Abstract
Dendrobium has been widely used not only as ornamental plants but also as food and medicines. The identification and evaluation of the genetic diversity of Dendrobium species support the conservation of genetic resources of endemic Dendrobium species. Uniquely identifying Dendrobium species used as medicines helps avoid misuse of medicinal herbs. However, it is challenging to identify Dendrobium species morphologically during their immature stage. Based on the DNA barcoding method, it is now possible to efficiently identify species in a shorter time. In this study, the genetic diversity of 76 Dendrobium samples from Southern Vietnam was investigated based on the ITS (Internal transcribed spacer), ITS2, matK (Maturase_K), rbcL (ribulose-bisphosphate carboxylase large subunit) and trnH-psbA (the internal space of the gene coding histidine transfer RNA (trnH) and gene coding protein D1, a polypeptide of the photosystem I reaction center (psaB)) regions. The ITS region was found to have the best identification potential. Nineteen out of 24 Dendrobium species were identified based on phylogenetic tree and Indel information of this region. Among these, seven identified species were used as medicinal herbs. The results of this research contributed to the conservation, propagation, and hybridization of indigenous Dendrobium species in Southern Vietnam.
Keywords: Dendrobium, ITS, ITS2, matK, rbcL, trnH-psbA, southern Vietnam, molecular identification, genetic diversity, DNA barcoding
1. Introduction
Dendrobium is among the most abundant genera of flowering plants with over 1148 known species, which ranks second in the orchid family, after the Bulbophyllum genus [1]. Dendrobium is diverse in shapes, colors, and sizes, and is hence considered as a favorite ornamental plant. Some Dendrobium species are also used as medicinal herbs, such as D. densiflorum and D. chrysotoxum [2]. Many studies on diverse Dendrobium species by geographic regions have been published for Australia [3,4], mainland Asia [5,6], China [7], Thailand [8,9], etc. These studies again confirm the rich diversity of the beautiful orchids.
The living environment of indigenous Dendrobium species in Vietnam is declining due to climate change and over-exploitation. An evaluation of genetic diversity and identification of Dendrobium species in Vietnam is critical for prompt conservation of this valuable genus. Morphology of Dendrobium species is similar at non-flowered stages, and hence misidentification often happens between conspecific species [10].
DNA barcoding is an effective method used in the identification of species, especially orchids. Many works have proved that the ITS region (Internal Transcribed Spacer) contains many genetic differences, so it is used to classify species and study relationships [10,11], particularly in Dendrobium [8,12]. The ITS2 region has been assessed as being able to clearly distinguish between Dendrobium species [13,14]. Two matK and rbcL regions have also been identified as being able to identify species of the genus Dendrobium [4,10].
Tran (2015) conducted a diversity examination of indigenous Dendrobium species in Vietnam, mostly from Northern Vietnam, using ITS sequences [15]; 23 of 32 samples of Dendrobium were identified, among which four of nine unidentified samples were confirmed as Dendrobium parishii [15]. Nguyen et al. (2017) constructed a phylogenetic tree for the ITS region, and separated 12 samples of wild Dendrobium species collected in Southern Vietnam and 11 samples of imported Dendrobium from Thailand divided into two distinct groups. Those results corresponded to the classification by the traditional identification method [16]. Nguyen (2018) continued to evaluate ITS on the identification of 15 samples belonging to Dendrobium thyrsiflorum, which were delineated on single branches [17].
A large number of Dendrobium species in Southern Vietnam were evaluated for genetic diversity to improve conservation efforts in the current work. The identification capability of different sequences was also investigated. The results of our work contribute to the enrichment of the sequences in GenBank and have applications in practical conservation and management of genetic resources.
2. Materials and Methods
2.1. DNA Extraction and Amplification
The total DNA of 76 samples was isolated from fresh leaves by the Isolate II Plant DNA kit BIO-52069 (TBR Company, Ho Chi Minh City, Vietnam). Primers and thermocycling conditions used for the amplification of 4 regions, ITS, matK, rbcL, trnH-psbA, are presented in Table 1. Components of the amplification reaction included 12.5 μL Taq DNA pol 2x-premix, 1 μL forward primer (5 μM–10 µM), 1 μL reverse primer (5 µM–10 μM), 1 μL DNA template and water to make 25 μL. PCR products were sequenced bi-directionally at Macrogen Company, Seoul, Korea.
Table 1.
Primer sequences and the thermal cycles for amplification reactions of the ITS, matK, rbcL, trnH-psbA regions.
| Barcode | Primer Name | Primer Sequence | Thermal Cycle | Source |
|---|---|---|---|---|
| ITS | ITS1F | 5′CTTGGTCATTTAGAGGAAGTAA3′ | Denaturing: 94 °C/30 sec Annealing: 55 °C/40 sec Extending: 72 °C/1 min |
[17,18] |
| ITS4R | 5′TCCTCCGCTTATTGATATGC3′ | |||
| matK | 390F | 5′CGATCTATTCATTCAATATTTC3′ | Denaturing: 94 °C/1 min Annealing: 48 °C/30 sec Extending: 72 °C/1 min |
[6,19] |
| 1326R | 5′TCTAGCACACGAAAGTCGAAGT3′ | |||
| rbcL | aF | 5′ATGTCACCACAAACAGAGACTAAAGC3′ | Denaturing: 94 °C/30 sec Annealing: 55 °C/1 min Extending: 70 °C/1 min |
[20] |
| aR | 5′CTTCTGCTACAAATAAGAATCGATCTCTCCA3′ | |||
| trnH-psbA | trnHF_05 | 5′CGCGCATGGTGGATTCACAATCC3′ | Denaturing: 95 °C/30 sec Annealing: 5 °C/20 sec Extending: 72 °C/20 sec |
[21] |
| psbA3′f | 5′GTTATGCATGAACGTAATGCTC3′ |
2.2. Data Analysis
FinchTV software [22] was used to read and adjust nucleotide sequences. Forward and reverse sequences were combined into consensus sequences and aligned using Seaview 4.0 [23]. The ITS2 sequence was then extracted from the ITS sequence (Based on accession number JN388570.1) for analyses. The phylogenetic tree and variable parameters were calculated in MEGA 7.0 software [24] by using the Maximum Likelihood algorithm, following the 2-parameter Kimura model. The sequence of orchid species Paphiopedilum delenatii was used as an outgroup to root the tree.
3. Results
3.1. Sample Collection, Amplification, and Sequencing
The 76 Dendrobium samples (Appendix A) were collected and divided into two groups: the collection of Biotechnology Center Ho Chi Minh (coded as TT) and the commercial samples (coded DT, PN). For ITS and matK, all 76 collected samples were amplified. Since rbcL is a conserved region, only 35 samples from 30 species were amplified.
The PCR results in both ITS and matK regions achieved success rates of 94.73% and 97.26%, respectively. Notably, the rbcL area had the best rate of 100%. Particularly in the trnH-psbA region, the PCR success rate was 82.19%. However, the amplification and sequencing of trnH-psbA were at low levels. Therefore, the data from the trnH-psbA region was not included in further analyses in the study.
3.2. Genetic Diversity Based on Nucleotide Polymorphism and Phylogenetic Analyses
Seventy-six samples of 30 collected Dendrobium species were included in the survey (Appendix A). For phylogenetic analysis, sequences of Dendrobium species from our study were compared with GenBank accessions (Accession numbers of GenBank sequences are shown in Appendix B). Based on the phylogenetic tree, individuals of the same species should cluster in the same branch that separates from the other species. In general, there was no conflict among the three constructed trees. However, the ITS gave the most separated branches. The ITS2 trees showed the same clusters as the ITS trees. Hence the ITS region was representatively analyzed for the divergence of Dendrobium species in Southern Vietnam.
On the ITS tree, samples of some species were grouped with their conspecific accessions from GenBank without mixing with other different species, i.e., D. aloifolium, D. amabile, D. capillipes, D. chrysotoxum, D. crumenatum, D. crystallinum, D. densiflorum, D. farmeri, D. intricatum, D. parishii, D. secundum, D. sulcatum, and D. venustum. D. superbum was the synonym name of D. anosmum. Hence their sequences were mixed up for both our samples and GenBank accessions and closely related to their sister D. parishii. As a result, the hybrid samples of D. anosmum × parishii and D. anosmum × D. aphyllum were also included in the phylogenetic branch of these species. D. anosmum × parishii is named D. nestor, and D. anosmum × D. aphyllum is named Adastra. The separation of D. parishii from D. anosmum was also reported by Tran et al. (2018) [15].
In both ITS and matK phylogenetic trees, our sample of D. salaccense was not clustered with a group of the species accessions from GenBank. Interestingly, after searching other similar sequences from GenBank using the BLAST tool, our sample 24DT was homologous with D. hancockii at 99.71% in ITS data and 100% in matK data (data not show). These two species have the same Vietnamese name, “Hoang Thao Truc”. Hence species confusion might happen during the sampling process. The scientific name of sample 24DTwas then corrected to D. hancockii.
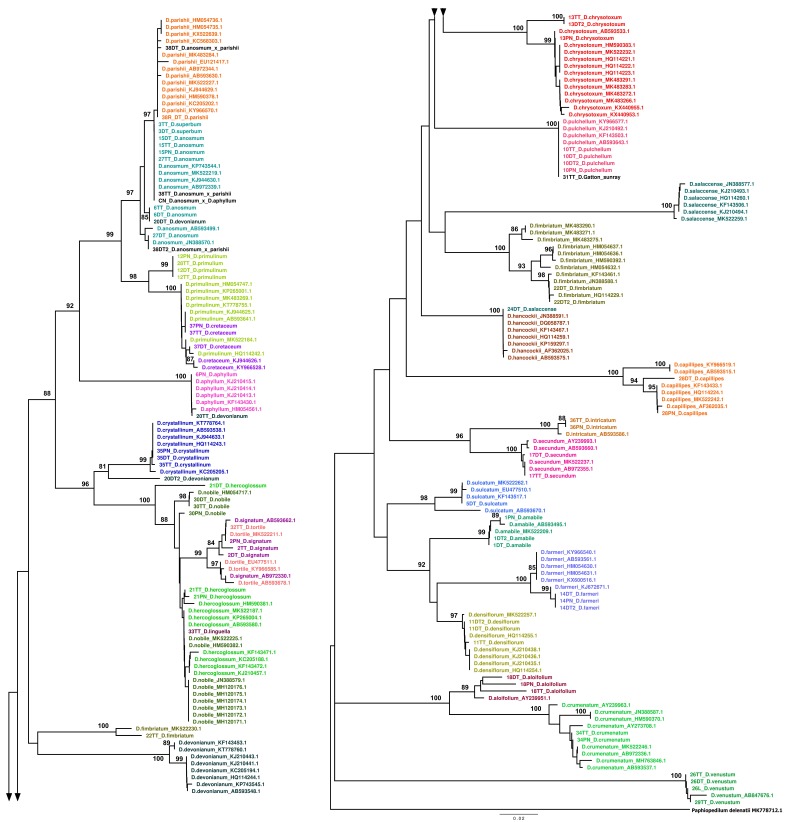
Among three samples of D. fimbriatum, two samples, 22DT and 22DT2, were grouped with other D. fimbriatum accessions from GenBank but sample 22TT was totally separated from this group. However, when compared to GenBank sequences, the remaining sample 22TT was also matched with another conspecific accession D. fimbriatum (MK522230.1) and was closely related to D. devonianum species (Figure 1). A further observation on the original alignment of these accessions showed that sequences of 22TT and D. fimbriatum (MK522230.1) were highly similar throughout the length and were fractionated into different regions, in which some fragments were similar to other D. fimbriatum accessions, some were similar to D. devonianum sequences, and some were distinct from all of others. This result proposed the conclusion that the 22TT sample was a hybrid of D. fimbriatum and D. devonianum as these two species share the same local habitat (Appendix A). Otherwise, D. fimbriatum might be diverted into different directions of the evolution process.
Figure 1.
ITS tree is constructed base on the Maximum likelihood for Dendrobium collected at Southern Vietnam.
The variety D. gatton sunray was located in the same branch of D. pulchellum in both ITS and matK trees. D. pulchellum was crossed with D. chrysotoxum forming D. illustre. Then, D. illustre was crossed back with D. pulchellum to create D. gatton sunray. As a result, the hybrid, which contains lots of genetic characters from D. pulchellum, was grouped with its parent in phylogenetic trees.
Sequences of two species, D. signatum and D. tortile, were mixed up on the same branch. In terms of sexual morphology, their flowers are remarkably similar except that petals of species D. tortile are non-yellowed, more purple, and more twisted. Hence the molecular result was consistent with morphological features. D. signatum is sometimes called by the synonym scientific name D. tortile var. hildebrandi (Rolfe) T. Tang and F.T. Wang (1951). As a result, they had a very close genetic relationship. D. hercoglossum and D. linguella, are two synonym names of one species. On all phylogenetic trees, this species was closely related to D. nobile, D. signatum, and D. tortile and could not be completely distinguished.
Two species, D. primulinum and D. cretaceum, which have similar morphological features, were also close in genetic characters. The same situation also happened for two species, D. primulinum and D. cretaceum. The most divergent species was D. devonianum within our three conspecific samples, and even sequences of this species from GenBank were significantly separated into different branches on all ITS, matK, and rbcL trees. Although there was not enough data to clarify this issue, the results suggested a hypothesis of breeding between D. devonianum and other species in nature.
Briefly, there is a diversity of 28 species of Dendrobium in Southern Vietnam, including three hybrid species, which were investigated in this study. Among conspecific variations, there was also divergence, shown in different lengths of branches on the same cluster, i.e., species D. amabile, D. secundum, D. capillipes, D. chrysotoxum, and D. crystallinum (Figure 1).
3.3. Potential Sequences for Identification of Dendrobium Species in Southern Vietnam
Investigating genetic diversity of Dendrobium populations not only provides information for species management but also helps distinguish herbals and their adulterants, and significantly supports conservation by identifying and limiting trade of valuable and endangered species illegally. In this study, we assayed the potential of using sequences in species identification for practical conservation. In this analysis, 24 original species were included, except for three hybrids and the undetermined species D. devonianum. Twenty-three species were analyzed using matK and rbcL data since D. parishii could not be amplified. The most critical measurement for evaluation was the species resolution of each region. Therefore, tree-based methods and indel information were combined to optimize achievement (Appendix C). Criteria such as variable sites, informative parsimony sites, and singleton sites were also recorded.
Both the ITS (56.65%) and ITS2 (52.89%) regions showed significantly high results in nucleotide polymorphism (variable sites) in comparison with matK (10.21%) and rbcL (6.58%) and trnH-psbA (8.31%) (Table 2). ITS2 was even more divergent than the full ITS region. This result was consistent with previous studies [25,26,27]. Based on the phylogenetic tree, the species identification by ITS2 (17 out of 24 species) was as effective as ITS (17 species).
Table 2.
Comparison parameters of ITS (internal transcribed spacer), ITS2, matK, rbcL, and trnH-psbA markers for identification of Dendrobium species.
| Region | Length | Number of Samples | Number of Species | Variable Site (%) | Parsimony (%) | Single-ton (%) | Indel | Identified Species Based on the Phylogenetic Tree | Identified Species Based on the Phylogenetic Tree and Indel Information |
|---|---|---|---|---|---|---|---|---|---|
| ITS | 639 | 68 | 24 | 362 (56.65) |
338 (52.89) |
24 (3.75) |
15 | 17/24 | 19/24 |
| ITS2 | 253 | 68 | 24 | 167 (66.00) |
152 (60.07) |
15 (5.92) |
12 | 17/24 | 17/24 |
| matK | 822 | 65 | 23 | 84 (10.21) |
53 (6.44) |
31 (3.77) |
3 | 12/23 | 12/23 |
| rbcL | 501 | 34 | 21 | 26 (6.58) |
16 (4.59) |
10 (1.99) |
0 | 5/23 | 5/23 |
| trnH-psbA | 782 | 56 | 24 | 65 (8.31) |
46 (5.88) |
17 (2.17) |
13 | 5/24 | 5/24 |
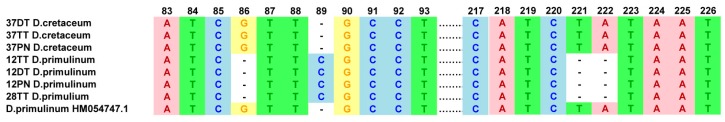
From both ITS trees, three pairs of species were not separated, i.e., D. cretaceum and D. primulinum; D. hercoglossum and D. nobile; D. tortile and D. signatum. Our examination of insertion and deletion information from their full ITS sequences indicated the differences between D. cretaceum and D. primulinum at sites 86, 89, 221–222 (aligned with the complete ITS of Dendrobium primulinum HM054747.1) (shown in Figure 2), which did not exist in short version, ITS2. D. primulinum in this study had three deletions at sites 86, 221, 222, and 1 insertion at site 89. Therefore, these two species were distinguished, and ITS could identify 19 out of 24 species (79.16%). Although less divergent, the long ITS (15) contained more indel sites than the short ITS2 (12) and was proven to be useful in previous studies [28,29]. The combination of multiple loci as a single marker did not provide more species resolution. Finally, 19 out of 24 species were clearly identified, including D. aloifolium, D. amabile, D. aphyllum, D. capillipes, D. chrysotoxum, D. cretaceum, D. crumenatum, D. crystallinum, D. densiflorum, D. farmeri, D. fimbriatum, D. intricatum, D. parishii, D. primulinum, D. pulchellum, D. hancockii, D. secundum, D. sulcatum, and D. venustum.
Figure 2.
Insertion-deletion (indel) sites in sequences of D. creatceum and D. primulinum accessions.
In terms of best match/best close match methods in the evaluation of potential sequences for species identification, ITS2 gave the best results of the correct match, following by ITS and matK. rbcL gave the lowest effect (Table 3).
Table 3.
The identification results of the “best match/ best close match” method.
| Barcode | No Sequences | Best Match (%) | Best Close Match (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | ||
| ITS | 68 | 55 (80.88) | 2 (2.94) | 11 (16.17) | 51 (75.00) | 2 (2.94) | 5 (7.35) | 10 (14.70) |
| ITS2 | 68 | 57 (83.82) | 4 (5.88) | 7 (10.29) | 52 (76.47) | 3 (4.41) | 4 (5.88) | 9 (13.23) |
| matK | 65 | 44 (67.69) | 16 (24.61) | 5 (7.69) | 44 (67.69) | 15 (23.07) | 5 (7.69) | 1 (1.53) |
| rbcL | 34 | 7 (20.58) | 24 (70.58) | 3 (8.82) | 7 (20.58) | 24 (70.58) | 3 (8.82) | 0 (0.00) |
| trnH-psbA | 56 | 38 (67.85) | 6 (10.71) | 12 (21.42) | 38 (67.85) | 6 (10.71) | 12 (21.42) | 0 (0.00) |
Correct: identified; ambiguous; incorrect: unidentified; no match: under threshold. Above number: numbers of sequences; below number: percentage of sequences out of total sequences.
The “best match/best close match” methods [30] are based on comparing the genetic distance of the analyzed sequences. The sequences that achieve intra-value are the smallest when compared to the order of the same species classified as correct. If this intra-value is also present when compared to other species, the sequence is classified as ambiguous. The sequences with intra-distances greater than inter-distances are categorized as incorrect. For the “best close match” method, a threshold value (%) is calculated based on all intra-distances, to determine the similarity of sequences. The sequences that do not meet this value (no match) will be deleted before being identified.
Both the matK and rbcL regions are quite conserved sequence areas [31], and there was a similarity level higher than 97%, so when the threshold (3%) was set, no sequence was classified as “no match”. Meanwhile, the ITS and ITS2 sequences are sequences of high diversity, so the results (50 and 53, respectively) were higher than matK and rbcL. When using the “best close match” with a threshold of 3% of the ITS2 region, the highest results were obtained (48 sequences), indicating that ITS2 was the most likely area of determination in the studied regions. Therefore, the ITS and ITS2 sequence regions were identified as potential barcodes.
In general, the results derived from best match/best close match methods (Appendix D) were consistent with branch forming of each sample on phylogenetic trees. For instance, on the tree (Figure 1), 30PN was separated in another branch from the group of 30DT and 30TT. The best match calculation from ITS data also reported sample 30PN D. nobile as incorrect while the two remain samples of that species, 30DT and 30TT, were correct. However, for this method, the relationship among species was not visualized as well as the tree-based method. For instance, we could not recognize that D. anosmum and D. superbum were clustered on the same branch as they are synonymous names of the same species, or D. primulinum with D. cretaceum. Hence, best match/best close match methods were used just for general evaluation of identification potential of a sequence.
4. Discussion
ITS was also used in previous studies on identification of Dendrobium species, among which some studies focused on medicinal species for distinguishing herbals and their adulterants [13,14]. In a previous study of Tran et al. (2018) [15], 19 out of 23 Vietnamese Dendrobium species (82.61%) were identified using the ITS marker (Appendix E). In our study, 28 species were considered in which 19 species (67.86%) were identified using the same marker ITS. Some species were identified in study of by Tran et al. (2018) but not in ours, i.e., D. anosmum and D. nobile. In contrast, two species, D. amabile and D. fameri, were clearly separated on monophyletic branches in our study but not in the previous research. Unidentified species were species with their sequences grouped with sequences of other species, forming paraphyletic or polyphyletic branches [28]. In the two studies, ITS could not resolve 100% of Dendrobium species. However it was the best in comparison with matK and rbcL markers in our study. The difference of resolution effectiveness actually much depends on component of sample data. Sixteen species from our study were not included in study of Tran et al. (2018) and, vice versa, 11 species in their study were not in our collection. Tran and his colleagues collected samples from the whole of Vietnam and mostly from the northern areas, while our study collected species from southern regions. Besides, in the study of Tran et al. (2018), the sample size was small, with 32 specimens, and most of the sampled species (15 out of 23) were examined with only one representative sample. Therefore genetic diversity among conspecific individuals was not investigated in their study. In our study, 2 to 3 samples for each species, except for five species, D. aphyllum, D. parishii, D. salaccense, D. sulcatum, and D. tortile, were included for intra- and inter-specific genetic analyses. In short, our study results and the report of Tran et al. do not contradict each other but both gave a remarkable contribution to the sequence library of Vietnamese native Dendrobium diversity.
The intergenic spacer trnH-psbA was recommended by Yao et al. (2009) for the identification of 15 Dendrobium species [32] due to high divergence of sequences. In our study, this region was more difficult to amplify than other regions. The amplification rate was just 82.19% after repetition. This problem was consistent with the previous report of Gigot et al. (2007). trnH-psbA is supposed to contain too many tandem mononucleotide repeats which results in high levels of length variation and causes problem in amplification, bidirectional sequencing, and alignment [33].
The matK and rbcL markers were used for this orchid group by Asahina et al. (2010) [10] and Moudi et al. (2013) [34]. Sigh et al. (2012) proposed the combination of three regions, matK, rpoB, and rpoC1 [35]. Among those barcoding regions, ITS was the most commonly used. [2,8,9,14,15,25,36,37,38,39]. Our results again confirmed the effect of ITS in the evaluation of genetic diversity and the identification of Dendrobium species not only in Southern Vietnam but also in other habitats.
5. Conclusions
The ITS2 region has the highest level of genetic diversity among the surveyed areas. In particular, the ITS region has more indels to help increase the ability to identify species. In general, both ITS and ITS2 have the most potential for assessment of genetic diversity and identification of Dendrobium species in Southern Vietnam. In this study, 19 Dendrobium species were recognized, many of which have high levels of diversity within the same species. Some species with easily confused morphological characteristics have also been redefined for accuracy based on molecular sequences. Research has contributed to increasing data in the library of Dendrobium of Vietnam and the world. Also, the two species with very similar morphologies can be distinguished, D. primulinum (used as medicinal herbs) and D. creatceum, to avoid confusion when using these species as medicinal herbs.
Acknowledgments
We would like to thank the Biotechnology Center of Ho Chi Minh City and HCMC University of Education, and Nguyen-Tat-Thanh University for supporting the facilities to do this research.
Appendix A
Table A1.
List: code, and location collection of the sample vouchers.
| Scientific Name | IUCN 2019 | Herbal | Sample Voucher | Collect Location | |
|---|---|---|---|---|---|
| 1 | D. aloifolium (Bl.) Rchb. f. | LC | 18TT | the collection of Biotechnology Center Ho Chi Minh | |
| 18DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 18PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 2 | D. amabile (Lour.) O’ Brien | 1DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 1DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 1PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 3a | D. anosmum Lindl. | 27TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 27DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 6TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 6DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 3b | 15TT | the collection of Biotechnology Center Ho Chi Minh | |||
| 15DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 15PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 4 | D. aphyllum (Roxb.) C. Fisch. 1928 | LC | X | 6PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam |
| 5 | D. capillipes Rchb.f. | X | 28DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | |
| 28PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 6 | D. chrysotoxum Rchb.f; | X | 13TT | the collection of Biotechnology Center Ho Chi Minh | |
| 13DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 13PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 7 | D. cretaceum Lindl. 1847. | 37TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 37DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 37PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 8 | D. crumenatum Sw. | 34TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 34DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 9 | D. crystallinum Rchb. f. (1868) | X | 35TT | the collection of Biotechnology Center Ho Chi Minh | |
| 35DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 35PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 10 | D. densiflorum Wall. ex Lindl | 11TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 11DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 11DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 11 | D. devonianum Paxton (1840) | X | 20TT | the collection of Biotechnology Center Ho Chi Minh | |
| 20DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 20DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 12 | D. farmeri Paxton Lindl.f.Rchb.f. | 14DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 14DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 14PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 13 | D. fimbriatum Hook (1823) | X | 22TT | the collection of Biotechnology Center Ho Chi Minh | |
| 22DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 22DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 14 | D. hercoglossum Rchb. f. 1886 | X | 21TT | the collection of Biotechnology Center Ho Chi Minh | |
| 21DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 21PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 15 | D. intricatum Gagnep (1930) | 36TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 36DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 16 | D. linguella Rchb. f. 1882 | 33TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 17 | D. nobile Lindl. | X | 30TT | the collection of Biotechnology Center Ho Chi Minh | |
| 30DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 30PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 18 | D. parishii Rchb. f 1863 | 38R-DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 19 | D. primulinum Lindl | X | 28TT | the collection of Biotechnology Center Ho Chi Minh | |
| 12TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 12DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 12PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 20 | D. pulchellum Roxb. ex Lindl. | 10TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 10DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 10DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 10PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 21 | D. salaccense (Bl.) Lindl. | X | 24DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | |
| 22 | D. secundum (Bl.) Lindl. | 17TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 17DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 23 | D. signatum Rchb. f. 1884 | 2DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 2PN | the collection in Phu Nhuan District, Ho Chi Minh City, Vietnam | ||||
| 2TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 24 | D. sulcatum Lindl. (1838) | 5DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 25 | D. superbum Rchb.f. | 3TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 3DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 26 | D. tortile Lindl | 32TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 27 | D. venustum Teijsm. & Binn. 1864 | 26TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 26DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 26L | the collection in Long An Province, Vietnam | ||||
| 29TT | the collection of Biotechnology Center Ho Chi Minh | ||||
| 28 | D. anosmum × D. parishi | 38TT | the collection of Biotechnology Center Ho Chi Minh | ||
| 38DT | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 38DT2 | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||||
| 29 | D. anosmum × D. aphyllum | CN | the collection in Duc Trong District, Lam Dong Province, Vietnam | ||
| 30 | D. Gatton Sunray | 31TT | the collection of Biotechnology Center Ho Chi Minh |
Appendix B
Table A2.
List of accession numbers of sequences obtained by this study and from Genbank for phylogenetic analysis.
Appendix C
Table A3.
Species resolution results based on phylogenetic trees and nucleotide polymorphism.
| No | Species | ITS | ITS2 | matK | rbcL | trnH-psbA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tree-Based | Indel-Based | Tree-Based | Indel-Based | Tree-Based | Indel-Based | Tree-Based | Indel-Based | Tree-Based | Indel-Based | ||
| 1 | D. aloifolium | + | + | + | + | + | |||||
| 2 | D. amabile | + | + | + | ― | ― | |||||
| 3 |
D. anosmum (synonym name D. superbum) |
― | ― | ― | ― | ― | |||||
| 4 | D. aphyllum | + | + | + | ― | ― | |||||
| 5 | D. capillipes | + | + | + | ― | ― | |||||
| 6 | D. chrysotoxum | + | + | + | ― | ― | |||||
| 7 | D. cretaceum | ― | + | ― | ― | ― | ― | ||||
| 8 | D. crumenatum | + | + | + | + | + | |||||
| 9 | D. crystallinum | + | + | + | ― | ― | |||||
| 10 | D. densiflorum | + | + | ― | ― | ― | |||||
| 11 | D. farmeri | + | + | ― | ― | ― | |||||
| 12 | D. fimbriatum | + | + | ― | ― | ― | |||||
| 13 |
D. hercoglossum (synonym name D. linguella) |
― | ― | ― | ― | ― | |||||
| 14 | D. intricatum | + | + | + | ― | ― | |||||
| 15 | D. nobile | ― | ― | ― | ― | ― | |||||
| 16 | D. parishii | + | + | not available | not available | ― | |||||
| 17 | D. primulinum | ― | + | ― | ― | ― | ― | ||||
| 18 | D. pulchellum | + | + | + | ― | + | |||||
| 19 | D. hancockii (previously named D. salaccense) |
+ | + | + | + | + | |||||
| 20 | D. secundum | + | + | ― | + | ― | |||||
| 21 | D. signatum | ― | ― | ― | ― | ― | |||||
| 22 | D. sulcatum | + | + | + | ― | ― | |||||
| 23 | D. tortile | ― | ― | ― | ― | ― | |||||
| 24 | D. venustum | + | + | + | + | + | |||||
| Identified species | 17/24 | 2 | 17/24 | 0 | 12/23 | 0 | 5/23 | 0 | 5/24 | 0 | |
| 19/24 | 17/24 | ||||||||||
Appendix D
Table A4.
Sequence identification results based on best match/best close match methods.
| Species | Voucher | ITS | ITS2 | matK | rbcL | trnH-psbA | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | Best Match (%) | Best Close Match (%) | |||||||||||||||||||||||||||
| Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | ||
| D. aloifolium | 18TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 18DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| 18PN | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. amabile | 1DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 1DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 1PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. anosmum | 27TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 27DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 6TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 6DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 15TT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 15DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 15PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. superbum | 3TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 3DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. aphyllum | 6PN | x | x | X | X | X | X | X | X | |||||||||||||||||||||||||||
| D. capillipes | 28DT | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| 28PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. chrysotoxum | 13TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 13DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 13PN | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. cretaceum | 37TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 37DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 37PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. crumenatum | 34TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 34PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. crystallinum | 35TT | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| 35DT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 35PN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| D. densiflorum | 11TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 11DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 11DT2 | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. farmeri | 14DT | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| 14DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 14PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. fimbriatum | 22TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 22DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 22DT2 | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. hercoglossum | 21TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 21DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 21PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. linguella | 33TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. intricatum | 36TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 36DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. nobile | 30TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 30DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 30PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. parishii | 38R-DT | X | X | X | X | X | X | |||||||||||||||||||||||||||||
| D. primulinum | 28TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 12TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 12DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| 12PN | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| D. pulchellum | 10TT | X | X | X | X | |||||||||||||||||||||||||||||||
| 10DT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 10DT2 | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 10PN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| D. salaccense | 24DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. secundum | 17TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 17DT | X | X | X | X | X | X | ||||||||||||||||||||||||||||||
| D. signatum | 2DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 2PN | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| 2TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
| D. sulcatum | 5DT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. tortile | 32TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| D. venustum | 26TT | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||||||||||
| 26DT | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 26L | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||||
| 29TT | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||||||||||||
Appendix E
Table A5.
Comparison of identification species between our study and the study of Tran et al. (2018) [25].
| No | Species | Identified Species Uisng ITS | |
|---|---|---|---|
| Our Study | Tran et al. (2018) | ||
| 1 | D. aloifolium | + | not included |
| 2 | D. amabile | + | ― |
| 3 |
D. anosmum (synonym name D. superbum) |
― | + |
| 4 | D. aphyllum | + | + |
| 5 | D. capillipes | + | + |
| 6 | D. chrysotoxum | + | + |
| 7 | D. cretaceum | + | not included |
| 8 | D. crumenatum | + | not included |
| 9 | D. crystallinum | + | not included |
| 10 | D. densiflorum | + | not included |
| D.devonianum | ― | not included | |
| 11 | D. farmeri | + | ― |
| 12 | D. fimbriatum | + | + |
| 13 |
D. hercoglossum (synonym name D. linguella) |
― | not included |
| 14 | D. intricatum | + | not included |
| 15 | D. nobile | ― | + |
| 16 | D. parishii | + | + |
| 17 | D. primulinum | + | + |
| 18 | D. pulchellum | + | not included |
| 19 |
D. hancockii (previously named D. salaccense) |
+ | + |
| 20 | D. secundum | + | not included |
| 21 | D. signatum | ― | not included |
| 22 | D. sulcatum | + | not included |
| 23 | D. tortile | ― | ― |
| 24 | D. venustum | + | not included |
| 25 | D. anosmum × D. parishi | ― | not included |
| 26 | D. anosmum × D. aphyllum | ― | not included |
| 27 | D. Gatton Sunray | ― | not included |
| 28 | D. findlayanum | not included | + |
| 29 | D. moschatum | not included | + |
| 30 | D. chrysanthum | not included | + |
| 31 | D. thyrsiflorum | not included | + |
| 32 | D. wattii | not included | + |
| 33 | D. jenkinsii | not included | + |
| 34 | D. haveyanum | not included | ― |
| 35 | D. aduncum | not included | + |
| 36 | D.brymerianum | not included | + |
| 37 | D. draconis | not included | + |
| 38 | D. christyanum | not included | + |
| 28 species | 23 species | ||
| 19 identified species | 19 identified species | ||
Author Contributions
Conceptualization, H.-D.T. and H.-X.D.; methodology, H.-D.T.; formal analysis, N.-D.L. and T.-D.N.; investigation, N.-H.N.; resources, H.-X.D.; data curation, N.-H.N., N.-D.L., and T.-D.N.; writing—original draft preparation, N.-H.N. and H.-T.V.; writing—review and editing, H.-D.T.; supervision, H.-D.T. and H.-X.D.; project administration, H.-D.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Leitch I.J., Kahandawala I., Suda J., Hanson L., Ingrouille M.J., Chase M.W., Fay M.F. Genome size diversity in orchids: Consequences and evolution. Ann. Bot. 2009;104:469–481. doi: 10.1093/aob/mcp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takamiya T., Wongsawad P., Tajima N., Shioda N., Lu J.F., Wen C.L., Wu J.B., Handa T., Iijima H., Kitanaka S., et al. Identification of Dendrobium species used for herbal medicines based on ribosomal DNA internal transcribed spacer sequence. Biol. Pharm. Bull. 2011;34:779–782. doi: 10.1248/bpb.34.779. [DOI] [PubMed] [Google Scholar]
- 3.Burke J.M., Bayly M.J., Adams P.B., Ladiges P.Y. Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification. Aust. Sys. Bot. 2008;21:1–14. doi: 10.1071/SB07038. [DOI] [Google Scholar]
- 4.Yukawa T., Kita K., Handa T. DNA phylogeny and morphological diversification of Australian Dendrobium (Orchidaceae) In: Wilson K.L., Morrison D.A., editors. Monocots: Systematics and Evolution. CSIRO Publishing; Collingwood, Australia: 2000. pp. 465–471. [Google Scholar]
- 5.Xiang X.G., Schuiteman A., Li D.Z., Huang W.C., Chung S.W., Li J.W., Zhou H.L., Jin W.T., Lai Y.J., Li Z.Y., et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 2013;69:950–960. doi: 10.1016/j.ympev.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Xu S., Li D., Li J., Xiang X., Jin W., Huang W., Jin X., Huang L. Evaluation of the DNA barcodes in Dendrobium (Orchidaceae) from mainland Asia. PLoS ONE. 2015;10:e0115168. doi: 10.1371/journal.pone.0115168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng S.-G., Lu J.-J., Gao L., Liu J.-J., Wang H.-Z. Molecular phylogeny analysis and species identification of Dendrobium (Orchidaceae) in China. Biochem. Genet. 2014;52:127–136. doi: 10.1007/s10528-013-9633-6. [DOI] [PubMed] [Google Scholar]
- 8.Peyachoknagul S., Mongkolsiriwatana C., Srikulnath S., Huehne P., Srikulnath K. Identification of native Dendrobium species in Thailand by PCR-RFLP of rDNA-ITS and chloroplast DNA. ScienceAsia. 2014;40:113–120. doi: 10.2306/scienceasia1513-1874.2014.40.113. [DOI] [Google Scholar]
- 9.Srikulnath K., Sawasdichai S., Jantapanon T.K., Pongtongkam P., Peyachoknagul S. Phylogenetic relationship of Dendrobium species in Thailand inferred from chloroplast matK gene and nuclear rDNA ITS region. Hort J. 2015;84:243–252. doi: 10.2503/hortj.MI-028. [DOI] [Google Scholar]
- 10.Asahina H., Shinozaki J., Masuda K., Morimitsu Y., Satake M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J. Nat. Med. 2010;64:133–138. doi: 10.1007/s11418-009-0379-8. [DOI] [PubMed] [Google Scholar]
- 11.Hollingsworth P., Forrest L., Spouge J., Hajibabaei M., Ratnasingham S., Bank M., Chase M., Cowan R., Erickson D., Fazekas A. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay P., Banerjee G., Banerjee N. Distinguishing orchid species by DNA barcoding: Increasing the resolution of population studies in plant biology. Omics. 2017;21:711–720. doi: 10.1089/omi.2017.0131. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Shi L.L., Zhou J., Zhu G.P. DNA barcoding identification of Dendrobium huoshanense and its adulterants. Zhongguo Zhong Yao Za Zhi. 2018;43:4055–4061. doi: 10.19540/j.cnki.cjcmm.2018.0107. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Fang C., Zhang T., Guo L., Ye Q. Molecular authentication and differentiation of Dendrobium species by rDNA ITS region sequence analysis. AMB Express. 2019;9:53. doi: 10.1186/s13568-019-0767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran D.D., Khuat H.T., La T.N., Nguyen T.T.T., Pham B.H., Nguyen T.K., Tran H.D., Do M.T. Identification of Vietnamese native Dendrobium species based on ribosomal DNA internal transcribed spacer sequence. Adv. Stud. Biol. 2018;10:1–12. [Google Scholar]
- 16.Nguyen N.H., Tran H.D., Duong H.X., Huynh H.D. Phylogenetic relationship between several Dendrobium strains based on ITS sequence. Vietnam J. Sci. Technol. Agri. 2017;12:41–45. (In Vietnamese) [Google Scholar]
- 17.Nguyen N.H. ITS sequence of several strains of Dendrobium thyrsiflorum. J. Sci. Univ. Educ. 2018;15:149–153. (In Vietnamese) [Google Scholar]
- 18.Waud M., Busschaert P., Ruyters S., Jacquemyn H., Lievens B. Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol. Ecol. Resour. 2014;14:679–699. doi: 10.1111/1755-0998.12229. [DOI] [PubMed] [Google Scholar]
- 19.Kyndt T., Van B., Romeijn E., Romero P., Scheldeman X., Goetghebeur P., Van Damme P., Gheysen G. Molecular phylogeny and evolution of Caricaceae based on rDNA internal transcribed spacers and chloroplast sequence data. Mol. Phylogenet. Evol. 2005;37:442–459. doi: 10.1016/j.ympev.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Yao P.-C., Gao H.-Y., Wei Y.-N., Zhang J.-H., Chen X.-Y., Li H.-Q. Evaluating sampling strategy for DNA barcoding study of coastal and inland halo-tolerant Poaceae and Chenopodiaceae: A case study for increased sample size. PLoS ONE. 2017;12:e0185311. doi: 10.1371/journal.pone.0185311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabelin V.L., Alejandro G.J. Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to molecularly authenticate philippine ethnomedicinal apocynaceae through DNA barcoding. Pharmacogn. Mag. 2016;12:S384–S388. doi: 10.4103/0973-1296.185780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geospiza Inc. FinchTV. [(accessed on 10 January 2020)]; Available online: http://www.geospiza.com/Products/finchtv.shtml.
- 23.Manolo G., Stéphane G., Olivier G. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng S., Jiang Y., Wang S., Jiang M., Chen Z., Ying Q., Wang H. Molecular identification of Dendrobium species (Orchidaceae) based on the DNA barcode ITS2 region and its application for phylogenetic study. Int. J. Mol. Sci. 2015;16:21975–21988. doi: 10.3390/ijms160921975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao H., Song J., Liu C., Luo K., Han J., Li Y., Pang X., Xu H., Zhu Y., Xiao P., et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Yao H., Han J., Liu C., Song J., Shi L., Zhu Y., Ma X., Gao T., Pang X., et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vu H.-T., Huynh P., Tran H.-D., Le L. In silico study on molecular sequences for identification of paphiopedilum species. Evol. Bioinform. 2018;14 doi: 10.1177/1176934318774542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vu H.-T., Vu Q.-L., Nguyen T.-D., Tran N., Nguyen T.-C., Duong T., Nguyen T.-K., Le L. Genetic Diversity and Identification of Vietnamese Paphiopedilum Species Using DNA Sequences. Biology. 2019;9:9. doi: 10.3390/biology9010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y.-Y., Huang L.-Q., Liu Z.-J., Wang X.-Q. Promise and challenge of DNA barcoding in venus slipper (Paphiopedilum) PLoS ONE. 2016;11:e0146880. doi: 10.1371/journal.pone.0146880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wattoo J., Saleem Z., Shahzad M.S., Arif A., Hameed A., Saleem A. DNA Barcoding: Amplification and sequence analysis of rbcl and matK genome regions in three divergent plant species. Adv. Life Sci. 2016;4:3–7. [Google Scholar]
- 32.Yao H., Song J.Y., Ma X.Y., Liu C., Li Y., Xu H.X., Han J.P., Duan L.S., Chen S.L. Identification of Dendrobium species by a candidate DNA barcode sequence: The chloroplast psbA-trnH intergenic region. Planta Med. 2009;75:667–669. doi: 10.1055/s-0029-1185385. [DOI] [PubMed] [Google Scholar]
- 33.Gigot G., Van Alphen-Stahl J., Bogarin D., Warner J., Chase M.W., Savolainen V. Finding a suitable DNA barcode for Mesoamerican orchids. Lankesteriana Int. J. Orchid. 2007;7:200–203. doi: 10.15517/lank.v7i1-2.19502. [DOI] [Google Scholar]
- 34.Moudi M. Phylogenetic analysis among four sections of Genus Dendrobium Sw. (Orchidaceae) in Peninsular Malaysia using rbcL sequence data. Int. J. Bioassays. 2013;2:932–937. [Google Scholar]
- 35.Singh H.K., Parveen I., Raghuvanshi S., Babbar S.B. The loci recommended as universal barcodes for plants on the basis of floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Res. Notes. 2012;5:42. doi: 10.1186/1756-0500-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D., Li Z., Mao P., Yan X., Chun Z., Ma X. Phylogenetic analysis and identification of Dendrobium species based on ribosomal DNA internal transcribed spacer (ITS) sequence. Acta Hortic. Sin. 2012;39:1539–1550. [Google Scholar]
- 37.Liu Y., Chen R., Lin S., Chen Y., Chin S., Chen F., Lee C. Analysis of sequence diversity through internal transcribed spacers and simple sequence repeats to identify Dendrobium species. Genet. Mol. Res. 2014;13:2709–2717. doi: 10.4238/2014.April.8.15. [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Chen X. Barcoding the Dendrobium (Orchidaceae) species and analysis of the intragenomic variation based on the internal transcribed spacer 2. BioMed Res. Int. 2017;2017:2734960. doi: 10.1155/2017/2734960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu S., Huang J., Li P.-X., Yang H.-J., Zhang Y.-Q., Zhang G.-Q., Chen L.-J., Niu Y.-X., Luo Y.-B. Morphological type identification of self-incompatibility in dendrobium and its phylogenetic evolution pattern. Int. J. Mol. Sci. 2018;19:2595. doi: 10.3390/ijms19092595. [DOI] [PMC free article] [PubMed] [Google Scholar]