Abstract
Background
We evaluated the functional capacity of plantaricin-producing Lactobacillus plantarum SF9C and S-layer-carrying Lactobacillus brevis SF9B to withstand gastrointestinal transit and to compete among the gut microbiota in vivo. Considering the probiotic potential of Lb. brevis SF9B, this study aims to investigate the antibacterial activity of Lb. plantarum SF9C and their potential for in vivo colonisation in rats, which could be the basis for the investigation of their synergistic functionality.
Results
A plantaricin-encoding cluster was identified in Lb. plantarum SF9C, a strain which efficiently inhibited the growth of Listeria monocytogenes ATCC® 19111™ and Staphylococcus aureus 3048. Homology-based three-dimensional (3D) structures of SF9C plantaricins PlnJK and PlnEF were predicted using SWISS-MODEL workspace and the helical wheel representations of the plantaricin peptide helices were generated by HELIQUEST. Contrary to the plantaricin-producing SF9C strain, the S-layer-carrying SF9B strain excluded Escherichia coli 3014 and Salmonella enterica serovar Typhimurium FP1 from the adhesion to Caco-2 cells. Finally, PCR-DGGE analysis of the V2–V3 regions of the 16S rRNA gene confirmed the transit of the two selected lactobacilli through the gastrointestinal tract (GIT). Microbiome profiling via the Illumina MiSeq platform revealed the prevalence of Lactobacillus spp. in the gut microbiota of the Lactobacillus-treated rats, even on the 10th day after the Lactobacillus application, compared to the microbiota of the healthy and AlCl3-exposed rats before Lactobacillus treatment.
Conclusion
The combined application of Lb. plantarum SF9C and Lb. brevis SF9B was able to influence the intestinal microbiota composition in rats, which was reflected in the increased abundance of Lactobacillus genus, but also in the altered abundances of other bacterial genera, either in the model of healthy or aberrant gut microbiota of rats. The antibacterial activity and capacity to withstand in GIT conditions contributed to the functional aspects of SF9C and SF9B strains that could be incorporated in the probiotic-containing functional foods with a possibility to positively modulate the gut microbiota composition.
Keywords: Antibacterial activity, Gut colonisation, Lactobacillus, Microbiota, Plantaricin, S-layer
Background
Lactobacillus strains are omnipresent in different ecological niches. The representative members dominate the microbiota of the sauerkraut and are under constant competition with other strains for nutrients and space [13]. The antibacterial activity of Lactobacillus strains is an important factor for the pathogen elimination in the complex microbial communities. Bacteriocin-producing Lactobacillus strains may achieve a competitive advantage in the surrounding microenvironment, which represents an attractive approach in terms of food biopreservation [13]. Their application expands even to the aspect of health since bacteriocin production is recognised as an important probiotic trait and bacteriocins have even been proposed as alternatives to antibiotics [12, 39]. Bacteriocinogenic activity may contribute to the functionality of probiotics through direct inhibition of the pathogens. Moreover, bacteriocins aid the survival of the producing strain and may act as quorum sensing molecules in the intestinal environment.
Previously, we monitored lactic acid bacteria (LAB) population during spontaneous fermentation of the Brassica oleracea var. capitata cultivar Varaždinski [6–8]. At the onset of the spontaneous fermentation, LAB diversity was present, including Leuconostoc mesenteroides strains, while a restricted number of Lactobacillus species, mainly Lactobacillus plantarum, dominated in the later stages [7]. Lb. brevis SF9B was also isolated from this fermentation. This strain showed desirable functional and technological properties largely influenced by surface (S)-layer proteins (Slps), which were detected by SDS-PAGE [7] and identified by 2D electrophoresis followed by LC–MS analysis. Slps have a functional role in conveying increased survival of the respective SF9B strain under simulated GIT conditions and during freeze-drying. Moreover, the results indicated a prominent role of Slps in adhesion of SF9B strain to mucin, extracellular matrix (ECM) proteins, and particularly to Caco-2 cells [6]. Besides SF9B, at the final stage of spontaneous fermentation, we isolated an autochthonous strain SF9C. This strain was identified as Lb. plantarum, which is a prevalent species in sauerkraut fermentation, probably due to its competitiveness with autochthonous microbiota. Turbidimetric method had previously revealed antibacterial activity of Lb. plantarum SF9C against some common pathogens [7]. Therefore, the aim of this study is to evaluate the competitive advantage potential of bacteriocin-producing Lb. plantarum SF9C and S-layer-carrying Lb. brevis SF9B against pathogens by in vitro and in vivo investigations. It also tests possible bacteriocinogenic activity of SF9C strain against Gram-positive pathogens Listeria monocytogenes ATCC® 19111™ and Staphylococcus aureus 3048. Coculturing of LAB strain with common Gram-positive food pathogens stimulates its bacteriocinogenic activity. The stimulation of bacteriocinogenic activity in SF9C strain was performed by its coculturing with common Gram-positive food pathogens. To observe whether the examined Lactobacillus strains have a broader spectrum of antimicrobial activity, antagonistic activity against Gram-negative Escherichia coli 3014 and Salmonella Typhimurium FP1 was also evaluated. Since preclinical evidence indicates that probiotic Lactobacillus strains may positively influence gut microbiota composition in different disorders followed by microbiota disturbance [11, 16], this study also aims to assess colonisation potential and the capacity of SF9C and SF9B strains to affect microbiome alterations in vivo, when applied together, either in healthy or AlCl3-exposed rats as a model of disturbed microbiota. PCR-DGGE (polymerase chain reaction-denaturing gradient gel electrophoresis) and the Illumina MiSeq sequencing analysis served to investigate if Lb. brevis SF9B and Lb. plantarum SF9C have the potential for in vivo colonisation and to influence the microbiota composition in the intestinal tract (IT) of rats.
Results
Plantaricin-related genes and whole genome sequencing (WGS) of Lb. plantarum SF9C
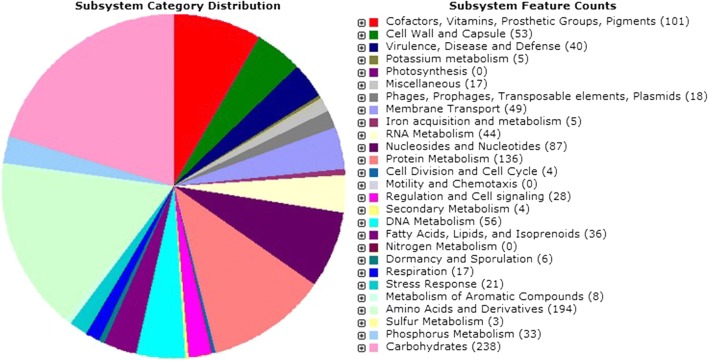
PCR gave positive result for plantaricin-related genes plnA, plnE and plnJ, suggesting that SF9C genome could harbour a pln locus (Additional file 1: Fig. S1), but it did not detect amplicons for plnNC8, plnS and plnW genes. Rapid Annotations using Subsystems Technology (RAST) of sequences obtained by Illumina MiSeq platform, and tblastn v. 2.2.27 comparison of the assembled contigs with the sequences deposited in NCBI employed for WGS identified SF9C strain as Lb. plantarum. This Whole Genome Shotgun project was deposited at DDBJ/ENA/GenBank under the accession RHLZ0000000. The version described in this paper is RHLZ01000000. The genome sequence contained 3.26 million bp (Mb) divided into 14 contigs. The size of Lb. plantarum SF9C genome of 3.2 Mb was similar to that of the other members of the species. The number of coding sequences was 3229 and the number of RNAs was 68. According to the comparative genomic studies, the estimated number of predicted protein-coding genes in Lactobacillus strains ranges from 1700 to around 3000 [47]. The G + C content of the Lb. plantarum SF9C genome was 44.4%, which is similar to the other Lb. plantarum strains, e.g. Lb. plantarum WCFS1 (44.5%) and Lb. plantarum ATCC 14917 (44.5%) [3]. Figure 1 shows the subsystem category distribution of major protein encoding genes (PEGs) for Lb. plantarum SF9C annotated by RAST server. The pie chart depicts the percentage distribution of 27 most abundant subsystem categories in SF9C strain. While most of the PEGs were related to universal cell functions such as DNA replication, transcription, translation, ribosomal structure and biogenesis, protein turnover and chaperones, and transport and metabolism of carbohydrates and nucleotides, certain PEGs were associated with the specific categories of cellular defence mechanisms and secondary metabolite biosynthesis, transport and catabolism, which may be responsible for the antimicrobial phenotype of SF9C strain.
Fig. 1.
Distribution of Lb. plantarum SF9C subsystem gene functions. The complete genome sequence of Lb. plantarum SF9C was annotated using the RAST server. The pie chart shows the count of each subsystem feature and the subsystem coverage
Given that SF9B and SF9C originate from the same microenvironment, their whole genomes were compared and the cluster dendrogram that reflects the diversity among strains was constructed. Single-nucleotide polymorphism (SNP) hierarchical clustering based on a similarity of whole genome sequences revealed that S-layer-carrying Lb. brevis SF9B is grouped with another S-layer-expressing Lb. brevis ATCC 367 (Fig. 2). Given that the phylogenetic distance between the strains was small, sauerkraut isolate Lb. plantarum SF9C was grouped with SF15C strain, another Lb. plantarum strain isolated from the same fermentation batch (Fig. 2).
Fig. 2.
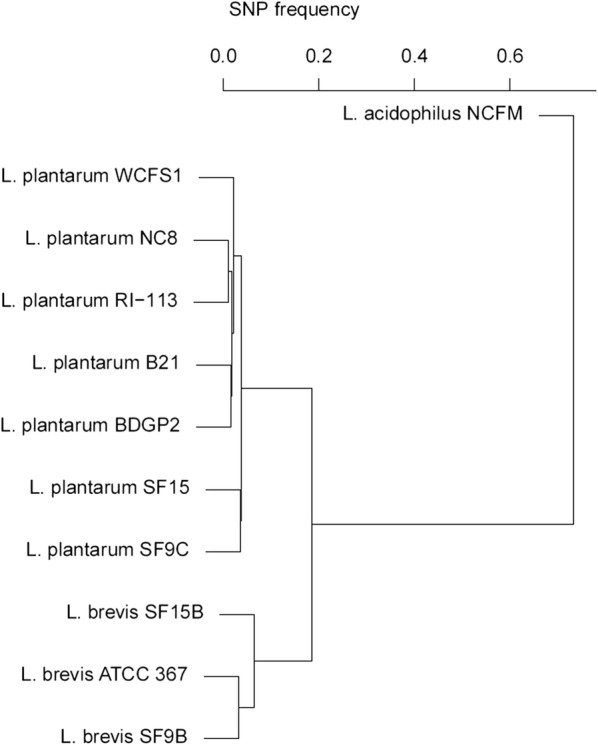
Hierarchical clustering of multiple Lactobacillus genomes based on single-nucleotide polymorphism (SNP) frequency. SNP frequency is “number of bases divided by bases aligned”. Origins and resources of the respected strains: WCFS1 [29], NC8 [4], RI-113 [28], B21 [23], BDGP2 [49], SF15C, SF9C, SF15B and SF9B [8], ATCC367 [36] and NCFM [1]
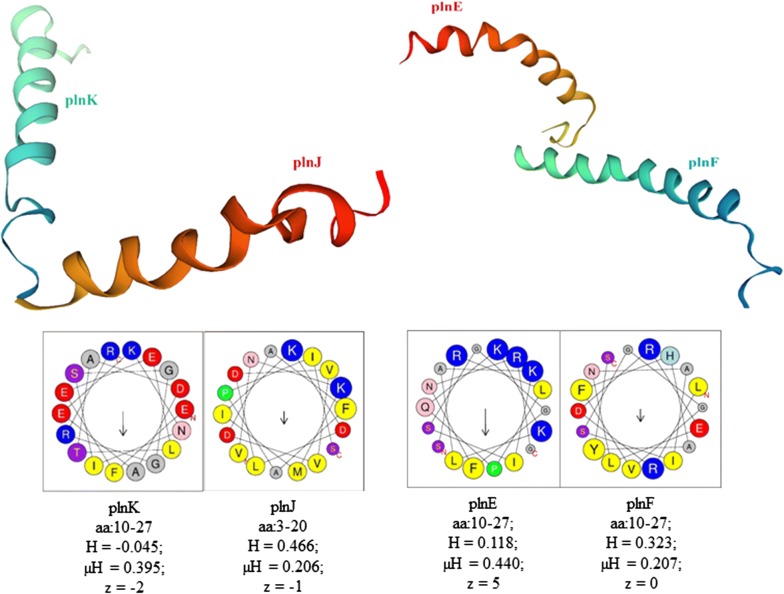
Next, WGS data were exploited to identify potential genomic triggers that may be responsible for the antibacterial phenotype. The assembled contigs were compared with the bacteriocins identified so far in the NCBI database using the tblastn v. 2.2.27. Through functional annotation and analysis of the high-coverage contigs obtained through Illumina sequencing, the genes involved in plantaricin production were predicted for Lb. plantarum SF9C and compared with the genes of other Lb. plantarum-bacteriocin-producing strains (Additional file 2: Table S1). The genome sequence of Lb. plantarum SF9C contains a cluster for biosynthesis of a putative plantaricin. In silico BAGEL4 analysis identified one area of interest (AOI) located at contig 13. The pln locus of SF9C contains genes encoding their cognate immunity proteins, whose location is just downstream of the bacteriocin genes, as well as ABC transporters, probably involved in the export of peptides with a double glycine leader (Fig. 3). Finally, the SWISS-MODEL predicted homology-based three-dimensional (3D) structures of SF9C two-peptide plantaricins PlnJK and PlnEF. Properties of the chosen amino acid (aa) residues that form a helix of each of the two plantaricins, PlnJK and PlnEF, were calculated by HeliQuest web server (Fig. 4).
Fig. 3.
Genetic map of the plantaricin gene cluster of Lb. plantarum SF9C strain
Fig. 4.
3D structures of PlnJK and PlnEF plantaricin peptides of SF9C strain predicted by the homology modelling and their helical wheel projections analysed by HeliQuest. aa—the amino acid residues (the one-letter code for amino acids is used); yellow—hydrophobic residues; purple—serine and threonine residues; dark blue—basic residues; red—acidic residues, pink—asparagine and glutamine residues; grey—alanine and glycine residues, light blue—histidine residues; green—proline residues; H—hydrophobicity; μH—hydrophobic moment; z—net charge (calculated at pH = 7.4, under the assumption that histidine is neutral and that the N-terminal amino group and the C-terminal carboxyl group of the sequence are uncharged)
Antimicrobial activity of Lb. plantarum SF9C after the cocultivation with pathogens
Preliminary results regarding the antagonistic activity of the two naturally coexisting strains Lb. plantarum SF9C and Lb. brevis SF9B, clearly demonstrated the difference in the spectrum of antibacterial activity (Tables 1, 2). Grown cultures of both strains, SF9C and SF9B, demonstrated antibacterial activity against L. monocytogenes ATCC® 19111™, S. aureus 3048, E. coli 3014 and S. Typhimurium FP1 (Tables 1, 2). Cell-free supernatant (CFS) of the strain SF9C exerted the antibacterial activity against the selected pathogens, while CFS of the strain SF9B did not inhibit the examined pathogens (Table 2). Furthermore, the combined CFSs of both strains, SF9C and SF9B, in equal ratio, showed decreased antibacterial activity against L. monocytogenes ATCC® 19111™, S. aureus 3048, E. coli 3014 and S. Typhimurium FP1, which was expected since the CFS of S-layer-carrying SF9B strain did not demonstrate antibacterial activity against these pathogens, and therefore alleviated cumulative inhibitory effect of combined CFSs (Table 2).
Table 1.
Inhibition of Listeria monocytogenes ATCC® 19111™, Staphylococcus aureus 3048, Escherichia coli 3014 and Salmonella Typhimurium FP1 by grown cultures of Lb. plantarum SF9C and Lb. brevis SF9B, separately and combined, evaluated by agar spot-test method and expressed as the diameter of inhibition zones around the grown cultures (cm)
| Lactobacillus strains | Diameter of inhibition zone (cm) | |||
|---|---|---|---|---|
| L. monocytogenes ATCC® 19111™ | S. aureus 3048 | E. coli 3014 | S. Typhimurium FP1 | |
| Lb. plantarum SF9C | 3.50 ± 0.10az | 3.32 ± 0.13azx | 3.05 ± 0.05ay | 3.07 ± 0.06ayx |
| Lb. brevis SF9B | 1.93 ± 0.40bz | 1.35 ± 0.05by | 1.75 ± 0.31bzy | 1.52 ± 0.08bzy |
| Combined SF9C + SF9B | 2.57 ± 0.06bz | 1.50 ± 0.10bx | 2.17 ± 0.06by | 2.60 ± 0.10cz |
abcDifferent letter means statistically significant difference (p < 0.05) within the same column among the used Lactobacillus strains
xyzDifferent letter means statistically significant difference (p < 0.05) within the same row among the used pathogens. Statistical analysis was carried out using ANOVA and the results are reported as mean value ± SD of three independent experiments
Table 2.
Comparison of the antimicrobial activity of cell-free supernatants (CFSs) of Lb. plantarum SF9C and Lb. brevis SF9B, separately and combined, before and after the treatment with proteinase K and high temperature heating, against L. monocytogenes ATCC® 19111™, S. aureus 3048, E. coli 3014 and S. Typhimurium FP1, determined by agar well-diffusion assay and expressed as the diameter of inhibition zones around the wells (cm)
| Treatment of CFS | Lactobacillus strains | Diameter of inhibition zone (cm) | |||
|---|---|---|---|---|---|
| L. monocytogenes ATCC® 19111™ | S. aureus 3048 | E. coli 3014 | S. Typhimurium FP1 | ||
| Before treatment | Lb. plantarum SF9C | 1.92 ± 0.03az | 1.57 ± 0.06ay | 1.50 ± 0.05ayw | 1.38 ± 0.03aw |
| Lb. brevis SF9B | 0.00 ± 0.00ez | 0.00 ± 0.00ez | 0.00 ± 0.00ez | 0.00 ± 0.00fz | |
| Combined SF9C + SF9B | 1.72 ± 0.03bz | 0.98 ± 0.03dx | 1.13 ± 0.06cy | 1.00 ± 0.00cx | |
| proteinase K | Lb. plantarum SF9C | 1.10 ± 0.00dy | 1.17 ± 0.03cyz | 1.23 ± 0.03bz | 1.20 ± 0.00bxz |
| Lb. brevis SF9B | 0.00 ± 0.00ez | 0.00 ± 0.00ez | 0.00 ± 0.00ez | 0.00 ± 0.00fz | |
| Combined SF9C + SF9B | 1.02 ± 0.03dz | 0.90 ± 0.00dy | 0.98 ± 0.03dz | 0.85 ± 0.00ey | |
| 100 °C/30 min | Lb. plantarum SF9C | 1.68 ± 0.03bz | 1.52 ± 0.03by | 1.30 ± 0.00bx | 1.23 ± 0.03bx |
| Lb. brevis SF9B | 0.00 ± 0.00ez | 0.00 ± 0.00ez | 0.00 ± 0.00ez | 0.00 ± 0.00fz | |
| Combined SF9C + SF9B | 1.53 ± 0.06cz | 0.93 ± 0.06dx | 1.03 ± 0.06cdy | 0.92 ± 0.03dx | |
abcdefDifferent letter means statistically significant difference (p < 0.05) within the same column among the treatments of CFSs and used Lactobacillus strains
wxyzDifferent letter means statistically significant difference (p < 0.05) within the same row among the used pathogens. Statistical analysis was carried out using ANOVA and the results are reported as mean value ± SD of three independent experiments
Lb. plantarum SF9C strain showed the strongest antibacterial activity against the closely related Gram-positive pathogen L. monocytogenes ATCC® 19111™, implying the potential bacteriocinogenic activity (Tables 1, 2). Therefore, CFSs of both examined Lactobacillus strains, separately and combined, were treated with proteinase K and exposed to high temperature in order to inactivate the potentially present bacteriocin. According to the obtained results, the antibacterial activity of the CFS of SF9C strain as well as combined CFSs of both Lactobacillus strains was partially inactivated after the treatment with proteinase K and after its exposure to high temperature of 100 °C for 30 min, compared to the CFSs that were not treated with proteinase K or high temperature (Table 2). These findings confirmed the presence of a substance with a proteinaceous nature in CFS of Lb. plantarum SF9C.
Similarly, the evaluation of the antibacterial activity of SF9C and SF9B against closely related LAB strains by agar well-diffusion method showed that SF9C was more effective in the inhibition of the examined LAB strains than SF9B, with the strongest effect observed against Enterococcus, moderate against Lactococcus and the weakest against Lactobacillus strains (data not shown).
To assess the possibility to enhance the bacteriocin activity of Lb. plantarum SF9C, this strain was cocultivated with S. aureus 3048 and L. monocytogenes ATCC® 19111™, since the activity of bacteriocins from LAB is mostly directed towards Gram-positive bacteria. The growth of Lb. plantarum SF9C strain was not impaired by the cocultivation with Gram-positive pathogens, but the log CFU/mL values of S. aureus 3048 and L. monocytogenes ATCC® 19111™ were reduced to non-detectable levels after 48 and 24 h of cocultivation, respectively (Fig. 5). Additionally, the antibacterial effect of Lb. plantarum SF9C, obtained by agar spot test, was significantly higher after 8, 10 and 22 h of incubation in coculture with S. aureus 3048, as well as after 22, 24 and 48 h of incubation in coculture with L. monocytogenes ATCC® 19111™ (Additional file 3: Fig. S2). The obtained results indicate the possibility to enhance antibacterial activity of Lb. plantarum SF9C by incubation in the presence of the sensitive Gram-positive pathogens.
Fig. 5.
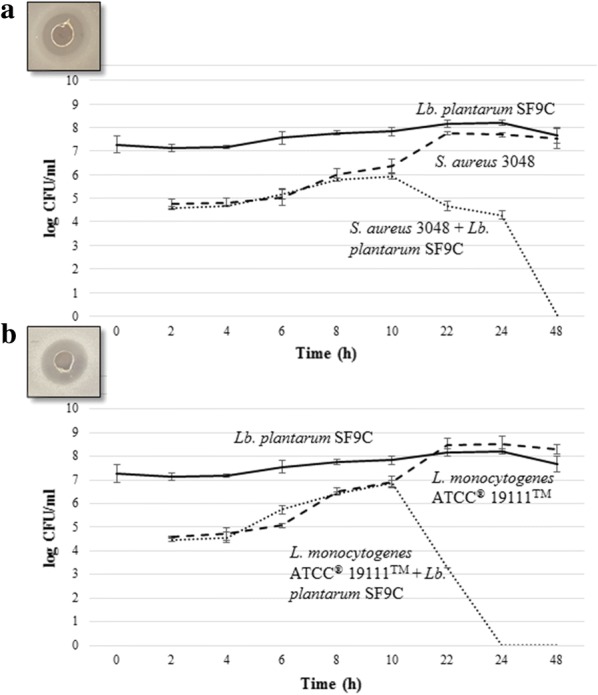
The growth curves of the test microorganisms: a) S. aureus 3048 and b) L. monocytogenes ATCC® 19111™ during cocultivation with (···) or without (▬ ▬) Lb. plantarum SF9C. Growth curve of Lb. plantarum SF9C (▬)
Inhibition of pathogen adherence to Caco-2 cells by Lb. brevis SF9B
Competitive pathogen exclusion assays by Lb. plantarum SF9C and Lb. brevis SF9B on Caco-2 human intestinal cells were performed. Since Lb. plantarum SF9C strain exhibited stronger antimicrobial activity against Gram-positive than against Gram-negative pathogens, as revealed by agar spot-test and agar well-diffusion assays, the possibility of SF9C to exclude targeted L. monocytogenes ATCC® 19111™ and S. aureus 3048 from the Caco-2 cell line was investigated. After 1 h of incubation, the number of adhered pathogens exposed to Lb. plantarum SF9C strain reached the values of (5.916 ± 0.527) log CFU/mL of L. monocytogenes ATCC® 19111™ and (7.167 ± 0.168) log CFU/mL of S. aureus 3048, respectively, which was not significantly reduced (p < 0.05) compared to the control ((5.823 ± 0.204) log CFU/mL of L. monocytogenes ATCC® 19111™ and (7.448 ± 0.181) log CFU/mL of S. aureus 3048, respectively).
While SF9C strain failed in preventing Caco-2 adhesion of the tested pathogens, Lb. brevis SF9B significantly reduced the adhesion of Gram-negative pathogens S. Typhimurium FP1 (p < 0.05) and E. coli 3014 (p < 0.01) in competitive exclusion assays. Namely, in exclusion assay, when Caco-2 cells were exposed to Lb. brevis SF9B before S. Typhimurium FP1, significantly fewer (p < 0.01) Salmonella cells adhered to them ((4.708 ± 0.014) log CFU/mL) than after exposure of Caco-2 cells to S. Typhimurium FP1 alone ((6.825 ± 0.099) log CFU/mL). In competition assay, when Caco-2 cells were incubated simultaneously with Lb. brevis SF9B and Salmonella, significantly fewer (p < 0.05) Salmonella cells adhered to them ((5.613 ± 0.135) log CFU/mL) than to Caco-2 cells infected with Salmonella alone ((6.825 ± 0.099) log CFU/mL). The competitive exclusion effect of Lb. brevis SF9B was even stronger against E. coli 3014, where the significant inhibition (p < 0.01) of an invasion of Caco-2 cells by E. coli 3014 in comparison to the control (without the addition of SF9B cells) was evident, with reduced values of 2.209 Δlog CFU/mL when pathogen cells were added after SF9B cells (exclusion), and of 2.117 Δlog CFU/mL when Lb. brevis SF9B and pathogen cells were added simultaneously (competition).
Influence of Lb. brevis SF9B and Lb. plantarum SF9C on gut microbiome composition
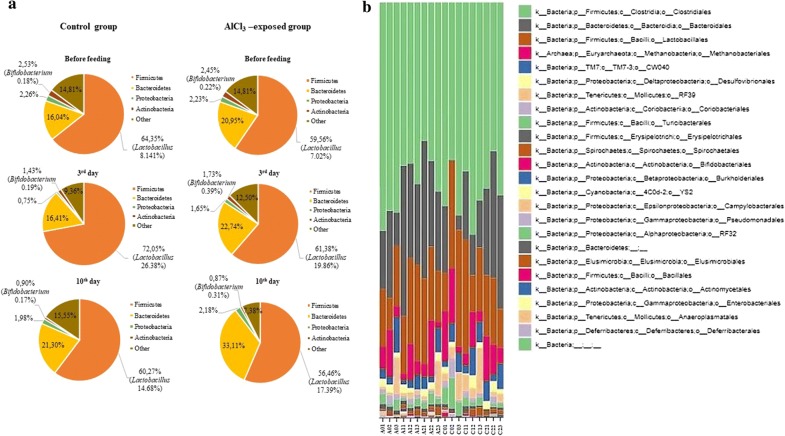
To assess the capacity of the two Lactobacillus strains to survive transit and eventually colonize the GIT, the gut microbiome composition after their transit through the GIT was monitored in vivo. Since the aberrant microbiota differs from the microbiota of healthy subjects in the prevalence of undesirable species, the AlCl3-exposed rats were chosen as an animal model for microbiome dysbiosis because previous research had shown that toxic metals like aluminium had a negative impact directly on the gut microbiota in humans and animals. Additionally, Lactobacillaceae, particularly Lb. plantarum, Lb. rhamnosus and Lb. brevis, were able to bind and remove toxic metals [50]. A recent study by Tian et al. [43] also suggests the potential of the Lb. plantarum strain to alleviate the aluminium-induced brain injuries in mice. Therefore, the potential of Lb. plantarum SF9C and Lb. brevis SF9B to compete among microbiota of AlCl3-exposed rats was investigated. The acetylcholinesterase (AChE) activity was assessed in the brain tissue homogenates to monitor the possible influence of AlCl3 treatment in the rats. The AChE activity and histopathological and immunohistochemical analyses of the brain showed that the number of plaques and the AChE activity were significantly higher in the brains of the AlCl3-exposed group than in the control group (p < 0.05) [34]. According to the obtained results, the neuropathological changes were observed in the AlCl3-exposed rats. Diffuse plaques, also called benign plaques, occurred much earlier than the neuritic plaques in the cerebellum. In the treatments, cerebellum of the AlCl3-exposed rats was negative on the AT8 marker, but positive on 4G8 and Iba1 markers (Additional file 4: Fig. S3). The value of AChE in the control group was lower (from 1.2 to 1.58 mol of hydrolyzed substrate/min/mg protein) than of the AlCl3-exposed group (from 1.38 to 2.4 mol of hydrolyzed substrate/min/mg protein). Furthermore, the analysis of faecal microbiota of rats by Illumina MiSeq sequencing revealed that across all, control or AlCl3-exposed groups (calculated as mean values of all the experiments), the dominant phyla were Firmicutes and Bacteroidetes, which respectively made up 63% (62.35 ± 5.40%) and 22% (21.76 ± 6.20%) of total abundance, with lower contributions from Actinobacteria (1.65 ± 0.73%) and Proteobacteria (1.84 ± 0.58%) (Fig. 6a). The Firmicutes and Bacteroidetes phyla accounted for more than 85% of total sequences, similar to previous findings in the gut microbiota of rats. However, phylum- through genus-wide differences in bacterial abundance were observed between these two groups. In the microbiome of AlCl3-exposed rats the abundance of Firmicutes and Actinobacteria decreased, while the abundance of Bacteroidetes increased compared to the control group. At the class level, the most abundant in all groups were Bacilli, Clostridia and Bacteroidia (Fig. 6b). Bifidobacterium was also consistently detected through the samples (Fig. 6a). Since our main goal was to evaluate the survival and colonisation potential of the two Lactobacillus strains in the model of healthy, but also in animals with disturbed microbiota, the focus was on the evaluation of Lactobacillus abundance. The abundance in Lactobacillus sp. was observed in all treated rat groups, implying the ability of SF9B and SF9C to adapt to the GIT, especially since these two strains are not of an intestinal, but sauerkraut origin. The gut microbiome analysis revealed taxonomic differences in gut microbiota composition influenced by Lactobacillus treatments. The culture-independent PCR-DGGE verified the presence of lactobacilli in the gut microbiota of faecal samples among rats before and after the treatment with SF9B and SF9C strains (Fig. 7). The cultivation on selective agar plates revealed the presence of presumptive Lactobacillus in the faeces of the control group at 5.6 × 107 CFU/mL and AlCl3-exposed rats at 1.99 × 108 CFU/mL on the 10th day after Lactobacillus treatment, which was in correlation with the results of microbiome analysis obtained by the Illumina MiSeq sequencing. PCR-DGGE analysis verified which Lactobacillus strains were potentially responsible for the observed higher Lactobacillus spp. abundance levels in the microbiota of Lactobacillus-treated rats. The PCR-DGGE of DNA fragments obtained by PCR amplification of the V2–V3 regions of the 16S rRNA gene suggested the presence of both Lactobacillus strains in the faeces of treated rats since their DNA fragment coincided with the 16S DNA fragment generated from the pure culture of Lb. brevis SF9B and Lb. plantarum SF9C (Fig. 7). The inoculation of the healthy rats with Lactobacillus strains led to the appearance of a new 16S DNA fragment in the PCR-DGGE profile of the sample on the 3rd day after Lactobacillus treatment that corresponded to Lactobacillus reuteri. Interestingly, the results of microbiota analysis at the species level showed the presence of Lb. reuteri and Lb. brevis as well. Furthermore, in a PCR-DGGE profile of the healthy rat, an intensive band was consistently detected, assigned after the sequencing and BLAST search to Lb. animalis, while on the 3rd day after Lactobacillus treatment, a faint band appeared corresponding to Lactobacillus intestinalis strain (Fig. 7).
Fig. 6.
a The four most abundant phyla detected in the faecal microbiota of control and AlCl3-exposed rats, both fed with Lb. plantarum SF9C and Lb. brevis SF9B. b The distribution of the bacterial classes in the faeces of control (C) and AlCl3-exposed rats (A), before application (0), and on the 3rd day (1), and 10th day (2) after the application of SF9B and SF9C strains. The second number represents the ordinal number of the rat
Fig. 7.
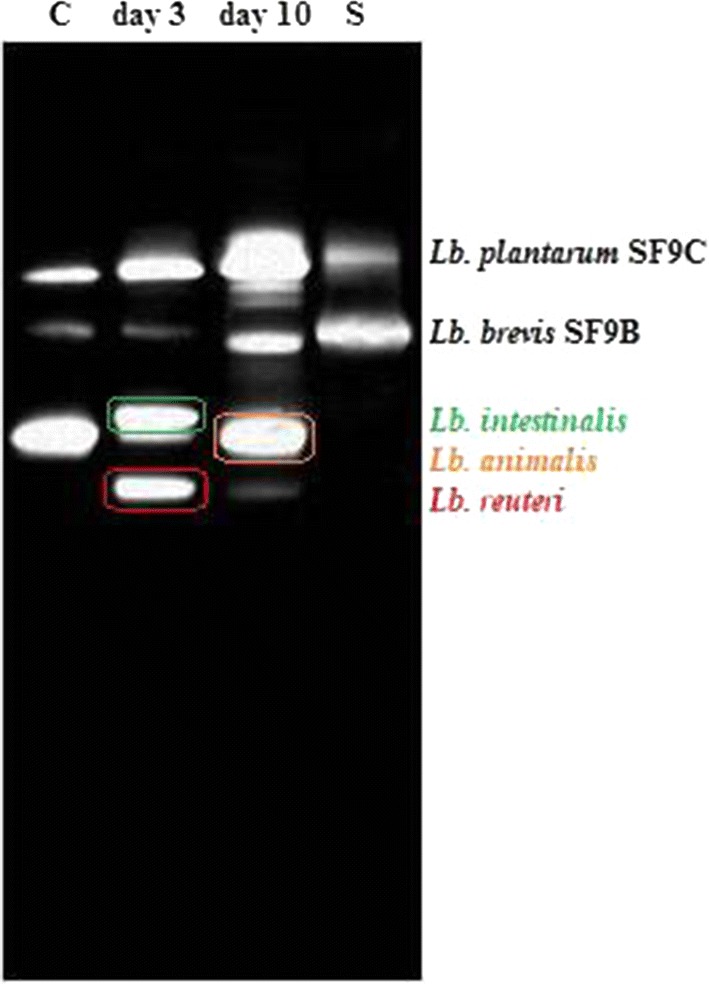
PCR-DGGE analysis of 16S DNA fragments generated with the universal bacterial primers HDA1 and HDA2 from the pooled DNA samples of the Lactobacillus species, isolated on MRS agar from faecal samples of rats fed with Lb. plantarum SF9C and Lb. brevis SF9B. Lanes: C—before application of SF9C and SF9B strains; day 3—3rd day after the application of SF9C and SF9B strains; day 10—10th day after application of SF9C and SF9B strains, S—the ladder of sequences from the pure cultures of SF9C and SF9B strains, respectively. Bands indicated by the symbols were excised and after amplification sequenced
Discussion
Functional genomics in probiotic research has facilitated the characterisation of candidate Lactobacillus strains. Bacteriocin production is a desirable trait of probiotic strains [25]. Herein, Lb. plantarum SF9C genome sequence was determined using a WGS assembly approach, with a focus on the characterisation of the plantaricin locus. The WGS confirmed the presence of the plantaricin (pln) loci in SF9C strain. The plnE and plnF genes that encode for bacteriocin precursor peptide, the plnA which encodes induction factor and individual gene plnJ were also detected by PCR (Additional file 1: Fig. S1). Plantaricin EF (PlnEF) and plantaricin JK (PlnJK) have already been described in certain Lb. plantarum strains as two-peptide bacteriocins. These compounds are biosynthesised as prepeptides and are cleaved off during the transport to the cell surface to become active peptides whose activity depends on the complementary action of the two peptides PlnE/PlnF, i.e. PlnJ/PlnK [15]. Predicted 3D structures of two Lb. plantarum SF9C plantaricins by SWISS homology modelling showed sequence similarity with the structures of PlnJK and PlnEF described by Rogne et al. [41] and Fimland et al. [19], respectively.
Bacteriocin activity of the particular strain together with its ability to compete for limited nutrients, competitive exclusion, and the stimulation of mucosal immunity could contribute to intestinal health [17]. Herein the antibacterial activity of the Lb. plantarum SF9C against Gram-positive L. monocytogenes ATCC® 19111™ and S. aureus 3048, and Gram-negative pathogens E. coli 3014 and S. Typhimurium FP1 was determined. Lb. plantarum SF9C drastically decreased the pH value (3.86 ± 0.04) after overnight growth due to the lactic acid production (2.25 ± 0.24% v/v), which creates unfavourable microenvironment for the pathogenic bacteria. The mechanisms of the antibacterial activity of SF9C strain are multifactorial and include the inhibition by the produced lactic acid, but also the potential antibacterial activity of plantaricin, especially since the inhibition was alleviated after the treatment with proteinase K and boiling of CFS of Lb. plantarum SF9C. L. monocytogenes ATCC® 19111™ and S. aureus 3048 possess several mechanisms to combat the challenges posed by acidic environments and therefore can tolerate low pH values. This claim was supported by the finding that Lb. plantarum SF9C strain demonstrated antibacterial activity against L. monocytogenes ATCC® 19111™ and S. aureus 3048, unlike Lb. brevis SF9B, which failed to inhibit respective pathogens, even though it is an effective lactic acid producer. Additionally, L. monocytogenes and S. aureus were deliberately chosen since these foodborne Gram-positive pathogens contaminate a wide range of fermented foods, although the pH value in these food matrices is low due to the metabolic activity of a spontaneously present population of LAB. Therefore, it was hypothesized that the potential plantaricin antibacterial activity was involved in the growth inhibition of Gram-positive pathogens, L. monocytogenes and S. aureus. Since one strategy to achieve the expression of otherwise silenced bacteriocins is the stimulation of their biosynthesis by growth in the coculture [10, 30, 38], the potential to enhance plantaricin antibacterial activity by cocultivation of Lb. plantarum SF9C with S. aureus 3048 and L. monocytogenes ATCC® 19111™ was studied. Antibacterial activity was initially detected in the early exponential phase of the pathogen growth after 10 h of incubation. The highest antibacterial activity was observed after 24 h in the late exponential phase of L. monocytogenes ATCC® 19111™ and after 48 h of S. aureus 3048. This is supported by the results of Maldonado-Barragán et al. [38], who suggested that the induction of bacteriocin production by means of coculturing with specific bacterial strains is a common feature among Lb. plantarum species. Since L. monocytogenes tolerates a broad pH range, it can be speculated that the obtained enhanced antilisterial effect of SF9C during cocultivation with L. monocytogenes ATCC® 19111™ is attributed to the potential enhanced plantaricin production. This is in agreement with the feature of Lactobacillus bacteriocins whose activity is mostly related towards Gram-positive bacteria.
Contrary to Lb. plantarum SF9C, S-layer-carrying Lb. brevis SF9B showed competitive exclusion of pathogens on the Caco-2 cells. Slps may act as mediators of bacterial adhesion and as such may contribute to the antagonism against the pathogens with which the S-layer-carrying strain competes for the same adhesion sites [27, 42, 45]. In our previous paper, SF9B strain exhibited the strongest coaggregation with E. coli 3014 and S. Typhimurium FP1 and the removal of Slps negatively affected its coaggregation ability. This study revealed that S-layer-carrying SF9B strain demonstrated significant levels of exclusion (p < 0.01) and competitive capacity (p < 0.05) against S. Typhimurium FP1, but was more effective in both competitive exclusion experiments against E. coli 3014 (p < 0.01). Nevertheless, Lb. brevis SF9B strain efficiently prevented the adhesion of S. Typhimurium FP1 under in vivo conditions [6], probably owing to the considerable coaggregation capacity, as well as longer in vivo coincubation period than the 1 h of coincubation tested in the respective experiment. The coaggregation enables lactobacilli to manipulate a microenvironment around the pathogenic bacteria and inhibit their growth in the gut by secreting antimicrobial substances at their very close proximity. The results suggest that Lb. brevis SF9B competed more efficiently with E. coli 3014 than S. Typhimurium FP1 since the mechanisms of competition and exclusion differ and are highly specific for each pathogen. Combining plantaricin-producing Lb. plantarum SF9C with S-layer-carrying Lb. brevis SF9B offers an effective strategy to suppress undesirable bacteria such as L. monocytogenes ATCC® 19111™, S. aureus 3048, E. coli 3014 and S. Typhimurium FP1 since the joint application of these potential probiotic strains could result in a broader spectrum of antibacterial activity. Furthermore, in contrast to the application of the sole purified bacteriocin, which could be degraded in the GIT, the application of bacteriocin-producer strain assures its continual production and persistence in the gut [24]. Here we suggested the application of the plantaricin-producing Lb. plantarum SF9C synergistically with Lb. brevis SF9B to eliminate common pathogens, having in mind that Lb. brevis SF9B, as a non-bacteriocin producing strain, was found to possess plnI gene which encodes for the bacteriocin immunity protein [6]. Both strains, SF9B and SF9C, have shown tolerance of the harsh conditions of the GIT because their relative survival rate decreased only by 2- and 3-log CFU/mL, respectively, under the conditions mimicking the GIT [6, 7]. The cooperation of coexisting Lactobacillus strains can also be exploited to control bacterial infection for the reestablishment of the disturbed gut microbiota associated with certain diseases [14]. Therefore, the potential of plantaricin-producing SF9C and S-layer-carrying SF9B strains to compete within healthy or disturbed gut microbiota was examined after their application to the healthy and AlCl3-exposed rats. The exposure to AlCl3 can cause a variety of adverse physiological effects in humans and animals, including the disturbance of gut microbiota [11, 34]. After Lactobacillus treatment of rats, the changes in intestinal microbiota composition were evident, not only in the abundance of Lactobacillus genus, but also in the abundance of other bacterial genera. According to the microbiome analysis, Blautia genus was not detected in healthy rats but was identified in the AlCl3-exposed rats in which its ratio decreased on the 3rd and 10th day after the Lactobacillus application. The ratio of Bacteroides and Phascolarctobacterium genera before the Lactobacillus treatment was higher in the AlCl3-exposed rats than in the healthy rats, but on the 3rd day after the Lactobacillus administration the ratio of these genera was reduced in AlCl3-exposed rats while it increased in healthy rats, compared to the ratio of these genera before Lactobacillus application. Furthermore, the abundance of the Bifidobacterium genus remained unchanged before and after the Lactobacillus treatment in both healthy and the AlCl3-exposed rats. Clostridium and Adlercruetzia genera were evenly present in both groups before the Lactobacillus application, while on the 3rd and 10th day after Lactobacillus addition the ratio of Adlercruetzia genus decreased in both groups and the ratio of Clostridium genus decreased only in AlCl3-exposed rats. An abundant prevalence of Lactobacillus spp. was observed in the microbiota of the Lactobacillus treated rats even 10 days after the Lactobacillus application compared to the microbiota of the healthy and AlCl3-exposed rats. This increased abundance of the Lactobacillus genus possibly reflected an adaptation of Lb. plantarum SF9C and Lb. brevis SF9B in GIT as demonstrated by PCR-DGGE. However, besides allochthone lactobacilli SF9B and SF9C, PCR-DGGE indicated a presence of other commensal lactobacilli, suggesting the possible impact of the applied Lactobacillus strains on the competitive ability of autochthonous strains. The obtained results emphasised the influence of the applied Lactobacillus strains on rat microbiota composition, which will be valuable for further experiments on more experimental animals to investigate interactions of specific features of Lactobacillus strains, such as Slps or bacteriocin production, with the commensal members of gut microbiota. Further studies are needed to better understand the probiotic effects of these two strains on a healthy and disturbed gut microbiome composition and function, and the possible impacts on other parameters important in alleviating AlCl3-induced toxicity in host.
Conclusion
The results of this research support an enhanced functionality potential of the joined application of Lb. plantarum SF9C and Lb. brevis SF9B strains in vivo. The cooperation between the two strains could result in a facilitated adhesion of Lb. plantarum SF9C due to the competitive pathogen exclusion by the coexisting Lb. brevis SF9B. Simultaneously, SF9B strain could benefit from the pathogen inhibition due to plantaricin production by SF9C strain, resulting in a broader spectrum of antibacterial activity. The plantaricin- and S-layer-expressing Lactobacillus strains could be promising probiotic candidates for combined application in functional food and for the treatment of different disorders linked with a dysbiosis of gut microbiota, which require further investigation.
Materials and methods
Bacterial strains, culture media and cultivation conditions
Table 3 shows bacterial strains and cultivation conditions used in this study.
Table 3.
Bacterial strains used in this study
| Bacterial strain | Cultivation conditions | References |
|---|---|---|
| Lb. brevis SF9B | MRS, 37 °C, microaerophilic | Banić et al. [6] |
| Lb. plantarum SF9C | MRS, 37 °C, microaerophilic | This study |
| E. coli 3014 | BHI broth, 37 °C, aerobic | CIM-FFTB |
| S. Typhimurium FP1 | BHI broth, 37 °C, aerobic | CIM-FFTB |
| L. monocytogenes ATCC® 19111™ | BHI broth, 37 °C, aerobic | ATCC |
| S. aureus 3048 | BHI broth, 37 °C, aerobic | CIM-FFTB |
CIM-FFTB—Culture collection of the Laboratory for Antibiotic, Enzyme, Probiotic and Starter Culture Technologies, Faculty of Food Technology and Biotechnology, University of Zagreb
ATCC American Type Culture Collection
Banić et al. [6] had already characterized S-layer-carrying Lb. brevis SF9B. Strains are deposited in the Culture Collection of the Laboratory for Antibiotic, Enzyme, Probiotic and Starter Culture Technologies, Faculty of Food Technology and Biotechnology, University of Zagreb (CIM-FFTB) and are maintained as frozen stock at -80 °C in appropriate medium supplemented with 15% (v/v) glycerol.
Human cell line, culture medium and cultivation conditions
Enterocyte-like Caco-2 cells (Ruđer Bošković Institute, Zagreb, Croatia) were grown as monolayer cultures in RPMI 1640 medium (GIBCO, Carlsbad, CA, USA), supplemented with 15% fetal bovine serum (GIBCO, Carlsbad, CA, USA) and 4500 mg/L glucose. Cells were grown up to confluence at 37 °C and 5% CO2 in T-flasks, trypsinised and seeded into 24-multiwell plates. Prior to experiments, cells reached sub-confluence.
DNA isolation and PCR analysis
Total genomic DNA, both for PCR analysis of the bacteriocin genes or WGS, was extracted according to the method of Leenhouts et al. [35] with minor modifications. The purity and concentration of the extracted DNA were then determined by using a BioSpec-Nano spectrophotometer (Shimadzu, Kyoto, Japan) and the extracted DNA was stored at − 20 °C. PCR screening of the bacteriocin structural genes was performed with primers listed in Table 4. Amplification of DNA fragments was performed in 50-µL reaction mixtures containing 25 µL Emerald Amp MAX HS PCR Mastermix Premix (TaKaRa, Ohtsu, Japan), 200 nmol/L each oligonucleotide primer, 300 ng DNA template and EmeraldAmp dH2O. A negative control, which contained all reagents except the DNA template, was used to detect contamination or non-specific amplification. The amplification was carried out in an Eppendorf Mastercycler personal thermal cycler (Eppendorf, Hamburg, Germany) using the conditions described by the authors cited in Table 4. PCR-amplified products were separated by electrophoresis in a 2% agarose gel, stained with ethidium bromide (0.5 μg/mL) and visualised on a MiniBIS Pro transilluminator (DNR Bio-Imaging Systems Ltd, Jerusalem, Israel) at 254 nm and images were captured by the GelCapture software v. 7.1 (DNR Bio-Imaging Systems Ltd, Jerusalem, Israel). The λ DNA HindIII (Fermentas, Waltham, MA, Canada) and 100 bp DNA Ladder (Invitrogen, Carlsbad, CA, USA) were used as molecular size standards.
Table 4.
PCR primers used for the amplification of the plantaricin-related genes
| Target gene | Bacteriocin | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon size (bp) | References |
|---|---|---|---|---|---|
| plnA | Plantaricin A | GTACAGTACTAATGGGAG | CTTACGCCAATCTATACG | 450 | Ben Omar et al. [9] |
| plnEF | Plantaricin EF | GGCATAGTTAAAATTCCCCCC | CAGGTTGCCGCAAAAAAAG | 428 | Ben Omar et al. [9] |
| plnJ | Plantaricin J | TAACGACGGATTGCTCTG | AATCAAGGAATTATCACATTAGTC | 475 | Ben Omar et al. [9] |
| plnNC8 | Plantaricin NC8 | GGTCTGCGTATAAGCATCGC | AAATTGAACATATGGGTGCTTTAAATTC | 207 | Maldonado et al. [37] |
| plnS | Plantaricin S | GCCTTACCAGCGTAATGCCC | CTGGTGATGCAATCGTTAGTTT | 320 | Ben Omar et al. [9] |
| plnW | Plantaricin W | TCACACGAAATATTCCA | GGCAAGCGTAAGAAATAAATGAG | 165 | Holo et al. [26] |
WGS and identification of genes encoding bacteriocins
Genomic DNA was prepared according to Frece et al. [20]. Genome sequencing was done using a paired-end approach as essentially described in Banić et al. [6]. Briefly, the Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, USA) was used to construct a library. The library was processed with the Illumina cBot and sequenced on the MiSeq 2500 (Illumina, San Diego, CA,USA) pair-end with 300 cycles per read. Contigs were classified as belonging to Lb. plantarum when obtaining the best blastn v. 2.2.27 hit [2] in the NCBI nt database. RAST server, which identifies protein-encoding, rRNA and tRNA genes, assigns functions to the genes and predicts which subsystems are represented in the genome [5], was used for the annotation, distribution and categorization of all sequenced genes. The assembled contigs were compared with the so far identified bacteriocins in the NCBI database using the tblastn v. 2.2.27. To further supplement the annotation, BAGEL4 software was used to predict genes related to bacteriocin synthesis [46]. The input file was the genome sequence of Lb. plantarum SF9C in a fasta file. Conserved genes associated with the bacteriocin synthesis were retrieved using the RAST server [5]. Additionally, whole genome sequences were pairwise aligned with’run-mummer3‘to detect alignments and SNPs. The plot was computed with R package hclust, based on SNP frequency. SNP hierarchical clustering was created based on a high similarity of whole genome sequences available in the NCBI microbial genome database. The closest whole genome sequences of the 11 Lactobacillus strains in respect to the symmetrical and gapped identities among the draft and the complete genome sequences were selected for comparison. The references of the respected Lactobacillus strains isolated from different sources are as follows: WCFS1 [29], NC8 [4], RI-113 [28], B21 [23], BDGP2 [49], SF15C, SF9C, SF15B and SF9B [8], ATCC367 [36] and NCFM [1].
Furthermore, the 3D structure homology was modelled using the SWISS-MODEL server (https://swissmodel.expasy.org/) based on the alignment of the amino acid sequences of the core peptides, generated from BAGEL4 software. Additionally, helix properties of the plantaricins were calculated using heliQuest web server [22].
In vitro assays
Testing of antimicrobial activity
The antimicrobial activity of the overnight grown culture of Lb. plantarum SF9C and Lb. brevis SF9B strains was tested against four different test microorganisms: L. monocytogenes ATCC® 19111™, S. aureus 3048, E. coli 3014 and S. Typhimurium FP1 by agar spot test and well-diffusion method respectively. The agar spot test was performed according to Leboš Pavunc et al. [32]. The ratio of the inhibition diameter (ID) to the spot culture diameter (CD) was calculated to determine the effective inhibition ratio (EIR) of SF9C and SF9B strains: ((ID-CD)/CD). Furthermore, antimicrobial activity of the CFS of SF9C and SF9B strains was examined by the agar well-diffusion method, previously described by Kos et al. [31]. CFS was recovered by centrifugation, filtered through a 0.22-µm sterile filter (Millipore Corporation, Billerica, MA, USA) and concentrated up to fivefold in an Amicon cell concentrator (Amicon, Beverly, MA, USA) equipped with a selective (10 kDa) membrane. The proteinaceous nature of potential inhibitory compounds in CFS was examined by treatment with 1 mg/mL proteinase K (Invitrogen, Carlsbad, CA, USA) for 2 h at 37 °C and by heating the samples at 100 °C/30 min, according to Elayaraja et al. [18].
Evaluation of the antibacterial activity after cocultivation with the targeted pathogens
A slightly modified method of Kos et al. [31] served to determine the influence of cocultivation of Lb. plantarum SF9C with L. monocytogenes ATCC® 19111™ and S. aureus 3048 on bacteriocin activity of SF9C strain. The number of viable cells was determined by spot-plate method using the corresponding selective media for each strain: MRS for lactobacilli, Baird-Parker (Oxoid, Hampshire, UK) for S. aureus and ChromoBio (Biolab Diagnostic Laboratory, Budapest, Hungary) for L. monocytogenes in 2-h intervals during the first 10 h, and after 22, 24 and 48 h of incubation. Plates were incubated for 24 h at 37 °C and the number of viable cells was expressed as log CFU/mL. Also, during the experiment, the antibacterial activity of SF9C strain in monoculture and coculture was tested by agar spot test as described above.
Pathogen competition and exclusion assay by Lb. brevis SF9B and Lb. plantarum SF9C on Caco-2 cell line
For exclusion and competition assay experiments, Caco-2 cells were routinely grown in 24-well culture plates until confluent differentiated monolayers were obtained. Cellular monolayers were carefully rinsed three times with PBS (pH = 7.4) before the addition of the bacterial cells. Two separate protocols were followed to assess the ability of viable strains of lactobacilli to inhibit E. coli 3014 and S. Typhimurium FP1 adhesion to Caco-2 cells. For both assays, Lactobacillus strains and pathogens were routinely cultivated; the cells were harvested and prepared in PBS (pH = 7.4) to reach A620 nm = 1 (approximately 109 CFU/mL). The competition assay was performed according to the procedure described by Uroic et al. [45] with some modifications. Lactobacilli and pathogens were co-incubated with Caco-2 monolayer for 1 h. For exclusion assays, Lactobacillus strains were cultured with Caco-2 monolayer for 1 h. Following a 1-h incubation, Caco-2 monolayers were gently washed three times with PBS (pH = 7.4), then pathogens were added and incubated for another 1 h. The 1.0-mL aliquots of the monospecies cultures of pathogenic bacteria together with 1.0 mL of EMEM per well were used as the controls in both assays. In all the above treatments, after the incubation, the non-adhered bacterial cells were removed by washing the Caco-2 monolayers three times with PBS (pH = 7.4). The Caco-2 cells were then lysed by the addition of 0.25% (v/v) Triton X-100 (AppliChem, Darmstadt, Germany) solution at 37 °C for 10 min in order to collect the adherent bacterial cells, and the total numbers of viable adhering Lactobacillus, E. coli and S. Typhimurium were determined by spot-plate method on MRS, Rapid (Biorad, Dubai, United Arab Emirates) and XLD (Biolife, Milano, Italy) agar plates, respectively. The efficiency of pathogen exclusion by Lactobacillus strains was assayed in three biologically independent experiments each with three replicates.
In vivo animal trial
Preparation of Lb. brevis SF9B and Lb. plantarum SF9C strains and administration to rats
Bacterial cultures Lb. brevis SF9B and Lb. plantarum SF9C were grown in 5 mL of MRS broth at 37 °C under anaerobic conditions until the absorbance value reached 1.0 at 620 nm. Thus prepared cultures were mixed in 1:1 (v/v) ratio and inoculated (4%) in 50 mL of MRS broth. After overnight incubation under optimal conditions, the cells were harvested by centrifugation at 5000×g for 10 min, suspended in saline solution and the presence of both strains was microscopically examined. The bacterial suspensions were prepared daily to ensure viability and the CFU was controlled to maintain their constant number administered to a rat as it is described in the next chapter.
Experimental animals
Three-month-old male highly inbred Y59 strain rats, weighing 200 to 250 g (http://www.informatics.jax.org/external/festing/rat/docs/Y59.shtml), obtained from our breeding within the Department of Animal Physiology, Faculty of Science, University of Zagreb, were used in this study. The animals were maintained under a 12/12-h light–dark cycle with free access to food and water and standard housing conditions (room temperature around 25 °C and 60% humidity). They were fed a standard laboratory diet (4 RF 21, Mucedola, Settimo Milanese, Italy) and tap water ad libitum. Maintenance and care of all experimental animals were carried out according to the guidelines of the Republic of Croatia (Law on the Welfare of Animals, NN135/06 and NN37/13) and in accordance with EU Directive 2010/63/EU for animal experiments [40] and in compliance with the Guide for the Care and Use of Laboratory Animals, DHHS Publ. # (NIH) 86-123. The experimental procedure was approved by the Bioethics Committee of the Faculty of Science, University of Zagreb, Croatia (No. HR-POK-012).
Rat study design and sample collection
Male rats belonging to the Y59 inbred strain were randomly divided into 2 equally sized trial groups and housed three per cage in stainless-steel cages, under the same controlled conditions. The rats were treated daily for five consecutive days with a single dose (3 × 109 CFU/mL) of Lb. brevis SF9B and Lb. plantarum SF9C strains suspended in saline solution, starting 24 h after the last treatment as follows: (a) first trial group represented a model of induced aluminium toxicity which was established by intraperitoneally injecting AlCl3 (10 mg/kg) and d-galactose (60 mg/kg) as described by Ulusoy et al. [44] and (b) second group served as healthy (control) group and was injected comparatively with saline solution in the same manner. No side effects were reported following Lactobacillus administration. In order to evaluate the AChE activity, which requires a brain sample, rats had to be sacrificed. Before the sacrifice, the rats were anaesthetized using a mixture of ketamine (75 mg/kg, Narketan®10, Vetoquinol AG, Belp Bern, Switzerland) with xylazine (10 mg/kg, Xylapana® Vetoquinol Biowet Sp., Gorzow, R. Poland). The intestinal mucosal content from each sacrificed rat was scraped and specimens were kept frozen at − 80 °C until the analysis. The brain was removed and frozen at − 80 °C or kept in buffered formaldehyde until the analysis. The brain tissue homogenates were used to assess AChE activity by colorimetric method. AChE activity is expressed in mol/min/g tissue. The brain samples were prepared according to standard paraffin procedure. Changes related to early-stage Alzheimer’s disease were also (un)confirmed by immunohistochemistry using primary antibodies Purified-β-Amyloid, 17-24 Antibody (4G8) diluted 1:2000 (BioLegend, San Diego, CA, USA), Phospho-PHF-Tau (pSer202 + Thr205) Monoclonal Antibody (AT8) diluted 1:500 (Thermo Fisher Scientific, Waltham, MA, USA) and Iba1 diluted 1:250 (Wako Pure Chemical Industries, Osaka, Japan). Photomicrographs were recorded using a digital camera (AxioCam ERc5s, Zeiss, Germany) and processed by a computer program morphometric image analysis (AxioCam ERc5s-ZEN2). The faecal samples were collected from the cages before starting the treatment and on the 3rd and 10th day following the last Lactobacillus administration in triplicates, and stored at − 80 °C until analysis as described in the next chapter.
Bacterial 16S rRNA sequencing and processing using QIIME
Rat faecal samples were collected at the end of the study and the total genomic DNA was extracted using a Maxwell DNA Tissue Kit with automated extraction platform, Maxwell® 16 Research System instrument (Promega, Madison, USA). The final equimolar pool was sequenced on the Illumina MiSeq platform using 341F (5′-CCTACGGGNGGCWGCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′) primers. PCR reactions and 16S sequencing were performed at the Molecular Research LP (MRDNA, Shallowater, Texas, USA). The MiSeq instrument (Illumina) was used for sequencing the 16S amplicons following the manufacturer’s instructions at MRDNA described by Garcia-Mazcorro et al. [21] with slight modifications. Raw 16S data were obtained from Illumina’s basespace as FASTQ files and analysed with the QIIME 2 pipeline using the procedure as described in the ‘moving pictures’ tutorial (https://qiime2.org/).
PCR–DGGE analysis
PCR-DGGE analysis was performed according to Leboš Pavunc et al. [33] with slight modifications in order to check the presence of the Lb. plantarum SF9B and Lb. brevis SF9C in the faeces of Lactobacillus-fed rats. DNA was extracted directly from faecal samples of healthy rats for culture-independent PCR-DGGE analysis, as well as from the bacterial colonies, isolated on MRS agar plates for culture-dependent PCR-DGGE analysis, from faeces of healthy rats sampled before feeding (control), and on the 3rd and 10th day after the application of Lactobacillus SF9B and SF9C strains. In both cases, DNA was isolated using Maxwell DNA Cell Kit with automated extraction platform, Maxwell® 16 Research System instrument (Promega, Madison, USA). The V2–V3 regions of the 16S ribosomal DNA gene of bacteria in the faeces contents or from pure cultures of lactobacilli were amplified with primers HDA1-GC (5′-ACTCCTACGGGAGGCAGCAGT-3′) and HDA2 (5′-GTATTACCGCGGCTGCTGGCAC-3′) [48]. To identify the lactobacilli recovered from rat faeces, the V2–V3 regions of the 16S rRNA gene of the strains were amplified. The amplicons were sequenced using ABI PRISM® 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). A search of sequences deposited in the GenBank DNA database was conducted by using the BLAST algorithm. The identities of the isolates were determined based on the highest score.
Statistical analysis
All the experiments were repeated three times and the results were expressed as mean value of three independent trials ± standard deviation (SD). Statistical significance was appraised by one-way analysis of variance. Pairwise differences between the mean values of groups were determined by the Tukey’s honestly significant difference (HSD) test for post-analysis of variance pairwise comparisons (http://vassarstats.net). Statistical differences between groups were considered significant when p values were less than 0.05.
Supplementary information
Additional file 1: Fig. S1 Plantaricin-related genes of bacteriocinogenic strain Lactobacillus plantarum SF9C detected by PCR with a plantaricin structural gene-specific primers. S—standard (in bp).
Additional file 2: Table S1. Genes of Lb. plantarum SF9C involved in plantaricin production and their known or putative biochemical functions.
Additional file 3: Fig. S2 Effective Inhibition Ratio (EIR) of test microorganisms, resulting from the antimicrobial activity of Lb. plantarum SF9C after the growth in coculture with: a) S. aureus 3048 (▓) and b) L. monocytogenes ATCC® 19111™ (▓), and after the growth of SF9C alone (░), obtained by agar spot test. Each shown value is the mean ± SD. Asterisks indicate statistically significant differences of EIR of test microorganisms obtained by the Lb. plantarum SF9C after the growth in coculture with test microorganisms and alone, at the same incubation time: *p < 0.05, **p < 0.01.
Additional file 4: Fig. S3 Photomicrograph of the sagittal section in a rat cerebellum; a control group (C) and AlCl3-exposed group. Morphological profile of the rat Purkinje cells (stained with Bielschowsky silver staining), diffuse plaques (4G8, scale bar 10 × = 100 µm) and expression of microglia cells markers Iba1 (scale bar 10 × = 100 µm; scale bar 40 × = 20 µm) in the molecular layer (ML) and granular layer (GL) of the cerebellum.
Authors’ contributions
KB, MB, JN, ALP, KU, KD, NO, MK, SR, SS, JZ, AS, JS and BK authors performed the analyses, prepared the manuscript, and contributed to editing and critical reviewing. All authors read and approved the final manuscript.
Funding
This work was supported by Croatian Science Foundation through projects IP-2014-09-7009 and IP-2019-04-2237. Authors also acknowledge financial support of the University of Zagreb, Croatia. The authors declare that there is no conflict of interest.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Animal experiments were carried out in accordance with the guidelines of the Republic of Croatia (Law on the Welfare of Animals, NN135/06 and NN37/13) and in accordance with EU Directive 2010/63/EU for animal experiments [40] and in compliance with the Guide for the Care and Use of Laboratory Animals, DHHS Publ. # (NIH) 86-123. The protocol was approved by the Bioethics Committee of the Faculty of Science, University of Zagreb, Croatia (No. HR-POK-012). All procedures were performed under the anaesthesia, and made to minimize suffering.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12934-020-01365-6.
References
- 1.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci USA. 2005;102(11):3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–404. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Anukam KC, Macklaim JM, Gloor GB, Reid G, Boekhorst J, Renckens B, et al. Genome sequence of Lactobacillus pentosus KCA1: vaginal isolate from a healthy premenopausal woman. PLoS ONE. 2013;8(3):e59239. doi: 10.1371/journal.pone.0059239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aukrust T, Blom H. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res Int. 1992;25:253–261. doi: 10.1016/0963-9969(92)90121-K. [DOI] [Google Scholar]
- 5.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banić M, Uroić K, Leboš Pavunc A, Novak J, Zorić K, Durgo K, et al. Characterization of S layer proteins of potential probiotic starter culture Lactobacillus brevis SF9B isolated from sauerkraut. LWT-Food Sci Technol. 2018;93:257–267. doi: 10.1016/j.lwt.2018.03.054. [DOI] [Google Scholar]
- 7.Beganović J, Kos B, Leboš Pavunc A, Uroić K, Jokić M, Šušković J. Traditionally produced sauerkraut as source of autochthonous functional starter cultures. Microbiol Res. 2014;169(7):623–632. doi: 10.1016/j.micres.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Beganović J, Leboš Pavunc A, Gjuračić K, Špoljarec M, Šušković J, Kos B. Improved sauerkraut production with probiotic strain Lactobacillus plantarum L4 and Leuconostoc mesenteroides LMG 7954. J Food Sci. 2011;76(2):M124–M129. doi: 10.1111/j.1750-3841.2010.02030.x. [DOI] [PubMed] [Google Scholar]
- 9.Ben Omar N, Abriouel H, Keleke S, Sánchez Valenzuela A, Martínez-Cañamero M, Lucas López R, et al. Bacteriocin-producing Lactobacillus strains isolated from poto poto, a Congolese fermented maize product, and genetic fingerprinting of their plantaricin operons. Int J Food Microbiol. 2008;127(1–2):18–25. doi: 10.1016/j.ijfoodmicro.2008.05.037. [DOI] [PubMed] [Google Scholar]
- 10.Chanos P, Mygind T. Co-culture-inducible bacteriocin production in lactic acid bacteria. Appl Microbiol Biot. 2016;100(10):4297–4308. doi: 10.1007/s00253-016-7486-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen P, Miah MR, Aschner M. Metals and neurodegeneration. F1000 Res 5. 2016 doi: 10.12688/f1000research.7431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LMT. Functions and emerging applications of bacteriocins. Curr Opin Biotech. 2017;49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins FWJ, Mesa-Pereira B, O’Connor PM, Rea MC, Hill C, Ross RP. Reincarnation of bacteriocins from the Lactobacillus Pangenomic graveyard. Front Microbiol. 2018;9:1298. doi: 10.3389/fmicb.2018.01298t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dicks LMT, Dreyer L, Smith C, van Staden AD. A review: the fate of bacteriocins in the human gastro-intestinal tract: do they cross the gut-blood barrier? Front Microbiol. 2018;9:2297. doi: 10.3389/fmicb.2018.02297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diep DB, Straume D, Kjos M, Torres C, Nes IF. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides. 2009;30:1562–1574. doi: 10.1016/j.peptides.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Distrutti E, Reilly JA, McDonald C, Ciprian S, Renga B, Lynch MA, et al. Modulation of intestinal microbiota by the probiotic VSL#3 resets brain gene expression and ameliorates the age-related deficit in LTP. PLoS ONE. 2014;9(9):e106503. doi: 10.1371/journal.pone.0106503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobson A, Cotter PD, Ross RP, Hill C. Bacteriocin production: a probiotic trait? Appl Environ Microb. 2012 doi: 10.1128/AEM.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elayaraja S, Annamalai N, Mayavu P, Balasubramanian T. Production, purification and characterization of bacteriocin from Lactobacillus murinus AU06 and its broad antibacterial spectrum. Asian Pac J Trop Biomed. 2014;4:305–311. doi: 10.12980/APJTB.4.2014C537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fimland N, Rogne P, Fimland G, Nissen-Meyer J, Kristiansen PE. Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF. Biochim Biophys Acta. 2008;1784(11):1711–1719. doi: 10.1016/j.bbapap.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Frece J, Kos B, Svetec IK, Zgaga Z, Beganović J, Leboš A, Šušković J. Synbiotic effect of Lactobacillus helveticus M92 and prebiotics on the intestinal microflora and immune system of mice. J Dairy Res. 2009;76:98–104. doi: 10.1017/S0022029908003737. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Mazcorro JF, Lage NN, Mertens-Talcott S, Talcott S, Chew B, Dowd SE, et al. Effect of dark sweet cherry powder consumption on the gut microbiota, short-chain fatty acids, and biomarkers of gut health in obese db/db mice. PeerJ. 2018;6:e4195. doi: 10.7717/peerj.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific a-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 23.Golneshin A, Adetutu E, Ball AS, May BK, Van Hao TT, Smith AT. Complete Genome Sequence of Lactobacillus plantarum Strain B21, a bacteriocin-producing strain isolated from vietnamese fermented sausage Nem Chua. Microbiol Resour Announc. 2015;3(2):e00055–e00115. doi: 10.1128/genomeA.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartmann HA, Wilke T, Erdmann R. Efficacy of bacteriocin-containing cell-free culture supernatants from lactic acid bacteria to control Listeria monocytogenes in food. Int J Food Microbiol. 2011;146:192–199. doi: 10.7717/peerj.4195. [DOI] [PubMed] [Google Scholar]
- 25.Hegarty JW, Guinane CM, Ross RP, Hill C, Cotter PD. Version 1 acteriocin production: a relatively unharnessed probiotic trait? F1000Res. 2016;5:2587. doi: 10.12688/f1000research.9615.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology. 2001;147(Pt 3):643–651. doi: 10.1099/00221287-147-3-643. [DOI] [PubMed] [Google Scholar]
- 27.Hynönen U, Kant R, Lähteinen T, Pietilä TE, Beganović J, Smidt H, et al. Functional characterization of probiotic surface layer protein-carrying Lactobacillus amylovorus strains. BMC Microbiol. 2014;14:199. doi: 10.1186/1471-2180-14-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inglin RC, Meile L, Klumpp J, Stevens MJA. Complete and assembled genome sequence of Lactobacillus plantarum RI-113 isolated from Salami. Genome Announc. 2017;5:e00183–e00217. doi: 10.1128/genomeA.00183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA. 2003;100(4):1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kos B, Beganović J, Jurašić L, Švađumović M, Leboš Pavunc A, Habjanič K, et al. Coculture-inducible bacteriocin biosynthesis of different probiotic strains by dairy starter culture Lactococcus lactis. Mljekarstvo. 2011;61(4):273–282. [Google Scholar]
- 31.Kos B, Šušković J, Beganović J, Gjuračić K, Frece J, Iannaccone C, et al. Characterization of the three selected probiotic strains for the application in food industry. World J Microbiol Biotechn. 2008;24:699–707. doi: 10.1007/s11274-007-9528-y. [DOI] [Google Scholar]
- 32.Leboš Pavunc A, Kos B, Beganović J, Uroić K, Bučan D, Šušković J. Antibiotic susceptibility and antimicrobial activity of autochthonous starter cultures as safety parameters for fresh cheese production. Mljekarstvo. 2013;63(4):185–194. [Google Scholar]
- 33.Leboš Pavunc A, Beganović J, Kos B, Uroić K, Blažić M, Šušković J. Characterization and application of autochthonous starter cultures for fresh cheese production. Food Technol Biotechnol. 2012;50(2):1412–2151. [Google Scholar]
- 34.Ledinski M, Oršolić N, Kukolj M, Odeh D, Mojzeš A, Uroić K, Gačina, L. Analysis of intestine microbiome in the Alzheimer’s disease rat model. In: Book of abstracts of the annual meeting of the croatian immunological society with EFIS on Tour Zagreb, Zagreb, 2017, p. 40.
- 35.Leenhouts K, Kok J, Venema G. Stability of integrated plasmids in the chromosom Lactococcus lactis. Appl Environ Microb. 1990;56(9):2726–2735. doi: 10.1128/AEM.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maldonado A, Ruiz-Barba JL, Jiménez-Díaz R. Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of gram-positive bacteria. Arch Microbiol. 2004;181:8–16. doi: 10.1007/s00203-003-0606-8. [DOI] [PubMed] [Google Scholar]
- 38.Maldonado-Barragán A, Caballero-Guerrero B, Lucena-Padrós H, Ruiz-Barba JL. Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 2013;33:40e47. doi: 10.1016/j.fm.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Mills S, Ross RP, Hill C. Bacteriocins and bacteriophage; a narrow-minded approach to food and gut microbiology. FEMS Microbiol Rev. 2017;022(41):S129–S153. doi: 10.1093/femsre/fux022. [DOI] [PubMed] [Google Scholar]
- 40.OJEU Directive 2010/63/EU of the European Parliament and of the Council on the protection of animals used for scientific purposes. Off J Eur Union. 2010;276:33–79. [Google Scholar]
- 41.Rogne P, Haugen C, Fimland G, Nissen-Meyer J, Kristiansen PE. Three-dimensional structure of the two-peptide bacteriocin plantaricin JK. Peptides. 2009;30(9):1613–1621. doi: 10.1016/j.peptides.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, et al. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl Environ Microb. 2013;79:1221–1231. doi: 10.1128/AEM.03056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian F, Yu L, Zhai Q, Xiao Y, Shi Y, Jiang J, Liu X, Zhao J, Zhang H, Chen W. The therapeutic protection of a living and dead Lactobacillus strain against aluminum-induced brain and liver injuries in C57BL/6 mice. PLoS ONE. 2017;12(4):e0175398. doi: 10.1371/journal.pone.0175398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulusoy HB, Sonmez MF, Kilic E, Caliskan K, Bkaraca B, Kara M, et al. Intraperitoneal administration of low dose aluminium in the rat: how good is it to produce a model for Alzheimer disease. Arch Ital Biol. 2015;153:266–278. doi: 10.12871/00039829201543. [DOI] [PubMed] [Google Scholar]
- 45.Uroić K, Novak J, Hynӧnen U, Pietilä TE, Leboš Pavunc A, Kant R, Šušković J. The role of S-layer in adhesive and immunomodulating properties of probiotic starter culture Lactobacillus brevis D6 isolated from artisanal smoked fresh cheese. LWT-Food Sci Technol. 2016;69:625–632. doi: 10.1016/j.lwt.2016.02.013. [DOI] [Google Scholar]
- 46.van Heel AJ, de Jong A, Song C, Viel JH, Kok J, Kuipers OP. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46(W1):W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VanPijkeren JP, Tolle PW. Comparative and functional genomics of the genus Lactobacillus. In: Ljungh Å, Wadström T, editors. Lactobacillus molecular biology: from genomics to probiotics. Norfolk: Caister Academic Press; 2013. pp. 59–82. [Google Scholar]
- 48.Walter J, Tannock GW, Tilsala-Timisjarvi A, Rodtong S, Loach DM, Munro K, Alatossava T. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl Environ Microb. 2000;66:297–303. doi: 10.1128/aem.66.1.297-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan KH, Yu C, Park S, Hammonds AS, Booth BW, Celniker SE. Complete genome sequence of Lactobacillus plantarum Oregon-R-modENCODE strain BDGP2 isolated from Drosophila melanogaster gut. Microbiol Resour Announc. 2017;5(41):e01155–e01217. doi: 10.1128/genomeA.01155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu L, Zhai Q, Liu X, Wang G, Zhang Q, Zhao J, Narbad A, Zhang H, Tian F, Chen W. Lactobacillus plantarum CCFM639 alleviates aluminium toxicity. Appl Microbiol Biotechnol. 2016;100:1891–1900. doi: 10.1007/s00253-015-7135-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1 Plantaricin-related genes of bacteriocinogenic strain Lactobacillus plantarum SF9C detected by PCR with a plantaricin structural gene-specific primers. S—standard (in bp).
Additional file 2: Table S1. Genes of Lb. plantarum SF9C involved in plantaricin production and their known or putative biochemical functions.
Additional file 3: Fig. S2 Effective Inhibition Ratio (EIR) of test microorganisms, resulting from the antimicrobial activity of Lb. plantarum SF9C after the growth in coculture with: a) S. aureus 3048 (▓) and b) L. monocytogenes ATCC® 19111™ (▓), and after the growth of SF9C alone (░), obtained by agar spot test. Each shown value is the mean ± SD. Asterisks indicate statistically significant differences of EIR of test microorganisms obtained by the Lb. plantarum SF9C after the growth in coculture with test microorganisms and alone, at the same incubation time: *p < 0.05, **p < 0.01.
Additional file 4: Fig. S3 Photomicrograph of the sagittal section in a rat cerebellum; a control group (C) and AlCl3-exposed group. Morphological profile of the rat Purkinje cells (stained with Bielschowsky silver staining), diffuse plaques (4G8, scale bar 10 × = 100 µm) and expression of microglia cells markers Iba1 (scale bar 10 × = 100 µm; scale bar 40 × = 20 µm) in the molecular layer (ML) and granular layer (GL) of the cerebellum.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.