Abstract
Acetylcholinesterase (AChE) inhibitors (AChEIs) still remain the leading therapeutic options for the symptomatic treatment of cognitive deficits associated with mild-to-moderate Alzheimer’s disease. The search for new AChEIs benefits from well-established knowledge of the molecular interactions of selective AChEIs, such as donepezil and related dual binding site inhibitors. Starting from a previously disclosed coumarin-based inhibitor (±)-cis-1, active as racemate in the nanomolar range toward AChE, we proceeded on a double track by (i) achieving chiral resolution of the enantiomers of 1 by HPLC and (ii) preparing two close achiral analogues of 1, i.e., compounds 4 and 6. An eudismic ratio as high as 20 was observed for the (−) enantiomer of cis-1. The X-ray crystal structure of the complex between the (−)-cis-1 eutomer (coded as MC1420) and T. californica AChE was determined at 2.8 Å, and docking calculation results suggested that the eutomer in (1R,3S) absolute configuration should be energetically more favored in binding the enzyme than the eutomer in (1S,3R) configuration. The achiral analogues 4 and 6 were less effective in inhibiting AChE compared to (±)-cis-1, but interestingly butylamide 4 emerged as a potent inhibitor of butyrylcholinesterase (BChE).
Keywords: Dual binding site acetylcholinesterase inhibitors, Alzheimer’s disease, coumarin derivatives, X-ray diffraction, molecular docking, chiral separation
The increased life expectancy in developed countries represents an unprecedented challenge for health systems and caregivers, due to the widening incidence of age-related pathologies. Among these, neurodegenerative diseases (NDs), most notably, Alzheimer’s disease (AD), are considered as a true emergency, because of their increasing incidence and accompanying social and economic costs.1 AD is a progressive and fatal neurological disease, involving degeneration of brain areas of the frontal cortex and basal forebrain nuclei, which evolves from memory disorders in its early stages, through progressive behavioral alterations that culminate in a total inability in the later stages of the disease.
Despite decades of intensive research, no disease-modifying therapy is yet available, and therapeutic approaches include solely symptomatic treatments based on acetylcholinesterase (AChE) inhibitors (AChEIs) and memantine, an N-methyl-d-aspartate (NMDA) receptor antagonist. AChE (EC 3.1.1.7) is the enzyme principally responsible for the termination of nerve impulse transmission at cholinergic synapses, by rapid hydrolysis of the neurotransmitter acetylcholine (ACh). Cholinergic innervation abounds in regions most affected by neurodegeneration in AD, e.g., the hypothalamus and entorhinal neo-cortex, and accordingly selective inhibition of AChE may help slow the progress of cognitive alteration in the early stages of the disease. Furthermore, it is commonly accepted that impairment of cholinergic innervation from the nucleus basalis and septal diagonal band to the cerebral cortex and hippocampus are involved in producing the cognitive changes.2 While effectiveness decreases along with the progression of AD, this approach is to date the only option for alleviating symptoms.3 In the central nervous system (CNS), AChE activity is complemented by that of butyrylcholinesterase (BChE), a related enzyme characterized by a larger active site, and thus capable of accommodating larger substrates. BChE is present in the serum and in the CNS, where its concentration increases with the progression of AD, colocalizing with neuritic plaques.4 These observations have supported the hypothesis that BChE might serve as a target for the symptomatic treatment of late-stage AD.5,6 The observation that cymserine analogues, selectively targeting BChE, can partially restore AChE brain levels and cognitive functions in aged rats7 provides support for this hypothesis.
3D structures of AChE from several species, e.g., Torpedo, electric eel, mouse, and human,8 in the presence or absence of inhibitors, have been solved by X-ray crystallography, demonstrating the existence of two binding sites at the top and bottom of the active-site gorge, termed the peripheral anionic site (PAS) and the catalytic anionic site (CAS), respectively.9 Among AChEIs, donepezil (Figure 1) is considered as a reference drug because of its potency and high therapeutic index. Its peculiar inhibitory mechanism involves a dual binding site (DBS) reversible interaction with both the CAS and the PAS of the enzyme, thus resulting in mixed, i.e. neither completely competitive nor noncompetitive, inhibition kinetics.10,11
Figure 1.
Ligand-based design of dual binding site inhibitors of AChE.
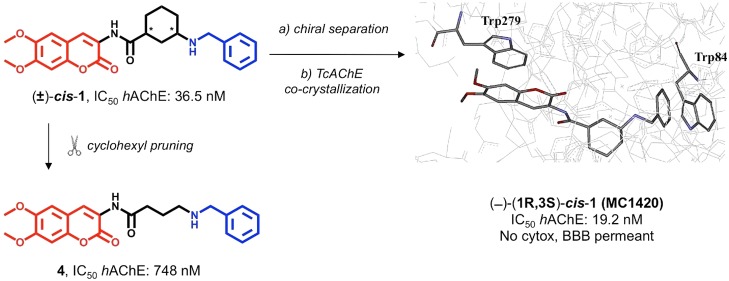
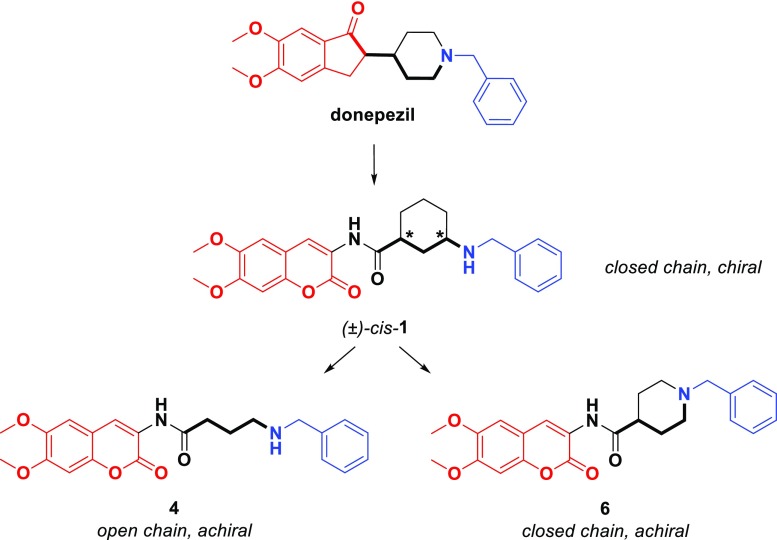
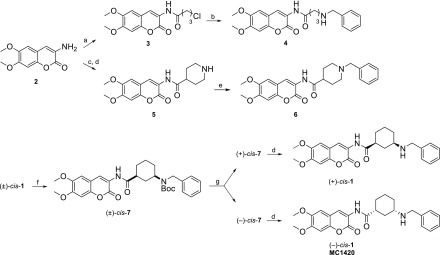
Many DBS inhibitors of AChE have been described in the literature,12 often displaying potent, reversible, and selective inhibition.13 Many of them exhibit the archetypal structural features of donepezil, i.e., a protonatable N-benzylamine moiety able to interact with the aromatic amino acids of the CAS, and a planar, aromatic, lipophilic terminal portion making hydrophobic interactions (mainly π–π stacking) within the PAS. These structural features were indeed displayed by the coumarin-based racemic compound (±)-cis-1 (Figure 1), which has been studied by some of the authors of the current study.14 To assess the stereochemical contribution to AChE inhibition of the cis-3-amino-1-ciclohexanecarboxylic acid used as the spacer in (±)-cis-1, we performed its chiral separation by HPLC and tested the two enantiomers. In parallel, we undertook the design of two new achiral analogues 4 and 6, also shown in Figure 1. Compound 4 includes a 3-atom linear open chain, and 6 encloses a piperidine ring, both joined to the donepezil-like N-benzyl moiety. By employing docking-assisted crystallographic studies, we determined the crystal structure of compound (−)-cis-1 (coded as MC1420), established as the eutomer after chiral separation, complexed with Torpedo californica AChE (TcAChE). Scheme 1 shows the synthetic and experimental procedure for obtaining 4, 6, and the two chiral forms of (±)-cis-1. Permeability and cytotoxicity of the coumarin derivative (±)-cis-1 were also evaluated in vitro as an early assessment of its potential as an AChE inhibitor for alleviating symptoms in AD-associated cognitive impairments.
Scheme 1.
Reagents and conditions: (a) 4-chlorobutyryl chloride, THF, triethylamine, rt; (b), benzylamine, KI, acetone, rt; (c), N-Boc protected isonipecotic acid, HOBt, DIC, CH2Cl2, rt; (d), TFA, CH2Cl2, 0 °C. (e), benzyl bromide, K2CO3, acetone, rt; (f) Boc2O, THF, rt; (g) semipreparative chiral HPLC.
Previous data on coumarin-donepezil hybrid (±)-cis-1 highlighted a good inhibitory potency on electric eel AChE (eeAChE) with very high selectivity over BChE.14 In order to obtain data comparable with published works in which the inhibition of human isoforms (hAChE, hBChE) were studied, we first investigated the inhibition of hAChE and hBChE by (±)-cis-1. The IC50 value for (±)-cis-1 against hAChE in Table 1 (36.5 nM) is in fair agreement with that previously determined for eeAChE (7.6 nM).14 The hAChE kinetic inhibition constant (Ki) of (±)-cis-1 was 4-fold higher than that measured for donepezil (46.6 vs. 12.7 nM, Table 1). The kinetic profile correlated well with a mixed-mode inhibition, typical for putative DBS inhibitors.
Table 1. Inhibition Data of Title Compoundsa.
| IC50 (nM) |
|||
|---|---|---|---|
| Entry | hAChE | hBChE | KihAChE (nM) |
| (±)-cis-1 | 36.5 ± 5.4 | 6250 ± 906 | 46.6 ± 3.6 (mixed) |
| (+)-cis-1 | 380 ± 45 | 2330 ± 219 | 93.4 ± 3.2 (mixed) |
| (−)-cis-1 (MC1420) | 19.2 ± 3.0 | 14100 ± 205 | 19.1 ± 1.4 (mixed) |
| 4 | 748 ± 24 | 181 ± 7 | n.d. |
| 6 | 223 ± 5 | 21 ± 2%b | n.d. |
| Donepezil | 16.1 ± 2.7 | 2900 ± 500 | 12.7 ± 1.0 (mixed) |
Values are mean ± SEM of three independent experiments; n.d.: not determined.
% inhibition at 10 μM.
A preliminary assessment of safety profile was obtained from a MTT-based cellular test15 of (±)-cis-1, which was incubated with HepG2 human liver cancer cells in the 20–100 μM concentration range (Figure 2). Cell viability after 2 h was always 80–90% even at the higher concentrations, thus revealing a low intrinsic cytotoxicity.
Figure 2.
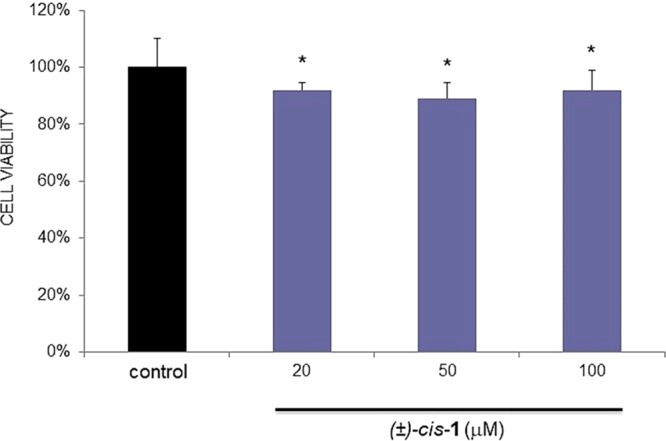
HepG2 cells viability, measured by the MTT assay, in the absence (black bar) and presence (gray bars) of (±)-cis-1. The percentage of MTT reduction observed is relative to control cells (DMEM). Values are expressed as mean ± SEM from six replicates, being significantly different from the control (untreated cells) as estimated by the Student’s t test (*p < 0.01).
The potential of (±)-cis-1 as a hit compound for pharmacological profiling was further explored by assessing its permeability in a well-validated cell membrane model, which utilizes the MDCK-MDR1 cell line expressing the efflux system P-gp. This cell line is widely considered to reliably mimic blood–brain barrier permeability, accounting for both transcellular and paracellular pathways.16 The MTT assay of cell viability, performed after 24 and 72 h of coincubation with 100 μM (±)-cis-1, showed lower cell survival compared with the control HepG2 samples, with cell viability dropping to 60% and 37%, respectively (Supporting Information, Table S1). However, the IC50 at this last time point was 30 μM, a value 3 orders of magnitude higher than the IC50 measured for AChE. Following a previously reported approach,17 the apparent permeabilities Papp were measured both from the apical to basolateral (Papp AP) and from the basolateral to apical (Papp BL) compartments. Diazepam and FD-4 were used as markers of transcellular and paracellular pathways, respectively. The permeability values shown in Table 2 are comparable to those of reference compounds, while the efflux ratio lower than 2 that was found indicates that the compound is not a substrate for P-gp.
Table 2. Permeability Assay Dataa.
| Entry | Papp AP (×10 –5 cm/s) | Papp BL (×10 –5 cm/s) | ER (Papp BL/Papp AP) |
|---|---|---|---|
| (±)-cis-1 | 3.7 ± 1.2 | 0.84 ± 0.20 | 0.22 |
| Diazepam | 2.0 ± 0.2 | 1.4 ± 0.2 | 0.70 |
| FD-4 | 0.69 ± 0.10 | 0.65 ± 0.12 | 0.93 |
Values are mean ± SEM of three independent experiments.
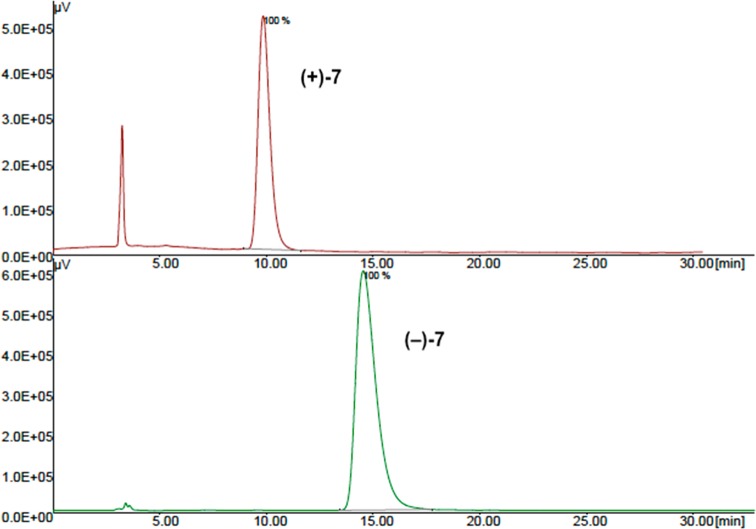
With this information in hand, we proceeded with the enantiomeric separation of (±)-cis-1. Due to the presence of a secondary N-benzylamine group, we performed a preliminary N-Boc protection, leading to the lipophilic derivative 7 (Scheme 1). Resolution of racemate (±)-cis-7 into the single enantiomers was achieved by semipreparative chiral HPLC (Figure 3), using a Kromasil 5-AmyCoat chiral stationary phase with isopropanol/n-hexane 1:1 v/v as the mobile phase, followed by Boc deprotection.
Figure 3.
Chromatograms of enantiopure samples obtained by chiral resolution of (±)-cis-7.
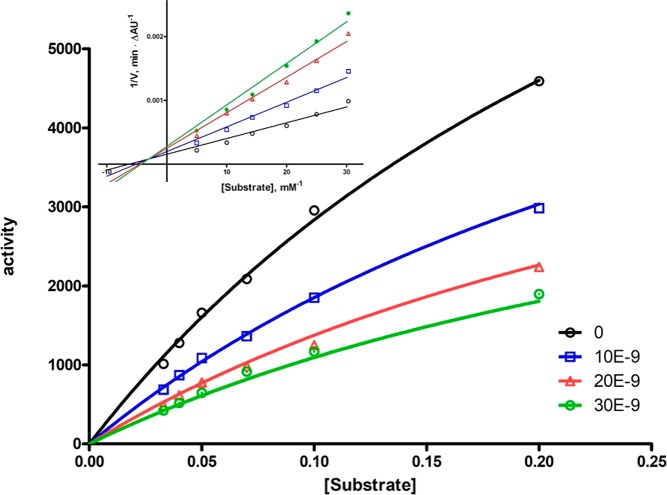
Inhibition data reported in Table 1 show a eudismic ratio of ca. 20 for the (−) enantiomer (coded as MC1420), with IC50 for hAChE very close to that of donepezil. Interestingly, an opposite eudismic ratio was found for hBChE inhibition, leading to high (730-fold) AChE/BChE selectivity of MC1420. The Ki for MC1420 was 19.1 nM, close to the value of 12.7 nM found for donepezil, and the kinetic data fitted a Michaelis–Menten model of a mixed-type inhibition, with very low variance (residuals <±1%; r2 = 0.996; Figure 4).
Figure 4.
Michaelis–Menten plot for inhibition of hAChE by MC1420 at various inhibitor concentrations. The inset displays the corresponding Lineweaver–Burk plot.
As shown in Table 1, the newly synthesized derivatives 4 and 6 (Scheme 1) were unable to reproduce the good inhibitory capacity of racemate 1. As far as compound 4 is concerned, we replaced the conformationally constrained cyclohexyl spacer with a linear open chain. This structural variation turned out to be detrimental, resulting in a 20-fold activity drop in inhibition of hAChE. The second structural variation was designed to keep the six-atom ring spacer, while incorporating the basic nitrogen into an N-benzylpiperidine fragment, which is a typical pharmacophore motif of donepezil and related structures. The isonipecotamide derivative 6 showed a 6-fold drop in hAChE inhibition compared to rac-1 while retaining fair AChE inhibition and very high AChE/BChE selectivity. It is noteworthy that the butylamide 4 showed strong inhibition of hBChE, thus resulting as a good and fairly selective inhibitor of this isozyme. The decrease in activity returned by these achiral analogues, irrespective of the limited variation in distance between the basic nitrogen and the coumarin moiety, interacting at the CAS and PAS, respectively, prompted us to elucidate the interactions of MC1420 with its target protein at the molecular level.
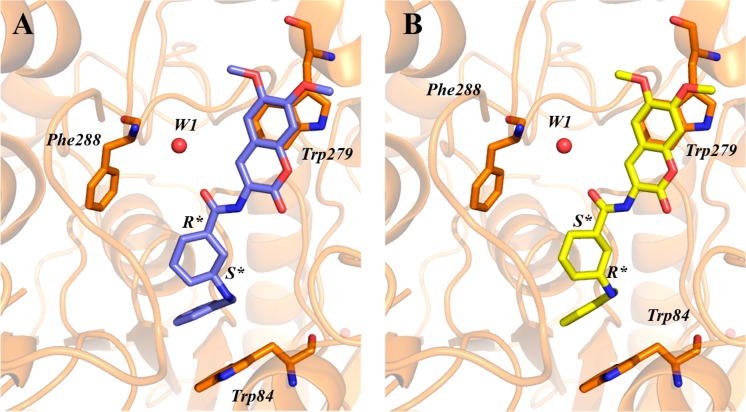
We thus determined the crystal structure of the complex of eutomer MC1420 with TcAChE. MC1420 was soaked into trigonal crystals of TcAChE, obtained as described earlier,18 and the structure of the TcAChE/MC1420 complex (PDB ID: 6TT0) was solved at 2.8 Å resolution from data collected at 100 K at a synchrotron beamline, following cryoprotection and flash-cooling of crystals. At this stage, the absolute configuration of MC1420 at the 1,3-cis-cyclohexyl ring spacer, namely, either (1R,3S) or (1S,3R), was unknown. Therefore, the structural refinement was performed assuming both the configurations of the spacer. In both cases, it was observed that the ligand molecule binds to the CAS through its N-benzyl moiety, with the coumarin group anchored at the PAS (Figure 5). This binding mode is driven mainly by two π–π stacking interactions, that of the aromatic ring of the N-benzyl group of the ligand with the indole of Trp84 (Trp86 in hAChE) at the CAS and of the coumarin ring with the indole moiety of Trp279 (Trp286 in hAChE) at the PAS. Regardless of the ligand’s absolute configuration, the rings involved in these stacking interactions are almost parallel (interplanar angle <15°) and display the typical parallel-displaced geometry. Interestingly, the interplanar distance between the coumarin group and Trp279 is about 0.5 Å shorter than that between Trp84 in the CAS and the N-benzyl group of the ligand.
Figure 5.
X-ray structure of the MC1420/TcAChE complex (PDB ID: 6TT0). Data refined for the 1R,3S-cis- (A) and 1S,3R-cis- (B) configurations. The ligands and relevant amino acid residues are rendered as sticks, the water molecule W1 responsible for a water-mediated interaction with Phe288 is shown as a red sphere, while protein is represented as a cartoon.
The experimental electron density map revealed the presence of a water molecule (W1 in Figure 5) at 2.8 Å from the oxygen atom of the amide group of MC1420 and 2.7 Å from the nitrogen atom of the Phe288 backbone, distances both compatible with H-bond interactions. This water-mediated interaction, along with the two stacking interactions described above, comprises the whole set of significant protein–ligand interactions. The oxygen of W1 has a thermal factor (59.4 Å2) that agrees very well with the average B-factor of atoms within 5 Å of W1 (60 Å2). Importantly, the B-factor is obtained by using W1 at full crystallographic occupancy, suggesting that this water molecule and related water-mediated interactions are present in each unit of the crystal. By performing hydration analysis of the ligand by wet script, two water molecules were identified close to the one observed experimentally (Figure S1 in Supporting Information). Interestingly, W1 corresponds to a conserved water as identified by Koellner et al.,19 and it therefore preexists at this position before binding of the compound. The structure offers an illustration that this structural water is indeed fully part of the gorge, where it helps to accommodate (and determine the binding affinity of) MC1420 in the gorge.
Despite the ligand being clearly visible in the experimental Fo–Fc electron density map, the absolute configuration of the (−)-cis-1 ligand used in the crystal preparation could not be unambiguously determined at the achieved resolution. We additionally performed molecular docking simulations, followed by binding free energy calculations for both configurations. Given the importance of W1 for ligand binding, this water molecule was included in the docking calculations performed.
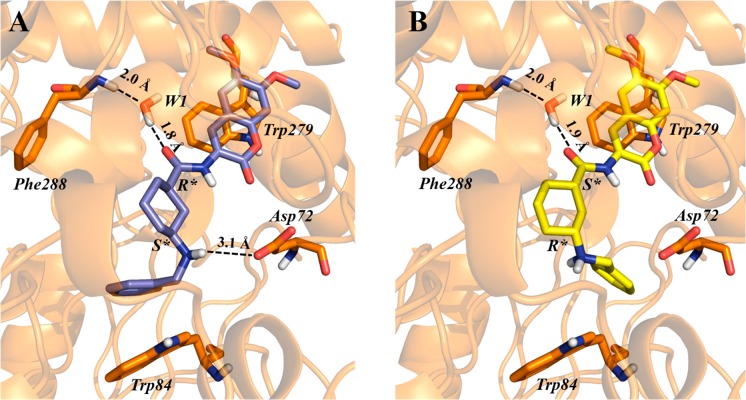
(1R,3S)-cis-1 returned not only a better docking score (−13.56 kcal/mol) and a better binding free energy (−99.28 kcal/mol) than (1S,3R)-cis-1 (−11.74 kcal/mol and −81.00 kcal/mol respectively), but also a more plausible pose, as shown by comparing Figures 5 and 6. We may postulate that this difference can be accounted for by an additional interaction with the PAS, established only by the (1R,3S) configuration (Figure 6). The S configuration of the carbon atom at position 3 of the cyclohexane should more likely permit an orientation of the charged amine adjacent to the benzyl ring, prone to form a salt bridge with Asp72. Taken together, these data suggest that MC1420 in the (1R,3S)-cis configuration should be more favored in binding compared to the (1S,3R)-cis configuration. It is noteworthy that the eutomer in both configurations should form a water-bridged H-bond with Phe288 in the acyl pocket.
Figure 6.
Top-scored docking poses for the (1R,3S)-cis- (A) and (1S,3R)-cis- (B) configurational isomers of 1 within the binding site of TcAChE. The ligand itself, relevant amino acid residues, and the water molecule W1 responsible for water-mediated interaction with Phe288, are all rendered as sticks, while protein is represented as a cartoon. H-bonds are depicted by dotted lines.
In agreement with the experimental findings, even the best solutions for achiral compounds 4 and 6 returned poorer docking scores (−11.44 kcal/mol and −12.34 kcal/mol, respectively) as well as poorer binding free energies (−79.49 kcal/mol and −78.70 kcal/mol, respectively) relative to (1R,3S)-cis-1. The obtained top-scored docking poses are reported in the Supporting Information (Figure S2).
In conclusion, our efforts to design selective and reversible AChE/BChE inhibitors led us to the synthesis of the AChE-selective hit compound (±)-cis-1 (Figure 1),14 which showed in vitro good safety and capacity to cross the BBB as assessed by HepG2 and MDCKII-MDR cell-based assays, respectively. After chiral resolution, the (−)-cis-1 enantiomer MC1420 resulted in the eutomer in hAChE inhibition, thus justifying the use of X-ray crystallography to resolve its binding mode in complex with TcAChE. The structure confirmed the dual binding mode of interaction predicted for MC1420, whereas docking calculations suggested that MC1420 should more favorably bind the enzyme in (1R,3S) absolute configuration than in (1S,3R).
Furthermore, in an attempt to overcome stereoisomeric limitations in a possible pharmacological evaluation, two achiral congeners of (±)-cis-1, i.e. 4 and 6, were synthesized and tested. These compounds did not replicate the strong inhibitory potency of 1. However, the butylamide 4 displayed good inhibition of BChE (IC50 = 181 nM) and the piperidine derivative 6 a high (about 2 orders of magnitude) AChE/BChE selectivity, suggesting that both of them could deserve further consideration.
Materials and Methods
Structural Determination of the MC1420/TcAChE Complex
After chiral separation of (±)-cis-1 racemate, the (−)-cis-1 enantiomer MC1420 was soaked for 2 h at 1 mM concentration in hanging drops containing trigonal crystals of TcAChE, obtained as described previously.18 Crystals were then flash-cooled, in situ, at 100 K under the gaseous nitrogen stream of a cryo-cooler (Oxford Cryosystems, Oxford, United Kingdom). Data collection was carried out on beamline ID29, at the European Synchrotron Facility (ESRF) at a wavelength of 1.074 Å. Initial phases were determined by rigid-body refinement using as a model the native TcAChE structure (PDB ID 2VT7). The Fo–Fc difference map showed continuous positive electron density at σ > 3.5 within the active site gorge of the enzyme (CAS and PAS). Both enantiomers compatible with MC1420 in cis conformation (1R,3S and 1S,3R of the cyclohexane ring) were tested in the refinement procedure. The restrain description file required for the refinement procedure was generated by using the PRODRG server20 and modified to ensure planarity of the coumarin and amide groups. Molecular geometry was optimized by eLBOW,21 under the crystallographic suite PHENIX.22 The ligand was fitted into the positive Fo–Fc Fourier difference map by using COOT.23 Water molecules were then added to the protein–ligand complex, and the resulting crystal structure was refined using phenix.refine,24 included in the Phenix crystallographic software suite.22 The structural model was validated using the Phenix implementation of MolProbity.25
Docking Simulations
Both enantiomers of (±)-cis-1 were docked into the refined X-ray structure of the complex 6TT0. The protein structure was prepared using Protein Preparation Wizard26 for adding missing hydrogen atoms, reconstructing incomplete side chains and loops, and assigning ambiguous protonation states. The ligand was prepared using LigPrep26 in order to properly generate all the possible tautomers and ionization states at a pH value of 7.0 ± 2.0. The files thus obtained were used for docking simulations performed by Grid-based ligand docking with energetics (GLIDE).26,27 During the docking process, the protein was held fixed, whereas full conformational flexibility was allowed for the ligand. The default Force Field OPLS_2005,28 and all the default settings of the extra precision (XP) protocol were used. A cubic grid was used that was centered on the refined structure of the cognate ligand, having an edge of 10 Å for the inner box and 30 Å for the outer box. Finally, a water molecule (referred to as W1) was kept in the binding site during docking simulations. Indeed, W1 arises from experimental electron density indicating a water-mediated H-bond involving the carbonyl group of the ligand and the backbone of Phe288.
MM-GBSA Calculations
The binding free energies (ΔG) between protein and ligands were computed by applying the molecular mechanics/generalized Born surface area on the obtained top-scored docking poses.29 More specifically, Prime30 was the software used, and the following eq 1 was applied:
| 1 |
where ΔEMM, ΔGsolv, and ΔGSA represent the difference between the contribution made by the ligand–protein complex and the sum of those made by the ligand and the protein taken alone, in terms of minimized energy, solvation energy, and surface area energy, respectively. Flexibility was allowed for all residues having at least one atom within a distance of 3 Å from the ligand.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.9b00656.
Syntheses of compounds 4–6, procedures for chiral separation of (±)-cis-1, cell viability assays (Table S1), hydration calculations for MC1420 (Figure S1), docking of compounds 4 and 6 (Figure S2), and crystallographic data (Table S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- 2019 Alzheimer’s disease facts and figures. Alzheimer's Dementia 2019, 15, 321–387. 10.1016/j.jalz.2019.01.010. [DOI] [Google Scholar]
- Mufson E. J.; Counts S. E.; Perez S. E.; Ginsberg S. D. Cholinergic System during the Progression of Alzheimer’s Disease: Therapeutic Implications. Expert Rev. Neurother. 2008, 8, 1703–1718. 10.1586/14737175.8.11.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham W. V.; Bonito-Oliva A.; Sakmar T. P. Update on Alzheimer’s Disease Therapy and Prevention Strategies. Annu. Rev. Med. 2017, 68, 413–430. 10.1146/annurev-med-042915-103753. [DOI] [PubMed] [Google Scholar]
- Macdonald I. R.; Maxwell S. P.; Reid G. A.; Cash M. K.; DeBay D. R.; Darvesh S. Quantification of Butyrylcholinesterase Activity as a Sensitive and Specific Biomarker of Alzheimer’s Disease. J. Alzheimer's Dis. 2017, 58, 491–505. 10.3233/JAD-170164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig N. H.; Utsuki T.; Yu Q.-S.; Zhu X.; Holloway H. W.; Perry T. A.; Lee B.; Ingram D. H.; Lahiri D. K. A New Therapeutic Target in AD Treatment: Attention to Butyrylcholinesterase. Curr. Med. Res. Opin. 2001, 17, 159–165. 10.1185/03007990152673800. [DOI] [PubMed] [Google Scholar]
- de Candia M.; Zaetta G.; Denora N.; Tricarico D.; Majellaro M.; Cellamare S.; Altomare C. D. New Azepino[4,3-b]indole Derivatives as Nanomolar Selective Inhibitors of Human Butyrylcholinesterase Showing Protective Effects against NMDA-induced Neurotoxicity. Eur. J. Med. Chem. 2017, 125, 288–298. 10.1016/j.ejmech.2016.09.037. [DOI] [PubMed] [Google Scholar]
- Greig N. H.; Utsuki T.; Ingram D. K.; Wang Y.; Pepeu G.; Scali C.; Yu Q. S.; Mamczarz J.; Holloway H. W.; Giordano T.; Chen D.; Furukawa K.; Sambamurti K.; Brossi A.; Lahiri D. K. Selective Butyrylcholinesterase Inhibition Elevates Brain Acetylcholine, Augments Learning and Lowers Alzheimer Beta-amyloid Peptide in Rodent. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 17213–17218. 10.1073/pnas.0508575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J.; Rudolph M. J.; Burshteyn F.; Cassidy M. S.; Gary E. N.; Love J.; Franklin M. C.; Height J. J. Structures of Human Acetylcholinesterase in Complex with Pharmacologically Important Ligands. J. Med. Chem. 2012, 55, 10282–10286. 10.1021/jm300871x. [DOI] [PubMed] [Google Scholar]
- Greenblatt H. M.; Dvir H.; Silman I.; Sussman J. L. Acetylcholinesterase: a Multifaceted Target for Structure-based Drug Design of Anticholinesterase Agents for the Treatment of Alzheimer’s Disease. J. Mol. Neurosci. 2003, 20, 369–383. 10.1385/JMN:20:3:369. [DOI] [PubMed] [Google Scholar]
- Nochi S.; Asakawa N.; Sato T. Kinetic Study on the Inhibition of Acetylcholinesterase by1-Benzyl-4-((5,6-dimethoxy-1-indanon)-2-yl)methylpiperidine Hydrochloride (E2020). Biol. Pharm. Bull. 1995, 18, 1145–1147. 10.1248/bpb.18.1145. [DOI] [PubMed] [Google Scholar]
- Kryger G.; Silman I.; Sussman J. L. Structure of Acetylcholinesterase Complexed with E2020 (Aricept): Implications for the Design of New Anti-Alzheimer Drugs. Structure 1999, 7, 297–307. 10.1016/S0969-2126(99)80040-9. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Wang H.; Chen H. Z. AChE Inhibition-based Multi-target-directed Ligands, a Novel Pharmacological Approach for the Symptomatic and Disease-modifying Therapy of Alzheimer’s Disease. Curr. Neuropharmacol. 2016, 14, 364–375. 10.2174/1570159X14666160119094820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani L.; Catto M.; De Palma A.; Farina R.; Cellamare S.; Altomare C. D. Discovery of Potent Dual Binding Site Acetylcholinesterase Inhibitors via Homo- and Heterodimerization of Coumarin-Based Moieties. ChemMedChem 2017, 12, 1349–1358. 10.1002/cmdc.201700282. [DOI] [PubMed] [Google Scholar]
- Catto M.; Pisani L.; Leonetti F.; Nicolotti O.; Pesce P.; Stefanachi A.; Cellamare S.; Carotti A. Design, Synthesis and Biological Evaluation of Coumarin Alkylamines as Potent and Selective Dual Binding Site Inhibitors of Acetylcholinesterase. Bioorg. Med. Chem. 2013, 21, 146–152. 10.1016/j.bmc.2012.10.045. [DOI] [PubMed] [Google Scholar]
- Berridge M. V.; Tan A. S. Characterization of the Cellular Reduction of 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT): Subcellular Localization, Substrate Dependence, and Involvement of Mitochondrial Electron Transport in MTT Reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- Franchini S.; Manasieva L. I.; Sorbi C.; Battisti U. M.; Fossa P.; Cichero E.; Denora N.; Iacobazzi R. M.; Cilia A.; Pirona L.; Ronsisvalle S.; Aricò G.; Brasili L. Synthesis, Biological Evaluation and Molecular Modeling of 1-Oxa-4-thiaspiro- and 1,4-Dithiaspiro[4.5]decane Derivatives as Potent and Selective 5-HT1AReceptor Agonists. Eur. J. Med. Chem. 2017, 125, 435–452. 10.1016/j.ejmech.2016.09.050. [DOI] [PubMed] [Google Scholar]
- Pisani L.; Farina R.; Soto-Otero R.; Denora N.; Mangiatordi G. F.; Nicolotti O.; Mendez-Alvarez E.; Altomare C. D.; Catto M.; Carotti A. Searching for MultitargetingNeurotherapeutics against Alzheimer’s: Discovery of Potent AChE-MAO B Inhibitors through the Decoration of 2H-Chromen-2-one Structural Motif. Molecules 2016, 21, 362. 10.3390/molecules21030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe S. N.; De la Mora E.; Chan S.; Kantham S.; McColl G.; Miles J. A.; Veliyath S. K.; Sreenivas S. K.; Nassar Z. D.; Silman I.; Sussman J. L.; Weik M.; McGeary R. P.; Parat M. O.; Brazzolotto X.; Ross B. P. Rivastigmine and Metabolite Analogues with Putative Alzheimer’s Disease-modifying Properties in a Caenorhabditis elegans Model. Commun. Chem. 2019, 2, 35. 10.1038/s42004-019-0133-4. [DOI] [Google Scholar]
- Koellner G.; Kryger G.; Millard C. B.; Silman I.; Sussman J. L.; Steiner T. Active-site Gorge and Buried Water Molecules in Crystal Structures of Acetylcholinesterase from Torpedo californica. J. Mol. Biol. 2000, 296, 713–735. 10.1006/jmbi.1999.3468. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf A. W.; van Aalten D. M. F. PRODRG: A Tool for High-Throughput Crystallography of Protein-Ligand Complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 1355–1363. 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Moriarty N. W.; Grosse-Kunstleve R. W.; Adams P. D. Electronic Ligand Builder and Optimization Workbench (eLBOW): A Tool for Ligand Coordinate and Restraint Generation. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65, 1074–1080. 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: A Comprehensive Python-based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V.; Grosse-Kunstleve R. W.; Echols N.; Headd J. J.; Moriarty N. W.; Mustyakimov M.; Terwilliger T. C.; Urzhumtsev A.; Zwart P. H.; Adams P. D. Towards Automated Crystallographic Structure Refinement with phenix.refine. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. 3rd; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. MolProbity: All-atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Suite 2018–4 Protein Preparation Wizard; Schrödinger, LLC: New York, NY, 2018. [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra Precision GLIDE: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Robertson M. J.; Tirado-Rives J.; Jorgensen W. L. Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J. Chem. Theory Comput. 2015, 11, 3499–3509. 10.1021/acs.jctc.5b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S.; Ryde U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discovery 2015, 10, 449–461. 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PrimeX; Schrödinger, LLC: New York, NY, 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.