Abstract
Background:
Gastric cancer is one of the most common malignancies worldwide with high mortality. Therefore, identifying cancer-related biomarkers for predicting prognosis and progression of gastric cancer is essential. This study aimed to investigate the clinical value and functional role of microRNA-3196 in gastric cancer.
Methods:
The relative expression levels of microRNA-3196 in gastric cancer tissues and adjacent normal tissues were detected by quantitative reverse transcription-polymerase chain reaction. In this study, quantitative reverse transcription-polymerase chain reaction, cell proliferation assay, and Transwell migration and invasion assays were performed to explore microRNA-3196 expression level and its effects on cell proliferation, migration, and invasion in gastric cancer cells. The Kaplan-Meier method and multivariate Cox regression analyses were used to explore the prognostic significance of microRNA-3196 in gastric cancer. Dual-luciferase report assay was performed to validate the potential target gene regulated by microRNA-3196 in gastric cancer.
Results:
The expression of microRNA-3196 was downregulated in gastric cancer tissues and cell lines. Downregulation of microRNA-3196 was associated with lymph node metastasis and Tumor Node Metastasis (TNM) stage. The Kaplan-Meier curve analysis indicated that patients with low expression of microRNA-3196 had a poor prognosis, and the Cox regression analysis results showed microRNA-3196 expression was an independent prognostic factor of gastric cancer. Moreover, overexpression of microRNA-3196 inhibited cell proliferation, migration, and invasion, while knockdown of microRNA-3196 promoted these cellular behaviors in AGS and MKN45 cells. OTX1 may be a potential target gene regulated by microRNA-3196 in gastric cancer.
Conclusions:
These results suggested that microRNA-3196 might not only a tumor suppressor in gastric cancer cells by modulating OTX1 but also might be an independent prognostic biomarker and therapeutic target of gastric cancer.
Keywords: gastric cancer, miR-3196, prognosis, proliferation, migration, invasion
Introduction
Gastric cancer is one of the most frequently occurring malignant tumors and remains the second leading cause of cancer-related death worldwide.1,2 Although the incidence of gastric cancer has a dropping trend globally, the incidence is still high in China, in which the incidence and mortality ranked second and third for all types of cancers, respectively.3 Currently, surgical resection is still the main choice for the treatment of gastric cancer combined with chemotherapy and radiotherapy.4 Although the early diagnosis and treatment of gastric cancer have been improved, the prognosis of gastric cancer is still very poor because of the lymphatic metastasis and invasion.5,6
MicroRNAs (miRNAs or miRs) are short small noncoding RNA molecules and regulate gene expression via binding the 3′-untranslated region (3′-UTR) of the target genes at the posttranscriptional level, leading to the translation inhibition or degradation of messenger RNAs.7,8 It reported that miRNAs play a pivotal role in the initiation and progression of human tumors and may serve as diagnostic or prognostic biomarkers.9-11 Importantly, aberrant expression of various miRNAs has been demonstrated in various types of cancers and involved in multiple cellular processes, including metabolism, proliferation, differentiation, migration, invasion, and apoptosis.12,13 Many miRNAs involved in the progression of gastric cancer have been reported in recent years, such as miR-150-5p,14 miR-1284,15 and miR-524-5p.16 Therefore, identify more effective cancer-related biomarkers for the treatment and prognosis of gastric cancer is important. In recent years, studies have found that miR-3196 is downregulated in several cancers, such as papillary thyroid carcinoma17 and basal cell carcinoma.18 A miRNA profile study of gastric cancer reported various abnormal expression of miRNAs in gastric cancer tissues, including miR-3196.19 However, the potential role of miR-3196 in gastric cancer has not been explored.
In the present study, we investigated the expression of miR-3196 and its clinicopathological implication, as well as prognostic value in gastric cancer. We further explored the function of miR-3196 in gastric cancer in vitro.
Materials and Methods
Patients and Tissues Collection
A number of 127 gastric cancer tissues and adjacent normal tissues were collected from the patients who underwent gastrectomy at Jining Hospital of Traditional Chinese Medicine from January 2010 and December 2013. None of the patients had received preoperative therapies, including chemotherapy and radiotherapy. The tissue samples were all pathologically confirmed and promptly frozen in liquid nitrogen until use. This study was approved by the Jining Hospital of Traditional Chinese Medicine Ethical Committee (approval no. JNZYY200906). All patients provided written informed consent prior to enrollment in the study. The clinical characteristics of all patients are collected and recorded in Table 1. The 5-year follow-up overall survival information was also recorded for survival analysis.
Table 1.
Relationship Between MiR-3196 Expression and Clinical Characteristics in Patients With Gastric Cancer.
| Parameters | Cases (n = 127) | miR-3196 expression | P | |
|---|---|---|---|---|
| Low (n = 67) | High (n = 60) | |||
| Gender | .675 | |||
| Male | 78 | 40 | 38 | |
| Female | 49 | 27 | 22 | |
| Age | .199 | |||
| <60 | 58 | 27 | 31 | |
| ≥60 | 69 | 40 | 29 | |
| Tumor size (cm) | .078 | |||
| <5 | 70 | 32 | 38 | |
| ≥5 | 57 | 35 | 22 | |
| Differentiation | .691 | |||
| Well moderate | 76 | 39 | 37 | |
| Poor | 51 | 28 | 23 | |
| Lymph node metastasis | .029 | |||
| Negative | 74 | 33 | 41 | |
| Positive | 53 | 34 | 19 | |
| TNM stage | .005 | |||
| I-II | 66 | 27 | 39 | |
| III-IV | 61 | 40 | 21 | |
Abbreviation: miRNA, microRNA; TNM, Tumor Node Metastasis.
Cell Culture and Transfection
The gastric cancer cell lines, HGC-27, MKN45, AGS, and NCI-N87, as well as normal human gastric mucosa cells GES-1 were purchased from the Cell Resource Center of the Chinese Academy of Sciences. All cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) at 37 °C and 5% CO2.
The miR-3196 mimic (5′-CGGGGCGGCAGGGGCCUC-3′), mimic negative control (mimic NC; 5′-UUUGUACUACACAAAAGUACUG-3′), miR-3196 inhibitor (5′-GAGGCCCCUGCCGCCCCG-3′), and inhibitor NC (5′-UCUACUCUUUCUAGGAGGUUGUGA-3′) were synthesized by GenePharma. And mimics, mimic NC, inhibitors, or inhibitor NC at a final concentration of 50 nM were transfected into gastric cancer cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc) according to the manufacturer’s instructions. miR-3196 mimic was used to upregulate miR-3196 expression, while miR-3196 inhibitor was used to downregulate miR-3196 expression in gastric cancer cells. Cells treated with transfection reagent were used as control. Transfection efficiency was assessed 24 hours after transfection.
RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA was isolated from tissue samples and cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA was then reverse transcribed using the miScript Reverse Transcription kit (QIAGEN). The quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was carried out on the ABI7500 Real-Time PCR System (Applied Biosystems) using Green PCR Mixture (Applied Biosystems) to estimate the expression of miR-3196. The relative expression of miR-3196 was calculated using the 2−ΔΔCt method and normalized to U6.
Cell Counting Kit-8 Assay
A total of 3000 cells transfected with miR-3196 mimic, miR-3196 inhibitor, or corresponding NCs were seeded into 96-well plates. Cell counting kit-8 (CCK-8; Dojindo) was added to the plates after being cultivated for 0, 24, 48, 72 hours. After 2 hours further incubation at 37 °C, the cell proliferation was evaluated by determining the absorbance at 450 nm with the microplate reader (BioRad Laboratories, Inc).
Transwell Assays
To investigate cell migratory or invasive ability of gastric cancer cells, Transwell assays were performed using Transwell chamber without or precoated with Matrigel (BD Bioscience), respectively. In brief, cell suspension with 5 × 104 transfected cells in the serum-free medium was seeded into the upper chamber, while the lower chamber was filled with complete medium supplemented with 10% FBS as a chemoattractant. After incubated for 16 hours, remanent cells on the upper surface of the membrane were cleared, while the migration and invasive cells on the bottom surface of the membrane were fixed and stained. Then, the number of these cells was counted in 5 random fields under a light microscope.
Dual-Luciferase Reporter Assay
The Targetscan (www.Targetscan.org) and miRDB (www.mirdb.org) algorithms were used to predict the putative target genes of miR-3196. MicroRNA-3196 was predicted to have binding sites of the 3′-UTR of OTX1. And the dual-luciferase reporter was used to explore the findings. The wild-type (Wt) and mutated (Mut) putative miR-3196-binding sites in the 3′-UTR region of OTX1 were cloned into the pmirGLO Dual-Luciferase miRNA target expression vector for dual-luciferase reporter assay (Promega Corporation). According to the protocol, the MKN45 cells were cotransfected with the miR-3196 mimics, mimic NC, miR-3196 inhibitors, or inhibitor NC and the Wt or Mut OTX1 3′-UTR-luciferase reporter plasmids using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc) according to the manufacturer’s instructions. The activities of firefly luciferase were detected using a dual-luciferase reporter assay system (Promega) that was normalized to the Renilla luciferase activities.
Statistical Analysis
Data were presented as mean ± standard deviation and analyzed with the statistical software SPSS 20.0 (IBM SPSS Inc) and GraphPad Prism 5.0 (GraphPad Software, Inc). Comparisons were determined for 2 groups using paired Student t test and for multiple comparisons using 1-way analysis of variance followed by Tukey post hoc test. The association between miR-3196 expression and clinicopathological factors was analyzed using the χ2 test. The prognostic value of miR-3196 was evaluated using the Kaplan-Meier method and multivariate Cox analysis. The value of P less than .05 was considered to be statistically significant.
Results
MicroRNA-3196 Was Downregulated in Gastric Cancer Tissues and Cell Lines
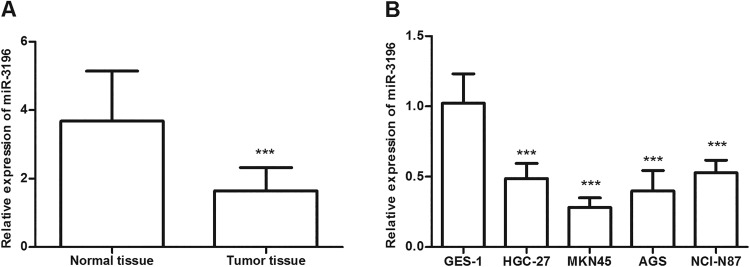
The expression of miR-3196 in gastric cancer tissues and cell lines was detected using qRT-PCR analysis. The results revealed that the miR-3196 level was significantly reduced in gastric cancer tissues in contrast to corresponding adjacent normal tissues (P < .001, Figure 1A). What’s more, a similar result was observed in gastric cancer cell lines, showing that the level of miR-3196 in gastric cancer cells was significantly lower than in normal human gastric mucosa cells GES-1 (P < .001, Figure 1B). Among these cell lines, MKN45 and AGS exhibited relatively lower miR-3196 expression levels, which were chosen for subsequent experiments.
Figure 1.
MicroRNA-3196 (miR-3196) expression levels in gastric cancer tissues and cell lines were detected using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. A, Relative expression levels of miR-3196 in gastric cancer tissues and normal tissues. B, The relative expression level of miR-3196 in cell lines. ***P < .001.
Expression of miR-3196 Is Associated With Clinical Characteristics of Patients With Gastric Cancer
Patients with gastric cancer were divided into a low miR-3196 expression group and a high miR-3196 expression group according to the mean miR-3196 expression level (1.641) in tissues. We further analyzed the association between miR-3196 expression and various clinical characteristics of patients with gastric cancer. We found that miR-3196 expression was associated with lymph node metastasis (P = .029) and Tumor Node Metastasis (TNM) stage (P = .005). Gender, age, tumor size, and differentiation were not associated with miR-3196 expression in patients with gastric cancer (P > .05; Table 1).
The Prognostic Value of miR-3196 in Gastric Cancer
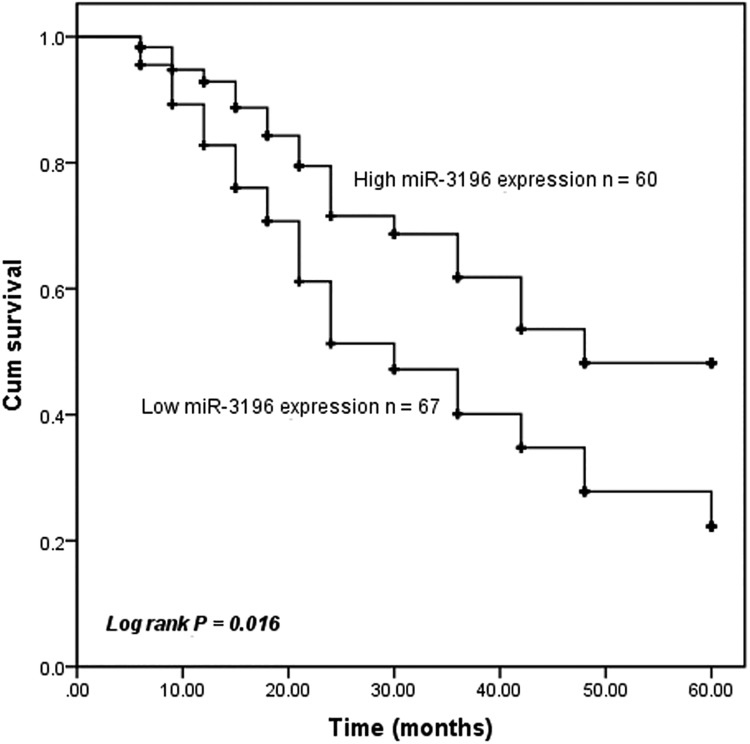
Then, we evaluated the effect of the miR-3196 expression on the overall survival of patients with gastric cancer using Kaplan-Meier curve methods. The results revealed that patients with low miR-3196 expression exhibited a shorter overall survival time compared with those with high miR-3196 expression (P = .016, Figure 2). Using multivariate Cox proportional hazard model analysis, advanced TNM stage and low miR-3196 expression were independent risk factors for overall survival (P < .05, Table 2). Collectively, these results suggested that miR-3196 may be a potential prognostic biomarker for patients with gastric cancer.
Figure 2.
Kaplan-Meier survival curves of microRNA-3196 (miR-3196) in the prognosis of patients with gastric cancer. Patients with low miR-3196 expression exhibited a shorter overall survival rate (log-rank P = .016). Cutoff point: mean value of the miR-3196 expression.
Table 2.
Multivariate Cox Regression Analysis for MiR-3196 in Patients With Gastric Cancer.
| Characteristics | Multivariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P | |
| miR-3196 | 1.884 | 1.046-3.394 | .035 |
| Gender | 1.452 | 0.754-2.797 | .264 |
| Age | 1.347 | 0.786-2.308 | .278 |
| Tumor size | 0.586 | 0.332-1.036 | .066 |
| Differentiation | 0.570 | 0.306-1.063 | .077 |
| Lymph node metastasis | 1.676 | 0.949-2.957 | .075 |
| TNM stage | 1.852 | 1.031-3.329 | .039 |
Abbreviation: miRNA, microRNA; TNM, Tumor Node Metastasis.
Effect of miR-3196 on Cell Proliferation, Migration, and Invasion in Gastric Cancer Cells
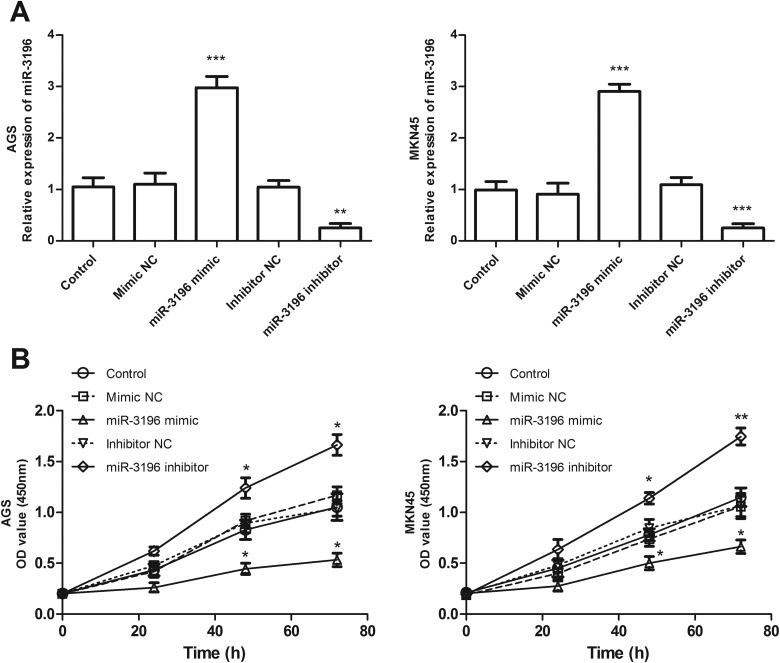
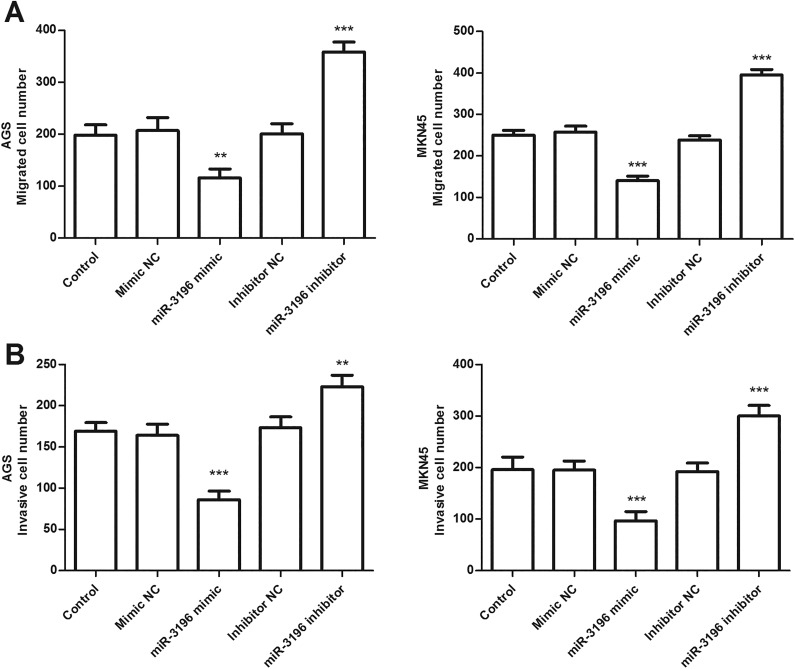
To evaluate the effect of miR-3196 on cell proliferation, migration, and invasion, miR-3196 mimic, mimic NC, miR-3196 inhibitor, or inhibitor NC was, respectively, transfected into AGS and MKN45 cells to achieve miR-3196 upregulation or downregulation. The qRT-PCR analysis confirmed the transfection efficiency, showing that miR-3196 expression was successfully increased or decreased by miR-3196 mimic or inhibitor in AGS and MKN45 cells (P < .01, Figure 3A). Then, we evaluated the effect of miR-3196 on AGS and MKN45 cell proliferative abilities using CCK-8 assays. As shown in Figure 3B, overexpression of miR-3196 inhibited the proliferation, while downregulation of miR-3196 accelerated the proliferation of AGS and MKN45 cells, compared with control (P < .05). Significant differences were observed at 48 and 72 hours (Figure 3B). The Transwell migration (Figure 4A) and invasion (Figure 4B) assays result exhibited that upregulation of miR-3196 apparently decreased the numbers of migrated and invaded cells, while downregulation of miR-3196 increased the numbers of migrated and invaded cells, compared with the corresponding control (P < .01). These data suggested that miR-3196 may function as a suppressor gene in gastric cancer in vitro.
Figure 3.
The role of microRNA-3196 (miR-3196) in cell proliferative function using AGS and MKN45 cells. A, Results of transfection efficacies of miR-3196 mimic or inhibitor. B, Analysis of cell proliferation using the cell counting kit-8 (CCK-8) assay. *P < .05, **P < .01, ***P < .001.
Figure 4.
MicroRNA-3196 (miR-3196) inhibited gastric cancer cell migration and invasion of AGS and MKN 45 cells. Transwell migration (A) and invasion (B) assay suggested that miR-3196 overexpression inhibited cell migration and invasion, while miR-3196 knockdown promoted cell migration and invasion, compared with control. ** P < .01, ***P < .001.
OTX1 May be a Target Gene Regulated by miR-3196
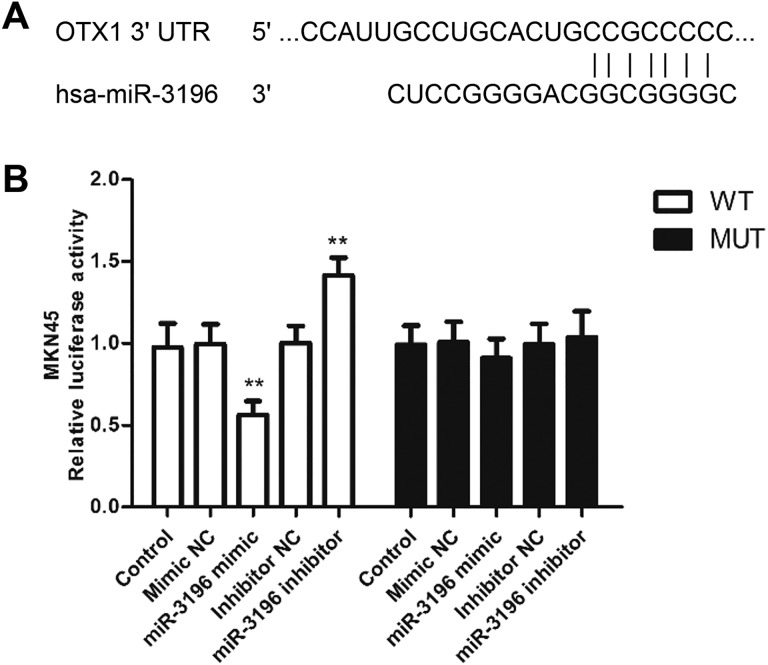
To explore the biological mechanisms underlying the effects of miR-3196 on the proliferation, migration, and invasion of gastric cancer cells, we investigated the potential targets of miR-3196. The OTX1 was identified as a target gene and had a miR-3196 binding site in the 3′-UTR, as shown in Figure 5A. To validate this interaction, a dual-luciferase reporter assay was conducted. Results revealed that the relative luciferase activity in MKN45 cells transfected with miR-3196 mimics and Wt-OTX1 was significantly decreased but that was increased in MKN45 cells transfected with miR-3196 inhibitors and Wt-OTX1, when compared with control (P < .01, Figure 5B). Inversely, the luciferase activity of the Mut group could not be affected by miR-3196 expression level (Figure 5B). Above all, OTX1 may be a direct target gene regulated by miR-3196 in gastric cancer.
Figure 5.
Validation of OTX1 as a direct target of microRNA-3196 (miR-3196). A, The binding site of miR-3196 and OTX1-3′-UTR was predicted by TargetScan and miRDB database. B, The luciferase activity of MKN45 cells was examined by a dual-luciferase reporter assay. **P < .01. UTR, untranslated region.
Discussion
Gastric cancer is one of the most common malignant tumors in the digestive system with high mortality. Following the continuous improvement in surgery, radiotherapy, and chemotherapy, as well as the implementation of new adjuvant therapy, the 5-year overall survival has improved but still unsatisfactory.20,21 Increasing evidence has indicated that miRNAs may play a crucial role in the initiation and development of gastric cancer.22-24 Thus, exploring cancer-related molecular markers associated with the development of gastric cancer is essential for the treatment of cancer.
MicroRNA-3196, a novel miRNA, has been reported to be aberrantly expressed in several human cancers. For instance, miR-3196 is downregulated in basal cell carcinoma.18 In patients with breast cancer with lymph node metastasis, miR-3196 is also one of the downregulated miRNAs.25 Circulating miR-3196 is also found downregulated in patients with lung adenocarcinoma with EGFR exon 19 deletion compared with wild type.26 Moreover, miR-3196 is reported to be downregulated in gastric cardia adenocarcinoma using miRNA expression profiles and is associated with differentiation.27 Consistently, we observed that miR-3196 was significantly downregulated in gastric cancer tissues and cell lines in the present study. In addition, the downregulation of miR-3196 was associated with positive lymph node metastasis and advanced TNM stages. These results suggested that miR-3196 may act as a tumor suppressor gene in gastric cancer.
MicroRNAs have obtained much attention in cancer research. Numerous studies have identified substantial miRNAs may function as diagnostic or prognostic biomarkers in gastric cancer.22,28,29 For instance, the miR-25 level was significantly upregulated in patients with gastric cancer, and serum levels of miR-25 could improve gastric cancer screening, and as the diagnostic and prognostic biomarker of gastric cancer.28 In the present study, results of the Kaplan-Meier survival curve revealed that the patients with gastric cancer with high miR-3196 expression levels would experience a relatively better overall survival. In addition, multivariate Cox regression analysis results showed that miR-3196 expression was an independent prognostic factor for gastric cancer. These data suggested that miR-3196 may be an independent prognostic biomarker for gastric cancer. Furthermore, we also observed that overexpression of miR-3196 inhibited cell proliferation, migration, and invasion, while knockdown of miR-3196 promoted these cellular behaviors in gastric cancer cells.
The functional role of miR-3196 in tumor progression has been demonstrated in several cancers. A study by Xu et al showed that miR-3196 was inhibited by H2AX phosphorylation and inhibited VP-16 induced lung cancer cell apoptosis by downregulating p53 upregulated modulator of apoptosis (PUMA).30 Another study in breast cancer provided miR-3196 was downregulated in breast cancer tissues, and overexpression of miR-3196 could suppress cell proliferation and induce cell apoptosis through targeting ERBB3, suggesting miR-3196 could serve as a potential biomarker and therapeutic target for breast cancer.31 A recent study showed ADPGK-AS1 acted as a miR-3196 sponge to release OTX1 in breast cancer cells, and high expression of ADPGK-AS1 predicted poor prognosis for breast cancer and promoted tumor cell proliferation, migration, induced epithelial–mesenchymal transition process, and suppressed cell apoptosis.32 In our present study, we observed that miR-3196 was downregulated in patients with gastric cancer and involved in the progression of gastric cancer. Moreover, in this study, miR-3196 could interact with OTX1 through predicted binding sites. And luciferase reporter assays revealed that miR-3196 directly targeted the 3′-UTR of OTX1. Interestingly, OTX1 was reported to be highly expressed in gastric cancer tissues and knockdown of OTX1 inhibited gastric cancer cell proliferation, migration, and invasion.33 According to the previous evidence and our present results, we speculate that miR-3196 functions as a suppressor gene in gastric cancer and may suppress tumor cell proliferation, migration, and invasion by regulating the expression of OTX1.
In conclusion, the results of the present study suggested that miR-3196 was downregulated in gastric cancer tissues and cells. OTX1 may be a direct target of miR-3196. Overexpression of miR-3196 may suppress cell proliferation, migration, and invasion by targeting OTX1. Additionally, a low level of miR-3196 predicted poor prognosis for patients with gastric cancer. These results indicated that miR-3196 may act as a novel prognostic biomarker and therapeutic target in the treatment of gastric cancer. More detailed investigation will carry out to further explore the precise molecular mechanism of miR-3196 in gastric cancer.
Acknowledgments
The authors deeply thank Ying Li, Yusong Liu, and Jie Yao (all from Department of Oncology, Jining Hospital of Traditional Chinese Medicine), for their assistance with the experiments.
Abbreviations
- CCK-8
cell counting kit-8
- FBS
fetal bovine serum
- miRNA
microRNAs
- Mut
mutated
- NC
negative control
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- Wt
wild-type
- 3′-UTR
3′-untranslated region
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by Binzhou Medical University Hospital (BY2015KJ25).
ORCID iD: Xiaocheng Fan  https://orcid.org/0000-0003-0852-2260
https://orcid.org/0000-0003-0852-2260
References
- 1. Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 4. Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626. [DOI] [PubMed] [Google Scholar]
- 5. Min C, Zhang A, Qin J. Increased expression of miR-601 is associated with poor prognosis and tumor progression of gastric cancer. Diagn Pathol. 2019;14(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bu Z, Zheng Z, Li Z, et al. Lymphatic vascular invasion is an independent correlated factor for lymph node metastasis and the prognosis of resectable T2 gastric cancer patients. Tumour Biol. 2013;34(2):1005–1012. [DOI] [PubMed] [Google Scholar]
- 7. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379. [DOI] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 9. Luo H, Xu R, Chen B, et al. MicroRNA-940 inhibits glioma cells proliferation and cell cycle progression by targeting CKS1. Am J Transl Res. 2019;11(8):4851–4865. [PMC free article] [PubMed] [Google Scholar]
- 10. Plum PS, Warnecke-Eberz U, Drebber U, et al. Upregulation of miR-17-92 cluster is associated with progression and lymph node metastasis in oesophageal adenocarcinoma. Sci Rep. 2019;9(1):12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y, Huang H, Zhang Y, Liao N. Combined detection of serum MiR-221-3p and MiR-122-5p expression in diagnosis and prognosis of gastric cancer. J Gastric Cancer. 2019;19(3):315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kapora E, Feng S, Liu W, et al. MicroRNA-505-5p functions as a tumor suppressor by targeting cyclin-dependent kinase 5 in cervical cancer. Biosci Rep. 2019;39(7):6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Dan B, Luo J, Li K, Chen S. Prognostic value of miR-375 for survival outcomes in various cancers: a systematic review and meta-analysis. Oncol Res Treat. 2018;41(1-2):47–50. [DOI] [PubMed] [Google Scholar]
- 14. Quan X, Chen D, Li M, Chen X, Huang M. MicroRNA-150-5p and SRC kinase signaling inhibitor 1 involvement in the pathological development of gastric cancer. Exp Ther Med. 2019;18(4):2667–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei W, Cao W, Zhan Z, Linhai Y, Yubo X, Qiang X. MiR-1284 suppresses gastric cancer progression by targeting EIF4A1. Onco Targets Ther. 2019;12:3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu CY, Meng FQ, Liu J. MicroRNA-524-5p suppresses cell proliferation and promotes cell apoptosis in gastric cancer by regulating CASP3. Eur Rev Med Pharmacol Sci. 2019;23(18):7968–7977. [DOI] [PubMed] [Google Scholar]
- 17. Qiu ZL, Shen CT, Song HJ, Wei WJ, Luo QY. Differential expression profiling of circulation microRNAs in PTC patients with non-131I and 131I-avid lungs metastases: a pilot study. Nucl Med Biol. 2015;42(5):499–504. [DOI] [PubMed] [Google Scholar]
- 18. Sand M, Skrygan M, Sand D, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167(4):847–855. [DOI] [PubMed] [Google Scholar]
- 19. Liu D, Hu X, Zhou H, Shi G, Wu J. Identification of aberrantly expressed miRNAs in gastric cancer. Gastroenterol Res Pract. 2014;2014:473817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milano AF. 20-year comparative survival and mortality of cancer of the stomach by age, sex, race, stage, grade, cohort entry time-period, disease duration & selected ICD-O-3 oncologic phenotypes: a systematic review of 157,258 cases for diagnosis years 1973-2014: (SEER*Stat 8.3.4). J Insur Med. 2019;48(1): 5–23. [DOI] [PubMed] [Google Scholar]
- 22. Li W, Cui X, Qi A, Lihui Y, Tong W, Bin L. miR-183-5p acts as a potential prognostic biomarker in gastric cancer and regulates cell functions by modulating EEF2. Pathol Res Pract. 2019;215(11): 152636. [DOI] [PubMed] [Google Scholar]
- 23. Su H, Ren F, Jiang H, Chen Y, Fan X. Upregulation of microRNA-520a-3p inhibits the proliferation, migration and invasion via spindle and kinetochore associated 2 in gastric cancer. Oncol Lett. 2019;18(3):3323–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, He C, Wang X, et al. MicroRNA-183 affects the development of gastric cancer by regulating autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR signals. Artif Cells Nanomed Biotechnol. 2019;47(1):3163–3171. [DOI] [PubMed] [Google Scholar]
- 25. Wang B, Li J, Sun M, Sun L, Zhang X. miRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life. 2014;66(5):371–377. [DOI] [PubMed] [Google Scholar]
- 26. Ju L, Han M, Li X, Zhao C. MicroRNA signature of lung adenocarcinoma with EGFR Exon 19 deletion. J Cancer. 2017;8(7):1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao S, Zhou F, Zhao C, et al. Gastric cardia adenocarcinoma microRNA profiling in Chinese patients. Tumour Biol. 2016;37(7):9411–9422. [DOI] [PubMed] [Google Scholar]
- 28. Kong Y, Ning L, Qiu F, Yu Q, Cao B. Clinical significance of serum miR-25 as a diagnostic and prognostic biomarker in human gastric cancer. Cancer Biomark. 2019;24(4):477–483. [DOI] [PubMed] [Google Scholar]
- 29. Huang ZS, Guo XW, Zhang G, Liang LX, Nong B. The diagnostic and prognostic value of miR-200c in gastric cancer: a meta-analysis. Dis Markers. 2019;2019:8949618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu C, Zhang L, Duan L, Lu C. MicroRNA-3196 is inhibited by H2AX phosphorylation and attenuates lung cancer cell apoptosis by downregulating PUMA. Oncotarget. 2016;7(47):77764–77776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ji ZC, Han SH, Xing YF. Overexpression of miR-3196 suppresses cell proliferation and induces cell apoptosis through targeting ERBB3 in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8383–8390. [DOI] [PubMed] [Google Scholar]
- 32. Yang J, Wu W, Wu M, Ding J. Long noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and EMT process through regulating miR-3196/OTX1 axis in breast cancer. In Vitro Cell Dev Biol Anim. 2019;55(7):522–532. [DOI] [PubMed] [Google Scholar]
- 33. Qin SC, Zhao Z, Sheng JX, et al. Dowregulation of OTX1 attenuates gastric cancer cell proliferation, migration and invasion. Oncol Rep. 2018;40(4):1907–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]