi: 10.1046/j.1365-201X.2003.01182.x. [DOI] [PubMed] [Google Scholar]
- 60.Spratt V.T., Guillemette J.G. Calcium-deficient calmodulin binding and activation of neuronal and inducible nitric oxide synthases. Biochim. Biophys. Acta. 1774 doi: 10.1016/j.bbapap.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 61.Sorokin Nitric oxide synthase and cyclooxygenase pathways: a complex interplay in cellular signaling. 2016. https://www.ingentaconnect.com/content/ben/cmc/2016/00000023/00000024/art00002 [DOI] [PubMed]
- 62.Iqbal E.G.H., Hong C., Stokum J.A., Kurland D.B., Gerzanich V., Simard J.M. Inducible nitric oxide synthase (NOS-2) in subarachnoid hemorrhage: regulatory mechanisms and therapeutic implications. Brain Circ. 2016;2:8–19. doi: 10.4103/2394-8108.178541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao C.N., Sriram S. nNOS mediated mitochondrial injury in LPS stimulated oligodendrocytes. Mitochondrion. 2012;12(2):336–344. doi: 10.1016/j.mito.2012.01.002. 2012. [DOI] [PubMed] [Google Scholar]
- 64.Wu Regulation of endothelial nitric oxide synthase activity and gene expression. Ann. N. Y. Acad. Sci. 2002 doi: 10.1111/j.1749-6632.2002.tb04062.x. [DOI] [PubMed] [Google Scholar]
- 65.Spratt D.E., Taiakina V., Guillemette J.G. Calcium-deficient calmodulin binding and activation of neuronal and inducible nitric oxide synthases. Biochim. Biophys. Acta. 2007:1774. doi: 10.1016/j.bbapap.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 66.Salim T., Sershen C.L., May E.E. Investigating the role of TNF-α and IFN-γ activation on the dynamics of iNOS gene expression in LPS stimulated macrophages. PloS One. 2016;11 doi: 10.1371/journal.pone.0153289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tötemeyer S., Sheppard M., Lloyd A., Roper D., Dowson C., Underhill D., Murray P., Maskell D., Bryant C. IFN-gamma enhances production of nitric oxide from macrophages via a mechanism that depends on nucleotide oligomerization domain-2. J. Immunol. 2006;176:4804–4810. doi: 10.4049/jimmunol.176.8.4804. [DOI] [PubMed] [Google Scholar]
- 68.Hagen W.V.V., Heremans H., Bakker‐Woudenberg I.A.J.M. Differential nitric oxide and TNF-α production of murine Kupffer cell subfractions upon priming with IFN-γ and TNF-α. Liver. 1998;18:299–305. doi: 10.1111/j.1600-0676.1998.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 69.MacMicking C.N., Hom G., Chartrain N., Fletcher D.S., Trumbauer M. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell. 1995;81(4):641–650. doi: 10.1016/0092-8674(95)90085-3. 1995. [DOI] [PubMed] [Google Scholar]
- 70.Kelleher Z.T., Matsumoto A., Stamler J.S., Marshall H.E. NOS2 regulation of NF-kappaB by S-nitrosylation of p65. J. Biol. Chem. 2007;282:30667–30672. doi: 10.1074/jbc.M705929200. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura S.A.L. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxidants Redox Signal. 2012;18:239–249. doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinrich R.S. da S., Miranda K.M., Switzer C.H., Wink D.A., Fukuto J.M. Biological nitric oxide signalling: chemistry and terminology. Br. J. Pharmacol. 2013;169:1417–1429. doi: 10.1111/bph.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsikas A.B. S-Transnitrosation reactions of hydrogen sulfide (H2S/HS−/S2−) with S-nitrosated cysteinyl thiols in phosphate buffer of pH 7.4: results and review of the literature. Nitric Oxide. 2017;65:22–36. doi: 10.1016/j.niox.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Wu A.M.P., Fu C., Liu T., Marino S.M., Gladyshev V.N., Jain M.R., Baykal A.T., Li Q., Oka S., Sadoshima J., Beuve A., Simmons W.J., Li H. Thioredoxin 1-mediated post-translational modifications: reduction, transnitrosylation, denitrosylation, and related proteomics methodologies. Antioxidants Redox Signal. 2011;15:2565–2604. doi: 10.1089/ars.2010.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez-Ruiz I.M.A., Izquierdo-Alvarez A., Hernansanz-Agustin P., Lamas S., Serrador J.M. Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxid Redox Signal. 2013;19(11):1220–1235. doi: 10.1089/ars.2012.5066. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batchelor A.M., Bartus K., Reynell C., Constantinou S., Halvey E.J., Held K.F., Dostmann W.R., Vernon J., Garthwaite J. Exquisite sensitivity to subsecond, picomolar nitric oxide transients conferred on cells by guanylyl cyclase-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22060–22065. doi: 10.1073/pnas.1013147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hunt N.L. Heme-nitrosyls: electronic structure implications for function in biology. Acc. Chem. Res. 2015;48:2117–2125. doi: 10.1021/acs.accounts.5b00167. [DOI] [PubMed] [Google Scholar]
- 78.Korkmaz S., Loganathan S., Mikles B., Radovits T., Barnucz E., Hirschberg K., Li S., Hegedüs P., Páli S., Weymann A., Karck M., Szabó G. Nitric oxide- and heme-independent activation of soluble guanylate cyclase attenuates peroxynitrite-induced endothelial dysfunction in rat aorta. J Cardiovasc Pharmacol Ther. 2013;18:70–77. doi: 10.1177/1074248412455696. [DOI] [PubMed] [Google Scholar]
- 79.Diers A.R., Keszler A., Hogg N. Detection of S-nitrosothiols. Biochim. Biophys. Acta. 2014;1840:892–900. doi: 10.1016/j.bbagen.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carvajal A.M.G., Huidobro‐Toro J.P., Weiner C.P. Molecular mechanism of cGMP-mediated smooth muscle relaxation. J. Cell. Physiol. 2000;184:409–420. doi: 10.1002/1097-4652(200009)184:3. [DOI] [PubMed] [Google Scholar]
- 81.Carnegie B.T.B. A-kinase anchoring proteins that regulate cardiac remodeling. J. Cardiovasc. Pharmacol. 2011;58:451–458. doi: 10.1097/FJC.0b013e31821c0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carnegie C.K.M., Scott J.D. A-kinase anchoring proteins: from protein complexes to physiology and disease. IUBMB Life. 2009;61:394–406. doi: 10.1002/iub.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Connelly M.P.-C., Ameixa C., Moncada S., Hobbs A.J. Biphasic regulation of NF-kappa B activity underlies the pro- and anti-inflammatory actions of nitric oxide. J. Immunol. 2001;166:3873–3881. doi: 10.4049/jimmunol.166.6.3873. [DOI] [PubMed] [Google Scholar]
- 84.Yang V.A.P., Cornfield D.N., Milla C., Panoskaltsis-Mortari A., Blazar B.R., Haddad I.Y. Effects of oxidant stress on inflammation and survival of iNOS knockout mice after marrow transplantation. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;281:922–930. doi: 10.1152/ajplung.2001.281.4.L922. [DOI] [PubMed] [Google Scholar]
- 85.Kalns J.S., Millenbaugh N., Vivekananda J., Shealy D., Eggers J. TNF receptor 1, IL-1 receptor, and iNOS genetic knockout mice are not protected from anthrax infection. Biochem. Biophys. Res. Commun. 2002;292(1):41–44. doi: 10.1006/bbrc.2002.6626. 2002. [DOI] [PubMed] [Google Scholar]
- 86.Bratt K.W., Rabowsky M.F., Last M.S., Franzi L.M., Last J.A., Kenyon N.J. Nitric oxide synthase enzymes in the airways of mice exposed to ovalbumin: NOS2 expression is NOS3 dependent. Mediat. Inflamm. 2010:321061. doi: 10.1155/2010/321061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gobeil F., Jr., Bernier S.G., Vazquez-Tello A., Brault S., Beauchamp M.H., Quiniou C. Modulation of pro-inflammatory gene expression by nuclear lysophosphatidic acid receptor type-1. J. Biol. Chem. 2003;278 doi: 10.1074/jbc.M212481200. [DOI] [PubMed] [Google Scholar]
- 88.Sun L.J.D., Zweier J.L. Reactive oxygen and nitrogen species regulate inducible nitric oxide synthase function shifting the balance of nitric oxide and superoxide production. Arch. Biochem. Biophys. 2010;494(2):130–137. doi: 10.1016/j.abb.2009.11.019. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oleson B.J., Broniowska K.A., Yeo C.T., Flancher M., Naatz A., Hogg N., Tarakanova V.L., Corbett J.A. The role of metabolic flexibility in the regulation of the DNA damage response by nitric oxide. Mol. Cell Biol. 2019;39 doi: 10.1128/MCB.00153-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang C.-F., Diers A.R., Hogg N. Cancer cell metabolism and the modulating effects of nitric oxide. Free Radic. Biol. Med. 2015;79:324–336. doi: 10.1016/j.freeradbiomed.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu I.G.C., Moncada S. Nitric oxide: orchestrating hypoxia regulation through mitochondrial respiration and the endoplasmic reticulum stress response. Cell Res. 2005;15:63–65. doi: 10.1038/sj.cr.7290267. [DOI] [PubMed] [Google Scholar]
- 92.Brown V.B. Nitric oxide inhibition of mitochondrial respiration and its role in cell death. Free Radic. Biol. Med. 2002;33:1440–1450. doi: 10.1016/S0891-5849(02)01112-7. [DOI] [PubMed] [Google Scholar]
- 93.O'Dell T.J., Huang P.L., Dawson T.M., Dinerman J.L., Snyder S.H., Kandel E.R., Fishman M.C. Endothelial NOS and the blockade of LTP by NOS inhibitors in mice lacking neuronal NOS. Science. 1994;265:542–546. doi: 10.1126/science.7518615. [DOI] [PubMed] [Google Scholar]
- 94.Ota M.S.M., Wu M.S., Young G.J., Schafe G.E. Synaptic plasticity and NO-cGMP-PKG signaling coordinately regulate ERK-driven gene expression in the lateral amygdala and in the auditory thalamus following Pavlovian fear conditioning. Learn. Mem. 2010;17:221–235. doi: 10.1101/lm.1592510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schafe G.E., Nader K., Blair H.T., LeDoux J.E. Memory consolidation of Pavlovian fear conditioning: a cellular and molecular perspective. Trends Neurosci. 2001;24:540–546. doi: 10.1016/S0166-2236(00)01969-X. [DOI] [PubMed] [Google Scholar]
- 96.Koriyama R.Y., Homma K., Mawatari K., Nagashima M., Sugitani K., Matsukawa T., Kato S. Nitric oxide-cGMP signaling regulates axonal elongation during optic nerve regeneration in the goldfish in vitro and in vivo. J. Neurochem. 2009;110:890–901. doi: 10.1111/j.1471-4159.2009.06182.x. [DOI] [PubMed] [Google Scholar]
- 97.Calabrese C.M., Calvani M., Rizzarelli E., Butterfield D.A., Stella A.M.G. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat. Rev. Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 98.Liu J.L., Zhao F., Wang H., Qu Y., Mu D. Nitric oxide synthase in hypoxic or ischemic brain injury. Rev. Neurosci. 2015;26:105–117. doi: 10.1515/revneuro-2014-0041. [DOI] [PubMed] [Google Scholar]
- 99.Kudlow D.S.C., Carvalho A.F., McIntyre R.S. Nitric oxide and major depressive disorder: pathophysiology and treatment implications. 2016. https://www.ingentaconnect.com/content/ben/cmm/2016/00000016/00000002/art00009 [DOI] [PubMed]
- 100.Chiueh T.A., Lai A.R., Lai E., Krishna G. Neuroprotective strategies in Parkinson's disease: protection against progressive nigral damage induced by free radicals. Neurotox. Res. 2000;2(2–3):293–310. doi: 10.1007/BF03033799. 2006. [DOI] [PubMed] [Google Scholar]
- 101.Deckel Nitric oxide and nitric oxide synthase in Huntington's disease. J. Neurosci. Res. 2001;64(2):99–107. doi: 10.1002/jnr.1057. 2001. [DOI] [PubMed] [Google Scholar]
- 102.Siow T.Y., Chen C.-C.V., Wan N., Chow K.-P.N., Chang C. In vivo evidence of increased nNOS activity in acute MPTP neurotoxicity: a functional pharmacological MRI study. BioMed Res. Int. 2013;2013:964034. doi: 10.1155/2013/964034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang Z.D., Nie X., Xi H., Li A., Guo A., Wu Q., Jiang S., Zhao J., Chen G. Activation of neuronal nitric oxide synthase (nNOS) signaling pathway in 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced neurotoxicity. Environ. Toxicol. Pharmacol. 2014;38:119–130. doi: 10.1016/j.etap.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 104.Hu L.-S.G., Dang X.-B., Ren P.-Y., Zhang Y.-L. Small-molecule inhibitors at the PSD-95/nNOS interface attenuate MPP+-induced neuronal injury through Sirt3 mediated inhibition of mitochondrial dysfunction. Neurochem. Int. 2014;79:57–64. doi: 10.1016/j.neuint.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 105.Qu T.N., Holland E.A., McKercher S.R., Lipton S.A. S-nitrosylation of Cdk5, prion. 2012. [DOI] [PMC free article] [PubMed]
- 106.Levecque A.E., Clavel J., Richard F., Vidal J.-S., Amouyel P., Tzourio C., Alpérovitch A., Chartier-Harlin M.-C. Association between Parkinson's disease and polymorphisms in the nNOS and iNOS genes in a community-based case–control study. Hum. Mol. Genet. 2003;12:79–86. doi: 10.1093/hmg/ddg009. [DOI] [PubMed] [Google Scholar]
- 107.Sorarù L.V., Fedrizzi L., D'Ascenzo C., Polo A., Bernazzi B., Angelini C. Activities of mitochondrial complexes correlate with nNOS amount in muscle from ALS patients. Neuropathol. Appl. Neurobiol. 2007;33:204–211. doi: 10.1111/j.1365-2990.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- 108.Gao X.R.Q., Zhao J., Balesar R., Bao A.M., Swaab D.F. Decreased NOS1 expression in the anterior cingulate cortex in depression. Cereb Cortex. 2013;23(12):2956–2964. doi: 10.1093/cercor/bhs285. 2012. [DOI] [PubMed] [Google Scholar]
- 109.Wang A.M., Poole B., Falk S., Lucia M.S., Tayal S., Schrier R. Endothelial nitric oxide synthase-deficient mice exhibit increased susceptibility to endotoxin-induced acute renal failure. Am. J. Physiol. Ren. Physiol. 2004;287:1044–1048. doi: 10.1152/ajprenal.00136.2004. [DOI] [PubMed] [Google Scholar]
- 110.Bonini K.S., Silva S.O., Corbett J., Dore M., Petranka J. Constitutive nitric oxide synthase activation is a significant route for nitroglycerin-mediated vasodilation. Proc. Natl. Acad. Sci. U. S. A. 2008;105(25):8569–8574. doi: 10.1073/pnas.0708615105. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mao M., Varadarajan S., Fukai T., Bakhshi F.R., Chernaya O., Jr S.C.D., Minshall R.D., Bonini M.G. Nitroglycerin tolerance in caveolin-1 deficient mice. PloS One. 2014;9 doi: 10.1371/journal.pone.0104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Steudel Wolfgang, Fumito Ichinose, Huang Paul L., Hurford William E., Jones Rosemary C., Bevan John A., Fishman Mark C., Zapol Warren M. Pulmonary vasoconstriction and hypertension in mice with targeted disruption of the endothelial nitric oxide synthase (NOS 3) gene. Circ. Res. 1997;81:34–41. doi: 10.1161/01.RES.81.1.34. [DOI] [PubMed] [Google Scholar]
- 113.Lorenz L.K., Baumann G., Stangl K., Stangl V. Endothelial NO production is mandatory for epigallocatechin-3-gallate–induced vasodilation: results from eNOS knockout (eNOS: −/−, mice. J. Cardiovasc. Pharmacol. 2015;65:607–610. doi: 10.1097/FJC.0000000000000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang Z.H., Mashimo H., Bloch K.D., Moskowitz M.A., Bevan J.A. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature. 1995;377(6546):239–242. doi: 10.1038/377239a0. 1995. [DOI] [PubMed] [Google Scholar]
- 115.van de Sandt A.M., Windler R., Gödecke A., Ohlig J., Zander S., Reinartz M., Graf J., van Faassen E.E., Rassaf T., Schrader J., Kelm M., Merx M.W. Endothelial NOS (NOS3) impairs myocardial function in developing sepsis. Basic Res. Cardiol. 2013;108:330. doi: 10.1007/s00395-013-0330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zubair K.A., Bexis S., Docherty J.R. Relaxations to β-adrenoceptor subtype selective agonists in wild-type and NOS-3-KO mouse mesenteric arteries. Eur. J. Pharmacol. 2008;587:216–223. doi: 10.1016/j.ejphar.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 117.Perez-Rojas K.M.K., Beierwaltes W.H., Garvin J.L., Herrera M. Nitric oxide produced by endothelial nitric oxide synthase promotes diuresis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298:1050–1055. doi: 10.1152/ajpregu.00181.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Buga G.M., Griscavage J.M., Rogers N.E., Ignarro L.J. Negative feedback regulation of endothelial cell function by nitric oxide. Circ. Res. 1993;73:808–812. doi: 10.1161/01.RES.73.5.808. [DOI] [PubMed] [Google Scholar]
- 119.Ravi L.A.B., Levic S., Ross P.A., Black S.M. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Straub A.W.L., Billaud M., Johnstone S.R., Dwyer S.T., Lee M.Y., Bortz P.S., Best A.K., Columbus L., Gaston B., Isakson B.E. Endothelial cell expression of haemoglobin α regulates nitric oxide signalling. Nature. 2012;491:473–477. doi: 10.1038/nature11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen F.R.B., Shajahan A.N., Sharma T., Mao M., Trane A., Bernatchez P., Nieuw Amerongen G.P., Bonini M.G., Skidgel R.A., Malik A.B., Minshall R.D. Nitric oxide–dependent Src activation and resultant caveolin-1 phosphorylation promote eNOS/caveolin-1 binding and eNOS inhibition. MBoC. 2012;23:1388–1398. doi: 10.1091/mbc.e11-09-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang C.Z.J., Jang J.H., Wang Y. Molecular mechanisms of neuronal nitric oxide synthase in cardiac function and pathophysiology. J Physiol. 2014;592(15):3189–3200. doi: 10.1113/jphysiol.2013.270306. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goryacheva O.L.T., Abramochkin D.V., Budanova O.P., Belkina L.M., Smirin B.V. Patol Fiziol Eksp Ter; 2016. Effect of Adaptation to Hypoxia on Expression of NO Synthase Isoforms in Rat Myocardium. [PubMed] [Google Scholar]
- 124.Danson J.K.C., Paterson D.J. Cardiac nitric oxide: emerging role for nNOS in regulating physiological function. Pharmacol. Ther. 2005;106(1):57–74. doi: 10.1016/j.pharmthera.2004.11.003. 2005. [DOI] [PubMed] [Google Scholar]
- 125.Sears S.M.B., Ashley E.A., Lygate C.A., Rakovic S., Wallis H.L. Cardiac neuronal nitric oxide synthase isoform regulates myocardial contraction and calcium handling. Circ. Res. 2003;92(5):e52–e59. doi: 10.1161/01.RES.0000064585.95749.6D. 2003. [DOI] [PubMed] [Google Scholar]
- 126.Capettini S.F.C., Lemos V.S. Relative contribution of eNOS and nNOS to endothelium-dependent vasodilation in the mouse aorta. Eur. J. Pharmacol. 2010;643(2–3):260–266. doi: 10.1016/j.ejphar.2010.06.066. 2010. [DOI] [PubMed] [Google Scholar]
- 127.Sato T.Y., Ichioka S., Shibata M., Takeda S. Vasodilation of intramuscular arterioles under shear stress in dystrophin-deficient skeletal muscle is impaired through decreased nNOS expression. Acta Myol. 2008;27:30–36. [PMC free article] [PubMed] [Google Scholar]
- 128.Jurzik M.F., Straub R.H., Scholmerich J., Wiest R. Up-regulation of nNOS and associated increase in nitrergic vasodilation in superior mesenteric arteries in pre-hepatic portal hypertension. J. Hepatol. 2005;43(2):258–265. doi: 10.1016/j.jhep.2005.02.036. 2005. [DOI] [PubMed] [Google Scholar]
- 129.Gericke E.G., Sniatecki J.J., Steege A., Wojnowski L., Pfeiffer N. Contribution of nitric oxide synthase isoforms to cholinergic vasodilation in murine retinal arterioles. Exp. Eye Res. 2013;109:60–66. doi: 10.1016/j.exer.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 130.Huang P.L.H., Panahian N., Dalkara T., Fishman M.C., Moskowitz M.A. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- 131.Galougahi C.C.L., Gentile C., Kok C., Nunez A., Garcia A. Glutathionylation mediates angiotensin II-induced eNOS uncoupling, amplifying NADPH oxidase-dependent endothelial dysfunction. J Am Heart Assoc. 2014;3(2) doi: 10.1161/JAHA.113.000731. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gielis J.Y.L., Wingler K., Schil P.E.V., Schmidt H.H., Moens A.L. Pathogenetic role of eNOS uncoupling in cardiopulmonary disorders. Free Radic. Biol. Med. 2011;50(7):765–776. doi: 10.1016/j.freeradbiomed.2010.12.018. 2010. [DOI] [PubMed] [Google Scholar]
- 133.Dijkstra G., van Goor H., Jansen P.L.M., Moshage H. Targeting nitric oxide in the gastrointestinal tract. Curr. Opin. Invest. Drugs. 2004;5:529–536. [PubMed] [Google Scholar]
- 134.Salzman Nitric oxide in the gut. New Horiz. 1995;3(1):33–45. 1995. [PubMed] [Google Scholar]
- 135.Bagyanszki P.T., Krecsmarik M., Fekete E., Adriaensen D., Nassauw L.V. Chronic alcohol consumption induces an overproduction of NO by nNOS- and iNOS-expressing myenteric neurons in the murine small intestine. Neuro Gastroenterol. Motil. 2011;23(6):e237–e248. doi: 10.1111/j.1365-2982.2011.01707.x. 2011. [DOI] [PubMed] [Google Scholar]
- 136.Petersson O.S., Steege A., Patzak A., Hellsten A., Phillipson M. eNOS involved in colitis-induced mucosal blood flow increase. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293(6):G1281–G1287. doi: 10.1152/ajpgi.00357.2007. 2007. [DOI] [PubMed] [Google Scholar]
- 137.Eisenreich U.B., Poller W., Schultheiss H.P., Rauch U. Effects of the Cdc2-like kinase-family and DNA topoisomerase I on the alternative splicing of eNOS in TNF-alpha-stimulated human endothelial cells. Biol. Chem. 2008;389(10):1333–1338. doi: 10.1515/BC.2008.152. 2008. [DOI] [PubMed] [Google Scholar]
- 138.Lorenz B.H., Hui J., Zepp A., Baumann G., Bindereif A., Stangl V., Stangl K. Alternative splicing in intron 13 of the human eNOS gene: a potential mechanism for regulating eNOS activity. Faseb. J. 2007;21:1556–1564. doi: 10.1096/fj.06-7434com. [DOI] [PubMed] [Google Scholar]
- 139.Wang D.C.N., Marsden P.A. Neuronal NOS: gene structure, mRNA diversity, and functional relevance. CRN. 1999;13 doi: 10.1615/CritRevNeurobiol.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- 140.Hall H.A., Wang Y., Cheung A.H., Arbus A.M., Olson S.L., Lu W.C., Kau C.L., Marsden P.A. Structural organization of the human neuronal nitric oxide synthase gene. Biol. Chem. 1994;269:33082–33090. [PubMed] [Google Scholar]
- 141.Baldelli D.L.B., Tatulli G., Aquilano K., Ciriolo M.R. The role of nNOS and PGC-1alpha in skeletal muscle cells. J. Cell Sci. 2014;127(Pt 22):4813–4820. doi: 10.1242/jcs.154229. 2014. [DOI] [PubMed] [Google Scholar]
- 142.Aquilano S.B., Ciriolo M.R. Nuclear recruitment of neuronal nitric-oxide synthase by α-syntrophin is crucial for the induction of mitochondrial biogenesis. J. Biol. Chem. 2014;289:365–378. doi: 10.1074/jbc.M113.506733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Silvagno H.X., Bredt D.S. Neuronal nitric-oxide synthase-mu, an alternatively spliced isoform expressed in differentiated skeletal muscle. J. Biol. Chem. 1996;271(19):11204–11208. doi: 10.1074/jbc.271.19.11204. 1996. [DOI] [PubMed] [Google Scholar]
- 144.Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 145.Xu J.P.E., Meissner G., Stamler J.S. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279(5348):234–237. doi: 10.1126/science.279.5348.234. 1998. [DOI] [PubMed] [Google Scholar]
- 146.MacMicking J., Xie Q.W., Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 147.Nathan, Role of iNOS in human host defense. Science. 2006;312(5782):1874–1875. doi: 10.1126/science.312.5782.1874b. 2006) 5. [DOI] [PubMed] [Google Scholar]
- 148.Bronte V., Serafini P., Mazzoni A., Segal D.M., Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003;24:302–306. doi: 10.1016/s1471-4906(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 149.Bogdan Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36(3):161–178. doi: 10.1016/j.it.2015.01.003. 2015. [DOI] [PubMed] [Google Scholar]
- 150.Zolini G.K.L., Lucinda N., Silva M.A., Dias M.F., Pessoa N.L. Defense against HSV-1 in a murine model is mediated by iNOS and orchestrated by the activation of TLR2 and TLR9 in trigeminal ganglia. J. Neuroinflammation. 2014;11 doi: 10.1186/1742-2094-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zeidler P.C., Roberts C., JR., V C., F B., L A., M E. Response of alveolar macrophages from inducible nitric oxide synthase knockout or wild-type mice to an in vitro lipopolysaccharide or silica exposure. J. Toxicol. Environ. Health. 2003;66(11):995–1013. doi: 10.1080/15287390306395. 2003. [DOI] [PubMed] [Google Scholar]
- 152.Adler A.S., Graulich E., Habermeier A., Bacher N., Friebe A. Neuronal nitric oxide synthase modulates maturation of human dendritic cells. J. Immunol. 2010;184(11):6025–6034. doi: 10.4049/jimmunol.0901327. 2010. [DOI] [PubMed] [Google Scholar]
- 153.Chakrabarti C.K.C., Jiang Y., Davidge S.T. Neuronal nitric oxide synthase regulates endothelial inflammation. J. Leukoc. Biol. 2012;91(6):947–956. doi: 10.1189/jlb.1011513. 2012. [DOI] [PubMed] [Google Scholar]
- 154.Huang F.W.H., Fay J.D., Hashimoto A.C., Chapagain M.L., Kaufusi P.H. Stimulation of unprimed macrophages with immune complexes triggers a low output of nitric oxide by calcium-dependent neuronal nitric-oxide synthase. J. Biol. Chem. 2012;287(7):4492–4502. doi: 10.1074/jbc.M111.315598. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Iijima M.K.T., Duguet A., Shan J., Carbonara P., Hamid Q. NOS 1 is required for allergen-induced expression of NOS 2 in mice. Int. Arch. Allergy Immunol. 2005;138(1):40–50. doi: 10.1159/000087356. 2005. [DOI] [PubMed] [Google Scholar]
- 156.Komatsu D.D.I., Chen N., Reiss C.S. Neuronal expression of NOS-1 is required for host recovery from viral encephalitis. Virology. 1999;258(2):389–395. doi: 10.1006/viro.1999.9734. 1999. [DOI] [PubMed] [Google Scholar]
- 157.Strebovsky P.W., Lang R., Dalpke A.H. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. Faseb. J. 2011;25(3):863–874. doi: 10.1096/fj.10-170597. 2010. [DOI] [PubMed] [Google Scholar]