Abstract
Interactions are reported of three representative naphthalenediimide derivatives with three quadruplex targets, from the promoter region of the telomerase (hTERT) gene, a human telomeric DNA quadruplex, and a telomeric RNA quadruplex (TERRA). Thermal melting studies showed that these compounds strongly stabilize the quadruplexes, with weak stabilization of a duplex DNA. Binding studies by surface plasmon resonance and fluorescence spectroscopy found that the compounds bind to the quadruplexes with nanomolar equilibrium dissociation constants. Plausible topologies for the quadruplex complexes were deduced from CD spectra, which together with the surface plasmon resonance data indicate that the quadruplexes with parallel quadruplex folds are preferred by two compounds, which was confirmed by qualitative molecular modeling.
Keywords: Quadruplex, naphthalene diimides, surface plasmon resonance, fluorescence, modeling
Quadruplex (G4) nucleic acids are higher-order four-stranded arrangements formed around guanine quartets.1,2 Their occurrence in the human genome at telomere ends3 and within promoter sequences4 of cancer initiation and progression genes1,5−7 is currently the focus of efforts to develop G4 therapeutics.8−11 One approach involves tandem telomeric DNA repeats of the sequence TTAGGG, which can be folded into G4s by appropriate G4-binding small molecules, which then inhibit telomerase enzyme complex activity.12,13 Telomerase plays a major role in tumorigenesis with up-regulated expression in the majority of human cancers.14 Promoter mutations of hTERT, the catalytic subunit gene of telomerase, occur in several human cancers.15 The G4-forming sequences in this promoter16−19 can be the target of G4-selective small molecules,20 inhibiting hTERT expression. Both telomere and hTERT G4 targeting can produce cancer cell growth inhibition and antitumor activity.20,21
Bioinformatics22,23 and sequencing-based6,24 estimates of the number of putative G4 structures present in the human genome suggest widespread occurrence. An experimental study on active chromatin6 suggesting ca. 10,000 occurrences is the most realistic to date. However, there is no current methodology to reliably predict the folding of all G4s. A topology with parallel G4 strands is consistent with the presence of short loops in a G4.23,25 Many such G4s encoded in cancer-related gene promoter sequences may form parallel-stranded G4s, as do the majority of RNA G4s, including those formed by human telomeric (TERRA) RNA sequences.26−28 Human telomeric DNA G4 sequences can fold into a variety of topologies, with parallel,29 antiparallel, and mixed strand orientations.30,31 Crystallographic and/or NMR structural data are available for some of these G4s, though fewer on G4 complexes with small molecules.32−35 These show a consistent pattern of binding geometry, with the planar groups of a typical G4 ligand being stacked onto a terminal G-quartet of a G4 and the ligand side-chains (typically cationic-charged), residing in grooves or loops of the G4. Since much of the binding energy of a G4 ligand is driven by π–π stacking to a terminal G-quartet, the accessibility to its planar surface is an important determinant of overall ligand binding. Accessibility is dependent on G4 topology,36 with the human telomeric parallel G429 providing a greater solvent-accessible surface area than (3 + 1) mixed parallel–antiparallel arrangements.
A number of substituted naphthalenediimide (ND) derivatives have been reported, that can target G4s with high affinity and induce a range of biological effects in cells.37−43 The present study focuses on a group of three bioactive ND compounds from successive generations of ND development (Figure 1).41−43 All have cellular and in vivo antitumor activity ascribable to G4 targeting. Two, CM03 and SOP1812, have been shown to target multiple cancer-related G4s.41,42 G4-specific antibody immunofluorescence staining has suggested that both CM03 and SOP1812 increase cellular G4 occurrence. We report here on the G4-binding properties of these compounds and implications for G4-ND drug design.
Figure 1.
Structures of the naphthalenediimide derivatives used in this study.
Results
Three G4-forming sequences were used:
hTERT promoter sequence 5′-AGG GGA GGG GCT GGG AGG GC-3′ (G4-hTERT);
telomeric RNA 5′-GGG UUA GGG UUA GGG UUA GGG-3′ (G4-TERRA);
telomeric DNA 5′-GGG TTA GGG TTA GGG TTA GGG-3′ (G4-HTR).
Experimental details are in the Supporting Information.
Thermal Stability Studies
UV melting44 (Figure S1) was used to estimate the ability of the compounds to stabilize DNA G4s and a control duplex DNA (a TERT hairpin duplex containing the hTERT sequence plus the complementary strand and a terminal hairpin). Ligand:DNA ratios beyond 1:1 result in inconsistent melting curves (data not shown), possibly caused by aggregation—MM41 and G4-hTERT also gave inconsistent results at a 1:1 ratio. Since MM41 and SOP1812 have similar structures, the former’s ΔTm (Table 1) value is likely to be closer to that of SOP1812. All compounds are selective for the G4 DNA sequences, with minimal changes in ΔTm with the hairpin duplex. All show a preference for stabilizing G4-hTERT, with SOP1812 having the greatest ΔTm, of 18 °C.
Table 1. UV Thermal Melting Changes (ΔTm) for Small Compound-Quadruplex Complexes and a Control Duplex Sequencea.
| G4-hTERT, ΔTm(°C)(Tm= 67 °C) | G4-HTR, ΔTm(°C)(Tm= 62 °C) | Duplex DNA, ΔTm(°C)(Tm= 85 °C) | |
|---|---|---|---|
| SOP1812 | 18 | 10 | 2 |
| CM03 | 13 | 5 | 2 |
| MM41 | >16b | 11 | <2 |
The thermal melting results are averages of two independent trials and are reproducible within ±1 °C. Tm values of these sequences without compounds are listed above.
Thermal melting changes of MM41with G4-hTERT resulted in a nonstandard melting curve. We therefore predicted the ΔTm value based on the similarity in structures between MM41 and SOP1812.
Surface Plasmon Resonance Analyses
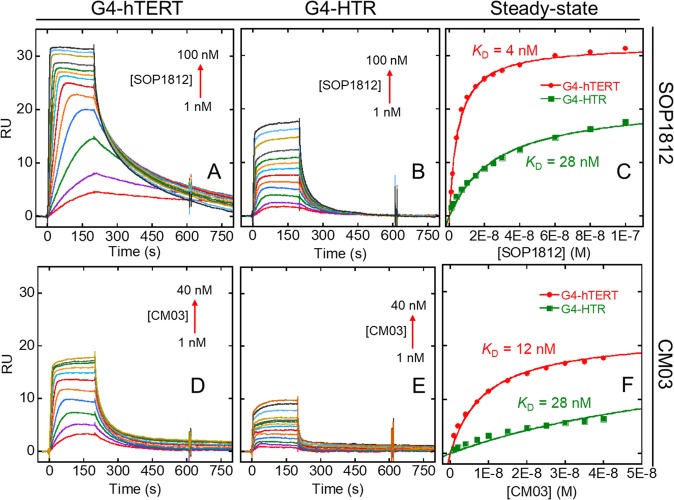
SPR methodology45 provided quantitative binding data for the compounds with the G4s (Figures 2 and 3). All have consistently low nM KD values with G4-hTERT. For SOP1812 with G4-hTERT, the sensorgrams show slow dissociation (Figure 2A), compared with faster association/dissociation with G4-HTR (Figure 2B). Sensorgram results with CM03 are suggestive of rapid association/dissociation for G4-hTERT and G4-HTR sequences. MM41 also shows nanomolar binding affinity toward both G4 DNAs. The sensorgrams with MM41 (Figure S3) do not show a regular dose–response pattern, possibly as a result of nonspecific interactions and/or adsorption of the compound at high concentrations on the sensor chip surface, although the data was fitted to a steady-state plot giving reliable KD values.
Figure 2.
Representative sensorgrams and steady state equilibrium binding of (A–C) SOP1812 and (D–F) CM03 to G4-hTERT and G4-HTR. The concentrations of each compound are shown, and the arrows show the direction of increased response in the sensorgrams with increased concentration. The steady state shows the binding plot at equilibrium. The steady-state response unit (RU) at lower concentration in A was predicted by kinetic estimation. Binding constants were obtained for a single-site binding model (Table 2).
Figure 3.
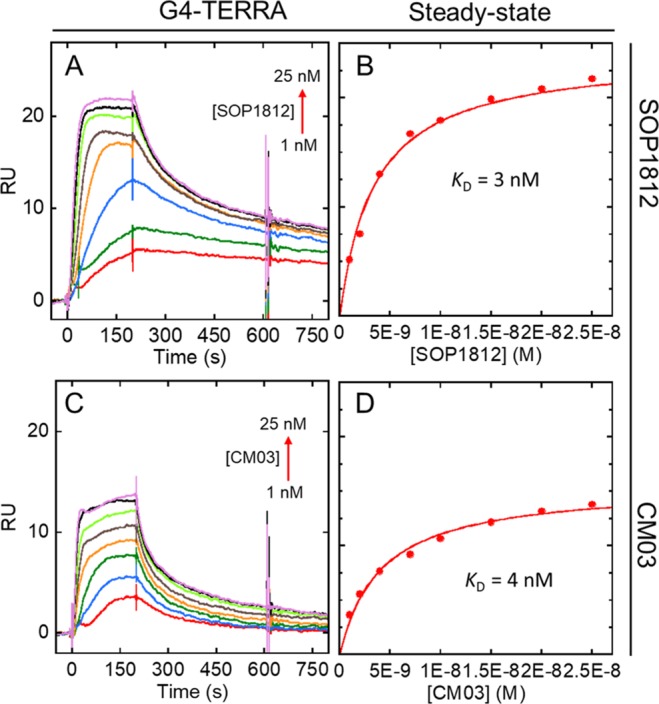
Representative sensorgrams and steady state equilibrium binding of (A, B) SOP1812 and (C, D) CM03 to G4 RNA G4-TERRA. Plots are as in Figure 2. Binding constants were obtained for a single-site binding model (Table 2).
The dissociation of CM03 from G4-hTERT is nanomolar (Table 2) although that to G4-HTR is weaker. The KD is 82 nM, i.e. about 7-fold lower. The results for SOP1812 show greater dissociation from G4-hTERT (KD = 4 nM), also compared to G4-HTR (KD = 28 nM). The differences in RUbound for these two compounds binding to G4-hTERT compared to G4-HTR might indicate higher stoichiometry (>1:1 binding ratio) for interaction with G4-hTERT. The steady-state fit does not improve significantly with a 2:1 binding model, with the dissociation (i.e., KD1 and KD2) within 5-fold range of the KD values obtained by single-site fitting (data not shown). This result suggests there is an additional equivalent binding site available on G4-hTERT, but not on G4-HTR.
Table 2. Dissociation Equilibrium Binding Constant KD for Compounds CM03, MM41, and SOP1812 with Quadruplex Sequencesa.
|
G4-hTERT,KD(nM) |
G4-HTR,KD(nM) |
G4 TERRA,KD(nM) |
||||
|---|---|---|---|---|---|---|
| SPR | Fluorescence | SPR | Fluorescence | SPR | Fluorescence | |
| CM03 | 12 ± 2 | 16 ± 2 | 82 ± 6 | 94 ± 5 | 4 ± 1 | 3 ± 1 |
| MM41 | 2 ± 1 | 4 ± 1 | 5 ± 1 | 10 ± 2 | 3 ± 1 | 2 ± 1 |
| SOP1812 | 4 ± 1 | 4 ± 1 | 28 ± 2 | 29 ± 2 | 3 ± 1 | 2 ± 1 |
The steady-state binding result obtained as a function of free ligand concentration was fitted to a one-site binding model to obtain KD values.
SPR showed weak duplex DNA interactions for SOP1812 and CM03 (Figure S2). The sensorgrams suggest that generic nonspecific interactions take place between compounds and duplex, with rapid dissociation/association. A steady-state was not reached at saturation even at high compound concentrations (≤100 nM). This could be due to electrostatic interactions between the positively charged compounds with negative duplex DNA. Approximate KD values are estimated to be >100 nM for both compounds.
Interactions with G4-TERRA RNA G4 were also studied by SPR (Figure 3). Sensorgrams with CM03 and SOP1812 gave low nM KD values. Steady-state analysis (Figures 3B, D) suggests that the observed RUbound indicates that higher stoichiometries may exist. Analogous to G4-hTERT, a steady-state fit with a 2:1 binding model resulted in two similar low nM KD values for SOP1812 and CM03, indicating a possible secondary independent binding site on G4-TERRA. Since hTERT and TERRA adopt parallel quadruplex structures, it is possible that the additional binding sites in hTERT and TERRA are the consequence of two similar terminal quartets, as both compounds prefer to stack on the end terminal of G4 structures (vide infra).
Fluorescence Titration and Anisotropy
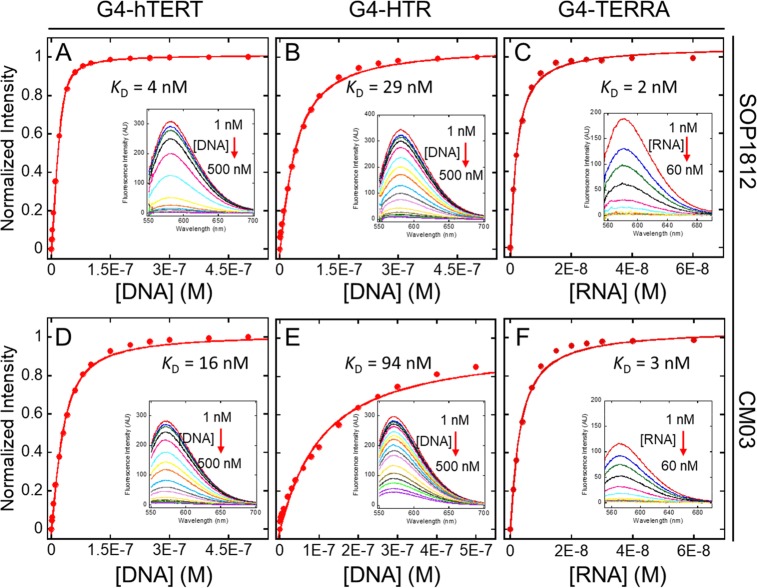
CM03 and SOP1812 have strong fluorescence emission signals (λex = 525 nm and λem = 580 nm for SOP1812; λex = 515 nm and λem = 570 nm for CM03) at nanomolar concentrations, making them suitable for determining binding affinities by fluorescence spectroscopy. The fluorescence titrations of CM03 and SOP1812 with the three G4s (Figure 4) show significant quenching of intensity on ligand binding; almost full fluorescence intensity is quenched in the titration. KD values were obtained (Table 2) by fitting the change in the total intensity at the maxima λem = 570 and 580 nm using eq 1 in Supporting Information for a single binding site. Those for CM03 and SOP1812 are consistently in good agreement with KD data from the SPR analyses.
Figure 4.
Intrinsic fluorescence titration of SOP1812 (A, B, C) and CM03 (D, E, F) to G4s. (A, D) For G4-hTERT; (B, E) for G4-HTR; (C, F) for G4-TERRA. Insets shown the total change in fluorescence spectrum with maximum λem = 580 nm (SOP1812) and λem = 570 nm (CM03). Red arrows show the decrease in total fluorescence intensity upon titration with G4 concentrations to 500 nM, compounds at a constant concentration of 25 nM for G4 DNA titrations and 10 nM for G4 RNA titrations. Triplicate readings were averaged.
MM41 has a strong fluorescence emission at the far end of visible light (λex = 630 nm and λem = 670 nm). Unlike CM03 and SOP1812, the intrinsic fluorescence intensity of MM41 did not significantly quench upon G4 titration, making it suitable for a fluorescence anisotropy study: reasons for this are unclear. The interaction between MM41 and G4 can be observed since MM41 molecular size is small compared to the G4s (∼600 Da versus ∼6000 Da for the G4s). Figure 5 shows the binding curves of MM41 with G4-HTR, G4-TERT, and G4-TERRA, from fluorescence anisotropy measurements. MM41 shows strong binding with all three G4s, with nanomolar KD values (Table 2), close to those from SPR, apart from that for G4-HTR, which differ by 5σ. This may be a real effect, possibly reflecting aggregation in these particular SPR experiments. Overall the SPR and fluorescent data for MM41 agree that it has less selectivity compared to CM03 and SOP1812.
Figure 5.
Fluorescence titration of MM41 to quadruplex DNA (A) with G4-hTERT, (B) with G4-HTR, and (C) with G4-TERRA. The change in anisotropy (r value) was calculated based on the change in MM41 polarized fluorescence intensity upon increasing G4 concentration. MM41 concentration was constant at 50 nM with the total change between final and initial volume <10%.
Circular Dichroism (CD) Reveals Ligand-Bound G4 Topologies
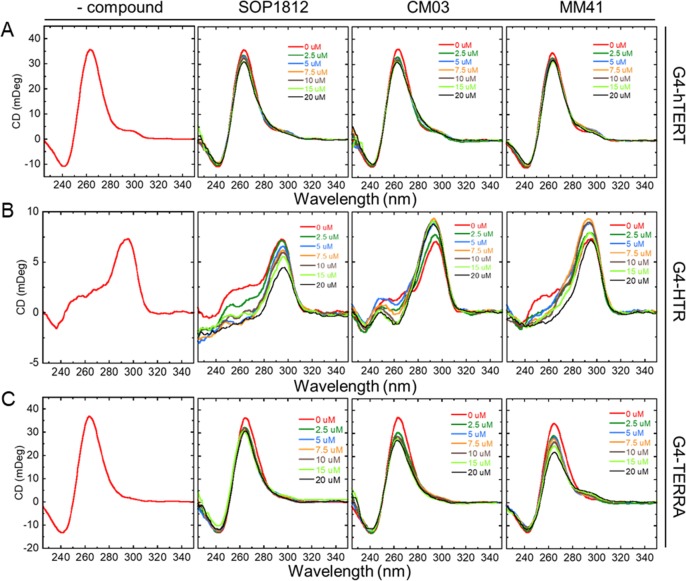
The CD spectra for each native G4 (Figure 6) show unambiguously that G4-hTERT and G4-TERRA adopt a parallel topology in the conditions used, with peak maxima around 265 nm and minima around 245 nm.36 The G4-HTR CD signal is consistent with a hybrid (3 + 1) form having three parallel and one antiparallel strands, with a maximum at 295 nm, a shoulder around 260 nm, and a negative peak at 240 nm.
Figure 6.
CD spectra of compounds titrated with G4s. (Row A) G4-hTERT. (Row B) G4-HTR. (Row C) G4-TERRA. CD spectra of native G4s are in the far-left column. Titrations with each compound are indicated. Each G4 concentration was constant at 5 μM with increasing compound concentrations up to 20 μM. Duplicate readings were averaged.
The effects of titration of each compound on the CD spectra (Figure 6) do not show significant ICDs, up to a 4:1 ratio of compound:G4 (i.e., 20 μM of compounds in 5 μM G4) and at any wavelength. This suggests that all three bind to the two parallel G4s by stacking at G-quartet terminals, without inducing any G4 topology changes. Titration of the compounds to G4-HTR shows a decrease in G4 peak intensity at 265 nm while other peaks at 295 nm and 240 remained unchanged. This suggests that the compounds might drive the (3 + 1) hybrid structure of G4-HTR to an antiparallel structure at higher ligand concentrations, although no significant ICDs were apparent, consistent with end-quartet binding. In summary, all ligands bind to these G4s with minimal structural perturbation.
Potential Ligand-G4 Structural Arrangements
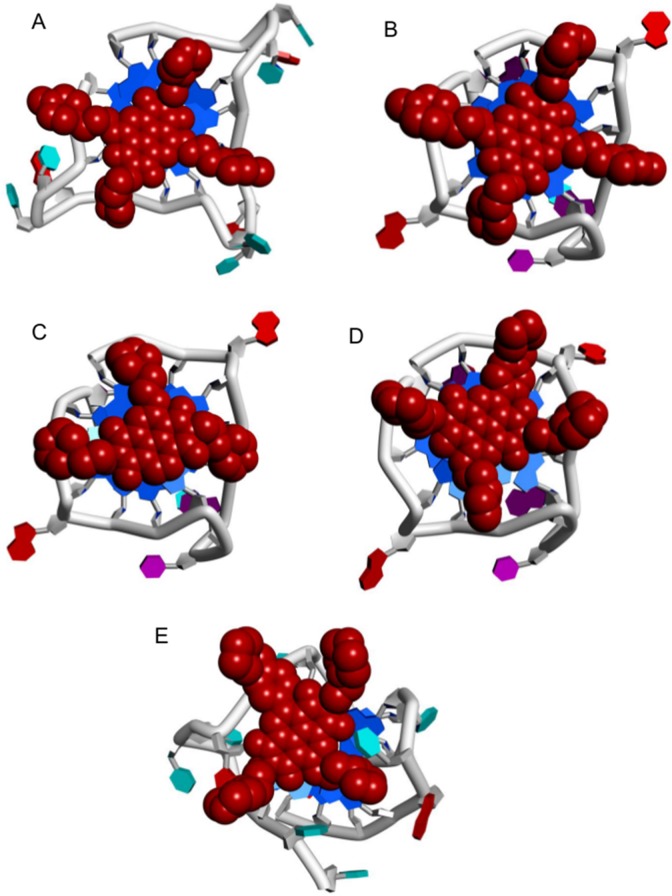
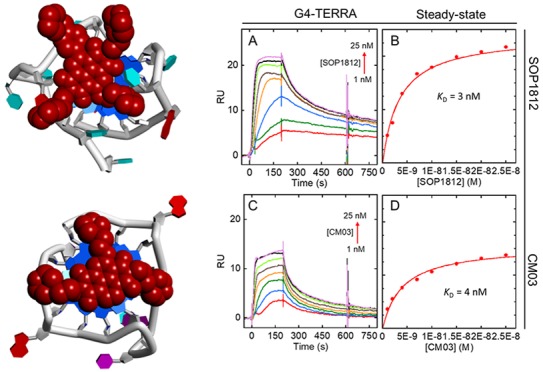
Two crystal structures of MM41 are available, bound to a human telomeric G4 (PDB ids 3UYH, 4DA3).40 The MM41 molecules (ND cores and substituents) in both structures are in closely related positions, with ⟨rmsd⟩ of 0.76 Å. 3UYH, with the G4-HTR, is at the higher resolution (1.95 Å) and has (Figure 7A) the ND core end-stacked onto a terminal G-quartet and extensive overlap between core and guanine bases. MM41 substituents each reside in a G4 groove. These features were also observed in other cocrystal structures of NDs with human telomeric G4s,39,40 demonstrating the consistency in binding mode of this family of ND compounds. This consensus binding pose suggests plausible structures for the complexes of all three compounds MM41, CM03, and SOP1812 bound to a parallel topology G4-hTERT (PDB id 2KZE).16 G4-TERRA has a parallel topology, with UUA trinucleotide loops.27,28 We have assumed that the overall structural features of the G4-TERRA complexes with the ND compounds would not vary significantly from those of G4-hTERT and have not been explored in detail. The G4-HTR sequences in these cocrystal structures adopt an all-parallel topology identical to that observed in the native crystal structure.
Figure 7.
Views of G4-ND complexes, projected onto a terminal G-quartet. NDs are dark red CPK solids. G4s are in cartoon form with guanines colored blue. (A) MM41-G4-HTR crystal structure (PDB id 3UYH). (B–D) Superposition of MM41, CM03, and SOP1812 onto the equivalent binding pose in the G4-hTERT structure (PDB id 2KZE). (E) SOP1812 in the ligand binding site of the (3 + 1) human telomeric G4 telomestatin derivative complex (PDB id 2MB3).
The plausible G4-hTERT complexes with the compounds (Figures 7B–D) retain the ND chromophore-G-quartet overlap seen in the MM41 crystal structures. The positioning of MM41 and SOP1812, each with four substituents, on a terminal G-quartet of the G4-hTERT parallel structure, is severely restrained by the geometry of the loops, and the arrangements closely resemble that found experimentally for MM41 with G4-HTR (Figures 7B, D). The side-chains reside at the mouths of the grooves, but none are sufficiently long to penetrate deeply into the grooves: The crystal structures show side-chains held in place by hydrogen bonds to water molecules populating the grooves. The arrangement for CM03 (Figure 7C), with three substituent arms, resembles these first three, albeit with the loss of potential contacts in a fourth groove. This design concept, of side-chains residing in grooves and in contact with loops, is analogous to that used in the design of the trisubstituted acridine compound BRACO19, as well as in the more recent design of several potent substituted macrocyclic G4 binding compounds.47−49
The solution CD data indicates that G4-HTR adopts a (3 + 1) hybrid mixed parallel-antiparallel strand topology,31 which is invariant on ligand binding, in the concentrations examined here. The NMR structure of a human telomeric G4-telomestatin analogue complex35 (PDB id 2MB3), has been used to examine how ND compounds might interact with the (3 + 1) G4-HTR, with SOP1812 as the exemplar (Figure 7E). One side of the G-quartet binding site is bounded by a lateral TTA loop, which hinders the ND chromophore from fully stacking onto the quartet surface. Thus, the ND-quartet overlap (Figure 7E) is less than that in the all-parallel structures (Figure 7A–D).
Conclusions
We report here biophysical studies on three structurally related ND compounds, each having cationic side-chains, which have been previously shown to possess potent ability to inhibit proliferation in cancer cell lines and antitumor activity in animal models.41−43 It is shown here that they also have high affinity for a small group of human G4 structures. Results from SPR are in consistently good agreement with those from fluorescence binding studies, so any effects due to aggregation or adsorption onto the SPR chip can be discounted, apart from some data with MM41. All three compounds have low nM KD values with G4-hTERT and G4-TERRA. CM03 and SOP1812 bind 5-fold more weakly to G4-HTR compared to G4-hTERT and G4-TERRA, in contrast to MM41. All bind quite weakly to a DNA duplex. A future goal will be to determine their cellular G4 affinities.
CD can define the overall conformation of a G4 and provides suggestive data on plausible binding modes for ligands upon complex formation using ICD signals. The absence of, or a weak ICD signal indicates that a compound induces little or no structural perturbation in the target G4, with end-stacking the most plausible binding mode, in accordance with crystallographic and NMR studies. A strong ICD signal indicates significant induced structural perturbation, such as that from groove changes or intercalation between G-quartets. The CD data shows that G4-hTERT and G4-TERRA adopt parallel topologies, in accord with NMR and crystallographic findings.16,27,28 Data from this small panel of G4s suggests that G4 topology plays an important role in the strength of the interactions of these ND compounds. The preference for G4 parallel topology is supported by models of ND binding on to the 5′ and 3′ terminal G-quartets of G4-hTERT and G4-TERRA (Figure 7).
The concept, supported here by (limited) evidence from two human G4s, that high-affinity G4-binding small molecules may exhibit a preference for binding to multiple G4s with parallel topology, is in agreement with recent cellular studies indicative of multiple targeting.37,42,50 This may well find a role in the treatment of those complex human cancers characterized by the dysregulation of multiple G4-containing genes and pathways. Some small molecules can target individual driver genes via G4 formation,41−46 although full transcriptome analyses have not been reported on these systems. Some G4 ligands can be selective51in vitro and maybe in vivo, as shown with hTERT,19 c-KIT, KRAS, BCL2, and MYC G4s.52−56 Since some of these genes are established drivers of particular cancers, targeting them may be useful in those instances, and perhaps also in early stage disease prior to multiple gene dysregulation. All three compounds show antitumor activity in vivo against human cancer xenograft models. CM03 and SOP1812 show activity against the demanding KPC model for pancreatic cancer.42,43 Pharmacological properties of these two compounds are surprisingly favorable,43 given their relatively high molecular weight and burden of positive charge. This suggests that their combination of ND scaffold and particular side-chain substitution may confer sufficient efficacy for consideration in the future for clinical evaluation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00041.
Supplementary figures and extended descriptions of the methodology for the biophysical experiments (PDF)
Author Present Address
∥ (T.V.) National Cancer Institute, National Institute of Health, Bethesda, MD 20892, USA.
Author Contributions
W.D.W. and S.N. devised this project. C.M., S.A.O., S.O., and R.A. devised the synthetic routes and synthesized the compounds. W.D.W. designed the biophysical studies, which were carried out by T.V. and analyzed by W.D.W. and T.V. The manuscript was written by W.D.W., T.V., and S.N.
S.N. thanks the Medical Research Council, the Wellcome Trust and the UCL Technology Fund for financial support. T.V. was supported by the Molecular Basis of Disease (MBD) fellowship from the College of Arts and Sciences, Georgia State University. Work in Atlanta was supported by the US National Institutes of Health (NIH) (Grant No. GM111749 to W.D.W. and D. W. Boykin).
The authors declare no competing financial interest.
Dedication
In memory of Professor Maurizio Botta, a superb scientist and wonderful friend.
Supplementary Material
References
- Spiegel J.; Adhikari S.; Balasubramanian S. The structure and function of DNA G-quadruplexes. Trends in Chem. 2020, 2, 123–136. 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S.; Parkinson G. N.; Hazel P.; Todd A. K.; Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist W. I.; Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.; Lipps H. J. G-quadruplexes and their regulatory roles in cancer. Nucleic Acids Res. 2015, 43, 8627–8637. 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- Zyner K. G.; et al. Genetic interactions of G-quadruplexes in humans. eLife 2019, 8, e46793 10.7554/eLife.46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Hurley L. H.; Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discovery 2011, 10, 261–275. 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo R.; Palumbo M.; Sissi C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta, Gen. Subj. 2017, 1861, 1399–1413. 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nature Rev. Chem. 2017, 1, 0041 10.1038/s41570-017-0041. [DOI] [Google Scholar]
- Asamitsu S.; Obata S.; Yu Z.; Bando T.; Sugiyama H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, E429 10.3390/molecules24030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997, 40, 2113–2116. 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- Islam M. K.; Jackson P. J.; Rahman K. M.; Thurston D. E. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016, 8, 1259–1290. 10.4155/fmc-2015-0017. [DOI] [PubMed] [Google Scholar]
- Shay J. W.; Wright W. E. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- Vinagre J.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- Lim K. W.; Lacroix L.; Yue D. J.; Lim J. K.; Lim J. M.; Phan A. T. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010, 132, 12331–12342. 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- Yu Z.; et al. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J. Am. Chem. Soc. 2012, 134, 5157–5164. 10.1021/ja210399h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires J. B.; et al. An improved model for the hTERT promoter quadruplex. PLoS One 2014, 9, e115580 10.1371/journal.pone.0115580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsen R. C.; et al. The hTERT core promoter forms three parallel G-quadruplexes. Nucleic Acids Res. 2020, 10.1093/nar/gkaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. H.; et al. Small-molecule-targeting hairpin loop of hTERT promoter G-quadruplex induces cancer cell death. Cell Chem. Biol. 2019, 26, 1110–1121. 10.1016/j.chembiol.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A. M.; Dai F.; Schultes C. M.; Reszka A. P.; Moore M. J.; Double J. A.; Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd A. K.; Johnston M.; Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers V. S.; Marsico G.; Boutell J. M.; Di Antonio M.; Smith G. P.; Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- Hazel P.; Huppert J. H.; Balasubramanian S.; Neidle S. Loop-length dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- Randall A.; Griffith J. D. Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J. Biol. Chem. 2009, 284, 13980–13986. 10.1074/jbc.M900631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martadinata H.; Phan A. T. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009, 131, 2570–2578. 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- Collie G. W.; Haider S. M.; Neidle S.; Parkinson G. N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010, 38, 5569–5580. 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson G. N.; Lee M. P. H.; Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Lim K. W.; et al. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Luu K. N.; Patel D. J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell N. H.; Parkinson G. N.; Reszka A. P.; Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- Dai J.; Carver M.; Hurley L. H.; Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011, 133, 17673–17680. 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie G. W.; Promontorio R.; Hampel S. M.; Micco M.; Neidle S.; Parkinson G. N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- Chung W. J.; Heddi B.; Tera M.; Iida K.; Nagasawa K.; Phan A. T. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J. Am. Chem. Soc. 2013, 135, 13495–13501. 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- Haider S. M.; Autiero I.; Neidle S. Surface area accessibility and the preferred topology of telomeric DNA quadruplex-ligand complexes. Biochimie 2011, 93, 1275–1279. 10.1016/j.biochi.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Pirota V.; Nadai M.; Doria F.; Richter S. N. Naphthalene diimides as multimodal G-quadruplex-selective ligands. Molecules 2019, 24, E426 10.3390/molecules24030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recagni M.; Greco M. L.; Milelli A.; Minarini A.; Zaffaroni N.; Folini M.; Sissi C. Distinct biological responses of metastatic castration resistant prostate cancer cells upon exposure to G-quadruplex interacting naphthalenediimide derivatives. Eur. J. Med. Chem. 2019, 177, 401–413. 10.1016/j.ejmech.2019.05.066. [DOI] [PubMed] [Google Scholar]
- Micco M.; et al. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013, 56, 2959–2974. 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- Collie G. W.; Promontorio R.; Hampel S. M.; Micco M.; Neidle S.; Parkinson G. N. Structural basis for telomeric G-quadruplex naphthalene diimide ligand targeting. J. Am. Chem. Soc. 2012, 134, 2723–2731. 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- Ohnmacht S. A.; et al. A G-quadruplex-binding compound showing anti-tumor activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015, 5, 11385. 10.1038/srep11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti C.; et al. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J. Med. Chem. 2018, 61, 2500–2517. 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.; et al. , to be published.
- Rachwal P. A.; Fox K. R. Quadruplex melting. Methods 2007, 43, 291–301. 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Paul A.; Musetti C.; Nanjunda R.; Wilson W. D. Biosensor-surface plasmon resonance: label-free method for investigation of small molecule-quadruplex nucleic acid interactions. Methods Mol. Biol. 2019, 2035, 63–85. 10.1007/978-1-4939-9666-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Villar-Guerra R.; Trent J. O.; Chaires J. B. G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem., Int. Ed. 2018, 57, 7171–7175. 10.1002/anie.201709184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M.; et al. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 4844–4849. 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby T.; et al. Disabling c-Myc in childhood medulloblastoma and atypical teratoid/rhabdoid tumor cells by the potent G-quadruplex interactive agent S2T1–6OTD. Mol. Cancer Ther. 2010, 9, 167–179. 10.1158/1535-7163.MCT-09-0586. [DOI] [PubMed] [Google Scholar]
- Ma Y.; et al. Development of G-quadruplex ligands for selective induction of a parallel-type topology. Org. Biomol. Chem. 2018, 16, 7375–7382. 10.1039/C8OB01702F. [DOI] [PubMed] [Google Scholar]
- Amato J.; et al. Tailoring a lead-like compound targeting multiple G-quadruplex structures. Eur. J. Med. Chem. 2019, 163, 295–306. 10.1016/j.ejmech.2018.11.058. [DOI] [PubMed] [Google Scholar]
- Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016, 59, 5987–6011. 10.1021/acs.jmedchem.5b01835. [DOI] [PubMed] [Google Scholar]
- Brown R. V.; Danford F. L.; Gokhale V.; Hurley L. H.; Brooks T. A. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 2011, 286, 41018–41027. 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. B.; Elsayed M. S. A.; Wu G.; Deng N.; Cushman M.; Yang D. Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J. Am. Chem. Soc. 2019, 141, 11059–11070. 10.1021/jacs.9b02679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelliccia S.; et al. Bio-inspired dual-selective BCL-2/c-MYC G-quadruplex binders: design, synthesis, and anticancer activity of drug-like imidazo[2,1-i]purine derivatives. J. Med. Chem. 2020, 63, 2035. 10.1021/acs.jmedchem.9b00262. [DOI] [PubMed] [Google Scholar]
- See for example:Gunaratnam M.; et al. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J. Med. Chem. 2009, 52, 3774–3783. 10.1021/jm900424a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrado J.; et al. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci. Rep. 2015, 5, 9696. 10.1038/srep09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.