Abstract
The multi-drug resistance of the opportunistic pathogen Acinetobacter baumannii is of growing concern, with many clinical isolates proving to be resistant to last resort as well as front line antibiotic treatments. The use of bacteriophages is an attractive alternative to controlling and treating this emerging nosocomial pathogen. In this study, we have investigated bacteriophages collected from hospital wastewater in Thailand and we have explored their activity against clinical isolates of A. baumannii. Bacteriophage vB_AbaM_PhT2 showed 28% host range against 150 multidrug resistant (MDR) isolates and whole genome sequencing did not detect any known virulence factors or antibiotic resistance genes. Purified vB_AbaM_PhT2 samples had endotoxin levels below those recommended for preclinical trials and were not shown to be directly cytotoxic to human cell lines in vitro. The treatment of human brain and bladder cell lines grown in the presence of A. baumannii with this bacteriophage released significantly less lactate dehydrogenase compared to samples with no bacteriophage treatment, indicating that vB_AbaM_PhT2 can protect from A. baumannii induced cellular damage. Our results have also indicated that there is synergy between this bacteriophage and the end line antibiotic colistin. We therefore propose bacteriophage vB_AbaM_PhT2 as a good candidate for future research and for its potential development into a surface antimicrobial for use in hospitals.
Keywords: Acinetobacter baumannii, antibiotic resistance, Thailand, opportunistic, nosocomial, bacteriophage, antibiotic alternative, phage therapy
1. Introduction
Acinetobacter baumannii infection has become a research priority due to its exceptional ability to acquire antimicrobial resistance (AMR) [1,2]. Strains of this Gram negative bacterium have been shown to have high genetic diversity and large genetic ‘resistance islands’ [3], and there have been cases of pan-drug resistant isolates that are resistant to all known clinical antibiotics [4]. This has made A. baumannii a formidable pathogen in the nosocomial environment, particularly against immunocompromised and intensive care unit (ICU) patients. Often targeting moist tissues such as mucous membranes [5], A. baumannii infection will commonly result in urinary tract infections, meningitis, pneumonia, and wound site infections. Forming strong biofilms, A. baumannii is also commonly connected with the use of catheters and ventilators [6], the latter of which is associated with particularly high mortality rates [7].
Current therapies against A. baumannii infections include the beta-lactamase inhibitor sulbactam, carbapenems, aminoglycosides, and tetracyclines [8]. However, there is no longer an empirical antimicrobial agent treatment [8] and the extensive use of the broad spectrum carbapenems [9] has led the World Health Organization to highlight carbapenem resistant A. baumannii as a Class 1: Priority Critical pathogen for the focus of research and development of novel antibiotics in 2017 [1]. End line treatment options include tigecycline and colistin [10], both of which have been associated with severe side effects, including hepatotoxicity, nephrotoxicity, and neurotoxicity [6].
A. baumannii infections are of particular concern in Thailand [11] where this pathogen is the most common cause of death by AMR associated nosocomial infection [7]. According to NARST (National Antimicrobial Resistance Surveillance Centre, Thailand), 98% of A. baumannii isolates were susceptible to the carbapenem imipenem in 1998 [1,12], but by 2002, this had dropped to 79%, and by 2008 only 43% of isolates screened were susceptible. Data collected for 2018 indicated that just over 32% of A. baumannii isolates in Thailand were susceptible to imipenem and 31.8% to meropenem. In the same year, only 30.7% of 42,212 isolates were susceptible to sulbactam, 39.1% to the aminoglycoside gentamycin, and 18.5% to tetracycline [12]. Currently, 97.1% of isolates are recorded to be susceptible to the last resort treatment, colistin. However, resistance genes to this antibiotic were discovered in 2011 [13] and if the trends follow those seen for the carbapenems, the number of colistin susceptible A. baumannii isolates could be reduced by two thirds within the next 20 years.
Bacteriophages are an attractive alternative to treating this drug resistant pathogen in the upcoming ‘post antibiotic era’ [14]. The application of lytic bacteriophages could be prepared as a cocktail or as a combinatorial therapy, thereby reducing the chances of novel resistances being developed [15]. Phage therapy [16] has been shown to be successful in treating mouse models for A. baumannii pneumonia [17] and wound infections [18], with the first human trial of bacteriophages being used against A. baumannii published in 2017 [19]. Gaining regulatory approval for phage therapy is complex and slow however, and as yet, there are no approved mainstream bacteriophage medicines in the EU or USA. In 2007 though, the Food and Drug Agency (FDA) approved the use of bacteriophages as a food additive to protect consumers against Listeria monocytogenes in meat [20], and over time, the regulation of phage therapy may become more straight forward. Until this time, bacteriophages still have the potential to be utilised for the benefit of human health in hand sanitizers, surface disinfectants, or antimicrobials [21], and should not be overlooked as a powerful disease control tool. We are only just beginning to touch on the scope of how bacteriophages could be used in the pursuit of substantial human health benefits.
This study initially investigated five bacteriophages, isolated from hospital wastewater treatment plants in the lower northern Thai province of Phitsanulok, due to their good host range against 150 multidrug resistant (MDR) A. baumannii isolates [11,22]. A preliminary investigation using 24-h growth curves indicated that bacteriophage vB_AbaM_PhT2 not only had the highest relative host range of those screened, but also maintained low A. baumannii turbidity for the longest period of the five isolates during growth curves. This bacteriophage was therefore carried forward for a more detailed investigation, with the aim of potentially developing a surface disinfectant. Upon sequencing of the vB_AbaM_PhT2 genome, there were no known virulence factors or antibiotic resistance genes found. The treatment of human brain and bladder cell lines with this bacteriophage produced a significant reduction in relative cytotoxicity caused to the human cells by being grown in the presence of A. baumannii. There was also an indication of vB_AbaM_PhT2 synergy with colistin, potentially allowing the clinical dosage of this nephrotoxic drug to be reduced. We therefore propose vB_AbaM_PhT2 as a candidate for future research and development into a tailored surface antimicrobial for use in hospitals.
2. Results
2.1. Acinetobacter baumannii Bacteriophages Present Broad Host Range
Bacteriophages, previously isolated from wastewater at Buddhachinaraj and Bang Rakam hospitals, were screened against multidrug resistant (MDR) A. baumannii strains [22] from five tertiary hospitals (HA-HE, see methods) (where MDR-AB is defined as strains resistant to at least three classes from penicillins/cephalosporins, fluroquinolones, and aminoglycosides [23,24]). The five of these bacteriophages with the highest host range were carried forward for further investigation in this study (Table 1 and Figure 1). All five bacteriophages had been previously partially characterised and were all confirmed to belong to the lytic non-enveloped dsDNA Caudovirales order of viruses via electron microscopy [25,26,27,28,29] (Supplementary Figure S2). Bacteriophages vPhT2, 4, and 44 were all Myoviridaes, whereas vPhT29 and 39 were the smaller Podoviridaes. Formerly, these bacteriophages were published under a different, less standardised nomenclature, but have now been renamed to reflect the Kropinski system [30] (see methods) for clarity and consistency with other bacteriophage research (Table 1). These bacteriophages are now called vB_AbaM_PhT2, vB_AbaM_PhT4, vB_AbaP_PhT29, vB_AbaP_PhT39, and vB_AbaM_PhT44 (previously ØAB02, ØAB04, ØAB29, ØAB39 and ØAB44), but for the purposes of the flow of the text in this paper, these names have also been abbreviated to vPhT2, vPhT4, vPhT29, vPhT39, and vPhT44 respectively [30].
Table 1.
Overview of properties of Acinetobacter baumannii bacteriophages with broad host ranges.
| Bacteriophage | Abbreviated Name | Previously Published Name | Host Strain | Family | Plaque Size |
|---|---|---|---|---|---|
| vB_AbaM_PhT2 | vPhT2 | ØAB02 [27,29] | A. baumannii AB183 | Myoviridae | 1 mm |
| vB_AbaM_PhT4 | vPhT4 | ØAB04 [27] | A. baumannii AB22 | Myoviridae | 12 mm |
| vB_AbaP_PhT29 | vPhT29 | ØAB29 [29] | A. baumannii AB20 | Podoviridae | 2–3 mm |
| vB_AbaP_PhT39 | vPhT39 | ØAB39 [29] | A. baumannii AB22 | Podoviridae | 12 mm |
| vB_AbaM_PhT44 | vPhT44 | ØAB44 [29] | A. baumannii AB20 | Myoviridae | 2–3 mm |
See also Figures S1 and S2.
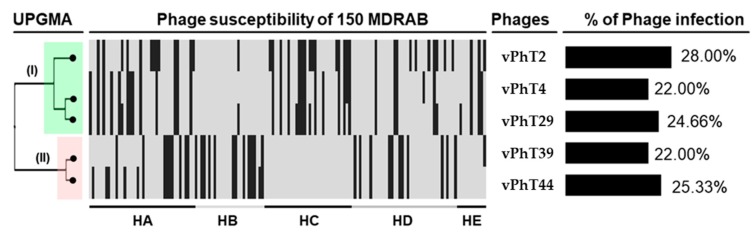
Figure 1.
Information on host ranges of Thai bacteriophages against 150 multidrug resistant Acinetobacter baumannii (MDR-AB) strains from five hospitals. From left to right are presented: a UPGMA (Unweighted Pair Group Method with Arithmetic mean) dendrogram, host susceptibility to isolates from different hospitals (black = A. baumannii isolates susceptible to bacteriophages (clear plaque), grey = A. baumannii isolates resistant to the bacteriophages (no plaque), HA–HE = hospitals A–E) and bacteriophage names and percentage host ranges.
This study showed host ranges of 28.00%, 22.00%, 22.00%, 24.66%, and 25.33% when screened against 150 MDR A. baumannii for vPhT2, vPhT4, vPhT29, vPhT39, and vPhT44, respectively. Ninety (60%) of the 150 isolates were killed by at least one of the five bacteriophages. An unweighted pair group method with arithmetic mean (UPGMA) dendrogram (Figure 1), which was created to represent the binary clustering of susceptibility of the 150 MDR-AB isolates to the bacteriophages, revealed that the five bacteriophages could be grouped into two distinct clusters (clusters I and II), based on similarities in host range. Cluster I consisted of three bacteriophages (vPhT2, vPhT4, and vPhT39), whereas vPhT29 and vPhT44 belonged to cluster II.
2.2. Bacteriophage Activity against Their Bacterial Host
2.2.1. Bacteriophages Reduce A. baumannii Turbidity over 24 Hours
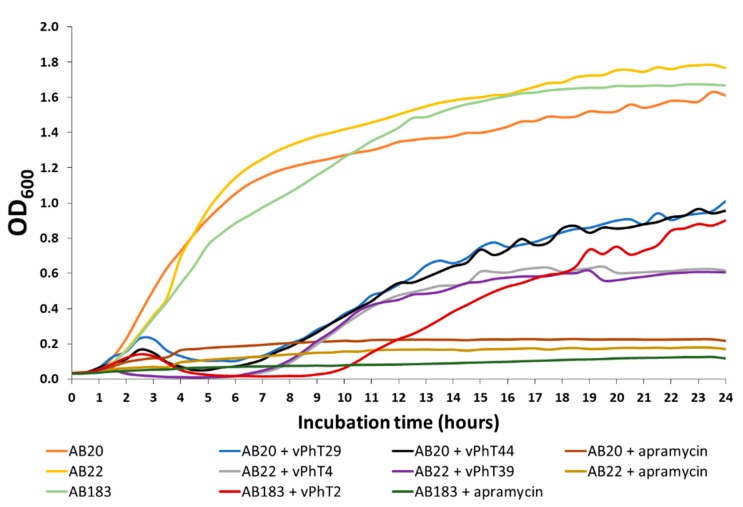
In a 24-h growth curve of bacterial samples treated with each bacteriophage at an MOI of 0.01, there was a brief rise in OD600 (up to ~1.5 to 2.0 h incubation) before a drop in turbidity compared to the untreated controls (Figure 2). For bacteriophage vPhT44, the OD600 remained low until around 5.5 h, when it gradually increased again for the rest of the time course. This gradual increase was also seen for vPhT4, vPhT29, and vPhT39 from around 6.5 h, but for vPhT2, the increase was not until around nine hours. The OD600 of all samples being treated with bacteriophages remained well below that of the untreated controls throughout the 24-h incubation period (Table 2). The lowest OD600 readings over the 24-h period (not including lag phase readings during the first hour) after treatment with vPhT2, vPhT4, vPhT29, vPhT39, and vPhT44 were observed after 7.0, 4.5, 6.0, 4.5, and 4.5 h respectively (Figure 2).
Figure 2.
Twenty-four-hour growth curve for Acinetobacter baumannii strains grown with and without bacteriophages or apramycin (500 µg/mL). Data displayed as OD600 (N = 3).
Table 2.
Relative metabolic endpoint activity and turbidity of Acinetobacter baumannii samples treated for 24 h with bacteriophages or apramycin.
| Sample | Relative Metabolic Activity (OD540) 1 | Relative Turbidity (OD600) | ||||
|---|---|---|---|---|---|---|
| Average (OD540 ± SD) | Normalised (% ± SD) | Significance of Results 2 | Average (OD600 ± SD) | Normalised (% ± SD) | Significance of Results 2 | |
| AB20 | 1.579 ± 0.422 | 100.00 ± 0.00 | N/A | 1.863 ± 0.264 | 100.00 ± 0.00 | N/A |
| AB20 + apramycin | 0.006 ± 0.001 | 0.41 ± 0.110 | *** | 0.240 ± 0.144 | 13.47 ± 8.57 | *** |
| AB20 + vPhT29 | 1.030 ± 0.368 | 64.89 ± 11.28 | *** | 1.055 ± 0.338 | 56.87 ± 16.74 | *** |
| AB20 + vPhT44 | 1.027 ± 0.326 | 65.22 ± 11.15 | *** | 1.119 ± 0.319 | 59.53 ± 11.51 | *** |
| AB20 + vPhT29 + vPhT44 | 1.151 ± 0.443 | 71.52 ± 13.82 | ** | 1.156 ± 0.262 | 60.84 ± 8.07 | *** |
| AB22 | 1.520 ± 0.188 | 100.00 ± 0.00 | N/A | 1.811 ± 0.127 | 100.00 ± 0.00 | N/A |
| AB22 + apramycin | 0.009 ± 0.003 | 0.60 ± 0.28 | *** | 0.245 ± 0.127 | 13.56 ± 7.35 | *** |
| AB22 + vPhT4 | 0.888 ± 0.151 | 58.96 ± 11.08 | *** | 0.592 ± 0.170 | 32.96 ± 10.01 | *** |
| AB22 + vPhT39 | 0.984 ± 0.198 | 64.82 ± 11.28 | *** | 0.695 ± 0.240 | 38.60 ± 13.92 | *** |
| AB22 + vPhT4 + vPhT39 | 0.794 ± 0.186 | 56.19 ± 12.42 | * | 0.673 ± 0.165 | 38.16 ± 9.67 | ** |
| AB183 | 1.562 ± 0.186 | 100.00 ± 0.00 | N/A | 1.671 ± 0.071 | 100.00 ± 0.00 | N/A |
| AB183 + apramycin | 0.009 ± 0.007 | 0.560 ± 0.370 | *** | 0.134 ± 0.061 | 7.93 ± 3.39 | *** |
| AB183 + vPhT2 | 0.978 ± 0.306 | 62.94 ± 20.13 | ** | 0.832 ± 0.247 | 49.79 ± 14.19 | *** |
* p < 0.05, ** p < 0.01, *** p < 0.001, SD = standard deviation, N/A = not applicable. Apramycin concentration = 500 µg/mL. 1 Relative metabolic activity was calculated from OD540 readings after application of triphenyl tetrazolium chloride (TTC). 2 Significance was calculated from a two tailed paired t-test compared to the untreated, bacteria only controls.
Samples of A. baumannii were also treated with apramycin to produce a comparison of changes in cell turbidity when treated with known antibiotics versus bacteriophages. The turbidity of the samples treated with apramycin remained low throughout the time course with the lowest OD600 being produced much earlier than the bacteriophage samples, at around 1.5 h, after which there was a slow but gradual increase.
2.2.2. Bacteriophages Reduce Relative A. baumannii Metabolic Activity over 24 Hours
As a further assessment of relative bacteriophage activity against their host, an endpoint triphenyl tetrazolium chloride (TTC) assay was performed on growth curve cultures, where the colorimetric TTC product was a measure of dehydrogenase activity and cell respiration, and was proportional to bacterial metabolic activity (measured at OD540) (Table 2). A significant reduction in metabolic activity (as well as turbidity) was observed for all bacteriophages at 24 h compared to the untreated controls (where significance was calculated using a two-tailed, paired t-test with a p-value of less than 0.05 being defined as significant). The percentage reduction in relative A. baumannii metabolic activity (OD540) ranged between 26.27% and 41.04% and turbidity (OD600) was reduced between 39.16% and 67.04%. Bacteriophage vPhT4 produced the greatest reductions in both metabolic activity and turbidity by 24 h. There was a positive correlation coefficient between turbidity and metabolic activity for all samples at 24 h (r = 0.95, p < 0.00001).
2.2.3. Pairs of Bacteriophages Do Not Act Synergistically
Growth curves were also produced using mixes of pairs of bacteriophages that had the same host (vPhT4 and vPhT39 with AB22 and vPhT29 and vPhT44 with AB20). As before, the relative endpoint metabolic activity (TTC at OD540) and turbidity (OD600) of the host A. baumannii were analysed (Table 2). No synergy was detected and so the use of these combinations was not replicated in future testing.
2.2.4. Bacteriophage vPhT2 Presents the Best Performance in all the Parameters Tested
No single bacteriophage performed best in every single assay performed. The relative performance of bacteriophages across all assays was compared in order to select one for further characterisation (Table 3). Bacteriophage vPhT2 performed consistently well across all experiments: it maintained a low A. baumannii OD600 for the longest period in the 24-h growth curves (Figure 2) and had the widest host range (Figure 1). Bacteriophage vPhT2 was therefore selected for an investigation of its suitability as a surface antimicrobial or as a potential candidate to be used for phage therapy.
Table 3.
Ranking of bacteriophage efficacy and performance across all assays from this study.
| Variable | vPhT2 | vPhT4 | vPhT29 | vPhT39 | vPhT44 |
|---|---|---|---|---|---|
| Host range 1 | 1 | 4 | 4 | 3 | 2 |
| Host metabolism 2 | 2 | 1 | 4 | 3 | 5 |
| Host turbidity 3 | 3 | 1 | 4 | 2 | 5 |
| Maintaining low host turbidity 4 | 1 | 2 | 3 | 3 | 5 |
Bacteriophages that performed equivalently have been given the same ranking for that variable. 1 Host range (1 = broadest host range, 5 = narrowest host range). 2 Reduction in relative endpoint host metabolism (OD540 of TTC assay) after 24 h incubation with the bacteriophage (1 = lowest metabolism, 5 = highest metabolism). 3 Reduction in endpoint host turbidity (OD600) after 24 h incubation with the bacteriophage (1 = lowest turbidity, 5 = highest turbidity). 4 Length of time maintaining a low host OD600 during 24-h growth curves (1 = longest, 5 = shortest).
2.3. Genomic Analysis of vPhT2
2.3.1. Sequencing of the Genome of vPhT2 Phage
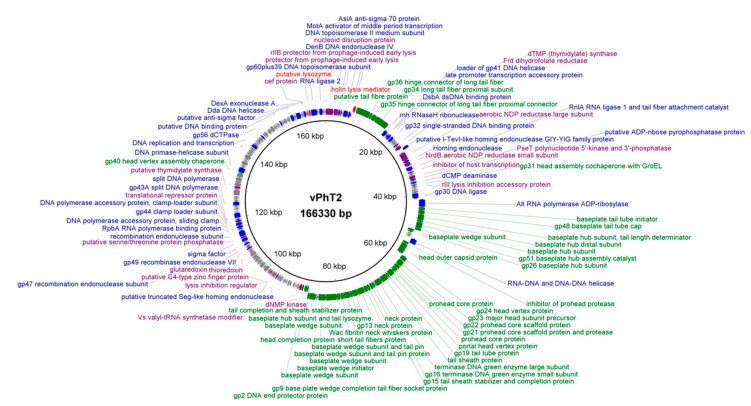
Sequencing of the vPhT2 genome revealed a genome size of 166,330 bp, with 246 predicted protein coding sequences and 9 tRNAs in a single chromosome with an average coverage of 596x (Figure 3, Table 4) (NCBI accession number: MN864865). The genome was determined to be double-stranded DNA with a G + C content of 36.4%. Of the 246 predicted open reading frames (ORFs), the majority genes encoded for proteins with no known function (144 genes) and 102 putative gene products (41%) were assigned to known functions. Putative proteins encoded by vPhT2 can be categorised into four groups: structural and morphogenesis (43 proteins), DNA replication/transcription (37 proteins), host lysis (2 proteins), and ‘other’ (20 proteins) (Figure 3). The major capsid proteins, major and minor tail proteins, baseplate hub subunit protein, tail fiber proteins and prohead core proteins involved in structure and morphogenesis were identified, as were the genes involved in DNA replication and transcription, such as endodeoxyribonucleases, DNA helicases, DNA polymerases, transcriptional regulators, and RNA ligases. With regards to host lysis by vPhT2, an endolysin and holin lysis mediator were found. Moreover, a gene coding for a tail-associated lysozyme was found (locus tag: vB_AbaM_PhT2_173). Additional genes identified coded for a predicted thymidylase synthase, DNA adenine methylase, acetyltransferase, thymidylate synthase and dihydrofolate reductase (Figure 3). No known genes involved in antibiotic-resistance and virulence factors were detected in the vPhT2 genome when searched in the virulence factor database (VFDB) [31] and in the antibiotic resistance genes database (ARDB) [32].
Figure 3.
Graphic circular map of vPhT2 genome and its predicted open reading frames (ORFs) (NCBI accession number: MN864865). From inside to outside: the scale units in kilo base pairs (kbp), predicted genes on forward strand and predicted genes on reverse strand. Predicted genes are coloured according to their function (green = structural and morphogenesis proteins, blue = DNA replication and transcription, red = host lysis, purple = other functions and grey = hypothetical proteins with unknown function). The genome map was drawn using the BLAST Ring Image Generator (BRIG) software, version 0.95, The University of Queensland, Brisbane, Australia.
Table 4.
Comparison between vPhT2 and its most similar genomes in the NCBI database including pairwise genome comparison ANI-tree similarity matrix compare and gene structure of vPhT2.
| Characteristic | vPhT2 | AbTZA1 | AM101 | KARL-1 | vB_ApiM_fHyAci03 | ZZ1 |
|---|---|---|---|---|---|---|
| vPhT2 ANI (%) | N/A | 95.34 | 78.16 | 77.45 | 77.35 | 71.31 |
| Accession number | MN864865 | MK278860 | MH165274 | MH713599 | MH460829 | HQ698922 |
| Genome size (bp) | 166,330 | 168,223 | 166,487 | 166,560 | 165,975 | 166,687 |
| GC content (%) | 36.4 | 36.3 | 36.7 | 36.8 | 36.8 | 34.4 |
| Number of ORFs | 246 | 253 | 250 | 253 | 247 | 256 |
| Number of tRNAs | 9 | 6 | 8 | 0 | 8 | 8 |
| Year uploaded | 2019 | 2019 | 2019 | 2018 | 2018 | 2012 |
| Isolate location | Thailand | Israel | Russia | Germany | Finland | China |
| Family | Myoviridae | Myoviridae | Myoviridae | Myoviridae | Myoviridae | Myoviridae |
| Reference | This study | Unpublished | Unpublished | Jansen et al. [34] | Pulkkinen et al. [35] | Jin et al. [36] |
ANI = average nucleotide identity, N/A = not applicable, bp = base pairs, ORF = open reading frame.
2.3.2. Comparison of vPhT2 Genome against the NCBI Database
A BLASTn [33] search against the NCBI database using vPhT2 genome as a query resulted in the identification of the five most closely related genomes, AbTZA1, AM101, KARL-1 [34], vB_ApiM_fHyAci03 [35] and ZZ1 [36] (Table 4), all of which were Acinetobacter infecting bacteriophages. At the nucleotide level, the vPhT2 genome was only genetically very similar to Israeli isolate AbTZA1, where an average nucleotide identity (ANI) of 95.34% was calculated (with a cut-off of >95% to represent being of the same species). A low ANI (~71–78%) was seen with the next four most similar bacteriophages, namely AM101, KARL-1, vB_ApiM_fHyAci03, and ZZ1.
2.4. Preparations of Bacteriophage vPhT2 Shows Good Stability in Long Term Studies in Lysogeny Broth, but Not in SM Buffer II
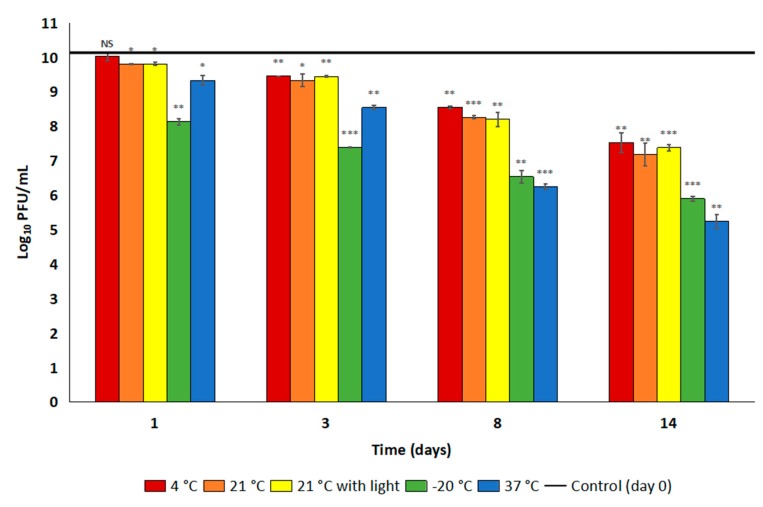
Aliquots of bacteriophage vPhT2 stored in lysogeny broth (LB) or SM buffer II (see methods) at a number of temperatures and in the presence and absence of light were titred at multiple recovery points in order to establish the laboratory stability of the suspensions. Bacteriophage suspensions stored in LB in the fridge (4 °C) and at room temperature (21 ± 2 °C), both protected from and exposed to light, did not show a significant reduction in titre during the first 14 days of storage (Table 5 and Supplementary Table S2). When the samples were stored in SM buffer II however, a reduction in titre was seen almost immediately with reductions of 1.57 ± 0.03, 1.87 ± 0.04 and 1.92 ± 0.21 log10PFU/mL after only eight days at 4 °C, 21 °C without light, and 21 °C with light respectively (Figure 4). This degradation continued over the rest of the time course, with a greater than 2.5 log reduction being seen after 14 days storage for all three samples. When stored in LB in the fridge during an extended study over 10 months, only a 1.30 ± 0.08 log10PFU/mL reduction in bacteriophage titre was seen in this time (data not shown here). When comparing the room temperature samples (21 °C) kept in the dark with those exposed to light, there was no significant difference in titre of samples stored in SM buffer II seen at any time point for 14 days (p > 0.05).
Table 5.
Logarithmic reduction in bacteriophage vPhT2 after storage in SM buffer II or LB for 14 days in different temperature and light conditions.
| Storage Location | Storage Reagent | Reduction in Titre (log10PFU/mL ± SD) | |||
|---|---|---|---|---|---|
| 1 Day | 3 Days | 8 Days | 14 Days | ||
| Fridge (4 °C) | SMII | NR | 0.66 ± 0.00 | 1.57 ± 0.03 | 2.60 ± 0.29 |
| Room temperature (21 °C) | SMII | 0.32 ± 0.01 | 0.79 ± 0.18 | 1.87 ± 0.04 | 2.94 ± 0.33 |
| Room temperature with light (21 °C) | SMII | 0.32 ± 0.04 | 0.68 ± 0.03 | 1.92 ± 0.21 | 2.75 ± 0.10 |
| Freezer (−20 °C) | SMII | 1.99 ± 0.09 | 2.74 ± 0.01 | 3.59 ± 0.18 | 4.23 ± 0.06 |
| Biological incubator (37 °C) | SMII | 0.79 ± 0.14 | 1.58 ± 0.06 | 3.88 ± 0.07 | 4.88 ± 0.21 |
| Fridge (4 °C) | LB | NR | NR | NR | NR |
| Room temperature (21 °C) | LB | NR | NR | NR | NR |
| Room temperature with light (21 °C) | LB | NR | NR | NR | NR |
PFU = plaque forming units, SD = standard deviation, NR = no significant reduction (p > 0.05), LB = lysogeny broth, SMII = SM buffer II (see methods).
Figure 4.
Logarithmic titre of vPhT2 after storage in SM buffer II or LB for 14 days in different temperature and light conditions compared to day zero. (NS = no significance, * p < 0.05, ** p < 0.01, *** p < 0.001).
For samples stored in SM buffer II in the freezer (−20 °C), there was already a 1.99 ± 0.09 log10PFU/mL reduction in titre after only 24 h, compared to time zero (p < 0.05) (Table 5). This then followed with log reductions of 2.74 ± 0.01, 3.59 ± 0.18 and 4.23 ± 0.06 log10PFU/mL by three (3), eight (8) and fourteen (14) days storage compared to time zero. Storage at 37 °C in SM buffer II resulted in the greatest degradation of vPhT2, with a significant reduction in titre of 0.79 ± 0.14 log10PFU/mL seen after 24 h (p < 0.05). By three days this was a 1.58 ± 0.06 log10PFU/mL reduction and by eight and 14 days this was 3.88 ± 0.07 and 4.88 ± 0.21 log10PFU/mL, respectively.
2.5. vPhT2 Activity in the Human Cell Environment
2.5.1. CsCl Purified vPhT2 Preparations Had Low Endotoxin Levels
Bacteriophage vPhT2 was purified via a caesium chloride gradient prior to testing in the human cell environment (and stability testing trials) in order to minimise the number of endotoxins in the preparation. A 1 × 108 PFU/mL sample of CsCl purified vPhT2 in SM buffer II was found to have 10.92 endotoxin units per millilitre (EU/mL) (Table 6). Crude lysates containing vPhT2 in LB had endotoxin levels above the threshold of the assay.
Table 6.
Data on units of endotoxins measured in bacteriophage preparations.
| Sample | Sample Concentration | EU/mL |
|---|---|---|
| Crude vPhT2 in LB | 5 × 105 PFU/mL | Above threshold of standard curve |
| Purified vPhT2 in SM buffer II | 1 × 108 PFU/mL | 10.92 |
| LB medium | N/A | 0.09 |
| SM buffer II | N/A | 0.03 |
| Leibovitz medium | N/A | 0.05 |
PFU = plaque forming units, EU = endotoxin unit, LB = lysogeny broth.
2.5.2. vPhT2 Presents No Cytotoxicity to Human Cell Lines
Lactate dehydrogenase is an enzyme found in nearly all living cells and catalyses the production of pyruvate from lactate and back again, producing NAD+ and NADH [37]. As an intracellular enzyme, the extracellular presence of LDH in a supernatant is an indicator of cytotoxicity, human cell line death and damage. Treatment of T24 and hCMEC/D3 cell lines with bacteriophage vPhT2 (in the absence of A. baumannii) produced no significant difference (p > 0.05) in LDH release compared to cell lines that had not been treated, indicating that this bacteriophage was not cytotoxic (Table 7). All human cell lines had a significant increase (p < 0.05) in levels of LDH released after treatment with A. baumannii (in the absence of bacteriophages) compared to no bacterial infection (Table 7), indicating that the presence of A. baumannii was cytotoxic.
Table 7.
Raw data for lactate dehydrogenase levels (OD490–OD680) released by different human cell lines treated with Acinetobacter baumannii or vPhT2 compared to no treatment.
| Human Cell Line | Average Relative LDH Release (OD490–680 ± SD) | ||
|---|---|---|---|
| Human Cells Only | Human Cells and vPhT2 | Human Cells and AB183 | |
| T24 | 0.185 ± 0.012 | 0.204 ± 0.011 | 2.444 ± 0.057 |
| hCMEC/D3 | 0.017 ± 0.002 | 0.022 ± 0.004 | 0.486 ± 0.125 |
LDH = lactate dehydrogenase, SD = standard deviation.
2.5.3. vPhT2 Reduces LDH Production by Human Cell Lines Grown in the Presence of A. baumannii
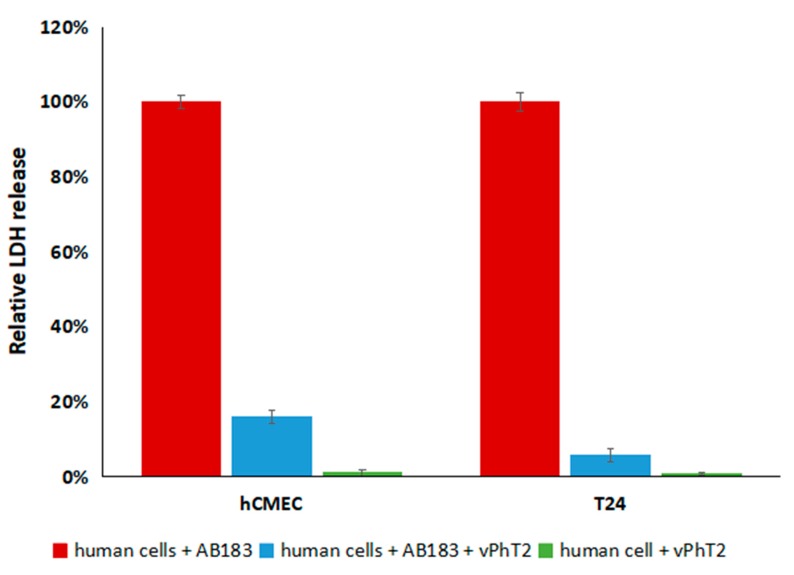
When vPhT2 was applied to human cell lines that had been exposed to A. baumannii for an hour, there was a 90.98 ± 0.49% and 84.01 ± 1.74% reduction in LDH production over the next 23 h by T24 and hCMEC/D3 cells, respectively (p < 0.001) (Figure 5).
Figure 5.
Normalised lactate dehydrogenase levels (LDH) (OD490–OD680) released by T24 and hCMEC/D3 human cell lines treated with Acinetobacter baumannii AB183 and or vPhT2 over 24 h (N = 3). Treatment of human cells with only AB183 represent 100% relative cytotoxicity. LDH levels produced by human cell negative controls with no treatment are representative of 0% cytotoxicity.
2.5.4. vPhT2 Reduces A. baumannii Activity in the Presence of Human Cell Lines
As a marker of bacterial inhibition, the endpoint turbidity (OD600) and titres of A baumannii from samples of the LDH assay were measured. The reduction in OD600 for the supernatant of the human cell lines exposed to A. baumannii and vPhT2 was 85.19% ± 4.85% and 89.35% ± 1.72% for T24 and hCMEC/D3 cells respectively, compared to samples only treated with A. baumannii (Table 8) (p < 0.001). There were also measured to be log reductions in A. baumannii titre of 3.34 ± 0.11 and 3.08 ± 0.13 log10CFU/mL when treated with vPhT2 in the presence of T24 and hCMEC/D3 cells respectively (Table 8). Unsurprisingly, there was a positive correlation between relative LDH levels produced by the human cell lines and the reduction in A. baumannii OD600, (r = 0.52, p < 0.00001) or endpoint titre (r = 0.67, p < 0.001). As A. baumannii growth was reduced, so were the levels of LDH released by human cell lines.
Table 8.
Raw data for Acinetobacter baumannii turbidity (OD600) and titre (log10CFU/mL) from samples grown in the presence of human cell lines.
| Human Cell Line | Average OD600 (±SD) | Average log10CFU/mL (±SD) | ||
|---|---|---|---|---|
| Human Cells and AB183 | Human Cells, AB183 and vPhT2 | Human Cells and AB183 | Human Cells, AB183 and vPhT2 | |
| T24 | 1.051 ± 0.040 | 0.156 ± 0.051 | 9.47 ± 0.01 | 6.13 ± 0.11 |
| hCMEC/D3 | 0.989 ± 0.073 | 0.105 ± 0.017 | 9.46 ± 0.07 | 6.38 ± 0.13 |
CFU = colony forming units, SD = standard deviation.
2.6. vPhT2 Works Synergistically with Colistin
Host strain A. baumannii AB183 was shown to be susceptible to colistin (EUCAST clinical breakpoint >2 µg/mL [38]) (Supplementary Table S3 and Figure S4) with a measured MIC of 1 µg/mL. Previous studies have indicated that endolysins from A. baumannii bacteriophages can work synergistically with colistin [27]. When colistin and bacteriophage vPhT2 were tested in combination against AB183, a fractional inhibitory concentration index (FICI) value of 0.35 was calculated (Table 9), indicating synergy between this bacteriophage and colistin (where the FICI was interpreted as follows: FICI ≤ 0.5, synergy; <2 and >0.5, indifference; ≥2, antagonism [39]).
Table 9.
Susceptibility of Acinetobacter baumannii host strain AB183 to colistin in combination with bacteriophage vPhT2.
| Treatment | Independent MIC Value | Combined MIC Value | FIC Values |
|---|---|---|---|
| Colistin | 1 µg/mL | 0.25 µg/mL | 0.25 |
| vPhT2 | 500 PFU/mL | 50 PFU/mL | 0.1 |
| FICI | 0.35 |
MIC = minimum inhibitory concentration, FIC = fractional inhibitory concentration, FICI = fractional inhibitory concentration index (calculated FICI = combined MIC/independent MIC), PFU = plaque forming units.
3. Discussion
The five bacteriophages in this study had a host range of between 22% and 28% of 150 MDR A. baumannii isolates. When analysing host range data and discussing the ‘broadness’ of the range, the diversity of location sources of the hosts should be taken into consideration [40]. Bacteriophages isolated from hospital waste-water are more likely to be efficacious against clinical isolates from the same hospital or region than independent sources, due to coevolution [28,29]. The A. baumannii isolates screened in this study were from five hospitals spread across Thailand, a country of over 500,000 km2 with a population of around 70 million inhabitants [41]. Hospitals were chosen to be representative of a range of different environments including industrial and rural areas, international borders, the presence of tourists and foreign workers, as well as geographic locations and regional climates. All five bacteriophages were from two hospitals in lower northern Thailand. Thus, the bacteriophages appear to have the potential to be used in a range of Thai hospital locations and possibly the rest of the world. In fact, a screen of the NCBI database [33] showed vPht2 to be of the same species (% ANI > 95) as a bacteriophage isolated from Israel, AbTZA1 (accession number: MK278860). Groupings produced by the UPGMA dendogram, also revealed that there are two groups of bacteriophage host ranges within the five bacteriophages screened. If applied as a cocktail of cluster I (vPhT2, vPhT4, vPhT39) and II (vPhT29 and vPhT44), the overall host ranges could be expected to be higher [29]. In fact, 60% (90 out of 150) of the clinical isolates were killed by at least one of the five bacteriophages.
Over the course of a 24-h growth curve, the OD600 of samples treated with the bacteriophages did not increase past 60.84% of the negative controls. Although there was a significant reduction in host turbidity by all bacteriophages at all-time points analysed, bacteriophage vPhT4 produced the greatest relative reductions in A. baumannii metabolic activity (TTC assay) and turbidity (OD600), whereas bacteriophage vPhT2 maintained the low OD600 for the longest period (nine hours compared to 5.5–6.5 h for all others). Bacteriophage vPhT2 also had the highest host range and for this reason was selected for further investigation and characterisation.
The production of sequencing data on vPhT2 means there is now the option for the genetic engineering of this bacteriophage, which could potentially lead to the optimisation of its use for multiple commercial applications. An analysis of the vPhT2 genome showed that it contained the gene to carry a lysozyme domain fused to its tail, suggesting that this bacteriophage encodes enzymes capable of degrading host’s cell wall at the phage’s first contact with the cell surface and the last step of the phage lytic cycle [42]. As the genome did not harbour any known virulence and antibiotic resistance genes that could potentially confer to a host, it shows promise as a suitable candidate for further applications in phage therapy.
Stability and a sensible shelf-life are important for the accessibility and usefulness of an antibacterial product in the clinical environment. Any specific storage conditions could complicate existing hospital procedures and possibly lead to incorrect usage by untrained individuals. It is promising therefore that the vPhT2 titre was stable at room temperature, the most likely routine storage temperature for a disinfectant, when stored in LB (both in the presence and absence of light) for 14 days. After 10 months in the fridge in this same diluent, there was just over a one log reduction in vPhT2 titre, indicating that this is a possible long-term storage option. However, lysogeny broth is not a common commercial storage medium for bacteriophages and it is unclear why the bacteriophage was less stable in SM buffer II. Further investigation is therefore required to find a more suitable, commercially relevant diluent for vPhT2. We also suggest the use of an opaque storage vessel in order to protect the long-term efficacy of bacteriophage suspensions.
Endotoxins are part of the outer membrane of the cell wall of Gram negative bacteria and are a known pyrogenic contaminant of injectable drugs [42]. Endotoxin levels for purified vPhT2 (1 × 108 PFU/mL) were shown to be below the threshold of 200 EU/mL as proposed by Brito et al. for endotoxin levels in live attenuated vaccines going through preclinical trials, based on United States Pharmacopoeia (USP) advice [42,43]. Should a higher dose or volume be required, endotoxin levels for dosages up to almost 20 times higher would still be below these guideline amounts and well within the USP recommendation of 5 EU/kg/hr [44].
The treatment of human T24 bladder and hCMEC/D3 brain cell lines with A. baumannii and vPhT2 resulted in a significant reduction in cytotoxicity (as measured by relative LDH levels) compared to when no bacteriophage was present. In addition, neither human cell line grown in the presence of vPhT2 (with no A. baumannii) released significantly more LDH than controls grown without exposure to bacteriophages indicating that vPhT2 is not cytotoxic.
Previous trials with purified A. baumannii bacteriophage endolysins showed broad range host activity and synergy with colistin [27]. The polymyxin colistin, with all its associated side effects, is one of the end line treatments for A. baumannii infection. With an FICI of 0.35, the indication that colistin can work synergistically with bacteriophage vPhT2 is very promising. Developing vPhT2 for use in the clinical environment, for example through developing an encapsulated delivery system [45] or further stability trials are therefore to be considered as the next steps for bacteriophage vPhT2.
4. Materials and Methods
4.1. Collection of Samples and Host Range Analysis
Seventeen bacteriophages were isolated from wastewater at Buddhachinaraj and Bang Rakam hospitals in Phitsanulok province, Thailand in 2010 and were screened against 150 MDR A. baumannii strains collected from five hospitals [11,22,27,29] using a spot test method as described previously [27]. For additional information on the chosen bacteriophages and MDR-AB, please refer to the supplement file: ‘Selected-MDRAB-150-isolates’. The first hospital (HA) was located in central Thailand, another one was in the lower northern region (HB), another one was on the northern tourist border with Myanmar (HC), one was located in an eastern industrial area with a high population of foreign workers (HD) and the final hospital was in northern Thailand (HE). Each hospital had between 405 and 1000 beds at the time of the study and isolates were collected from a range of different departments and specimen types.
Colonies of A. baumannii were suspended in 0.85% NaCl to an equivalent of a 0.5 McFarland standard (1 × 108 CFU/mL). The suspension was then swabbed on to Trypticase Soy Agar (TSA) to create a bacterial lawn. Phage suspensions (2 µL at 1 × 108 PFU/mL) were dropped into the lawn before plates were incubated at 37 °C for eight (8) hours to allow cell lysis. Bacterial clearance at the site of bacteriophage inoculation implied that the host was sensitive to that particular bacteriophage. All experiments were performed in duplicate.
Of the five bacteriophages with the highest host range, bacteriophages vPhT4 and vPhT39 were tested against the chosen A. baumannii host strain AB22, vPhT29 and vPhT44 against the host strain AB20 and vPhT2 against the host strain AB183 (Table 1, Supplementary Figure S1 and Table S1) due to the good titres seen for these bacteriophage-host pairs in initial screening studies. Bacteriophage vPhT2 also showed some specificity against host strain AB22. Bacteriophages vPhT2, 4, 39 and 44 were all isolated from Buddhachinaraj hospital whereas vPhT29 was from Bang Rakam hospital. Hosts for propagating these bacteriophages were isolated from two different hospitals, one in northern Thailand and another in central Thailand (Supplementary Table S1) and cultivated in LB and LBA (lysogeny broth agar) (1.5% agar) as described previously [29].
4.1.1. Analysis of Host Range Data
A UPGMA dendrogram was created using DendroUPGMA: a dendrogram construction utility website using the default settings [46]. The input query was for a binary outcome (yes or no). If a particular bacteriophage could infect an A. baumannii host, then it was given a rating equal to one (yes), and if it could not infect, it was given a rating equal to zero (no).
4.1.2. Naming of the Bacteriophages
The five bacteriophages were renamed (Table 1) for this study according to Kropinski system [30] to make their referencing and entry into open access databases more consistent with contemporary and progressive research. The prefix of vB_AbaM_PhT or vB_AbaP_PhT was used to indicate that these were bacterial viruses (vB), infecting A. baumannii (Aba), were either Myoviridaes or Podoviridaes (M or P) and were isolated from Phitsanulok province in Thailand (PhT). This nomenclature system was not used in previous publications with these bacteriophages as they were first isolated only a year after the Kropinski system was first suggested in 2009 and knowledge of this nomenclature was not widespread or commonly practiced.
4.2. Bacteriophage Characterisation
4.2.1. Viral Enrichment—Propagation of Bacteriophages of Interest
To propagate the bacteriophage isolates, the A. baumannii hosts were grown overnight in LB (Sigma-Aldrich: Lennox—10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) at 37 °C and 130 rpm. In the morning, 1 mL of the overnight liquid culture was used to inoculate 50 mL fresh LB. This was then incubated at 37 °C and 130 rpm until an OD600 of 0.3 was reached. At this point, 100 μL of bacteriophage stock was added to each flask and samples were incubated for a further four hours. Bacterial debris was pelleted by centrifugation at 3220 g for 10 min before the supernatant was passed through a 0.2 μm pore size membrane filter. The prepared phage stocks in LB were stored at 4 °C. Bacteriophage samples stored in LB were used for initial 24-h growth curve assays, stability trials, and other bacterial studies.
4.2.2. Caesium Chloride Purification of Bacteriophages
Bacteriophages were propagated as described above, with the assay being scaled up to a 250 mL sample. Sodium chloride was added to samples to a final concentration of 1 M. After incubation on ice for one hour, samples were centrifuged at 3220× g and the supernatant passed through a 0.2 μm pore size membrane filter before PEG8000 was added to a final concentration of 10% w/v. This was then left overnight at 4 °C, before the samples were centrifuged at 25,000× g for 60 min. The phage pellet was then resuspended in 6–7 mL SM buffer I (1 M NaCl, 8 mM MgSO4 · 7H2O, 25 mM Tris-HCl pH 7.5) and passed through a 0.2 μm pore size membrane filter before undergoing concentration and further purification in a CsCl gradient for 20 h at 150,000× g and 4 °C. The extracted phage band was then dialysed first in SM buffer I and then twice with SM buffer II (100 mM NaCl, 8 mM MgSO4 · 7H2O, 25 mM Tris-HCl pH 7.5) in order to remove the CsCl. The purified phage was then stored at 4 °C (see stability trials, Section 2.4). Bacteriophage samples stored in SM buffer II were used in human cell assays and stability trials. Samples were processed promptly due to their instability in SM buffer II.
4.2.3. Plaque Assay—Quantification of Bacteriophages of Interest
Bacteriophage titre was determined via a soft agar plaque assay, using 0.7% agar top LBA [47]. One hundred microliters of serially diluted bacteriophage were incubated with 100 μL host cells (1 × 108 CFU/mL) at room temperature for 15 min before 3 mL molten top agar was added and poured over a 90 mm 1.5% agar LBA plate. Plaques were quantified after overnight incubation at 37 °C. For spot/drop tests, 100 μL host cells (1 × 108 CFU/mL) were added to molten 0.7% LBA, poured over a 1.5% agar LBA plate, and allowed to set before 5 μL bacteriophage (1 × 109 PFU/mL) or 5 or 10 µL antibiotic were spotted onto the surface and allowed to dry.
4.2.4. Classification of Bacteriophage Family
Bacteriophage families were classified as previously described, using electron microscopy [27,29,48].
4.3. Bacterial Host Characterisation
Antibiotic Susceptibility and MIC Testing
An initial screen of antibiotic susceptibilities was done using a drop test, where 10 µL of different antibiotics were spotted onto a lawn of A. baumannii on LBA. To determine if the three A. baumannii host strains were multi, extensively or pan drug resistant, the more quantitative disc diffusion method was carried out on selected clinically relevant antibiotics according to CLSI guidelines [49]. A further investigation into the colistin, apramycin, tigecycline, and meropenem resistance profiles of the A. baumannii host strains, was performed according to broth microdilution methods outlined by EUCAST [50] using cation adjusted Mueller Hinton broth 2 (CA-MHB) (Sigma-Aldrich—17.5 g/L acid hydrolysate of casein, 3 g/L beef extract, 1.5 g/L starch) and an initial inoculum of 5 × 105 CFU/mL. The CA-MHB used with tigecycline was either used fresh (<12 h) or frozen and used later, in order to optimise the activity of this antibiotic, which can be negatively impacted by media that has been stored for too long [10].
4.4. Bacteriophage Activity against the Bacterial Host
4.4.1. Twenty-Four-Hour A. baumannii Growth Curves
A total load of 1 × 107 CFU A. baumannii hosts were added to each of the wells of a 96 well plate (5 × 107 CFU/mL). Bacteriophages were added to a MOI of 0.01 (final load of 5 × 105 CFU/mL) (total volume = 200 µL). Samples were grown in LB at 37 ± 2 °C shaking in a plate reader with measurements of the optical density (OD600) taken every five minutes over a 24-h period. Additional MOIs were also tested in the MOI optimisation growth curve assay (Supplementary Figure S3a–e).
4.4.2. Triphenyl Tetrazolium Chloride Assay
After the 24 h growth curve, OD600 readings were completed, 50 µL of each sample was added to 150 µL water, and 50 µL 0.1% triphenyl tetrazolium chloride (TTC, Sigma-Aldrich) was diluted in LB and samples were incubated statically at 37 °C for three hours [48]. At this point the OD540 was measured, with the red coloured product being used as an indicator of relative metabolism. All tests were carried out with biological triplicates, with triplicate technical replicates assayed each time. Apramycin (final concentration of 500 µg/mL) was used as a positive control during this assay (a non-clinically relevant antibiotic was selected due to the host strains being resistant to most clinically relevant antibiotics trialled) (Supplementary Figure S4). A statistical analysis of results was carried out with Microsoft Excel using a two-tailed paired t-test, with a p-value of less than 0.05 being used as the threshold for significance.
4.5. Whole Genome Sequencing and Bioinformatics Analysis
Extraction and purification of bacteriophage genomic DNA was performed from on a high titre phage lysate (109 PFU/ml) using a phenol:chloroform method as previously described [47]. An Illumina sequencing library was generated with the Nextera XT DNA library preparation kit, following the manufacturer’s instructions. Sequencing was performed on an Illumina MiSeq (250 bp paired-end). The resultant reads were trimmed with Sickle version 1.33 using default settings [51], prior to being assembled with SPAdes version 3.6.0 using the “--only-assembler” option [52]. The resulting single contig was annotated with Prokka version 1.11 using a custom database constructed from proteins of all current viral genomes [53]. The genome sequence of the selected bacteriophage was deposited in GenBank [33] under accession number MN864865. The annotated ORFs were searched by using BLASTn to detect virulence and antibiotic resistance genes in the virulence factor database (VFDB) [31] and antibiotic resistance genes database (ARDB) [32]. Hits with more than 75% coverage and 50% identity were considered as positive results.
To compare the vPhT2 with other bacteriophages, a BLASTn search with the vPhT2 genome against the NCBI database was performed and the most similar genomes were selected for further study [33]. The average nucleotide identity (ANI) percentages, based on BLAST pairwise sequence alignments, of these bacteriophage genomes were calculated using JSpeciesWS version 3.2.6 [54] with the default settings. An ANI of above 95% was classed as an indicator of two genome being from the same species [55]. The circular map of the bacteriophage genome was generated through the BLAST Ring Image Generator (BRIG) program, version 0.95, written by Alikhan et al., Australian Infectious Diseases Research Centre, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, QLD 4072, Australia [56].
4.6. Stability Trials
Aliquots of propagated bacteriophage were stored in the fridge (4 °C), at room temperature (21 °C, both protected and exposed to light), at 37 °C and in the freezer (−20 °C) in LB or SM buffer II for 14 days. Samples were taken after one, three, eight and 14 days storage and were quantified using the plaque assay described above and tested in duplicate. Aliquots stored in the freezer only underwent one freeze/thaw cycle each. Additional aliquots of samples suspended in LB were also kept in the fridge for 10 months to provide data on longer term storage in this location. Samples were plated in duplicate and reductions in titre were calculated as PFU/mL and log10PFU/mL compared to samples from time zero.
4.7. Bacteriophage Activity in the Human Cell Environment
4.7.1. Endotoxin Testing
The presence of endotoxins in purified bacteriophage samples was measured using the Limulus Amebocyte Lysate (LAL) Chromogenic Endpoint Assay Kit from Hycult Biotech, according to the protocols supplied with the kit.
4.7.2. Lactate Dehydrogenase Cytotoxicity Assay
Human cell lines for this study were T24 urinary bladder epithelial cells (ATCC® HTB-4™) and hCMEC/D3 blood brain barrier endothelial cells (Sigma Aldrich SCC066). Cytotoxicity to human cells was measured using the Invitrogen™ CyQUANT™ LDH Cytotoxicity Assay Kit, according to the protocols supplied with the kit.
Human cell lines were grown up at 37 °C in their own cell specific media with 5% CO2 until they reached a suitable level of confluency. Cells were then trypsinised and resuspended in Leibovitz media (Sigma-Aldrich) with 5% FBS (Gibco) to a final concentration of 400,000 cells/mL. One millilitre of this suspension was then added to the wells of six well plates and grown overnight at 37 °C (no CO2 supplementation). On the same day, an overnight culture of A. baumannii was started in Leibovitz media + 5% FBS. Bacteriophages were diluted in LB to a final concentration of 1 × 109 PFU/mL. After 24 h growth, 800 µL of A. baumannii overnight culture (with refreshed medium) or 800 µL Leibovitz medium + 5% FBS were added to the human cell lines and incubated at 37 °C for one hour. After an hour, 200 µL host specific bacteriophage or 200 µL LB medium was added to relevant samples (final bacteriophage concentration of 1 × 108 PFU/mL). The total volume per well of the six well plates was 2 mL. Samples were then incubated for a further 23 h at 37 °C. After this incubation, aliquots were taken from each sample and the OD600 measured, as an indicator of A. baumannii turbidity. A second set of aliquots was taken in order to titre the CFU/mL of A. baumannii present, as a measure of host bacteria viability. A third set of aliquots was also taken and centrifuged at 17,000× g in a microfuge for five minutes to produce a cell pellet. Fifty microliters of the supernatant were then transferred to the wells of a 96 well plate in triplicate before adding 50 µL LDH assay test solution. Samples were mixed by tapping the side of the plate and then covered with foil to protect them from light during a 30-min incubation at room temperature. As a control, 200 µL lysis buffer supplied with the kit was added to relevant samples 45 min prior to adding LDH test solution. Fifty microliters of stop solution were added to all samples before measuring the optical density (OD) at 490 and 680 nm. Relative cytotoxicity to human cell lines was calculated from OD490 minus OD680. All tests were carried out with triplicate biological replicates. A statistical analysis of results was carried out with Microsoft Excel using a two-tailed un-paired t-test, with a p-value of less than 0.05 being used as the threshold for significance.
4.8. Synergy Trials
Synergy trials between colistin and bacteriophage vPhT2 were carried out using A. baumannii AB183 in a checker-board assay, following the same set up methods as the MIC testing, according to the EUCAST broth microdilution methods [50]. Synergy of colistin with the bacteriophages was determined from fractional inhibitory concentration (FIC) values, where a FIC is the MIC for an agent working in combination divided by the MIC of an agent when working alone. If the sum of the FIC values for both agents is ≤0.5, then this indicates synergy, whereas a ΣFIC > 0.5 and <2 shows indifference, and a value ≥2 indicates the antagonism of two agents against each other [39].
5. Conclusions
Understanding the in vivo activity of bacteriophages against their host bacteria is the first step in the successful development of a commercial or clinical product. In this study, the performance of a broad host range bacteriophage was analysed against its A. baumannii host and in the presence of human cell lines through the use of growth curves, bacterial viability assays, cytotoxicity studies, whole genome sequencing, stability testing, and assaying for synergy with colistin. Bacteriophage vPhT2 has shown potential to be made into a safe hand sanitizer or antimicrobial for use in hospitals, showing promise for development into a phage therapy tool in the future.
Acknowledgments
With thanks also to The British Council Newton Fund for funding this project, to Robert Spooner for his suggestions on analysing the data and EPSRC Bridging the Gaps AMR funding EP/M027503/1 for the establishment of the Warwick Antimicrobial Testing Facility.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/4/200/s1, Figure S1: Morphology of plaques formed by bacteriophages on a lawn of Acinetobacter baumannii. Figure S2: Transmission Electron Micrographs of A. baumannii bacteriophages. Table S1: Details on clinical sources of Acinetobacter baumannii host strains. Figure S3a–e: Eight-hour growth curve for Acinetobacter baumannii strains grown with bacteriophages added at different multiplicity of infections (MOIs). Table S2: Logarithmic titer of bacteriophage vPhT2 after storage for 30 days in different temperature and light conditions. Table S3: Minimum inhibitory concentrations (MICs) of antibiotics against Acinetobacter baumannii host strains. Figure S4: Photo of drop test screen for zones of inhibition caused by a range of antibiotics against Acinetobacter baumannii strains. Supplement File: Selected-MDRAB-150-isolates.
Author Contributions
Growth curve assays, CsCl purification, endotoxin testing, human cell line assays, synergy testing, K.M.S.; MOI optimization, G.S.C., K.M.S. and S.E.S.; stability trials, K.M.S. and S.E.S.; whole genome sequencing, A.M. and R.T.; host range analysis, R.T. and U.L.; MIC testing, C.G.D. and J.M.; writing—original draft preparation, K.M.S.; writing—review and editing, A.M., A.P.S., and S.S.; funding acquisition, A.P.S. and S.S. with E.M.H.W. and A.M. as collaborators; supervision, A.P.S. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an Institutional Links grant, ID 332371796, under the UK—Thailand Research and Innovation Partnership Fund. The grant is funded by the UK Department for Business, Energy and Industrial Strategy and delivered by the British Council. For further information, please visit www.newtonfund.ac.uk.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Van de Sande-Bruinsma N., Lahra M., Patel J. Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation. [(accessed on 7 October 2019)]; Available online: http://apps.who.int/iris/bitstream/10665/188783/1/9789241549400_eng.pdf?ua=1.
- 2.Tacconelli E., Magrini N., Carmeli Y., Harbarth S., Kahlmeter G., Kluytmans J., Mendelson M., Pulcini C., Singh N., Theuretzbacher U. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organization; Geneva, Switzerland: 2017. [Google Scholar]
- 3.Fournier P.-E., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., et al. Comparative Genomics of Multidrug Resistance in Acinetobacter baumannii. PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas M.E., Bliziotis I.A. Pandrug-resistant Gram-negative bacteria: The dawn of the post-antibiotic era? Int. J. Antimicrob. Agents. 2007;29:630–636. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Howard A., O’Donoghue M., Feeney A., Sleator R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence. 2012;3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maragakis L.L., Perl T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 7.Phodha T., Riewpaiboon A., Malathum K., Coyte P.C. Annual relative increased in inpatient mortality from antimicrobial resistant nosocomial infections in Thailand. Epidemiology Infect. 2019;147:e133. doi: 10.1017/S0950268818003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishbain J.T., Peleg A. Treatment of Acinetobacter Infections. Clin. Infect. Dis. 2010;51:79–84. doi: 10.1086/653120. [DOI] [PubMed] [Google Scholar]
- 9.Santimaleeworagun W., Thathong A., Samret W., Preechachuawong P., Sae-lim W., Jitwasinkul T. Identification and characterization of carbapenemase genes in clinical isolates of carbapenem-resistant Acinetobacter Baumannii from general hospital in Thailand. Southeast Asian J. Trop. Med. Public Health. 2014;45:874–880. [PubMed] [Google Scholar]
- 10.Peleg A., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niumsup P.R., Boonkerd N., Tansawai U., Tiloklurs M. Carbapenem-resistant Acinetobacter baumannii producing OXA-23 in Thailand. Jpn. J. Infect. Dis. 2009;62:152–154. [PubMed] [Google Scholar]
- 12.National Antimicrobial Resistance Surveillance Centre Thailand (NARST) AMR Antibiograms 1998–2018. [(accessed on 12 October 2019)]; Available online: http://narst.dmsc.moph.go.th/antibiograms.html.
- 13.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 14.Bragg R.R., Meyburgh C.M., Lee J.-Y., Coetzee M. Potential Treatment Options in a Post-antibiotic Era. Adv. Exp. Med. Biol. 2018;1052:51–61. doi: 10.1007/978-981-10-7572-8_5. [DOI] [PubMed] [Google Scholar]
- 15.Cisek A., Dąbrowska I., Gregorczyk-Zboroch K.P., Wyżewski Z. Phage Therapy in Bacterial Infections Treatment: One Hundred Years After the Discovery of Bacteriophages. Curr. Microbiol. 2016;74:277–283. doi: 10.1007/s00284-016-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulakvelidze A., Alavidze Z., Morris J.G. Bacteriophage Therapy. Antimicrob. Agents Chemother. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Y., Luo T., Yang Y., Dong N., Wang R., Wang Y., Xu M., Guo X., Hu F., He P. Phage Therapy as a Promising New Treatment for Lung Infection Caused by Carbapenem-Resistant Acinetobacter baumannii in Mice. Front. Microbiol. 2018;8:2659. doi: 10.3389/fmicb.2017.02659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regeimbal J.M., Jacobs A.C., Corey B.W., Henry M.S., Thompson M.G., Pavlicek R.L., Quinones J., Hannah R.M., Ghebremedhin M., Crane N.J., et al. Personalized Therapeutic Cocktail of Wild Environmental Phages Rescues Mice from Acinetobacter baumannii Wound Infections. Antimicrob. Agents Chemother. 2016;60:5806–5816. doi: 10.1128/AAC.02877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateczun A.J., Lancaster J., Quinones J., Strathdee S.A., Picel A.C., Henry M.S., Kumaraswamy M., Regeimbal J.M., Segall A.M., Hamilton T., et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017;61:1–14. doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bren L. Bacteria-eating virus approved as food additive. FDA Consum. 2007;41:20–22. doi: 10.1037/e589942007-003. [DOI] [PubMed] [Google Scholar]
- 21.Yang H., Liang L., Lin S., Jia S. Isolation and Characterization of a Virulent Bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010;10:131. doi: 10.1186/1471-2180-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leungtongkam U., Thummeepak R., Wongprachan S., Thongsuk P., Kitti T., Ketwong K., Runcharoen C., Chantratita N., Sitthisak S. Dissemination of blaOXA-23, blaOXA-24, blaOXA-58, and blaNDM-1 Genes of Acinetobacter baumannii Isolates from Four Tertiary Hospitals in Thailand. Microb. Drug Resist. 2018;24:55–62. doi: 10.1089/mdr.2016.0248. [DOI] [PubMed] [Google Scholar]
- 23.Toledo S., An A., Takemoto F., Norman D., Rubenstein L., Wieland D. Improving Care in the Nursing Home: Comprehensive Reviews of Clinical Research. Volume 2. SAGE Publications, Inc.; Thousand Oaks, CA, USA: 2014. Infections and Infection Control; pp. 65–101. [DOI] [Google Scholar]
- 24.Magiorakos A.-P., Srinivasan A., Carey R., Carmeli Y., Falagas M.E., Giske C., Harbarth S., Hindler J., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Ackermann H.-W. Tailed bacteriophages: The order caudovirales. Adv. Appl. Microbiol. 1998;51:135–201. doi: 10.1016/s0065-3527(08)60785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulo C., De Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I., Le Mercier P. ViralZone: A knowledge resource to understand virus diversity. Nucleic Acids Res. 2010;39:D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitti T., Thummeepak R., Thanwisai A., Boonyodying K., Kunthalert D., Ritvirool P., Sitthisak S. Characterization and Detection of Endolysin Gene from Three Acinetobacter baumannii Bacteriophages Isolated from Sewage Water. Indian J. Microbiol. 2014;54:383–388. doi: 10.1007/s12088-014-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koskella B., Meaden S. Understanding Bacteriophage Specificity in Natural Microbial Communities. Viruses. 2013;5:806–823. doi: 10.3390/v5030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitthisak S., Kitti T., Thummeepak R., Leungtongkam U. Efficacy of Acinetobacter baumannii bacteriophage cocktail on Acinetobacter baumannii growth. African J. Microbiol. Res. 2015;9:2159–2165. [Google Scholar]
- 30.Kropinski A.M., Prangishvili D., Lavigne R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009;11:2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 31.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2004;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B., Pop M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2008;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;45:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansen M., Wahida A., Latz S., Krüttgen A., Häfner H., Buhl E.M., Ritter K., Horz H.-P. Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Sci. Rep. 2018;8:14140. doi: 10.1038/s41598-018-32344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulkkinen E., Wicklund A., Oduor J.M.O., Skurnik M., Kiljunen S. Characterization of vB_ApiM_fHyAci03, a novel lytic bacteriophage that infects clinical Acinetobacter strains. Arch. Virol. 2019;164:2197–2199. doi: 10.1007/s00705-019-04284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin J., Li Z.-J., Wang S.-W., Wang S., Huang D.-H., Li Y.-H., Ma Y., Wang J., Liu F., Chen X.-D., et al. Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 2012;12:156. doi: 10.1186/1471-2180-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams M.J., Buehner M., Chandrasekhar K., Ford G.C., Hackert M.L., Liljas A., Rossmann M.G., Smiley I.E., Allison W.S., Everse J., et al. Structure-Function Relationships in Lactate Dehydrogenase. Proc. Natl. Acad. Sci. USA. 1973;70:1968–1972. doi: 10.1073/pnas.70.7.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.EUCAST The European Committee on Antimicrobial Susceptibility Testing. [(accessed on 9 September 2019)];2019 Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. Available online: http://www.eucast.org.
- 39.Moellering R.C. Antimicrobial Combinations. Rinsho Yakuri/Jpn. J. Clin. Pharmacol. Ther. 1993;24:293–300. doi: 10.3999/jscpt.24.293. [DOI] [Google Scholar]
- 40.Ross A., Ward S., Hyman P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016;7:1. doi: 10.3389/fmicb.2016.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.United Nations, Department of Economic and Social Affairs, Population Division World Population Prospects 2019, Custom Data Acquired Via Website. [(accessed on 12 October 2019)]; Available online: https://population.un.org/wpp/dataquery.
- 42.Daneshian M., Guenther A., Wendel A., Hartung T., Von Aulock S. In vitro pyrogen test for toxic or immunomodulatory drugs. J. Immunol. Methods. 2006;313:169–175. doi: 10.1016/j.jim.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Brito L.A., Singh M. COMMENTARY: Acceptable Levels of Endotoxin in Vaccine Formulations During Preclinical Research. J. Pharm. Sci. 2011;100:34–37. doi: 10.1002/jps.22267. [DOI] [PubMed] [Google Scholar]
- 44.Malyala P., Singh M. Endotoxin Limits in Formulations for Preclinical Research. J. Pharm. Sci. 2008;97:2041–2044. doi: 10.1002/jps.21152. [DOI] [PubMed] [Google Scholar]
- 45.Malik D.J., Sokolov I.J., Vinner G.K., Mancuso F., Cinquerrui S., Vladisavljević G.T., Clokie M.R., Garton N.J., Stapley A.G., Kirpichnikova A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017;249:100–133. doi: 10.1016/j.cis.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Vallvé S., Puigbo P. DendroUPGMA: A dendrogram Construction Utility. [(accessed on 22 October 2019)]; Available online: http://genomes.urv.es/UPGMA/
- 47.Guttman B., Raya R., Kutter E., Carlson K., Boyd E.F. In: BACTERIOPHAGES Biology and Applications. 1st ed. Kutter E., Sulakvelidze A., editors. CRC Press; Boca Raton, FL, USA: 2005. [Google Scholar]
- 48.Thummeepak R., Kitti T., Kunthalert D., Sitthisak S. Enhanced Antibacterial Activity of Acinetobacter baumannii Bacteriophage ØABP-01 Endolysin (LysABP-01) in Combination with Colistin. Front. Microbiol. 2016;7:113. doi: 10.3389/fmicb.2016.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinstein M.P. M02-Performance Standards for Antimicrobial Disk Susceptibility Tests. 13th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 50.Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 51.Joshi N.A., Fass J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software] 2011. [(accessed on 20 March 2019)]; Available online: https://github.com/najoshi/sickle.
- 52.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 54.Richter M., Rossello-Mora R., Glöckner F.O., Peplies J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2015;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adriaenssens E.M., Brister J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses. 2017;9:70. doi: 10.3390/v9040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alikhan N.-F., Petty N.K., Ben Zakour N., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.