Abstract
Members of the aldehyde dehydrogenase 1A family are commonly acknowledged as hallmarks of cancer stem cells, and their overexpression is significantly associated with poor prognosis in different types of malignancies. Accordingly, treatments targeting these enzymes may represent a successful strategy to fight cancer. In this work we describe a novel series of imidazo[1,2-a]pyridines, designed as aldehyde dehydrogenase inhibitors by means of a structure-based optimization of a previously developed lead. The novel compounds were evaluated in vitro for their activity and selectivity against the three isoforms of the ALDH1A family and investigated through crystallization and modeling studies for their ability to interact with the catalytic site of the 1A3 isoform. Compound 3f emerged as the first in class submicromolar competitive inhibitor of the target enzyme.
Keywords: Aldehyde dehydrogenases; aldehyde dehydrogenase inhibitors; ALDH1A1; ALDH1A2; ALDH1A3; imidazo[1,2-a]pyridines
Aldehyde dehydrogenases (ALDHs) comprise a superfamily of 19 enzymatic NAD(P+) dependent isoforms characterized by distinctive subcellular localization, chemical structure, and substrate binding. They all catalyze the irreversible oxidation of both endogenous and exogenous aldehydes to the corresponding carboxylic acids and CoA esters, thus playing a key role in a wide spectrum of biological functions.1 The ALDH1A subfamily, including the ALDH1A1, ALDH1A2, and ALDH1A3 isoforms, turns the vitamin A metabolite retinal into all-trans retinoic acid (ATRA), 9-cis retinoic acid (9-cis-RA), and 13-cis retinoic acid (13-cis-RA), having a major role in regulating gene expression. Indeed, to date, more than 530 genes have been reported as regulatory targets of RAs, being modulated by these ligands either directly, thanks to the RA response element (RARE) sequence in their promoter, or indirectly, through transcriptional intermediaries.2 Such a massive modulation gives the sense of RA as a crucial mediator and, above all, testifies the functional role of the ALDH1A family in regulating cell morphogenesis and development. Actually, these enzymes are highly expressed in embryonal tissues and in cells showing a stem-like phenotype, including both normal stem cells and cancer stem cells (CSCs).3
Besides acting as a marker for the identification of colonic stem cells, members of the ALDH1A family are able to affect stem cell vitality regulating self-renewal, differentiation, and expansion. In the case of CSCs, ALDH activity is decisive for tumor progression and invasion, so much that a high expression of the different ALDH1A isoforms often has a prognostic value for poor clinical outcomes.4,5 ALDH1A1 activation has been observed in the stem-like population of multiple myeloma and acute myeloid leukemia.6 Moreover, high levels of this isoenzyme predict negative prognosis in patients with esophageal squamous cell carcinoma7 and prostate cancer8 and are reported as responsible for platinum resistance of ovarian cancer cells.9 ALDH1A3 overexpression is associated with worse prognosis in aggressive and/or triple-negative breast cancer,10,11 and it is also responsible for tumorigenicity in nonsmall cell lung cancer.12 In addition, increased expression of this isoform endorses high grade gliomas.13
Accordingly, targeting these enzymes with suitably designed inhibitors represents a successful strategy to fight CSCs, resulting in an innovative and groundbreaking molecular treatment of cancer.14
We recently developed a novel series of ALDH inhibitors bearing the 2,6-diphenyl-imidazo[1,2-a]pyridine core structure, developed as prototypical drug candidates to fight glioma stem-like cells (GSCs). Pursuing a rational optimization of the representative hit GA1115 (Chart 1) and making use of distinctive substitution patterns on the two pendant phenyl rings, we succeeded in modulating the functional profile of the synthesized compounds obtaining fully selective derivatives able to target either ALDH1A1 isoenzyme or the parent ALDH1A3.16 Besides an unquestionable significance as pharmacological tools, the novel compounds displayed also a relevant functional efficacy. Indeed, when tested against well-characterized patient-derived GSCs, representative examples of ALDH1A3 inhibitors displayed antiproliferative efficacy in the nanomolar/picomolar range, thus imposing themselves as novel prototypes of drug candidates deserving further investigation.
Chart 1.
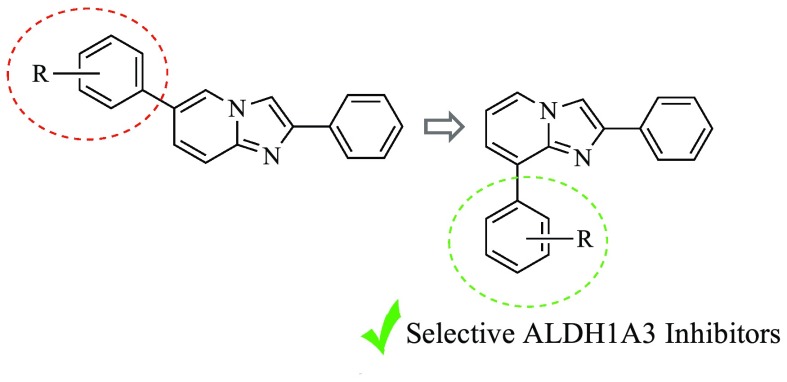
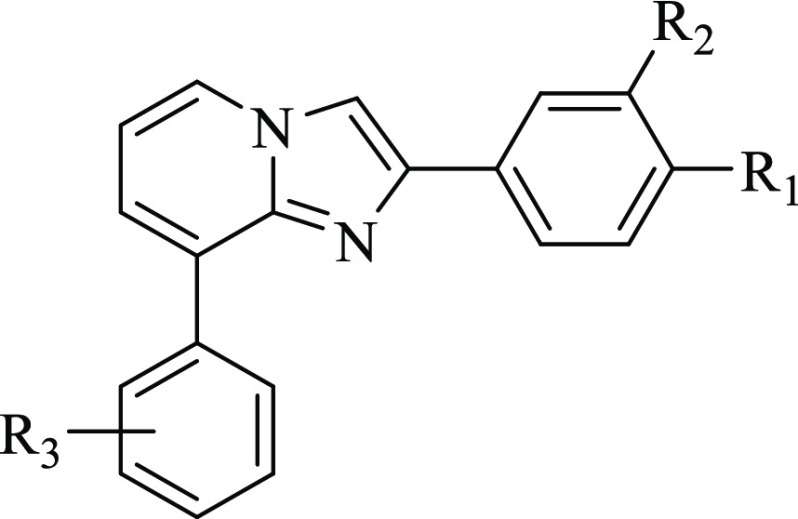
A close inspection of the complex obtained cocrystallizing GA11 with the human recombinant ALDH1A3 revealed that the reference compound gets settled at the entrance of the catalytic tunnel of the enzyme.16 Arranged in this pose, GA11 forecloses access to the natural substrate retinal, thus allowing inhibition of the catalytic activity of the enzyme but was proved to occupy only partly the enzyme site, thus offering interesting clues for the rational optimization of this class of compounds. In particular, the obtained binding mode lead to speculation that shifting the 6-phenyl ring to the 8-position could come in very handy to cast these derivatives into the deepest part of the enzyme binding site, in close proximity to the key catalytic residue C314. Prompted by this speculation, we embarked in the synthesis of a novel series of 8-substituted derivatives, 3a–t (Chart 1), which were investigated for their functional efficacy and mode of interaction against the human recombinant isoforms of the ALDH1A family.
Results and Discussion
Chemistry
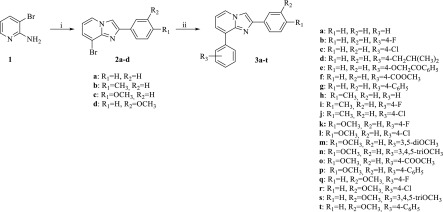
The synthesis of the target inhibitors 3a–t was performed as depicted in Scheme 1. Reaction of the commercially available 2-amino-3-bromo pyridine 1 with the suitably substituted 2-bromo-1-phenylethan-1-one, in the presence of potassium carbonate, gave the corresponding key intermediates 8-bromo-2-phenylimidazo[1,2-a]pyridines 2a–d, which provided the desired inhibitors 3a–t by coupling reactions with differently substituted phenylboronic acids, performed in the presence of Pd(OAc)2 and PPh3 as the catalysts.
Scheme 1. Synthesis of 8-Substituted-Imidazo[1,2-a]pyridine Derivatives 3a–t.
Reagents and conditions: (i) 2-Bromo-1-(4-(3)substituted phenyl)ethan-1-one, K2CO3, Δ; (ii) Substitued-phenylboronic acid, Pd(OAc)2, PPh3, K2CO3, Δ.
Functional Evaluation of the Novel Imidazo[1,2-a]pyridine Derivatives
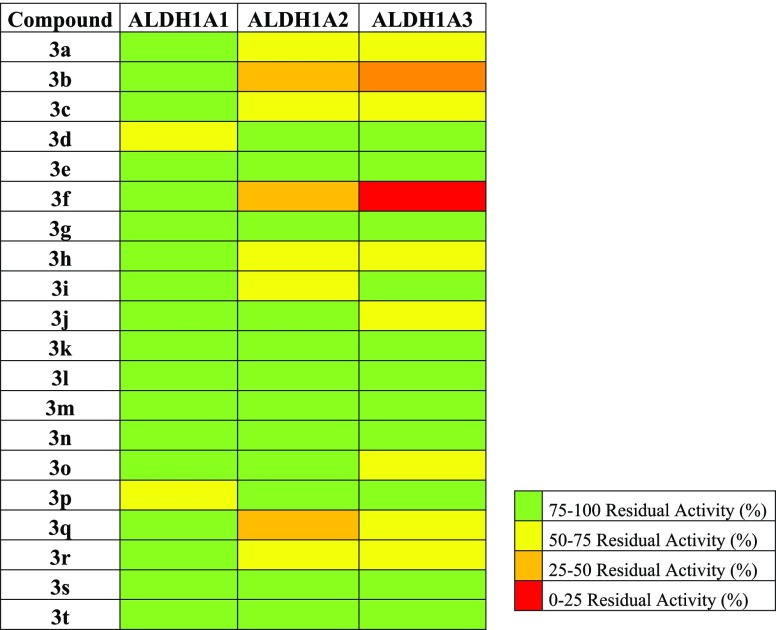
All the synthesized compounds, characterized by different substitution patterns on both the 2- and 8-phenyl rings, were tested in vitro for their efficacy against the human recombinant ALDH1A3 and selectivity toward the parent isoforms ALDH1A1 and ALDH1A2. They were preliminarily screened at a single concentration of 25 μM (Figure 1). Compounds showing the best inhibitory efficacy were further investigated for their kinetic parameters, including IC50 and Ki values. Results obtained allowed us to sketch some structure–activity relationships. In particular, moving from the unsubstituted 3a (IC50 80.6 ± 1.8 μM, Table 1), the most significant increase in the inhibitory efficacy of the novel series was obtained through the insertion of electron-withdrawing substituents in the para position of the 8-phenyl ring. Actually, both compound 3b (IC50 6.0 ± 1.3 μM, Table 1), carrying a fluoro atom, and compound 3c (IC50 5.5 ± 1.4 μM, Table 1), carrying a chloro atom, turned out to be almost ten times more potent than the lead 3a. An even better inhibitory profile was obtained through the insertion of a methoxycarbonyl fragment as in derivative 3f (IC50 0.66 ± 1.3 μM, Table 1), whose activity in the submicromolar range increased a hundredfold compared to the unsubstituted 3a. Interesting functional results were also obtained when additional substituents, like methyl and methoxyl groups, were added on the 2-phenyl ring. Among the investigated combinations, the one including a methoxy fragment in the meta position of the 2-phenyl ring, as in compounds 3q (IC50 7.2 ± 1.5 μM, Table 1) and 3r (IC50 4.3 ± 1.6 μM, Table 1), turned out to be the most effective one, helping to keep the inhibitory activity in the low micromolar range.
Figure 1.
Heat map of ALDH1As residual activities in the presence of 8-substituted-imidazo[1,2-a]pyridine derivatives 3a–t.
Table 1. ALDH1As Inhibitory Activity of 8-Substituted-imidazo[1,2-a]pyridine Derivatives 3a–c,f,q,r.
| IC50 (μM)a |
||||||
|---|---|---|---|---|---|---|
| N | R1 | R2 | R3 | ALDH1A1 | ALDH1A2 | ALDH1A3 |
| 3a | H | H | H | n.a.b | 61.4 ± 1.6 | 80.6 ± 1.8 |
| 3b | H | H | 4-F | n.a. | n.a. | 6.0 ± 1.3 |
| 3c | H | H | 4-Cl | 168.3 ± 2.6 | 27.6 ± 1.2 | 5.5 ± 1.4 |
| 3f | H | H | 4-COOCH3 | n.a. | n.a. | 0.66 ± 1.3 |
| 3q | H | OCH3 | 4-F | n.a. | n.a. | 7.2 ± 1.5 |
| 3r | H | OCH3 | 4-Cl | n.a. | n.a. | 4.3 ± 1.6 |
IC50 values represent the concentration required to produce 50% enzyme inhibition.
Not active. No significant enzyme inhibition was observed at 25 μM.
An in depth biochemical investigation carried out on the most effective derivatives, 3c and 3f, revealed the fully competitive inhibitory profile of the novel compounds. They both were highly selective against the human isoenzyme ALDH1A3 (3c, Ki 8.7 ± 0.9 μM, 3f, Ki 0.12 ± 0.01 μM) with respect to the parent 1A1 (3c, Ki 168.3 ± 2.6 μM, 3f, Ki 79.5 ± 1.8 μM), but not completely toward the 1A2 isoform (3c, Ki 27.6 ± 1.2 μM, 3f, Ki 0.92 ± 1.5 μM).
Crystallization and Molecular Modeling Studies
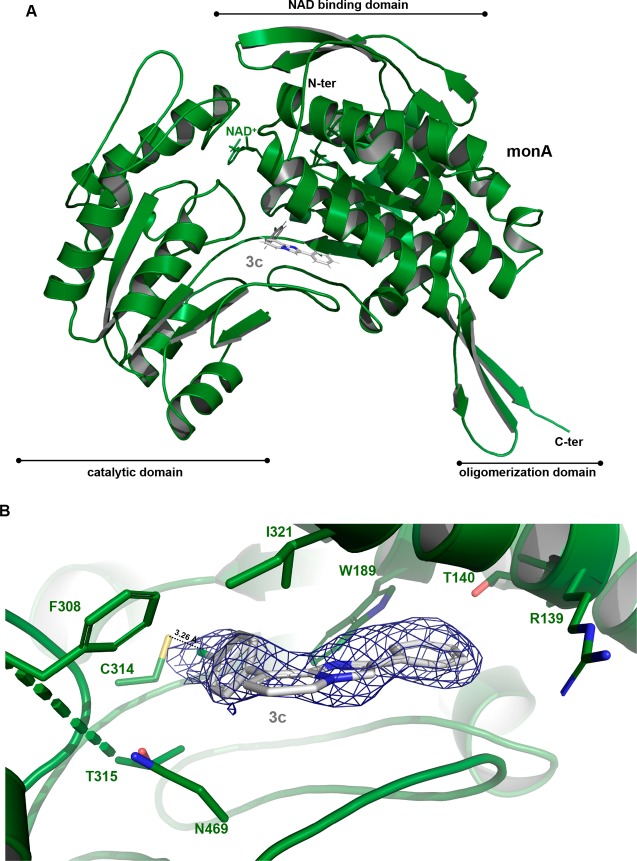
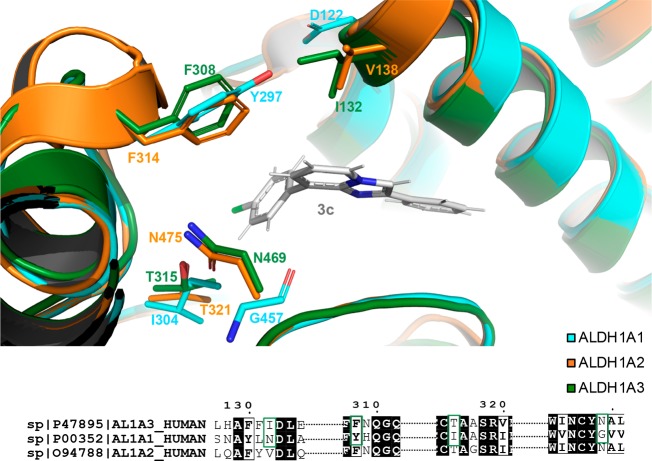
Compounds 3c and 3f, showing the best inhibitory profile of the whole series, were then investigated further, to clarify their mode of interaction with the ALDH1A3 isoform. The three-dimensional structure of human ALDH1A3 in complex with 3c has been obtained and solved by molecular replacement, using the human ALDH1A3 atomic coordinates as a search model (PDB code: 5FHZ(17)), and refined at a resolution of 2.9 Å. The final human ALDH1A3 model contains two identical chains per asymmetric unit, arranged as a dimer, and a total of 11 solvent molecules (Table 1-SI, Supporting Information). The structure analysis confirms that the obtained 3c–ALDH1A3 complex folds into 13 α-helices, 19 β-strands, and the connecting loops, arranged into three functional domains. Both the NAD+ binding domain and the catalytic domain are built on a topologically related βαβ type polypeptide fold (Figure 2A). Our crystallographic data revealed that, in all monomers, 3c enters the catalytic tunnel projecting toward the catalytic cysteine 314 (C314). In particular, the chloride atom (Cl01) on the 8-phenyl ring contacts directly C314 through an H-bond interaction (3.26 Å) (Figure 2B). By adopting this pose, 3c casts the 8-phenyl ring toward the side chain of the aromatic F308, I321, and N469, thus allowing π–π stacking interactions which make the binding more effective. In addition, T315 stabilizes 3c bonding through van der Waals contact. Notably, the four mentioned residues are specific of ALDH1A3 isoform and are not conserved in the other two isoenzymes (Figure 3). As they significantly contribute to define the binding pocket hosting 3c, they give reason to the inhibitory selectivity displayed by the compound against the three isoenzymes. Moreover, the phenyl ring in position 2 of the nucleus establishes additional contacts with the protein backbone, including π–π stacking interaction and van der Waals contacts with W189, T140, and R139 (Figure 2B). The mentioned residues are conserved in each of the 1A1, 1A2, and 1A3 isozymes, with this enzymatic area being the one stabilizing retinoic acid binding in the whole members of the 1A subfamily, as previously demonstrated.17
Figure 2.
(A) ribbon representation of the overall structure of the monomer B ALDH1A3 (PDB code: 6TRY) with 3c and NAD+. Each monomer consists of three domains: the NAD binding-domain, the catalytic-domain and the oligomerization domain, all highlighted with rows. The ligands NAD+ and 3c are shown as green and gray sticks, respectively. (B) electron density map (2mFo-dFc) in blue, countered at 1.0 sigma, used to determine the presence of 3c into the enzyme binding site.
Figure 3.
Superposition of inhibitor binding site between ALDH1A3 in forest green, ALDH1A1 (PDB_code: 4WB9(24)) in cyan and ALDH1A2 (PDB_code: 6ALJ(25)) in orange. The three structures maintain the same structure arrangement. Side chains of key residues, involved in the 3c–ALDH1A3 binding (I132, F308, T315, and N469), are shown in forest green sticks. The correspondent not conserved residues of ALDH1A2 and of ALDH1A1 are represented in orange sticks and cyan sticks, respectively.
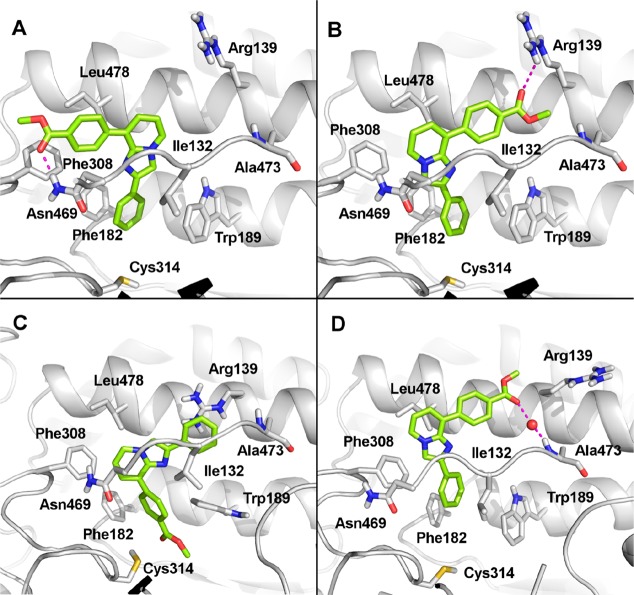
Different attempts to cocrystallize 3f in complex with human recombinant ALDH1A3 were fruitless. Therefore, in the absence of a reliable crystal structure, the binding mode of 3f within the catalytic site of ALDH1A3 has been assumed by molecular docking simulations, carried out using the GOLD program.18 To assess the reliability of the docking protocol, the cocrystallized ligand 3c was self-docked into the catalytic site of ALDH1A3, providing a very good agreement with the crystallographic pose (RMSD < 0.5 Å). Molecular docking simulations identified two possible, although alternative, binding modes for 3f. The replacement of the chlorine atom with the ester moiety significantly modified the binding mode of the imidazopyridine scaffold. In the first pose (Figure 4A), the phenyl ring in position 2 is oriented toward the catalytic C314 and establishes a sandwich-like and a parallel-displaced π–π stacking interaction with F182 and with W189, respectively. The imidazopyridine core is accommodated into the hydrophobic pocket flanked by I132, F308, and L478, at the entrance of the catalytic tunnel, while the ester group establishes a H-bond with N469. In the second pose (Figure 4B), interactions of the phenyl group at position 2 and the imidazopyridine group remain unchanged, while the ester moiety is H-bonded to R139. It is worth mentioning that, regardless of the binding poses proposed by the GOLD program, 3f is able to establish an additional H-bond interaction with key ALDH1A3 residues that are not present in the crystallographic binding mode of 3c, and this gives reason for the stronger ALDH1A3 inhibitory activity of 3f.
Figure 4.
Stick representations of the proposed docking poses (A and B boxes) and representative frames from MD simulations (C and D boxes) of compound 3f into the binding site of ALDH1A3 (PDB code: 6TRY), represented as gray ribbons. Residues relevant for the inhibitor binding have been labeled, and their side chains are shown as gray sticks.
The obtained docking poses were further investigated by molecular dynamics (MD) simulations using Amber18.19 Each 3f–ALDH1A3 complex was simulated for 300 ns of unrestrained MD, while the most representative conformations were extracted from MD trajectories by cluster analysis. The docking pose shown in Figure 4A is highly destabilized in MD and converges to a new binding mode (MD simulation video, Supporting Information) in which the ester group is oriented toward the catalytic C314, while the unsubstituted phenyl ring establishes a cation−π interaction with R139 (Figure 4C). Notably, this pose remains stable along the MD trajectory (Figure 1-SI, Supporting Information) and is highly comparable to the crystallographic pose of 3c (Figure 2-SI, Supporting Information). The pose shown in Figure 4B, in which the ester group is H-bonded to R139, keeps the same orientation for the whole MD simulation. In the MD trajectory, 3f loses the interaction with R139 and establishes a water-bridged H-bond with A473 (Figure 4D). The theoretical affinity of 3f to ALDH1A3 was computed by the molecular mechanics/generalized Born surface area (MMGBSA) approach.20 Notably, the two poses are almost isoenergetic (delta energies of binding are −33.07 ± 0.26 kcal/mol and −35.88 ± 0.47 kcal/mol for the pose shown in Figure 4C and 4D, respectively).
Overall, both crystallographic and molecular modeling studies, carried out on the representative compounds 3c and 3f, evidence that the novel 2,8-diphenylimidazopyridines are able to bind ALDH1A3 protruding toward the catalytic C314 residue. Results obtained confirm the soundness of our initial working hypothesis, based on the suitable relocation of the 6-phenyl ring of GA11 to increase the structural complementarity of this class of compounds with the enzyme binding site.
Antiproliferative Study of Derivative 3c
Derivative 3c, chosen as the representative compound among the most effective inhibitors, was investigated for its antiproliferative efficacy against the U87-MG cell line, used as a model of human glioblastoma overexpressing ALDH1A321 and, as reported in Human Atlas, silencing the isoforms ALDH1A1 and ALDH1A2. As displayed in Figure 3-SI (Supporting Information), compound 3c proved to reduce U87-MG vitality, although not significantly, when tested at concentrations in the high micromolar range. To rationalize the observed discrepancy between the cell-free and the cell-based assay results, a prediction of the physiochemical properties of the test compound was carried out using the QikProp program.22 Despite an estimated good cell permeability (QPPCaco, 8020 nm/s), 3c was predicted to have a weak water solubility (QPlogS, −6.482) and a relatively high logP (QPlogPo/w, 5.666), although within the range of 95% of current drugs. Accordingly, the low antiproliferative activity observed might be ascribed in principle to the limited water solubility and bioavailability of the compound, which certainly deserves an in depth investigation and optimization.
Concluding Remarks
It is now clear that the ALDH1A enzyme subfamily, and especially the 1A1 and 1A3 isoforms, play a crucial role in tumor progression and invasion by modulating self-renewing differentiation and expansion of CSCs. Accordingly, compounds able to block their catalytic activity may represent an effective strategy to fight cancer. Pursuing our interest in the field of ALDH inhibitors, and taking advantage of a structure-based optimization, we succeeded in improving the functional activity of a previously developed hit compound (GA11, IC50 4.7 ± 1.7 μM15) obtaining the novel lead 3f, methyl 4-(2-phenylimidazo[1,2-a]pyridin-8-yl)benzoate (IC50 0.66 ± 1.3 μM). Characterized by a distinctive molecular geometry, 3f proved to bind the catalytic site of ALDH1A3 protruding toward the key C314. Moreover, thanks to the metoxycarbonyl fragment on the 8-phenyl ring, it is able to H-bond the protein backbone, thus settling firmly into the binding site of the target protein. Such a structural complementarity gave reason of its significant functional efficacy. Actually, showing both IC50 and Ki values in the sub-micromolar range, 3f stands out as the most effective ALDH1A3 inhibitor ever described to date. Clearly, thorough investigations in complex structures like cell lines and animal models are now necessary, to prove its robustness as a novel prototype of an anticancer agent.
Experimental Procedures
Chemistry
Melting points were determined using a Reichert Köfler hot-stage apparatus and are uncorrected. Routine 1H NMR and 13C NMR spectra were recorded in DMSO-d6 solution on a Bruker 400 spectrometer operating at 400 MHz. Evaporation was performed in vacuo (rotary evaporator). Analytical TLCs were carried out on Merck 0.2 mm precoated silica gel aluminum sheets (60 F-254). Purity of the target inhibitors was determined by HPLC analysis, using a Shimadzu LC-20AD liquid chromatograph (PDA, 250–500 nm) and a Luna C18 column (250 mm × 4.6 mm, 5 μm, Phenomenex), with a gradient of 30% water and 70% acetonitrile and a flow rate of 1.0 mL/min. All the compounds showed percent purity values ≥95%. HRMS were obtained with a Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). 3-Bromopyridin-2-amine, the suitably substituted 2-bromo-1-phenylethan-1-one, and the appropriate boronic acids, used to obtain the target inhibitors, were from Alfa Aesar, Aldrich, and Fluka. 8-Bromo-2-phenylimidazo[1,2-a]pyridine 2a and 8-bromo-2-(3-methoxyphenyl)imidazo[1,2-a]pyridine 2d were synthesized following reported procedures.23
General Procedure for the Synthesis of 6-Bromo-2-(substituted)phenylimidazo[1,2-a]pyridines, 2b,c
A mixture of 5-bromopyridin-2-amine 1 (1.00 mmol), the suitably substituted 2-bromo-1-phenylethan-1-one (1.00 mmol), and potassium carbonate (138 mg, 1.00 mmol) in 20.0 mL of EtOH was refluxed under stirring until the disappearance of the starting material (TLC analysis). After cooling, the crude obtained was evaporated to dryness. Water was then added to the residue and the desired heterocycle, 2b,c, separated as a white solid, was collected by filtration, purified through recrystallization from the suitable solvent, and characterized with physio-chemical and spectroscopic data (Table 2-SI, Supporting Information).
General Procedure for the Synthesis of 2,8-(Substituted)diphenylimidazo[1,2-a]pyridines, 3a–t
A solution of the appropriate imidazo[1,2-a]pyridine derivative 2a–d (1.00 mmol), Pd(OAc)2 (0.10 mmol) and PPh3 (0.20 mmol) in toluene was added with the suitable phenyl boronic acid (1.50 mmol), dissolved in ethanol, and 2 mL of Na2CO3 2 M. The resulting mixture was refluxed under stirring until the disappearance of the starting material (TLC analysis). After cooling, the crude obtained was evaporated to dryness and the residue was purified by column chromatography (silica gel, ethyl acetate/petroleum ether). The pure product was then recrystallized from the suitable solvent and characterized by physiochemical and spectroscopic data (Table 3-SI, Supporting Information).
Acknowledgments
The authors acknowledge Beatrice Muscatello, CISUP (Centre for Instrumentation Sharing - University of Pisa) for HRMS analysis, and the European Synchrotron Radiation Facility (ESRF, Grenoble, France) for data collections at the beamlines ID29. S.G. and D.M.F. acknowledge the University of Piemonte Orientale (grant Ricerca Ateneo FAR_2016) and ROCHE per la Ricerca 2017, for financial support. F.P. acknowledges a MIUR (Ministero dell’Istruzione, dell’Università e della Ricerca) grant “Dipartimento di Eccellenza 2018–2022”.
Glossary
Abbreviations
- ALDH
aldehyde dehydrogenase
- ALDH1A1
aldehyde dehydrogenase subtype 1A1
- ALDH1A2
aldehyde dehydrogenase subtype 1A2
- ALDH1A3
aldehyde dehydrogenase subtype 1A3
- RA
retinoic acid
- ATRA
all-trans retinoic acid
- 9-cis-RA
9-cis retinoic acid
- 13-cis-RA
13-cis retinoic acid
- RARE
retinoic acid (RA) response element
- GSCs
glioma stem-like cells.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.9b00686.
Data collection and refinement statistics of the human ALDH1A3 model. Rearrangement of compound 3f within the catalytic site of ALDH1A3, extrapolated from MD simulation. Conformational stability of the 3f–ALDH1A3 complex in MD simulations. Overlap of the binding mode of the crystallized compound 3c and the representative poses of compound 3f extrapolated from MD trajectories. Antiproliferative activity of compounds 3c against the U87-MG cell line. Physiochemical and spectroscopic data of compounds 2b,c and 3a–t. Experimental procedures of biochemical and antiproliferative assays. Experimental procedures of crystallization and computational studies. PDB ID Code. 6TRY. Authors will release the atomic coordinates and experimental data upon article publication. (PDF)
MD simulation video (MPG)
Author Contributions
∥ L.Q. and E.L.M.G. contributed equally to the work.
The authors declare no competing financial interest.
Supplementary Material
References
- Black W.; Vasiliou V. The aldehyde dehydrogenase gene superfamily resource center. Hum. Genomics 2009, 4, 136–42. 10.1186/1479-7364-4-2-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer J. E.; Blomhoff R. Gene expression regulation by retinoic acid. J. Lipid Res. 2002, 43, 1773–808. 10.1194/jlr.R100015-JLR200. [DOI] [PubMed] [Google Scholar]
- Moreb J. S. Aldehyde dehydrogenase as a marker for stem cells. Curr. Stem Cell Res. Ther. 2008, 3, 237–246. 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- Xu X.; Chai S.; Wang P.; Zhang C.; Yang Y.; Yang Y.; Wang K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015, 369, 50–57. 10.1016/j.canlet.2015.08.018. [DOI] [PubMed] [Google Scholar]
- Ginestier C.; Hur M. H.; Charafe-Jauffret E.; Monville F.; Dutcher J.; Brown M.; Jacquemier J.; Viens P.; Kleen C.; Liu S.; Schott A.; Hayes D.; Birnbaum D.; Wicha M. S.; Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D. J.; Taussig D.; Simpson C. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells 2005, 23, 752–760. 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- Yang L.; Ren Y.; Yu X.; Quian F.; Bian B.-S.-J.; Xiao H.-L.; Wang W.-G.; Xu S.-L.; Yang J.; Cui W.; Liu Q.; Wang Z.; Guo W.; Xiong G.; Yang K.; Quian C.; Zhang X.; Zhang P.; Chi Y.-H.; Bian X.-W. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod. Pathol. 2014, 27, 775–783. 10.1038/modpathol.2013.189. [DOI] [PubMed] [Google Scholar]
- Li T.; Su Y.; Mei Y.; Leng Q.; Leng B.; Liu Z.; Stass S. A.; Jiang F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’outcome. Lab. Invest. 2010, 90, 234–244. 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng E.; Mitra A.; Tripathi K.; Finan M. A.; Scalici J.; McClellan S.; Madeira da Silva L.; Reed E.; Shevde L. A.; Palle K.; Rocconi R. P. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One 2014, 9, e107142 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcato P.; Dean C. A.; Liu R.-Z.; Coyle K. M.; Bydoun M.; Wallace M.; Clemens D.; Turner C.; Mathenge E. G.; Gujar S. A.; Giacomantonio C. A.; Mackey J. R.; Godbout R.; Lee P. W. K. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol. Oncol. 2015, 9, 17–31. 10.1016/j.molonc.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita D.; Minata M.; Ibrahim A. N.; Yamaguchi S.; Coviello V.; Bernstock J. D.; Harada S.; Cerione R. A.; Tannous B. A.; La Motta C.; Nakano I. Identification of ALDH1A3 as a viable therapeutic target in breast cancer metastasis-initiating cells. Mol. Cancer Ther. 2020, molcanther.0461.2019. 10.1158/1535-7163.MCT-19-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C.; Sullivan J. P.; Girard L.; Augustyn A.; Yenerall P.; Rodriguez-Canales J.; Liu H.; Behrens C.; Shay J. W.; Wistuba I. I.; Minna J. D. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin. Cancer Res. 2014, 20, 4154–4166. 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P.; Joshi K.; Li J.; Kim S. H.; Li P.; Santana-Santos L.; Luthra S.; Chandran U. R.; Benos P. V.; Smith L.; Wang M.; Hu B.; Cheng S. Y.; Sobol R. W.; Nakano I. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 8644–8649. 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinavahi S. S.; Bazewicz C. G.; Gowda R.; Robertson G. P. Aldehyde dehydrogenase inhibitors for cancer therapeutics. Trends Pharmacol. Sci. 2019, 40, 774–789. 10.1016/j.tips.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Cheng P.; Wang J.; Waghmare I.; Sartini S.; Coviello V.; Zhang Z.; Kim S. H.; Mohyeldin A.; Pavlyukov M. S.; Minata M.; Valentim C. L.; Chhipa R. R.; Bhat K. P.; Dasgupta B.; La Motta C.; Kango-Singh M.; Nakano I. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016, 76, 7219–7230. 10.1158/0008-5472.CAN-15-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrini L.; Gelardi E. L. M.; Coviello V.; Sartini S.; Ferraris D. M.; Mori M.; Nakano I.; Garavaglia S.; La Motta C.. Imidazo[1,2-a]pyridine Derivatives as Aldehyde Dehydrogenase Inhibitors: Novel Chemotypes to Target Glioblastoma Stem Cells. J. Med. Chem.. Manuscript under revision. [DOI] [PubMed] [Google Scholar]
- Moretti A.; Li J.; Donini S.; Sobol R. W.; Rizzi M.; Garavaglia S. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid. Sci. Rep. 2016, 19, 3571. 10.1038/srep35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.; Willett P.; Glen R. C.; Leach A. R.; Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Case D. A.; Ben-Shalom I. Y.; Brozell S. R.; Cerutti D. S.; Cheatham T. E. III; Cruzeiro V. W. D.; Darden T. A.; Duke R. E.; Ghoreishi D.; Gilson M. K.; Gohlke H.; Goetz A. W.; Greene D.; Harris R.; Homeyer N.; Izadi S.; Kovalenko A.; Kurtzman T.; Lee T. S.; LeGrand S.; Li P.; Lin C.; Liu J.; Luchko T.; Luo R.; Mermelstein D. J.; Merz K. M.; Miao Y.; Monard G.; Nguyen C.; Nguyen H.; Omelyan I.; Onufriev A.; Pan F.; Qi R.; Roe D. R.; Roitberg A.; Sagui C.; Schott-Verdugo S.; Shen J.; Simmerling C. L.; Smith J.; Salomon-Ferrer R.; Swails J.; Walker R. C.; Wang J.; Wei H.; Wolf R. M.; Wu X.; Xiao L.; York D. M.; Kollman P. A.. AMBER 2018; University of California, San Francisco, 2018. [Google Scholar]
- Miller B. R.; McGee T. D.; Swails J. M.; Homeyer N.; Gohlke H.; Roitberg A. E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Liu Y.; Hu H.; Huang H.; Bao Z.; Yang P.; Wang Y.; You G.; Yan W.; Jiang T.; Wang J.; Zhang W. ALDH1A3: a marker of mesenchymal phenotype in gliomas associated with cell invasion. PLoS One 2015, 10, e0142856 10.1371/journal.pone.0142856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2019-4; QikProp, Schrödinger, LLC: New York, NY, 2019. [Google Scholar]
- Bischoff F.; Berthelot D.; De Cleyn M.; MacDonald G.; Minne G.; Oehlrich D.; Pieters S.; Surkyn M.; Trabanco A. A.; Tresadern G.; Van Brandt S.; Velter I.; Zaja M.; Borghys H.; Masungi C.; Mercken M.; Gijsen H. J. M. Design and synthesis of a novel series of bicyclic heterocycles as potent γ-secretase modulators. J. Med. Chem. 2012, 55, 9089–9106. 10.1021/jm201710f. [DOI] [PubMed] [Google Scholar]
- Morgan C. A.; Hurley T. D. Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1. Chem.-Biol. Interact. 2015, 234, 29–37. 10.1016/j.cbi.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Zhu J.-Y.; Hong K. H.; Mikles D. C.; Georg G. I.; Goldstein A. S.; Amory J. K.; Schönbrunn E. Structural basis of ALDH1A2 inhibition by irreversible and reversible small molecule inhibitors. ACS Chem. Biol. 2018, 13 (3), 582–590. 10.1021/acschembio.7b00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.