Abstract
Objective
Beclin1 is a core molecule of the macroautophagy machinery. Although dysregulation of macroautophagy is known to be involved in metabolic disorders, the function of Beclin1 in adipocyte metabolism has not been investigated. In the present study, we aimed to study the role of Beclin1 in lipolysis and mitochondrial homeostasis of adipocytes.
Methods
Autophagic flux during lipolysis was examined in adipocytes cultured in vitro and in the adipose tissue of mice. Adipocyte-specific Beclin1 knockout (KO) mice were used to investigate the activities of Beclin1 in adipose tissues.
Results
cAMP/PKA signaling increased the autophagic flux in adipocytes differentiated from C3H10T1/2 cells. In vivo autophagic flux was higher in the brown adipose tissue (BAT) than that in the white adipose tissue and was further increased by the β3 adrenergic receptor agonist CL316243. In addition, surgical denervation of BAT greatly reduced autophagic flux, indicating that sympathetic nerve activity is a major regulator of tissue autophagy. Adipocyte-specific KO of Beclin1 led to a hypertrophic enlargement of lipid droplets in BAT and impaired CL316243-induced lipolysis/lipid mobilization and energy expenditure. While short-term effects of Beclin1 deletion were characterized by an increase in mitochondrial proteins, long-term Beclin1 deletion led to severe disruption of autophagy, resulting in mitochondrial loss, and dramatically reduced the expression of genes involved in lipid metabolism. Consequently, adipose tissue underwent increased activation of cell death signaling pathways, macrophage recruitment, and inflammation, particularly in BAT.
Conclusions
The present study demonstrates the critical roles of Beclin1 in the maintenance of lipid metabolism and mitochondrial homeostasis in adipose tissues.
Keywords: Beclin1, Lipophagy, Mitophagy, Lipolysis, Brown adipose tissue, White adipose tissue
Highlights
-
•
β3 adrenergic receptor stimulation induced autophagy in adipose tissue.
-
•
Beclin1 in adipocytes is required for lipolysis and lipid utilization.
-
•
Adipocyte-specific Beclin1 KO reduced CL316243-induced thermogenic gene expression.
-
•
Adipocyte-specific Beclin1 KO results in defective autophagy, loss of mitochondria, and inflammation.
1. Introduction
Adipose tissue is a central metabolic organ that is specialized for lipid storage and mobilization [1,2]. Excess energy is stored as neutral lipids in lipid droplets (LDs) in adipocytes, and energy deprivation stimulates lipolysis and LD breakdown to provide free fatty acids as an energy source via the systemic blood circulation [1,2]. Catecholamines are one of the primary activators of lipolysis in adipocytes through the beta-adrenergic receptor activation. In this process, the cAMP-dependent protein kinase A- (PKA-) mediated downstream signaling activates cytosolic lipases, including adipose triglyceride lipase (ATGL) [[3], [4], [5]] and hormone-sensitive lipase (HSL) [1].
In addition to lipolysis that depends on cytosolic lipases, there is an emerging concept of autophagic lipolysis, which is the lysosomal breakdown of LDs by macroautophagy, termed lipophagy [6]. LDs contain mainly triglycerides; thus, triglyceride hydrolysis must be cooperatively regulated during LD degradation. In this regard, previous studies have demonstrated a significant contribution of the Rab7-mediated lipophagy to lipolysis in adipocytes in vitro [7]. However, the role of autophagy in lipolysis and LD breakdown in adipocytes in vivo has not been elucidated completely.
Autophagy plays a crucial role in the regulation of adipogenesis and mitochondrial dynamics in adipocytes. For instance, the adipocyte-specific ATG7 knockout (KO) increases the brown adipose tissue (BAT) mass and enhances insulin sensitivity [8]. The transition from beige adipocytes to the white adipocytes requires increased autophagic activity, and ATG5 and ATG12 KO enhance thermogenic capacity and protect mice from diet-induced obesity [9]. In addition, KO mouse studies have demonstrated critical roles of ATG4b, ATG7, and ATG5 in adipogenesis [[10], [11], [12]].
Autophagy can be categorized into three major types: macroautophagy, chaperon-mediated autophagy, and microautophagy [13]. During the macroautophagy process, phagophore nucleation and expansion lead to autophagosome formation [13]. Subsequently, autophagosomes mature to fuse with lysosomes to generate autolysosomes [13]. Beclin1 is one of the major molecules in class III phosphatidylinositol 3-kinase (PI3KC3) VPS34 complex, which is required for nucleation of the phagophore. Thus, the Beclin1-VPS34 complex provides a platform to recruit the macroautophagy machinery and plays an essential role in autophagosome maturation [14,15]. Beclin1 also regulates cell survival responses and crosstalk between apoptosis and autophagy by the interaction with BCL-2 [14,16]. In relation to metabolic function, acute exercise activates autophagy in skeletal and cardiac muscle by disrupting Beclin1-BCL-2 interactions [17]. The impaired dissociation of Beclin1 from the BCL-2 complex reduces exercise-induced autophagy and its beneficial effects on muscle insulin sensitivity [17]. Although dysregulation of macroautophagy is known to be involved in metabolic disorders, the metabolic function of Beclin1 in adipocytes is not fully understood.
In the present study, we investigated the role of Beclin1 in lipid metabolism by using adipocyte-specific Beclin1 KO mice. We found that the lack of Beclin1 in adipocytes led to defects in the β3 adrenergic receptor agonist-induced lipolysis, energy expenditure, and fatty acid utilization. In addition, the long-term effect of Beclin1 KO resulted in defective autophagy, loss of mitochondria, and a reduction in the expression levels of genes involved in both anabolic and catabolic lipid metabolisms. Consequently, adipose tissue underwent increased activation of cell death signaling pathways, macrophage recruitment, and inflammation. This study demonstrates that Beclin1-dependent processes are critical for maintaining synthetic and catabolic lipid metabolism and mitochondrial function, particularly in BAT.
2. Material and methods
2.1. Animals
All of the protocols related to animal experiments were approved by the Institutional Animal Care and Use Committees of Yonsei University (IACUC-A-201803-704-01) and Seoul National University (SNU-191025-2). Mice were fed a standard chow diet and were kept on a 12-hour light/12-hour dark cycle at 22 ± 1 °C with free access to food and water. Male mice were used for experiments. Wild type C57BL/6 mice (5–6 weeks old) were purchased from Central Lab. Animal Inc. Mice were treated with CL316243 (CL) (Sigma, 1 mg/kg mouse) via intraperitoneal injection for up to 3 days, as indicated. Serum glycerol levels were measured using glycerol reagent (Sigma), as described previously [18]. Energy expenditure was estimated by indirect calorimetry (PhenoMaster, TSE Systems, Bad Homburg, Germany) [18]. Body composition was measured by nuclear magnetic resonance (NMR) scanning EchoMRI-700 (Echo Medical Systems). For cold tolerance test, mice were placed in a 4 °C cold room where body temperature was measured by rectal thermometry, as described previously [19].
To determine the effects of sympathetic neuronal input on autophagic flux, unilateral surgical denervation of interscapular BAT was performed, as previously described [19].
Adipoq-CreER (B6.129-Tg(Adipoq-cre/Esr1∗)1Evdr/J: stock# 024671) mice were purchased from The Jackson Laboratory. Beclin1-floxed mice were a gift from Dr. H. W. Lee (Yonsei University), which were generated by crossing Becn1tm1a(KOMP)Wtsi mice [20] with ACTB-FLPe (B6.Cg-Tg(ACTFLPe)9205Dym/J: JAX stock #005703). Adipoq-CreER mice were crossed with Becn1-floxed mice to generate adipocyte-specific Becn1 KO mice [21]. For Cre recombination, transgenic mice and wild type (WT) controls were treated with tamoxifen dissolved in sunflower oil (Sigma, 75 mg/kg) by oral gavage on each of five consecutive days. Also, we included vehicle (oil)-treated control groups to confirm specific induction of Cre recombinase activity upon tamoxifen treatment in adipose tissue. The analyses were performed three to four days after the last dose of tamoxifen for the short-term effect of Beclin1 deletion. For long-term experiments, the analyses were performed two weeks after the last dose of tamoxifen. An intraperitoneal glucose tolerance test was performed as previously described [22]. To examine the adipocyte-specific effects, BAT was dissociated by collagenase (Type 1, Worthington) digestion, and floating adipocyte fractions were collected by centrifugation, as previously described [23].
2.2. Quantitative PCR and western blot analyses
Quantitative PCR and western blot analyses were performed as described previously [23]. Primers used for qPCR are listed in Table S1. The primary antibodies used for western blot analysis are listed in Table S2.
2.3. RNA-seq
Trizol reagent (Invitrogen) was used for tissue total RNA extraction according to the manufacturer's protocol. RNA integrity number (RIN), rRNA ratio, and concentration of samples were verified on an Agilent Technologies 2100 Bioanalyzer (Agilent Technology, Santa Clara, CA, USA) using a DNA 1000 chip. For RNA-seq analysis, cDNA libraries were constructed with the TruSeq mRNA Library Kit using 1 μg of total RNA. The total RNA was sequenced by the NovaSeq 6000 System (Macrogen, Seoul, Republic of Korea). The raw RNA-seq data have been deposited in Gene Expression Omnibus (GEO) (GSE 148275).
The Cytoscape plugin iRegulon (version 1.3) [24] was used to predict the upstream transcription factors of the top upregulated and downregulated genes in mouse BAT with Beclin1 KO. iRegulon performs a gene-based motif enrichment analysis for cis-regulatory regions within 20 kb centered around the transcription start site using more than 9,000 position weight matrices linked to candidate-binding transcription factors [24]. These transcription factors are ranked according to their maximal normalized enrichment scores (NESs), which quantifies the extent to which the identified motif(s) recover(s) the associated set of input genes. iRegulon regulator prediction was run for the top fifty genes that were upregulated by Beclin1 KO and fifty of the highest-expressed genes in controls that were downregulated by Beclin1 KO. Species and gene nomenclature were set to “Mus musculus, MGI symbols”, and the program was run with default parameters, including a NES cutoff of 3.0.
2.4. Relative mitochondrial DNA copy number analysis
The analysis of relative mitochondrial DNA (mtDNA) copy number was performed as described previously [25]. Briefly, DNA was extracted from adipose tissue using Genomic DNA Extraction Kit (Bioneer). qPCR was performed using the primers for nuclear 18S DNA and mtDNA listed in Table S1. The mtDNA copy number was normalized to the results of nuclear 18S DNA.
2.5. Cell culture
C3H10T1/2 (American Type Culture Collection (ATCC), Manassas, VA, USA) cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) at 37 °C in a humidified atmosphere with 5% CO2. Adipogenic differentiation was performed as previously described [22]. Earle's Balanced Salt Solution (EBSS, Thermo Fisher) was used for nutrient starvation. 8-Bromoadenosine 3′5′-cyclic monophosphate (8Br-cAMP) (Biolog, B007) was used for PKA activation of differentiated C3H10T1/2 adipocytes. For the inhibition of autophagy, chloroquine (an inhibitor of lysosomal degradation, 50 μM, Sigma, C6628) and 3-methyl adenine (3-MA, an inhibitor of early phase, 5 mM, Sigma, M9281) were used. Adipocytes were treated with inhibitors for 30 min before 8Br-cAMP treatment.
For autophagic flux analysis, C3H10T1/2 cells transduced with pBABE-puro mCherry-EGFP-LC3B (a gift from Jayanta Debnath; Addgene plasmid #22418, http://n2t.net/addgene:22418; RRID:Addgene_22418) [26] were differentiated into adipocytes and then monitored during 8Br-cAMP (1 mM) treatment. The pH-sensitive decrease in green fluorescent protein (GFP) intensity over red fluorescent protein (mCherry) intensity was quantified to indicate autolysosome formation. Annexin V Apoptosis Detection Kit with PI (BioLegend, 640914) was used for flow cytometric analysis of apoptosis, according to the manufacturer's instruction. Flow cytometry was performed using LSRFortessa X-20 (BD Biosciences) and FlowJo software (Tree Star). Live-cell imaging was performed with IncuCyte ZOOM Live-Cell Imaging equipment (Essen Bioscience), and fluorescence intensities of the images were analyzed using ImageJ (https://imagej.nih.gov).
To examine the cell-autonomous effects of Beclin1 KO in adipocytes, we analyzed primary cultures of adipocytes that were differentiated from brown adipocyte precursors obtained from Beclin1adipoqKO mice, as previously described [21]. Fully differentiated adipocytes were cultured with 10% FBS DMEM containing 1 μM 4-hydroxytamoxifen (Sigma, H7904) for 3 days and maintained in 10% FBS DMEM without 4-hydroxytamoxifen for 2 days [27]. LIVE/DEAD Viability/Cytotoxicity Kit (Thermo Fisher Scientific, L3224) was used, according to the manufacturer's instructions. For mitochondrial membrane potential analysis, adipocytes were stained with 2 μM JC-1 (Sigma, T4069) for 30 min, and fluorescence intensities were measured, according to the manufacturer's instruction.
2.6. Histology and transmission electron microscopy (TEM)
Hematoxylin and eosin (H&E) staining and immunohistochemical analysis were performed using paraffin sections, as described previously [22]. TEM was performed as described previously [22]. Briefly, small pieces of minced adipose tissue (1∼2 mm3) were immersed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2). Tissues were postfixed in 1% osmium tetroxide (OsO4) for 2 h, dehydrated using ethanol gradient, and embedded into Spurr's Resin (Electron Microscopy Sciences). Samples were sectioned at 60 nm with Ultramicrotome (Leica) and subsequently transferred on nickel grids. Images were collected under Talos L120C cryo-TEM (FEI) or JEM-1010 (JEOL) transmission electron microscope at the Nanobio Imaging Center, Seoul National University.
TUNEL Assay Kit-HRP-DAB (Abcam, ab206386) was used to detect apoptotic cells in paraffin sections of BAT, according to the manufacturer's instructions.
2.7. Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA). Data are presented as mean ± standard errors of the mean (SEM). Statistical significance between two groups was determined by unpaired t-test. Comparisons among multiple groups were performed using a one-way or two-way analysis of variance (ANOVA), with Bonferroni post hoc tests to determine P values. Heatmap was generated by the PermutMatrix program, as previously described [23].
3. Results
3.1. β3 adrenergic receptor stimulation induces autophagy in adipose tissue
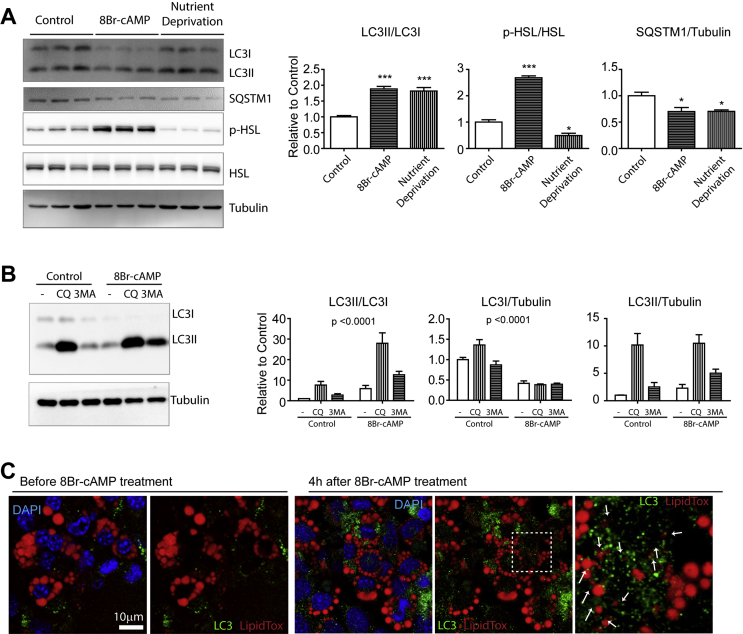
To examine the adipocyte-specific roles of autophagy during lipolysis, we examined autophagic flux in adipocytes during PKA-dependent lipolysis. A ratio of cytosolic LC3 (LC3I) and lipidated, autophagosome-recruited LC3 (LC3II) was used to detect autophagic flux [28]. The 8Br-cAMP (cell-permeable cAMP analog) treatment induced autophagy in differentiated C3H10T1/2 adipocytes, which was similar in magnitude to the autophagy induced by the nutrient deprivation status, as indicated by an increase in LC3II/LC3I ratio and a decrease in SQSTM1 levels (Figure 1A). To capture the dynamic process of autophagy, we inhibited autophagy with chloroquine, which prevents lysosomal acidification and inhibits the lysosomal degradation of the autophagosomes [28], and 3-MA, which inhibits class III PI3K and early steps of autophagosome formation [29]. Chloroquine treatment further increased the accumulation of LC3II relative to LC3I in the adipocytes; the LC3II/LC3I ratio was higher in the cells that were treated with 8Br-cAMP than in the vehicle controls, indicating an increase in the autophagic flux by PKA signaling (Figure 1B). Interestingly, 3-MA had no significant effects on LC3II levels under control conditions but induced LC3II accumulation with 8Br-cAMP cotreatment. To assess autophagic flux in the absence of pharmacological inhibition, we examined the effects of 8Br-cAMP using the LC3-GFP-RFP reporter [26], in which green fluorescence from acid-sensitive GFP was suppressed as the LC3 reporter entered autolysosomes (Figure S1A). qPCR analysis indicated that 8Br-cAMP treatment upregulated the expression of several autophagy-related genes (Figure S1B). As lipophagy is involved in lipolysis, we examined the association between LC3 and LD via immunofluorescence analysis. Results suggested that 8Br-cAMP treatment facilitated lipophagy, which was indicated by increased punctate staining of LC3 surrounding the micro-LDs (Figures 1C and S1C, arrow in magnified view).
Figure 1.
Autophagy is induced by PKA signaling in adipocytes in vitro. (A–B) Immunoblot analysis of LC3 and HSL in adipocytes differentiated from C3H10T1/2 cells. Earle's Balanced Salt Solution (EBSS) was used for nutrient deprivation. Cells were treated with 1 mM 8Br-cAMP for 4 h (n = 6 per condition, mean ± SEM, ∗P < 0.05, ∗∗∗P < 0.001). Cells were pretreated with chloroquine (CQ: 50 μM) or 3-methyl adenine (3 MA: 5 mM) for 30 min before 8Br-cAMP or vehicle treatment. (C) Representative images of adipocytes differentiated from C3H10T1/2 cells treated with 8Br-cAMP for 4 h and stained for LC3. LipidTox and DAPI were used for neutral lipid and nucleus staining, respectively.
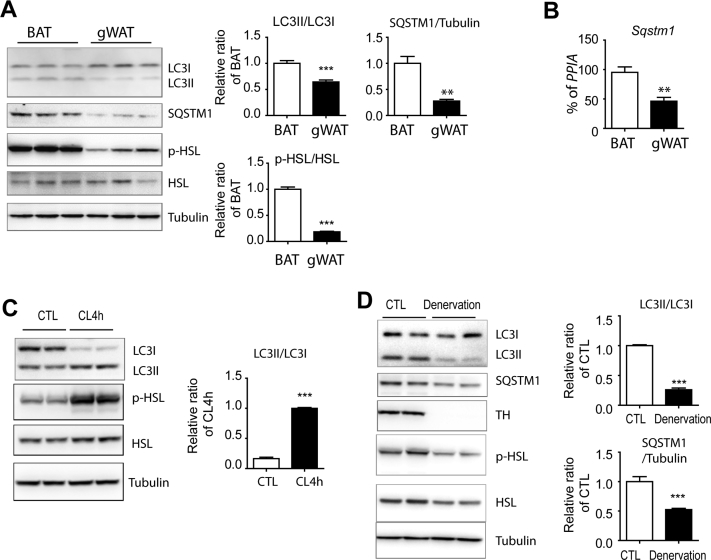
We also examined in vivo autophagic flux in the interscapular BAT and white adipose tissue (WAT). We found higher levels of LC3II/LC3I ratio in BAT than in the gonadal white adipose tissue (gWAT) (Figure 2A). Interestingly, sequestosome-1 (SQSTM1) protein levels were higher in BAT compared to gWAT (Figure 2A). The expression of Sqstm1 mRNA was also higher in BAT compared to gWAT (Figure 2B), suggesting differential transcriptional regulation of Sqstm1 levels in BAT and gWAT. This is in line with the previously reported metabolic roles of SQSTM1 (p62) in mitochondrial function and thermogenesis in BAT [30]. The β3 adrenergic receptor agonist CL increased autophagic responses in BAT, indicated by an increase in the LC3II/LC3I ratio (Figure 2C). This increase in autophagy was accompanied by phosphorylation of HSL (Figure 2B), indicating the activation of cAMP/PKA downstream signaling. Similarly, cold exposure increased the LCII/LC3I ratio in BAT (Figure S2). In addition, surgical denervation of BAT, which reduced phosphorylation of HSL, strongly reduced autophagic flux (Figure 2D), indicating that basal levels of sympathetic nerve activity contribute to autophagic flux in vivo. Denervation also reduced SQSTM1 protein levels in BAT (Figure 2D). In vitro 8Br-cAMP treatment increased Sqstm1 transcript levels in adipocytes (Figure S1B), suggesting that Sqstm1 expression levels are transcriptionally controlled by sympathetic nerve activity and PKA signaling. The correlation between the activation of lipolysis and the increase in autophagic flux underscored the potential role of autophagy in the PKA-dependent lipolysis.
Figure 2.
Autophagy is induced by beta-adrenergic stimulation in adipose tissue in vivo. (A) Immunoblot analysis of LC3, SQSTM1, and HSL in BAT and gWAT of mice. (B) qPCR analysis of Sqstm1 in BAT and gWAT. (C) Immunoblot analysis of LC3 and HSL in BAT of mice treated with CL316243 for 4 h and controls. (D) Immunoblot analysis of LC3, SQSTM1, tyrosine hydroxylase (TH), and HSL of denervated BAT and sham-surgery controls (n = 8 per condition, mean ± SEM, ∗∗∗P < 0.001).
3.2. Beclin1 in adipocytes is required for lipolysis and lipid utilization
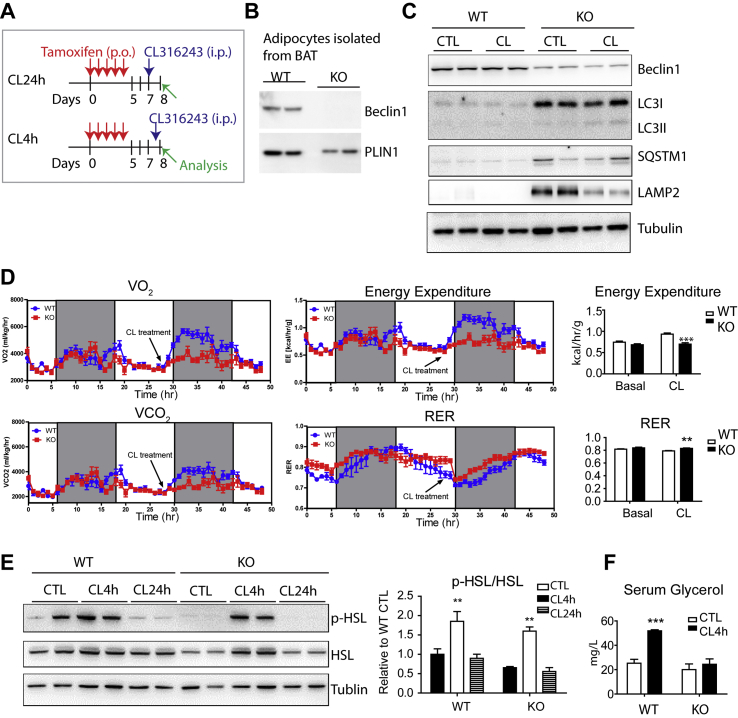
To determine the in vivo effects of autophagy inhibition in the adipocyte lipid metabolism, we examined the effects of adipocyte-specific KO of Beclin1, a core molecule of autophagosome formation. To avoid the potential developmental effects and to assess the temporal effects of autophagy inhibition, we deleted Beclin1 in adipocytes in an inducible manner using Adipoq-CreER/Becn1floxed/floxed mice treated with tamoxifen (Beclin1adipoq KO mice). Becn1floxed/floxed mice treated with tamoxifen were used as WT controls. Genetic inactivation of Beclin1 was confirmed by western blot analysis of the isolated adipocytes from BAT of Beclin1adipoq KO mice four days after the final dose of tamoxifen (Figure 3B). Tamoxifen treatment also reduced Beclin1 protein in the whole BAT, with nonadipocyte stromal cells accounting for residual levels (Figure 3C).
Figure 3.
Adipocyte-specific Beclin1 KO attenuates CL316243-induced lipolysis and energy expenditure. (A) Schematic diagram illustrating the experimental schedule for tamoxifen and CL316243 (CL) treatment (single injection). (B) Immunoblot analysis of Beclin1 expression in adipocytes isolated from BAT of WT and Beclin1adipoqKO mice (KO). (C) Immunoblot analysis of BAT of WT and KO mice treated with CL or vehicle for 24 h. (D) Indirect calorimetry analysis of WT and KO mice before and after CL treatment. (E) Immunoblot analysis of BAT of WT and KO mice. (F) Serum glycerol of WT and KO mice 4 h after CL treatment (n = 6–8 per condition, mean ± SEM, ∗∗P < 0.01, ∗∗∗P < 0.001).
We next performed indirect calorimetry analysis to assess the basal metabolic rate and thermogenesis in response to β3-adrenergic receptor activation. Adipocyte-specific KO of Beclin1 did not affect the normal daily energy expenditure. However, whereas CL treatment sharply increased energy expenditure in WT mice, this activation was strongly attenuated in the Beclin1adipoqKO mice (Figure 3D). Moreover, the Beclin1adipoqKO mice demonstrated relatively higher levels of respiratory exchange ratio (RER) after CL treatment, indicating a reduction in the CL-induced fatty acid oxidation (Figure 3D). Western blot analysis indicated that Beclin1adipoqKO significantly increased the accumulation of intermediates in autophagy (LC3I, SQSTM1) (Figure 3C), which indicates disrupted autophagy (“frustrated autophagy [31]”). Induction of lysosome-associated membrane glycoprotein 2 (LAMP2) was observed, indicating possible compensation by chaperone-mediated autophagy (Figure 3C). Phosphorylation of HSL by CL appeared to be largely intact Beclin1adipoq KO mice, indicating that PKA-mediated signaling was not impaired at this time (Figure 3E). Nonetheless, CL treatment failed to increase circulating levels of glycerol, indicating a defect in adipocyte lipolysis (Figure 3F).
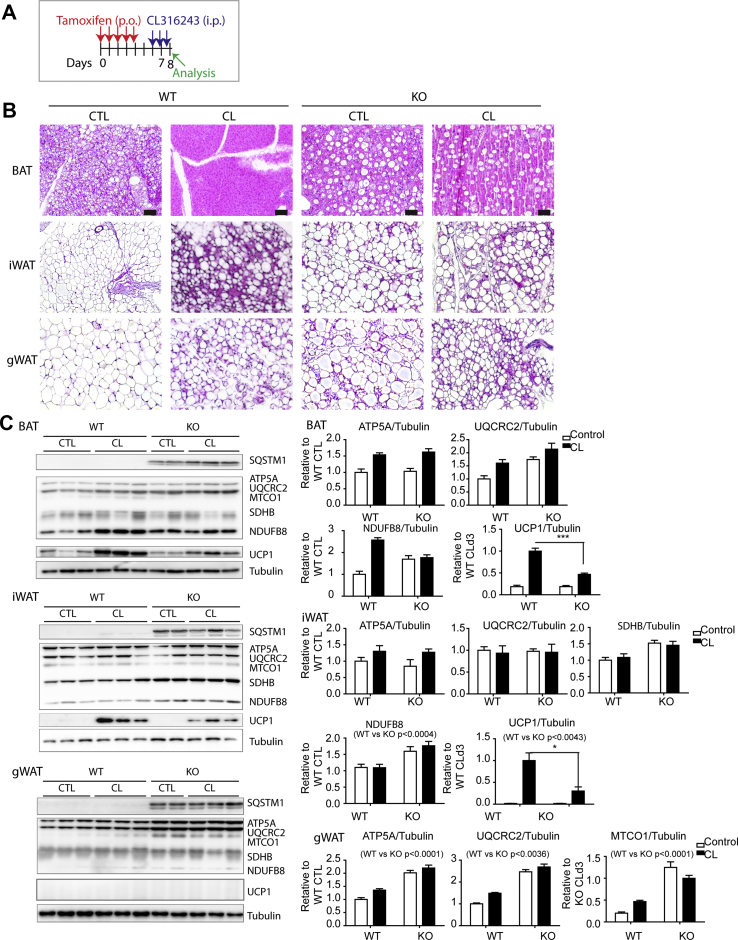
Histological observation by H&E staining of the paraffin-embedded sections demonstrated enlarged hypertrophic LD in BAT of the KO mice while some of the brown adipocytes lost LD, indicating pronounced heterogeneity in LD homeostasis induced by Beclin1 KO; the CL-induced shrinkage of LD size was blocked by Beclin1 KO (Figure 4B). Morphological changes by CL treatment further emphasized the heterogeneity in LD size (Figure 4B). Collectively, these data indicate that Beclin1adipoqKO mice are defective in the CL-induced lipolysis.
Figure 4.
Effects of adipocyte-specific Beclin1 KO on lipid droplets and mitochondrial content in BAT and WAT. (A) Schematic diagram illustrating the experimental schedule for tamoxifen (5 days) and CL316243 (3 days) treatment. B. H&E staining of paraffin sections of BAT, iWAT, and gWAT of WT and Beclin1adipoqKO mice (size bar = 40 μm). (C) Immunoblot analysis of BAT, iWAT, and gWAT of WT and Beclin1adipoqKO mice treated with CL for 3 days (n = 6–8 per condition, mean ± SEM, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
3.3. Adipocyte-specific Beclin1 deletion reduced CL-induced thermogenic gene expression
Previous studies have reported that the deletion of genes encoding certain autophagy-related proteins (ATGs) or pharmacological inhibition of autophagy increases the mitochondrial content of adipose tissue [9,[32], [33], [34]]. Therefore, we examined the mitochondrial content of BAT and WAT in KO mice using western blot analysis. The levels of mitochondrial enzymes involved in oxidative phosphorylation in BAT of KO mice were slightly higher than (ubiquinol-cytochrome-c reductase complex core protein 2 (UQCRC2), NADH dehydrogenase [ubiquinone]1 beta subcomplex subunit 8 (NDUFB8)), or comparable to (ATP synthase F1 subunit alpha (ATP5A), succinate dehydrogenase [ubiquinone] iron-sulfur subunit (SDHB)) those in WT controls, although each subunit responded differentially (Figure 4C). However, uncoupling protein 1 (UCP1) induction by CL treatment was attenuated in BAT and inguinal white adipose tissue (iWAT) of Beclin1 KO mice (Figure 4C). Several mitochondrial enzymes (ATP5A, UQCRC2, and cytochrome c oxidase subunit 1 (MTCO1)) were significantly increased in gWAT; however, UCP1 protein was not detected in the basal condition or following CL treatment (Figure 4C). This is consistent with the results of the qPCR analysis, which indicates that CL-induced increases in brown adipocyte markers were reduced in KO mice (Figure S3). In addition, KO mice were more sensitive to cold exposure compared to WT control mice (Figure S4).
3.4. Long-term deletion of Beclin1 results in defective autophagy, loss of mitochondria, and inflammation
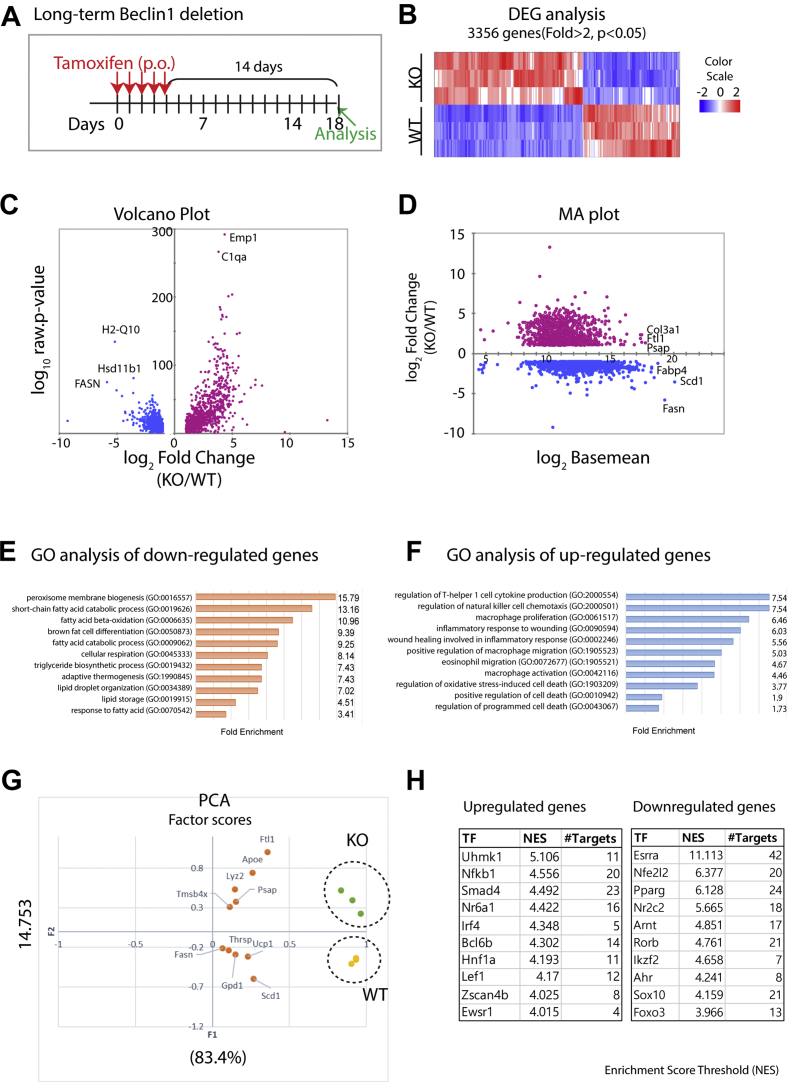
We also examined the long-term effects of Beclin1 deletion on adipose tissue. In contrast to the phenotypes obtained three days after the final tamoxifen dose, BAT mitochondrial content was markedly reduced two weeks after the tamoxifen-induced deletion (Figure S5). The drastic change in the BAT phenotype prompted us to perform RNA-seq profiling of BAT of control and KO mice 2 weeks after tamoxifen induction (long-term deletion) (Figure 5). A total of 3356 genes were differentially regulated in BAT between Beclin1 KO and WT. GO analysis of genes upregulated by Beclin1 KO demonstrated an elevated immune response and cell death signaling pathways (Figure 5C–F). The upregulated genes included epithelial membrane protein 1 (Emp1; 15.5-fold) and NADPH oxidase 4 (Nox4, 12-fold upregulation), which have been involved in oxidative stress-induced ferroptotic death (Figure 5C) [35]. qPCR analysis confirmed the upregulation of Emp1 and Nox4 in BAT of Beclin1 KO mice (Figure. S6). Principle component analysis (PCA) indicated that factor 2 (F2) clearly separated WT and KO clusters (Figure 5G). The genes with the top 5 contributing factor scores to F2 toward transcriptomic profiles of KO included lysozyme 2 (Lyz2), which is highly expressed in the monocyte-macrophage system, indicating monocyte/macrophage infiltration in BAT of Beclin1 KO mice. In contrast, the downregulated genes in BAT of KO mice were involved in the fatty acid synthesis and thermogenesis (stearoyl-CoA desaturase 1 (Scd1), Fasn, and Ucp1) (Figure 5C,D, and G). Consistently, GO analysis indicated that Beclin1 KO reduced the expression of genes involved in the core functions of brown adipocyte lipid metabolism, including genes involved in thermogenesis, lipogenesis, and mitochondrial oxidative phosphorylation (Figure 5E). Western blot analysis confirmed the protein expression levels of several differentially regulated genes (FASN, HSL, UCP1, and enzymes involved in mitochondrial oxidative phosphorylation) identified from RNA-seq analysis (Figure 6B).
Figure 5.
Transcriptomic characterization of BAT of adipocyte-specific Beclin1 KO mice. (A) Schematic diagram illustrating the experimental schedule. (B) Heatmap of 3356 mRNA differentially regulated in BAT of adipocyte-specific Beclin1 KO mice from RNA sequencing data (|fold change| ≥ 2, P < 0.05). (C–D) Volume plot and MA-plot of differentially expressed genes in BAT of Beclin1 KO mice (|fold change| ≥ 2, P < 0.05). (E–F) GO analysis of differentially regulated genes. (G–H) Principle component analysis (G) and transcription factor enrichment (H) of differentially regulated genes in BAT of adipocyte-specific Beclin1 KO mice from RNA sequencing data.
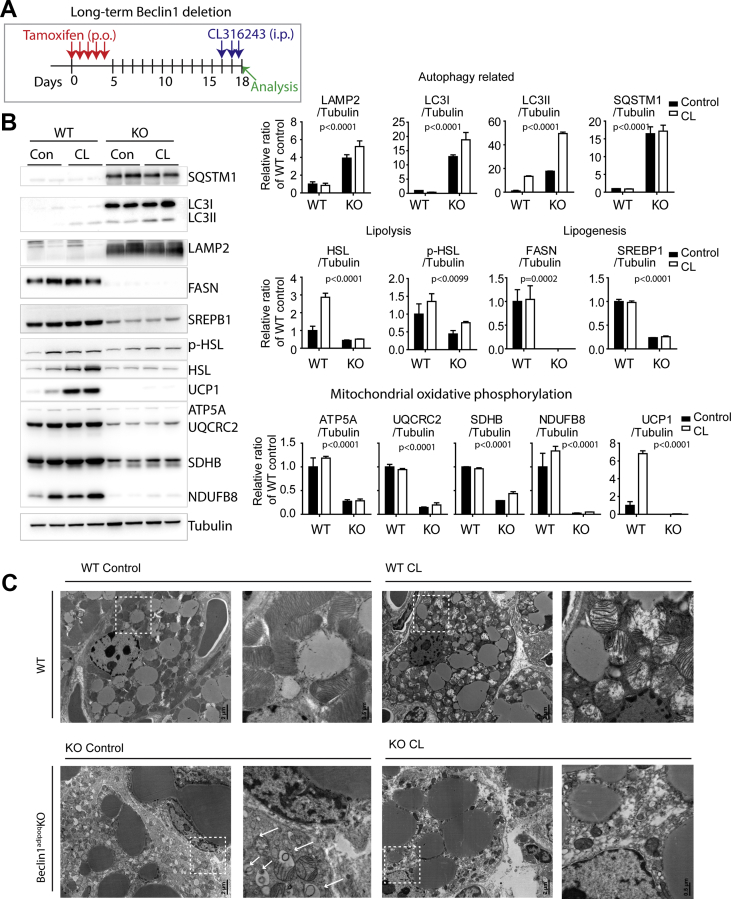
Figure 6.
Long-term adipocyte-specific Beclin1 KO impairs autophagy and mitochondrial integrity in BAT. (A) Schematic diagram illustrating the experimental schedule for tamoxifen (5 days) and CL316243 treatment. (B) Immunoblot analysis of BAT of Beclin1adipoq KO mice and WT controls (n = 8 per condition, mean ± SEM; two-way ANOVA; the significance of Beclin1 KO effects is indicated). (C) Representative electron micrographs of BAT of Beclin1adipoq KO mice and WT controls. Magnified views of boxed regions are presented. The arrows indicated the accumulation of autophagosomes.
Transcription factor enrichment analysis by iRegulon analysis predicted that potential transcription networks control upregulated and downregulated gene sets in BAT of KO mice (Figure 5H and Table S3). Beclin1 KO induced massive upregulation of proinflammatory network involving TNFα/NFκB (Tnfsf11, Rela, and Spp1) and TGFb/Smad4 (Saa3 and Dlx4) signaling. Importantly, the core transcriptional regulators of BAT catabolic (Esrra, Esrrb, Ppara, and Ppargc1b) and anabolic (Pparg, Cebpa, Srebf, and Mlxipl) functions were strongly suppressed 2 weeks after Beclin1 inactivation (Figure 5H and Table S3). Interestingly, the key elements of the β-adrenergic/PKA signaling pathway (Adrb1/3, Gnas, Adcy6, and Prkaca) and lipolysis/fat oxidation (Cidea, Plin1, Pnpla2, Cpt1b, and Acadm) pathways were strongly suppressed, which likely contribute to the appearance of unilocular adipocytes in BAT (Figure S10A).
As lipolysis activates thermogenic gene programs and de novo lipogenesis [36], these genetic signatures also supported the defective lipolysis in the BAT of the Beclin1adipoqKO mice. As hypothesized, the Beclin1adipoqKO was also defective in autophagy, indicated by increased levels of SQSTM1 and LC3I, indicating the accumulation of early autophagosomes. Moreover, electron-microscopic (EM) analysis indicated an impaired autophagy/mitophagy with numerous vacuoles and disruption of mitochondrial integrity caused by a lack of Beclin1 (Figure 6C). Indeed, Beclin1 KO reduced the levels of the outer (MFF, MFN2, and TOM20) and inner (OPA1) mitochondrial membrane proteins, as well as the matrix protein citrate synthase (Figure S7). In addition, adipocyte-specific Beclin1 KO reduced the relative mtDNA copy number in BAT (Figure S8). Beclin1 KO reduced the levels of certain enzymes involved in mitochondrial oxidative phosphorylation in iWAT (Figure S9A), and TEM analysis confirmed the accumulation of autophagosome/autolysosomes and presence of irregular vacuoles (Figure S9C). Although mitochondrial protein (ATP5A and UQCRC2) was increased in gWAT (Figure S9B), the mitochondrial integrity was impaired (Figure S9C).
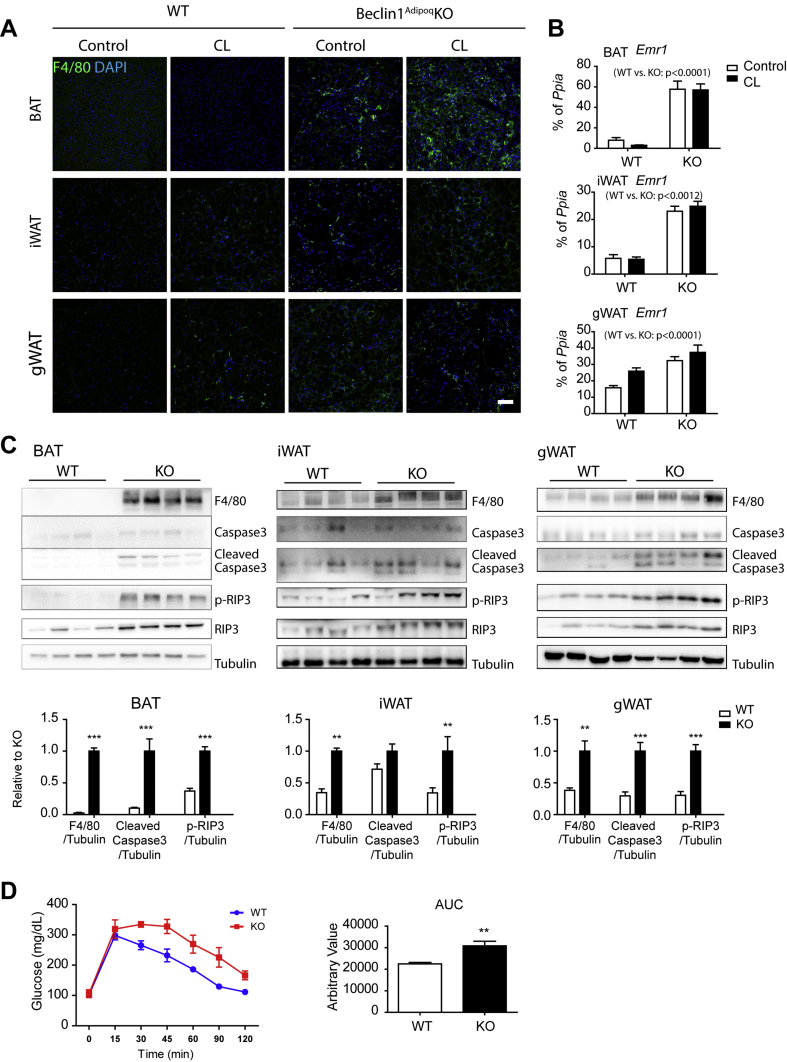
As predicted by RNA-seq data, macrophage recruitment was prominent in the BAT of the KO mice (Figure 7A), which may explain the increase in inflammatory gene expression. H&E staining (Figure S10A) indicated that long-term KO increased the appearance of adipocytes with large unilocular LD and stromal cells lacking visible LD. Adipocytes in BAT of KO mice exhibited reduced UCP1 expression, as indicated by UCP1 immunostaining (Figure S10B) and western blot analysis of isolated adipocytes (Figure S10C). Consistently, qPCR analysis and western blot analysis indicated increased expression F4/80 and Emr1 (gene encoding F4/80) (Figure 7B,C). Indeed, cell death-related markers of necroptosis (receptor-interacting serine/threonine-protein kinase 3 (RIP3), p-RIP3) and apoptosis (cleaved caspase 3) were increased in Beclin1 KO in BAT and WAT (Figure 7C). Furthermore, immunostaining of p-RIP3 and TUNEL assay confirmed that Beclin1 KO induced necroptosis and apoptosis in BAT (Figure S11). Due to adipocyte death, adipose tissue mass was significantly reduced by long-term Beclin1 deletion, while body weight was not significantly altered (Figure S12). Long-term adipocyte-specific Beclin1 deletion impaired glucose tolerance (Figure 7D), potentially due to dysfunctional adipose tissue and inflammation.
Figure 7.
Long-term adipocyte-specific Beclin1 KO increases macrophage recruitment and immune responses in adipose tissue. (A) F4/80 staining of paraffin sections of BAT, iWAT, and gWAT of Beclin1 KO and WT mice (size bar = 100 μm). (B) qPCR analysis of Emr1 expression in BAT, iWAT, and gWAT of Beclin1 KO and WT mice. (C) Immunoblot analysis of F4/80, caspase 3, and RIP3 in BAT, iWAT, and gWAT of Beclin1 KO and WT mice. (D) Intraperitoneal glucose tolerance test of Beclin1 KO and WT mice (n = 8 per condition, mean ± SEM, ∗∗P < 0.01, ∗∗∗P < 0.001).
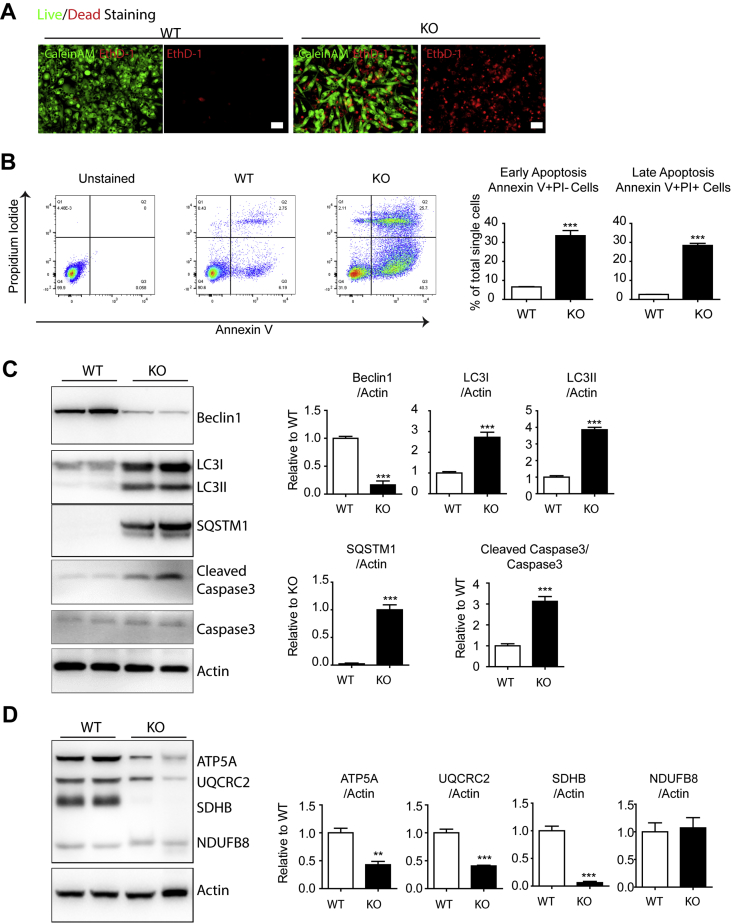
To test whether the defective autophagy induces cell death in a cell-autonomous fashion, we examined the primary cultures of adipocytes that were differentiated from brown adipocyte precursors obtained from Beclin1adipoqKO mice (Figure 8). Beclin1 deletion was confirmed by western blot 5 days after 4-hydroxytamoxifen exposure (Figure 8C). Beclin1 KO induced cell death as demonstrated by ethidium homodimer-1 (EthD-1) staining (Figure 8A) and by flow cytometric detection of Annexin V (Figure 8B). Also, western blot analysis indicated that Beclin1 KO reduced the levels of enzymes involved in mitochondrial oxidative phosphorylation and increased caspase 3 cleavage (Figures 8C and D). In addition, Beclin1 KO reduced mitochondrial membrane potential, measured by JC-1 analysis (Figure S13). Pharmacological inhibition of autophagy by chloroquine treatment induced cell death responses, indicated by increased levels of cleaved caspase 3 and phospho-RIP3 (Figure S14).
Figure 8.
Beclin1 KO in cultured brown adipocytes induces cell death and impairs autophagy-related and mitochondrial protein expression. Cell death analysis of adipocytes differentiated from precursors obtained from BAT of Beclin1adipoqKO mice and WT controls. Adipocytes differentiated from the precursor cells were treated with 4-hydroxyl-tamoxifen (1 μM) to delete Beclin1. (A) LIVE/DEAD staining by CaleinAM (green) and EthD-1 (red). (B) FACS analysis of Annexin V. (C–D) Immunoblot analysis of autophagy-related and mitochondrial proteins (n = 6 per condition, mean ± SEM, t-test, ∗∗P < 0.01, ∗∗∗P < 0.001).
4. Discussion
The dysregulation of autophagy is known to be involved in the development of metabolic disease [37]. Clinical and experimental evidence suggests that increased autophagy in WAT is correlated with obesity and diabetes [38]. Moreover, the inhibition of mitochondrial clearance has been recognized as a mechanism in the browning of WAT and in the metabolic plasticity of adipose tissue. Thus, we investigated the roles of adipocyte Beclin1, a core molecule in autophagosome formation, in adipose tissue lipolysis and mitochondrial homeostasis.
Adipocytes are a specialized cell type that store excess energy in the form of neutral lipids in LDs to maintain energy homeostasis. The mechanisms of LD degradation have been investigated in relation to the LD-selective autophagic process. For example, recent studies indicate that chaperone-mediated autophagy regulates neutral lipid metabolism via degradation of the LD proteins PLIN2 and PLIN3 in hepatocytes [39]. In addition, the contribution of the Rab7-mediated lipophagy to adipocyte lipolysis has been reported [7]. Consistent with previous reports, the current study demonstrates that cAMP/PKA signaling activates adipocyte lipophagy and cytosolic lipolysis simultaneously in vitro and in vivo. In addition, we demonstrated the defective lipolysis of the adipocyte-specific Beclin1 KO mice, suggesting that the simultaneous activation of lipophagy with cytosolic lipolysis is required for an efficient lipid catabolism and LD homeostasis in vivo.
By utilizing a tamoxifen-inducible KO system, we were able to discriminate the time-dependent effects of Beclin1 deletion on adipose tissue homeostasis. Three days after tamoxifen induction, mitochondrial protein levels were increased in BAT and WAT of KO mice relative to WT controls. It is likely that defective Beclin1-mediated autophagy/mitophagy acutely increased mitochondrial proteins. However, loss of Beclin1 for two weeks (i.e., long-term effect) reduced mitochondrial content and integrity in BAT and WAT, partially due to dysregulation of autophagy. Our findings are in line with the recent studies of Atg3 and Atg16L1 KO mice [40]; however, this result seems to contradict previous reports indicating that the inhibition of autophagy by ATG5 and ATG12 KO prevents mitochondrial clearance and increases the mitochondrial content and thermogenic capacity of adipose tissue [9]. These studies suggest that the disruption of specific components produces different outcomes in metabolic phenotypes. It was clearly shown that Beclin1 KO accumulated damaged mitochondria in adipose tissue, which led to adipocyte death responses. Furthermore, we demonstrate that the effects of Beclin1 deletion on cell survival and mitochondrial function are cell autonomous.
Failure of mitochondrial quality control leads to apoptosis, inflammation, or necrosis, which has been proposed as one of the major pathological stimuli in degenerative diseases [41]. While Beclin1 has been investigated as a key regulator of macroautophagy [15], Beclin1 has been involved in the control of mitophagy [[42], [43], [44]]. For instance, interactions between PINK1 and Beclin1 on mitochondria-associated membranes facilitate autophagosome formation during mitophagy [42,44]. E3 ubiquitin-protein ligase parkin (PARK2)-Beclin1 interactions are required for PARK2 translocation to mitochondria [43]. In relation to mitochondrial quality control, cardiac-specific Beclin1 overexpression reduces mitochondrial damage and improves heart function during lipopolysaccharide- (LPS-) induced sepsis [45]. As the current study was focused on the effects of Beclin1 KO in mitochondrial integrity and cell death responses, the roles of Beclin1 in mitophagy in adipocytes deserve further investigation. In addition, future investigations on autophagy-independent roles of Beclin1 in adipocytes will be informative to understand the mechanisms of Beclin1-mediated adipocyte death.
Long-term Beclin1 deletion resulted in the accumulation of defective autophagosomes/autolysosomes in adipocytes that resembled “frustrated autophagy” [31]. As Beclin1 is responsible for lysosomal fusion, as well as autophagosome formation [46], further investigation on the activity of Beclin1 in individual autophagy steps in adipocytes will be required.
Beclin1 inactivation increased the levels of LAMP2, which is a well-known factor in chaperon-mediated autophagy, macroautophagy [47], and lipophagy [39]. The elevation of LAMP2 protein levels might indicate compensatory induction of LAMP2-mediated lipophagy in KO mice. LAMP2 plays a critical role in the degradation of LD proteins, PLIN2 and PLIN3 in hepatocytes, and NIH3T3 fibroblasts [39], facilitating lipophagy [39]. Whether LAMP2-mediated lipophagy contributes to PKA/cAMP-induced lipolysis involving phosphorylation of PLIN1 in adipocytes deserves further investigation.
Several studies have reported altered autophagic activity in obese individuals [37,38]. As adipose tissue comprises multiple cell types, further research is necessary to carefully determine which of these are important contributors to alterations in the autophagic activity. In addition, understanding the specific function of each of the autophagy-related proteins in the various individual steps of the autophagic process is vital for developing novel strategies of treating obesity-related metabolic diseases via the regulation of autophagy.
One interesting finding in our study is that brown adipocytes are particularly sensitive to Beclin1 KO compared to white adipocytes, which have comparatively low autophagic/catabolic activity. These results suggest that Beclin1-dependent autophagic functions are critical in maintaining the brown adipocyte phenotype during the activation of the catabolism via β-adrenergic signaling. Indeed, lipolysis engages Beclin1-dependent autophagic flux, which is critical for maintaining the phenotypic character of brown adipocytes.
Collectively, the present study characterized the role of Beclin1 in adipose tissue, in relation to β-adrenergic stimulation of adipose tissue. Although our study focused on the role of Beclin1 in lipolysis and mitochondrial turnover, future investigations into cell type-specific regulation of Beclin1-dependent autophagy will be required for developing therapeutic applications that treat metabolic diseases by improving lipid metabolism.
Authors' contributions
YHL conceived and designed the study. YHS, YKC, AS, JHP, MSK, YSJ, HJK, JKS, RBB, JGG, and YHL conducted the experiments and analyzed the results. YHL, YKC, AS, HJK, JKS, and YHS performed in vivo mouse experiments. YKC, AS, and YHS performed in vitro experiments. YHL, YHS, YKC, and JGG wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
We thank Dr. H. W. Lee (Yonsei University) for providing Becn1-floxed mice. This research was supported by the National Research Foundation of Korea (NRF) grants (NRF-2019R1C1C1002014, NRF-2018R1A5A2024425, NRF-2013M3A9D5072550) funded by the Korean government (MSIT). RBB and JGG were supported by NIH grants F31DK116536 and RO1DK062292.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101005.
Conflicts of interest
The authors declare no conflicts of interest
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Granneman J.G., Moore H.-P.H. Location, location: protein trafficking and lipolysis in adipocytes. Trends in Endocrinology and Metabolism: Trends in Endocrinology and Metabolism. 2008;19(1):3–9. doi: 10.1016/j.tem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Duncan R.E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H.S. Regulation of lipolysis in adipocytes. Annual Review of Nutrition. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306(5700):1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 4.Sanders M.A., Madoux F., Mladenovic L., Zhang H., Ye X., Angrish M. Endogenous and synthetic ABHD5 ligands regulate ABHD5-perilipin interactions and lipolysis in fat and muscle. Cell Metabolism. 2015;22(5):851–860. doi: 10.1016/j.cmet.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granneman J.G., Moore H.-P.H., Krishnamoorthy R., Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) Journal of Biological Chemistry. 2009;284(50):34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cingolani F., Czaja M.J. Regulation and functions of autophagic lipolysis. Trends in Endocrinology and Metabolism. 2016;27(10):696–705. doi: 10.1016/j.tem.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lizaso A., Tan K.-T., Lee Y.-H. β-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9(8):1228–1243. doi: 10.4161/auto.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A.M., Luu Y.K. Autophagy regulates adipose mass and differentiation in mice. Journal of Clinical Investigation. 2009;119(11):3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altshuler-Keylin S., Shinoda K., Hasegawa Y., Ikeda K., Hong H., Kang Q. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metabolism. 2016;24(3):402–419. doi: 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L., Huang J.-X., Liu Y., Li X., Zhou S.-R., Qian S.-W. Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Molecular and Cellular Biology. 2013;33(16):3180–3190. doi: 10.1128/MCB.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proceedings of the National Academy of Sciences. 2009;106(47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baerga R., Zhang Y., Chen P.-H., Goldman S., Jin S.V. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5(8):1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nature Reviews Molecular Cell Biology. 2018;19(6):349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 14.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death & Differentiation. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirawan E., Lippens S., Vanden Berghe T., Romagnoli A., Fimia G.M., Piacentini M. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8(1):6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y., Miao H., Wu S., Yang W., Zhang Y., Xie G. ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2. Autophagy. 2016;12(11):2167–2182. doi: 10.1080/15548627.2016.1217380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C., Bassik M.C., Moresi V., Sun K., Wei Y., Zou Z. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S.-N., Jung Y.-S., Kwon H.-J., Seong J.K., Granneman J.G., Lee Y.-H. Sex differences in sympathetic innervation and browning of white adipose tissue of mice. Biology of Sex Differences. 2016;7(1):67. doi: 10.1186/s13293-016-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y.-H., Petkova A.P., Konkar A.A., Granneman J.G. Cellular origins of cold-induced brown adipocytes in adult mice. The FASEB Journal. 2015;29(1):286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noguchi S., Honda S., Saitoh T., Matsumura H., Nishimura E., Akira S. Beclin 1 regulates recycling endosome and is required for skin development in mice. Communications Biology. 2019;2:37. doi: 10.1038/s42003-018-0279-0. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y.H., Petkova A.P., Mottillo E.P., Granneman J.G. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metabolism. 2012;15(4):480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S.-N., Kwon H.-J., Im S.-W., Son Y.-H., Akindehin S., Jung Y.-S. Connexin 43 is required for the maintenance of mitochondrial integrity in brown adipose tissue. Scientific Reports. 2017;7(1):7159. doi: 10.1038/s41598-017-07658-y. 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y.H., Kim S.N., Kwon H.J., Granneman J.G. Metabolic heterogeneity of activated beige/brite adipocytes in inguinal adipose tissue. Scientific Reports. 2017;7:39794. doi: 10.1038/srep39794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janky R., Verfaillie A., Imrichová H., Van de Sande B., Standaert L., Christiaens V. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Computational Biology. 2014;10(7) doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redmann M., Benavides G.A., Wani W.Y., Berryhill T.F., Ouyang X., Johnson M.S. Methods for assessing mitochondrial quality control mechanisms and cellular consequences in cell culture. Redox Biology. 2018;17:59–69. doi: 10.1016/j.redox.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.N'Diaye E.N., Kajihara K.K., Hsieh I., Morisaki H., Debnath J., Brown E.J. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Reports. 2009;10(2):173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harms M.J., Ishibashi J., Wang W., Lim H.W., Goyama S., Sato T. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metabolism. 2014;19(4):593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y.T., Tan H.L., Shui G., Bauvy C., Huang Q., Wenk M.R. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. Journal of Biological Chemistry. 2010;285(14):10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller T.D., Lee S.J., Jastroch M., Kabra D., Stemmer K., Aichler M. p62 Links β-adrenergic input to mitochondrial function and thermogenesis. Journal of Clinical Investigation. 2013;123(1):469–478. doi: 10.1172/JCI64209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb R.A., Mentzer R.M. Autophagy during cardiac stress: joys and frustrations of autophagy. Annual Review of Physiology. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armani A., Cinti F., Marzolla V., Morgan J., Cranston G.A., Antelmi A. Mineralocorticoid receptor antagonism induces browning of white adipose tissue through impairment of autophagy and prevents adipocyte dysfunction in high-fat-diet-fed mice. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2014;28(8):3745–3757. doi: 10.1096/fj.13-245415. [DOI] [PubMed] [Google Scholar]
- 33.Cairó M., Villarroya J., Cereijo R., Campderrós L., Giralt M., Villarroya F. Thermogenic activation represses autophagy in brown adipose tissue. International Journal of Obesity (2005) 2016;40(10):1591–1599. doi: 10.1038/ijo.2016.115. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh A.K., Mau T., O'Brien M., Yung R. Novel role of autophagy-associated Pik3c3 gene in gonadal white adipose tissue browning in aged C57/Bl6 male mice. Aging. 2018;10(4):764–774. doi: 10.18632/aging.101426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W.-H., Ding C.-K.C., Sun T., Rupprecht G., Lin C.-C., Hsu D. The hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Reports. 2019;28(10):2501–2508.e2504. doi: 10.1016/j.celrep.2019.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mottillo E.P., Balasubramanian P., Lee Y.H., Weng C., Kershaw E.E., Granneman J.G. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. The Journal of Lipid Research. 2014;55(11):2276–2286. doi: 10.1194/jlr.M050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maixner N., Bechor S., Vershinin Z., Pecht T., Goldstein N., Haim Y. Transcriptional dysregulation of adipose tissue autophagy in obesity. Physiology. 2016;31(4):270–282. doi: 10.1152/physiol.00048.2015. [DOI] [PubMed] [Google Scholar]
- 38.Kovsan J., Blüher M., Tarnovscki T., Klöting N., Kirshtein B., Madar L. Altered autophagy in human adipose tissues in obesity. Journal of Clinical Endocrinology & Metabolism. 2011;96(2):E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 39.Kaushik S., Cuervo A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nature Cell Biology. 2015;17(6):759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai J., Pires K.M., Ferhat M., Chaurasia B., Buffolo M.A., Smalling R. Autophagy ablation in adipocytes induces insulin resistance and reveals roles for lipid peroxide and Nrf2 signaling in adipose-liver crosstalk. Cell Reports. 2018;25(7):1708–1717.e1705. doi: 10.1016/j.celrep.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green D.R., Galluzzi L., Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333(6046):1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelmetti V., De Rosa P., Torosantucci L., Marini E.S., Romagnoli A., Di Rienzo M. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy. 2017;13(4):654–669. doi: 10.1080/15548627.2016.1277309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choubey V., Cagalinec M., Liiv J., Safiulina D., Hickey M.A., Kuum M. BECN1 is involved in the initiation of mitophagy: it facilitates PARK2 translocation to mitochondria. Autophagy. 2014;10(6):1105–1119. doi: 10.4161/auto.28615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michiorri S., Gelmetti V., Giarda E., Lombardi F., Romano F., Marongiu R. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death & Differentiation. 2010;17(6):962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 45.Sun Y., Yao X., Zhang Q.J., Zhu M., Liu Z.P., Ci B. Beclin-1-Dependent autophagy protects the heart during sepsis. Circulation. 2018;138(20):2247–2262. doi: 10.1161/CIRCULATIONAHA.117.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature Cell Biology. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 47.Issa A.-R., Sun J., Petitgas C., Mesquita A., Dulac A., Robin M. The lysosomal membrane protein LAMP2A promotes autophagic flux and prevents SNCA-induced Parkinson disease-like symptoms in the Drosophila brain. Autophagy. 2018;14(11):1898–1910. doi: 10.1080/15548627.2018.1491489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.