Abstract
Background and Purpose
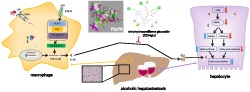
Regulating macrophage–hepatocyte crosstalk through P2X7 receptors has led to new pharmacological strategies to reverse alcoholic hepatosteatosis. We investigated how tetrahydroxystilbene glucoside (2354glu), isolated from Polygonum multiflorum, modulates macrophage–hepatocyte crosstalk during alcoholic hepatosteatosis.
Experimental Approach
A model of alcoholic hepatosteatosis was established by giving ethanol intragastrically to C57BL/6 mice. HepG2 cells were incubated in conditioned medium from LPS+ATP‐activated THP‐1 human macrophages with silenced or overexpressed P2X7 receptors. THP‐1 macrophages or mouse peritoneal macrophages were pretreated with 2354glu for 1 hr prior to LPS+ATP stimulation. Western blots, RT‐PCR and immunohistochemical analysis were used, along with over‐expression and silencing of P2X7 receptors.
Key Results
Knockdown or overexpression of P2X7 receptors in THP‐1 macrophages affected release of mature IL‐1β and, subsequently, modulated lipid metabolism in HepG2 cells via the LKB–AMPK pathway. 2354glu ameliorated alcoholic hepatosteatosis in mice by regulating LKB1–AMPK–SREBP1 pathway and its target genes. Suppression of P2X7 receptor activation by 2354glu inhibited IL‐1β release and reduced macrophage and neutrophil infiltration. In macrophages stimulated with LPS+ATP, expression of P2X7 receptors, caspase‐1 and NF‐κB, release of IL‐1β, calcium influx and PI uptake were reduced by 2354glu. SIRT1–LKB1–AMPK–SREBP1 axis‐mediated lipid accumulation in HepG2 cells was reduced when they were cultured with conditioned media from LPS+ATP‐activated THP‐1 macrophages pretreated with 2354glu.
Conclusion and Implications
Modulation of P2X7 receptors in macrophages regulated lipid accumulation in hepatocytes during alcoholic hepatosteatosis. 2354glu might be a promising candidate that targets P2X7 receptors in macrophages interacting with hepatocytes during alcoholic hepatosteatosis.
Abbreviations
- 2345glu
tetrahydroxystilbene glucoside, 2,3,5,4′‐tetrahydroxy‐stilbene‐2‐O‐β‐d‐glucoside
- AMPK
AMP‐activated protein kinase
- LKB1
liver kinase B1
- NLRP3
nucleotide‐binding oligomerization domain‐like receptor protein 3
- SIRT1
sirtuin 1
- SREBP1
sterol regulatory element‐binding protein 1
- TLRs
toll like receptors
What is already known
P2X7 receptor blockade exhibited notable therapeutic value in the treatment of alcoholic liver disease
What this study adds
The natural product, tetrahydroxystilbene glucoside (2354glu) ameliorated a mouse model of acute alcoholic hepatosteatosis.
This effect of 2345glu results from blockade of P2X7 receptors in macrophages crosstalking to hepatocytes.
What is the clinical significance
2354glu is a promising candidate for novel antagonists of P2X7 receptors in macrophages.
Targeting P2X7 receptors in macrophages interacting with hepatocytes provides new approaches to treating alcoholic hepatosteatosis
1. INTRODUCTION
Alcohol‐associated health issues, including alcoholic liver disease (ALD), have become a major threat to human health worldwide (World Health Organization, 2018). ALD is caused by excessive alcohol use or binge drinking over a prolonged period of time. The histological spectrum of ALD includes hepatosteatosis, steatohepatitis, hepatic fibrosis, cirrhosis, and, ultimately, hepatocellular carcinoma (Gao & Bataller, 2011). To date, prolonged alcohol abstinence is the most effective therapy to attenuate ALD, and this approach even significantly decreases mortality and morbidity. However, alcohol abstinence cannot reverse advanced ALD (Frazier, Stocker, Kershner, Marsano, & McClain, 2011; Seitz et al., 2018; Singal, Bataller, Ahn, Kamath, & Shah, 2018). Various treatments, including nutritional therapy, pharmacological therapy, psychotherapy, and surgery, have not been proven to improve the survival of ALD patients. For patients with end‐stage liver disease, the most effective therapeutic option is liver transplantation (European Association for the Study of the Liver, 2018; Lackner et al., 2017). Thus, the control of ALD at the hepatic steatosis stage, an early stage of ALD, could be of great significance in delaying the development of ALD. Although the pathogenesis of ALD is not yet fully elucidated, research has shown that the potential mechanisms of ALD are associated with inflammation, endotoxin and lipid metabolism imbalance (Rocco, Compare, Angrisani, Sanduzzi Zamparelli, & Nardone, 2014; Seitz et al., 2018).
Excessive alcohol consumption increases the levels of its metabolite acetaldehyde (the major product of alcohol metabolism), which activates the adaptive immune system and recruits lymphocytes to the damaged liver (Altamirano & Bataller, 2011). Furthermore, alcohol abuse not only results in changes in the colonic microbiota but also increases gut permeability to bacterial endotoxin, leading to elevated serum levels of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5019 (Altamirano & Bataller, 2011; Gao & Bataller, 2011). Gut‐derived LPS, as the first signal to up‐regulate inflammatory mediators, engages with https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1754 (TLR4) and activates the NF‐κB signalling pathway, leading to the accumulation of pro‐inflammatory cytokines, including https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4974 (Hritz et al., 2008). Sensitized hepatocytes respond to inflammatory cytokines leading to cellular damage (Nagy, Ding, Cresci, Saikia, & Shah, 2016). Additionally, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1713 is released from damaged hepatocytes in response to ethanol and acts as a danger signal in alcohol‐induced liver inflammation (Petrasek et al., 2012). The purinergic https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=484 receptor is an ATP‐gated ion channel that mediates activation of the https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1770) inflammasome in response to extracellular ATP. Upon activation, NLRP3 forms a complex with the inflammasome‐adaptor protein ASC and pro‐caspase‐1. Assembly of the NLRP3 inflammasome complex leads to the cleavage of pro‐caspase‐1 to its active form (cleaved https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1617), which subsequently causes the maturation of IL‐1β (Abderrazak et al., 2015). Previously, we reported that the inhibition of the P2X7 receptor–NLRP3 inflammasome axis could prevent extracellular matrix deposition and liver fibrosis (Jiang et al., 2017). Additionally, inhibition of IL‐1β secretion by macrophages using low MW compounds might be beneficial to the steatotic phenotype of hepatocytes (Li et al., 2018). However, whether and how P2X7 receptors affect the crosstalk between hepatocytes and macrophages during alcoholic hepatosteatosis is not known. Therefore, elucidating the role of P2X7 receptors in the interactions between macrophages and hepatocytes may provide new pharmacological strategies to reverse alcoholic hepatosteatosis.
Tetrahydroxystilbene glucoside (2,3,5,4′‐tetrahydroxy‐stilbene‐2‐O‐β‐d‐glucoside, 2354glu; Figure 4a; PubChem CID: 5321884), a resveratrol analogue of glucoside, is the main characteristic monomer of stilbene purified from the traditional Chinese herbal medicine prepared from Polygonum multiflorum (Lin et al., 2015; Wu et al., 2017). 2354glu is also a reference material used in the Chinese Pharmacopoeia to evaluate the quality of P. multiflorum (Wu et al., 2017). This glucoside exhibits a wide range of pharmacological effects, including anti‐aging (Cheung, Leung, Liu, & Che, 2014), anti‐oxidation (Wang, Zhao, Han, Chen, & Wang, 2008), and anticancer (Lin et al., 2016) effects, without toxic side effects. Although 2354glu is known to attenuate the non‐alcoholic fatty liver disease induced by a methionine and choline‐deficient diet (Li et al., 2018; Lou et al., 2017), whether and how 2354glu could regulate hepatic steatosis and inflammation induced by binge alcohol consumption had not been studied. Previously, we reported the potential reversal of alcoholic hepatosteatosis via regulation of P2X7 receptors (Li et al., 2018).
Figure 4.
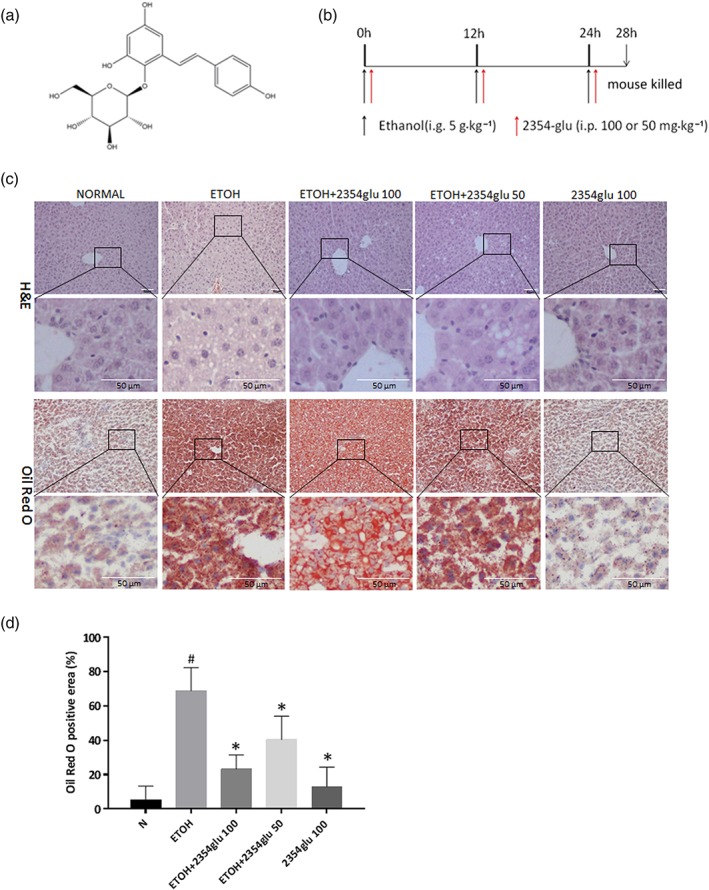
2354glu alleviates lipid accumulation in acute alcoholic hepatosteatosis. C57BL/6 mice were gavaged with ethanol (5 g·kg−1) every 12 hr for a total of three doses to induce acute hepatic steatosis. 2354glu (100 and 50 mg·kg−1) was intraperitoneally injected simultaneously with ethanol administration. (a) Chemical structure of 2354glu. (b) Animal experimental procedure. (c) HE staining, Oil red O staining (200× original magnification). (d) Positive area of Oil Red O staining was analysed with Image Pro‐Plus 6.0. All histograms represent the mean ± SD of six independent assays with at least three replicates. # P < .05, significantly different from normal group; *P < .05, significantly different from ethanol group
Our current research differs from the studies already cited, as our goal was (a) to assess the role of P2X7 receptors in macrophages when crosstalking with hepatocytes and (b) to find out if changes in the P2X7 receptor–NLRP3 signalling in macrophages, due to exposure to 2354glu, would regulate lipid accumulation in hepatocytes, in a model of alcoholic hepatosteatosis. Our results suggested that 2354glu might be a promising candidate targeting the P2X7 receptors in macrophages that interact with hepatocytes during alcoholic hepatosteatosis.
2. MATERIALS AND METHODS
2.1. Animals
All animal care and experimental procedures conformed to the criteria of the “Guide for the Care and Use of Laboratory Animals” published by the U.S. National Institutes of Health (National Research, 2011) and were approved by the Animal Research Committee of Yanbian University with the approval number 20170303.
Male C57BL/6 mice (8–10 weeks old, 20–22 g) were purchased from Changchun Yisi Laboratory Animal Technology Co., Ltd. [(SPF, SCXK [JI] 2018‐0007), Jilin, China]. The mice were housed in an animal facility at Yanbian University and were fed a standard chow diet and given tap water ad libitum. All mice were maintained at a temperature of 23 ± 2°C, with a relative humidity of 55 ± 5% and 12/12‐hr light/dark cycles. The animals were treated humanely, and all efforts were made to minimize the animal suffering and numbers of experimental animals. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2011; McGrath & Lilley, 2015) and with the recommendations made by the British Journal of Pharmacology.
After 1 week of acclimatization to the environment, the mice were randomly divided into the following five groups (six mice per group): the normal group, ethanol group, ethanol plus 2354glu groups (50 or 100 mg·kg−1 body weight), and 2354glu alone group (100 mg·kg−1 body weight). The murine model of acute alcoholic hepatosteatosis was prepared as follows. Mice were given 50% (v/v) ethanol three times at a dose of 5 g·kg−1 (body weight) every 12 hr by gastric gavage, except for the normal group and the 2354glu alone group. The animal protocol was previously proved to establish a significant acute hepatosteatosis according to previously published protocols from our previous work and others (Hamarneh et al., 2017; Li et al., 2018; Lian et al., 2010). The mice in the normal group and the 2354glu alone group were intragastrically treated with isocaloric maltose dextran (Figure 4b). Four hours after the last dose, all mice were killed by isoflurane inhalation. The blood was collected by direct cardiac puncture, and the whole liver was collected and stored at −80°C for subsequent use in the assays, as indicated.
2.2. Cell culture
The human monocyte cell line, THP‐1 (RRID:CVCL_0006), was a generous gift from Prof. Xuejun Jin (Molecular Medicine Research Center, College of Pharmacy, Yanbian University, Yanji, China) and was cultured in an RPMI‐1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified, 5% CO2/95% air incubator, at 37°C. Phorbol 12‐myristate 13‐acetate (PMA)‐differentiated THP‐1 macrophages were utilized for the subsequent studies. The human hepatoma cell line, HepG2 (RRID:CVCL_0027), was a generous gift from Professor Jung Joon Lee (Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea). HepG2 cells were cultured in DMEM supplemented with 10% FBS, 100 units·ml−1 penicillin G, and 100 mg·ml−1 streptomycin at 37°C in 5% CO2. The cells were passaged by trypsinization every 2 or 3 days, and the experiments were performed using cells from the fourth to the seventh passages.
2.3. Isolation and culture of murine peritoneal macrophages
Murine peritoneal macrophages were isolated from 8‐ to 10‐week‐old male C57BL/6 mice (20–22 g) according to the regulations of Animal Research Committee of Yanbian University (project number 20170303). Mice were anaesthetized with isoflurane, followed by an immediate cervical dislocation. Murine peritoneal macrophages were isolated as previously described (Tilija Pun & Park, 2018). Briefly, cells were elicited by injection of 1 ml of 3.8% sterile thioglycollate medium (BD Biosciences, San Jose, CA, USA) into the peritoneal cavity. After 4 days, the peritoneal cavity was flushed with 5 ml of sterile ice‐cold PBS containing 3% FBS, and the peritoneal fluid was dispensed into 50‐ml conical centrifuge tubes. After centrifugation, the peritoneal exudate cells were resuspended in DMEM with 10% FBS and 1% penicillin G/streptomycin and were incubated at 37°C and 5% CO2. Purified, adherent macrophages were obtained by washing three times with PBS to remove the unattached cells. The cells were treated with LPS for 4 hr and were subsequently stimulated for 30 min with 3‐mM ATP.
2.4. Transfection of siRNA for P2X7 receptors in vitro
THP‐1 cells were transfected with scrambled control siRNA (2 μM) or P2X7 receptor‐siRNA (2 μM) using the HiPerfect transfection reagent (Qiagen, Valencia, CA, USA) according to the manufacturer's instruction. The THP‐1 cells were treated with 100‐nM PMA for 24 hr before transfection. The sequences for the human P2X7 receptor‐siRNA were as follows: sense, 5′‐AGAGCAAAGUGACCUGGUU‐3′; antisense, 5′‐AACCAGGUCACUUUGCUCU‐3′.
2.5. Construction of P2X7 receptor plasmids
THP‐1 cells were transfected with the negative control plasmid (cytomegalovirus, CMV) or the P2X7 receptor overexpression plasmid (CMV‐P2X7R, GeneChem, Shanghai, China) using the Lipofectamine™ 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). Expression of P2X7 receptors was evaluated using western blotting and RT‐PCR. The plasmids were effective and suitable for the study.
2.6. Western blotting
The antibody‐based procedures used comply with the recommendations made by the British Journal of Pharmacology (Alexander et al., 2018). The total proteins of cells and liver samples were extracted using lysis buffer (Abcam, Cambridge, MA, USA) supplemented with protease and phosphatase inhibitors (Roche Diagnostics, Mannheim, Germany). Equal quantities of protein (20 μg) were loaded onto an 8–12% gel, separated by SDS‐PAGE, and transferred to PVDF membranes (Amersham Hybond, GE Healthcare Bio‐Sciences, Pittsburgh, PA, USA). Next, the membranes were blocked with 5% skim milk in PBS containing 0.05% Tween 20 and were incubated at 4°C overnight with the primary antibodies (listed in Table S2). Thereafter, the membranes were incubated with the appropriate HRP‐conjugated secondary antibodies (listed in Table S2), and the signals were visualized using the Amersham ECL Prime Western Blotting Detection Reagent (RPN2232, GE Healthcare, UK). The intensity of the protein bands was quantified using Quantity One software (RRID:SCR_014280, Bio‐Rad, CA, USA).
2.7. Real‐time PCR and RT‐PCR
Total RNA was isolated from the liver or cells using the SV Total RNA Isolation System (LS1040, Promega, Madison, WI, USA) according to the manufacturer's instructions. Total RNA was reverse transcribed into cDNA using 1 μg of Oligo (dT)15 Primer (Promega), 20 U of RNasin Ribonuclease Inhibitors (Promega), and 10 U of AMV reverse transcriptase (Promega) according to the manufacturers' instructions. The relative mRNA expression levels were measured using the real‐time PCR with Power SYBR® Green PCR Master Mix (Life Technologies, Carlsbad, CA, USA) on an Agilent Mx3000P QPCR System, and the relative fold difference was quantified using the comparative threshold cycle (ΔΔCt) method. GAPDH was used as a housekeeper gene for normalization. The mouse‐specific primer sequences are listed in Table S1. RT‐PCR was performed as previously described (Jin et al., 2014) using primers specific for human P2x7r and Gapdh. The specific primer sequences are listed in Table S1. The PCR products were subjected to electrophoresis and were stained with ethidium bromide. GAPDH was used as the internal standard calibrator.
2.8. ELISA
The concentrations of human or murine IL‐1β were measured using the appropriate species‐specific ELISA kits (BD Biosciences or R&D Systems, Minneapolis, MN, USA, respectively) according to the manufacturers' instructions.
2.9. Liver histological analysis
For haematoxylin and eosin (H&E) staining, the liver sections were fixed in 10% neutral buffered formalin, dehydrated in descending grades of ethanol, and embedded in paraffin. Sections (5 μm) of the liver were deparaffinized and stained with haematoxylin and eosin in sequence. For Oil Red O staining, liver cryosections (5 μm) were stained with Oil Red O and were counterstained with haematoxylin. The stained slides were visualized with a Nikon TI‐E fluorescence microscope (Nikon, Tokyo, Japan). All analysis was carried out in a blinded manner.
2.10. Immunohistochemistry
The liver sections were immunostained with a primary antibody (listed in Table S3) as previously described (Li et al., 2018). Briefly, the sections were deparaffinized, rehydrated, and heated in a microwave oven in citrate buffer for antigen retrieval. Next, the slides were successively incubated in 3% hydrogen peroxide and 5% normal goat serum, Avidin/Biotin Blocking solution (Vector Laboratories, Inc., Burlingame, CA, USA), primary antibody, which was incubated overnight at 4°C, and then with the specific secondary antibody (listed in Table S3). The bound antibodies were visualized with 3,3‐diamino benzidine (DAB; Thermo Fisher Scientific, Fremont, CA, USA). All the visual inspections were performed by a light microscopy (Nikon TI‐E). All the analyses were carried out in a blinded manner. The immunohistochemical procedures in our study also conform to the BJP guidelines (Alexander et al., 2018).
2.11. Single/double immunofluorescence staining
The cells were fixed with 4% paraformaldehyde solution and were incubated in 0.5% Triton X‐100 (v/v) in PBS for 20 min. Five‐micrometre cryostat sections were fixed in a 1:1 acetone–methanol mixture. Next, frozen sections or cells were incubated in a blocking solution with 5% goat serum, followed by incubation with primary antibodies (listed in Table S4) overnight at 4°C and then with appropriate secondary antibodies (listed in Table S4). The stained slides were mounted with Fluoroshield Mounting Medium with DAPI (Abcam). Fluorescence was visualized using a Nikon TI‐E fluorescence microscope. The immunofluorescence intensity was analysed using Image Pro‐Plus 6.0 (RRID:SCR_016879, Media Cybernetics, Inc., MD, USA). All the analyses were carried out in a blinded manner.
2.12. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). All studies were designed to generate groups of equal size, randomized to condition, and blinded to analysis. Group size is the number of independent values, and statistical analysis was undertaken only for studies with independent values of n ≥ 5. To reduce unwanted sources of variation, we normalized experimental data values to control group values. The normalized data used fold‐matched control values and not fold mean of control values. The data are presented as the means ± SD. All the data were analysed by Kolmogorov–Smirnov normality test and followed a Gaussian distribution. The statistical significance of the differences was determined by one‐way ANOVA, followed by Turkey's multiple comparison tests. Post hoc tests were run only if F achieved P < .05, and there was no significant variance inhomogeneity. P < .05 was deemed to indicate statistical significance. Statistical analysis was performed using GraphPad Prism 6.0 (RRID:SCR_002798, GraphPad Software, Inc., San Diego, CA, USA). No data were excluded from any study.
2.13. Materials
2354glu (tetrahydroxystilbene glucoside) was obtained from Push Bio‐Technology (>99% purity; Chengdu, China) and was dissolved in saline. Ultrapure LPS from Escherichia coli O111:B4 was purchased from InvivoGen (San Diego, CA, USA), and ATP disodium salt was purchased from Sigma Chemical Co. (St. Louis, MO, USA). The P2X7 receptor‐selective antagonist https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4118 (PubChem CID: 11673921) was purchased from Abcam. The TLR4 signalling inhibitor, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=9036 (TAK‐242, PubChem CID: 11703255), was purchased from InvivoGen. HRP‐conjugated goat anti‐mouse, goat anti‐rabbit, and donkey anti‐goat antibodies were purchased from Abcam. The BCA Protein Assay Kit was obtained from Beyotime (Nantong, Jiangsu, China).
2.14. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org/, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Cidlowski et al., 2019; Alexander, Fabbro et al., 2019; Alexander, Mathie et al., 2019).
3. RESULTS
3.1. P2X7 receptors in macrophages are necessary to regulate lipid accumulation in hepatocytes, through cell–cell crosstalk
We previously reported that ethanol bingeing in mice induced increased circulating ATP levels (Zhang et al., 2018), a finding that was consistent with previous studies (Petrasek et al., 2012). Here, we treated HepG2 cells with ethanol and then measured the extracellular and intracellular ATP levels. As shown in Figure S1, after ethanol exposure, the extracellular ATP level was greatly elevated in a concentration‐dependent manner, while the intracellular ATP level was not affected by 50‐mM ethanol but did increase when HepG2 cells were exposed to higher concentrations of ethanol (100 and 200 mM).
To evaluate the regulatory function of P2X7 receptors on the release of IL‐1β from activated macrophages, THP‐1 macrophages were transfected with specific siRNA to silence P2X7 receptors or with a CMV plasmid to overexpress P2X7 receptors (Figure 1a,f); the cells were then stimulated with LPS plus ATP (Figure 1c,h). As shown in Figure 1a–e, knockdown of P2X7 receptors markedly decreased the intracellular levels of pro‐IL‐1β and the extracellular levels of mature IL‐1β. After P2X7 receptor overexpression, a significant increase in IL‐1β release from THP‐1 cells was observed (Figure 1f–j). These results demonstrated that the genetic alteration of P2X7 receptors influenced the release of IL‐1β, indicating that these receptors were essential to trigger IL‐1β maturation and secretion.
Figure 1.
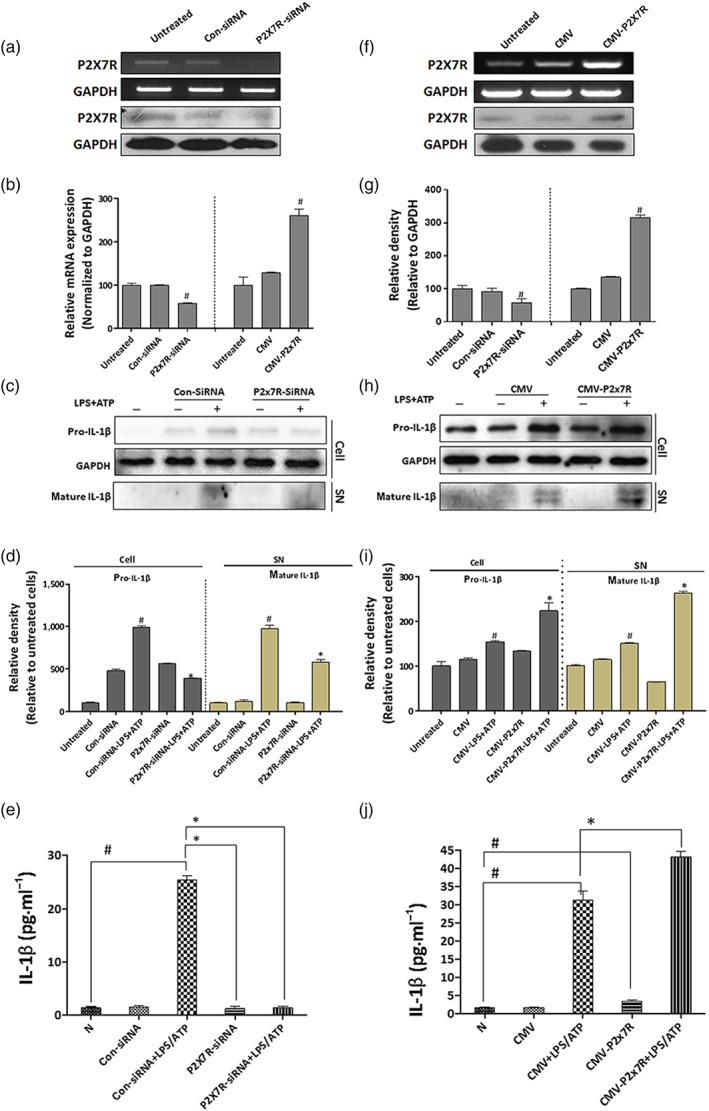
P2X7 receptors (P2X7R) are essential in triggering IL‐1β release from LPS+ATP‐activated macrophages. PMA‐differentiated THP‐1 macrophages were transfected with specific siRNA to silence P2X7 receptors or CMV plasmid to overexpress P2X7 receptors, and subsequently treated with LPS (1 μg·ml−1) for 4 hr, followed by 30 min of ATP (3 mM) stimulation. (a, f) Expression of P2X7 receptors in THP‐1 macrophages was confirmed by RT‐PCR analysis and western blot analysis. (c, h) Intracellular pro‐IL‐1β and extracellular mature IL‐1β were detected by western blotting. (b, g, d, i) Each immunoreactive band from RT‐PCR or western blot was normalized against GAPDH and relative to normal to control for unwanted sources of variation. (e, j) Protein expression of IL‐1β was determined by ELISA assay. The data are expressed as the mean ± SD of five independent assays with at least three replicates. # P < .05, significantly different from untreated THP‐1 macrophages; *P < .05, significantly different from THP‐1 macrophages transfected with siRNA or CMV plasmid specific for P2X7 receptors and then treated with LPS+ATP
Previously, we reported that IL‐1β from activated macrophages accelerates lipid accumulation in hepatocytes (Li et al., 2018). To further assess the essential role of P2X7 receptors in the effect of macrophages on hepatocyte lipid accumulation, HepG2 cells were cultured in a conditioned medium from THP‐1 macrophages with knocked down or overexpressed P2X7 receptors that had been treated with LPS plus ATP. The conditioned medium from P2X7 receptor‐deficient THP‐1 macrophages treated with LPS+ATP increased the protein levels of total and phosphorylated LKB1 and AMPKα as shown by western blotting (Figure 2a) and decreased the intensity of the sterol regulatory element‐binding protein 1 (SREBP1) in cytoplasm and nucleus of HepG2 detected by immunofluorescence staining (Figure 3a). Furthermore, when HepG2 cells were cultured with the conditioned medium from P2X7 receptor‐overexpressing THP‐1 macrophages stimulated by LPS+ATP, the levels of total and phosphorylated LKB1 and AMPKα were reduced, and the intensity of SREBP1 in cytoplasm and nucleus of HepG2 was greatly increased (Figure 2c,b). Our results indicated that macrophage‐derived IL‐1β might interfere with the AMPKα and LKB1 signalling in hepatocytes by affecting NLRP3‐mediated IL‐1β release from macrophages.
Figure 2.
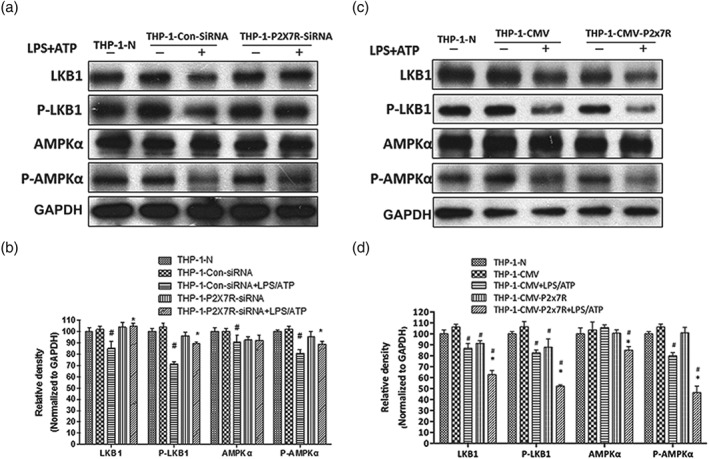
P2X7 receptor‐dependent IL‐1β released from activated macrophages accelerates lipid accumulation via LKB1–AMPK in hepatocytes. THP‐1 macrophages were transfected with specific siRNA to silence P2X7 receptors (P2X7R) or CMV plasmid to overexpress P2X7 receptors, followed by LPS+ATP stimulation. Then HepG2 cells were incubated with different conditioned medium from above THP‐1 macrophages. (a, c) Total and phosphorylated AMPKα and LKB1 protein levels in HepG2 cells were determined using western blot analysis. (b, d) Each immunoreactive band was normalized against GAPDH and relative to normal to control for unwanted sources of variation. The data are expressed as the mean ± SD of five independent assays with at least three replicates. # P < .05, significantly different from HepG2 cells cultured with medium from untreated THP‐1 macrophages; *P < .05, significantly different from HepG2 cells cultured with medium from THP‐1 macrophages transfected with siRNA or CMV plasmid specific for P2X7 receptors and then treated with LPS+ATP
Figure 3.
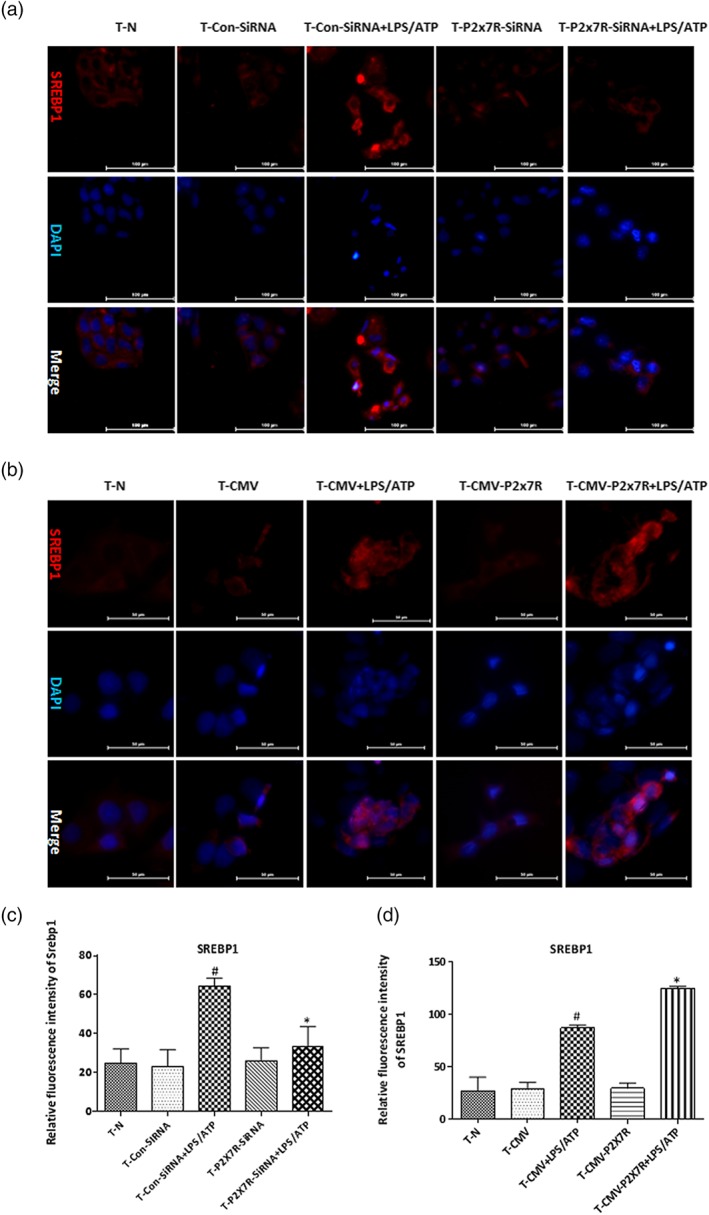
P2X7 receptors located on macrophages influence lipid accumulation of hepatocytes through regulating SREBP1. HepG2 cells were incubated with conditioned medium from LPS+ATP‐stimulated THP‐1 macrophages, followed by transfection with specific siRNA to silence P2X7 receptors (P2X7R) or CMV plasmid to overexpress P2X7 receptors. (a, b) Immunofluorescence staining for SREBP1. Images were taken at 200× magnification. SREBP1 staining (red) and nuclei with DAPI (blue) are shown. Representative images were shown. (c, d) Relative fluorescence intensity of SREBP1 was analysed with Image Pro‐Plus 6.0. The data are expressed as the mean ± SD of five independent assays with at least three replicates. # P < .05, significantly different from HepG2 cells cultured with medium from untreated THP‐1 macrophages; *P < .05, significantly different from HepG2 cells cultured with medium from THP‐1 macrophages transfected with siRNA or CMV plasmid specific for P2X7 receptors and then treated with LPS+ATP
3.2. 2354glu alleviates lipid accumulation in acute alcoholic hepatosteatosis
After exposure to three doses of alcohol and treatment with various concentrations of 2354glu, staining with HE and Oil Red O showed obvious lipid droplets in the livers of ethanol‐treated mice, while 2354glu effectively relieved the lipid accumulation caused by ethanol exposure. Treatment with 2354glu alone exhibited no effect on liver function (Figure 4c).
The regulatory effects of 2354glu treatment on the levels of LKB1 and AMPK in the liver were investigated by western blot analysis. Ethanol feeding significantly suppressed the protein level of total and phosphorylated LKB1 and AMPK, while 2354glu restored the activation of AMPK–LKB1 (Figure 5a,c). Additionally, the ratio of phosphorylated to total protein of AMPK and LKB1 was decreased after ethanol exposure, while 2354 administration enhanced the ratio of phosphorylated to total protein of AMPK and LKB1 (Figure 5b). Activated SREBP1 is involved in lipid synthesis, while activated PPAR can promote the oxidation of lipid acids, both of which can regulate lipid metabolism in the liver. The expression of SREBP1 was increased both in the cytoplasm and nucleus in the livers of mice after acute alcohol exposure, and the increase in SREBP1 was inhibited by 2354glu administration (Figure 5d). Moreover, acute alcohol intake up‐regulated the mRNA expression of SREBP1 target genes (Fasn and Acly) and down‐regulated the expression of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=593 target genes (Cpt2 and Abcd2), while 2354glu modulated lipid metabolism‐related changes induced by alcohol (Figure 5e). These data suggest that 2354glu might prevent ethanol‐induced acute hepatic steatosis through the inhibition of lipogenesis and the elevation of lipid oxidation.
Figure 5.
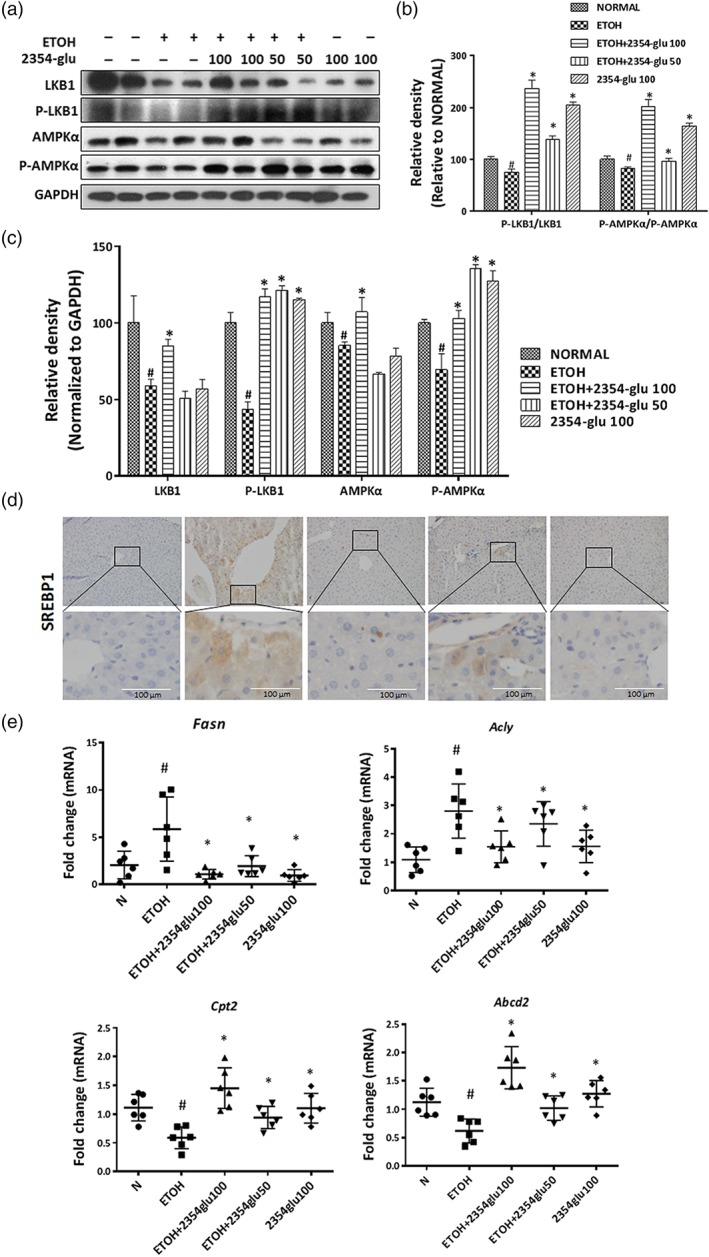
2354glu regulates LKB1–AMPK–SREBP1 in acute alcoholic hepatosteatosis. (a) Total and phosphorylated AMPKα and LKB1 protein levels in mouse livers were determined using western blot analysis. (b) Each immunoreactive band of phosphorylated and total protein of AMPK or LKB1 was normalized against GAPDH. Then the ratio of phosphorylated to total protein of AMPK or LKB1 was calculated. (c) Each immunoreactive band was normalized against GAPDH and relative to normal to control for unwanted sources of variation. (d) Immunohistochemistry for SREBP1 (200× original magnification). (e) Relative mRNA expression of SREBP1 targeting genes‐Fasn, Acly, and PPARα targeting genes Cpt2 and Abcd2. All histograms represent the mean ± SD of six independent assays with at least three replicates. # P < .05, significantly different from normal group; *P < .05, significantly different from ethanol group
3.3. 2354glu suppresses NLRP3 inflammasome activation via inhibition of P2X7 receptors in ethanol‐induced acute hepatic steatosis
Alcohol consumption induced activation of P2X7 receptors and NLRP3 inflammasomes in both hepatocytes and macrophages in mouse liver, as shown by double immunohistochemistry staining (Figure 6a,b). Consistent with the immunohistochemistry staining data, the expression of P2X7 receptor protein was increased by alcohol intake (Figure 7a). However, 2354glu administration blocked IL‐1β production and reduced NLRP3 and P2X7 receptor expression compared with the group treated with ethanol only.
Figure 6.
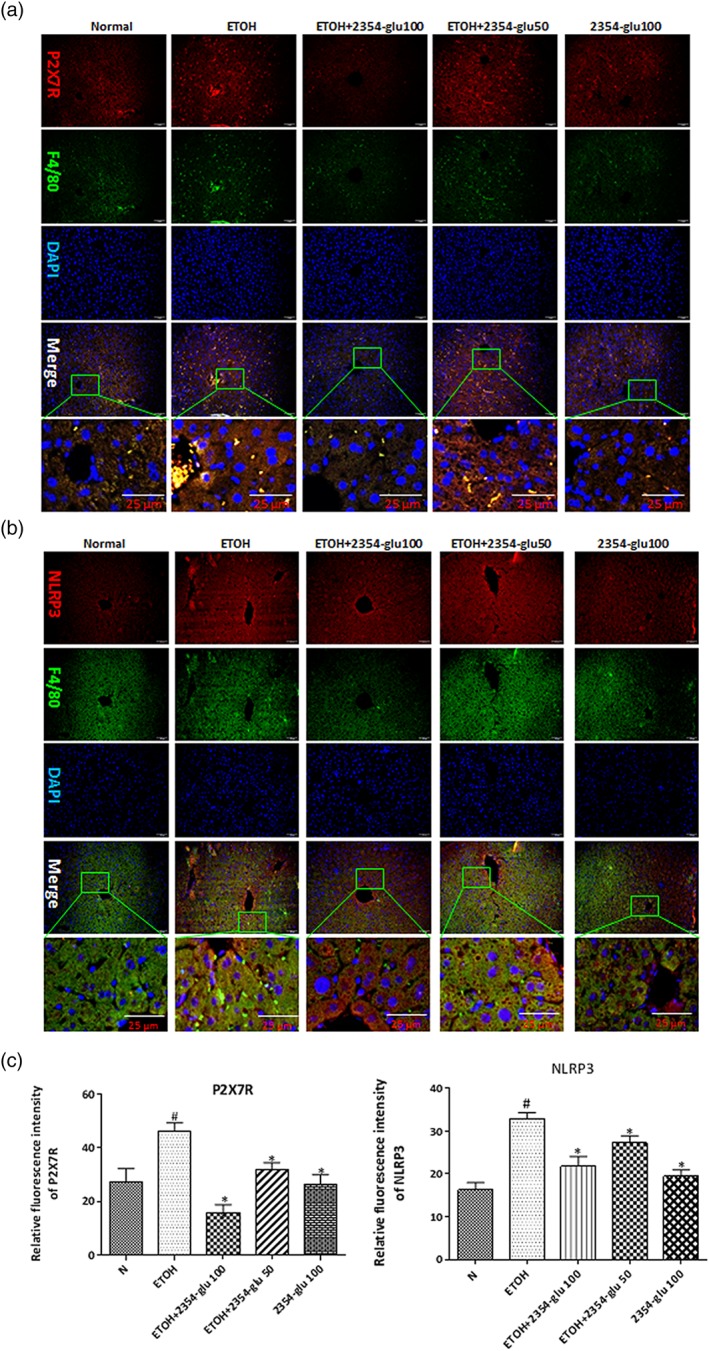
2354glu suppresses NLRP3 inflammasome activation via blockade of P2X7 receptors in acute alcoholic hepatic steatosis. Expression of P2X7 receptors (P2X7R; a, red) and NLRP3 (b, red) in mouse livers was evaluated by dual immunofluorescence staining combined with F4/80 (green, 200× original magnification). (c) Relative fluorescence intensity of P2X7 receptors and NLRP3 was analysed with Image Pro‐Plus 6.0. All histograms represent the mean ± SD of six independent assays with at least three replicates. # P < .05, significantly different from normal group; *P < .05, significantly different from ethanol group
Figure 7.
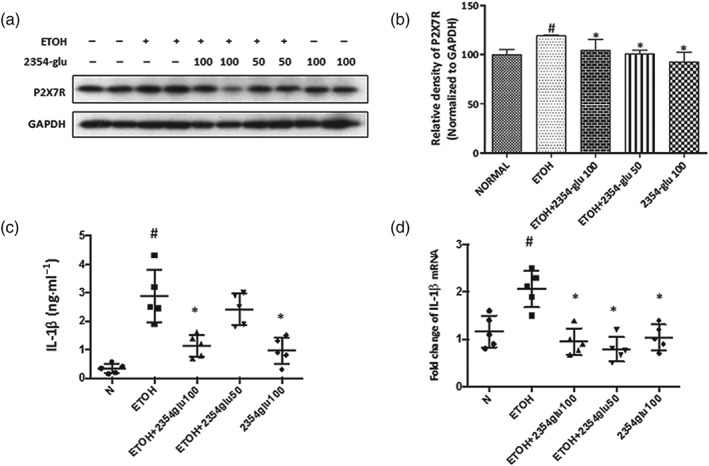
2354glu suppresses IL‐1β released via blockade of P2X7 receptors in acute alcoholic hepatic steatosis. (a) Protein expression of P2X7 receptors (P2X7R) was determined by western blotting. (b) Each immunoreactive band was normalized against GAPDH and relative to normal to control for unwanted sources of variation. IL‐1β protein levels in serum (c) and IL‐1β mRNA expression in the liver (d) were determined by ELISA assay or real‐time PCR. All histograms represent the mean ± SD of six independent assays with at least three replicates. # P < .05, significantly different from normal group; *P < .05, significantly different from ethanol group
To further assess the effects of 2354glu on release of inflammatory cytokines in the liver and serum of ethanol‐treated acute steatotic mice, we used real‐time PCR and ELISA to examine the mRNA and protein expression levels of IL‐1β respectively. We found that the mRNA level of IL‐1β in the liver and IL‐1β released into the serum was greatly increased in ethanol‐treated mice, compared with the levels in normal mice, and was decreased to the normal level after 2354glu administration (Figure 7d,c).
3.4. 2354glu blocks P2X7 receptor‐dependent IL‐1β cleavage in macrophages
As shown in panels a and b of Figures 6 and 7, 2354glu could inhibit alcohol‐induced P2X7 receptor–NLRP3 inflammasome activation in hepatocytes and macrophages, while 2354glu pretreatment markedly reduced the number of macrophages, suggesting that the observed immune responses in ethanol‐fed mice were likely to be mediated by macrophage recruitment. We therefore used thioglycollate‐elicited peritoneal macrophages (Figure 8) and naïve‐resident peritoneal macrophages (Figure S3) to investigate whether the action of 2354glu against alcoholic hepatosteatosis involved activation of P2X7 receptors and NLRP3 inflammasomes in macrophages. First, we evaluated the viability of murine peritoneal macrophages and the cytotoxicity of different concentrations of 2354glu using MTT and LDH activity assays. 2354glu did not affect the viability of murine peritoneal macrophages and exhibited little cellular cytotoxicity (Figure S2A, C). As shown in Figures 8a and S3C, when thioglycollate‐elicited peritoneal macrophages and naïve‐resident peritoneal macrophages were treated with LPS for 4 hr and were subsequently stimulated for 30 min with 3‐mM ATP, the expression of P2X7 receptor protein was greatly increased. As shown in Figure S3A, with LPS plus ATP stimulation, the two types of mouse peritoneal macrophages produced very large amounts of pro‐IL‐1β and secreted a large amount of mature IL‐1β into culture media, a finding consistent with the in vivo results (Figure 7c,d). LPS plus ATP stimulation also increased caspase‐1 cleavage compared with that in two types of mouse peritoneal macrophages when left untreated (Figures 8a and S3C). When the macrophages were directly treated with ethanol, no IL‐1β release was induced (Figure S3A), a finding that agreed with our previous finding in THP‐1 macrophages (Li et al., 2018). A P2X7 receptor‐selective antagonist, A438079, and a TLR4 antagonist, CLI‐095, were added to murine peritoneal macrophages to block activation of P2X7 receptors and TLR4 respectively. CLI‐095, but not A438079, decreased the level of pro‐IL‐1β, while both CLI‐095 and A438079 decreased the levels of mature IL‐1β, cleaved caspase‐1, and P2X7 receptors in the extracellular and intracellular fractions. 2354glu also significantly reduced LPS+/ATP‐induced IL‐1β cleavage and mature IL‐1β release in a concentration‐dependent manner but only slightly altered the pro‐IL‐1β levels (Figures 8a,e and S3C). Furthermore, 2354glu decreased pro‐caspase‐1, cleaved caspase‐1 and P2X7 receptors, suggesting that 2354glu might be a specific inhibitor of these receptors. To confirm a specific inhibitory effect on P2X7 receptors by 2354glu, we determined the intracellular calcium concentration [Ca2+]i in LPS+ATP‐stimulated mouse peritoneal macrophages. LPS+ATP stimulation increased the intracellular calcium concentration, while 2354glu and A438079 suppressed the [Ca2+]i in macrophages, in a concentration‐dependent manner (Figure S4A, C). The IC50 of 2354glu for [Ca2+]i was 148 μM. ATP is known to elicit membrane pore dilatation by activating P2X7 receptors followed by cytolysis to trigger IL‐1β maturation (Faria, Reis, Ferreira, Cezar‐de‐Mello, & Moraes, 2016; Takenouchi et al., 2011; Wewers, 2004). Thus, we examined whether 2354glu prevented P2X7 receptor‐mediated pore formation in THP‐1 macrophages using PI uptake. The fluorescence images confirmed that the P2X7 receptor pore was activated by LPS+ATP, while it was blocked by 2354glu, A438079, and CLI‐095 in THP‐1 macrophages (Figure S4B, D). The positive control group was treated with Triton X‐100 (0.1%), a widely used non‐ionic surfactant to permeabilize the living cell membrane (Koley & Bard, 2010). These results provided further evidence to support the finding that 2354glu blocked the activation of P2X7 receptors.
Figure 8.
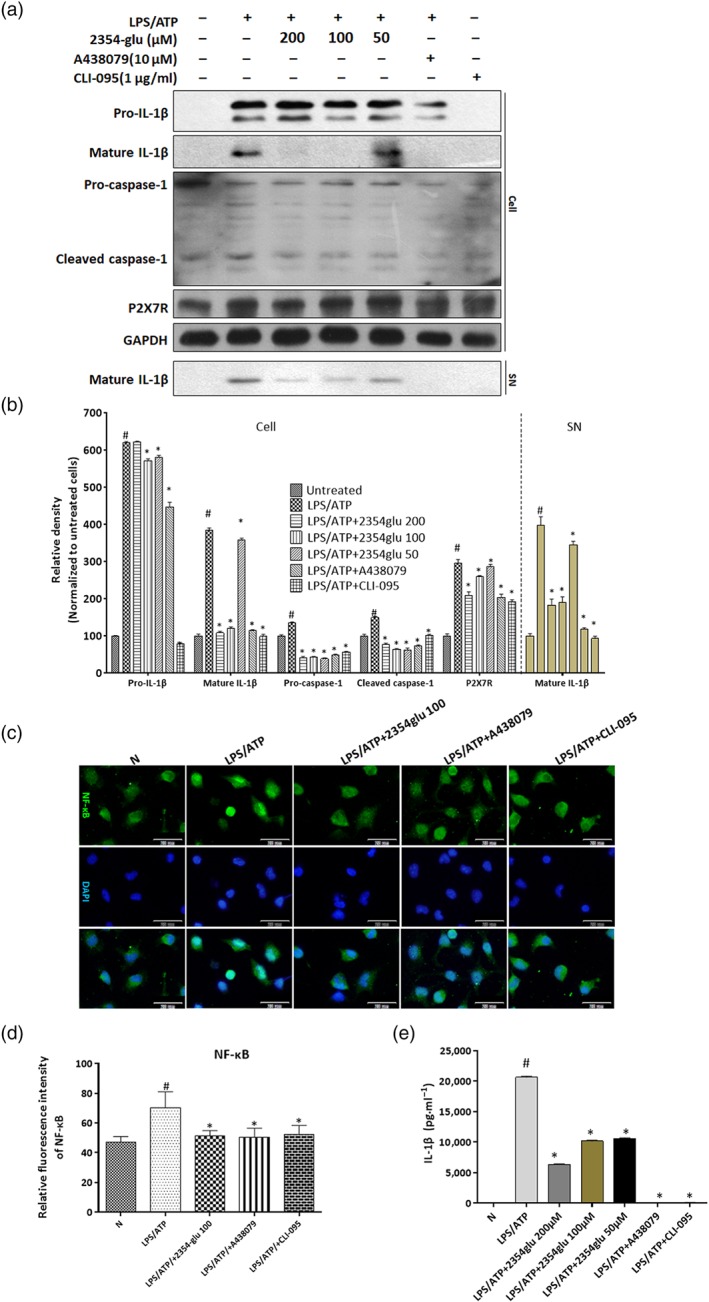
2354glu blocks P2X7 receptor‐dependent IL‐1β cleavage in murine peritoneal macrophages. Murine peritoneal macrophages were treated with the indicated concentrations of 2354glu (200, 100, and 50 μM), A438079 (10 μM), or CLI‐095 (1 μg·ml−1) for 1 hr and subsequently stimulated with LPS (1 μg·ml−1) for another 4 hr followed by 30 min of ATP (3 mM) stimulation. (a) Protein expression of P2X7 receptors (P2X7R) and caspase‐1, and extracellular and intracellular pro‐IL‐1β and mature IL‐1β in murine peritoneal macrophages was verified by western blot analysis. GAPDH was used as the loading control. (b) Each immunoreactive band was digitized and expressed as a ratio of untreated cells. (c) Immunofluorescence staining for NF‐κB. Images were taken at 200× magnification. NF‐κB staining (green) and nuclei with DAPI (blue) are shown. Representative images were shown. (d) Relative fluorescence intensity of NF‐κB was analysed with Image Pro‐Plus 6.0. (e) Protein expression of extracellular IL‐1β in murine peritoneal macrophage was determined by ELISA. All histograms represent the mean ± SD of five independent assays with at least three replicates. # P < .05, significantly different from untreated cells; *P < .05, significantly different from cells stimulated with LPS + ATP
To explore the direct interaction of 2354glu or the selective P2X7 receptor inhibitor, A438079, with P2X7 receptors, we investigated the binding modes using a molecular docking study (Gonzaga et al., 2017; Homerin et al., 2019) (Figure S5). We found that all parts of 2354glu were in the P2X7 receptor binding pocket consisting of Lys174, Ile109, Thr111, Glu171, Gln143, Lys145, Lys311, and Val173, while part of A438079 was exposed outside the P2X7 receptor binding pocket. The oxygen atom of the phenolic alcohol in 2354glu formed a hydrogen bond with the hydrogen atom of the hydroxyl group on the side chain of Thr111. The hydrogen atom of the primary alcohol at the 6‐position of the glycoside in 2354glu formed a hydrogen bond with the carbonyl atom of α‐amino acid of Glu171. The oxygen atom of secondary alcohols at the 4‐ and 5‐position of glycoside of 2354glu formed a hydrogen bond with the hydrogen atom of the amine on the side chain of Lys145. The amine group at the 4‐position of the tetrazole moiety in A438079 formed a hydrogen bond with the hydrogen atom of amino group on the side chain of Lys145. According to the above analysis results, 2354glu appeared to be more favourably accommodated in the binding pocket of P2X7 receptors than A438079, a well‐known selective P2X7 receptor antagonist (Figure S5B, D).
As shown in Figure S6, P2X7 receptor knockdown in PMA‐differentiated THP‐1 macrophages successfully abolished mature IL‐1β release. Although pretreatment with 2354glu decreased the level of mature IL‐1β release in LPS + ATP‐stimulated THP‐1 macrophages, 2354glu did not further regulate the release of mature IL‐1β in P2X7 receptor‐silenced THP‐1 macrophages. These results suggested that P2X7 receptors might be a major target of 2354glu in macrophages. Taken together, our research indicated that 2354glu might be a potential P2X7 receptor antagonist.
As shown in the immunohistochemistry staining data (Figure 8c), after LPS+ATP stimulation of murine peritoneal macrophages, a substantial amount of NF‐κB was translocated to the nucleus, while pretreatment with 2354glu, A438079, and CLI‐095 significantly down‐regulated the level of NF‐κB in the nuclear fraction, indicating that 2354glu could regulate the inflammatory response induced by LPS + ATP through the NF‐κB signalling pathway in macrophages.
3.5. 2354glu ameliorates lipid accumulation in hepatocytes induced by IL‐1β released from activated macrophages
Based on the above results, we wondered whether the inhibition by 2354glu of the release of mature IL‐1β from macrophages could prevent lipid accumulation in hepatocytes. By itself, 2354glu was not cytotoxic to HepG2 cells and THP‐1 macrophages (Figure S2B, D, E). As shown by western blotting data (Figure 9a), when HepG2 cells were cultured with the conditioned medium from LPS+ATP‐activated THP‐1 macrophages, the medium from THP‐1 macrophages pretreated with 2354glu up‐regulated total and phosphorylated LKB1 and AMPK in a dose‐dependent manner. As shown by immunohistochemical staining for SREBP1 and DAPI (Figure 9c), after incubation of HepG2 cells with the conditioned medium from LPS+ATP‐activated THP‐1 macrophages, SREBP1 was up‐regulated both in the cytoplasm and the nucleus of HepG2 cells, while such increased SREBP1 in HepG2 cells was reversed by incubation with the conditioned medium from 2354glu‐pretreated, LPS+ATP‐activated, THP‐1 macrophages.
Figure 9.
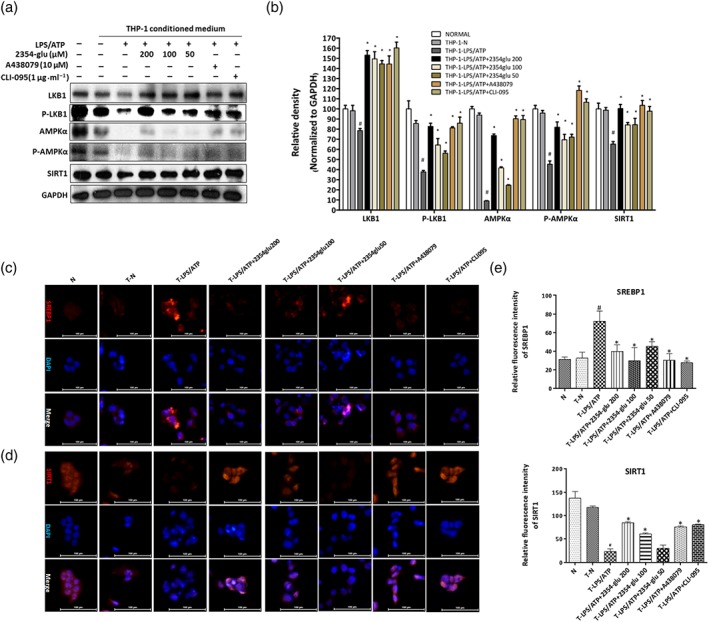
2354glu ameliorates lipid accumulation in hepatocytes induced by IL‐1β from activated macrophages. HepG2 cells were incubated 24 hr with conditioned medium from LPS+ATP‐stimulated THP‐1 macrophages pretreated with 2354glu (200, 100, and 50 μM), A438079 (10 μM), or CLI‐095 (1 μg·ml−1) for 1 hr. (a) Total and phosphorylated AMPKα and LKB1 and SIRT1 protein levels were determined using western blot analysis with HepG2 cells lysates. (b) Each immunoreactive band was normalized against GAPDH and relative to normal to control for unwanted sources of variation. Immunofluorescence staining of SREBP1 (c) and SIRT1 (d). Original images were taken at 200× magnification. SREBP1 (red) and SIRT1 (red) staining and nuclei with DAPI (blue) are shown. Representative images were shown. (e) Relative fluorescence intensity of SREBP1 and SIRT1 was analysed with Image Pro‐Plus 6.0. All histograms represent the mean ± SD of five independent assays with at least three replicates. # P < .05, significantly different from untreated HepG2 cells; *P < .05, significantly different from HepG2 cells cultured with conditioned medium from LPS+ATP‐activated THP‐1 macrophages
Figure 10.
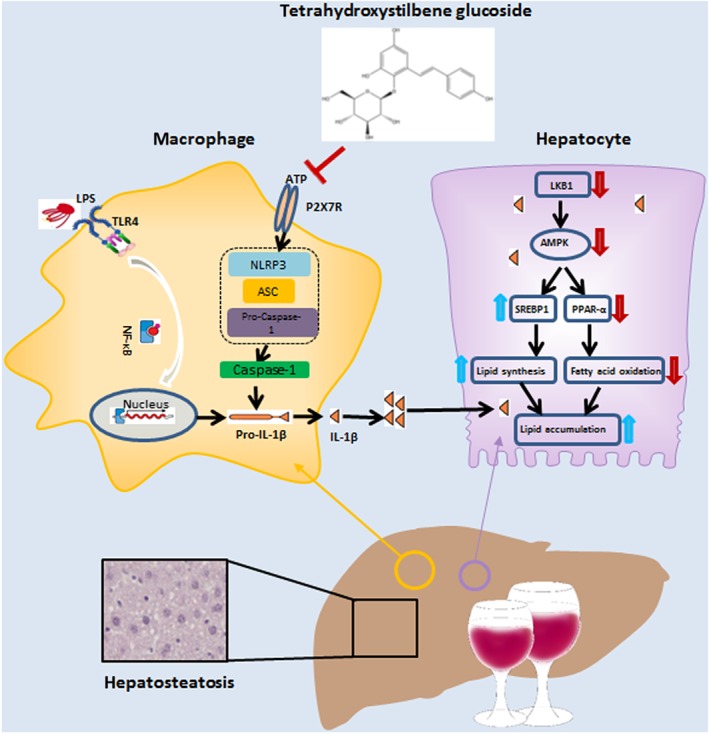
Diagram of the possible mechanisms underlying the amelioration of alcoholic hepatosteatosis by tetrahydroxystilbene glucoside (2345glu). The amelioration of this murine model of alcoholic hepatosteatosis by 2354glu appears to depend on the blockade of the P2X7 receptor–NLRP3 pathway in macrophages, leading to decreased secretion of IL‐1β. In hepatocytes, this decreased level of extracellular IL‐1β leads to a suppression of lipid accumulation
SIRT1, as a nuclear protein, is predominately localized in nucleus of cells. After incubation of HepG2 cells with the conditioned medium from LPS+ATP‐activated THP‐1 macrophages, nuclear levels of SIRT1 were diminished. Additionally, the nuclear SIRT1 of HepG2 cells was greatly activated in response to conditioned medium from 2354glu pretreated THP‐1 macrophages (Figure 9a,d). Treatment of THP‐1 macrophages with both A438079 and CLI‐095 increased the SIRT1 levels of HepG2 cells. Inhibition of IL‐1β release from macrophages, by treatment with 2354glu, significantly ameliorated lipid accumulation in hepatocytes, indicating that blockade of IL‐1β secretion from macrophages, by targeting P2X7 receptors, might be beneficial in controlling alcoholic hepatosteatosis.
4. DISCUSSION
Long‐term excessive alcohol intake causes a fatty liver, which can further deteriorate into ALD. Acute and chronic inflammatory conditions have been recognized to play important roles in the pathogenesis of ALD (Wang, Gao, Zakhari, & Nagy, 2012). Previous research had suggested that the regulation of cell‐to‐cell interactions between Kupffer cells and hepatocytes during alcohol‐induced inflammation can ameliorate hepatocyte steatosis (Louvet et al., 2011). Therefore, it is necessary to seek low MW inhibitors to regulate the interactions between Kupffer cells and hepatocytes and that such inhibitors might provide a potential therapeutic strategy to treat or prevent ALD.
The low MW compound 2354glu is a stilbenoid compound from P. multiflorum, which had been reported to reverse the occurrence and development of non‐alcoholic fatty liver disease (Lou et al., 2017). Moreover, previous research (Wu et al., 2017) and our data (Figure S2) showed that 2354glu exhibits little or no cell toxicity. These findings encouraged us to evaluate whether 2354glu could ameliorate alcohol‐induced hepatic steatosis and inflammation by regulating the crosstalk of macrophages and hepatocytes. The present work provided evidence that regulation of the P2X7 receptor–NLRP3 axis by 2354glu in macrophages could ameliorate alcoholic hepatic steatosis by modulating the crosstalk of macrophages and hepatocytes to affect lipid accumulation.
The infiltration of monocytes and macrophages into the damaged tissue in the liver represents an important pathophysiological feature in patients with ALD (Ju & Mandrekar, 2015). Our data demonstrated that macrophages are recruited to the liver in response to alcohol exposure (Figure 6), suggesting that alcohol consumption triggers an inflammatory response. Additionally, we clearly observed that 2354glu significantly suppressed the infiltration of immune cells. Therefore, we utilized murine peritoneal macrophages to investigate whether the P2X7 receptor–NLRP3 inflammasome pathway was involved in this inhibitory effect of 2354glu. Alcohol consumption increases gut permeability and the rate of bacterial endotoxin translocation, which is increasingly recognized as a major factor in ALD (Szabo, 2015). Thus, translocated LPS provides the first signal for TLR4‐mediated up‐regulation of inflammatory mediators, such as pro‐IL‐1β. In alcohol‐fed mice, ATP is released from alcohol‐damaged hepatocytes, and serum ATP levels are elevated, indicating that high levels of ATP might be a danger signal in ALD progression (Petrasek et al., 2012). The combination of LPS and ATP provides two signals, which activate the NLRP3 inflammasome through the activation of TLR4 and P2X7 receptors. Upon stimulation of P2X7 receptors, the inactive precursor of IL‐1β is cleaved into mature IL‐1β, causing an inflammatory response. As shown in our earlier work (Shang et al., 2018), LPS by itself does not induce the macrophages to secrete IL‐1β but, as a first signal, triggers accumulation of pro‐IL‐1β in macrophages. When LPS is combined with a high concentration of extracellular ATP, a powerful second signal, the cleavage of pro‐IL‐1β into mature IL‐1β is promoted, followed by the release of mature IL‐1β from the macrophage (Gombault, Baron, & Couillin, 2012). Therefore, we used LPS plus ATP combination to obtain conditioned medium containing mature IL‐1β from cultures of macrophages. We compared A438079, a selective P2X7 receptor antagonist, and CLI‐095, a TLR4 inhibitor, with 2354glu. A438079 and CLI‐095 were added to naïve and activated murine peritoneal macrophages to inhibit activation of P2X7 receptors and TLR4 respectively (Figure 8a and Figure S3C). CLI‐095 completely inhibited the production of pro‐IL‐1β, while A438079 only inhibited the release of the mature IL‐1β to the extracellular medium. Compared with CLI‐095 and A438079, 2354glu also significantly reduced LPS+ATP‐induced IL‐1β cleavage and mature IL‐1β release in a concentration‐dependent manner but only slightly affected levels of the pro‐IL‐1β form. Those data might indirectly indicate that 2354glu is a specific inhibitor of P2X7 receptors. A high concentration of ATP disrupts the calcium influx, and activation of P2X7 receptors by ATP leads to the elevation of intracellular Ca2+. Therefore, we measured calcium fluxes in naïve murine peritoneal macrophages to confirm the specific inhibitory capacity of 2354glu against P2X7 receptors and found that 2354glu suppressed the intracellular calcium concentration [Ca2+]i (Figure S4A). Together with blockade of P2X7 receptor‐mediated pore formation by 2354glu (Figure S4B) and docking conformation of 2354glu in the ATP binding site of the P2X7 receptor (Figure S5), our overall results confirmed that 2354glu might be a specific inhibitor of P2X7 receptors, resulting in decreased IL‐1β secretion in macrophages.
Previous research has shown that the activation of P2X7 receptors is associated with tissue injury by promoting fibrosis, inflammation, and oxidative stress (Das et al., 2013; Huang et al., 2014). P2X7 receptors are ATP‐gated ion channels that are mainly involved in IL‐1β maturation and these receptors also recruit the NLRP3 inflammasome–caspase‐1 complex (Giuliani, Sarti, Falzoni, & Di Virgilio, 2017). IL‐1β, as a potent pro‐inflammatory cytokine, is crucial to the pathogenesis of ALD (Petrasek et al., 2012). In the current study, IL‐1β release from macrophages was assessed after the knockdown or overexpression of P2X7 receptors in LPS‐primed and ATP‐stimulated THP‐1 macrophages (Figure 1). The results indicated that P2X7 receptors are essential in triggering IL‐1β maturation and secretion in macrophages.
Macrophages respond to numerous hepatic inflammatory stimuli by producing specific cytokines, such as IL‐1β, influencing the function of hepatocytes through intercellular interactions (Silva, Peccerella, Mueller, & Rausch, 2019; Stienstra et al., 2010). The accumulation of lipids in hepatocytes influenced by IL‐1β released from macrophages is accompanied by increased lipid synthesis or decreased lipid oxidation through SREBP‐1 and PPARα respectively (Han et al., 2015; Pellicoro, Ramachandran, Iredale, & Fallowfield, 2014; Sozio & Crabb, 2008). Moreover, the up‐regulation of SREBP‐1 following alcohol exposure results from the inactivation of https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1540), which phosphorylates and subsequently inactivates target enzymes involved in lipid metabolism, leading to the activation of genes involved in lipid synthesis (Gao & Bataller, 2011; Hu et al., 2012; You, Matsumoto, Pacold, Cho, & Crabb, 2004). Moreover, the https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2212) is the key upstream activator of AMPK. https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2707), as a central signalling molecule, controls the regulators mentioned above in the presence of ethanol (You, Jogasuria, Taylor, & Wu, 2015). Alcohol treatment suppresses the phosphorylation of AMPK via LKB1, and SIRT1 deacetylates and phosphorylates LKB1. Clearly, the activation of the SIRT1–LKB1–AMPK pathway provides a feasible strategy to regulate lipogenesis and lipid accumulation during ALD. As shown in Figure 5, ethanol intake or 2354glu modulated both the phosphorylated and the total protein of AMPK and LKB1, but the magnitude of the effects on phosphorylation of AMPK and LKB1 by ethanol intake or 2354glu was greater than that on the total protein. We further observed that AMPK and LKB1 phosphorylation was decreased in HepG2 cells cultured with medium from LPS+ATP‐stimulated THP‐1 macrophages. As expected, 2354glu activated the phosphorylation of LKB1 and AMPK and inhibited the transcriptional activity of SREBP1 in alcohol‐induced steatotic mouse liver (Figures 5). Based on the elevated hepatic and circulating levels of LPS and ATP in alcoholic steatotic mice, the restoration of AMPK and LKB1 by 2354glu in alcohol‐exposed mouse livers, especially in hepatocytes, might be connected with the amount of IL‐1β released from LPS+ATP‐stimulated THP‐1 macrophages, to a certain extent.
Those results implied that 2354glu might provide a new strategy towards macrophage–hepatocyte crosstalk in alcoholic hepatosteatosis. To strengthen this possibility, we mimicked the indirect interaction of hepatocytes and macrophages by using conditioned medium. The knockdown or overexpression of P2X7 receptors in THP‐1 macrophages altered the release of mature IL‐1β into the medium, and the subsequent changes in IL‐1β release accompanying P2X7 receptor gene alteration on macrophages resulted in the regulation of the SIRT1–AMPK–LKB1–SREBP1 axis in HepG2 cells incubated with the conditioned medium from the LPS+ATP‐stimulated THP‐1 macrophages (Figures 2 and 3). These results indicate that P2X7 receptors located on macrophages regulate the secretion of mature IL‐1β, and then the released mature IL‐1β from activated macrophages regulates LKB1–AMPK‐mediated lipid accumulation in hepatocytes during alcoholic hepatosteatosis. Consistent with in vivo findings, when HepG2 cells were incubated with the culture medium from LPS+ATP‐stimulated THP‐1 macrophages, 2354glu pretreatment of macrophages suppressed hepatocyte lipid synthesis via the SIRT1–LKB1–AMPK–SREBP1 axis (Figures 9). These results suggest that the ability of 2354glu to ameliorate alcoholic hepatosteatosis might critically depend on the effects of 2345glu on the P2X7 receptor–NLRP3 axis in macrophages. The consequent reduction of mature IL‐1β released from macrophages inhibited lipid accumulation in hepatocytes through the crosstalk between macrophages and hepatocytes.
In conclusion, our data suggest that the regulation of alcoholic hepatosteatosis by 2354glu is the result of modulating the crosstalk between macrophages and hepatocytes. As illustrated in Figure 10, the mature IL‐1β released by activation of P2X7 receptors in macrophages blocks the SIRT1–LKB1–AMPK pathway in hepatocytes, leading to the acceleration of lipid accumulation in hepatocytes. The low MW compound, 2354glu, inhibits P2X7 receptor–NLRP3 signalling in macrophages, decreasing the secretion of IL‐1β and the consequent blockade of the SIRT1–LKB1–AMPK pathway in hepatocytes, thus suppressing lipid accumulation in these cells. Our research suggested that the natural product, 2354glu, might be a promising candidate to target P2X7 receptors in macrophages and the subsequent interaction with hepatocytes, during alcoholic hepatosteatosis.
AUTHOR CONTRIBUTIONS
Y.Z. and M.J. are the primary investigators in this study. B.‐W.C., Y.S., and M.W. participated in part of animal experiments. H.‐X.Y., J.L., C.‐Y.Q., Z.‐Y.Z., and H.Y. participated in part of culture cell experiments. C.H.J. performed molecular docking. Y.‐L.W., G.‐H.Z., and Q.J. participated in part of data analysis. L.‐H.L. and J.‐X.N. designed this study and wrote the manuscript as corresponding authors.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for https://doi.org/10.1111/bph.14207, https://doi.org/10.1111/bph.14208, and https://doi.org/10.1111/bph.14206, and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Table S1 Primers sequences used in Real‐Time PCR and RT‐PCR.
Table S2. Primary and secondary antibodies used in Western blotting
Table S3. Primary and secondary antibodies used in liver histological analysis
Table S4. Primary and secondary antibodies used immunofluorescence staining
Fig. S1 The ATP content of ethanol exposure in HepG2 cells. HepG2 cells were incubated with or without ethanol (50, 100 and 200 mM) for 24 h. The extracellular (A) and intracellular (B) ATP levels were determined using Enhanced ATP Assay Kit as mentioned in Method section. All histograms represent the mean ± SD of at least five independent assays with three replicates. # p<0.05, significantly different when compared with untreated cells.
Fig. S2. Effects of 2354glu on cell viability and cytotoxicity. Cell viability of murine peritoneal macrophage cells (A) and HepG2 cells (B) was detected by MTT assay within 24 h after treatment with various 2354glu concentrations. The LDH release of murine peritoneal macrophage cells (C), HepG2 cells (D) and THP‐1 macrophages (E) was quantified by LDH assay within 24 h after treatment with various concentrations of 2354glu. The data are expressed as the mean ± SD of at least five independent assays with three replicates.
Fig. S3. 2354glu blocks P2X7R‐dependent IL‐1β cleavage in murine Naïve peritoneal macrophages. (A) Murine Naïve peritoneal macrophages and thioglycollate‐elicited peritoneal macrophages were primed with LPS (1 μg/ml) for 4 h then continuously stimulated ATP (3 mM) for additional 30 min or incubated with ethanol (50 mM) for 4 h. Intracellular and extracellular IL‐1β were detected by Western blotting. (C) Murine Naïve peritoneal macrophages were treated with the indicated concentrations of 2354glu (50, 100 and 200 μM), A438079 (10 μM) or CLI‐095 (1 μg/ml) for 1 h, and subsequently stimulated with LPS (1 μg/ml) for another 4 h followed by 30 min of ATP (3 mM) stimulation. Protein expression of P2X7R and caspase‐1, and extracellular and intracellular IL‐1β was verified by Western Blotting analysis. GAPDH was used as the loading control. (B, D) Each immunoreactive band was digitized and expressed as a ratio of untreated cells. All histograms represent the mean ± SD of at least five independent assays with three replicates. # p<0.05, significantly different when compared with untreated cells; * p<0.05, significantly different when compared with cells stimulated with LPS plus ATP.
Fig. S4. ATP‐induced P2X7R‐mediated membrane pore formation was inhibited by 2354glu. Murine Naïve peritoneal macrophages and THP‐1 macrophages were pretreated with the indicated concentrations of 2354glu (50, 100 and 200 μM), A438079 (10 μM) or CLI‐095 (1 μg/ml) for 1 h, and subsequently stimulated with LPS (1 μg/ml) for another 4 h followed by 30 min of ATP (3 mM) stimulation. Cells were incubated in the presence of 5 μM Fluo‐4 AM for 30 min or 5 μM PI for 15 min in the condition of 37°C, respectively. Before incubating PI, THP‐1 macrophages were treated with Triton 100 (0.1%) for 30 min, served as the positive control permeabilized the living cell membrane. (A) Changes of intracellular calcium measured through Immunofluorescence staining. Original images were taken at 400 × magnification. Representative images were shown. [Ca2+] (green) staining are shown. (B) PI uptake was measured by Immunofluorescence staining. Original images were taken at 600 × magnification. Representative images were shown. PI uptake (red) are shown. (C) Relative fluorescence intensity of intracellular calcium was analyzed with Image Pro‐Plus 6.0. (D) Relative fluorescence intensity of PI uptake was analyzed with Image Pro‐Plus 6.0. All histograms represent the mean ± SD of at least five independent assays with three replicates. # p<0.05, significantly different when compared with untreated cells; * p<0.05, significantly different when compared with cells stimulated with LPS plus ATP.
Fig. S5. Docking conformation of 2354glu and A438079 in the P2X7R ATP binding site (PDB: 5u1x). (A) 2D binding mode of tetrahydroxystilbene glucoside. (B) Proposed conformation of tetrahydroxystilbene glucoside in the binding pocket of P2X7R. (C) 2D binding mode of A438079 (3‐((5‐(2,3‐Dichlorophenyl)‐1H‐tetrazol‐1‐yl)methyl)pyridine). (D) Proposed conformation of A438079 in the binding pocket of P2X7R. The ligand is shown in yellow.
Fig. S6. Inhibition of IL‐1β cleavage in THP‐1 macrophages by 2354glu involves P2X7 receptors. (A) Extracellular mature IL‐1β were detected by western blotting. (B) Each immunoreactive band was digitized and expressed as a ratio of untreated cells. All histograms represent the mean ± SD of at least five independent assays with three replicates. # P < 0.05, significantly different from control siRNA‐transfected untreated cells; * P < 0.05, significantly different from control siRNA‐transfected cells stimulated with LPS plus ATP; a P < 0.05, significantly different from control siRNA‐transfected cells pretreated with 2354glu then stimulated with LPS plus ATP.
ACKNOWLEDGEMENTS
This study was supported by a grant from the National Natural Science Foundation of China (81960677, 81560597, 81660689, and 81860751) and partially by Science and Technology Planning Projects from the Science and Technology Department of Jilin Province (20180414048GH, 20180201065YY, and 20180519010JH) and Science and Technology Planning Project of the Jilin Provincial Education Department (JJKH20191155KJ).
Zhang Y, Jiang M, Cui B‐W, et al. P2X7 receptor‐targeted regulation by tetrahydroxystilbene glucoside in alcoholic hepatosteatosis: A new strategy towards macrophage–hepatocyte crosstalk. Br J Pharmacol. 2020;177:2793–2811. 10.1111/bph.15007
Yu Zhang and Min Jiang contributed equally to this work.
Contributor Information
Li‐Hua Lian, Email: lhlian@ybu.edu.cn.
Ji‐Xing Nan, Email: jxnan@ybu.edu.cn.
REFERENCES
- Abderrazak, A. , Syrovets, T. , Couchie, D. , El Hadri, K. , Friguet, B. , Simmet, T. , & Rouis, M. . (2015). NLRP3 inflammasome: From a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biology, 4, 296–307. 10.1016/j.redox.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Cidlowski, J. A. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators (2019). The Concise Guide to PHARMACOLOGY 2019/20: Nuclear hormone receptors. British Journal of Pharmacology, 176, S229–S246. 10.1111/bph.14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators (2019). The Concise Guide to PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , … CGTP Collaborators (2019). The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology, 176, S142–S228. 10.1111/bph.14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology . British Journal of Pharmacology, 175, 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamirano, J. , & Bataller, R. (2011). Alcoholic liver disease: Pathogenesis and new targets for therapy. Nature Reviews. Gastroenterology & Hepatology, 8, 491–501. 10.1038/nrgastro.2011.134 [DOI] [PubMed] [Google Scholar]
- Cheung, F. W. , Leung, A. W. , Liu, W. K. , & Che, C. T. (2014). Tyrosinase inhibitory activity of a glucosylated hydroxystilbene in mouse melan‐a melanocytes. Journal of Natural Products, 77, 1270–1274. 10.1021/np4008798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S. , Seth, R. K. , Kumar, A. , Kadiiska, M. B. , Michelotti, G. , Diehl, A. M. , & Chatterjee, S. (2013). Purinergic receptor X7 is a key modulator of metabolic oxidative stress‐mediated autophagy and inflammation in experimental nonalcoholic steatohepatitis. American Journal of Physiology. Gastrointestinal and Liver Physiology, 305, G950–G963. 10.1152/ajpgi.00235.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Association for the Study of the Liver (2018). EASL clinical practice guidelines: Management of alcohol‐related liver disease. Journal of Hepatology, 69, 154–181. 10.1016/j.jhep.2018.03.018 [DOI] [PubMed] [Google Scholar]
- Faria, R. X. , Reis, R. A. , Ferreira, L. G. , Cezar‐de‐Mello, P. F. , & Moraes, M. O. (2016). P2X7R large pore is partially blocked by pore forming proteins antagonists in astrocytes. Journal of Bioenergetics and Biomembranes, 48, 309–324. 10.1007/s10863-016-9649-9 [DOI] [PubMed] [Google Scholar]
- Frazier, T. H. , Stocker, A. M. , Kershner, N. A. , Marsano, L. S. , & McClain, C. J. (2011). Treatment of alcoholic liver disease. Therapeutic Advances in Gastroenterology, 4, 63–81. 10.1177/1756283X10378925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, B. , & Bataller, R. (2011). Alcoholic liver disease: Pathogenesis and new therapeutic targets. Gastroenterology, 141, 1572–1585. 10.1053/j.gastro.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani, A. L. , Sarti, A. C. , Falzoni, S. , & Di Virgilio, F. (2017). The P2X7 receptor‐interleukin‐1 liaison. Frontiers in Pharmacology, 8, 123 10.3389/fphar.2017.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombault, A. , Baron, L. , & Couillin, I. (2012). ATP release and purinergic signaling in NLRP3 inflammasome activation. Frontiers in Immunology, 3, 414 10.3389/fimmu.2012.00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga, D. T. G. , Ferreira, L. B. G. , Moreira Maramaldo Costa, T. E. , von Ranke, N. L. , Anastacio Furtado Pacheco, P. , Sposito Simoes, A. P. , … da Silva, F. C. (2017). 1‐Aryl‐1H‐ and 2‐aryl‐2H‐1,2,3‐triazole derivatives blockade P2X7 receptor in vitro and inflammatory response in vivo. European Journal of Medicinal Chemistry, 139, 698–717. 10.1016/j.ejmech.2017.08.034 [DOI] [PubMed] [Google Scholar]
- Hamarneh, S. R. , Kim, B. M. , Kaliannan, K. , Morrison, S. A. , Tantillo, T. J. , Tao, Q. , … Hodin, R. A. (2017). Intestinal alkaline phosphatase attenuates alcohol‐induced hepatosteatosis in mice. Digestive Diseases and Sciences, 62, 2021–2034. 10.1007/s10620-017-4576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J. , Li, E. , Chen, L. , Zhang, Y. , Wei, F. , Liu, J. , … Wang, Y. (2015). The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature, 524, 243–246. [DOI] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR (2018). The IUPHAR/BPS Guide to pharmacology in 2018: Updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Research, 46, D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homerin, G. , Jawhara, S. , Dezitter, X. , Baudelet, D. , Dufrenoy, P. , Rigo, B. , … Ghinet, A. (2019). Pyroglutamide‐based P2X7 receptor antagonists targeting inflammatory bowel disease. Journal of Medicinal Chemistry. 10.1021/acs.jmedchem.9b00584 [DOI] [PubMed] [Google Scholar]
- Hritz, I. , Mandrekar, P. , Velayudham, A. , Catalano, D. , Dolganiuc, A. , Kodys, K. , … Szabo, G. (2008). The critical role of toll‐like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology, 48, 1224–1231. 10.1002/hep.22470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M. , Wang, F. , Li, X. , Rogers, C. Q. , Liang, X. , Finck, B. N. , … You, M. (2012). Regulation of hepatic lipin‐1 by ethanol: Role of AMP‐activated protein kinase/sterol regulatory element‐binding protein 1 signaling in mice. Hepatology, 55, 437–446. 10.1002/hep.24708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Yu, W. , Cui, H. , Wang, Y. , Zhang, L. , Han, F. , & Huang, T. (2014). P2X7 blockade attenuates mouse liver fibrosis. Molecular Medicine Reports, 9, 57–62. 10.3892/mmr.2013.1807 [DOI] [PubMed] [Google Scholar]
- Jiang, S. , Zhang, Y. , Zheng, J. H. , Li, X. , Yao, Y. L. , Wu, Y. L. , … Lian, L. H. (2017). Potentiation of hepatic stellate cell activation by extracellular ATP is dependent on P2X7R‐mediated NLRP3 inflammasome activation. Pharmacological Research, 117, 82–93. 10.1016/j.phrs.2016.11.040 [DOI] [PubMed] [Google Scholar]
- Jin, Q. , Jiang, S. , Wu, Y. L. , Bai, T. , Yang, Y. , Jin, X. , … Nan, J. X. (2014). Hepatoprotective effect of cryptotanshinone from Salvia miltiorrhiza in d‐galactosamine/lipopolysaccharide‐induced fulminant hepatic failure. Phytomedicine, 21, 141–147. 10.1016/j.phymed.2013.07.016 [DOI] [PubMed] [Google Scholar]
- Ju, C. , & Mandrekar, P. (2015). Macrophages and alcohol‐related liver inflammation. Alcohol Research: Current Reviews, 37, 251–262. [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill I, Emerson M, & Altman D (2011) Animal research: Reporting in vivo experiments: The ARRIVE guidelines. Vol. 31. [DOI] [PMC free article] [PubMed]
- Koley, D. , & Bard, A. J. (2010). Triton X‐100 concentration effects on membrane permeability of a single HeLa cell by scanning electrochemical microscopy (SECM). Proceedings of the National Academy of Sciences of the United States of America, 107, 16783–16787. 10.1073/pnas.1011614107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, C. , Spindelboeck, W. , Haybaeck, J. , Douschan, P. , Rainer, F. , Terracciano, L. , … Stauber, R. E. (2017). Histological parameters and alcohol abstinence determine long‐term prognosis in patients with alcoholic liver disease. Journal of Hepatology, 66, 610–618. 10.1016/j.jhep.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhang, Y. , Jin, Q. , Xia, K. L. , Jiang, M. , Cui, B. W. , … Nan, J. X. (2018). Liver kinase B1/AMP‐activated protein kinase‐mediated regulation by gentiopicroside ameliorates P2X7 receptor‐dependent alcoholic hepatosteatosis. British Journal of Pharmacology, 175, 1451–1470. 10.1111/bph.14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, L. H. , Wu, Y. L. , Song, S. Z. , Wan, Y. , Xie, W. X. , Li, X. , … Nan, J. X. (2010). Gentiana manshurica Kitagawa reverses acute alcohol‐induced liver steatosis through blocking sterol regulatory element‐binding protein‐1 maturation. Journal of Agricultural and Food Chemistry, 58, 13013–13019. 10.1021/jf103976y [DOI] [PubMed] [Google Scholar]
- Lin, C. L. , Hsieh, S. L. , Leung, W. , Jeng, J. H. , Huang, G. C. , Lee, C. T. , & Wu, C. C. (2016). 2,3,5,4′‐Tetrahydroxystilbene‐2‐O‐β‐d‐glucoside suppresses human colorectal cancer cell metastasis through inhibiting NF‐κB activation. International Journal of Oncology, 49, 629–638. 10.3892/ijo.2016.3574 [DOI] [PubMed] [Google Scholar]
- Lin, L. , Ni, B. , Lin, H. , Zhang, M. , Li, X. , Yin, X. , … Ni, J. (2015). Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb.: A review. Journal of Ethnopharmacology, 159, 158–183. 10.1016/j.jep.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, Z. , Xia, B. , Su, J. , Yu, J. , Yan, M. , Huang, Y. , & Lv, G. (2017). Effect of a stilbene glycoside‐rich extract from Polygoni Multiflori Radix on experimental non‐alcoholic fatty liver disease based on principal component and orthogonal partial least squares discriminant analysis. Experimental and Therapeutic Medicine, 14, 4958–4966. 10.3892/etm.2017.5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet, A. , Teixeira‐Clerc, F. , Chobert, M. N. , Deveaux, V. , Pavoine, C. , Zimmer, A. , … Lotersztajn, S. (2011). Cannabinoid CB2 receptors protect against alcoholic liver disease by regulating Kupffer cell polarization in mice. Hepatology, 54, 1217–1226. 10.1002/hep.24524 [DOI] [PubMed] [Google Scholar]
- McGrath, J. C. , & Lilley, E. (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): New requirements for publication in BJP. British Journal of Pharmacology, 172, 3189–3193. 10.1111/bph.12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, L. E. , Ding, W. X. , Cresci, G. , Saikia, P. , & Shah, V. H. (2016). Linking pathogenic mechanisms of alcoholic liver disease with clinical phenotypes. Gastroenterology, 150, 1756–1768. 10.1053/j.gastro.2016.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research, C. (2011). Guide for the care and use of laboratory animals (Eighth ed.). The National Academies Press: Washington, DC. [Google Scholar]
- Pellicoro, A. , Ramachandran, P. , Iredale, J. P. , & Fallowfield, J. A. (2014). Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nature Reviews. Immunology, 14, 181–194. 10.1038/nri3623 [DOI] [PubMed] [Google Scholar]
- Petrasek, J. , Bala, S. , Csak, T. , Lippai, D. , Kodys, K. , Menashy, V. , … Szabo, G. (2012). IL‐1 receptor antagonist ameliorates inflammasome‐dependent alcoholic steatohepatitis in mice. The Journal of Clinical Investigation, 122, 3476–3489. 10.1172/JCI60777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco, A. , Compare, D. , Angrisani, D. , Sanduzzi Zamparelli, M. , & Nardone, G. (2014). Alcoholic disease: Liver and beyond. World Journal of Gastroenterology, 20, 14652–14659. 10.3748/wjg.v20.i40.14652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz, H. K. , Bataller, R. , Cortez‐Pinto, H. , Gao, B. , Gual, A. , Lackner, C. , … Tsukamoto, H. (2018). Alcoholic liver disease. Nature Reviews. Disease Primers, 4, 16 10.1038/s41572-018-0014-7 [DOI] [PubMed] [Google Scholar]
- Shang, Y. , Li, X. F. , Jin, M. J. , Li, Y. , Wu, Y. L. , Jin, Q. , … Nan, J. X. (2018). Leucodin attenuates inflammatory response in macrophages and lipid accumulation in steatotic hepatocytes via P2x7 receptor pathway: A potential role in alcoholic liver disease. Biomedicine & Pharmacotherapy, 107, 374–381. 10.1016/j.biopha.2018.08.009 [DOI] [PubMed] [Google Scholar]
- Silva, I. , Peccerella, T. , Mueller, S. , & Rausch, V. (2019). IL‐1β‐mediated macrophage‐hepatocyte crosstalk upregulates hepcidin under physiological low oxygen levels. Redox Biology, 24, 101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal, A. K. , Bataller, R. , Ahn, J. , Kamath, P. S. , & Shah, V. H. (2018). ACG clinical guideline: Alcoholic liver disease. The American Journal of Gastroenterology, 113, 175–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozio, M. , & Crabb, D. W. (2008). Alcohol and lipid metabolism. American Journal of Physiology. Endocrinology and Metabolism, 295, E10–E16. 10.1152/ajpendo.00011.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra, R. , Saudale, F. , Duval, C. , Keshtkar, S. , Groener, J. E. , van Rooijen, N. , … Müller, M. (2010). Kupffer cells promote hepatic steatosis via interleukin‐1β‐dependent suppression of peroxisome proliferator‐activated receptor α activity. Hepatology, 51, 511–522. 10.1002/hep.23337 [DOI] [PubMed] [Google Scholar]
- Szabo, G. (2015). Gut‐liver axis in alcoholic liver disease. Gastroenterology, 148, 30–36. 10.1053/j.gastro.2014.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi, T. , Iwamaru, Y. , Sugama, S. , Tsukimoto, M. , Fujita, M. , Sekigawa, A. , … Kitani, H. (2011). The activation of P2X7 receptor induces cathepsin d‐dependent production of a 20‐kDa form of IL‐1β under acidic extracellular pH in LPS‐primed microglial cells. Journal of Neurochemistry, 117, 712–723. 10.1111/j.1471-4159.2011.07240.x [DOI] [PubMed] [Google Scholar]
- Tilija Pun, N. , & Park, P. H. (2018). Adiponectin inhibits inflammatory cytokines production by Beclin‐1 phosphorylation and B‐cell lymphoma 2 mRNA destabilization: Role for autophagy induction. British Journal of Pharmacology, 175, 1066–1084. 10.1111/bph.14144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. J. , Gao, B. , Zakhari, S. , & Nagy, L. E. (2012). Inflammation in alcoholic liver disease. Annual Review of Nutrition, 32, 343–368. 10.1146/annurev-nutr-072610-145138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zhao, L. , Han, T. , Chen, S. , & Wang, J. (2008). Protective effects of 2,3,5,4′‐tetrahydroxystilbene‐2‐O‐β‐d‐glucoside, an active component of Polygonum multiflorum Thunb, on experimental colitis in mice. European Journal of Pharmacology, 578, 339–348. 10.1016/j.ejphar.2007.09.013 [DOI] [PubMed] [Google Scholar]
- Wewers, M. D. (2004). IL‐1β: An endosomal exit. Proceedings of the National Academy of Sciences of the United States of America, 101, 10241–10242. 10.1073/pnas.0403971101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2018) Global status report on alcohol and health 2018. vol. Licence: CC BY‐NC‐SA 3.0 IGO.
- Wu, J. , Hu, W. , Gong, Y. , Wang, P. , Tong, L. , Chen, X. , … Huang, C. (2017). Current pharmacological developments in 2,3,4′,5‐tetrahydroxystilbene‐2‐O‐β‐d‐glucoside (TSG). European Journal of Pharmacology, 811, 21–29. 10.1016/j.ejphar.2017.05.037 [DOI] [PubMed] [Google Scholar]
- You, M. , Jogasuria, A. , Taylor, C. , & Wu, J. (2015). Sirtuin 1 signaling and alcoholic fatty liver disease. Hepatobiliary Surg Nutr, 4, 88–100. 10.3978/j.issn.2304-3881.2014.12.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, M. , Matsumoto, M. , Pacold, C. M. , Cho, W. K. , & Crabb, D. W. (2004). The role of AMP‐activated protein kinase in the action of ethanol in the liver. Gastroenterology, 127, 1798–1808. 10.1053/j.gastro.2004.09.049 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Jin, Q. , Li, X. , Jiang, M. , Cui, B. W. , Xia, K. L. , … Nan, J. X. (2018). Amelioration of alcoholic liver steatosis by dihydroquercetin through the modulation of AMPK‐dependent lipogenesis mediated by P2X7R‐NLRP3‐inflammasome activation. Journal of Agricultural and Food Chemistry, 66, 4862–4871. 10.1021/acs.jafc.8b00944 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers sequences used in Real‐Time PCR and RT‐PCR.
Table S2. Primary and secondary antibodies used in Western blotting
Table S3. Primary and secondary antibodies used in liver histological analysis
Table S4. Primary and secondary antibodies used immunofluorescence staining
Fig. S1 The ATP content of ethanol exposure in HepG2 cells. HepG2 cells were incubated with or without ethanol (50, 100 and 200 mM) for 24 h. The extracellular (A) and intracellular (B) ATP levels were determined using Enhanced ATP Assay Kit as mentioned in Method section. All histograms represent the mean ± SD of at least five independent assays with three replicates. # p<0.05, significantly different when compared with untreated cells.
Fig. S2. Effects of 2354glu on cell viability and cytotoxicity. Cell viability of murine peritoneal macrophage cells (A) and HepG2 cells (B) was detected by MTT assay within 24 h after treatment with various 2354glu concentrations. The LDH release of murine peritoneal macrophage cells (C), HepG2 cells (D) and THP‐1 macrophages (E) was quantified by LDH assay within 24 h after treatment with various concentrations of 2354glu. The data are expressed as the mean ± SD of at least five independent assays with three replicates.
Fig. S3. 2354glu blocks P2X7R‐dependent IL‐1β cleavage in murine Naïve peritoneal macrophages. (A) Murine Naïve peritoneal macrophages and thioglycollate‐elicited peritoneal macrophages were primed with LPS (1 μg/ml) for 4 h then continuously stimulated ATP (3 mM) for additional 30 min or incubated with ethanol (50 mM) for 4 h. Intracellular and extracellular IL‐1β were detected by Western blotting. (C) Murine Naïve peritoneal macrophages were treated with the indicated concentrations of 2354glu (50, 100 and 200 μM), A438079 (10 μM) or CLI‐095 (1 μg/ml) for 1 h, and subsequently stimulated with LPS (1 μg/ml) for another 4 h followed by 30 min of ATP (3 mM) stimulation. Protein expression of P2X7R and caspase‐1, and extracellular and intracellular IL‐1β was verified by Western Blotting analysis. GAPDH was used as the loading control. (B, D) Each immunoreactive band was digitized and expressed as a ratio of untreated cells. All histograms represent the mean ± SD of at least five independent assays with three replicates. # p<0.05, significantly different when compared with untreated cells; * p<0.05, significantly different when compared with cells stimulated with LPS plus ATP.
Fig. S4. ATP‐induced P2X7R‐mediated membrane pore formation was inhibited by 2354glu. Murine Naïve peritoneal macrophages and THP‐1 macrophages were pretreated with the indicated concentrations of 2354glu (50, 100 and 200 μM), A438079 (10 μM) or CLI‐095 (1 μg/ml) for 1 h, and subsequently stimulated with LPS (1 μg/ml) for another 4 h followed by 30 min of ATP (3 mM) stimulation. Cells were incubated in the presence of 5 μM Fluo‐4 AM for 30 min or 5 μM PI for 15 min in the condition of 37°C, respectively. Before incubating PI, THP‐1 macrophages were treated with Triton 100 (0.1%) for 30 min, served as the positive control permeabilized the living cell membrane. (A) Changes of intracellular calcium measured through Immunofluorescence staining. Original images were taken at 400 × magnification. Representative images were shown. [Ca2+] (green) staining are shown. (B) PI uptake was measured by Immunofluorescence staining. Original images were taken at 600 × magnification. Representative images were shown. PI uptake (red) are shown. (C) Relative fluorescence intensity of intracellular calcium was analyzed with Image Pro‐Plus 6.0. (D) Relative fluorescence intensity of PI uptake was analyzed with Image Pro‐Plus 6.0. All histograms represent the mean ± SD of at least five independent assays with three replicates. # p<0.05, significantly different when compared with untreated cells; * p<0.05, significantly different when compared with cells stimulated with LPS plus ATP.
Fig. S5. Docking conformation of 2354glu and A438079 in the P2X7R ATP binding site (PDB: 5u1x). (A) 2D binding mode of tetrahydroxystilbene glucoside. (B) Proposed conformation of tetrahydroxystilbene glucoside in the binding pocket of P2X7R. (C) 2D binding mode of A438079 (3‐((5‐(2,3‐Dichlorophenyl)‐1H‐tetrazol‐1‐yl)methyl)pyridine). (D) Proposed conformation of A438079 in the binding pocket of P2X7R. The ligand is shown in yellow.
Fig. S6. Inhibition of IL‐1β cleavage in THP‐1 macrophages by 2354glu involves P2X7 receptors. (A) Extracellular mature IL‐1β were detected by western blotting. (B) Each immunoreactive band was digitized and expressed as a ratio of untreated cells. All histograms represent the mean ± SD of at least five independent assays with three replicates. # P < 0.05, significantly different from control siRNA‐transfected untreated cells; * P < 0.05, significantly different from control siRNA‐transfected cells stimulated with LPS plus ATP; a P < 0.05, significantly different from control siRNA‐transfected cells pretreated with 2354glu then stimulated with LPS plus ATP.


