Abstract
Several cytochrome P450 (CYP) CYP3A polymorphisms were associated with reduced enzyme function. We aimed to evaluate the influence of these alleles on the pharmacokinetic parameters (PK) of several CYP3A substrates. We included 251 healthy volunteers who received a single dose of ambrisentan, atorvastatin, imatinib, aripiprazole, fentanyl, amlodipine, donepezil, olanzapine, fesoterodine, or quetiapine. The volunteers were genotyped for CYP3A4 and CYP3A5 polymorphisms by qPCR. To compare the PK across studies, measurements were corrected by the mean of each parameter for every drug and were logarithmically transformed. Neither CYP3A phenotype nor individual CYP3A4 or CYP3A5 polymorphisms were significantly associated with differences in PK. However, regarding the substrates that are exclusively metabolized by CYP3A, we observed a higher normalized AUC (p = 0.099) and a tendency of lower normalized Cl (p = 0.069) in CYP3A4 mutated allele carriers what was associated with diminished drug metabolism capacity. CYP3A4 polymorphisms did not show a pronounced influence on PK of the analysed drugs. If so, their impact could be detectable in a very small percentage of subjects. Although there are few subjects carrying CYP3A4 double mutations, the effect in those might be relevant, especially due to the majority of subjects lacking the CYP3A5 enzyme. In heterozygous subjects, the consequence might be less noticeable due to the high inducible potential of the CYP3A4 enzyme.
Keywords: CYP3A4, CYP3A5, pharmacokinetics
1. Introduction
Precision medicine uses the tools provided by molecular biology to improve the diagnosis and expand the treatment options of patients based on their biological and molecular profile. Pharmacogenetics, the study of genetic markers that influence the inter-individual variation in drug response, is one of the cornerstones of personalized medicine [1,2]. The role of heritable genetic variation in drug response has been studied since the 1950s [3]. In 1959, Vogel coined the term pharmacogenetics to define a new field that aimed to study the influence of inherited factors on drug response variability through genetic and pharmacological knowledge and methods [4].
Several genes are related to drug response. The U.S. Food and Drug Administration (FDA) describes four gene categories in drug development that can affect the benefit-risk profile of a certain drug: (1) genes relevant to the pharmacokinetics of the drug, (2) genes related to the pharmacological effect of the drug, (3) genes not directly related to the pharmacological effect of the drug, but may predispose to toxicities, and (4) genes that influence the susceptibility or progression of the disease [5]. To date, the most important genes that are related to the pharmacokinetics (PK) of drugs encode cytochrome P450 superfamily (CYP) enzymes.
At least 57 active CYP genes are present in the human genome, along with approximately the same number of pseudogenes. CYP enzymes are divided into 18 families and 44 subfamilies. CYP 1–3 families are particularly active in the detoxification of exogenous chemicals, such as drugs [6]. Every CYP protein sequence is designated as CYP (superfamily root symbol), followed by a number (gene family), then a capital letter (subfamily), and another number (protein). Family members should share >40%, while subfamily members should share >55% amino acid sequence identity [7].
CYP enzymes are responsible for about 75–80% of phase I drug metabolism [8]. Genetic polymorphisms in CYP enzymes which are associated with a characteristic effect in drug metabolism are translated into different metabolizer phenotypes: ultra-rapid metabolizers (UMs), normal metabolizers (NMs), intermediate metabolizers (IMs) and poor metabolizers (PMs). However, these categories give only a static idea about the functionality of an enzyme in an individual. In addition to genetics, variability in CYP activity may be explained by other factors, such as age, sex, morbidity, co-medication, food, or smoking [9].
1.1. CYP3A Subfamily
The human CYP3A subfamily consists of 4 isoforms, CYP3A4, CYP3A5, CYP3A7, and CYP3A43 [10]. These isoforms are present in variable proportion in the human liver, being about 30% of the total CYP enzyme content [11]. Genes encoding for CYP3A enzymes are located on the 7q21–q22.1 chromosome band [12] along with CYP3AP1 and CYP3AP2 pseudogenes, in a cluster of about 200kb [13]. All four CYP3A enzymes comprise 13 exons [14] and show high structural similarity (between 71.5% and 84.1%) [15].
The 5′ untranslated region (UTR) of each human CYP3A gene consists of an average of 101 nucleotides, which is below the mean human 5’-UTR length of 150 nucleotides. On the contrary, the size of the 3′-UTR is highly variable, being 111 nucleotides for CYP3A5, 463 for CYP3A7, 549 for CYP3A43 and 1152 for CYP3A4 [16]. The role of UTRs lies in the regulation of gene expression. Therefore, as CYP3A4 has the longest 3′UTR, it may be greatly regulated [17].
CYP3A43 is generally expressed at low levels in prostate, liver, kidney, and pancreas [10]. It is mainly involved in the metabolism of endogenous compounds, but not drugs [18].
CYP3A7 represents approximately 30–50% of the total CYP450 enzyme amount expressed in fetal liver, where CYP3A4 is not expressed [19]. CYP3A7 was detected also in placenta and endometrium. CYP3A7 is rarely present in adult livers at significant levels as it is gradually substituted by CYP3A4 after birth [18].
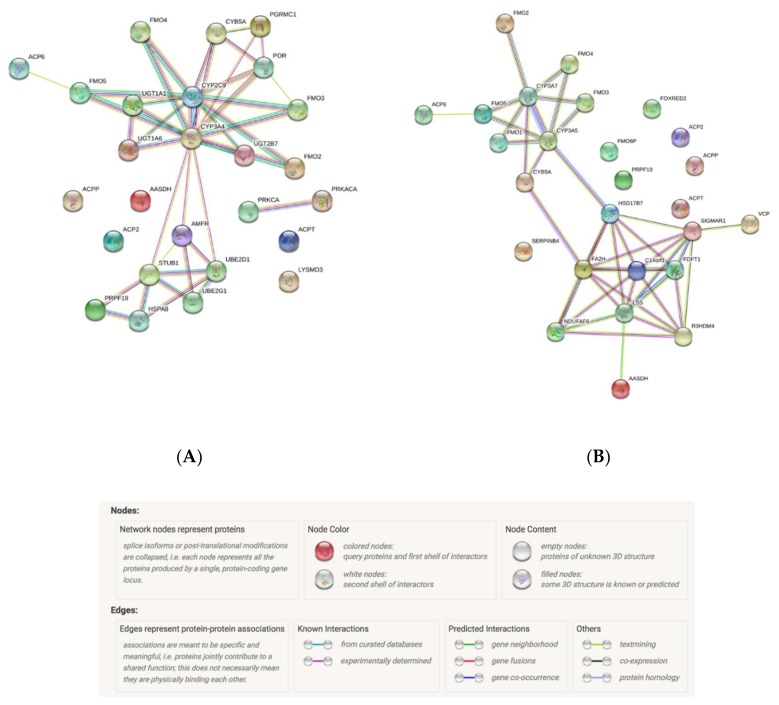
Enzyme activity and functional protein interactions are key to understanding the cellular machinery. In order to fully understand every biological phenomena, it is of importance to study the connectivity of the whole network [20]. CYP3A enzymes are involved in steroid hormone biosynthesis and in lipid and fatty acid metabolism pathways, while interacting with the main molecules described in Figure 1.
Figure 1.
Top CYP3A4 (A) and CYP3A5 (B) protein interactants. Data obtained from: STRING Interaction Network. Szklarczyk et al. Nucleic acids research 47.D1 (2018): D607-D613.2.
1.2. CYP3A4 and CYP3A5
Among the human CYP3A enzymes, CYP3A4 and CYP3A5 are considered the most important in drug metabolism [21,22]. Both are highly abundant in the adult liver and intestine [11,23].
In 1984, Kleinbloesem et al. reported a bimodal frequency distribution of the area under the concentration-time curve (AUC) of nifedipine and suggested that a polymorphism was related to its disposition kinetics [24]. Guengerich in 1985 identified and purified the enzyme responsible for the oxidation of nifedipine from rat and human livers, which he named P450 Nifedipine Oxidase. This enzyme is known as CYP3A4 today [25]. Since then, several studies aimed to characterize this enzyme more precisely.
CYP3A4 gene is located on the 7q22.1 band of the human karyotype [26] with a length of nucleotides [27]. Previously another CYP3A gene, CYP3A3, was thought to exist; however, it seems that it is a transcript variant of CYP3A4. To date, 8 splice variants encoding different isoforms have been identified. Six of these contain an Open Reading Frame (ORF) and code proteins with the size range between 153 and 534 amino acids [28].
CYP3A4 is expressed predominantly in the liver, where it is the most expressed CYP enzyme [29], ranging from 14.5% to 37% of the hepatic P450 pool [6]. Overall, CYP3A5 is expressed at lower levels than CYP3A4 (~10% of CYP3A4) [30]. Moreover, CYP3A4 is the most abundant CYP enzyme in the human intestinal epithelial cells [31]. Intestinal CYP3A4 plays a role in the bioavailability of some drugs, such as midazolam and cyclosporine via first-pass metabolism during liver transplantation [32,33].
One of the CYP3A4 most commonly studied reduced-function single nucleotide polymorphism (SNP) is *20, which gives rise to a truncated protein with complete loss of function of the enzymatic activity [34]. Similarly, CYP3A4*22 results in a reduced mRNA expression and a low CYP3A4 enzyme activity [17,35,36].
A previous study characterised CYP3A4 as a highly polymorphic enzyme as CYP3A4*20 allele was present in 1.2% of the Spanish population and up to 3.8% in specific regions [37]. This study described the CYP3A4*20 no function variant with such a high frequency for the first time [37].
Regarding the CYP3A4*22 variant, it is of especial interest to find relevant associations with several drugs, such as tacrolimus [38,39,40,41,42,43]. Briefly, it was linked to lower clearance, and therefore lower dose requirements. Moreover, CYP3A4*22 was also associated with a greater reduction in total and LDL cholesterol levels in an observational study with simvastatin users from a Caucasian cohort [44].
Another relevant variant which was predominantly studied in Caucasians is CYP3A4*1B (rs2740574G), a single base change in the 5’-UTR of CYP3A4. At first, this variant was studied related to prostate tumors [45,46]. It was associated with lower enzyme activity and reduced binding of nuclear proteins in vitro [47]. However, other studies contradicted these results [46,48]. In PK studies, CYP3A4*1B was associated to decreased dose-adjusted trough concentrations (C0/D) and higher dose requirements of tacrolimus [38,41,42,49,50,51], and cyclosporine [52,53,54] in transplanted patients. It was also associated with a lower risk of dose decrease or switching therapy during simvastatin treatment [55]. Usually when studied together with CYP3A5*3 or CYP3A4*22, it loses relevance against them [39,41,50]. On the other hand, a significant linkage disequilibrium between this variant and CYP3A5*1 was described, which could partly explain the underlying cause of the observed associations with CYP3A4*1B [56].
CYP3A4*1G variant is located in intron 10 of the CYP3A4 gene [57] and was predominantly studied in Asian populations [58]. In some studies, CYP3A4*1G carriers showed an increased clearance and a decreased C0/D of tacrolimus [59,60,61,62,63] and cyclosporine treatment [64,65]. CYP3A4*1G was associated with less dose requirements of fentanyl by patient-controlled analgesia (PCA) in the treatment of postsurgical pain [66,67,68,69,70]. However, further evidence is needed from independent studies to confirm these associations.
CYP3A4*2, CYP3A4*12, and CYP3A4*17 alleles have a very low frequency and their functional impact is not established clearly [15,57,71].
Finally, the CYP3A4*3 variant (rs4986910) is a missense polymorphism [71] with very little clinical evidence. In a study with 2735 individuals on statin therapy, CYP3A4*3 was associated with an increase in HDL-cholesterol levels with fluvastatin treatment [72].
To date, eight splice variants of CYP3A5 have been described. Three of these encode proteins with the size range between 1720 and 4473 amino acids [73]. The presence of the SNP 6986A>G (rs776746) in intron 3 defines CYP3A5*3 and results in a non-functional CYP3A5 protein in homozygous carriers (CYP3A5*3/*3) [74]. This defective variant is the most frequent among Caucasians and Asians. As a result, CYP3A5 is expressed in approximately 10–25% of individuals, depending on their ethnicity (Table 1). When expressed, CYP3A5 can represent about 50% of the total CYP3A hepatic content, which is equal to CYP3A4 activity [75]. On the other hand, CYP3A4 and CYP3A5 usually share substrate specificity. Therefore, a combined phenotype including both enzymes were proposed for several drugs, such as midazolam [76], everolimus [77], statins [78], or tamoxifen [79].
Table 1.
Allele frequencies of the most studied variants of CYP3A4 and CYP3A5.
| CYP3A Allele | Reference SNP Identifier | In Vitro Effect | PK Effect in CYP3A Metabolism | Minor Allele Frequencies | |||
|---|---|---|---|---|---|---|---|
| Europeans | Latin-Americans | Africans | East Asians | ||||
| CYP3A4*22 | rs35599367 | ↓ [35,36] | ↓ | 50/1006 (4.97%) | 18/694 (2.59%) | 1/1322 (0.08%) | 0/1008 (0%) |
| CYP3A4*1B | rs2740574 | (= [46], ↓ [47]?, ↑ [48]?) | (↑?) [38], [41,42,49,50,51,52,53,54] | 28/1006 (2.78%) | 73/694 (10.52%) | 1012/1322 (76.55%) | 4/1008 (0.4%) |
| CYP3A4*1G | rs2242480 | (↑?) [59], [60,61,62,63,64,65] | 82/1006 (8.15%) | 273/694 (39.34%) | 1124/1322 (85.02%) | 270/1008 (26.79%) | |
| CYP3A4*3 | rs4986910 | (↓?) [72] | 7/1006 (0.70%) | 5/694 (0.72%) | 1/1322 (0.08%) | 0/1008 (0%) | |
| CYP3A4*20 | rs67666821 | None | ↓ | 26/64600 (0.04%)~ | 22/13290 (0.17%)~ | 3/42021 (0.01%)~ | 0/3134 (0.00%)~ |
| CYP3A5*3 | rs776746 | ↓ | ↓ | 949/1006 (94.33%) | 553/694 (79.68%) | 238/1322 (18.00%) | 719/1008 (71.33%) |
| CYP3A5*6 | rs10264272 | (↓?) [74] | (↓?) [39], [40] | 3/1006 (0.30%) | 16/694 (2.31%) | 204/1322 (15.43%) | 0/1008 (0.00%) |
| CYP3A5*7 | rs41303343 | =/↓ | (↓?) [39], [40] | 0/942 (0.00%)^ | 27/1090 (2.48%)^ | 174/2014 (8.64%)^ | 0/480 (0.00%)^ |
| CYP3A5*8 | rs55817950 | ↓ | 0/ 113648 (0.00%) | 0/ 34590 (0.00%) | 0/ 16216 (0.00%) | 0/ 18391 (0.00%) | |
| CYP3A5*9 | rs28383479 | ↓ | 0/113434(0.00%) | 0/34340(0.00%) | 0/16226(0.00%) | 2/18358 (0.01%) | |
Minor allele frequencies obtained from Ensemble Genome Browser. PK effect in CYP3A metabolism obtained from PharmVar or cited studies. ~ Data from gnomAD browser v3. ^Data from CPIC guideline for Tacrolimus and CYP3A5: https://cpicpgx.org/guidelines/guideline-for-tacrolimus-and-cyp3a5/. ? Not enough evidence.
Other variants that result in a non-functional CYP3A5 protein are CYP3A5*6 (rs10264272), which causes an alternative splicing of CYP3A5 [74] and CYP3A5*7 (rs41303343), an insertion that results in a frameshift [80]. Both are greatly frequent in Latin American and African populations (Table 1). In a study with 140 Latin-American adult kidney transplant recipients under tacrolimus treatment, CYP3A5 defective haplotypes (*3, *6, *7) showed an increased C0/D [39,40].
Moreover, CYP3A5*8 (rs55817950) and CYP3A5*9 (rs28383479) are defective coding alleles that showed decreased CYP3A5 activity in in vitro models [81]. However, to date, these SNPs have been rarely studied in vivo.
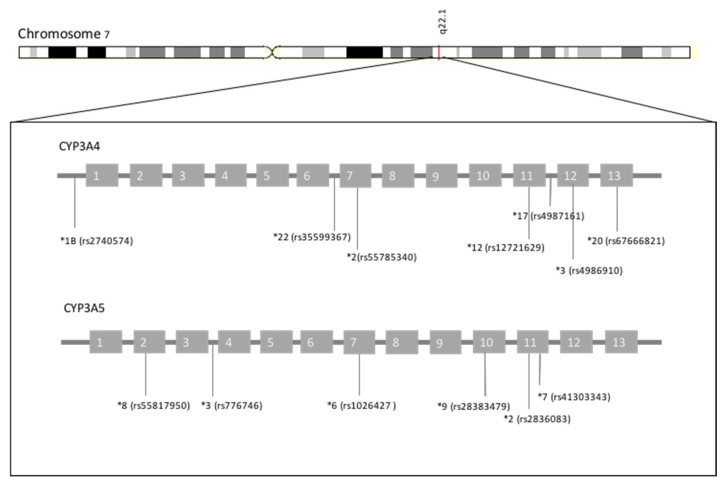
Figure 2 depicts the polymorphic context of CYP3A4 and CYP3A5, with the main polymorphisms described above.
Figure 2.
Polymorphic context of CYP3A4 and CYP3A5.
Tacrolimus is the only drug to which CYP3A-genotype guided recommendations are applied. In 2015, the Clinical Pharmacogenetics Implementation Consortium (CPIC) published the guideline for CYP3A5 genotype and tacrolimus dosing [82]. This association is well established; nevertheless, the variable frequency of CYP3A5*1 allele among different populations (Table 1) makes the utility of the genetic test variable.
In most of the studies that analyzed CYP3A4*22, CYP3A5*3 was also analyzed and in the majority of them the latter showed less associations [83,84]. This could be due to the different contribution of each gene to the metabolism of the different substrates, as it was demonstrated in vitro [85], or can alternatively be caused by a bias related to the low frequency of CYP3A5 expressers in some populations (Table 1).
CYP3A4 protein liver content and activity show an important inter-individual variation ranging from 10 to >100 fold [11,46,86]. In addition, CYP3A4 content was higher in female liver specimens compared to males [29,87]. Furthermore, a higher clearance of various CYP3A substrates was observed in females treated with different drugs, such as nifedipine [88], oxycodone [89], or cyclosporine [90]. CYP3A4 activity may also be affected by the phases of the menstrual cycle [91].
Drugs, herbals or alimentary products can also increase CYP3A4 metabolic activity by increasing enzyme expression. The mechanism of induction involves various nuclear receptors: pregnane X receptor (PXR), constitutive androstane receptor (CAR), glucocorticoid receptor (GR), vitamin D receptor (VDR), and hepatocyte nuclear factor—4 alpha (HNF4α). These nuclear receptors bind to DNA segments present in CYP3A promoter regions, such as PXR responsive element (prPXRE), xenobiotic-responsive enhancer module (XREM), or constitutive liver enhancer module (CLEM4), while increasing transcription and expression of CYP3A4 [6,92]. Some known CYP3A4 inducers are rifampicin, carbamazepine, phenytoin, phenobarbital, paclitaxel, cyclophosphamide, glucocorticoids, and herbs such as St. John’s Wort [92]. CYP3A4 is also susceptible to reversible and irreversible inhibition. Some of the most used drugs that inhibit CYP3A4 are macrolide antibiotics, protease inhibitors, anti-HIV agents, azole antifungals, fluoxetine, verapamil, diltiazem, and several herbal and dietary components [92]. Irreversible or mechanism-based inhibition is more relevant in in vivo drug interactions, specifically, nicotinamide adenine dinucleotide phosphate hydrogen (NADPH)-, time- and concentration-dependent [93].
Evidence of the effect of some CYP3A polymorphisms on the PK of some relevant drugs is scarce and often inconclusive; therefore, its inclusion in prescription guidelines is limited. Our aim was to verify and quantify the effect of the main CYP3A4 and CYP3A5 polymorphisms and a merged phenotype on the pharmacokinetic parameters of a large number of CYP3A substrates.
2. Materials and Methods
2.1. Study Population and Study Design
Our study population comprised 251 healthy volunteers who participated in one of 11 bioequivalence clinical trials (10 of them performed at the Clinical Trials Unit of Hospital Universitario de La Princesa, Madrid, Spain, and 1 at the Clinical Trials Unit of Universidad Autónoma de Madrid, Madrid, Spain) with CYP3A-substrates and agreed to participate in the pharmacogenetic study. The volunteers received a single oral dose of ambrisentan 10 mg (1 trial, n = 23), atorvastatin 80 mg (2 trials, n = 50), imatinib 400 mg (1 trial, n = 12), aripiprazole 10 mg (1 trial, n = 26), fentanyl 0.3 mg (1 trial, n = 35), amlodipine 10 mg (1 trial, n = 25), donepezil 10 mg (1 trial, n = 23), olanzapine 5 mg (1 trial, n = 25), fesoterodine 8 mg (1 trial, n = 13) or quetiapine 25 mg (1 trial, n = 19). Each study was a bioequivalence clinical trial in which a test formulation was compared to a reference formulation, after a single oral dose administration under fasting conditions.
All protocols met the terms of the current Spanish Legislation on human clinical research and were approved by the Research Ethics Committee, duly authorized by the Spanish Drugs Agency and under the guidelines of Good Clinical Practice. All participants signed a written informed consent for both the clinical trial and the pharmacogenetic study. Subjects were free to withdraw from the study at any time.
The inclusion criteria for each study were as follows: non-smoking male and female volunteers, age 18–55 years, body mass index within the 18.5–30.0 range, free from any organic or psychiatric conditions, with normal vital signs, electrocardiogram, medical records and physical examination.
Healthy volunteers were asked prior to study inclusion if they had taken any medications or supplements in the past month. Those volunteers who had taken any drugs or supplements were excluded from the clinical trial. Concomitant pharmacological treatments were not administered during none of the phases of the study. At the time of admission, the night prior to the administration of the study drug, volunteers were again asked if they had taken any concomitant medication during the time they had not been admitted to the Clinical Trials Unit.
2.2. Pharmacokinetic Analysis
Since the determination of every drug plasma concentration was outsourced, all samples were stored at −20 ± 5 °C or −75 ± 5 °C until their shipment to the external analytical laboratory. Each quantification was accomplished by a high-performance liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS) validated method, according to the European Medicines Agency guidelines. Pharmacokinetic parameters were calculated by means of a non-compartmental analysis with WinNonlin Professional software, Version 2.0 (Pharsight Corporation, Palo Alto, California), as described previously [94]. The AUC and the maximum concentration (Cmax) were corrected by dose/weight. Clearance (Cl) and volume of distribution (Vd) were divided by weight.
2.3. Genotyping
DNA was obtained from 1 mL of peripheral blood samples using MagNA Pure LC DNA Isolation Kit in an automatic DNA extractor (MagNa Pure® System, Roche Applied Science, Indianapolis, Indiana). CYP3A4*2 (rs55785340), CYP3A4*3 (rs4986910), CYP3A4*12 (rs12721629), CYP3A4*17 (rs4987161) and CYP3A4*22 (rs35599367) and CYP3A5*2 (rs28365083), CYP3A5*3(rs776746), CYP3A5*6 (rs10264272), CYP3A5*7 (rs41303343), CYP3A5*8 (rs55817950), and CYP3A5*9 (rs28383479) polymorphisms were genotyped by quantitative PCR (qPCR), in a QuantStudio 12k Flex instrument (Applied Biosystems, Foster City, CA, USA). We analysed these SNPs as they were the ones included in the commercial and predesigned TaqMan™ OpenArray™ PGx Express Panel (Applied Biosystems, Foster City, CA, USA).
The CYP3A4*20 (rs67666821) polymorphism call was assessed by KASPar SNP Genotyping System (LGC Genomics, Herts, UK) in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Darmstadt, Germany). All CYP3A4*20 carriers were confirmed by Sanger sequencing in an ABI PRISM 3700 DNA Analyser capillary sequencer (Applied Biosystems, Foster City, California, USA) [37].
The *1 allele, considered as wild-type, was assigned when the subject lacked all of the analyzed alleles.
2.4. Statistical Analysis
The influence of CYP3A4 and CYP3A5 genotypes were analysed separately. However, to assess the influence of both enzyme conjunctively, CYP3A4 and CYP3A5 genotypes were merged into a “CYP3A phenotype” as follows: subjects with at least one CYP3A4 reduced activity allele (i.e., CYP3A4 *1/*22, *1/*3, *22/*22 or *3/*22) and no CYP3A5 activity (CYP3A5 *3/*3,*3/*6 or *3/*7) were considered PM; subjects with normal CYP3A4 activity (CYP3A4 *1/*1) and no CYP3A5 activity (CYP3A5 *3/*3, *3/*6 or *3/*7) were considered IM; finally, subjects with normal CYP3A4 activity (CYP3A4*1/*1) and at least one CYP3A5 functional allele (CYP3A5 *1/*1 or *1/*3) were categorized as extensive metabolizers (EM), based on Sanchez Spitman et al. [79]
Statistical analyses were accomplished with the SPSS 22.0 software (SPSS Inc., Chicago, Illinois, USA). p values < 0.05 were considered statistically significant. Chi-square test was used to define sex-related differences in the genotypic frequencies. T-test or ANOVA were used to evaluate differences in PK among sexes and genotypes. To compare the PK parameters across studies, they were divided by the mean of each parameter for every drug and logarithmically transformed, thus termed as normalized parameter. The power analysis to know how many patients would be necessary to be evaluated in order to reveal a difference in PK parameters was performed with the G.power 3.1.9.7 software.
3. Results and Discussion
From all the volunteers included, 164 (65.3%) were Caucasians, 81 (32.3%) were Latin-Americans, four (1.6%) were Blacks, and two (0.8%) were Arabs. Regarding sex, 102 (40.6%) were women, while 149 (59.4%) were men. Mean age was 27.8 years old (ranging between 18 and 55). Mean weight was 69.6 kg (ranging between 44.4 and 98.5 kg) and mean height was 1.70 m (ranging between 1.49 and 1.97 m). Men were younger (26.5 ± 6.5 years vs. 29.8 ± 9.3 years), weighted more (76.0 ± 9.1 kg vs. 60.2 ± 8.1 kg) and were taller (1.76 ± 0.06 m vs. 1.61± 0.06 m) than women, p < 0.001.
Mean age was 25.8 ± 6.0, 32.0 ± 9.8, 32.0 ± 9.1 and 23.0 ± 0.0 years old (p < 0.001) in Caucasians, Latin-Americans, Blacks, and Arabs, respectively. Mean weight was 69.0 ± 11.3, 70.1 ± 12.1, 85.7 ± 5.2, 63.6 ± 17.0 kg (p = 0.033) in Caucasians, Latin-Americans, Blacks, and Arabs, respectively. Finally, mean height was 1.72 ± 0.08, 1.67 ± 0.11, 1.74 ± 0.06 and 1.70 ± 0.13 m (p = 0.001) in Caucasians, Latin-Americans, Blacks, and Arabs, respectively.
Table 2 shows the main CYP3A genotypes found in our study. None of the genotyped subjects carried CYP3A4*2, CYP3A4*12, CYP3A4*17, CYP3A5*2, CYP3A5*8, or CYP3A5*9 alleles. Our allele frequencies (AF) did not differ from those reported in the literature: CYP3A4*3 AF was 0.3% (Minor allele frequency, MAF: <1%) [95], CYP3A4*20 AF was 0.9% (MAF: <1%) [96], and CYP3A4*22 AF was 2.3% (Iberian MAF: 3.7%; Latin MAF: 2.6%) [97]. Additionally, CYP3A5*3 AF was 87.2% (Iberian MAF: 92.5%; Latin MAF: 79.7%) [98], CYP3A5*6 AF was 1.5% (Iberian MAF: 0.9%; Latin MAF: 2.3%) [99] and CYP3A5*7 AF was 0.2% (MAF: <1%) [100]. No statistically significant difference was found in CYP3A genotype frequencies among sexes.
Table 2.
CYP3A genotype and phenotype frequencies in our study population.
| Gene | Genotype | N | % |
| CYP3A4 | *1/*1 | 233 | 92.8 |
| *1/*20 | 5 | 2.0 | |
| *1/*22 | 11 | 4.4 | |
| *1/*3 | 1 | 0.4 | |
| *3/*22 | 1 | 0.4 | |
| CYP3A5 | *1/*1 | 5 | 2.0 |
| *1/*3 | 45 | 17.9 | |
| *3/*3 | 192 | 76.5 | |
| *3/*6 | 8 | 3.2 | |
| *3/*7 | 1 | 0.4 | |
| Phenotype | N | % | |
| CYP3A | PM | 17 | 6.8 |
| IM | 184 | 73.3 | |
| EM | 50 | 19.9 |
Abbreviation: CYP, cytochrome p450 oxidase.
Sex influenced the PK parameters since women showed higher normalized T1/2 (1.05 ± 0.34 vs. 0.96 ± 0.30, p = 0.03) and normalized Vd (1.06 ± 0.41 vs. 0.96 ± 0.32, p = 0.057), compared to men (Table 3). No statistically significant difference was found between men and women in normalized AUC, Cmax, Cl and Tmax. A higher T1/2 in women is the opposite of what is expected according to the greater CYP3A4 content in women [29,87]. The higher Vd could be due to the higher fat percentage in women. However, these differences are minor and possibly not sufficient to establish a dose adjustment based on sex.
Table 3.
Pharmacokinetic parameters (normalized by the mean of each drug) according to CYP3A genotypes and phenotype and sex in all the included drugs.
| Gene | Genotype/Phenotype | Pharmacokinetic Parameter | |||||
|---|---|---|---|---|---|---|---|
| Normalized AUC | Normalized Cmax | Normalized T1/2 | Normalized Tmax | Normalized Cl | Normalized Vd | ||
| CYP3A4 | *1/*1 (n = 233) | 0.99 (0.35) | 0.99 (0.42) | 0.99 (0.32) | 1.00 (0.56) | 0.99 (0.38) | 1.00 (0.37) |
| *1/*20 (n = 5) | 1.12 (0.24) | 0.96 (0.33) | 1.18 (0.24) | 0.66 (0.27) | 0.73 (0.18) | 0.90 (0.10) | |
| *1/*22 (n = 11) | 1.06 (0.46) | 0.98 (0.38) | 0.99 (0.18) | 1.00 (0.71) | 0.96 (0.43) | 0.98 (0.43) | |
| *1/*3 + *3/*22 (n = 2) | 1.24 (0.40) | 1.25 (0.10) | 1.07 (0.28) | 0.93 (0.03) | 0.75 (0.39) | 0.83 (0.18) | |
| p-value | 0.523 | 0.738 | 0.566 | 0.482 | 0.338 | 0.874 | |
| CYP3A4 | Wild-type (n = 233) | 0.99 (0.35) | 0.99 (0.42) | 0.99 (0.32) | 1.00 (0.56) | 0.99 (0.37) | 1.00 (0.37) |
| Mutated (n = 18) | 1.10 (0.39) | 1.00 (0.34) | 1.05 (0.20) | 0.90 (0.58) | 0.87 (0.37) | 0.94 (0.34) | |
| p-value | 0.240 | 0.853 | 0.276 | 0.351 | 0.146 | 0.428 | |
| CYP3A5 | *1/*1 (n = 5) | 0.90 (0.30) | 1.13 (0.48) | 0.88 (0.19) | 0.73 (0.26) | 1.20 (0.51) | 1.05 (0.27) |
| *1/*3 (n = 45) | 1.02 (0.32) | 1.02 (0.44) | 1.04 (0.36) | 0.90 (0.52) | 0.94 (0.38) | 0.96 (0.23) | |
| *3/*3 + *3/*6 + *3/*7 (n = 201) | 0.99 (0.37) | 0.99 (0.40) | 0.99 (0.31) | 1.03 (0.57) | 0.98 (0.38) | 1.00 (0.39) | |
| p-value | 0.710 | 0.626 | 0.731 | 0.257 | 0.486 | 0.833 | |
| CYP3A | EM (n = 50) | 1.01 (0.31) | 1.03 (0.44) | 1.02 (0.35) | 0.88 (0.50) | 0.97 (0.39) | 0.97 (0.23) |
| IM (n = 183) | 0.99 (0.37) | 0.99 (0.41) | 0.98 (0.31) | 1.03 (0.57) | 0.99 (0.37) | 1.01 (0.40) | |
| PM (n = 18) | 1.09 (0.39) | 1.00 (0.34) | 1.05 (0.21) | 0.93 (0.59) | 0.87 (0.37) | 0.94 (0.34) | |
| p-value | 0.408 | 0.692 | 0.501 | 0.155 | 0.324 | 0.720 | |
| Sex | Men (n = 149) | 1.00 (0.37) | 1.00 (0.43) | 0.96 (0.30) | 0.99 (0.58) | 0.99 (0.38) | 0.96 (0.32) |
| Women (n = 102) | 0.99 (0.35) | 1.00 (0.39) | 1.05 (0.34) | 1.01 (0.54) | 0.97 (0.37) | 1.06 (0.41) | |
| p-value | 0.765 | 0.832 | 0.030 | 0.426 | 0.889 | 0.057 | |
Abbreviation: AUC, area under the curve; Cmax, maximum plasma concentration; Tmax, time to reach the maximum plasma concentration; T1/2, half-life; Cl, total drug clearance adjusted for bioavailability; Vd, volume of distribution adjusted for bioavailability; CYP, cytochrome p450 oxidase.
Regarding the influence of CYP3A alleles or phenotype categories on normalized PK parameters, we performed a separate analysis for each compound (Supplementary Table S1) that showed no significant association of CYP3A polymorphisms for ambrisentan, aripiprazole, donepezil, atorvastatin and olanzapine. However, some associations were found for amlodipine (higher AUC and lower Cl in CYP3A5 PM), imatinib, and quetiapine (higher AUC in CYP3A4 mutated allele carriers). Although not significant, some tendencies were observed in CYP3A4 mutated alleles carriers for fentanyl (higher AUC and lower Cl) and fesoterodine (lower AUC and higher Cl). However, the limited number of individuals carrying mutations in each compound separately makes it difficult to reach any conclusion. Therefore, we merged the compounds for a conjunct analysis.
Regarding the influence of CYP3A genotypes on the PK parameters of the analysed compounds, we observed that neither CYP3A phenotype nor individual CYP3A4 or CYP3A5 polymorphisms were significantly associated with PK variability (Table 2). This fact could be explained by the influence of other metabolizing enzymes (i.e., CYP2D6 for aripiprazole [101]), whose polymorphisms might have a higher impact on PK parameters of some drugs.
Although it was not statistically significant, when analysing the substrates that are only metabolized by CYP3A (ambrisentan, atorvastatin, imatinib, fentanyl, amlodipine and quetiapine), some tendencies could be observed (Table 4). Subjects with CYP3A4 mutated alleles (*3, *20, *22) showed higher normalized AUC (p = 0.099) and almost significantly lower normalized Cl (p = 0.069), which could be a result of a diminished metabolism capacity. Moreover, the only subject carrying double mutation (CYP3A4*3/*22) had a 53% increase in AUC and a 53% lower Cl. However, the validity of this result need to be confirmed in other studies due to the limited sample size.
Table 4.
Pharmacokinetic parameters (normalized by the mean of each drug) according to CYP3A genotypes and phenotype and sex in pure CYP3A-substrates (ambisentran, atorvastatine, imatinib, fentanyl, amlodipine and quetiapine).
| Gene | Genotype/Phenotype | Pharmacokinetic Parameter | |||||
|---|---|---|---|---|---|---|---|
| Normalized AUC | Normalized Cmax | Normalized T1/2 | Normalized Tmax | Normalized Cl | Normalized Vd | ||
| CYP3A4 | *1/*1 (n = 150) | 0.98 (0.41) | 1.00 (0.49) | 1.00 (0.31) | 1.00 (0.56) | 1.00 (0.40) | 1.00 (0.41) |
| *1/*20 (n = 4) | 1.08 (0.26) | 0.91 (0.36) | 1.13 (0.25) | 0.74 (0.24) | 0.78 (0.16) | 0.89 (0.12) | |
| *1/*22 (n = 9) | 1.14 (0.46) | 1.02 (0.39) | 1.01 (0.19) | 1.01 (0.80) | 0.85 (0.39) | 0.91 (0.45) | |
| *3/*22 (n = 1) | 1.53 | 1.32 | 1.27 | 0.96 | 0.47 | 0.71 | |
| p-value | 0.317 | 0.796 | 0.626 | 0.822 | 0.194 | 0.673 | |
| CYP3A4 | Wild-type (n = 150) | 0.98 (0.41) | 1.00 (0.49) | 0.99 (0.31) | 1.00 (0.56) | 1.00 (0.40) | 1.00 (0.41) |
| Mutated (n = 14) | 1.15 (0.40) | 1.01 (0.37) | 1.06 (0.20) | 0.93 (0.64) | 0.81 (0.33) | 0.89 (0.36) | |
| p-value | 0.099 | 0.723 | 0.274 | 0.459 | 0.069 | 0.264 | |
| CYP3A5 | *1/*1 (n = 2) | 0.96 (0.49) | 1.24 (0.72) | 0.83 (0.19) | 0.75 (0.48) | 1.25 (0.81) | 0.99 (0.47) |
| *1/*3 (n = 23) | 1.02 (0.42) | 1.01 (0.59) | 1.03 (0.40) | 0.94 (0.63) | 0.97 (0.48) | 0.92 (0.28) | |
| *3/*3 + *3/*6 + *3/*7 (n = 139) | 1.00 (0.41) | 0.99 (0.45) | 1.00 (0.29) | 1.01 (0.56) | 0.98 (0.38) | 1.01 (0.42) | |
| p-value | 0.987 | 0.760 | 0.834 | 0.538 | 0.731 | 0.818 | |
| CYP3A | EM (n = 25) | 1.01 (0.42) | 1.03 (0.59) | 1.01 (0.39) | 0.92 (0.62) | 0.99 (0.49) | 0.93 (0.29) |
| IM (n = 126) | 0.98 (0.41) | 1.00 (0.47) | 0.99 (0.30) | 1.01 (0.55) | 0.99 (0.38) | 1.02 (0.43) | |
| PM (n = 13) | 1.14 (0.41) | 0.94 (0.28) | 1.06 (0.21) | 0.97 (0.65) | 0.82 (0.34) | 0.91 (0.37) | |
| p-value | 0.342 | 0.988 | 0.566 | 0.560 | 0.280 | 0.526 | |
| Sex | Men (n = 95) | 0.99 (0.43) | 0.98 (0.50) | 0.96 (0.27) | 0.96 (0.57) | 1.00 (0.41) | 0.96 (0.34) |
| Women (n = 69) | 1.01 (0.39) | 1.03 (0.44) | 1.05 (0.34) | 1.04 (0.56) | 0.96 (0.38) | 1.05 (0.48) | |
| p-value | 0.747 | 0.278 | 0.093 | 0.209 | 0.618 | 0.304 | |
Abbreviation: AUC, area under the curve; Cmax, maximum plasma concentration; Tmax, time to reach the maximum plasma concentration; T1/2, half-life; Cl, total drug clearance adjusted for bioavailability; Vd, volume of distribution adjusted for bioavailability; CYP, cytochrome p450 oxidase.
In a previous study with the CYP3A-substrate fentanyl, our group was able to demonstrate that carrying CYP3A4*22 polymorphism results in higher AUC and lower Cl [102]. Here, congruently, the same tendency was observed, yet the significance level was not reached as we used AUCt instead of AUCinf [102]. Moreover, the validity of our previous results need to be confirmed in other studies with similar settings due to the small sample size. Other CYP3A4 genotypes were not related to different exposure to fentanyl [103]. Regarding CYP3A5, the presence of *3 allele was linked to a two-fold increase in fentanyl systemic exposure [104]. However, we could not find any homozygous wild-type subject to compare with CYP3A5*3/*3 carriers.
CYP3A4*22 and CYP3A5*3 were not associated with lipid reductions in 105 children and adolescents with familial hypercholesterolemia treated with atorvastatin [105]. Moreover, CYP3A4*22 neither affected the lipid reduction response to atorvastatin and simvastatin treatment in 416 adults [106]. However, Rosales et al. described a better response to atorvastatin in Chilean patients who carried CYP3A4*1B [107]. Likewise, Peng et al. found an influence of the polymorphism CYP3A4*1G (rs2242480) on atorvastatin treatment in patients with ischemic stroke [108]. However, these two polymorphisms were not analysed in our study since they were not included in the commercial TaqMan™ OpenArray™ PGx Express Panel (Applied Biosystems, Foster City, CA, USA).
Indeed, no association with CYP3A4*1G was reported in tacrolimus concentrations in paediatric patients [109] or imatinib pharmacokinetics in Chinese patients [110]. Another CYP3A4 polymorphism, rs755828176, was significantly associated with dose-adjusted trough plasma concentrations of imatinib and its metabolite [111]. Nevertheless, this frameshift variant is reported not to be present in our population [112]. Another study in 112 patients with chronic myeloid leukaemia found no relationship between CYP3A4 rs2740574 and CYP3A5*3 and imatinib plasma levels and therapeutic response [113]. Neither did another study in 82 patients treated with imatinib [114]. Indeed, Petain et al. stated that morphologic and biological characteristics have a greater effect on imatinib PK than pharmacogenetics [115]. However, CYP3A5*3 was associated with higher imatinib trough plasma concentrations in Nigerian population [116] and a significant lower risk of acquiring imatinib resistance, while CYP3A4*18 had no statistically significant effect [117]. In our study, the only subject with the CYP3A4*20 allele showed higher imatinib levels (p = 0.046), although it is not conclusive due to the limited sample size in our study; the frequency of CYP3A4*20 previously found in Spanish population (1.2%) was not reached [37]. Moreover, in contrast to the study by Adeagbo et al. [116], CYP3A5 PMs showed lower AUC compared to the only IM subject, although, as before, it is not conclusive.
According to our knowledge, there is no study evaluating the effect of polymorphisms in CYP3A on the PK of ambrisentan. However, in our study, it does not appear that CYP3A4 or CYP3A5 polymorphisms influence its elimination.
Regarding amlodipine, Huang et al. described an altered antihypertensive efficacy of amlodipine related to CYP3A4*1G and CYP3A5*3 alleles in patients with hypertension following renal transplantation [118]. Moreover, in a study in 40 healthy volunteers, CYP3A5*3/*3 carriers exhibited lower plasma amlodipine concentrations [119]. In our study, although not significant, we found the lowest amlodipine plasma concentration in the only CYP3A5 *1/*1 subject. Indeed, we found a significantly higher Cl in this subject, but our sample size is not sufficient to arise any conclusion.
Regarding quetiapine, as shown in Supplementary Table S1, we found significant higher plasma levels in carriers of a CYP3A4 mutated allele (AUC 1107.7 ± 376.1 ng·h·Kg/mL·mg vs. 588.9 ± 272.6 ng·h·Kg/mL·mg in wild-type subjects, p = 0.024). A previous work from our laboratory showed that quetiapine AUC was influenced by polymorphisms in CYP1A2 [120]. According to our knowledge, a direct relationship between CYP3A4 polymorphisms and quetiapine was not described before. Notwithstanding, the association we found has to be interpreted with caution, given that only two subjects were carriers of mutations. Moreover, Shilbayeh et al. described in a population PK model a 29% increased Cl of quetiapine in CYP3A5 *1/*1 subjects compared to*1/*3 and *3/*3, but they stated that these simulations do not call for a genotype-based quetiapine dosing scheme [121]. Indeed, polymorphisms in CYP3A5 had no significant influence on steady-state serum concentrations of quetiapine in psychiatric patients [122]. In our study, we found no difference in any PK parameter of quetiapine according to CYP3A5 genotypes.
Finally, in regards to the other substrates that are not only metabolized by CYP3A, Noetzli et al. evaluated the effect of polymorphisms in CYP2D6 and CYP3A, finding that only CYP2D6 influenced donepezil clearance [123]. Moreover, another study in 54 patients did not find any statistically significant association between CYP3A4 or CYP3A5 genotypes and plasma donepezil concentrations or clinical response [124].
Moreover, in the current work, when analysing the combined effect of CYP3A4 and CYP3A5 into the CYP3A phenotype, the tendencies towards higher AUC and lower Cl disappeared. It is of great importance to increase our sample size and to analyse whether the influence of CYP3A4 polymorphisms alone is sufficient to have a clear effect on CYP3A-substrates PK parameters, since the majority of subjects lack CYP3A5 activity. With the 251 subjects included in the study the statistical power achieved was 0.81 for the ANOVA analysis of CYP3A phenotype and for an expected effect size of 0.20 for all the normalized PK parameters. The effect size observed was 0.074 for normalized AUC, 0.045 for normalized Cmax, 0.090 for normalized T1/2, 0.074 for normalized Tmax, 0.081 for normalized Cl, and 0.063 for normalized Vd. The sample size required for a smaller effect size of 0.09 (if clinically relevant) in an ANOVA analysis of normalized PK variables is 1.194.
Moreover, further approaches might shed light on this study field, such as the design of human recombinant microsomes with different CYP3A4 and CYP3A5 polymorphisms, which might allow the assessment of these polymorphisms on in vitro PK, and its comparison with the in vivo PK data. For instance, the study of recombinant CYP3A human liver microsomes showed that CYP3A5 expression influenced the metabolism of the antiviral asunaprevir, but not daclatasvir and beclabuvir [125].
Finally, it is important to note that CYP3A4 is an enzyme that is easily induced and inhibited by a wide variety of drugs, so the effect of polymorphisms, if any, could be masked in patients with multiple medications. Since this enzyme metabolizes many of the drugs actually on the market, it is likely that several CYP3A4 substrates are used concomitantly and that some of them are also CYP3A4 inhibitors or inducers of CYP3A4.
Study Limitations
Our study was performed after a single-dose administration of each drug to healthy volunteers, that does not allow the assessment of long-term effectiveness and safety. Our results might differ from patients under chronic treatment. Nevertheless, this study design allows the investigation of genetic polymorphisms affecting PK while avoiding the influence of other confounding factors, such as smoking or concomitant treatment. However, further research is needed to increase the statistical power of these results, especially to evaluate the influence in homozygote subjects for inactivating alleles. Finally, there is a possibility that other polymorphisms that were not included in the study could be functionally impactful, but we did not screen for them.
4. Conclusions
Our main conclusion is that CYP3A polymorphisms themselves do not have a pronounced influence on the PK parameters of the CYP3A-substrates analysed in this study. If so, the influence of CYP3A4 polymorphisms could be detectable in a very small percentage of subjects, so the evidence so far is scarce and does not support a dose adjustment in the majority of CYP3A-metabolized drugs. Indeed, it is of importance to highlight that, although there are few subjects carrying CYP3A4 double mutations, the effect in those might be relevant. The fact that the majority of subjects lack the CYP3A5 enzyme, might make the effect of CYP3A4 double mutations even more important when present. However, in CYP3A4 heterozygous subjects the consequence might be less noticeable, partly due to the high inducible potential of CYP3A4 enzyme.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2227-9059/8/4/94/s1.
Author Contributions
Conceptualization, M.S.-R., S.A. and F.A.-S.; methodology, M.N.-G., D.O. and M.R.; software, M.S.-R.; validation, M.S.-R., S.A. and F.A.-S.; formal analysis, M.N.-G, M.S., C.R.-A.; investigation, , M.S.-R., S.A., M.N.-G., D.O., M.R., P.Z., D.K., G.M., M.S., C.R.-A., A.M.B. and F.A.-S.; resources, D.O. and F.A.-S.; data curation, M.S.-R.; writing—original draft preparation, M.S.-R. and S.A.; writing—review and editing, M.S.-R., S.A. and F.A.-S.; visualization, M.S.-R. and S.A.; supervision, F.A.-S.; project administration, F.A.-S.; funding acquisition, F.A.-S., D.O. and C.R.-A. All authors have read and agreed to the published version of the manuscript.
Funding
F.A.S. and D.O. have been consultants or investigators in clinical trials sponsored by the following pharmaceutical companies: Abbott, Alter, Chemo, Cinfa, FAES, Farmalíder, Ferrer, GlaxoSmithKline, Galenicum, Gilead, Italfarmaco, Janssen-Cilag, Kern, Normon, Novartis, Servier, Silverpharma, Teva and Zambon. M.N.G is co-financed by the European Social Fund and the Youth European Initiative, grant number PEJ-2018-TL/BMD-11080. D.K. is co-financed by the H2020 Marie Sklodowska-Curie Innovative Training Network 721236 grant.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.König I.R., Fuchs O., Hansen G., von Mutius E., Kopp M.V. What is precision medicine? Eur. Respir. J. 2017;50 doi: 10.1183/13993003.00391-2017. [DOI] [PubMed] [Google Scholar]
- 2.Weinshilboum R., Wang L. Pharmacogenomics: Bench to bedside. Nat. Rev. Drug Discov. 2004;3:739–748. doi: 10.1038/nrd1497. [DOI] [PubMed] [Google Scholar]
- 3.Motulsky A.G. Drug reactions, enzymes, and biochemical genetics. J. Am. Med. Assoc. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 4.Moderne Probleme der Human Genetik Ergebn. [(accessed on 24 February 2020)];inn-Google Académico. Available online: https://scholar.google.es/scholar?hl=es&as_sdt=0%2C5&q=Moderne+Probleme+der+Human+genetik.+Ergebn.+inn&btnG=
- 5.US Food and Drug Administration . Clinical Pharmacogenomics. Premarket Evaluation in Early-Phase Clinical Studies and Recommendations for Labeling. Volume 26 Silver Spring; Rockville, MD, USA: 2013. [Google Scholar]
- 6.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharm. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Feyereisen R., Nelson D.R., Coon M.J., Estabrook R.W., Nebert D.W., Fujii-kuriyama Y., Gonzalez F.J., Guengerich F.P., Gunsalus I.C., Johnson E.F. The P450 superfamily: Update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991;10:1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- 8.Ingelman-Sundberg M., Rodriguez-Antona C. Pharmacogenetics of drug-metabolizing enzymes: Implications for a safer and more effective drug therapy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1563–1570. doi: 10.1098/rstb.2005.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rostami-Hodjegan A., Amin A.M., Spencer E.P., Lennard M.S., Tucker G.T., Flanagan R.J. Influence of dose, cigarette smoking, age, sex, and metabolic activity on plasma clozapine concentrations: A predictive model and nomograms to aid clozapine dose adjustment and to assess compliance in individual patients. J. Clin. Psychopharmacol. 2004;24:70–78. doi: 10.1097/01.jcp.0000106221.36344.4d. [DOI] [PubMed] [Google Scholar]
- 10.Domanski T.L., Finta C., Halpert J.R., Zaphiropoulos P.G. cDNA cloning and initial characterization of CYP3A43, a novel human cytochrome P450. Mol. Pharmacol. 2001;59:386–392. doi: 10.1124/mol.59.2.386. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T., Yamazaki H., Mimura M., Inui Y., Guengerich F.P. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: Studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 12.Brooks B.A., McBride O.W., Dolphin C.T., Farrall M., Scambler P.J., Gonzalez F.J., Idle J.R. The gene CYP3 encoding P450pcn1 (nifedipine oxidase) is tightly linked to the gene COL1A2 encoding collagen type 1 alpha on 7q21-q22. 1. Am. J. Hum. Genet. 1988;43:280. [PMC free article] [PubMed] [Google Scholar]
- 13.Finta C., Zaphiropoulos P.G. The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons. Gene. 2000;260:13–23. doi: 10.1016/S0378-1119(00)00470-4. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto H., Toide K., Kitamura R., Fujita M., Tagawa S., Itoh S., Kamataki T. Gene structure of CYP3A4, an adult-specific form of cytochrome P450 in human livers, and its transcriptional control. Eur. J. Bioch. 1993;218:585–595. doi: 10.1111/j.1432-1033.1993.tb18412.x. [DOI] [PubMed] [Google Scholar]
- 15.Gellner K., Eiselt R., Hustert E., Arnold H., Koch I., Haberl M., Deglmann C.J., Burk O., Buntefuss D., Escher S., et al. Genomic organization of the human CYP3A locus: Identification of a new, inducible CYP3A gene. Pharm. Genom. 2001;11:111–121. doi: 10.1097/00008571-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Plant N. The human cytochrome P450 sub-family: Transcriptional regulation, inter-individual variation and interaction networks. Biochim. Biophys. Acta. 2007;1770:478–488. doi: 10.1016/j.bbagen.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Elens L., Van Gelder T., Hesselink D.A., Haufroid V., Van Schaik R.H. CYP3A4* 22: Promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics. 2013;14:47–62. doi: 10.2217/pgs.12.187. [DOI] [PubMed] [Google Scholar]
- 18.Daly A.K. Significance of the minor cytochrome P450 3A isoforms. Clin. Pharm. 2006;45:13–31. doi: 10.2165/00003088-200645010-00002. [DOI] [PubMed] [Google Scholar]
- 19.Shimada T., Yamazaki H., Mimura M., Wakamiya N., Ueng Y.F., Guengerich F.P., Inui Y. Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs. Drug Metab. Dispos. 1996;24:515–522. [PubMed] [Google Scholar]
- 20.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams J.A., Ring B.J., Cantrell V.E., Jones D.R., Eckstein J., Ruterbories K., Hamman M.A., Hall S.D., Wrighton S.A. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab. Dispos. 2002;30:883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 22.Huang W., Lin Y.S., McConn D.J., Calamia J.C., Totah R.A., Isoherranen N., Glodowski M., Thummel K.E. Evidence of significant contribution from CYP3A5 to hepatic drug metabolism. Drug Metab. Dispos. 2004;32:1434–1445. doi: 10.1124/dmd.104.001313. [DOI] [PubMed] [Google Scholar]
- 23.Kivistö K.T., Bookjans G., Fromm M.F., Griese E.-U., Münzel P., KRoemer H.K. Expression of CYP3A4, CYP3A5 and CYP3A7 in human duodenal tissue. Br. J. Clin. Pharm. 1996;42:387–389. doi: 10.1046/j.1365-2125.1996.42615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinbloesem C.H., van Brummelen P., Faber H., Danhof M., Vermeulen N.P., Breimer D.D. Variability in nifedipine pharmacokinetics and dynamics: A new oxidation polymorphism in man. Biochem. Pharmacol. 1984;33:3721–3724. doi: 10.1016/0006-2952(84)90165-5. [DOI] [PubMed] [Google Scholar]
- 25.Guengerich F.P., Martin M.V., Beaune P.H., Kremers P., Wolff T., Waxman D.J. Characterization of rat and human liver microsomal cytochrome P-450 forms involved in nifedipine oxidation, a prototype for genetic polymorphism in oxidative drug metabolism. J. Biol. Chem. 1986;261:5051–5060. [PubMed] [Google Scholar]
- 26.Inoue K., Inazawa J., Nakagawa H., Shimada T., Yamazaki H., Guengerich F.P., Abe T. Assignment of the human cytochrome P-450 nifedipine oxidase gene (CYP3A4) to chromosome 7 at band q22. 1 by fluorescencein situ hybridization. Jpn. Hum. Genet. 1992;37:133–138. doi: 10.1007/BF01899734. [DOI] [PubMed] [Google Scholar]
- 27.The Nucleotide Database Nucleotide–NCBI. [(accessed on 24 February 2020)]; Available online: https://www.ncbi.nlm.nih.gov/nuccore/197313719?report=graph.
- 28.Ensembl Genome Browser. [(accessed on 24 February 2020)]; Available online: http://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000160868;r=7:99756960-99784248;t=ENST00000415003.
- 29.Achour B., Barber J., Rostami-Hodjegan A. Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: A meta-analysis. Drug Metab. Dispos. 2014;42:1349–1356. doi: 10.1124/dmd.114.058834. [DOI] [PubMed] [Google Scholar]
- 30.Michaels S., Wang M.Z. The Revised Human Liver Cytochrome P450 “Pie”: Absolute Protein Quantification of CYP4F and CYP3A Enzymes Using Targeted Quantitative Proteomics. Drug Metab. Dispos. 2014;42:1241–1251. doi: 10.1124/dmd.114.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q.Y., Dunbar D., Ostrowska A., Zeisloft S., Yang J., Kaminsky L.S. Characterization of human small intestinal cytochromes P-450. Drug Metab. Dispos. 1999;27:804–809. [PubMed] [Google Scholar]
- 32.Paine M.F., Shen D.D., Kunze K.L., Perkins J.D., Marsh C.L., McVicar J.P., Barr D.M., Gillies B.S., Thummel K.E. First-pass metabolism of midazolam by the human intestine. Clin. Pharm. Ther. 1996;60:14–24. doi: 10.1016/S0009-9236(96)90162-9. [DOI] [PubMed] [Google Scholar]
- 33.Kolars J.C., Watkins P.B., Merion R.M., Awni W.M. First-pass metabolism of cyclosporin by the gut. Lancet. 1991;338:1488–1490. doi: 10.1016/0140-6736(91)92302-I. [DOI] [PubMed] [Google Scholar]
- 34.Westlind-Johnsson A., Hermann R., Huennemeyer A., Hauns B., Lahu G., Nassr N., Zech K., Ingelman-Sundberg M., von Richter O. Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity. Clin. Pharmacol. Ther. 2006;79:339–349. doi: 10.1016/j.clpt.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Wang D., Guo Y., Wrighton S.A., Cooke G.E., Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharm. J. 2011;11:274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein K., Thomas M., Winter S., Nussler A.K., Niemi M., Schwab M., Zanger U.M. PPARA: A novel genetic determinant of CYP3A4 in vitro and in vivo. Clin. Pharm. Ther. 2012;91:1044–1052. doi: 10.1038/clpt.2011.336. [DOI] [PubMed] [Google Scholar]
- 37.Apellániz-Ruiz M., Inglada-Pérez L., Naranjo M.E.G., Sánchez L., Mancikova V., Currás-Freixes M., de Cubas A.A., Comino-Méndez I., Triki S., Rebai A., et al. High frequency and founder effect of the CYP3A4*20 loss-of-function allele in the Spanish population classifies CYP3A4 as a polymorphic enzyme. Pharm. J. 2015;15:288–292. doi: 10.1038/tpj.2014.67. [DOI] [PubMed] [Google Scholar]
- 38.Tavira B., Coto E., Diaz-Corte C., Alvarez V., López-Larrea C., Ortega F. A search for new CYP3A4 variants as determinants of tacrolimus dose requirements in renal-transplanted patients. Pharm. Genom. 2013;23:445–448. doi: 10.1097/FPC.0b013e3283636856. [DOI] [PubMed] [Google Scholar]
- 39.Santoro A.B., Struchiner C.J., Felipe C.R., Tedesco-Silva H., Medina-Pestana J.O., Suarez-Kurtz G. CYP3A5 genotype, but not CYP3A4*1b, CYP3A4*22, or hematocrit, predicts tacrolimus dose requirements in Brazilian renal transplant patients. Clin. Pharmacol. Ther. 2013;94:201–202. doi: 10.1038/clpt.2013.68. [DOI] [PubMed] [Google Scholar]
- 40.Santoro A., Felipe C.R., Tedesco-Silva H., Medina-Pestana J.O., Struchiner C.J., Ojopi E.B., Suarez-Kurtz G. Pharmacogenetics of calcineurin inhibitors in Brazilian renal transplant patients. Pharmacogenomics. 2011;12:1293–1303. doi: 10.2217/pgs.11.70. [DOI] [PubMed] [Google Scholar]
- 41.Kurzawski M., Dąbrowska J., Dziewanowski K., Domański L., Perużyńska M., Droździk M. CYP3A5 and CYP3A4, but not ABCB1 polymorphisms affect tacrolimus dose-adjusted trough concentrations in kidney transplant recipients. Pharmacogenomics. 2014;15:179–188. doi: 10.2217/pgs.13.199. [DOI] [PubMed] [Google Scholar]
- 42.de Jonge H., Elens L., de Loor H., van Schaik R.H., Kuypers D.R.J. The CYP3A4*22 C>T single nucleotide polymorphism is associated with reduced midazolam and tacrolimus clearance in stable renal allograft recipients. Pharm. J. 2015;15:144–152. doi: 10.1038/tpj.2014.49. [DOI] [PubMed] [Google Scholar]
- 43.Elens L., van Schaik R.H., Panin N., de Meyer M., Wallemacq P., Lison D., Mourad M., Haufroid V. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics. 2011;12:1383–1396. doi: 10.2217/pgs.11.90. [DOI] [PubMed] [Google Scholar]
- 44.Elens L., Becker M.L., Haufroid V., Hofman A., Visser L.E., Uitterlinden A.G., Stricker B.C., van Schaik R.H.N. Novel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin-mediated cholesterol reduction in the Rotterdam Study. Pharm. Genom. 2011;21:861–866. doi: 10.1097/FPC.0b013e32834c6edb. [DOI] [PubMed] [Google Scholar]
- 45.Rebbeck T.R., Jaffe J.M., Walker A.H., Wein A.J., Malkowicz S.B. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J. Natl. Cancer Inst. 1998;90:1225–1229. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 46.Westlind A., Löfberg L., Tindberg N., Andersson T.B., Ingelman-Sundberg M. Interindividual differences in hepatic expression of CYP3A4: Relationship to genetic polymorphism in the 5′-upstream regulatory region. Biochem. Biophys. Res. Commun. 1999;259:201–205. doi: 10.1006/bbrc.1999.0752. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez-Antona C., Sayi J.G., Gustafsson L.L., Bertilsson L., Ingelman-Sundberg M. Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem. Biophys. Res. Commun. 2005;338:299–305. doi: 10.1016/j.bbrc.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 48.Amirimani B., Ning B., Deitz A.C., Weber B.L., Kadlubar F.F., Rebbeck T.R. Increased transcriptional activity of the CYP3A4*1B promoter variant. Environ. Mol. Mutagen. 2003;42:299–305. doi: 10.1002/em.10199. [DOI] [PubMed] [Google Scholar]
- 49.Hesselink D.A., van Schaik R.H.N., van der Heiden I.P., van der Werf M., Gregoor P.J.H.S., Lindemans J., Weimar W., van Gelder T. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin. Pharmacol. Ther. 2003;74:245–254. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 50.Tavira B., Garciá E.C., Díaz-Corte C., Ortega F., Arias M., Torres A., Díaz J.M., Selgas R., López-Larrea C., Campistol J.M. Pharmacogenetics of tacrolimus after renal transplantation: Analysis of polymorphisms in genes encoding 16 drug metabolizing enzymes. Clin. Chem. Lab. Med. 2011;49:825–833. doi: 10.1515/CCLM.2011.143. [DOI] [PubMed] [Google Scholar]
- 51.Gervasini G., Garcia M., Macias R.M., Cubero J.J., Caravaca F., Benitez J. Impact of genetic polymorphisms on tacrolimus pharmacokinetics and the clinical outcome of renal transplantation. Transpl. Int. 2012;25:471–480. doi: 10.1111/j.1432-2277.2012.01446.x. [DOI] [PubMed] [Google Scholar]
- 52.Crettol S., Venetz J.-P., Fontana M., Aubert J.-D., Pascual M., Eap C.B. CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther. Drug Monit. 2008;30:689–699. doi: 10.1097/FTD.0b013e31818a2a60. [DOI] [PubMed] [Google Scholar]
- 53.Żochowska D., Wyzgał J., Pączek L. Impact of CYP3A4*1B and CYP3A5*3 polymorphisms on the pharmacokinetics of cyclosporine and sirolimus in renal transplant recipients. Ann. Transplant. 2012;17:36–44. doi: 10.12659/aot.883456. [DOI] [PubMed] [Google Scholar]
- 54.Gervasini G., García-Pino G., Vergara E., Mota-Zamorano S., García-Cerrada M., Luna E. CYP3A genotypes of donors but not those of the patients increase the risk of acute rejection in renal transplant recipients on calcineurin inhibitors: A pilot study. Eur. J. Clin. Pharmacol. 2018;74:53–60. doi: 10.1007/s00228-017-2353-9. [DOI] [PubMed] [Google Scholar]
- 55.Becker M.L., Visser L.E., van Schaik R.H.N., Hofman A., Uitterlinden A.G., Stricker B.H.C. Influence of genetic variation in CYP3A4 and ABCB1 on dose decrease or switching during simvastatin and atorvastatin therapy. Pharmacoepidemiol. Drug Saf. 2010;19:75–81. doi: 10.1002/pds.1866. [DOI] [PubMed] [Google Scholar]
- 56.Birdwell K.A., Grady B., Choi L., Xu H., Bian A., Denny J.C., Jiang M., Vranic G., Basford M., Cowan J.D., et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharm. Genom. 2012;22:32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai D., Tang J., Rose R., Hodgson E., Bienstock R.J., Mohrenweiser H.W., Goldstein J.A. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J. Pharm. Exp. Ther. 2001;299:825–831. [PubMed] [Google Scholar]
- 58.Fukushima-Uesaka H., Saito Y., Watanabe H., Shiseki K., Saeki M., Nakamura T., Kurose K., Sai K., Komamura K., Ueno K., et al. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum. Mutat. 2004;23:100. doi: 10.1002/humu.9210. [DOI] [PubMed] [Google Scholar]
- 59.Zuo X., Ng C.M., Barrett J.S., Luo A., Zhang B., Deng C., Xi L., Cheng K., Ming Y., Yang G., et al. Effects of CYP3A4 and CYP3A5 polymorphisms on tacrolimus pharmacokinetics in Chinese adult renal transplant recipients: A population pharmacokinetic analysis. Pharm. Genom. 2013;23:251–261. doi: 10.1097/FPC.0b013e32835fcbb6. [DOI] [PubMed] [Google Scholar]
- 60.Uesugi M., Hosokawa M., Shinke H., Hashimoto E., Takahashi T., Kawai T., Matsubara K., Ogawa K., Fujimoto Y., Okamoto S., et al. Influence of cytochrome P450 (CYP) 3A4*1G polymorphism on the pharmacokinetics of tacrolimus, probability of acute cellular rejection, and mRNA expression level of CYP3A5 rather than CYP3A4 in living-donor liver transplant patients. Biol. Pharm. Bull. 2013;36:1814–1821. doi: 10.1248/bpb.b13-00509. [DOI] [PubMed] [Google Scholar]
- 61.Li C.-J., Li L., Lin L., Jiang H.-X., Zhong Z.-Y., Li W.-M., Zhang Y.-J., Zheng P., Tan X.-H., Zhou L. Impact of the CYP3A5, CYP3A4, COMT, IL-10 and POR genetic polymorphisms on tacrolimus metabolism in Chinese renal transplant recipients. PLoS ONE. 2014;9:e86206. doi: 10.1371/journal.pone.0086206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J., Liu S., Fu Q., Zhang Y., Wang X., Liu X., Liu L., Wang C., Huang M. Interactive effects of CYP3A4, CYP3A5, MDR1 and NR1I2 polymorphisms on tracrolimus trough concentrations in early postrenal transplant recipients. Pharmacogenomics. 2015;16:1355–1365. doi: 10.2217/pgs.15.78. [DOI] [PubMed] [Google Scholar]
- 63.Liu M., He H., Zhang Y., Hu Y., He F., Luo J., Luo Z., Chen X., Liu Z., Zhou H., et al. IL-3 and CTLA4 gene polymorphisms may influence the tacrolimus dose requirement in Chinese kidney transplant recipients. Acta Pharm. Sin. 2017;38:415–423. doi: 10.1038/aps.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun B., Guo Y., Gao J., Shi W., Fan G., Li X., Qiu J., Qin Y., Liu G. Influence of CYP3A and ABCB1 polymorphisms on cyclosporine concentrations in renal transplant recipients. Pharmacogenomics. 2017;18:1503–1513. doi: 10.2217/pgs-2017-0127. [DOI] [PubMed] [Google Scholar]
- 65.Li T.-F., Hu L., Ma X.-L., Huang L., Liu X.-M., Luo X.-X., Feng W.-Y., Wu C.-F. Population pharmacokinetics of cyclosporine in Chinese children receiving hematopoietic stem cell transplantation. Acta Pharm. Sin. 2019;40:1603–1610. doi: 10.1038/s41401-019-0277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang W., Chang Y.-Z., Kan Q.-C., Zhang L.-R., Li Z.-S., Lu H., Wang Z.-Y., Chu Q.-J., Zhang J. CYP3A4*1G genetic polymorphism influences CYP3A activity and response to fentanyl in Chinese gynecologic patients. Eur. J. Clin. Pharm. 2010;66:61–66. doi: 10.1007/s00228-009-0726-4. [DOI] [PubMed] [Google Scholar]
- 67.Yuan R., Zhang X., Deng Q., Wu Y., Xiang G. Impact of CYP3A4*1G polymorphism on metabolism of fentanyl in Chinese patients undergoing lower abdominal surgery. Clin. Chim. Acta. 2011;412:755–760. doi: 10.1016/j.cca.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 68.Zhang W., Yuan J.-J., Kan Q.-C., Zhang L.-R., Chang Y.-Z., Wang Z.-Y., Li Z.-S. Influence of CYP3A5*3 polymorphism and interaction between CYP3A5*3 and CYP3A4*1G polymorphisms on post-operative fentanyl analgesia in Chinese patients undergoing gynaecological surgery. Eur. J. Anaesthesiol. 2011;28:245–250. doi: 10.1097/EJA.0b013e3283438b39. [DOI] [PubMed] [Google Scholar]
- 69.Dong Z.-L., Li H., Chen Q.-X., Hu Y., Wu S.-J., Tang L.-Y., Gong W.-Y., Xie G.-H., Fang X.-M. Effect of CYP3A4*1G on the fentanyl consumption for intravenous patient-controlled analgesia after total abdominal hysterectomy in Chinese Han population. J. Clin. Pharm. Ther. 2012;37:153–156. doi: 10.1111/j.1365-2710.2011.01268.x. [DOI] [PubMed] [Google Scholar]
- 70.Zhang J., Zhang L., Zhao X., Shen S., Luo X., Zhang Y. Association between MDR1/CYP3A4/OPRM1 gene polymorphisms and the post-caesarean fentanyl analgesic effect on Chinese women. Gene. 2018;661:78–84. doi: 10.1016/j.gene.2018.03.081. [DOI] [PubMed] [Google Scholar]
- 71.Sata F., Sapone A., Elizondo G., Stocker P., Miller V.P., Zheng W., Raunio H., Crespi C.L., Gonzalez F.J. CYP3A4 allelic variants with amino acid substitutions in exons 7 and 12: Evidence for an allelic variant with altered catalytic activity. Clin. Pharmacol. Ther. 2000;67:48–56. doi: 10.1067/mcp.2000.104391. [DOI] [PubMed] [Google Scholar]
- 72.Thompson J.F., Man M., Johnson K.J., Wood L.S., Lira M.E., Lloyd D.B., Banerjee P., Milos P.M., Myrand S.P., Paulauskis J., et al. An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharm. J. 2005;5:352–358. doi: 10.1038/sj.tpj.6500328. [DOI] [PubMed] [Google Scholar]
- 73.Ensembl Genome Browser. [(accessed on 24 February 2020)]; Available online: http://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000106258;r=7:99648194-99679998.
- 74.Kuehl P., Zhang J., Lin Y., Lamba J., Assem M., Schuetz J., Watkins P.B., Daly A., Wrighton S.A., Hall S.D. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 75.Lamba J.K., Lin Y.S., Schuetz E.G., Thummel K.E. Genetic contribution to variable human CYP3A-mediated metabolism. Adv. Drug Deliv. Rev. 2002;54:1271–1294. doi: 10.1016/S0169-409X(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 76.Miao J., Jin Y., Marunde R.L., Kim S., Quinney S., Radovich M., Li L., Hall S.D. Association of genotypes of the CYP3A cluster with midazolam disposition in vivo. Pharm. J. 2009;9:319–326. doi: 10.1038/tpj.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moes D.J.A.R., Swen J.J., den Hartigh J., van der Straaten T., van der Heide J.J.H., Sanders J.S., Bemelman F.J., de Fijter J.W., Guchelaar H.J. Effect of CYP3A4*22, CYP3A5*3, and CYP3A Combined Genotypes on Cyclosporine, Everolimus, and Tacrolimus Pharmacokinetics in Renal Transplantation. CPT Pharm. Syst. Pharmacol. 2014;3:e100. doi: 10.1038/psp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kitzmiller J.P., Sullivan D.M., Phelps M.A., Wang D., Sadee W. CYP3A4/5 combined genotype analysis for predicting statin dose requirement for optimal lipid control. Drug Metab. Drug Interact. 2013;28:59–63. doi: 10.1515/dmdi-2012-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanchez Spitman A.B., Moes D.J.A.R., Gelderblom H., Dezentje V.O., Swen J.J., Guchelaar H.J. Effect of CYP3A4*22, CYP3A5*3, and CYP3A combined genotypes on tamoxifen metabolism. Eur. J. Clin. Pharmacol. 2017;73:1589–1598. doi: 10.1007/s00228-017-2323-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hustert E., Haberl M., Burk O., Wolbold R., He Y.Q., Klein K., Nuessler A.C., Neuhaus P., Klattig J., Eiselt R., et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 81.Lee S.-J., Usmani K.A., Chanas B., Ghanayem B., Xi T., Hodgson E., Mohrenweiser H.W., Goldstein J.A. Genetic findings and functional studies of human CYP3A5 single nucleotide polymorphisms in different ethnic groups. Pharmacogenetics. 2003;13:461–472. doi: 10.1097/00008571-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 82.Birdwell K.A., Decker B., Barbarino J.M., Peterson J.F., Stein C.M., Sadee W., Wang D., Vinks A.A., He Y., Swen J.J. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharm. Ther. 2015;98:19–24. doi: 10.1002/cpt.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsamandouras N., Dickinson G., Guo Y., Hall S., Rostami-Hodjegan A., Galetin A., Aarons L. Identification of the effect of multiple polymorphisms on the pharmacokinetics of simvastatin and simvastatin acid using a population-modeling approach. Clin. Pharmacol. Ther. 2014;96:90–100. doi: 10.1038/clpt.2014.55. [DOI] [PubMed] [Google Scholar]
- 84.Luzum J.A., Theusch E., Taylor K.D., Wang A., Sadee W., Binkley P.F., Krauss R.M., Medina M.W., Kitzmiller J.P. Individual and Combined Associations of Genetic Variants in CYP3A4, CYP3A5, and SLCO1B1 With Simvastatin and Simvastatin Acid Plasma Concentrations. J. Cardiovasc. Pharmacol. 2015;66:80–85. doi: 10.1097/FJC.0000000000000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prueksaritanont T., Gorham L.M., Ma B., Liu L., Yu X., Zhao J.J., Slaughter D.E., Arison B.H., Vyas K.P. In vitro metabolism of simvastatin in humans [SBT] identification of metabolizing enzymes and effect of the drug on hepatic P450s. Drug Metab. Dispos. 1997;25:1191–1199. [PubMed] [Google Scholar]
- 86.Westlind-Johnsson A., Malmebo S., Johansson A., Otter C., Andersson T.B., Johansson I., Edwards R.J., Boobis A.R., Ingelman-Sundberg M. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab. Dispos. 2003;31:755–761. doi: 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 87.Wolbold R., Klein K., Burk O., Nüssler A.K., Neuhaus P., Eichelbaum M., Schwab M., Zanger U.M. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38:978–988. doi: 10.1002/hep.1840380424. [DOI] [PubMed] [Google Scholar]
- 88.Krecic-Shepard M.E., Park K., Barnas C., Slimko J., Kerwin D.R., Schwartz J.B. Race and sex influence clearance of nifedipine: Results of a population study. Clin. Pharm. Ther. 2000;68:130–142. doi: 10.1067/mcp.2000.108678. [DOI] [PubMed] [Google Scholar]
- 89.Andreassen T.N., Klepstad P., Davies A., Bjordal K., Lundström S., Kaasa S., Dale O. Influences on the pharmacokinetics of oxycodone: A multicentre cross-sectional study in 439 adult cancer patients. Eur. J. Clin. Pharmacol. 2011;67:493–506. doi: 10.1007/s00228-010-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kahan B.D., Kramer W.G., Wideman C., Flechner S.M., Lorber M.I., Van C.B. Demographic factors affecting the pharmacokinetics of cyclosporine estimated by radioimmunoassay. Transplantation. 1986;41:459–464. doi: 10.1097/00007890-198604000-00009. [DOI] [PubMed] [Google Scholar]
- 91.Zhu B., Liu Z.-Q., Chen G.-L., Chen X.-P., Ou-Yang D.-S., Wang L.-S., Huang S.-L., Tan Z.-R., Zhou H.-H. The distribution and gender difference of CYP3A activity in Chinese subjects. Br. J. Clin. Pharm. 2003;55:264–269. doi: 10.1046/j.1365-2125.2003.01728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou S.-F. Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr. Drug Metab. 2008;9:310–322. doi: 10.2174/138920008784220664. [DOI] [PubMed] [Google Scholar]
- 93.Zhou S., Chan S.Y., Goh B.C., Chan E., Duan W., Huang M., McLeod H.L. Mechanism-Based Inhibition of Cytochrome P450 3A4 by Therapeutic Drugs. Clin. Pharmacokinet. 2005;44:279–304. doi: 10.2165/00003088-200544030-00005. [DOI] [PubMed] [Google Scholar]
- 94.Saiz-Rodríguez M., Ochoa D., Belmonte C., Román M., Vieira de Lara D., Zubiaur P., Koller D., Mejía G., Abad-Santos F. Polymorphisms in CYP1A2, CYP2C9 and ABCB1 affect agomelatine pharmacokinetics. J. Psychopharmacol. (Oxf.) 2019;33:522–531. doi: 10.1177/0269881119827959. [DOI] [PubMed] [Google Scholar]
- 95.Ensembl Data CYP3A4 rs4986910. [(accessed on 25 February 2020)]; Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:99760401-99761401;v=rs4986910;vdb=variation;vf=17958388.
- 96.Ensembl Data CYP3A4 rs67666821. [(accessed on 25 February 2020)]; Available online: http://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:99757684-99758683;v=rs67666821;vdb=variation;vf=12903884.
- 97.Ensembl Data CYP3A4 rs35599367. [(accessed on 25 February 2020)]; Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:99768193-99769193;v=rs35599367;vdb=variation;vf=20923380.
- 98.Ensembl Data CYP3A5 rs776746. [(accessed on 25 February 2020)]; Available online: http://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:99672416-99673416;v=rs776746;vdb=variation;vf=550116.
- 99.Ensembl Data CYP3A5 rs10264272. [(accessed on 25 February 2020)]; Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:99664712-99665712;v=rs10264272;vdb=variation;vf=19096090.
- 100.Ensembl Data CYP3A5 rs41303343. [(accessed on 25 February 2020)]; Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:99652271-99653271;v=rs41303343;vdb=variation;vf=21091103.
- 101.Belmonte C., Ochoa D., Román M., Saiz-Rodríguez M., Wojnicz A., Gómez-Sánchez C.I., Martín-Vílchez S., Abad-Santos F. Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 Polymorphisms on Pharmacokinetics and Safety of Aripiprazole in Healthy Volunteers. Basic Clin. Pharmacol. Toxicol. 2018;122:596–605. doi: 10.1111/bcpt.12960. [DOI] [PubMed] [Google Scholar]
- 102.Saiz-Rodríguez M., Ochoa D., Herrador C., Belmonte C., Román M., Alday E., Koller D., Zubiaur P., Mejía G., Hernández-Martínez M., et al. Polymorphisms associated with fentanyl pharmacokinetics, pharmacodynamics and adverse effects. Basic Clin. Pharmacol. Toxicol. 2019;124:321–329. doi: 10.1111/bcpt.13141. [DOI] [PubMed] [Google Scholar]
- 103.Kuip E.J.M., Zandvliet M.L., Koolen S.L.W., Mathijssen R.H.J., van der Rijt C.C.D. A review of factors explaining variability in fentanyl pharmacokinetics; focus on implications for cancer patients. Br. J. Clin. Pharmacol. 2017;83:294–313. doi: 10.1111/bcp.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takashina Y., Naito T., Mino Y., Yagi T., Ohnishi K., Kawakami J. Impact of CYP3A5 and ABCB1 gene polymorphisms on fentanyl pharmacokinetics and clinical responses in cancer patients undergoing conversion to a transdermal system. Drug Metab. Pharmacokinet. 2012;27:414–421. doi: 10.2133/dmpk.dmpk-11-rg-134. [DOI] [PubMed] [Google Scholar]
- 105.Drogari E., Ragia G., Mollaki V., Elens L., Van Schaik R.H., Manolopoulos V.G. POR*28 SNP is associated with lipid response to atorvastatin in children and adolescents with familial hypercholesterolemia. Pharmacogenomics. 2014;15:1963–1972. doi: 10.2217/pgs.14.138. [DOI] [PubMed] [Google Scholar]
- 106.Ragia G., Kolovou V., Tavridou A., Elens L., Tselepis A.D., Elisaf M., Van Schaik R.H.N., Kolovou G., Manolopoulos V.G. No effect of CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) on lipid-lowering response to statins in Greek patients with primary hypercholesterolemia. [(accessed on 25 February 2020)];Drug Metab. Personal. Ther. 2015 30 doi: 10.1515/dmdi-2014-0021. Available online: https://www.degruyter.com/view/j/dmdi.2015.30.issue-1/dmdi-2014-0021/dmdi-2014-0021.xml. [DOI] [PubMed] [Google Scholar]
- 107.Rosales A., Alvear M., Cuevas A., Saavedra N., Zambrano T., Salazar L.A. Identification of pharmacogenetic predictors of lipid-lowering response to atorvastatin in Chilean subjects with hypercholesterolemia. Clin. Chim. Acta. 2012;413:495–501. doi: 10.1016/j.cca.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 108.Peng C., Ding Y., Yi X., Shen Y., Dong Z., Cao L., Li Q., Ren H., He L., Zhou D., et al. Polymorphisms in CYP450 Genes and the Therapeutic Effect of Atorvastatin on Ischemic Stroke: A Retrospective Cohort Study in Chinese Population. Clin. Ther. 2018;40:469–477. doi: 10.1016/j.clinthera.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 109.Liu H., Xu Q., Huang W., Zhao Q., Jiang Z., Kuang X., Li Z., Sun H., Qiu X. CYP3A5 and CYP3A7 genetic polymorphisms affect tacrolimus concentration in pediatric patients with nephrotic range proteinuria. Eur. J. Clin. Pharmacol. 2019;75:1533–1540. doi: 10.1007/s00228-019-02726-w. [DOI] [PubMed] [Google Scholar]
- 110.Qiu H., Zhuang W., Wang X., Huang M., Zhou Z. Association between genetic polymorphisms and variation of imatinib pharmacokinetics in gastrointestinal stromal tumors. Zhonghua Wei Chang Wai Ke Za Zhi. 2017;20:1031–1034. [PubMed] [Google Scholar]
- 111.Qian Y., Sun L.-N., Liu Y.-J., Zhang Q., Xu J.-H., Ma Z.-Q., Zhang X.-H., Xu H., Wang Y.-Q. Genetic Polymorphisms and Adverse Events on Unbound Imatinib and Its Active Metabolite Concentration in Patients With Gastrointestinal Stromal Tumors. Front. Pharmacol. 2019;10:854. doi: 10.3389/fphar.2019.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ensembl Data CYP3A4 rs755828176. [(accessed on 17 February 2020)]; Available online: https://www.ensembl.org/Homo_sapiens/Variation/Explore?r=6:160139351-160140352;v=rs755828176;vdb=variation;vf=213359709.
- 113.Belohlavkova P., Vrbacky F., Voglova J., Racil Z., Zackova D., Hrochova K., Malakova J., Mayer J., Zak P. The significance of enzyme and transporter polymorphisms for imatinib plasma levels and achieving an optimal response in chronic myeloid leukemia patients. Aoms. 2018;14:1416–1423. doi: 10.5114/aoms.2018.73538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seong S.J., Lim M., Sohn S.K., Moon J.H., Oh S.-J., Kim B.S., Ryoo H.M., Chung J.S., Joo Y.D., Bang S.M., et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann. Oncol. 2013;24:756–760. doi: 10.1093/annonc/mds532. [DOI] [PubMed] [Google Scholar]
- 115.Petain A., Kattygnarath D., Azard J., Chatelut E., Delbaldo C., Geoerger B., Barrois M., Séronie-Vivien S., LeCesne A., Vassal G., et al. Population pharmacokinetics and pharmacogenetics of imatinib in children and adults. Clin. Cancer Res. 2008;14:7102–7109. doi: 10.1158/1078-0432.CCR-08-0950. [DOI] [PubMed] [Google Scholar]
- 116.Adeagbo B.A., Bolaji O.O., Olugbade T.A., Durosinmi M.A., Bolarinwa R.A., Masimirembwa C. Influence of CYP3A5*3 and ABCB1 C3435T on clinical outcomes and trough plasma concentrations of imatinib in Nigerians with chronic myeloid leukaemia. J. Clin. Pharm. Ther. 2016;41:546–551. doi: 10.1111/jcpt.12424. [DOI] [PubMed] [Google Scholar]
- 117.Maddin N., Husin A., Gan S.H., Aziz B.A., Ankathil R. Impact of CYP3A4*18 and CYP3A5*3 Polymorphisms on Imatinib Mesylate Response Among Chronic Myeloid Leukemia Patients in Malaysia. Oncol. Ther. 2016;4:303–314. doi: 10.1007/s40487-016-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang Y., Wen G., Lu Y., Wen J., Ji Y., Xing X., Li Y., Wen J., Yuan H. CYP3A4*1G and CYP3A5*3 genetic polymorphisms alter the antihypertensive efficacy of amlodipine in patients with hypertension following renal transplantation. CP. 2017;55:109–118. doi: 10.5414/CP202559. [DOI] [PubMed] [Google Scholar]
- 119.Kim K., Park P., Lee O., Choi S., Min B., Shin K., Chun B., Shin J., Park J. Effect of CYP3A53 genotype on the pharmacokinetics and pharmacodynamics of amlodipine in healthy Korean subjects. Clin. Pharm. Ther. 2006;80:646–656. doi: 10.1016/j.clpt.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 120.Cabaleiro T., López-Rodríguez R., Román M., Ochoa D., Novalbos J., Borobia A., Carcas A., Abad-Santos F. Pharmacogenetics of quetiapine in healthy volunteers: Association with pharmacokinetics, pharmacodynamics, and adverse effects. Int. Clin. Psychopharmacol. 2015;30:82–88. doi: 10.1097/YIC.0000000000000047. [DOI] [PubMed] [Google Scholar]
- 121.Shilbayeh S.A.R., Sy S.K.B., Melhem M., Zmeili R., Derendorf H. Quantitation of the impact of CYP3A5 A6986G polymorphism on quetiapine pharmacokinetics by simulation of target attainment: Clinical Pharmacology in Drug Development. Clin. Pharm. Drug Dev. 2015;4:387–394. doi: 10.1002/cpdd.172. [DOI] [PubMed] [Google Scholar]
- 122.Bakken G.V., Molden E., Hermann M. Impact of Genetic Variability in CYP2D6, CYP3A5, and ABCB1 on Serum Concentrations of Quetiapine and N-desalkylquetiapine in Psychiatric Patients. Ther. Drug Monit. 2015;37:256–261. doi: 10.1097/FTD.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 123.Noetzli M., Guidi M., Ebbing K., Eyer S., Wilhelm L., Michon A., Thomazic V., Stancu I., Alnawaqil A.-M., Bula C., et al. Population pharmacokinetic approach to evaluate the effect of CYP2D6, CYP3A, ABCB1, POR and NR1I2 genotypes on donepezil clearance: Pharmacogenetic study on donepezil. Br. J. Clin. Pharmacol. 2014;78:135–144. doi: 10.1111/bcp.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Magliulo L., Dahl M.-L., Lombardi G., Fallarini S., Villa L.M., Biolcati A., Scordo M.G. Do CYP3A and ABCB1 genotypes influence the plasma concentration and clinical outcome of donepezil treatment? Eur. J. Clin. Pharmacol. 2011;67:47–54. doi: 10.1007/s00228-010-0883-5. [DOI] [PubMed] [Google Scholar]
- 125.Matsumoto J., San S.N., Fujiyoshi M., Kawauchi A., Chiba N., Tagai R., Sanbe R., Yanaka S., Sakaue H., Kato Y., et al. Effect of CYP3A5*3 genetic variant on the metabolism of direct-acting antivirals in vitro: A different effect on asunaprevir versus daclatasvir and beclabuvir. J. Hum. Genet. 2020;65:143–153. doi: 10.1038/s10038-019-0685-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.