Abstract
Background
Circular RNAs (circRNAs) have been proven to play important roles in tumorigenesis. However, the mechanism by which circRNAs act on gastric cancer (GC) through epithelial-to-mesenchymal transition (EMT) is unclear. In this study, we identified circ-OXCT1 and elucidated its function on EMT in GC.
Methods
Tissue circRNA microarray analysis and qRT-PCR were utilized to determine the expression level of circ-OXCT1 in GC. Luciferase reporter assay and FISH were employed to confirm the interaction between circ-OXCT1 and miR-136. CCK-8, cloning formation, transwell, wound healing, nude mice experiment, circ-OXCT1 overexpression and silencing were conducted to elucidate the function of circ-OXCT1 in vivo and in vitro. Western blot and rescue experiment were carried out to evaluate the changes of EMT-related proteins induced by circ-OXCT1 overexpression or silencing.
Results
Circ-OXCT1 was downregulated in GC tissues and cell lines. Its expression level was significantly associated with lymph node metastasis, pathologic stage and overall survival rate through clinicopathologic data analysis. Circ-OXCT1 silencing downregulated SMAD4 expression and accordingly regulated expression of E-cadherin, N-cadherin and vimentin through the transforming growth factor-beta (TGF-β)/Smad signaling pathway by a circ-OXCT1/miR-136/SMAD4 axis, resulting in enhancement of EMT and subsequent boost of cell migration, invasion and nude mice lung metastasis.
Conclusion
Our data showed that circ-OXCT1 suppresses gastric cancer EMT and metastasis through TGF-β/Smad signaling pathway. The clinicopathologic data analysis revealed that circ-OXCT1 overexpression could be a novel treatment for advanced GC especially with distant metastasis by targeting the circ-OXCT1/miR-136/SMAD4 axis.
Keywords: gastric cancer, GC, circ-OXCT1, miR-136, SMAD4, EMT
Introduction
GC is among the most frequent cancers around the world, making it a universal cancer burden.1,2 Its mortality ranks third worldwide.3 The prognostic prospect is still poor despite the profound development of surgical methods and medication,4 and most of the patients have been suffering from tumor recurrence and metastasis, resulting in a less than 30% of 5-year overall survival (OS).5 Hence, deciphering the potential molecular mechanisms of GC cell migration and invasion has always been the first priority so as to screen available biomarkers and develop novel effectively therapeutic methods.
CircRNAs (circular RNAs), highly resistant to RNase R,6 one of the pre-mRNA back-splicing productions, are widely distributed in zooblast.7 The vast majority of circRNAs shows highly conservative characters among species and also tissue- or development-specific expression patterns.8 Moreover, the specific structures of circRNAs determine their particular biological functions: some repress the biological functions of miRNAs as their sponges,9 some act as RNA-binding protein scaffolds or decoys10 and some even encode proteins.11,12 MiRNAs (microRNAs), normally 20–24 nt in length,13 are highly conserved among species and function in, for instance, apoptosis, proliferation, EMT and so on.14 miRNA regulates gene expression at the posttranscriptional level:15 it binds with messenger RNA (mRNA) at its 3ʹUTR (3ʹ untranslated region) and cuts it off, leading to mRNA degradation;16 it also binds with the coding sequence (CDS), blockading its translation.17
That circ-OXCT1 attenuating the GC cell migration and invasion was verified in our study. Circ-OXCT1 also reduced lung metastasis in nude mice experiment. Mechanically, circ-OXCT1 reversed SMAD4 expression by sponging miR-136, resulting in suppression of TGF-β/Smad-related EMT by regulating its relevant gene expression (E-CAD, N-CAD, and VIM) in GC cells.
Materials and Methods
Patients and Specimens
Seventy-four GC patients from the First Affiliated Hospital of Chongqing Medical University were randomly selected between March 2013 and November 2014. Cancerous and paired paracancerous specimens were collected accordingly. All patients underwent radical surgery and none of them took radiotherapy and/or systemic preoperative chemotherapy before surgery. All specimens were stored at −180°C in liquid nitrogen until RNA extraction. The final diagnosis was histopathologically confirmed and TNM-staged based on the seventh classification of the American Joint Committee on Cancer. Criteria as gender, age, tumor size, pathological type, cell differentiation grade, lymphatic metastasis, TNM stage and Borrmann classification are displayed in Table 1 as the clinicopathological features. The follow-up started at the moment of operation and ended until the last clinical visiting, loss to follow-up, disease progression or death. The mean follow-up time is 36.0 months. The overall survival analysis was carried out to study the survival value of circ-OXCT1. This study was supported by the Medical Ethics Review Committee of the First Affiliated Hospital of Chongqing Medical University. Moreover, every patient signed the informed consent before study.
Table 1.
Relationship Between Circ-OXCT1 Expression and Clinicopathological Features in GC Patients
| Clinicopathological Features | N (74) | Circ-OXCT1 | χ2 | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | 0.525 | 0.630 | |||
| <60 | 27 | 12 | 15 | ||
| ≥60 | 47 | 25 | 22 | ||
| Gender | 0.216 | 0.816 | |||
| Male | 38 | 18 | 20 | ||
| Female | 36 | 19 | 17 | ||
| Tumor size (cm) | 0.294 | 0.787 | |||
| <2 | 18 | 10 | 8 | ||
| ≥2 | 56 | 27 | 29 | ||
| Cell differentiation | 0.702 | 0.704 | |||
| Well | 19 | 8 | 11 | ||
| Moderately | 35 | 18 | 17 | ||
| Poorly | 20 | 11 | 9 | ||
| Borrmann classification | 0.704 | 0.872 | |||
| I | 11 | 6 | 5 | ||
| II | 36 | 18 | 17 | ||
| III | 19 | 8 | 11 | ||
| IV | 9 | 5 | 4 | ||
| Lymphatic node metastasis | 6.167 | 0.024* | |||
| N0 | 24 | 7 | 17 | ||
| N1–N3 | 50 | 30 | 20 | ||
| Pathological stages | 6.852 | 0.017* | |||
| I/II | 20 | 5 | 15 | ||
| III/IV | 54 | 32 | 22 | ||
Notes: P<0.05 indicates significance, χ2 test or Fisher’s exact test; *statistically significant, with median circ-OXCT1 values used as cut-off values.
Cell Culture and Transfection
AGS, MGC803 and MKN45 cell lines were purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences/Peking Union Medical College (CAMS/PUMC). HEK-293T, MKN28, BGC823 and GES-1 were donated by the Molecular Tumor and Epigenetics Laboratory of the First Affiliated Hospital of Chongqing Medical University and the use of the cell lines was approved by the Medical Ethics Review Committee of the First Affiliated Hospital of Chongqing Medical University. FBS (fetal bovine serum) (Biological Industries, Israel), penicillin-streptomycin sulfate (Hyclone, Shanghai, China) and RPMI-1640 (Gibco, USA) were used for cell culture under 5% CO2 at 37°C in an incubator (Thermo, USA). For overexpressing circ-OXCT1, a 1000 bp cDNA, including the manufactured liner form of circ-OXCT1, was constructed and inserted into the pHBLV-CMV vector (Hanbio Biotech, Shanghai, China). Then, lentivirus was utilized and transfected according to the instructions. MiR-136 mimics/inhibitors or negative control (RiboBio, Guangzhou, China) were transfected into AGS and MGC803 with Lipofectamine 2000 (Invitrogen, USA).
RNA Extraction and qRT-PCR
The TRIzol reagent (Takara, Dalian, China) was used in extracting total RNA from specimens and cells. cDNA was obtained through RNA reverse transcription using PrimeScriptTM RT Reagent Kit (RR037A, Takara) according to its protocol. SYBR Premix Ex TaqTM II (RR820A; Takara) was used in the two-step qRT-PCR on a CFX96 PCR equipment (Bio-Rad, USA). All primers (divergent primers for circ-OXCT1) were listed in Table 2. All data were measured by the ΔΔCT method. GAPDH was taken as an internal control.
Table 2.
Primers Used in qRT-PCR
| Circ-OXCT1 | Forward | 5′-GCACTGATTCAGAAAAGGTGTTA-3′ |
| Reverse | 5′-CCTGAAGGAGAATGTCTCTT-3′ | |
| SMAD2 | Forward | 5′-GTTTTCCTAGCGTGGCTTGC-3′ |
| Reverse | 5′-AGTCTCTTCACAACTGGCGG-3′ | |
| SMAD3 | Forward | 5′-CTAGAGTCAGGAGCAGGGACT-3′ |
| Reverse | 5′-ACTCTAGGAAGCCAAGGGGA-3′ | |
| SMAD4 | Forward | 5′-CCCATCCCGGACATTACTGG-3′ |
| Reverse | 5′-TGTGCAACCTTGCTCTCTCAA-3′ | |
| E-CAD | Forward | 5′-CTTCTCTCACGCTGTGTCATC-3′ |
| Reverse | 5′-CTCCTGTGTTCCTGTTAATGGT-3′ | |
| N-CAD | Forward | 5′-CGTGAAGGTTTGCCAGTGTGA-3′ |
| Reverse | 5′-CCTGGCGTTCTTTATCCCG-3′ | |
| VIM | Forward | 5′-TCAATGTTAAGATGGCCCTTG-3′ |
| Reverse | 5′-TGAGTGGGTATCAACCAGAGG-3′ | |
| GAPDH | Forward | 5′-CTTTGGTATCGTGGAAGGACTC-3′ |
| Reverse | 5′-GTAGAGGCAGGGATGATGTTCT-3′ |
Cell Counting Kit-8 and Cloning Formation Assay
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used for cell proliferation assay. Twenty-four hours after transfection, 2×103/well cells were planted into a 96-well plate. After 0, 24, 48, 72, or 96 h planting, CCK-8 reagent (20 μL) was added into each well, following incubation for another 1.5 h under 5% at 37°C. The OD value was then evaluated by a spectrophotometer (Tecan, Switzerland) at 450 nm before the growth curve plotting. The colony formation assay was performed using a 6-well plate seeded with 500/well and maintained in complete medium under 5% CO2 at 37°C. The cells were fixed and stained after 2 weeks’ incubation. Finally, cell colonies were imaged and counted.
Transwell and Wound Healing Assay
The 8 μm Transwells (BD, Bioscience) were used for cell migration assay and with an extra 100 μL of Matrigel matrix (Corning, NY, USA) coated at the base of upper chamber for the invasion assay. Two hundred microliter of medium with no serum suspended with 2×104 cells were evenly placed into the base of the upper chamber, meanwhile, 600 µL of complete medium was filled into the bottom chamber. After 24 h incubation for migration assay and 48 h for invasion assay under 5% CO2 at 37°C, the base of upper chamber was scrupulously cleaned with a cotton tip. The bottom chamber was fixed, stained and then pictured at 100× magnification under a microscope (Leica, Germany). A 6-well plate was seeded with 5×105/well for wound healing assay. After 24 h incubation, when the cells grew to 100% confluence, a 200 µL pipette tip was used for scratching linear wounds, PBS was used for washing off the exfoliated cells. Afterwards, 2 mL of RPMI-1640 containing 2% FBS was added into each well. Wound closures were respectively imaged at 0 and 24 h under a microscope (Leica), then the width of each wound was recorded.
Tissue circRNA Microarray Analysis
Five paired specimens were randomly selected for GC circRNA microarray analysis. Microarray hybridization was applied in accordance with instructions from Arraystar (Rockville, USA). First, linear RNA was eradicated from total RNA for enriching circRNAs using RNase R kit (Epicentre, Inc.). Then, fluorescent cRNAs were obtained from the processed circRNAs in accordance with the protocols of Arraystar Super RNA Labeling Kit (Arraystar). Afterwards, the Arraystar Human circRNA Array V2 (8×15 K, Arraystar) chip was applied to hybridize with the fluorescent-labeled cRNAs. Finally, the chip was scanned and imaged by an Agilent scanner G2565CA (Jamul, CA, USA). Agilent Feature Extraction software (v 10.7) was applied for extracting and processing the data. Differentiated circRNAs with fold change (FC) ≥2 and p < 0.05 were regarded as statistically significant.
Fluorescence in situ Hybridization (FISH)
Fluorescent in situ Hybridization Kit (RiboBio Guangzhou, China) was applied to perform FISH. Probe for circ-OXCT1 antagonizing its back-splice site was biotin (Bio)-labeled (Bio-5ʹ-TCCATCATCAAAACAAATGGAGT-3ʹ-Bio) and probe for miR-136 was digoxin (Dig)-labeled (Dig-5ʹ-CAGAGCAATAAAGACACAGAGACGGCG-3ʹ-Dig), separately. First, the bottom of each well (24-well plate) was placed by a coverslip. Second, 1×104/well cells were seeded and maintained under 5% CO2 at 37°C until growing to 60-70% confluence. The coverslips were permeabilized in 0.5% Triton X-100 after washing in PBS three times and fixing in paraformaldehyde. Third, a dark moist box was used for hybridization at 37°C for 24 h according to the protocol. Last, DAPI (6-diamidino-2-phenylindole) was used to stain the cell nucleus after washing with 4× SSC, 2× SSC, 1× SSC and PBS, gradually. Images were captured by a laser confocal microscope at 400× magnification (ZEISS LSM800, Germany).
Luciferase Reporter Assay
The mutual binding among circ-OXCT1, miR-136/miR-145/miR-665 and SMAD4 were implemented using pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega, USA). The wild-type or mutant of miR-136-, miR-145- or miR-665-binding sequences in circ-OXCT1 and those of the CDS in SMAD417,18 were separately subcloned into pmirGLO vector (circ-OXCT1-Wt/circ-OXCT1-Mut and SMAD4-Wt/SMAD4-Mut, GeneCreate Biological Engineering Co., Ltd., Wuhan, China). Cells (HEK-293T/AGS/MGC803) were incubated to 60-70% confluence in a 96-well plate under 5% CO2 at 37°C, then cotransfected the vector and miR-136/miR-145/miR-665 mimics or miR-NC (normal control) with Lipofectamine 2000. After 48 h incubation, the reporter assay was processed according to the protocol of the Dual-luciferase Reporter Gene Assay Kit (Beyotime, China) and then the luciferase intensity was detected by a Dual-luciferase Reporter Assay System (Promega, USA). The normalized firefly luciferase value was determined by the ratio of firefly to Renilla.
Western Blot
RIPA (Beyotime), protease inhibitors and phosphatase inhibitors were used for lysing cells on ice for 30 min. Total protein was extracted by centrifugation (12,000 rpm), and the Enhanced BCA Protein Assay Kit (Beyotime) was used to measure the protein concentration according to its protocol. Ten percent SDS-PAGE was used for electrophoresis to isolate target proteins. The proteins were then blotted onto PVDF membrane strips (Sigma-Aldrich, USA). Primary antibodies were listed as following: SMAD4 (1:1000, Cell Signaling Technology, USA), SMAD2 (1:1000, Cell Signaling Technology), SMAD3 (1:1000, Cell Signaling Technology), p-SMAD2 (1:1000, Ser465/467, Cell Signaling Technology), p-SMAD3 (1:1000, Ser423/425, Cell Signaling Technology), E-CAD (1:1000, Cell Signaling Technology), N-CAD (1:1000, Cell Signaling Technology), VIM (1:1000, Cell Signaling Technology), and GAPDH (1:10,000, Abcam, USA). GAPDH was taken as a loading control. Finally, all PVDF strips were incubated in a secondary antibody solution (1:5000, HRP-conjugated anti-rabbit IgG) (Invitrogen, USA). Bound antibodies were detected using BeyoECL plus (Beyotime) by a Fusion equipment (Vilber Lourmat, Paris, France).
In vivo Experiments
The BALB/c nude mice were purchased from the Laboratory Animal Centre Chongqing Medical University (SYXK2018-0003, Chongqing, China). Mice were divided into two groups: AGS-circ-OXCT1 and AGS-circ-NC (normal control). 0.1 mL of PBS suspended with 1×106 cells was subcutaneously injected into the right flank and tail vein of nude mice (BALB/c, male, 4 weeks old), respectively. Tumor volume was confirmed by half of length multiplied by width square every third day. The mice were euthanized 4 weeks later. Lungs were removed and the metastatic nodules were counted under hematoxylin-eosin staining. All animal protocols were carried out following the Guidelines for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006) and authorized by the Institutional Animal Ethics Care and Use Committee of the First Affiliated Hospital of Chongqing Medical University.
Statistical Analysis
All data were analyzed with SPSS 23.0 and GraphPad Prism 8. The Kaplan-Meier method and the Log-rank test were used to plot the overall survival curve for differentially expressed circ-OXCT1 and calculate the prognostic significance, respectively. Differences between two groups were determined by Student’s t-test. Every experiment was repeated thrice and data are displayed as the mean ± SD., p < 0.05 was considered as statistically significant.
Results
CircRNA Microarray Chip Expression Profiles in Gastric Cancer
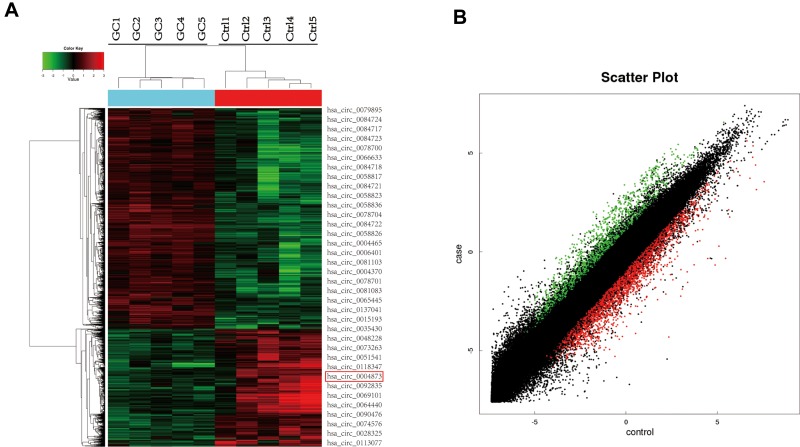
Five GC specimens (GC1-GC5) and paired paracancerous specimens (Ctrl1-Ctrl5) were applied for the circRNA microarray chip. After data processing, we distinguished 3594 downregulated and 1914 upregulated circRNAs with FC greater than or equal to 2. The top 24 upregulated and 12 downregulated circRNAs in GC are shown (Figure 1A). The scatter plot (Figure 1B) shows all the circRNAs variates. Hsa_circ_0004873, among the top 12 downregulated circRNAs, was found closely related to TGF-β-induced EMT in our study. Then, we confirmed hsa_circ_0004873 (located at chr5: 41794104–41807540) is derived from parental gene 3-oxoacid CoA-transferase 1 (OXCT1), which is located on chromosome 5p13.1 according to the human reference genome (GRCh37/hg19). Therefore, we named hsa_circ_0004873 “circ-OXCT1”.
Figure 1.
CircRNA microarray chip expression profiles in gastric cancer. (A) The top 24 upregulated (green) and 12 downregulated (red) circRNAs shown by heat map. Rows represent circRNAs, while columns represent specimens (GC-GC5, Ctrl1-Ctrl5). The red rectangle indicates hsa_circ_0004873 (circ-OXCT1). (B) Scatter plot demonstrating the differentially expressed circRNAs in the microarray analysis. Green dots represent the upregulated circRNAs, while red dots represent the downregulated circRNAs (FC >2 and p < 0.05).
Abbreviations: GC, gastric cancer; Ctrl, paracancerous specimens; FC, fold change.
Circ-OXCT1 Was Downregulated and Indicated Poor Prognosis
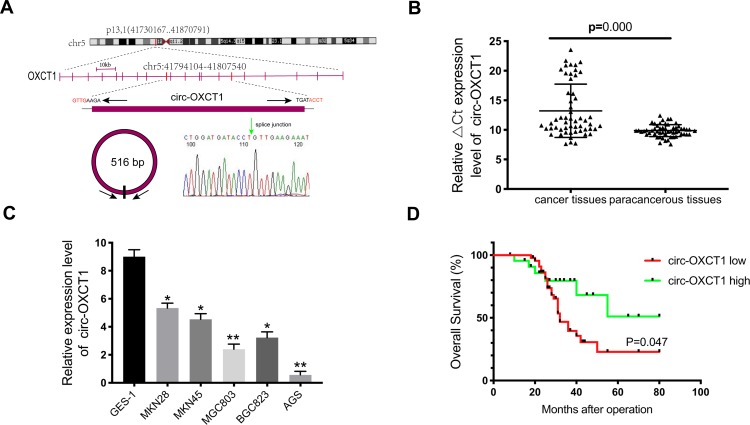
Given that circ-OXCT1 is a 516 bp-long exonic circRNA and has a strong connection with EMT, we did a series of experiments to explore the relationships between circ-OXCT1 and EMT in GC. In order to clarify its expression, divergent primers specific to the splice junction were designed and confirmed by Sanger sequencing on the PCR products after being treated with RNase R (Figure 2A). Circ-OXCT1 was then verified downregulated in the GC specimens comparing to the paracancerous specimens (Figure 2B). Six cell lines, MKN28, MKN45, MGC803, BGC823, AGS and GES-1 were applied in this study. Circ-OXCT1 was significantly downregulated in GC cell lines comparing to GES-1 (Figure 2C). The expression level of circ-OXCT1 were significantly correlated to lymph node metastasis (p = 0.024) and pathological stages (p = 0.017) despite no obvious correlations with tumor size, gender, age, cell classification and Borrmann classification according to the clinicopathological features (Table 1). Additionally, a lower circ-OXCT1 level indicated a shorter 5-year overall survival in GC patients, as shown by Kaplan–Meier survival curve analysis (p < 0.05) (Figure 2D).
Figure 2.
Circ-OXCT1 was downregulated and indicated poor prognosis. (A) Schematic illustration of the genomic locus of circ-OXCT1. The green arrow represents the back-splice junction of circ-OXCT1, confirmed by Sanger sequencing. (B, C) qRT-PCR determination of circ-OXCT1 expression in specimens and cell lines (*p < 0.05, **p < 0.01). (D) Patients with lower circ-OXCT1 level had a shorter overall survival rate than those with higher circ-OXCT1 level (p < 0.05).
Circ-OXCT1 Overexpression Suppressed Cell Migration and Invasion Except for Proliferation
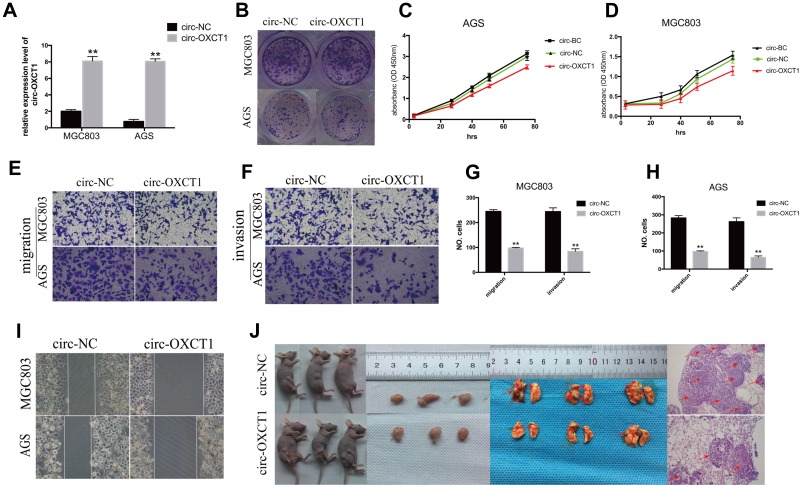
Since circ-OXCT1 expressed minimally in AGS and MGC803 among the five GC cell lines (Figure 2C), we used AGS and MGC803 to overexpress circ-OXCT by lentivirus transfection. Approximately fourfold higher circ-OXCT1 overexpression in MGC803 and 10-fold higher in AGS were consistently exhibited after transfecting with the lentivirus packaged with pHBLV-CMV-circ-OXCT1-GFP(T2A)-Puro plasmids (Figure 3A). The CCK-8 and colony formation assays suggested that circ-OXCT1 overexpression did not significantly suppress cell proliferation in neither MGC803 nor AGS (Figure 3B–D). Surprisingly, cell invasion and migration were remarkably inhibited by circ-OXCT1 overexpression through wound healing assay and Transwell in MGC803 and AGS (Figure 3E–I). Xenograft assay was implemented subsequently. By subcutaneously implanting AGS (stably transfected with circ-OXCT1 or circ-NC), we found that circ-OXCT1 overexpression did not inhibit tumor growth in nude mice as well. The tumor volumes showed no significant difference between the circ-OXCT1-overexpression group and the circ-NC group (Figure 3J), which agreed with the same outcome observed in vitro experiment. However, the lung metastatic nodules in the circ-OXCT1-overexpression mice were significantly much less than those in the circ-NC mice. Hematoxylin-eosin staining subsequently confirmed that circ-OXCT1 overexpression decreased lung metastasis (Figure 3J).
Figure 3.
Circ-OXCT1 overexpression suppressed cell migration and invasion except for proliferation. (A) Overexpression of circ-OXCT1 in MGC803 and AGS confirmed by qRT-PCR ( **p < 0.01). (B–D) Clone formation and CCK-8 assays showed that circ-OXCT1 overexpression did not significantly suppress the proliferation ability. (E–H) Transwell assay demonstrated that circ-OXCT1 overexpression dramatically reduced migration and invasion (**p < 0.01). (I) Representative images of wound healing assay in MGC803 and AGS. (J) Tumor volume between circ-NC and circ-OXCT1-overexpression group showed no significant difference in xenograft assay. However, hematoxylin-eosin staining showed that the lung metastatic nodules (red arrows) in the circ-OXCT1-overexpression group were significantly much less than that in the circ-NC group (**p < 0.01).
Abbreviations: GC, gastric cancer; BC, blank control; NC, negative control.
Circ-OXCT1 Bound Directly with miR-136 in GC Cells
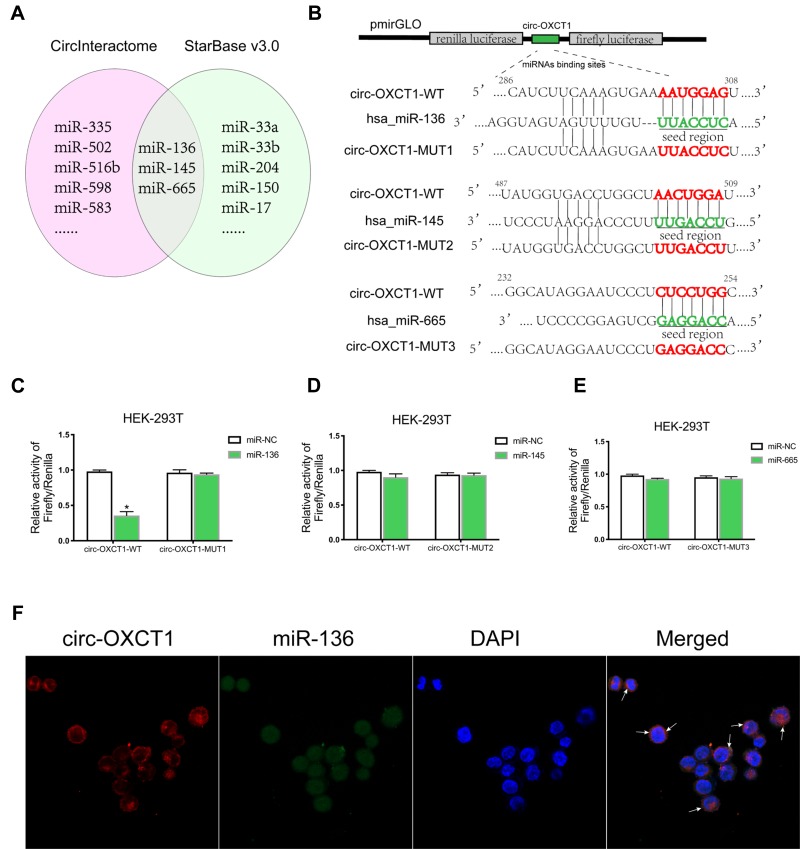
CircRNAs could suppress the biological functions of miRNAs by sponging them.19 Thus, we utilized CircInteractome (https://circinteractome.nia.nih.gov/index.html) and StarBase v3.0 (http://starbase.sysu.edu.cn/index.php) online software to predict potential miRNAs targeting circ-OXCT1 and identified 3 overlapped miRNAs (miR-136, 145 and 665) which might bind circ-OXCT1 with very high affinity (Figure 4A). Then, HEK-293T cells were utilized to verify which miRNAs could bind with circ-OXCT1. First, the putative miR-136-, miR-145-, or miR-665-binding site (wild-type/mutant) in circ-OXCT1 along with a certain length of flank sequence were separately subcloned into the pmirGLO luciferase reporter vectors (Figure 4B). Then the vectors were cotransfected with miR-136/miR-145/miR-665 mimics into HEK-293T cells, respectively. Among the three miRNAs, only miR-136 was verified could bind with circ-OXCT1 according to the luciferase activity (Figure 4C–E). Then, FISH assay was implemented and confirmed that circ-OXCT1 and miR-136 were preferentially colocalized in the cytoplasm (Figure 4F), providing another direct binding evidence for them. Above all, these results revealed that circ-OXCT1 bound directly with miR-136 in GC cells.
Figure 4.
Circ-OXCT1 bound directly with miR-136 in GC cells. (A) Schematic illustration exhibiting the overlapped miRNAs targeting circ-OXCT1 predicted by CircInteractome and StarBase v3.0. (B) Schematic description of the predicted miR-136-, miR-145- and miR-665-binding sites in circ-OXCT1 separately subcloned into the pmirGLO luciferase vector. (C–E) Relative luciferase activities of the wild-type/mutant circ-OXCT1 reporter in HEK-293T cotransfected with miR-136/miR-145/miR-665 or miR-NC (*p < 0.05). (F) Colocalization of circ-OXCT1 (red) and miR-136 (green) in AGS was observed (white arrows) by FISH. Cellular nuclei were counterstained with DAPI (blue).
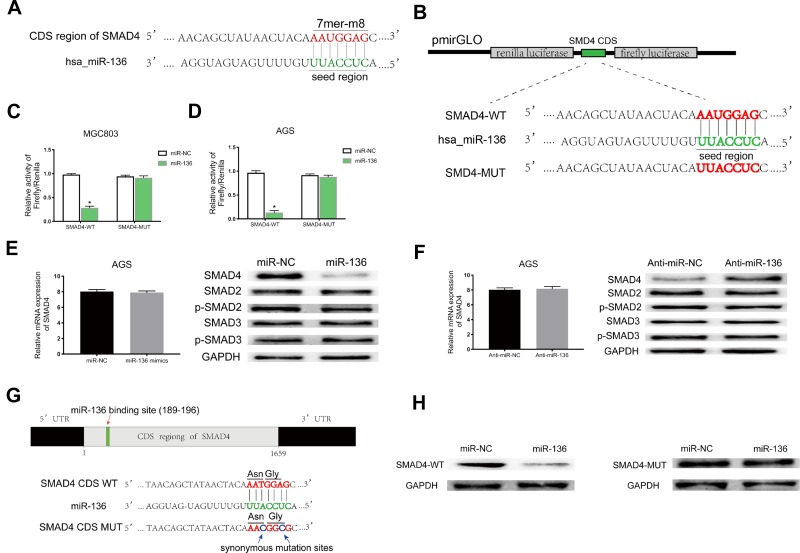
MiR-136 Repressed SMAD4 Expression by Targeting Its CDS
Based on StarBase v3.0 and miRanda prediction software, we found a 7mer-m8 binding location at the CDS of SMAD4 for miR-136 (Figure 5A). For validating whether miR-136 could directly target the CDS thus inhibit SMAD4 expression, we subcloned a fragment of the CDS (wild-type/mutant, including the miR-136-binding site) into the pmirGLO plasmid (Figure 5B) and transiently cotransfected with miR-136 mimics into MGC308 and AGS. The luciferase intensity was significantly weaker in the SMAD4-WT-miR-136 group than that in the SMAD4-MUT-miR-136 group in both MGC803 and AGS (Figure 5C and D), verifying that miR-136 bound with SMAD4 at its CDS and regulates SMAD4 expression by blockading its mRNA translation, which explained why its mRNA level stayed unchanged while its protein level significantly upregulated or downregulated accordingly (Figure 5E and F). Then, we constructed two synonymous mutations at the seed region of miR-136-binding location at the CDS of SMAD4 (Asn: AAT replaced by AAC, Gly: GGA replaced by GGG) (Figure 5G). MiR-136 mimics/miR-NC and pmirGLO-SMAD4-WT/MUT were cotransfected into HEK-293T cells, respectively. The SMAD4 protein level were significantly inhibited in the SMAD4-WT-miR-136 group (Figure 5H). In summary, the results validated miR-136 regulates SMAD4 expression via directly targeting its CDS.
Figure 5.
MiR-136 repressed SMAD4 expression by targeting its CDS. (A) Schematic illustration of the binding site between miR-136 and CDS of SMAD4 predicted by StarBase v3.0 and miRanda. (B) Schematic description about the predicted miR-136-binding locus in SMAD4. Targeting between wild-type/mutant SMAD4 and miR-136 are shown. (C, D) Dual-luciferase reporter assay demonstrated that miR-136 attenuated SMAD4 expression in MGC803 and AGS (*p < 0.05). (E, F) The SMAD4 protein level were decreased/increased significantly when transfected with miR-136 mimics/anti-miR-136 while it’s mRNA level remained nearly unchanged. (G) Schematic illustration of the synonymous mutations at the miR-136-binding location in CDS of SMAD4 (red arrows represent the miR-136-binding location, blue arrows represent the synonymous mutation sites. Asn: AAT replaced by AAC, Gly: GGA replaced by GGG). (H) SMAD4 protein level was dramatically downregulated when cotransfected with pmirGLO-SMAD4-WT and miR-136 mimics compared to the control (pmirGLO-SMAD4-MUT and miR-136 mimics).
Abbreviations: Asn, asparagine; Gly, glycine; CDS, coding sequence; Anti-miR-NC, miR-NC inhibitors; Anti-miR-136, miR-136 inhibitors.
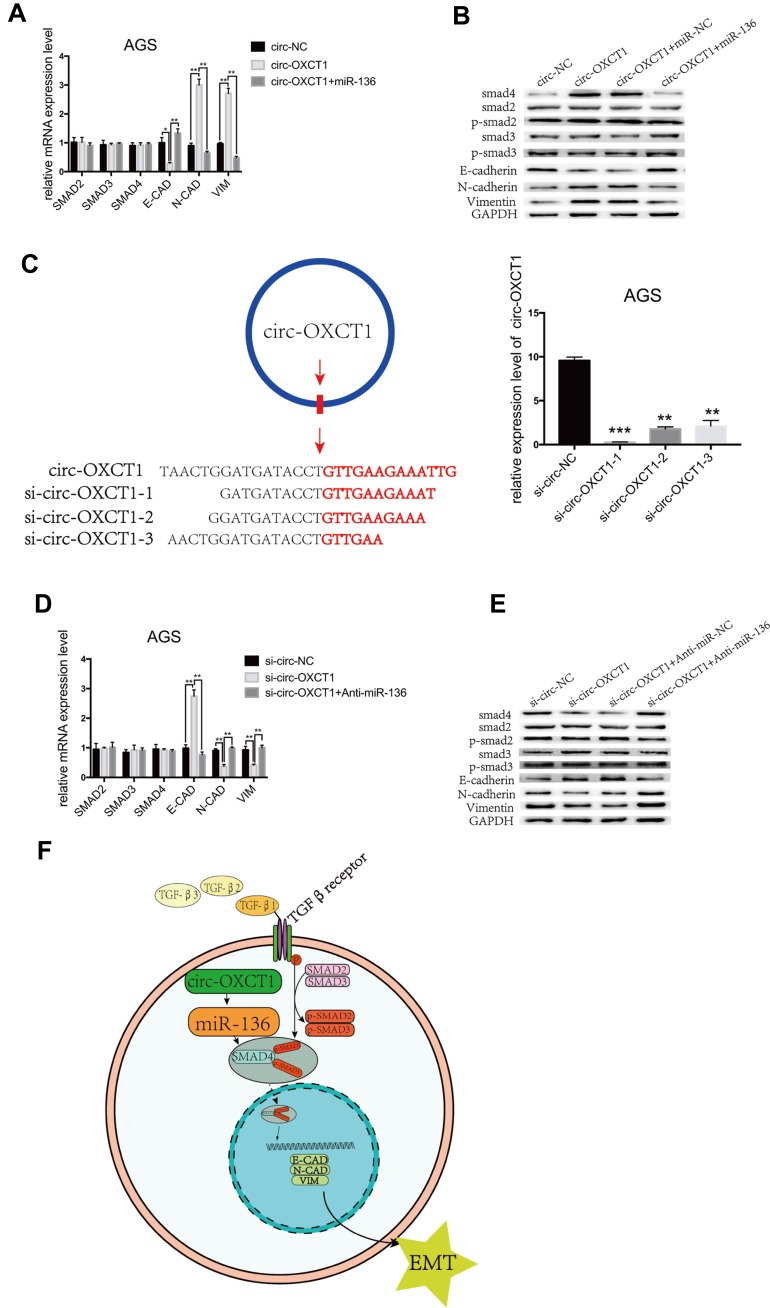
Circ-OXCT1 Suppressed SMAD4 Expression and EMT via the Circ-OXCT1/miR-136/SMAD4 Axis in GC Cells
We next examined whether miR-136 rescued the effect of circ-OXCT1 on SMAD4 expression and mediated EMT in GC cells. To validate our speculation, we carried out the following two rescue experiments. First, circ-OXCT1 overexpression significantly upregulated N-CAD, VIM, SMAD4 and downregulated E-CAD, their mRNA levels changed accordingly except for SMAD4 (miR-136 binding with its CDS) (Fig. 5G, 6A, 6B). Afterwards, we transfected miR-136 mimics into the circ-OXCT1-overexpressed AGS and verified that miR-136 rescued the effect caused by circ-OXCT1 overexpression (Figure 6B). Then, we constructed three short interfering RNAs (siRNA-1, siRNA-2 and siRNA-3) targeting the back-splice sequence of circ-OXCT1 (Figure 6C). qRT-PCR showed that siRNA-1 reduced circ-OXCT1 expression most efficiently, therefore, it was then applied in subsequent experiments. As estimated, circ-OXCT1 silencing significantly downregulated N-CAD, VIM, SMAD4 and upregulated E-CAD (Figure 6D), while anti-miR-136 (miR-136 inhibitors) cotransfection rescued the expression effect caused by circ-OXCT1 silencing (Figure 6E). In conclusion, our data suggested that circ-OXCT1 suppressed SMAD4 expression and EMT by sponging miR-136 through the circ-OXCT1/miR-136/SMAD4 axis in GC cells (Figure 6F).
Figure 6.
Circ-OXCT1 suppressed SMAD4 expression and EMT via the circ-OXCT1/miR-136/SMAD4 axis in GC cells. (A, B) mRNA levels of SMAD2, SMAD3, SMAD4, E-CAD, N-CAD, VIM and protein levels of SMAD2, p-SMAD2, SMAD3, p-SMAD3, SMAD4, E-CAD, N-CAD, VIM are shown after cotransfected with circ-OXCT1 and miR-136 mimics in AGS (*p < 0.05, **p < 0.01). (C) Schematic display of the targeting sequences of siRNAs (si-circ-OXCT1-1, si-circ-OXCT1-2 and si-circ-OXCT1-3) specific to the back-splice junction of circ-OXCT1. Si-circ-OXCT1-1 was verified the most effectively inhibitory siRNA (red dart represent the back-splice junction) (**p < 0.01, ***p=0.00). (D, E) Similar results were observed when cotransfected with si-circ-OXCT1-1 and anti-miR-136 (**p < 0.01). (F) A proposed mechanistic model in which circ-OXCT1 functions as a miR-136 sponge and regulates SMAD4/TGF-β-induced EMT in GC cells. GAPDH was used as a loading control.
Abbreviations: E-CAD, E-cadherin; N-CAD, N-cadherin; VIM, vimentin.
Discussion
GC is one of the most common cancers and has a low cure rate and a high recurrence rate.20 Limited by current treatment approaches and people’s cognition, the GC prognostic prospect is far below expected.21 In particular, there is still no effective treatment for advanced GC, especially with metastasis.22 In recent years, there is growing evidence showed that circRNAs play an important part in tumorigenesis, bringing new light to patient with GC. CircRNAs, as one of the endogenous RNAs, are commonly exist in eukaryotic cells and have gene regulatory functions.23 They distribute mostly in cytoplasm, with a little in nucleus.24 Previous studies unveiled that they are crucial in gene regulation, such as sponging miRNAs, regulating transcription factors, adsorbing RNA-binding proteins and even encoding proteins.25,26 Although circRNAs were discovered more than 26 years ago,27 Up to now, whether or how they influence on TGF-β-induced EMT is still unknown.
In this study, circ-OXCT1 was identified as a research candidate from the top 12 downregulated circRNAs in GC. We then collected 74 pairs of specimens (GC and paracancerous specimens) and inspected their clinicopathological features to assess the clinical value of circ-OXCT1. qRT-PCR ran on the specimens showed that circ-OXCT1 was significantly downregulated in GC. Moreover, the downregulation was strongly related to the lymph node metastasis, pathological stages (Table 1), and even a poor overall survival rate (Figure 2D), indicating that circ-OXCT1 downregulation might contribute to GC metastasis. Afterwards, we confirmed that circ-OXCT1 overexpression could suppress the lung metastasis in nude mice as well as migration and invasion in GC cell lines. Through dual-luciferase reporter assay and FISH, we also confirmed circ-OXCT1 sponged miR-136 in the cytoplasm, which also suggested that circ-OXCT1 and miR-136 may function at the posttranscriptional level. Finally, we verified that circ-OXCT1 could suppress the expression of SMAD4 by sponging miR-136 through binding SMAD4 at its CDS, indicating a circ-OXCT1/miR-136/SMAD4 axis.
TGF-β/Smad pathway is a latent initiator of EMT, a process ends in tumor invasion and metastasis.28 When SMAD2 and SMAD3 are phosphorylated in the cytoplasm, they form a trimer complex with SMAD4 before moving into nucleus. Then, the complex binds with the SMAD-binding element, initiating gene transcription. This process partly controls the switch of transcription for EMT-related proteins, resulting in upregulating of N-CAD, VIM and downregulating of E-CAD and so on.29,30 Through this study, circ-OXCT1 was first confirmed that could regulate SMAD4 expression and control EMT-related gene expression via sponging miR-136. Rescue experiments also showed that miR-136 mimics reversed the effect caused by circ-OXCT1 overexpression, and vice versa, miR-136 inhibitors reversed the effect caused by circ-OXCT1 silencing. All the data indicated that circ-OXCT1 suppressed TGF-β/Smad-induced EMT through sponging miR-136 in GC.
Overall, we validated the circ-OXCT1 expression pattern and its clinicopathological features in GC. Through sponging miR-136, circ-OXCT1 suppressed SMAD4 expression so as to manipulate the expression of EMT-related genes, for instance, E-CAD, N-CAD, VIM, resulting in GC EMT. In addition, whether the level of TGF-β/Smad-related downstream transcription factors (such as snail1, snail2, ZEB, etc.) have changed accordingly and what kind of relations between them and circ-OXCT1 required further study and the role of the parental gene OXCT1 in GC EMT need to be scrutinized as well. Unlike previous studies, our results provided a direct proof that overexpression of circ-OXCT1 could suppress GC EMT progress by sponging miR-136, suggesting a novel treatment for advanced GC with or without metastasis via targeting the circ-OXCT1/miR-136/SMAD4 axis and a novel biomarker for monitoring GC prognosis.
Acknowledgments
This work was supported by the National Key Clinical Specialties Construction Program of China (No [2012]0.649).
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Li Y, Huang S, Zheng Q. Response to “Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388(2017):208–219. doi: 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang J, Hong H, Xue X, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–232. doi: 10.1016/j.canlet.2018.10.040 [DOI] [PubMed] [Google Scholar]
- 5.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). (1474-547X (Electronic)). [DOI] [PMC free article] [PubMed]
- 6.Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi: 10.1080/15476286.2016.1220473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 8.Kong P, Yu Y, Wang L, et al. circ-Sirt1 controls NF-kappaB activation via sequence-specific interaction and enhancement of SIRT1 expression by binding to miR-132/212 in vascular smooth muscle cells. Nucleic Acids Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piwecka M, Glazar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357). [DOI] [PubMed] [Google Scholar]
- 10.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21 e27. doi: 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi F, Legnini I, Megiorni F, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Li Z, Yuan H, et al. Reciprocal regulatory mechanism between miR-214-3p and FGFR1 in FGFR1-amplified lung cancer. Oncogenesis. 2019;8(9):50. doi: 10.1038/s41389-019-0151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Bo X, Zheng Q, Xiao X, Wu L, Li B. miR-296 inhibits proliferation and induces apoptosis by targeting FGFR1 in human hepatocellular carcinoma. FEBS Lett. 2016;590(23):4252–4262. doi: 10.1002/1873-3468.12442 [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Wang Y, Lu Z, et al. miR-127 promotes EMT and stem-like traits in lung cancer through a feed-forward regulatory loop. Oncogene. 2017;36(12):1631–1643. doi: 10.1038/onc.2016.332 [DOI] [PubMed] [Google Scholar]
- 16.Ye YY, Mei JW, Xiang SS, et al. MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis. 2018;9(3):410. doi: 10.1038/s41419-018-0444-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23(4):604–615. doi: 10.1101/gr.139758.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Cai J, Fang C, et al. Mesenchymal glioblastoma constitutes a major ceRNA signature in the TGF-beta pathway. Theranostics. 2018;8(17):4733–4749. doi: 10.7150/thno.26550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 20.Shitara K, Ito S, Misawa K, et al. Genetic polymorphism of IGF-I predicts recurrence in patients with gastric cancer who have undergone curative gastrectomy. Ann Oncol. 2012;23(3):659–664. doi: 10.1093/annonc/mdr293 [DOI] [PubMed] [Google Scholar]
- 21.Boku N, Ryu MH, Kato K, et al. Safety and efficacy of nivolumab in combination with S-1/capecitabine plus oxaliplatin in patients with previously untreated, unresectable, advanced, or recurrent gastric/gastroesophageal junction cancer: interim results of a randomized, Phase II trial (ATTRACTION-4). Ann Oncol. 2019;30(2):250–258. doi: 10.1093/annonc/mdy540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Cutsem E, Boni C, Tabernero J, et al. Docetaxel plus oxaliplatin with or without fluorouracil or capecitabine in metastatic or locally recurrent gastric cancer: a randomized phase II study. Ann Oncol. 2015;26(1):149–156. doi: 10.1093/annonc/mdu496 [DOI] [PubMed] [Google Scholar]
- 23.Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi: 10.1007/s00018-017-2688-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao W, Huang X, Wang L, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38(1):169. doi: 10.1186/s13046-019-1136-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 26.Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 27.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 28.Hesling C, Fattet L, Teyre G, et al. Antagonistic regulation of EMT by TIF1gamma and Smad4 in mammary epithelial cells. EMBO Rep. 2011;12(7):665–672. doi: 10.1038/embor.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung MK, Jung YH, Lee JK, et al. CD271 confers an invasive and metastatic phenotype of head and neck squamous cell carcinoma through the upregulation of slug. Clin Cancer Res. 2018;24(3):674–683. doi: 10.1158/1078-0432.CCR-17-0866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Zhu D, Qin S. SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation. J Exp Clin Cancer Res. 2018;37(1):148. doi: 10.1186/s13046-018-0819-y [DOI] [PMC free article] [PubMed] [Google Scholar]