Abstract
Background
Cutaneous melanoma is one of the most aggressive and lethal skin cancers. It is greatly important to identify prognostic biomarkers to guide the clinical management. However, it is technically challenging for untrained researchers to process high dimensional profiling data and identify potential prognostic genes in profiling datasets.
Methods
In this study, we developed a webserver to analyze the prognostic values of genes in cutaneous melanoma using data from TCGA and GEO databases. The webserver is named Online consensus Survival webserver for Skin Cutaneous Melanoma (OSskcm) which includes 1085 clinical melanoma samples. The OSskcm is hosted in a windows tomcat server. Server-side scripts were developed in Java script. The database system is managed by a SQL Server, which integrates gene expression data and clinical data. The Kaplan–Meier (KM) survival curves, Hazard ratio (HR) and 95% confidence interval (95%CI) were calculated in a univariate Cox regression analysis.
Results
In OSskcm, by inputting official gene symbol and selecting proper options, users could obtain KM survival plot with log-rank P value and HR on the output web page. In addition, clinical characters including race, stage, gender, age and type of therapy could also be included in the prognosis analysis as confounding factors to constrain the analysis in a subgroup of melanoma patients.
Conclusion
The OSskcm is highly valuable for biologists and clinicians to perform the assessment and validation of new or interested prognostic biomarkers for melanoma. OSskcm can be accessed online at: http://bioinfo.henu.edu.cn/Melanoma/MelanomaList.jsp.
Keywords: Cutaneous melanoma, Survival, Prognosis, Biomarker
Background
Cutaneous melanoma (CM) is one of the most lethal malignancies of skin [1]. It was estimated that 287,700 new cases of melanoma and 60,700 deaths of melanomas occurred worldwide in 2018 [2]. Patients with metastatic melanoma have a shorter long-term survival time. Moreover, survival outcomes can vary widely among patients even within the same stage due to the biological heterogeneity of melanoma. At present, the methods commonly used in the treatment of melanoma include surgical resection, chemotherapy and immunotherapy. Only a few patients with advanced melanoma have a persistent response to surgical resection and chemotherapy. Some researchers have used mouse models to analyze the causes of drug resistance, possibly due to changes in metabolic levels in the state of obesity [3, 4]. Weight control can improve the effectiveness of medications and help reduce melanoma metastasis [5]. In addition, the combination of chemotherapy drugs may improve drug resistance [6, 7]. However, because of the molecular heterogeneity, not all the melanoma patients responded well to the treatments. Mutant BRAF has been shown to be significantly associated with worsen overall survival and metastasis free survival of melanoma [8], meanwhile mutant BRAF has been also proven to be a good therapeutic target for melanoma, but the resistance of small molecule drugs against mutant BRAF for melanoma is invariably observed [9]. Therefore, it is imperative to develop novel prognostic biomarkers for risk stratification and treatment optimization in melanoma patients. The specific and novel biomarker may provide the opportunities for guidance of personalized therapeutic interventions and new therapeutic target development.
High-throughput RNA-sequencing (RNA-Seq) has been shown to successfully measure gene expression, discover novel transcripts and identify differentially expressed genes [10]. BRAF and NRAS mutations have been used as molecular biomarkers in evaluating the clinical course of melanoma. Identification of novel molecular biomarkers becomes an area of interests to clinicians and researchers. Ideally, prognostic biomarkers are sensitive, specific, reliable, rapidly analyzable and cost effective. To date, a number of prognostic biomarkers have been proposed in melanoma [11]; however, most of these putative biomarkers lack independent validation in multiple cohorts. Mining available transcriptome data with appropriate clinical follow-up information offers opportunities to prescreen and validate new prognostic biomarkers [12]. Currently, there are several web-browsers, such as PRECOG [13], KM plotter [14] and CaPSSA [15], which have provided survival analysis based on gene expression. However, most of these prognostic analysis web servers only provide data from TCGA, without data from other sources such as GEO and published literatures. As we all know, the most important and difficult part of the biomarker development is to validate the performance of potential biomarker in multiple independent datasets, in this current study, we developed an Online consensus Survival webserver for Skin Cutaneous Melanoma, named OSskcm, which analyzes tumor gene expression profiles and clinical follow-up information of 1085 melanoma patients from multiple independent cohorts. The OSskcm webserver is registration-free and can assist biologists and clinicians to evaluate the prognostic potency of genes of interests and identify potential therapeutic targets.
Materials and methods
Expression profiling and clinical follow-up data used in OSskcm were collected from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) by searching with the keywords of “cutaneous melanoma” and “survival”. Only datasets containing mRNA expression profiling data, clinical survival information, and at least 20 cutaneous melanoma cases were included. The Kaplan–Meier (KM) survival curves were set up using a central server, Hazard ratio (HR) and 95% confidence interval (95%CI) were calculated in a univariate Cox regression analysis. Risk factors, including race, stage, gender, age and type of therapy, can be selected for a subgroup analysis. The OSskcm is hosted in a windows tomcat server. Server-side scripts were developed in Java script, which control the request of analysis and return the analysis results. The database system is managed by a SQL Server, which integrates gene expression data and clinical data. The central server for OSskcm can be accessed at http://bioinfo.henu.edu.cn/Melanoma/MelanomaList.jsp. More details of the methods of OSskcm development have been described [16–19].
Results
Clinical characteristics of cutaneous melanoma cohorts in OSskcm
We collected 1085 unique patients, including 615 patients from six GEO datasets and 470 patients from TCGA dataset. These melanoma samples include 221 primary cutaneous melanomas, 851 metastatic melanomas, and the tumor origin of 13 patients was unknown. (Table 1). The median age of patients is 59 years old. 762 patients have overall survival (OS) data, and the median overall survival is 39.3 months. In addition, 475 patients have progression-free survival (PFS) data, 665 patients have disease-specific survival (DSS) data, 470 patients have progression-free interval (PFI) data, and 150 patients have distant metastasis-free survival (DMFS) data.
Table 1.
Clinical properties of cutaneous melanoma patients in OSskcm
| GEO ID | References | Platform | No. of samples | Death event | Median overall survival (months) | Ages (years) | Gender (male/female) | Primary/metastatic | Stage (I/II/III/IV) |
|---|---|---|---|---|---|---|---|---|---|
| GSE17275 | [20] | GPL1930 | 60 | 41 | 64.00 (46.25–89.50) | NA | NA | 20/40 | 2/8/19/31 |
| GSE22155 | [21] | 70 | 60 | 7.27 (2.10–13.80) | 56.63 ± 14.58 | 39/31 | 0/70 | 0/0/3/67 | |
| GSE46517 | [22] | GPL96 | 84 | 40 | 71 (55–89)a | 77.03 ± 26.37 | 39/24c | 31/53 | 12/15/11/24 |
| GSE50509 | [23] | GPL10558 | 19 | 15 | 18.11 (8.63–26.53) | 57.68 ± 15.49 | 12/7 | 0/19 | NA |
| GSE65904 | [24] | GPL10558 | 214 | 102 | 17.80 (7.03–41.83)b | 62.35 ± 14.40 | 124/89‡ | 16/188‡ | NA |
| GSE98394 | [25] | GPL16791 | 51 | 18 | 93.50 (35.00–111.00) | NA | 31/20 | 51/0 | 12/22/10/0‡ |
| GSE19234 | [26] | GPL570 | 38 | 24 | 38.08 (23.57–65.90) | 62.66 ± 17.86 | 24/14 | 0/38 | 0/0/34/4 |
| GSE53118 | [27] | GPL6884 | 79 | 47 | 79.74 (28.81–120.05) | 55.49 ± 15.27 | 50/29 | 0/79 | 0/0/79/0 |
| TCGA | [28] |
Illumina HiSeqV2 |
470 | 216 | 34.45 (14.90–75.17) | 58.22 ± 15.73 | 290/180 | 103/364‡ | 77/140/171/23‡ |
| Total | 1085 | 563 | 39.30 (15.92–88.00) | 59.14 ± 15.55 | 609/394 | 221/851 | 131/215/268/149 |
NA not available
aThe survival endpoint was defined as event-free survival from resection until death
bThe survival endpoint was defined as disease-specific survival
cPartial data missing
The application of OSskcm webserver
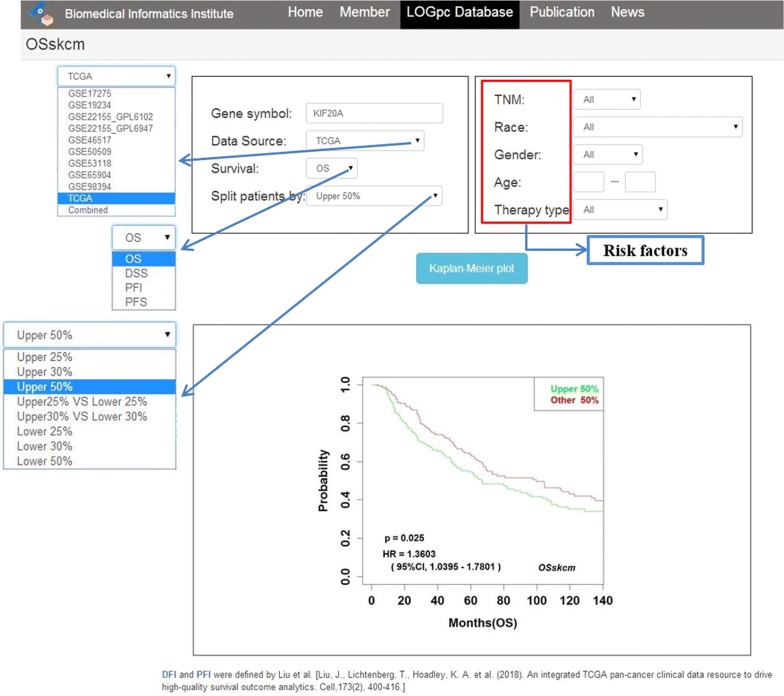
To apply OSskcm to determine the prognostic value of gene of interest, users only need to input an official gene symbol into “Gene symbol” dialog box, and choose “Data source” as either one individual dataset or combined datasets, then select one of the “Survival” terms such as OS, PFS, DSS or PFI, and select a appropriate cut-off value of gene expression stratification by “Split patients by”. After then click the ‘Kaplan–Meier plot’ button, the KM plots with log-rank P value and HR with 95%CI will be shown on the output web page (Fig. 1). If users are interested in the prognostic significance of biomarkers in a particular subgroup of patients, such as races, tumor stages and treatment methods, they may select corresponding risk factors to filter the patients prior to Kaplan–Meier analysis.
Fig. 1.
The usage and output web page of OSskcm webserver
Validation of previously published cutaneous melanoma biomarkers
A PubMed search was performed using keywords of ‘cutaneous melanoma’, ‘survival’, and ‘biomarker’ to identify genes previously reported as prognostic biomarkers for cutaneous melanoma in the literatures. In total, 30 such prognostic genes were validated in OSskcm (listed in Table 2). These biomarker candidates were generally detected by tissue-based immunohistochemistry or immunofluorescent staining.
Table 2.
Performance of previously published protein prognostic biomarker candidates in OSskcm
| Gene symbol | Literature data | Validation results | ||||||
|---|---|---|---|---|---|---|---|---|
| References | n | Survival endpoint | Prognostic significance of high expression | HR (95%CI) | Log-rank P value | Datasets | Cut off | |
| KLK7 | [20] | 45 | OS | Good | 2.65 (1.27–5.53)† | 0.0095 | GSE17275 | Upper 25% |
| 3.60 (1.48–8.80)† | 0.0049 | GSE19234 | Upper 25% | |||||
| 1.93 (1.40–2.65)† | < 0.0001 | TCGA | Upper 25% | |||||
| MITF | [29] | 200 | OS | Poor | 1.43 (1.09–1.87)† | 0.0104 | TCGA | Upper 50% |
| 3.46 (1.42–8.42)† | 0.0063 | GSE19234 | Upper 50% | |||||
| 3.33 (1.18–9.41)† | 0.0230 | GSE98394 | Upper 50% | |||||
| KIF20A | [30] | 61 | RFS | Poor | 2.17 (1.12–4.20)† | 0.0218 | GSE22155 | Upper 25% |
| 2.56 (1.20–5.47)† | 0.0151 | GSE50509 | Upper 25% | |||||
| 3.21 (1.26–8.20)† | 0.0147 | GSE98394 | Upper 25% | |||||
| 2.44 (1.02–5.83)† | 0.0454 | GSE19234 | Upper 25% | |||||
| CTHRC1 | [31] | 35 | OS | Poor | 3.41 (1.31–8.89)† | 0.0122 | GSE98394 | Upper 25% |
| TFAP2A | [32] | Nearly 600 | DSS | Poor | 1.59 (1.03–2.47)‡ | 0.0379 | GSE65904 | Upper 25% |
| ATF2 | [33] | 544 | OS | Poor | 3.05 (1.56–5.97)† | 0.0012 | GSE22155 | Upper 25% |
| NCOA3 | [34] | 343 | RFS and DSS | Poor | 1.79 (1.17–2.74)‡ | 0.0071 | GSE65904 | Upper 25% |
| BCL2 | [35] | 339 | OS | Good | 0.21 (0.04–0.97)† | 0.0458 | GSE22155 | Upper 25% |
| BIRC5 | [36] | 50 | DFS and OS | Poor | 3.73 (1.44–9.67)† | 0.0068 | GSE98394 | Upper 25% |
| MCAM | [37] | 76 | OS | Poor | 4.66 (1.78–12.18)† | 0.0017 | GSE19234 | Upper 25% |
| PLAT | [38] | 214 | DMFI and OS | Poor | 2.24 (1.16–4.34)† | 0.0164 | GSE22155 | Upper 25% |
| 3.88 (1.47–10.24)† | 0.0063 | GSE98394 | Upper 25% | |||||
| NOS2 | [39] | 132 | DSS and OS | Poor | 1.41 (1.07–1.85)† | 0.0131 | TCGA | Upper 50% |
| CDKN1B | [40] | 383 | DSS and OS | Poor | 0.48 (0.24–0.95)† | 0.0341 | GSE22155 | Upper 25% |
| 0.69 (0.50–0.95)† | 0.0235 | TCGA | Upper 25% | |||||
| BCL6 | [41] | 88 | 6-year OS | Poor | 0.57 (0.40–0.80)† | 0.0011 | TCGA | Upper 25% |
| FXYD5 | [42] | 115 | OS | Poor | 3.10 (1.24–7.76)† | 0.0159 | GSE19234 | Upper 25% |
| DDIT3 | [43] | 106 | OS | Good | 5.74 (2.18–15.13)† | 0.0004 | GSE98394 | Upper 25% |
| MCAT | [44] | 1270 | DFI and OS | Poor | 5.75 (1.26–26.10)† | 0.0236 | GSE22155 | Upper 25% |
| 4.51 (1.72–11.82)† | 0.0021 | GSE98394 | Upper 25% | |||||
| CTNNB1 | [45] | 202 | DSS | Good | 1.55 (1.02–2.37)‡ | 0.0412 | GSE65904 | Upper 25% |
| 1.75 (1.15–2.67)‡ | 0.0088 | GSE65904 | Upper 25% | |||||
| AKT1 | [46] | 222 | 5-year DSS or OS | Poor | 6.41 (2.39–17.23)† | 0.0002 | GSE98394 | Upper 25% |
| 1.53 (1.13–2.06)† | 0.0056 | TCGA | Upper 25% | |||||
| RUNX3 | [47] | 421 | 5-year OS | Good | 3.75 (1.36–10.33)† | 0.0107 | GSE50509 | Upper 25% |
| 5-year DSS | 1.81 (1.18–2.76)‡ | 0.0062 | GSE65904 | Upper 25% | ||||
| BBC3 | [48] | 158 | 5-year DSS or OS | Poor | 3.62 (1.38–9.52)† | 0.0092 | GSE98394 | Upper 25% |
| MMP2 | [49] | 157 | DSS and RFS | Poor | 1.41 (1.06–1.89)‡ | 0.0197 | TCGA | Upper 50% |
| SPP1 | [50] | 345 | RFS | Poor | 9.42 (3.46–25.67)† | < 0.0001 | GSE98394 | Upper 25% |
| TNC | [51] | 98 | DFS | Poor | 1.54 (1.01–2.34)‡ | 0.0434 | GSE65904 | Upper 25% |
| CCNA2 | [52] | 245 | RFS | Poor | 2.23 (1.02–4.88)† | 0.0437 | GSE50509 | Upper 25% |
| RGS1 | [53] | 40 | DSS | Poor | 2.74 (1.03–7.24)† | 0.0425 | GSE98394 | Upper 25% |
| 3.24 (1.31–8.00)† | 0.0110 | GSE19234 | Upper 25% | |||||
| 2.66 (1.07–6.65)† | 0.0357 | GSE19234 | Upper 25% | |||||
| SPARC | [54] | 112 | DFS | Poor | 2.78 (1.21–6.34)† | 0.0154 | GSE50509 | Upper 25% |
| CXCR4 | [55] | 71 | DFS and OS | Poor | 0.70 (0.51–0.97)† | 0.0315 | TCGA | Upper 25% |
| RBM3 | [56] | 246 | OS | Good | NS | NS | – | – |
| EPAS1 | [57] | 46 | DSS | Poor | 3.51 (1.56–7.91)† | 0.0024 | GSE50509 | Upper 25% |
NS not significance, RFS recurrence-free survival, DFS disease-specific survival, DFI disease-free interval, DMFI distant metastasis-free interval
†,‡HR (95%CI) and Log-rank P value of overall survival (OS) and disease-specific survival (DSS)
The analysis of these reported prognostic biomarkers in OSskcm showed that the prognostic roles of 22 genes were consistent with previous findings, RBM3 gene had no statistically significance on prognosis, and the other 7 genes (KLK7, CXCR4, CDKN1B, BCL6, CTNNB1, RUNX3 and DDIT3) had opposite prognostic trends compared to literatures. The analysis results were presented in Table 2.
Screening of new prognostic biomarkers for cutaneous melanoma
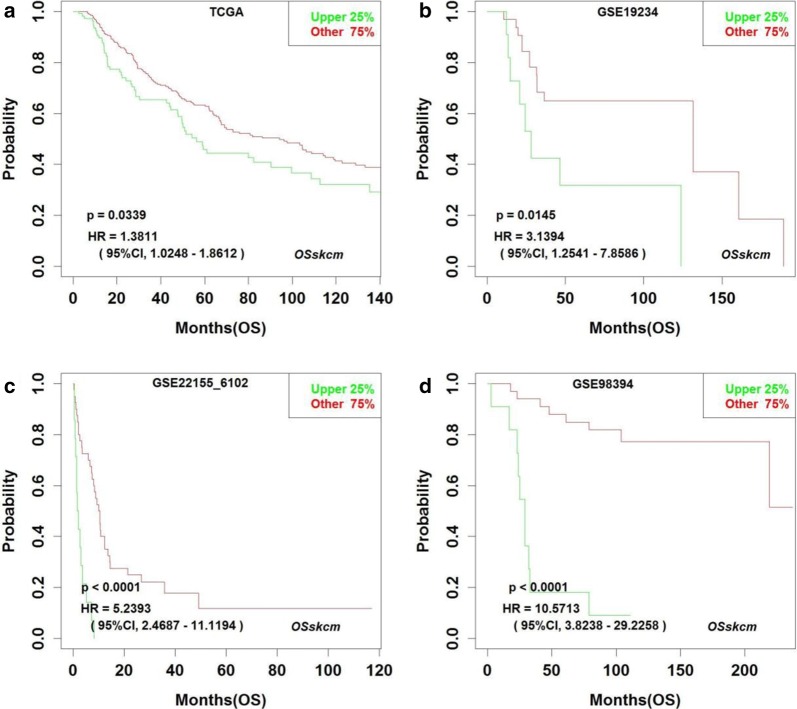
OSskcm can also be used to screen novel prognostic biomarkers for cutaneous melanoma, where OS, DSS, PFS, PFI and DMFS can be investigated. By OSskcm, we found that high expression of SAE1 gene is associated with poor prognosis of cutaneous melanoma (Fig. 2), and the prognostic potency of SAE1 gene has not been previously reported in cutaneous melanoma.
Fig. 2.
SAE1 is identified as an unfavorable prognostic biomarker in OSskcm. Overall survival (OS) curve of cutaneous melanoma patients based on TCGA (a), GSE19234 (b), GSE22155 (c) and GSE98394 (d) data. Upper 25%: the SKCM cases with ranked top 25% higher expression level for the inputted gene; Other 75%: the SKCM cases with ranked bottom 75% lower expression level of the inputted gene
Discussion
Due to the variant prognosis of cutaneous melanoma patients, the development of molecular prognosis biomarkers is significant. Here, we collected multiple large transcriptomic datasets to increase the statistical power for analyzing the association between the investigated marker and survival rate, and developed a freely accessible webserver OSskcm to estimate the prognostic value of any inputted gene in a large cohort of patients, by which KM survival curves as well as HR and log-rank P values could be outputted and presented. OSskcm is a webserver that can mutually validate prognostic biomarkers of cutaneous melanoma in multiple data sets. A total of 1085 patients of cutaneous melanoma with RNA-seq data from clinical tissues and clinical information were included in OSskcm. In addition, risk factors, including race, stage, gender, age and therapy type, can be selected for subgroup analysis. Clinical outcome data of OS, PFS, DSS, PFI, and DMFS was included in analysis.
We tested the performance of OSskcm using 30 previously reported cutaneous melanoma prognostic biomarkers. Among these, 22 genes were validated in OSskcm, but the prognostic significance of RBM3, KLK7, CXCR4, CDKN1B, BCL6, CTNNB1, RUNX3 and DDIT3 genes were inconsistent between literatures and OSskcm. It may be because the OSskcm utilizes mRNA expression data while all previously published biomarkers were studied based on the protein level. It is known that there is an inconsistency between the levels of mRNA and protein due to intracellular modifications, such as post-transcriptional regulation, protein translation and post-translational regulation. In addition, the prognostic significance of a protein may be determined by its subcellular localization. For example, loss of nuclear CDKN1B expression is correlated with a worse 5-year survival of primary melanoma patients in Kaplan–Meier analysis, but gain of cytoplasmic CDKN1B was associated with a poor 5-year survival of metastatic melanoma patients.
KIF20A and RGS1 genes have been reported to play critical roles in the development and progression of cancer, and promote the proliferation, migration and invasion of cancer cells [58, 59]. In OSskcm, KIF20A and RGS1 were found to be strongly associated with cutaneous melanoma prognosis. In addition, we found that SAE1 could be a new prognostic biomarker in cutaneous melanoma. SAE1 is dimeric SUMO Activating Enzyme E1, involves in SUMO conjugation [60]. Breast cancer patients with lower SAE1 expression have been reported to have significantly lower instances of metastatic cancer and increased survival compared to those that express a higher level of SAE1 [61]. Moreover, SAE1 was reported to have the strongest synthetic lethal interactions with K-Ras and can be used to evaluate the aggressiveness of mutated K-Ras-dependent malignancies [62]. It will be interesting to further verify by experiments whether SAE1 gene could be a new prognostic biomarker in cutaneous melanoma.
Conclusion
In summary, by utilizing genome-wide microarray datasets and RNAseq datasets, we built a prognosis webserver, OSskcm, which offer a platform for biologists and clinicians to identify prognostic biomarkers for cutaneous melanoma. Additional more research regarding how to better translate our web server and web server derived biomarkers for practice from local to global health is required [63].
Acknowledgements
Not applicable.
Abbreviations
- OSskcm
Online consensus Survival webserver for Skin Cutaneous Melanoma
- GEO
Gene Expression Omnibus
- TCGA
The Cancer Genome Atlas
- OS
Overall survival
- PFS
Progression-free survival
- DSS
Disease-specific survival
- PFI
Progression-free interval
- DMFS
Distant metastasis-free survival
- RFS
Recurrence-free survival
- DFS
Disease-specific survival
- DFI
Disease-free interval
- DMFI
Distant metastasis-free interval
Authors’ contributions
All authors materially participated in the study and manuscript preparation. XQG, LZ and QW participated in the design and the conception of the study. LJW, ZHL, XCZ, PPT and XZH collected and managed data. XQG, LZ and QW developed methods and performed data analysis. QW built the webserver. YQL provided technical support. LZ and QW wrote the original draft. LXX, YA, GSZ, WZ and XQG reviewed and edited the manuscript. XQG supervised the research process. All authors read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China (No. 81602362), Program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048), Kaifeng Science and Technology Major Project (18ZD008), Supporting grant of Bioinformatics Center of Henan University (No. 2018YLJC01; No. 2019YLXKJC01), Student Innovation and Entrepreneurship Training Program of Henan University (No. 2019101905).
Availability of data and materials
The datasets used and analyzed during the current study are available from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/).
Ethics approval and consent to participate
Not applicable.
Consent for publication
We have obtained consents to publish this paper from all participants of this study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lu Zhang and Qiang Wang contributed equally to this study
References
- 1.Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, et al. Melanoma. Lancet. 2018;392(10151):971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Malvi P, Chaube B, Singh SV, Mohammad N, Vijayakumar MV, Singh S, et al. Elevated circulatory levels of leptin and resistin impair therapeutic efficacy of dacarbazine in melanoma under obese state. Cancer Metab. 2018;6:2. doi: 10.1186/s40170-018-0176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malvi P, Chaube B, Pandey V, Vijayakumar MV, Boreddy PR, Mohammad N, et al. Obesity induced rapid melanoma progression is reversed by orlistat treatment and dietary intervention: role of adipokines. Mol Oncol. 2015;9(3):689–703. doi: 10.1016/j.molonc.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malvi P, Chaube B, Singh SV, Mohammad N, Pandey V, Vijayakumar MV, et al. Weight control interventions improve therapeutic efficacy of dacarbazine in melanoma by reversing obesity-induced drug resistance. Cancer Metab. 2016;4:21. doi: 10.1186/s40170-016-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaube B, Malvi P, Singh SV, Mohammad N, Meena AS, Bhat MK. Targeting metabolic flexibility by simultaneously inhibiting respiratory complex I and lactate generation retards melanoma progression. Oncotarget. 2015;6(35):37281–37299. doi: 10.18632/oncotarget.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammad N, Malvi P, Meena AS, Singh SV, Chaube B, Vannuruswamy G, et al. Cholesterol depletion by methyl-β-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol Cancer. 2014;13:204. doi: 10.1186/1476-4598-13-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreau S, Saiag P, Aegerter P, Bosset D, Longvert C, Helias-Rodzewicz Z, et al. Prognostic value of BRAF(V600) mutations in melanoma patients after resection of metastatic lymph nodes. Ann Surg Oncol. 2012;19(13):4314–4321. doi: 10.1245/s10434-012-2457-5. [DOI] [PubMed] [Google Scholar]
- 9.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508(7494):118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Yang C, Dong L, Zhu J, Wang J, Zhang S. Comparative transcriptome analyses of drought-resistant and—susceptible Brassica napus L. and development of EST-SSR markers by RNA-Seq. J Plant Biol. 2015;58:259–269. [Google Scholar]
- 11.Abbas O, Miller DD, Bhawan J. Cutaneous malignant melanoma: update on diagnostic and prognostic biomarkers. Am J Dermatopathol. 2014;36(5):363–379. doi: 10.1097/DAD.0b013e31828a2ec5. [DOI] [PubMed] [Google Scholar]
- 12.Xie L, Dang Y, Guo J, Sun X, Xie T, Zhang L, et al. High KRT8 expression independently predicts poor prognosis for lung adenocarcinoma patients. Genes. 2019;10(1):36. doi: 10.3390/genes10010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- .Jang Y, Seo J, Kim S, Lee S. CaPSSA: visual evaluation of cancer biomarker genes for patient stratification and survival analysis using mutation and expression data. Bioinformatics. 2019;35(24):5341–5343. doi: 10.1093/bioinformatics/btz516. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Xie L, Dang Y, Sun X, Xie T, Guo J, et al. OSlms: a web server to evaluate the prognostic value of genes in leiomyosarcoma. Front Oncol. 2019;9:190. doi: 10.3389/fonc.2019.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, Wang Q, Yang M, Yuan Q, Dang Y, Sun X, et al. OSblca: a web server for investigating prognostic biomarkers of bladder cancer patients. Front Oncol. 2019;9:466. doi: 10.3389/fonc.2019.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie L, Wang Q, Nan F, Ge L, Dang Y, Sun X, et al. OSacc: gene expression-based survival analysis web tool for adrenocortical carcinoma. Cancer Manag Res. 2019;11:9145–9152. doi: 10.2147/CMAR.S215586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Wang Q, Li N, Ge L, Yang M, An Y, et al. OSuvm: an interactive online consensus survival tool for uveal melanoma prognosis analysis. Mol Carcinog. 2020;59(1):56–61. doi: 10.1002/mc.23128. [DOI] [PubMed] [Google Scholar]
- 20.Martins WK, Esteves GH, Almeida OM, Rezze GG, Landman G, Marques SM, et al. Gene network analyses point to the importance of human tissue kallikreins in melanoma progression. BMC Med Genomics. 2011;4:76. doi: 10.1186/1755-8794-4-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson G, Busch C, Knappskog S, Geisler J, Miletic H, Ringner M, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16(13):3356–3367. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 22.Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS ONE. 2010;5(5):e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20(7):1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 24.Cirenajwis H, Ekedahl H, Lauss M, Harbst K, Carneiro A, Enoksson J, et al. Molecular stratification of metastatic melanoma using gene expression profiling: prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget. 2015;6(14):12297–12309. doi: 10.18632/oncotarget.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badal B, Solovyov A, Di Cecilia S, Chan JM, Chang LW, Iqbal R, et al. Transcriptional dissection of melanoma identifies a high-risk subtype underlying TP53 family genes and epigenome deregulation. JCI Insight. 2017 doi: 10.1172/jci.insight.92102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogunovic D, O’Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106(48):20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann GJ, Pupo GM, Campain AE, Carter CD, Schramm SJ, Pianova S, et al. BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J Investig Dermatol. 2013;133(2):509–517. doi: 10.1038/jid.2012.283. [DOI] [PubMed] [Google Scholar]
- 28.Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 29.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita J, Fukushima S, Jinnin M, Honda N, Makino K, Sakai K, et al. Kinesin family member 20A is a novel melanoma-associated antigen. Acta Derm Venereol. 2012;92(6):593–597. doi: 10.2340/00015555-1416. [DOI] [PubMed] [Google Scholar]
- 31.Eriksson J, Le Joncour V, Nummela P, Jahkola T, Virolainen S, Laakkonen P, et al. Gene expression analyses of primary melanomas reveal CTHRC1 as an important player in melanoma progression. Oncotarget. 2016;7(12):15065–15092. doi: 10.18632/oncotarget.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger AJ, Davis DW, Tellez C, Prieto VG, Gershenwald JE, Johnson MM, et al. Automated quantitative analysis of activator protein-2alpha subcellular expression in melanoma tissue microarrays correlates with survival prediction. Cancer Res. 2005;65(23):11185–11192. doi: 10.1158/0008-5472.CAN-05-2300. [DOI] [PubMed] [Google Scholar]
- 33.Berger AJ, Kluger HM, Li N, Kielhorn E, Halaban R, Ronai Z, et al. Subcellular localization of activating transcription factor 2 in melanoma specimens predicts patient survival. Cancer Res. 2003;63(23):8103–8107. [PubMed] [Google Scholar]
- 34.Rangel J, Torabian S, Shaikh L, Nosrati M, Baehner FL, Haqq C, et al. Prognostic significance of nuclear receptor coactivator-3 overexpression in primary cutaneous melanoma. J Clin Oncol. 2006;24(28):4565–4569. doi: 10.1200/JCO.2006.07.3833. [DOI] [PubMed] [Google Scholar]
- 35.Divito KA, Berger AJ, Camp RL, Dolled-Filhart M, Rimm DL, Kluger HM. Automated quantitative analysis of tissue microarrays reveals an association between high Bcl-2 expression and improved outcome in melanoma. Cancer Res. 2004;64(23):8773–8777. doi: 10.1158/0008-5472.CAN-04-1387. [DOI] [PubMed] [Google Scholar]
- 36.Piras F, Murtas D, Minerba L, Ugalde J, Floris C, Maxia C, et al. Nuclear survivin is associated with disease recurrence and poor survival in patients with cutaneous malignant melanoma. Histopathology. 2007;50(7):835–842. doi: 10.1111/j.1365-2559.2007.02695.x. [DOI] [PubMed] [Google Scholar]
- 37.Pacifico MD, Grover R, Richman PI, Daley FM, Buffa F, Wilson GD. Development of a tissue array for primary melanoma with long-term follow-up: discovering melanoma cell adhesion molecule as an important prognostic marker. Plast Reconstr Surg. 2005;115(2):367–375. doi: 10.1097/01.prs.0000148417.86768.c9. [DOI] [PubMed] [Google Scholar]
- 38.Ferrier CM, Suciu S, van Geloof WL, Straatman H, Eggermont AM, Koops HS, et al. High tPA-expression in primary melanoma of the limb correlates with good prognosis. Br J Cancer. 2000;83(10):1351–1359. doi: 10.1054/bjoc.2000.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekmekcioglu S, Ellerhorst JA, Prieto VG, Johnson MM, Broemeling LD, Grimm EA. Tumor iNOS predicts poor survival for stage III melanoma patients. Int J Cancer. 2006;119(4):861–866. doi: 10.1002/ijc.21767. [DOI] [PubMed] [Google Scholar]
- 40.Chen G, Cheng Y, Zhang Z, Martinka M, Li G. Prognostic significance of cytoplasmic p27 expression in human melanoma. Cancer Epidemiol Biomark Prev. 2011;20(10):2212–2221. doi: 10.1158/1055-9965.EPI-11-0472. [DOI] [PubMed] [Google Scholar]
- 41.Alonso SR, Ortiz P, Pollan M, Perez-Gomez B, Sanchez L, Acuna MJ, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am J Pathol. 2004;164(1):193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishizawa A, Nakanishi Y, Yoshimura K, Sasajima Y, Yamazaki N, Yamamoto A, et al. Clinicopathologic significance of dysadherin expression in cutaneous malignant melanoma: immunohistochemical analysis of 115 patients. Cancer. 2005;103(8):1693–1700. doi: 10.1002/cncr.20984. [DOI] [PubMed] [Google Scholar]
- 43.Korabiowska M, Cordon-Cardo C, Betke H, Schlott T, Kotthaus M, Stachura J, et al. GADD153 is an independent prognostic factor in melanoma: immunohistochemical and molecular genetic analysis. Histol Histopathol. 2002;17(3):805–811. doi: 10.14670/HH-17.805. [DOI] [PubMed] [Google Scholar]
- 44.Weinlich G, Eisendle K, Hassler E, Baltaci M, Fritsch PO, Zelger B. Metallothionein—overexpression as a highly significant prognostic factor in melanoma: a prospective study on 1270 patients. Br J Cancer. 2006;94(6):835–841. doi: 10.1038/sj.bjc.6603028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachmann IM, Straume O, Puntervoll HE, Kalvenes MB, Akslen LA. Importance of P-cadherin, beta-catenin, and Wnt5a/frizzled for progression of melanocytic tumors and prognosis in cutaneous melanoma. Clin Cancer Res. 2005;11(24 Pt 1):8606–8614. doi: 10.1158/1078-0432.CCR-05-0011. [DOI] [PubMed] [Google Scholar]
- 46.Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J Clin Oncol. 2005;23(7):1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Chen G, Cheng Y, Martinka M, Li G. Prognostic significance of RUNX3 expression in human melanoma. Cancer. 2011;117(12):2719–2727. doi: 10.1002/cncr.25838. [DOI] [PubMed] [Google Scholar]
- 48.Karst AM, Dai DL, Martinka M, Li G. PUMA expression is significantly reduced in human cutaneous melanomas. Oncogene. 2005;24(6):1111–1116. doi: 10.1038/sj.onc.1208374. [DOI] [PubMed] [Google Scholar]
- 49.Vaisanen AH, Kallioinen M, Turpeenniemi-Hujanen T. Comparison of the prognostic value of matrix metalloproteinases 2 and 9 in cutaneous melanoma. Hum Pathol. 2008;39(3):377–385. doi: 10.1016/j.humpath.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 50.Rangel J, Nosrati M, Torabian S, Shaikh L, Leong SP, Haqq C, et al. Osteopontin as a molecular prognostic marker for melanoma. Cancer. 2008;112(1):144–150. doi: 10.1002/cncr.23147. [DOI] [PubMed] [Google Scholar]
- 51.Ilmonen S, Jahkola T, Turunen JP, Muhonen T, Asko-Seljavaara S. Tenascin-C in primary malignant melanoma of the skin. Histopathology. 2004;45(4):405–411. doi: 10.1111/j.1365-2559.2004.01976.x. [DOI] [PubMed] [Google Scholar]
- 52.Florenes VA, Maelandsmo GM, Faye R, Nesland JM, Holm R. Cyclin A expression in superficial spreading malignant melanomas correlates with clinical outcome. J Pathol. 2001;195(5):530–536. doi: 10.1002/path.1007. [DOI] [PubMed] [Google Scholar]
- 53.Sun MY, Wang Y, Zhu J, Lv C, Wu K, Wang XW, et al. Critical role for non-GAP function of Gαs in RGS1-mediated promotion of melanoma progression through AKT and ERK phosphorylation. Oncol Rep. 2018;39(6):2673–2680. doi: 10.3892/or.2018.6341. [DOI] [PubMed] [Google Scholar]
- 54.Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Pollan M, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67(7):3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 55.Scala S, Ottaiano A, Ascierto PA, Cavalli M, Simeone E, Giuliano P, et al. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin Cancer Res. 2005;11(5):1835–1841. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- 56.Jonsson L, Bergman J, Nodin B, Manjer J, Ponten F, Uhlen M, et al. Low RBM3 protein expression correlates with tumour progression and poor prognosis in malignant melanoma: an analysis of 215 cases from the Malmo Diet and Cancer Study. J Transl Med. 2011;9:114. doi: 10.1186/1479-5876-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giatromanolaki A, Sivridis E, Kouskoukis C, Gatter KC, Harris AL, Koukourakis MI. Hypoxia-inducible factors 1alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res. 2003;13(5):493–501. doi: 10.1097/00008390-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 58.Liu SL, Lin HX, Qiu F, Zhang WJ, Niu CH, Wen W, et al. Overexpression of kinesin family member 20A correlates with disease progression and poor prognosis in human nasopharyngeal cancer: a retrospective analysis of 105 patients. PLoS ONE. 2017;12(1):e169280. doi: 10.1371/journal.pone.0169280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roh J, Shin SJ, Lee AN, Yoon DH, Suh C, Park CJ, et al. RGS1 expression is associated with poor prognosis in multiple myeloma. J Clin Pathol. 2017;70(3):202–207. doi: 10.1136/jclinpath-2016-203713. [DOI] [PubMed] [Google Scholar]
- 60.Mattoscio D, Chiocca S. SUMO pathway components as possible cancer biomarkers. Future Oncol. 2015;11(11):1599–1610. doi: 10.2217/fon.15.41. [DOI] [PubMed] [Google Scholar]
- 61.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malla C, Aylward P, Ward P. Knowledge translation for public health in low- and middle-income countries: a critical interpretive synthesis. Glob Health Res Policy. 2018;3:29. doi: 10.1186/s41256-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/).