Abstract
The immunomodulatory effects of Suppressor of Cytokine Signaling (SOCS) proteins, that control the JAK/STAT pathway, indicate them as attractive candidates for immunotherapies. Recombinant SOCS3 protein suppresses the effects of inflammation, and its deletion in neurons or in immune cells increases pathological blood vessels growth. Recently, on the basis of the structure of the ternary complex among SOCS3, JAK2, and gp130, we focused on SOCS3 interfacing regions and designed several interfering peptides (IPs) that were able to mimic SOCS3 biological role in triple negative breast cancer (TNBC) models. Herein, to explore other protein regions involved in JAK2 recognition, several new chimeric peptides connecting noncontiguous SOCS3 regions and including a strongly aromatic fragment were investigated. Their ability to recognize the catalytic domain of JAK2 was evaluated through MST (microscale thermophoresis), and the most promising compound, named KIRCONG chim, exhibited a low micromolar value for dissociation constant. The conformational features of chimeric peptides were analyzed through circular dichroism and NMR spectroscopies, and their anti-inflammatory effects were assessed in cell cultures. Overall data suggest the importance of aromatic contribution in the recognition of JAK2 and that SOCS3 peptidomimetics could be endowed with a therapeutic potential in diseases with activated inflammatory cytokines.
Keywords: Mimetic peptides, cytokine signaling, JAK-STAT, SOCS3, inflammation
The ability of Suppressor of Cytokine Signaling (SOCS) proteins to modulate Janus Kinases-Signal Transducer and Activator of Transcription (JAK/STAT) pathway effects suggests them as attractive templates for immunotherapeutics design.1,2 Traditionally SOCS are considered as negative feedback inhibitors of this pathway, but recent investigations pointed out a different regulation of their expression in dependence of transcriptional, translational, and post-translational conditions.3 The SOCS family consists of eight proteins sharing similar organization of domains constituted by an N-term region of variable length and sequence, a central SH2 domain, and a SOCS box in the C-term.4 Particularly, SOCS1 and 3 are the unique members presenting a kinase inhibitory region (KIR) and a small region called ESS (Extended SH2 Subdomain) in the N-term region of the SH2.5 These proteins regulate STAT signaling through different mechanisms: (a) the direct inhibition of Janus kinases, (b) the competition with STAT-SH2 domains for specific phosphotyrosine residues of intracellular portion of the receptor, and (c) an initial SH2-recruitment to the receptor cytoplasmic domain, followed by the inhibition of JAK activity, as shown by SOCS3.6 Diverse stimuli, including cytokines, growth factors, and bacterial and viral pathogen-associated molecular patterns, induce SOCS3’s expression.7 This protein is a physiological regulator in immune homeostasis, and its dysregulation can cause allergies, autoimmune diseases, inflammation, and cancer.8,9 SOCS3 suppression or epigenetic downregulation has been found in many types of solid tumors, and its functional deficiency is implicated in tumor development and metastases.10,11 Indeed, SOCS3 overexpression in NSCLC (Nonsmall cell lung cancer) cell lines inhibited tumor cell function, indicating that loss of SOCS3 is critical for tumorigenesis.12 SOCS3 acquired from alveolar macrophage-derived extracellular vesicles restrains epithelial tumorigenesis, while its deficiency at the mucosal surface contributes to epithelial cell transformation and allows growth and survival of mature tumor cells.13 Furthermore, SOCS3 regulates inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer (TNBC) models.14,15 A major role for SOCS3 has been outlined in spinal cord plasticity and mechanical allodynia associated with peripheral nerve injury16 and in the regulation of chondrocyte responses during inflammatory arthritis.17 In human atherosclerotic lesions, both vascular smooth muscle cells (VSMCs) and macrophages express SOCS3, indicating its key regulatory function in vascular cell responses.18−20 Importantly, SOCS3 regulates muscle homeostasis; indeed, muscle-specific deletion of SOCS3 increases the early inflammatory response21 and in muscles from SOCS3-knockout mice the SOCS3 mRNA response to lipopolysaccharide stimulation was significantly blunted, while STAT3 p-Tyr705 was exacerbated, clearly indicating that SOCS3 deficiency is tissue specific.22,23 SOCS3 protein acts as a feedback inhibitor of the JAK/STAT3 pathway24−26 indeed both SOCS3 and STAT3 restrain inflammation and induce the first level of protective immune response in different mouse models.2 In some cases, SOCS3 regulates the inflammatory responses positively through inhibiting STAT3. Numerous studies evidence abnormal expression levels of SOCS3/STAT3 in different myeloid and lymphoid cells as well as in various nonhematopoietic cells, due to its involvement in various infection and inflammatory diseases.22,23 Given the importance to maintain SOCS3 expression in opposing STAT-dependent responses, different approaches to modulate the STAT-SOCS3 axis are employed: (i) inhibitors of STAT activation (e.g., ruxolitinib); (ii) exogenous expression of SOCS3 via viral vectors12,18−20,26,27 or recombinant methods;28 and (iii) very recently, the administration of SOCS3 peptides.29,15 On the basis of similar studies carried out for SOCS1 protein30−32 and following a protein dissection structure-based approach33 we identified, for the first time to the best of our knowledge, a linear peptide spanning 22–45 residues of the protein, called KIRESS, endowed with a good helical content and able to bind to the JAK2 catalytic domain. This peptide mimicking the action of SOCS3 efficiently suppresses the IL(Interleukin)-22 molecular signaling in keratinocytes29 and that of IL-6 in TNBC models.15 Herein, to unveil a cooperative mechanism of action in the recognition of JAK2 among different protein fragments, we designed and investigated several chimeric peptides connecting noncontiguous SOCS3 regions and including a strongly aromatic fragment. Their abilities to recognize the catalytic domain of JAK2 were evaluated through MST (microscale thermophoresis), and the most promising compound, named KIRCONG chim, was conformationally investigated through CD (circular dichroism) and NMR (nuclear magnetic resonance) spectroscopies. The functionality and anti-inflammatory effects of designed sequences were evaluated in cells.
Results and Discussion
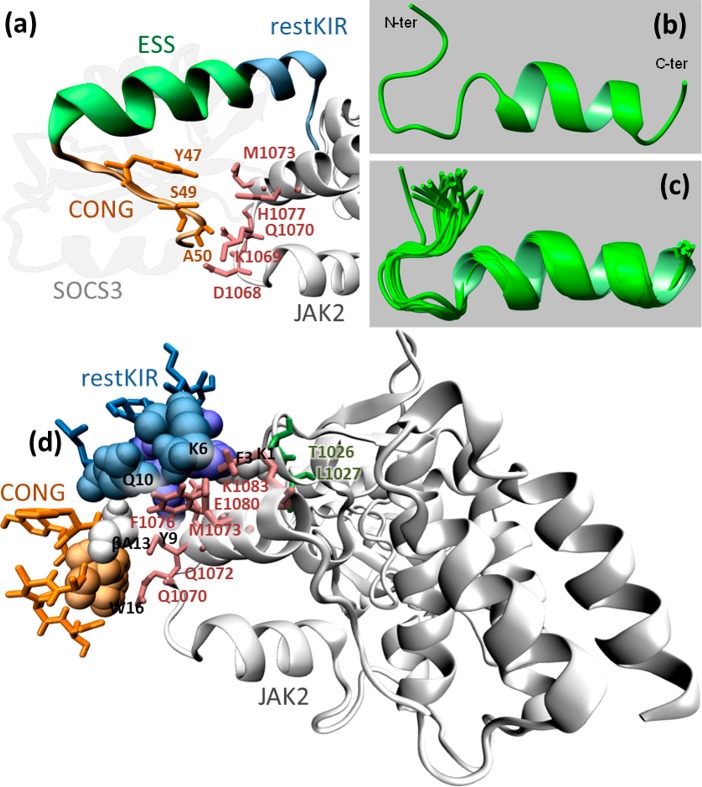
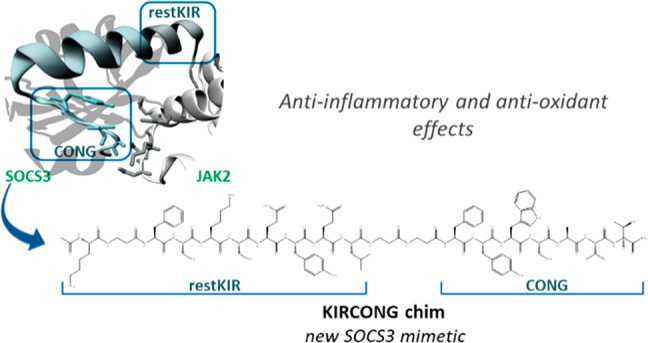
The crystal structure of the ternary complex among murine SOCS3/JAK2 kinase domain/gp130 phosphotyrosine-peptide was employed to design mimetics of SOCS3.34 From its inspection, SOCS3 appeared to interact with JAK2 and gp130 simultaneously through adjacent interfaces: KIR and ESS domains appeared mainly involved in the recognition of “GQM” and helix G motifs of JAK2 essentially through hydrophobic interactions. These considerations were fully confirmed by our previous investigations: peptides covering KIR (22–33) and ESS (34–45) domains separately and in conjunction, in the peptide KIRESS (22–45) (Table 1) demonstrated both in vitro and in vivo, to act as inhibitors of JAK2.29,15 In that study, SPR binding assays provided KD values in the low micromolar range for the complex KIRESS/biotin-JAK2. A deeper insight into the structure of SOCS3/JAK2/gp130 indicates that other contacts involving some residues in a “hinge” region between ESS and helix A (HA) of the SOCS3-SH2 domain are crucial for the formation of the complex. In particular, residues Tyr,47 Ser,49 and Ala50 of SOCS3 appeared to be located closer to Asp1068, Lys1069, Met 1073 (3.8 Å from Tyr47), His1077 (4.1 Å from Ser49), and Gln1070 (<3.7 Å from Ala50 and <3.4 Å from Thr52 which is the first residue of HA of SOCS3) of JAK2 (Figure 2A). Interestingly, the region spanning 46–52 residues, hereafter named CONG, contains three adjacent aromatic amino acids 46FYW48 out of seven residues. To unveil a potential contribution of this region on the ability of SOCS3 to recognize JAK2 we designed several new chimeric peptides: (a) ESSCONG (34–52) connecting contiguous ESS (34–45) and CONG (46–52) fragments, (b) restKIRESSCONG (spanning 25–52 residues) constituted by ESS (34–45), CONG (46–52) and a shorter version of KIR sequence (named restricted KIR, deleted of three residues at the N-termini) and (c) KIRCONG chim a chimeric peptide including two noncontiguous fragments restKIR (25–33) and CONG (46–52) connected by β-alanines as spacers (Table 1).
Table 1. Sequences and Names of Sequences Investigated in This Studya.
Figure 2.
Structural features of KIRCONG fragment in complex with JAK2. (a) SOCS3/JAK2 (PDB: 4GL9 41, chains A and E) where the fragment restKIRESSCONG and residues described in the text are highlighted; (b, c) NMR structure of the KIRCONG chim peptide calculated in H2O/TFE (60/40, v/v). (b) The first conformer of the NMR ensemble and (c) the best 20 conformers superimposed on the N, C, O, CA backbone atoms of residues 3–20 and CB atoms of residues 12–13 (RMSD = 0.59 Å). The NMR structure was generated from 204 upper distance limits (59 intraresidue, 85 short-range, and 60 medium-range) and 86 angular constraints (KIRCONG chim residues are numbered as reported in Figure S3). (d) Putative pose of KIRCONG chim peptide docked to JAK2: KIRCONG chim interacting residues are highlighted with their van der Waals sphere, while those on JAK2 are reported in pink. Thr1026 and Leu1027 forming a hydrogen bond with Lys1 are also highlighted (dark green). Color code: JAK2 (white), SOCS3 (gray), restKIR (blue), ESS (green), CONG (orange).
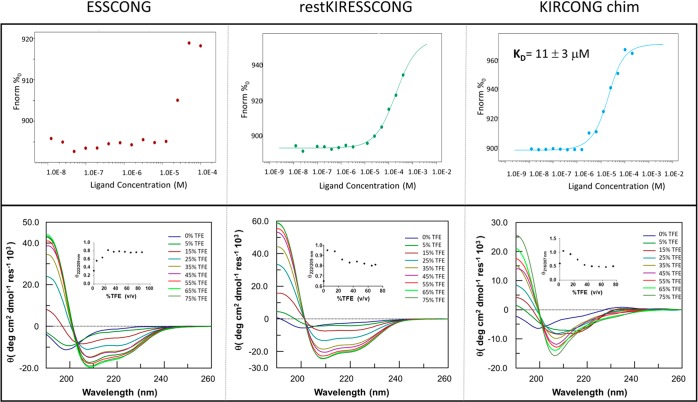
To evaluate the ability of newly derived SOCS3 peptides to recognize the JAK2 catalytic domain, in vitro MST experiments were carried out (Figure 1, upper panels). Microscale thermophoresis is based on the detection of a temperature-induced change in the fluorescence of a target as a function of the concentration of a nonfluorescent ligand, allowing the biophysical analysis of interactions between biomolecules.35 Its major advantages with respect to other in vitro binding assays are very low sample requirement and a substantially low maintenance for instrumentation. Signals exhibit a dose–response curve for all three peptides (thermophoretic traces are reported in Figure S1), but for ESSCONG signal variation was too small to be meaningful and for restKIRESSCONG, the signal did not reach saturation. The fitting of experimental data for KIRCONG chim provided a KD value of 11 ± 3 μM (Figure 1, upper panels) that is comparable with the previously reported value for KIRESS.15
Figure 1.
Upper panel: binding isotherms for MST signals versus peptide concentrations. SOCS3 derived peptides were used with a serial dilution (1:1) by preparing 14–16 samples on average of the following stock solutions: ESSCONG 404 μM, KIRCONG chim 409 μM, restKIRESSCONG 408 μM in labeling buffer at pH 7.5. Lower panel: overlay of CD spectra in TFE/H20 0–75% v/v. Spectra were acquired in 10 mM phosphate buffer at pH 7.4, at 25 °C, in mixtures with TFE (2,2,2-trifluoroethanol) and peptides concentration was 50 μM.
The conformational preferences of SOCS3 designed peptides were investigated through CD and NMR spectroscopies. CD spectra were recorded in the far UV region in 10 mM phosphate buffer, pH 7 and in the presence of TFE (2,2,2-trifluoroethanol), as structuring agent.36,37 In Figure 1, lower panels, the overlay of CD spectra, at different TFE percentages for each peptide, are reported. For all sequences, spectra in buffer present one minimum around 200 nm indicating a prevalent random content even if the presence of a shoulder at ∼215 nm suggests a small contribution of helical conformations and, in the case of KIRCONG chim, a positive slight band at ∼235 nm suggests a direct involvement of aromatic residues in the conformational features of the sequence.38 To evaluate the effects of TFE addition on the conformational features of designed sequences, we followed the variation of Θratio value at increasing concentration of the cosolvent, as reported as insets of Figure 1, lower panels. For ESSCONG and restKIRESSCONG we evaluated the ratio between ellipticity at 222 and 209 nm; and for KIRCONG chim, at 216 and 207 nm. The presence of TFE for all three sequences allows us to reach more ordered helical conformations, as expected. In detail, for restKIRESSCONG and KIRCONG chim the lowest TFE concentration, 5%, tends to partially induce coiled coil conformations, since the maximum value of Θratio is observed (∼1 for KIRCONG chim and 0.95 for restKIRESSCONG (Figure 1, lower panels). However, the addition of higher amounts of TFE stabilizes single helices, indeed Θratio values decrease until 20% and then remain constant. For ESSCONG the Θratio profile increases monotonically until 20%, when, similarly to other sequences, it reaches a constant value of ∼0.8 (Figure 1, lower panels).
To get further structural insights on the most potent ligand, KIRCONG chim, NMR studies were carried out in H2O and H2O/TFE mixtures (15% and 40% v/v).39
Initial studies performed in H2O revealed a flexible conformation lacking regular secondary structure elements. Comparison of 2D [1H, 1H] TOCSY40 and 2D [1H, 1H] ROESY41 spectra (Figure S2) allowed us to get almost complete proton resonance assignments42 (Table S1). The ROE pattern (Figure S3, blue panel) was dominated by strong Hαi-HNi+1 peaks in between sequential residues (i and i+1) which are canonical of extended and random coil species.42 A complete structure calculation further reinforced the coexistence of multiple conformations (Figures S4, S5 and Table S2); even if in the C-terminal peptide region, encompassing residues Tyr15-Ala,18 a tendency to form a bend is observed (Figure S4).
A similar investigation at 15% TFE demonstrated a slight decrease of the conformational freedom of the peptide. Proton resonance assignments were obtained from combined analysis of TOCSY40 and NOESY43 spectra (Figure S6 and Table S3). A complete 3D structure calculation indicated that conformers remain prevalently disordered (Table S4 and Figure S7): indeed the NOESY spectrum still contains mostly strong sequential contacts characteristic of a random coil status (Figure S3, red panel),42 although a few NOE correlations of the type HNi-HNi+2 point out a certain helical conformation even if distorted (Figure S3, red panel).42
Coherently, the inspection of calculated conformers (Table S4) with the software MOLMOL44 reveals the presence of a 3.10 helix at the C-terminus between residues Tyr15-Ser17 in 4 over 20 structures (Figures S5 and S7).
By increasing TFE from 15% to 40%, NMR spectra (Figure S8), in agreement with CD studies, indicate an increase of order. The NOE pattern (Figure S3, green panel) shows many inter-residue correlations indicative of α and 3.10 helices including Hαi-HNi+2, Hαi-HNi+3, Hαi-Hβi+3, and Hαi-HNi+442 (Figure S3, green panel). The analysis with MOLMOL44 of the structure (Table S6), identified, in most conformers, a α-helix encompassing two β-Ala residues 12 and 13, within the segment Gln10-Ser17 and extending in a few members of the NMR structure bundle from Gln8/Tyr9 until Val19 while, the N-terminal segment still lacks canonical secondary structure elements (Figures 2b, c and S3). Nevertheless, the presence of two β-alanines likely perturbs a regular α-helical organization since not all the backbone COi-NHi+4 H-bonds are observed in NMR conformers as required in a stable α-helical organization (Table S7). Noticeably the CO and NH backbone atoms of β-Ala at position 13 are involved in α-helical H-bonds different from those of β-Ala in position 12 (Table S7).
NMR results (Figure 2c) were used to model the peptide/JAK2 complex. Each NMR conformation of KIRCONG chim was docked to the SOCS3 binding site on JAK2 PDB: 4GL9,45 chain A, (Figure S9a). One pose was selected for each NMR conformation, and among the 20 identified poses, seven presented the root mean squared deviation (RMSD) of heavy atoms of KIRCONG chim peptide with respect to the same atoms of SOCS3 below 1.30 nm (Figure S9b,c). In particular, the closest pose (Figure S9d) had RMSD 0.88 nm for restKIR, 0.69 nm for CONG, and 0.80 nm for both fragments. In this pose some peptide contacts are maintained with respect to SOCS3 protein (Figure S10a,b and 2d): Phe,25 Lys,28 and Thr31 (Phe3, Lys6, and Thr9 in the peptide numeration). Novel interactions are also formed: the residue Lys1 forms H-bonds with both Thr1027 and Leu1026 and the β-Ala13 interacts with JAK2 through van der Waals contacts. In comparison with the structure, JAK2 in complex with KIRCONG chim maintains contacts with both the restKIR fragment through Leu1026 and Thr1027 and with the CONG fragment through Gln1070, Phe1076, and Glu1080, while new contacts involve Gln1072, Met 1073, and Lys 1083.
To get insights on potential cellular effects of SOCS3 derived peptides, on the basis of the primary role exerted by both SOCS1 and SOCS3 proteins in the formation of atherosclerotic plaque,19,20 we checked the activity of KIRCONG chim peptide in VSMC and macrophages, two major cellular constituents of atheroma plaques with an active role in atherogenesis. In these cellular assays with murine primary VSMC and macrophage-like cell line (RAW264.7), we also employed KIRESS peptide that, although demonstrated to mimic SOCS3’s cellular effects, was never tested in these cells. As negative control (NC) a peptide with the same length of KIRESS, bearing only alanines to avoid side chain interactions and five arginines to help aqueous solubility, was employed (Table 1).
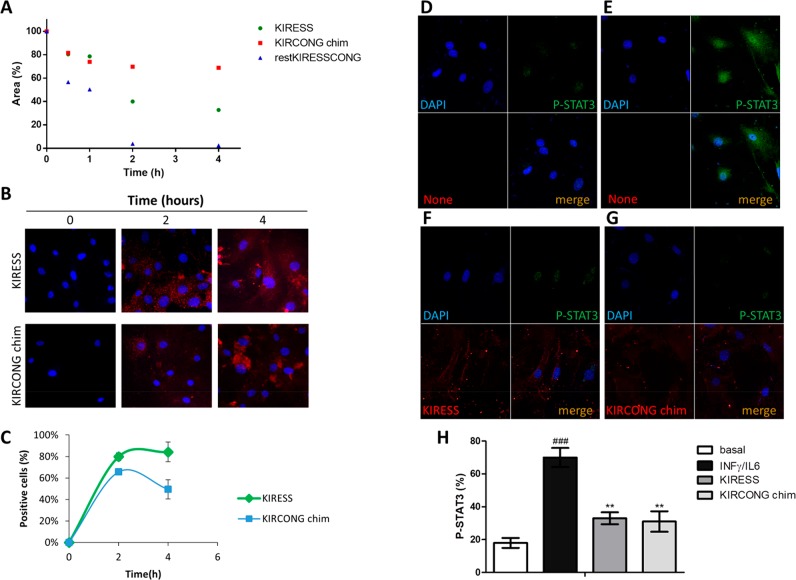
To check the stability of designed sequences during cellular experiments, the decrease of chromatographic peaks of pure SOCS3 derived peptides was followed during time. In Figure 3A, the area percentages of peptides versus time of analysis is reported. After 2 h of incubation in FBS (fetal bovine serum), restKIRESSCONG peptide appeared completely degraded while KIRESS and importantly KIRCONG chim peptides present a residual concentration of 40% and 70%, respectively. The greater stability shown by KIRCONG chim is likely due to the presence of three β-alanines as “non native” amino acid. On this basis, we analyzed only KIRESS and KIRCONG chim, excluding restKIRESSCONG that appeared too easily degraded in cellular media and ESSCONG that did not provide a significant binding in MST experiments (Figure 1, upper panel).
Figure 3.
Effects of SOCS3 derived peptides on the JAK/STAT pathway. Serum stability (A) and confocal images of cellular internalization (B) of peptides at different times, in VSMCs. (C) Quantification of peptides internalization on the basis of TAMRA (tetramethylrhodamine) fluorescence intensity. (D–G) Representative confocal images (n = 3 independent experiments) of cells under basal conditions (D), stimulated with IFNγ+IL-6 (E), stimulated with IFNγ+IL-6 in the presence of TAMRA-KIRESS (5 μM) (F), and stimulated with IFNγ+IL-6 in the presence of TAMRA-KIRCONG chim (5 μM) (G). (red, SOCS3 derived peptides; green, P-STAT3; blue, DAPI stained nuclei). (H) P-STAT3 fluorescence intensity percentages expressed as Mean ± SEM (###P < 0.001 vs basal; **P < 0.01 vs cytokines).
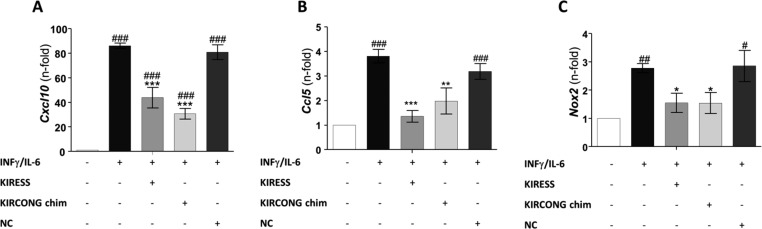
To perform cellular experiments, KIRESS, KIRCONG chim and NC were conjugated with the fragment 48–60 of the HIV Tat protein that allows cell penetration. Confocal microscopy images of mouse VSMCs treated with fluorescent-SOCS3 peptides46 are reported in Figure 3B–C: they indicated an efficient and similar uptake of KIRESS and KIRCONG chim peptides after 2 h, exhibiting a prevalent cytoplasmic distribution. Subsequently, immunofluorescence experiments in VSMCs revealed that both KIRESS and KIRCONG chim inhibited cytokine-induced STAT3 activation, as evidenced by a 65% reduction of STAT3 phosphorylation (P-STAT3) and nuclear translocation (Figure 3D–H). The effects of SOCS3 peptides were also investigated in RAW264.7 macrophages. Pretreatment of cells with KIRESS and KIRCONG chim peptides significantly reduced the mRNA expression of STAT-regulated pro-inflammatory genes such as Cxcl10 and Ccl5 chemokines47 (Figure 4A–B). Moreover, as reported in Figure 4C, both peptides inhibited the gene expression of NADPH oxidase 2 (Nox2), a superoxide-generating enzyme that can be modulated by SOCS3 expression.48 As expected, NC peptide was inactive in cellular assays. Overall data indicated anti-inflammatory and antioxidant effects of SOCS3 peptides and support further investigations of their potential in inflammatory diseases, including atherosclerosis.
Figure 4.
Real-time PCR analysis of STAT-dependent genes in macrophages. RAW264.7 cells were stimulated with IFNγ+IL-6 for 1 h in the absence or presence of 12.5 μM of SOCS3 peptidomimetics and NC peptide. The mRNA expression levels of Cxcl10 (A), Ccl5 (B), and Nox2 (C) were normalized to 18S and expressed as fold increases over basal condition (arbitrary set as 1). Mean ± SEM of three independent experiments (#P < 0.05, ##P < 0.01, ###P < 0.001 vs basal; *P < 0.05, **P < 0.01, ***P < 0.001 vs cytokines).
Conclusion
To investigate the molecular mechanisms of protein–protein interactions involved in the JAK/STAT pathway and to identify new interfering compounds of this signaling, we have already carried out several studies by assuming SOCS proteins as structural and functional templates.9,15,30−32,49 SOCSs are natural inhibitors of JAK/STAT, and the effects of their overexpression or exogenous administration suggested them as crucial pharmaceutical targets in several inflammation-related diseases such as rheumatoid arthritis,50 severe acute pancreatitis (SAP),51 and metabolic dysfunction induced by obesity.52 In particular, mimetics of SOCS1 and SOCS3 proteins have been demonstrated to be endowed with potential therapeutical applications.53,54,14,55−58 On the basis of our recent study focused on the design of SOCS3 mimetics and their potential application in TNBC disease, here we presented structural and functional investigations of another series of peptidomimetics of SOCS3 that include protein fragments neglected in the previous design. In detail, we engineered sequences spanning several SOCS3 fragments, contiguous and not, including the so-called CONG motif that in the protein architecture acts as a hinge between the KIR region and SH2 domain and appears to have hot spots of interaction with the JAK2 catalytic domain. All designed sequences were analyzed for their recognition ability to bind to JAK2 through MST assay. Indeed, here we preferred to employ a completely in solution binding assay since previous SPR experiments had highlighted unspecific signals on the chip, likely due to a limited water solubility of sequences containing the KIR motif. A chimeric peptide including noncontiguous protein fragments named KIRCONG chim was able to recognize JAK2 exhibiting a low micromolar dissociation constant value. This sequence demonstrated a good propensity to assume prevalent α-helical conformations, as outlined by NMR investigations and docking experiments. This observation further indicated that it is able to maintain several interactions with JAK2 with respect to SOCS3/JAK2 crystal structure and to create new points of connections.
To deepen the functional aspects of KIRCONG chim to mimic SOCS3, its anti-inflammatory properties were investigated in VSMC and RAW264.7 macrophages. The presence of KIRCONG chim as well as of KIRESS reduced the phosphorylation and nuclear translocation of STAT3, and in parallel, several dependent genes were repressed.
Reported data suggest the importance of helical content and aromatic contribution in the recognition of SOCS3 toward JAK2. In this interaction the KIR domain exerts the major role, but in the current study the importance of neighboring regions close to KIR has been outlined. These regions aid recognition in a helical context and both KIRESS and KIRCONG chim peptides exhibited low micromolar values of KD and similar anti-inflammatory properties inhibiting different cytokines (IL-6, IL-22, and INFγ) pathways. The limited water solubility and high molecular weights of KIRCONG chim, as well as of KIRESS sequences, make these compounds unsuitable to be used as they are as drugs. Their study however can pave the way for the design of new molecules, preferably macrocycles,59 through their structural and chemical modifications. Indeed, future structure–activity relationship (SAR) investigations will be done by reducing the conformational flexibility of these linear peptides through introduction of constraints at various positions and simultaneously, by enhancement in their water solubility. In this direction, different strategies can be followed combining iterative in silico and experimental optimization cycles through the appropriate introduction of small ionizable functional groups.60
Acknowledgments
We thank L. De Luca and L. Zona for helpful technical support. S.L.M. was supported by AIRC fellowship for Italy.
Glossary
Abbreviations
- CD
circular dichroism
- NMR
nuclear magnetic resonance
- SOCS
suppressors of cytokine signaling
- JAK
Janus kinases
- STAT
signal transducer and activator of transcription
- KIR
kinase inhibitory region
- ESS
extended SH2 subdomain
- INF-γ
Interferon gamma
- CXCL
C-X-C motif chemokine ligands
- CCL
C–C motif chemokine ligand
- IL
interleukin
- SH2
Src homology 2
- VSMCs
vascular smooth muscle cells
- MST
microscale thermophoresis
- TAMRA
tetramethylrhodamine
- PCR
polymerase chain reaction
- FBS
fetal bovine serum
- TOCSY
total correlation spectroscopy
- NOESY
nuclear Overhauser enhancement spectroscopy
- ROESY
rotating frame Overhauser enhancement spectroscopy
- NSCLC
nonsmall cell lung cancer
- TNBC
triple negative breast cancer
- Nox2
NADPH oxidase 2
- SAP
severe acute pancreatitis
- RMSD
root mean squared deviation.
- NSCLC
nonsmall cell lung cancer
- TFE
2,2,2-trifluoroethanol
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.9b00664.
Figure S1. Thermophoretic traces of MST assays. Figure S2. 2D [1H–1H] TOCSY and ROESY 250 spectra of KIRCONG chim peptide. Figure S3. ROE and NOE patterns in H2O. Figure S4. NMR structure of KIRCONG chim peptide in H2O. Figure S5. First conformers of KIRCONG chim peptide. Figure S6. 2D [1H–1H] TOCSY and NOESY 300 spectra of KIRCONG chim peptide in H2O/TFE. Figure S7. NMR structure of KIRCONG chim peptide in H2O/TFE 85/15. Figure S8. 2D [1H–1H] TOCSY and NOESY 300 spectra of KIRCONG chim peptide in H2O/TFE 60/40. Figure S9–S10. Schematic diagram of the interaction between restKIRESSCONG or KIRCONG chim and JAK2. Tables of 1H chemical shifts of KIRCONG chim peptide, structure statistics of KIRCONG chim peptide NMR ensemble, and H-bonds calculated with UCSF-Chimera. Complete experimental procedures. (PDF)
Author Contributions
S.L.M., L.L.S., F.A.M., and S.F. performed experiments, M.L. analyzed NMR investigations, C.G.G. and D.M. conceived the study and critically analyzed results. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was partially supported by POR CAMPANIA FESR 2014/2020 “Combattere la resistenza tumorale: piattaforma integrata multidisciplinare per un approccio tecnologico innovativo alle oncoterapie-Campania Oncoterapie” (Project N. B61G18000470007) and Spanish Government projects (MINECO/FEDER SAF2015-63696-R and MICINN/FEDER RTI2018-098788-B-I00 to C.G.G.).
The authors declare no competing financial interest.
Supplementary Material
References
- Keating N.; Nicholson S. E. SOCS-mediated immunomodulation of natural killer cells. Cytokine+ 2019, 118, 64–70. 10.1016/j.cyto.2018.03.033. [DOI] [PubMed] [Google Scholar]
- Carow B.; Rottenberg M. E. SOCS3, a Major Regulator of Infection and Inflammation. Front. Immunol. 2014, 5, 58. 10.3389/fimmu.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi J. U.; Kabir N. N.; Flores-Morales A.; Ronnstrand L. SOCS proteins in regulation of receptor tyrosine kinase signaling. Cell. Mol. Life Sci. 2014, 71 (17), 3297–310. 10.1007/s00018-014-1619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic S.; Sachithanandan N.; Kay T. W.; Steinberg G. R. Suppressor of cytokine signalling (SOCS) proteins as guardians of inflammatory responses critical for regulating insulin sensitivity. Biochem. J. 2014, 461 (2), 177–88. 10.1042/BJ20140143. [DOI] [PubMed] [Google Scholar]
- Yoshimura A.; Yasukawa H. JAK’s SOCS: a mechanism of inhibition. Immunity 2012, 36 (2), 157–9. 10.1016/j.immuni.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Croker B. A.; Kiu H.; Nicholson S. E. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008, 19 (4), 414–22. 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trengove M. C.; Ward A. C. SOCS proteins in development and disease. American journal of clinical and experimental immunology 2013, 2 (1), 1–29. [PMC free article] [PubMed] [Google Scholar]
- Yin Y.; Liu W.; Dai Y. SOCS3 and its role in associated diseases. Hum. Immunol. 2015, 76 (10), 775–80. 10.1016/j.humimm.2015.09.037. [DOI] [PubMed] [Google Scholar]
- La Manna S.; Di Natale C.; Florio D.; Marasco D., Peptides as Therapeutic Agents for Inflammatory-Related Diseases. Int. J. Mol. Sci. 2018, 19 ( (9), ), 2714. 10.3390/ijms19092714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikuma S.; Kanamori M.; Mise-Omata S.; Yoshimura A. Suppressors of cytokine signaling: Potential immune checkpoint molecules for cancer immunotherapy. Cancer science 2017, 108 (4), 574–580. 10.1111/cas.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki-Ohara K.; Kondo T.; Ito M.; Yoshimura A. SOCS, inflammation, and cancer. Jak-Stat 2013, 2 (3), e24053 10.4161/jkst.24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C.; Lin C. K.; Tsai Y. H.; Weng H. H.; Li Y. C.; You L.; Chen J. K.; Jablons D. M.; Yang C. T. Adenovirus-mediated SOCS3 gene transfer inhibits the growth and enhances the radiosensitivity of human non-small cell lung cancer cells. Oncol. Rep. 2010, 24 (6), 1605–12. 10.3892/or_00001024. [DOI] [PubMed] [Google Scholar]
- Speth J. M.; Penke L. R.; Bazzill J. D.; Park K. S.; de Rubio R. G.; Schneider D. J.; Ouchi H.; Moon J. J.; Keshamouni V. G.; Zemans R. L.; Lama V. N.; Arenberg D. A.; Peters-Golden M.. Alveolar macrophage secretion of vesicular SOCS3 represents a platform for lung cancer therapeutics. JCI insight 2019, 4 ( (20), ), 10.1172/jci.insight.131340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.; Ouzounova M.; Quraishi A. A.; Davis A.; Tawakkol N.; Clouthier S. G.; Malik F.; Paulson A. K.; D’Angelo R. C.; Korkaya S.; Baker T. L.; Esen E. S.; Prat A.; Liu S.; Kleer C. G.; Thomas D. G.; Wicha M. S.; Korkaya H. SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene 2015, 34 (6), 671–80. 10.1038/onc.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manna S.; Lee E.; Ouzounova M.; Di Natale C.; Novellino E.; Merlino A.; Korkaya H.; Marasco D. Mimetics of suppressor of cytokine signaling 3: Novel potential therapeutics in triple breast cancer. Int. J. Cancer 2018, 143 (9), 2177–2186. 10.1002/ijc.31594. [DOI] [PubMed] [Google Scholar]
- Dominguez E.; Mauborgne A.; Mallet J.; Desclaux M.; Pohl M. SOCS3-mediated blockade of JAK/STAT3 signaling pathway reveals its major contribution to spinal cord neuroinflammation and mechanical allodynia after peripheral nerve injury. J. Neurosci. 2010, 30 (16), 5754–66. 10.1523/JNEUROSCI.5007-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Croker B. A.; Campbell I. K.; Gauci S. J.; Alexander W. S.; Tonkin B. A.; Walsh N. C.; Linossi E. M.; Nicholson S. E.; Lawlor K. E.; Wicks I. P. Key role of suppressor of cytokine signaling 3 in regulating gp130 cytokine-induced signaling and limiting chondrocyte responses during murine inflammatory arthritis. Arthritis Rheumatol. 2014, 66 (9), 2391–402. 10.1002/art.38701. [DOI] [PubMed] [Google Scholar]
- Ortiz-Munoz G.; Lopez-Parra V.; Lopez-Franco O.; Fernandez-Vizarra P.; Mallavia B.; Flores C.; Sanz A.; Blanco J.; Mezzano S.; Ortiz A.; Egido J.; Gomez-Guerrero C. Suppressors of cytokine signaling abrogate diabetic nephropathy. J. Am. Soc. Nephrol. 2010, 21 (5), 763–72. 10.1681/ASN.2009060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Munoz G.; Martin-Ventura J. L.; Hernandez-Vargas P.; Mallavia B.; Lopez-Parra V.; Lopez-Franco O.; Munoz-Garcia B.; Fernandez-Vizarra P.; Ortega L.; Egido J.; Gomez-Guerrero C. Suppressors of cytokine signaling modulate JAK/STAT-mediated cell responses during atherosclerosis. Arterioscler., Thromb., Vasc. Biol. 2009, 29 (4), 525–31. 10.1161/ATVBAHA.108.173781. [DOI] [PubMed] [Google Scholar]
- Recio C.; Oguiza A.; Mallavia B.; Lazaro I.; Ortiz-Munoz G.; Lopez-Franco O.; Egido J.; Gomez-Guerrero C. Gene delivery of suppressors of cytokine signaling (SOCS) inhibits inflammation and atherosclerosis development in mice. Basic Res. Cardiol. 2015, 110 (2), 8. 10.1007/s00395-014-0458-1. [DOI] [PubMed] [Google Scholar]
- Swiderski K.; Thakur S. S.; Naim T.; Trieu J.; Chee A.; Stapleton D. I.; Koopman R.; Lynch G. S. Muscle-specific deletion of SOCS3 increases the early inflammatory response but does not affect regeneration after myotoxic injury. Skeletal Muscle 2016, 6, 36. 10.1186/s13395-016-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldow M. K.; Ham D. J.; Chee A.; Trieu J.; Naim T.; Stapleton D. I.; Swiderski K.; Lynch G. S.; Koopman R. Muscle-specific deletion of SOCS3 does not reduce the anabolic response to leucine in a mouse model of acute inflammation. Cytokine+ 2017, 96, 274–278. 10.1016/j.cyto.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Zhao H.; Wang P.; Wang J.; Zou L. The roles of SOCS3 and STAT3 in bacterial infection and inflammatory diseases. Scandinavian journal of immunology 2018, 88 (6), e12727 10.1111/sji.12727. [DOI] [PubMed] [Google Scholar]
- Nicholson S. E.; De Souza D.; Fabri L. J.; Corbin J.; Willson T. A.; Zhang J. G.; Silva A.; Asimakis M.; Farley A.; Nash A. D.; Metcalf D.; Hilton D. J.; Nicola N. A.; Baca M. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (12), 6493–8. 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A.; Ito M.; Chikuma S.; Akanuma T.; Nakatsukasa H. Negative Regulation of Cytokine Signaling in Immunity. Cold Spring Harbor Perspect. Biol. 2018, 10 (7), a028571. 10.1101/cshperspect.a028571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W. N.; Xia Z. Y.; Liu M.; Sun Q.; Lei S. Q.; Wu X. J.; Meng Q. T.; Leng Y. Protective effects of SOCS3 overexpression in high glucoseinduced lung epithelial cell injury through the JAK2/STAT3 pathway. Mol. Med. Rep. 2017, 16 (3), 2668–2674. 10.3892/mmr.2017.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. C.; Shi M.; Chueh F. Y.; Venkitachalam S.; Yu C. L. Enforced SOCS1 and SOCS3 expression attenuates Lck-mediated cellular transformation. Int. J. Oncol. 2010, 36 (5), 1201–8. 10.3892/ijo_00000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo D.; Liu D.; Yao S.; Collins R. D.; Hawiger J. Intracellular protein therapy with SOCS3 inhibits inflammation and apoptosis. Nat. Med. 2005, 11 (8), 892–8. 10.1038/nm1269. [DOI] [PubMed] [Google Scholar]
- Madonna S.; Scarponi C.; Morelli M.; Sestito R.; Scognamiglio P. L.; Marasco D.; Albanesi C. SOCS3 inhibits the pathological effects of IL-22 in non-melanoma skin tumor-derived keratinocytes. Oncotarget 2017, 8 (15), 24652–24667. 10.18632/oncotarget.15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doti N.; Scognamiglio P. L.; Madonna S.; Scarponi C.; Ruvo M.; Perretta G.; Albanesi C.; Marasco D. New mimetic peptides of the kinase-inhibitory region (KIR) of SOCS1 through focused peptide libraries. Biochem. J. 2012, 443 (1), 231–40. 10.1042/BJ20111647. [DOI] [PubMed] [Google Scholar]
- Madonna S.; Scarponi C.; Doti N.; Carbone T.; Cavani A.; Scognamiglio P. L.; Marasco D.; Albanesi C. Therapeutical potential of a peptide mimicking the SOCS1 kinase inhibitory region in skin immune responses. Eur. J. Immunol. 2013, 43 (7), 1883–95. 10.1002/eji.201343370. [DOI] [PubMed] [Google Scholar]
- La Manna S.; Lopez-Sanz L.; Leone M.; Brandi P.; Scognamiglio P. L.; Morelli G.; Novellino E.; Gomez-Guerrero C.; Marasco D. Structure-activity studies of peptidomimetics based on kinase-inhibitory region of suppressors of cytokine signaling 1. Biopolymers 2018, 110 (5), e23082 10.1002/bip.23082. [DOI] [PubMed] [Google Scholar]
- Viparelli F.; Cassese A.; Doti N.; Paturzo F.; Marasco D.; Dathan N. A.; Monti S. M.; Basile G.; Ungaro P.; Sabatella M.; Miele C.; Teperino R.; Consiglio E.; Pedone C.; Beguinot F.; Formisano P.; Ruvo M. Targeting of PED/PEA-15 molecular interaction with phospholipase D1 enhances insulin sensitivity in skeletal muscle cells. J. Biol. Chem. 2008, 283 (31), 21769–78. 10.1074/jbc.M803771200. [DOI] [PubMed] [Google Scholar]
- Kershaw N. J.; Murphy J. M.; Liau N. P.; Varghese L. N.; Laktyushin A.; Whitlock E. L.; Lucet I. S.; Nicola N. A.; Babon J. J. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 2013, 20 (4), 469–76. 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M.; Andre T.; Wanner R.; Roth H. M.; Duhr S.; Baaske P.; Breitsprecher D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. 10.1016/j.molstruc.2014.03.009. [DOI] [Google Scholar]
- La Manna S.; Roviello V.; Scognamiglio P. L.; Diaferia C.; Giannini C.; Sibillano T.; Morelli G.; Novellino E.; Marasco D. Amyloid fibers deriving from the aromatic core of C-terminal domain of nucleophosmin 1. Int. J. Biol. Macromol. 2019, 122, 517–525. 10.1016/j.ijbiomac.2018.10.210. [DOI] [PubMed] [Google Scholar]
- La Manna S.; Scognamiglio P. L.; Roviello V.; Borbone F.; Florio D.; Di Natale C.; Bigi A.; Cecchi C.; Cascella R.; Giannini C.; Sibillano T.; Novellino E.; Marasco D. The acute myeloid leukemia-associated Nucleophosmin 1 gene mutations dictate amyloidogenicity of the C-terminal domain. FEBS J. 2019, 286 (12), 2311–2328. 10.1111/febs.14815. [DOI] [PubMed] [Google Scholar]
- Andersson D.; Carlsson U.; Freskgard P. O. Contribution of tryptophan residues to the CD spectrum of the extracellular domain of human tissue factor: application in folding studies and prediction of secondary structure. Eur. J. Biochem. 2001, 268 (4), 1118–28. 10.1046/j.1432-1327.2001.01981.x. [DOI] [PubMed] [Google Scholar]
- Vincenzi M.; Mercurio F. A.; Leone M. About TFE: Old and New Findings. Curr. Protein Pept. Sci. 2019, 20 (5), 425–451. 10.2174/1389203720666190214152439. [DOI] [PubMed] [Google Scholar]
- Griesinger C.; Otting G.; Wuthrich K.; Ernst R. R. Clean TOCSY for proton spin system identification in macromolecules, 110, 7870–7872. J. Am. Chem. Soc. 1988, 110, 7870–7872. 10.1021/ja00231a044. [DOI] [Google Scholar]
- Bax A.; Davis D. G. Practical Aspects of Two-Dimensional Transverse Noe Spectroscopy. J. Magn. Reson. 1985, 63 (1), 207–213. 10.1016/0022-2364(85)90171-4. [DOI] [Google Scholar]
- Wuthrich K.NMR of Proteins and Nucleic Acids; Wiley: New YorK, 1986; p 320. [Google Scholar]
- Kumar A.; Ernst R. R.; Wuthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95 (1), 1–6. 10.1016/0006-291X(80)90695-6. [DOI] [PubMed] [Google Scholar]
- Koradi R.; Billeter M.; Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 1996, 14 (1), 51–5, 29–32. 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kershaw N. J.; Murphy J. M.; Liau N. P.; Varghese L. N.; Laktyushin A.; Whitlock E. L.; Lucet I. S.; Nicola N. A.; Babon J. J. SOCS3 binds specific receptor–JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 2013, 20 (4), 469. 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A.; Franceschini M.; Paiardini A.; Grottesi A.; Chiarella S.; Rocchio S.; Di Natale C.; Marasco D.; Vitagliano L.; Travaglini-Allocatelli C.; Federici L. Structural investigation of nucleophosmin interaction with the tumor suppressor Fbw7gamma. Oncogenesis 2017, 6 (9), e379 10.1038/oncsis.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H.; Niyongere S. A.; Lee S. J.; Baker B. J.; Benveniste E. N. Expression and functional significance of SOCS-1 and SOCS-3 in astrocytes. J. Immunol. 2008, 181 (5), 3167–76. 10.4049/jimmunol.181.5.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S.; Lowther S.; Stambas J. Inhibition of reactive oxygen species production ameliorates inflammation induced by influenza A viruses via upregulation of SOCS1 and SOCS3. J. Virol. 2015, 89 (5), 2672–83. 10.1128/JVI.03529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manna S.; Scognamiglio P. L.; Di Natale C.; Leone M.; Mercurio F. A.; Malfitano A. M.; Cianfarani F.; Madonna S.; Caravella S.; Albanesi C.; Novellino E.; Marasco D. Characterization of linear mimetic peptides of Interleukin-22 from dissection of protein interfaces. Biochimie 2017, 138, 106–115. 10.1016/j.biochi.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Nozaki Y.; Ri J.; Sakai K.; Niki K.; Kinoshita K.; Funauchi M.; Matsumura I., Inhibition of the IL-18 Receptor Signaling Pathway Ameliorates Disease in a Murine Model of Rheumatoid Arthritis. Cells 2020, 9 ( (1), ), 11. 10.3390/cells9010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M. Z.; Qin M. B.; Liang Z. H.; Tang G. D. Effect of SOCS3 on lung injury in rats with severe acute pancreatitis through regulating JAK2/STAT3 signaling pathway. European review for medical and pharmacological sciences 2019, 23 (22), 10123–10131. [DOI] [PubMed] [Google Scholar]
- Marin-Royo G.; Rodriguez C.; Le Pape A.; Jurado-Lopez R.; Luaces M.; Antequera A.; Martinez-Gonzalez J.; Souza-Neto F. V.; Nieto M. L.; Martinez-Martinez E.; Cachofeiro V. The role of mitochondrial oxidative stress in the metabolic alterations in diet-induced obesity in rats. FASEB J. 2019, 33 (11), 12060–12072. 10.1096/fj.201900347RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers L. O.; Johnson H. M.; Mujtaba M. G.; Ellis M. R.; Haider S. M.; Subramaniam P. S. Characterization of a peptide inhibitor of Janus kinase 2 that mimics suppressor of cytokine signaling 1 function. J. Immunol. 2004, 172 (12), 7510–8. 10.4049/jimmunol.172.12.7510. [DOI] [PubMed] [Google Scholar]
- Flowers L. O.; Subramaniam P. S.; Johnson H. M. A SOCS-1 peptide mimetic inhibits both constitutive and IL-6 induced activation of STAT3 in prostate cancer cells. Oncogene 2005, 24 (12), 2114–20. 10.1038/sj.onc.1208437. [DOI] [PubMed] [Google Scholar]
- He C.; Yu C. R.; Mattapallil M. J.; Sun L.; Larkin Iii J.; Egwuagu C. E. SOCS1 Mimetic Peptide Suppresses Chronic Intraocular Inflammatory Disease (Uveitis). Mediators Inflammation 2016, 2016, 2939370. 10.1155/2016/2939370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recio C.; Lazaro I.; Oguiza A.; Lopez-Sanz L.; Bernal S.; Blanco J.; Egido J.; Gomez-Guerrero C. Suppressor of Cytokine Signaling-1 Peptidomimetic Limits Progression of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2017, 28 (2), 575–585. 10.1681/ASN.2016020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recio C.; Oguiza A.; Lazaro I.; Mallavia B.; Egido J.; Gomez-Guerrero C. Suppressor of cytokine signaling 1-derived peptide inhibits Janus kinase/signal transducers and activators of transcription pathway and improves inflammation and atherosclerosis in diabetic mice. Arterioscler., Thromb., Vasc. Biol. 2014, 34 (9), 1953–60. 10.1161/ATVBAHA.114.304144. [DOI] [PubMed] [Google Scholar]
- Hernandez C.; Bogdanov P.; Gomez-Guerrero C.; Sampedro J.; Sola-Adell C.; Espejo C.; Garcia-Ramirez M.; Prieto I.; Egido J.; Simo R., SOCS1-Derived Peptide Administered by Eye Drops Prevents Retinal Neuroinflammation and Vascular Leakage in Experimental Diabetes. Int. J. Mol. Sci. 2019, 20 ( (15), ), 3615. 10.3390/ijms20153615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A.; Aiello C.; Grieco P.; Marasco D. Targeting ″Undruggable″ Proteins: Design of Synthetic Cyclopeptides. Curr. Med. Chem. 2016, 23 (8), 748–62. 10.2174/0929867323666160112122540. [DOI] [PubMed] [Google Scholar]
- Premnath P. N.; Craig S. N.; Liu S.; Anderson E. L.; Grigoroudis A. I.; Kontopidis G.; Perkins T. L.; Wyatt M. D.; Pittman D. L.; McInnes C. Iterative conversion of cyclin binding groove peptides into druglike CDK inhibitors with antitumor activity. J. Med. Chem. 2015, 58 (1), 433–42. 10.1021/jm5015023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.