Abstract
Circular RNAs, as hopeful diagnosis markers and therapeutic molecules, have been studied, probed and applied into several diseases, such as cardiovascular diseases, systemic lupus erythematosus, leukemia, pulmonary tuberculosis, and cancer especially. Recently, mounting evidence has supported that circRNAs play a key role in the tumorigenesis, progress, invasion and metastasis in lung cancer. Its special structure—3′–5′ covalent loop—allow it to execute several special functions in both normal eukaryotic cells and cancer cells. Our review summaries the latest studies on characteristics and biogenesis of circRNAs, and highlight the regulatory functions about miRNA sponge of lung-cancer-related circRNAs. In addition, the interaction of the circRNA-miRNA-mRNA regulatory network will also be elaborated in detail in this review. Therefore, this review can provide a new idea or strategy for further development and application in clinical setting in terms of early-diagnosis and better treatment.
Keywords: Circular RNAs, Lung neoplasms, ceRNA-miRNA-mRNA, miRNA sponge
Background
In the 21st century, cancer is still one of the most life threating diseases of the global health and wasting ever-increasing amounts of medical resources at the global level, and lung cancer is a highly heterogeneous malignance disease, which reaches the first in both the incidence and mortality of all-cancer (accounting for 11.6% of the overall total of 18.1 million new cases and 18.4% of the total number of 9.6 million cancer deaths respectively) in 2018 [1]. Thus, future curative strategies should be made to gain better understanding of cancer development, early diagnosis and effective treatment in oncology. In recent years, drug precise treatments targeting proteins encoding by EGFR, ALK, KRAS, mTOR, ErbB2 and PI3K, and anti-tumor immunity immune check point blockade, like PD-1, PD-L1 and CTLA4 inhibitors, emerge as promising treatments for human malignancies especially a more profound antitumor effect in patients with advanced lung carcinoma [2, 3]. By in-depth study, development of the new molecules therapeutic targets, including non-coding RNAs (ncRNAs), are the novel subjects of the research program. microRNA (miRNA), long non-coding RNA (lncRNA) and circular RNA (circRNA) are the top three members of ncRNAs in researches when search in the Pubmed. Arguably, increasing accumulating researches have been carried out to explicate the functions of circRNAs. Considered as the noisy transcripts for a long time at the beginning, circRNAs are kinds of single-stranded circular transcripts with a 3′–5′ covalently closed link [4]. Thanks to this special structure and to the fact that, unlike normal linear mRNA, circRNAs keep stable and resist to exonuclease. Derived from exons and (or) introns, circRNAs have certain evolutionary conservation and feature as tissue-specific expression.
Although Ma et al. have summarized the characteristics, biogenesis, function and progress of circRNA in lung cancer up to May 2018 [5], many published studies report new circRNAs involved in diagnosis biomarkers and therapeutic molecules in 2018. The current review was aimed to update the knowledge of circRNAs in lung cancer and focusing more on the effect of circRNA-miRNA-mRNA regulatory axes in lung cancer.
Characteristics of circRNA
CircRNAs, identified as brand-new star molecules, are abundantly expressed in a variety of eukaryotic cells, that are transcribed from a considerable number of genes both in human and animals [6]. Without the 5′-cap structure and 3′-ploy(A) tail, circRNAs with a structure of closed loop covalently links from 5′-head to 3′-tail [4]. In contrast with linear RNAs, a free 3′ overhang with at least 7 nucleotides required for exoribonuclease, circRNAs are more resistant to degradation by RNase or RNA exonucleases so that circRNAs are able to maintain a stable loop [7]. In 1970s, Sanger et al. confirmed the existence of circRNA firstly in viroid propagated in tomatoes and then 3 years later, circRNA was discovered in eukaryotic cells by electron microscope [8]. Due to the limitation of the technology, circRNAs are considered as nonsense RNAs or transcripts noises for a long time, which led to less attentions to its biological functions. Integrated with advanced technology of RNA sequencing (RNA-seq) and bioinformatics, it was supported that circRNAs exerted superior properties on diagnosis and treatment in clinical setting. It is reported that circRNAs are less abundant than their cognate linear transcripts. Some tissue-specific circRNAs, however, express more highly than their linear transcripts produced from the same gene, such as circTulp4, circRims2, circElf2, circphf21a, etc. in brain tissue [9, 10]. Four features of circRNA can be concluded below: (1) abundance and variety: Memczak and colleagues found that 1950 circRNA in HEK 293 cells, 1903 circRNA in mouse, 1028 circRNA in zebrafish and 724 circRNA in different stages of C. elegans through RNA-seq. High-throughput sequencing profiles revealed that circRNAs are widely expressed in genome transcriptional profiles of different types of normal and malignant human cells [6, 11]. Additionally, some circRNAs express tenfold as many as their associated linear mRNAs [12]; (2) stability: after being injected into fertilized eggs of Xenous laevis, the half-life of circRNAs are at least fivefold longer than linear message RNAs (mRNAs) with or without 5′-cap and 3′-poly(A) [13]; (3) conservation: circRNAs produced by mouse orthologous location of human genes are similar to the RNA products encoding by the same locus of human gene [12]; (4) clear location: most mature circRNAs are located in the cytoplasm, while some of circRNAs containing introns are usually retained in the nucleus [14]; (5) specificity as novel biomarkers, different circRNAs can be secreted into serum from different tissues and cells. There is much evidence to indicate that circRNAs with high specificity and sensitivity play potential roles on many human diseases with respect to early-diagnosis and effective therapy [15].
Classification of circRNA
In most eukaryotic cells, split genes encode RNAs with exons interrupted by introns, which are called precursor mRNAs (pre-mRNAs). Instead of canonical splicing using by linear mRNAs, circRNA transcripts generate themselves with a reverse mechanism, in which 3′ upstream donor connects with 5′ downstream acceptor, called head-to-tail back-splicing [16]. According to their constituent parts and sources, circRNAs can be briefly classified into nine types:
exonic circRNAs (ecircRNAs) EIciRNAs, which account for the main body of circRNAs (more than three fourths of the whole), circulate from head to tail with only one or several exons in the cytoplasm. The later are called exon skipping.
exonic-intronic circRNAs (EIciRNAs) EIciRNAs consist of both exons and introns which retain from back-splicing.
intronic circRNA (ciRNAs) CiRNAs prominently locate in nucleus and are generated from pre-mRNAs by removing the exons and closely connecting the intron’s head to its tail.
intergenic circRNAs These circRNAs are comprised of sequences between two intronic fragments flanked by GT-AG splicing signals, which locate in at least 1 kb away from two genes at both sides.
tRNA intronic circRNAs (tricRNAs) This kind of circRNAs is a special type of intronic circRNAs derived from pre-tRNAs. Intron-containing pre-tRNAs are cut by endonuclease complex resulting in forming intron transcripts and mature tRNAs, thereby tricRNAs were produced.
antisense circRNAs The formation mechanism of this circRNA is the same as ecircRNA, but happening in the opposite strand rather than sense strand.
overlapping circRNAs
circRNA rRNAs (circrRNAs)
intragenic circRNAs This type of circRNA is similar to intergenic circRNA, but the cyclization sequence is less than 1 kb away from either side between two genes [12, 17–21].
Biogenesis of circRNA
The formation of both linear RNA and circRNA are processed by splicing transcriptional sequence derived from pre-mRNA. However, the splicing mechanisms of generating themselves are totally different. Pre-mRNA splicing requires the involvement of spliceosome. As for the formation mechanism of linear mRNAs, pre-mRNAs are processed by spliceosomal machinery, where introns can be removed and exons can be selectively linked in juxtaposition, subsequently resulting in generation of 5′–3′ linked linear transcripts through constitutive splicing (CS) and alternative splicing (AS) mechanism [22]. However, back-splicing in reserve order of canonical splicing, which connects the 3′ upstream splice site (3′ ss) to the 5′ downstream splice site (5′ ss), is the main producing mechanism for circRNA. Indeed, it is well known that three splicing models can be found in the biogenesis of the circRNA: (1) circulation driven by lariat; (2) circulation driven by intron-pairing; (3) splicing intermediated by RNA binding protein (RBP). The detail information about these mechanisms can be concretely described as follow in terms of biogenesis of circRNA. Taking ecircRNAs, EIciRNAs and ciRNAs for representative examples.
EcircRNAs
Diverse ecircRNAs containing one or more exons are generally produced from splicing procedure of pre-mRNAs by the means of two models and three splicing mechanisms [23]. We can remember these as ‘3 + 2’. First, two models, namely, direct back-splicing and exon-skipping, can generate ecircRNAs with single one or several exons [4]. The main difference between two models is which pattern of splicing, canonical splicing or back-splicing, come first. Second, Lariat-driven cyclization, RBP-mediated cyclization and intron-pairing cyclization are the three mechanisms, which purpose to forming diverse ecircRNAs. Lariat-driven cyclization is the main mechanism in the splicing process of pre-mRNAs in the exon-skipping model, while RBP-mediated and intron-pairing cyclization are primarily responsible for the direct back-splicing [24]. For one thing, in the lariat-driven cyclization process, canonical splicing comes first, which will narrow the distance between upstream splice site and downstream splice site. Consequently, a linear mRNA with exons selective inclusion and introns exclusion, as well as a loop structure with one or more exons sequence flanking introns will be generated in the first step. And then spliceosome will cut the redundant introns and join 3′ hydroxyl of exon to 5′ phosphate, thus resulting in forming a 3′–5′ linked lariat-ecircRNA. Such process is called exon-skipping. For another thing, if trans- factors, like RBPs, bind to a pre-mRNA and cis-elements pairing brings the introns together, back-splicing will come first. Either RBPs or intron-pairing will dramatically promote back-splicing. Accordingly, ecircRNAs can be straightly generated at the very start [25]. Although the fact that back-splicing is much lower efficient than canonical splicing maybe attribute to the hypothesis that spliceosome unfavorably bind to pre-mRNA, RBP or introns-pairing can facilitate the splice process, complementing the disadvantage of efficiency of back-splicing [16, 26]. In addition, Memczak et al. identified that AG/GT flanking exons is a splice signal which plays a vital role for selecting breakpoint by spliceosome [6].
EIciRNAs
Introns-containing of EIciRNAs is the major distinction from ecircRNAs with respect to constituent parts. Notably, the mechanisms of generating EIciRNAs are fundamentally similar to ecircRNAs’ [24]. During the formation of ecircRNA, an intermediate production including exons flanking unspliced introns will be generated, which can be further processed into ecircRNA or EIciRNA by splicing or retaining introns. In RBP-binding cyclization, RBPs, such as Muscleblind (MBL/MBNL1), Quaking (QKI), Fused-in sarcoma (FUS), NF90/NF110, DHX9, HNRNRL, et al. have been reported to contribute a lot in the biogenesis of circRNA, through binding to pre-mRNA, getting the introns together and promoting the RNA splicing enzymes binding to RNA–protein complex [27–31]. MBL protein specifically binds to conserved muscleblind binding sites of pre-mRNA encoding by mbl gene and promotes the spliceosome to strongly bind to pre-mRNA, resulting in upregulating the production of circMbl in Drosophila. Furthermore, functional MBL binding sites in the introns of pre-mRNA encoding by mbl gene are required for the increase production of circMbl [32]. In contrast, nuclear RNA helicase DHX9 acting as an inhibitor regulatory factor interacts with inverted complementary sequence and downregulate the splice process of circRNA. Recently, research results of Yu show that unregulated DHX9 contributing to downregulation of cSMARCA5 mediates the promotion of growth, aggression and metastasis of hepatocellular carcinoma (HCC) [33].
Additionally, as cis elements, intron-pairing can also increase the rate of producing circRNAs. Exons flanking introns with complementary reserved sequences will form like a loop by intron pairing. This structure is expected to bring the distal splice site to proximity, which is an efficient way to facilitate the back-splicing of circRNA [16]. One of the best known are ALU repetitive elements, which occupy approximately 10% of the whole genome [34]. Interestingly, short inverted sequences (30–40 nucleotides) like ALU repeats can dramatically enhance the production of circRNA, while longer repeats can sometimes inhibit the back splicing of circRNA [35]. It is reported that competition of RNA pairing exists between ALU repeats within different introns [26, 34]. Besides circRNA, ALU repeats also involve in the biogenesis of miRNA. Di Ruocco et al. found that accumulation of ALU sequences is associated with both epithelial-to-mesenchymal transition (EMT) and cancer progression by targeting miR-566, which can lead to upregulation of DICER1 [36]. Surprisingly, Zhang et al. observed that not only the short repetitive sequences can promote the splicing process of circRNA, but also the non-repeated elements contribute to pre-mRNA cyclization [16].
CiRNAs
CiRNAs are identified as a sequence containing only introns and mainly perform their functions in the nucleus. Their sizes range from 200 to 3000 nucleotides [37]. It is generally believed that excised introns or removed remain introns will be easily and immediately degraded by RNase [38]. To generate a stable ciRNA after excised lariat introns are removed from spliceosome, some specific structures are usually necessary for during the splicing [39]. Zhang et al. reported that the reasons why ciRNAs fail to be debranched by enzymes are the existence of an special consensus motif that 7 nucleotides GU-rich sequences close to 5′ upstream splice site and 11 nucleotides C-rich sequence locate near in branch point of 3′ downstream splice site [40]. Treated by RNase on branch point of intron lariat, ciRNA is finally formed. Therefore, these elements play an important role in generating a stable ciRNA in the back splicing.
Functions of circRNAs in cancer
Giving that circRNAs possess some special structures which can interact with DNAs, RNAs and even proteins, circRNAs have potential to be new multifunctional small molecules. Regulatory networks can be established when circRNAs bind to other molecules forming circRNA-DNA, circRNA-RNA, circRNA-protein interactions. Recent years have witnessed a flood of researches which focus on the functions and mechanisms of how circRNAs work biologically. Multiple functions of circRNAs are demonstrated in Table 1. Detail information will be classified into three parts as follow.
Table 1.
Functions of circRNAs
| Function | Example | References |
|---|---|---|
| miRNA sponge |
circ-HIPK3 circ-PRKCI |
[41, 42] |
| Translation regulator | circ-PABPN1 | [43] |
| Alternative splicing | circ-Mbl | [44] |
| RNAP II elongation |
circ-EIF3J circ-PAIP2 |
[18] |
| RNA maturation | circ-ANRIL | [45] |
| Protein translation (including m6A-driven) | circ-ZNF609 | [46, 47] |
| Protein sponge | circ-Foxo3 | [48] |
| Histone methylation | Circ-ANRIL | [49] |
Transcription EIciRNAs such as circEIF3J or circPAIP2 naturally interact with U1 snRNP which is indispensable for the function of EIciRNAs where occur predominately in nucleus. EIciRNAs associate with RNA polymerase II and play a special role in promoting parental gene expression [18].
Translation Although circRNAs are identified as non-coding RNAs which canonically do not translate into proteins, some researchers found that circRNAs process a special function that can express translationally. Legnini et al. show that circ-ZNF609 contains a 753-nt open reading frame (ORF) and encode proteins in myogenesis [46]. Moreover, some initiation of circRNAs translation is driven by N6-methyladenosine (m6A). Yang et al. identified that circRNAs containing m6A motif can produce proteins and its translation efficiency can be control by regulation of m6A level. In-deep study shows that m6A motif recruit IF4G2 and YTHDF3 which requires for the initiation of translation [47]. Additionally, Abdelmohsen et al. identified that HuR proteins are able to bind to PABPN1 mRNA promoting mRNA translation. Targeting HuR, overexpressing circPABPN1 can be observed to upregulated PABPN1 abundance by keeping HuR from binding to its cognate transcript, eventually leading to low expression of PABPN1 [43].
Others Holdt found that in vascular smooth muscle cells and macrophages of human atherosclerosis, circANRIL is a member of a pre-ribosomal assembly complex by binding to pescadillo homologue 1 (PES1), which mediates the biogenesis and maturation of ribosome. This process leads to cell apoptosis and inhibits cell proliferation, which is independent on miRNA sponge [45]. CircRNAs not only can function as miRNA sponge, but also are able to sponge and interact with proteins. Du et al. reported that expression level of circFoxo3 in heart tissues of aging patients is higher than that of young patients. Of interest, experimentation using validated the fact that circFoxo3 interact with ID-1, E2F1, FAK and HIF-1α which increase cellular senescence by suppressing the effect of these proteins [48].
Most importantly, miRNA sponge will be described in detail below.
Function of miRNA sponge in human cancer
circRNAs identified as competing endogenous RNAs (ceRNAs) are the parts of small non-coding RNAs which play big parts in post-transcription and participate in genetic expression transcriptionally and translationally. It is well known that a part of circRNAs containing miRNA response elements (MREs) can act as competing molecules by interacting with diverse miRNAs and affect the efficiency of miRNAs to regulate the downstream mRNA expression [50].
circRNAs have been studied in many human diseases, such as neurodegenerative diseases, neural injuries, cardiac hypertrophy, myocardial infarction, atherosclerosis and diverse malignant carcinomas [51]. The prevention, diagnosis and therapy of cancers are the hot topics for human in a long time. There is no hesitation that the cure for cancer is the unremitting struggle of human. circRNAs as the star molecules in diagnosis and therapy have been investigated in a great deal of malignant tumors. Taking ciRS-7 for examples. Li et al. found that ciRS-7 upregulates in esophageal squamous cell carcinoma (ESCC) while miR-7 downregulates in the same cancer tissues compared with para-carcinoma tissues. Furthermore, they verified that elevated ciRS-7 can eliminate the effect of inhibitory miR-7 which inhibits the proliferation and metastasis of ESCC. Knockdown of HOXB13 in elevated ciRS-7/miR-7 cells can negatively affect colony formation and migration of cancer cells [52]. For breast cancer, ciRS-7 can also contribute to its tumorigenesis and metastasis. The expression level of ciRS-7 in triple negative breast cancer (TNBC) is higher than other types breast cancer tissues from who patients did not undergo chemotherapy and radiotherapy before surgery. Knockdown of ciRS-7 can suppress the metastasis of TNBC in vivo and in vitro. Mechanistically, ciR-7 acting as miR-1299 sponge mediates the EMT by upregulating matrix metalloproteinases (MMPs) [53].
With the extensive and in-depth research on cancer in the past decades, tumor microenvironment (TME) has been founded and researched. TME refers to the outcome of interaction among various tumor cell types, and act as an important component in fields of tumor cells progressing, growing and metastasizing [54]. Surprisingly, numerous studies have demonstrated that circRNAs can affect TME via circRNA-miRNA-mRNA axis [55]. To this end, the circRNA-miRNA-mRNA axis on TME has been regarded as a promising breakthrough in possible preventive strategies, molecular diagnosis, and therapy strategies on cancer.
-
Regulate tumor immunosuppression
During the past decades of TME research, programmed death 1/programmed death-ligand 1 (PD-1/PD-L1) has been identified as a crucial element to tumor immunosuppression [56]. Current research shows that due to the regulation of upstream circRNAs, the expression of PD-L1 can be affected by the corresponding circRNA-miRNA-PD-L1 axis. Subsequently, the interaction between PD-L1 and PD-1 can properly suppress the stimulator of effector T lymphocytes, eventually causing tumor cells to escape immune surveillance [57, 58]. The considerable roles of miRNAs in modulating PD-1/PD-L1 have been well investigated in the past few decades. However, the definite mechanism of circRNA regulates PD-1/PD-L1 through ceRNA is still unclear. One recent study revealed that circular RNA circ-0020397 was upregulated in colorectal cancer, circ-0020397 through circ-0020397 ↑-miR-138- ↓ PD-L1 ↑ to bind PD-1, inhibit T cell proliferation and activation, and induce T cell apoptosis, ultimately resulting in tumor immune escape [57]. As a consequence, the corresponding circRNAs through the ceRNA pathway to target and modulate PD-1/PD-L1 may become a novelty research direction for tumor immune escape and new treatment for cancer.
-
The cytotoxicity of natural killer cells
Natural killer (NK) cells, known as a vital component defense for human immune surveillance. Increasing experimental evidence has indicated that once the NK cells recognize and destroy the target cells since there is no major histocompatibility complex to prevent it, more and more immune activators can be generated to enhance the destruction of the target cells [59]. Previous studies elucidated the underlying mechanism by that circRNAs’ involve in NK-mediated immune responses by the ceRNA pathway. For instance, in pancreatic cancer cells under hypoxia, circ-0000977 can pass circ-0000977-miR-153-Hypoxia-inducible factor 1-α (HIF1α) axis to regulate HIF1α -mediated immune escape [60].
-
Angiogenesis
Tumor angiogenesis is an extremely complex process in biology [61]. Cancer cells can induce angiogenesis by affecting TME, thereby providing tumor cells with the oxygen and nutrients they need to pass through and provide a guarantee for tumor proliferation and invasion [60]. In recent decades, circRNAs have been reported to regulating tumor angiogenesis by playing as related downstream miRNA “sponges”. For example, a fascinating feedback loop namely FUS-circ-002136-miR-138-5p-SOX13 was discovered in glioma associated endothelial cells (GECs), and stimulating GECs angiogenesis [62]. Emerging research showed the expression of AGGF1 was increased by circSHKBP1-miR-379-FOXP2 or circSHKBP1-miR-544a/FOXP1 axis, which promoted the angiogenesis of glioma [63]. The above studies expound that circRNAs are important in regulating tumor angiogenesis through the mechanism of sponge miRNA. Therefore, we reasonably speculate that the ceRNA pathway of related circRNAs is a promising direction in the future research of tumor angiogenesis inhibitors.
-
Hypoxia regulates circRNA production
In TME, hypoxia can cause a series of complex biological changes, therefore affecting the occurrence and progression of cancer. HIFs, a type of transcriptional regulator, plays a crucial role in regulating TME responses and tumor cell proliferation and metastasis by stimulating downstream oncogene transcription of hypoxic response elements and regulating various relevant signaling pathways [64]. Mounting evidence demonstrates that hypoxic TME-related circRNAs involve in metastasis, invasion, angiogenesis, and radiation-resistance [65]. For example, the hypoxia-induced expression of circDENND4C and the circDENND4C/miR-200b, miR-200c axis could effectively modulate glycolysis, migration and invasion in breast cancer cells [66]. Interestingly, another study illustrated that circDENND2A was induced by hypoxia in glioma cells, and the circDENND2A-miR-625-5p axis promotes invasion and migration in glioma cells via the miR-625-5p interacted with HIF1α [67]. For this reason, the ceRNA mechanism of circRNA plays an important role in the case of tumor hypoxia, which may become the focus of our future research.
In short, researches about circRNAs can be found in various cancers, such as breast cancer, hepatocellular carcinoma, lung cancer, gastric cancer, colon cancer, neuro glioma, et al. [68]. In my review, we mainly focus on the effect of circRNAs in lung cancer. The contribution of circRNA-miRNA-mRNA regulatory network to lung cancer will be highlighted as follow.
circRNA-miRNA-mRNA regulatory axis in lung cancer
In order to fully understand the circRNAs on lung cancer, our team searched the related literatures on Pubmed, Embase and Web of Science. 53 literatures demonstrating the relationship or the application between circRNAs and lung cancer come into notice. However, taking into consideration that we shed light on circRNA-miRNA-mRNA regulatory network on lung cancer, only 43 individual researches can be included in this review (shown Table 2).
Table 2.
The summary of validated circRNA-miRNA-mRNA regulatory axes in lung cancer
| Cancer types | Sample | circRNA-miRNA-mRNA | Cell characteristics | Clinical characteristics | Clinical application | References |
|---|---|---|---|---|---|---|
| LAC |
Tissues Plasma Cells |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis 4. Cell migration |
1. TNM stage 2. Lymph node metastasis |
Diagnostic values AUC: 0.815 Sensitivity: 0.755 Specificity: 0.796 |
[69, 70] | |
| NSCLC | Cells | 1. Cell proliferation | – | – | [41] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis 4. Cell migration |
1. T stage 2. Lymph node metastases 3. TNM stage 4. Poor differentiation |
– | [71] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [72] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis 4. Cell migration |
1. TNM stage 2. Lymph node metastasis 3. Poor differentiation 4. Survival rate |
– | [73, 74] | |
| LAC |
Tissues Cells |
1. Cell proliferation | 1. Tumor size | – | [75] | |
| LAC |
Tissues Cells |
1. Cell proliferation 2. Cell apoptosis |
1. Poor differentiation 2. TNM stage 3. Lymph node metastasis |
– | [76] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. Histological grade 2. N stage 3. TNM stage |
– | [77] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion |
– | – | [80] | |
| LAC |
Tissues Cells |
1. Cell proliferation 2. Cell apoptosis |
1. TNM stage 2. Tumor size 3. T stage 4. Survival rate |
Prognostic value | [42] | |
| LAC | Tissues | – |
1. T stage 2. Distant metastasis |
Diagnostic values AUC: 0.815 |
[81] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [82] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [83] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion |
1. Tumor size 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [84] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. Poor differentiation 2. Lymph node metastasis 3. T stage and N stage 4. Survival rate |
Prognostic value | [85] | |
| NSCLC |
Tissues Cell |
1. Cell proliferation 2. Cell apoptosis |
– | – | [86] | |
| NSCLC |
Cells Mice |
1. Cell proliferation 2. Cell migration |
– | – | [87] | |
| LAC |
Tissues Cells Mice |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
4. TNM stage | – | [88] | |
| NSCLC |
Tissues cells |
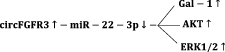 |
1. Cell proliferation 2. Cell invasion |
1. TNM stage 2. Lymph node metastasis 3. Tumor size 4. Poor differentiation 5. Survival rate |
Prognostic value | [89] |
| NSCLC |
Tissues Cells Mice |
1. Cell proliferation 2. Cell invasion |
1. Survival rate | Prognostic value | [90] | |
| NSCLC |
Tissues Blood Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. Distant metastasis |
Diagnostic values Tissues: AUC: 0.803 Sensitivity: 0.825 Specificity: 0.675 Serum: AUC: 0.794 Sensitivity: 0.711 Specificity: 0.800 |
[91] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell apoptosis |
1. Tumor size 2. TNM stage 3. Survival rate |
Prognostic value | [92] | |
| LAC |
Tissues Cells |
1. Cell proliferation 2. Cell apoptosis |
1. TNM stage 2. Tumor size 3. Survival rate |
Prognostic value | [93] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis |
– | [94] | |
| NSCLC |
Tissues Cells |
 |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis |
– |
Diagnostic values AUC: 0.643 Sensitivity: 0.725 Specificity: 0.575 |
[95] |
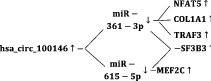
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion |
1. TNM stage 2. Distant metastasis 3. Poor differentiation |
– | [96] | |
| NSCLC |
Tissues Cells |
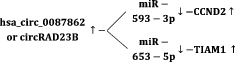 |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. Lymph node metastasis 2. Poor differentiation 3. Survival rate |
– | [97] |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [98] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis |
1. TNM stage 2. T stage 3. Poor differentiation 4. Survival rate |
Prognostic value | [99] | |
| NSCLC |
Tissues Cells |
1. Cell migration 2. Cell invasion |
1. Lymph node metastasis 2. Prognostic value |
Prognostic value | [100] | |
| NSCLC |
TISSUES Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
– | – | [101] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion 4. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis |
Prognostic value | [102] | |
| SCLC |
Tissues Blood |
1. Cell proliferation 2. Cell migration 3. Cell apoptosis |
1. Lymph node metastasis 2. Survival rate |
Prognostic value | [103] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation |
1. Patients’ age (particularly after the age of 60) 2. TNM stage |
– | [104] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell invasion 3. Cell apoptosis |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [105] | |
| NSCLC |
Tissues Cells |
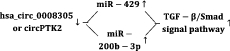 |
1. Cell invasion 2. Cell metastasis |
– | – | [106] |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [107] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion |
1. TNM stage 2. Lymph node metastasis 3. Survival rate |
Prognostic value | [108] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 3. Cell apoptosis |
– | – | [109] | |
| NSCLC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion |
– |
Diagnostic values AUC: 0.782 Sensitivity: 0.800 Specificity: 0.733 |
[110] | |
| LAC |
Tissues Cells |
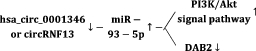 |
1. Cell migration 2. Cell invasion |
1. TNM stage 2. Lymph node metastasis |
– | [111] |
| LAC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion |
1. TNM stage 2. Lymph node metastasis 3. Tumor size 4. Survival rate |
Prognostic value | [112] | |
| LAC |
Tissues Cells |
1. Cell proliferation 2. Cell migration 3. Cell invasion |
1. T stage 2. Cell proliferation 3. Cell migration and invasion |
– | [113] |
CircRNAs are involved in the occurrence and progress of lung cancer, resulting in attracting considerable attention recently. Currently,Although a lot of literatures reported that circRNA can participate in the tumorigenesis and progress of lung cancer through various ways such as interaction with transcription factors and ceRNA mechanism, ceRNA mechanism is still the main focus among diverse circRNA functions in the tumorigenesis of lung cancer. In this review, we list all existing studies on circRNA-miRNA-mRNA axis in lung cancer, throwing light on further research on circRNAs in lung cancer, and even giving new ideas for early diagnosis and precise treatment of lung cancer.
It is reported that some circRNAs interacting with miRNA are overexpressed in lung cancer. Qu et al. found that hsa_circ_0020123, identified as the sponge of miR-144, is upregulated in NSCLC, thus leading to uprising of ZEB1/EZH2. The phenomenon that this regulatory axis may associate with the tumor differentiation, lymph node metastasis, TNM stage and survival rate in NSCLC can be found in the statistics data of a large collection of human cancer tissues. Simultaneously, hsa_circ_0020123 also has potential to give an easy access to early-diagnosis for NSCLC [85]. Almost all the researches about lung cancer focus on NSCLC, while only one literature about SCLC can be found in the database described above. Li et al. identified that circFLI1, as an ecircRNA, is involved in the occurrence of metastasis, which is dramatically up-regulated in the SCLC tissues compared with NSCLC tissues. Of interest, there are no significance between circFLI1 and cell proliferation, apoptosis and colony formation. They also found the regulatory network in circFLI1s that circFLI1 binds to miR-584-3p which further regulating its downstream targets-RhoA/ROCK1 signal pathway [103].
On the other hand, expression of some circRNAs in lung cancer significantly lower than in normal tissues or para-carcinoma tissues. Yang et al. reported that hsa_circ_0046264 is associated with lymph node metastasis and TNM stage in NSCLC, and it plays a vital role through hsa_circ_0046264↓-miR-1245↑- BRCA2↓ axis [105]. Moreover, Chen et al. found that low expression of hsa_circ_100395 in NSCLC can affect the TNM stage, metastasis and survival rate in NSCLC. Further study explores that hsa_circ_100395 modulating miR-1228 regulates lung cancer cells proliferation, migration and invasion. The hypothesis derived from RNA-seq profile that hsa_circ_100395 preforms its function by targeting miR-1228/TCF21 axis can be confirmed by luciferase reporter assay and overexpression experiment [107].
According to WHO cancer report, lung cancer has become the first killer among all cancers, leading to one out of five deaths of cancer-related mortality [114]. Although the chemotherapy is still available, it is noteworthy that once patients develop to chemotherapy resistance, the survival rate will be dramatically decreased, leading to higher mortality in the population of suffering lung cancer. It is reported that resistance of canonical chemotherapy like cisplatin and targeted therapy like TKI have occurred in clinical setting, which become the barriers to treatment [115, 116]. Strikingly, Zhou et al. shown that upregulation of hsa_circ_0004015 can remarkably increase the resistance of gefitinib in resistant cancer cells HCC827I/R, while downregulation can dramatically decrease the gefitinib IC50 of resistant cancer cells. Likewise, they also found that hsa_circ_0004015 functions through miR-1183/PDPK1 regulatory axis, which selectively promotes progression and invasion of lung cancer, but no significance can be found in the relationship between hsa_circ_0004015 and metastasis, no matter lymphatic metastasis or distant metastasis [99]. Therefore. the ceRNA mechanism of circRNA may play an indispensable role in chemotherapy resistance or susceptibility, which provides a brand-new perspective for this research.
After summarizing the literatures of circRNA-miRNA-mRNA regulatory network in lung cancer, we can clearly see that there are some interactions between different circRNAs regulatory axes. We describe this phenomenon as follow in terms of two parts.
Interactions of miRNAs in different circRNA-miRNA-mRNA network
miR-7 There are some different opinions about the relationship between miR-7 and lung cancer oncogenesis. Canonically, ciR-7 identified as a sponge of miR-7 acts on downstream targets such as NF-κB, EGRF, CCNE1, PIK3CD, eventually contributing to cell proliferation, migration and metastasis [71, 72]. Acting like an oncogene, miR-7 is downregulated in NSCLC tissues and also inhibits the cell proliferation, migration and invasion by suppressing ERK/MAPK signaling pathway [117]. Strikingly, however, Wan et al. reported that miR-7 sponged by circICTH enhances lung cancer cell proliferation. They also found that miR-7 can degrade the expression level of circITCH. Moreover, circITCH competitively inhibited miR-7 binding to ITCH remarkably enhances the expression of ITCH which promotes the oncogenesis of lung cancer by activating Wnt/β-catenin signal pathway [104].
miR-326 It is reported that that both hsa_circ_0003998 and circPUM1 are able to function as a sponge of miR-326. Differentially, hsa_circ_0003998 as a sponge of miR-326 modulate the downstream Notch1, while circPUM1 binding to miR-326 regulates targeted genes, cyclin D1 and Bcl-2 [84, 88]. Either hsa_circ_0003998 or circPUM1 considerably increases in lung cancer tissues and cells and contributes the tumor-promoting effect by different signal pathways. What’s more, Hang et al. also reported that circFARSA is upregulated in lung cancer and has predicted binding sites of miR-326. Experimentation of RPI and dual-luciferase reporter assay are needed to further confirm the relationship between cicrFARSA and miR-326.
miR-1179 miR-1179 sponged by both hsa_circ_0000735 and hsa_ circ_0003645 is downregulated in NSCLC tissues and cells [94, 98]. An et al. shown that circ_0003645 facilitates the promotion of cancer cell growth, apoptosis, and metastasis through miR-1179/TMEM14A regulatory axis [98].
Interactions of mRNAs in different circRNA-miRNA-mRNA network
Wnt/β-catenin signaling pathway: As is described in the above table, there are four circRNA regulatory networks which involved in the common downstream targeted pathway, Wnt/β-catenin signaling pathway. has_circ_0001946, has_circ_001569, has_circ_0006427 and circITCH will be drawn our attention due to their sharing signal pathway. has_circ_0001946 sponges miR-135a-5p and activates Wnt/β-catenin pathway by enhancing Sirtuin 1 (SIRT1) protein [93]. It is known that SIRT1 contributes its effect of tumor promotion to tumorigenesis of lung cancer. Recently, Lee et al. reported that Metformin and tenovin‐6 targeting downregulated SIRT1 synergistically induce apoptosis in lung cancer cell [118]. has_circ_001569 preforms its function by miRNA-148a, enhancing Wnt/β-catenin pathway associated gene expression [119]. has_circ_0006427 identified as miR-6783-3p sponge was found to be downregulated in NSCLC. The finding that, differently from has_circ_0001946, has_circ_0006427 activates its downstream pathway by increasing DKK1 protein [112]. Likewise, circITCH is also dramatically downregulated in NSCLC with binding sites of miR-7 and miR-214. Overexpression of circITCH can remarkably inhibits the activation of Wnt/β-catenin signal pathway through declining the vital downstream gene, c-Myc and cyclinD1 [104]. Shortly, although these four circRNAs share the common important signal pathway that significantly promote lung cancer cell in terms of growth, invasion and metastasis, they activate the pathway by targeting different proteins in the cell.
Cyclin D1 and bcl-2 Zhu et al. reported that hsa_circ_0013958 can promote LAC by regulating cyclin D1, while Chen et al. found that circPUM1 can also perform its function by targeting cyclin D1 and bal-2 [69, 88]. Additionally, hsa_circ_0000064 is reported to upregulate in NSCLC by modulating cyclin D1, bcl-2 and so on which is not described in our table [120]. Besides circPUM1 and hsa_circ_0000064, hsa_circ_0007385 and circPVT1 can also respectively bind to miR-181 and miR-497 and target the same downstream gene, bcl-2 [78, 91].
Briefly, both Wnt/β-catenin and Cyclin D1, bcl-2 are the common signal pathway affecting cell proliferation, apoptosis, invasion and migration in lung cancer. Some circRNAs binding to different miRNAs share the same downstream targets. In the era of precision medicine, blocking these common pathways maybe is a good idea to inhibit the growth and prevent the metastasis in lung cancer.
Clinical characteristics of circRNAs in lung cancer
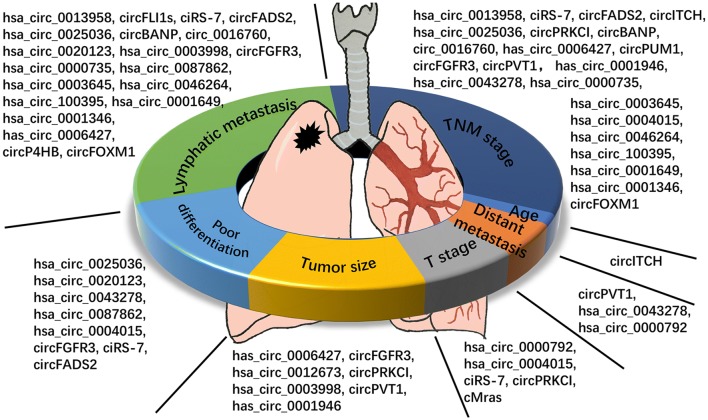
As we can see in the Table 2, circRNAs have multiple clinical characteristics in lung cancer. We summarize the clinical characteristics of circRNAs with ceRNA-miRNA-mRNA regulatory function in lung cancer (Figs. 1 and 2). It is clear that the most widely studied characteristics of circRNAs in lung cancer are TNM stage and lymphatic metastasis, both of which take up nearly 80% of all availably found studies. Meaningfully, we can find that there is a close relationship between circRNAs and TNM stage and lymphatic metastasis in lung cancer. We are delighted that a large amount of circRNAs have potential to develop into excellent biomarkers of lymphatic metastasis in lung cancer. Both distant metastasis and T stage are the characteristics which absorb least attention to researchers. Suggestion can be taken from these phenomena that maybe more circRNAs are significantly involved in distant metastasis and T stage in lung cancer if researchers play more attention to distant metastasis and T stage. Statistics of patients’ age can be found in almost all the circRNA studies of lung cancer, but only one circRNA is significantly related to patients’ age. Hence, if we can make good use of the significant relationship between circRNAs and TNM stage, lymphatic metastasis in lung cancer, maybe circRNA can be developed into an excellent prognostic biomarker of clinical characteristics.
Fig. 1.
The summary of clinical functions in lung cancer. All the circRNAs were concluded from validated circRNAs with circRNA-miRNA-mRNA regulatory network. CircRNAs included in our study were divided into 7 parts according to their clinical characteristics. As we can see in the picture, most of the circRNAs are significantly related to lymphatic metastasis and TNM stage
Fig. 2.
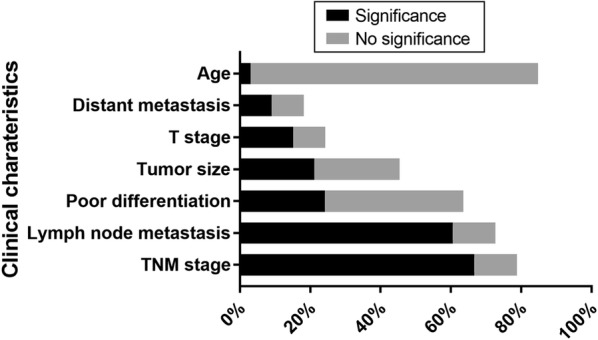
Summary of clinical characteristics of circRNAs in lung cancer. Percent in X axis means the studies involved in each clinical event take up in the whole studies about circRNA-miRNA-mRNA regulatory axis in lung cancer. Significance in each group means there is a significant relationship between clinical characteristics and certain circRNA. No significance in each group means no significance can be found between clinical characteristics and certain circRNA
Conclusion and perspective
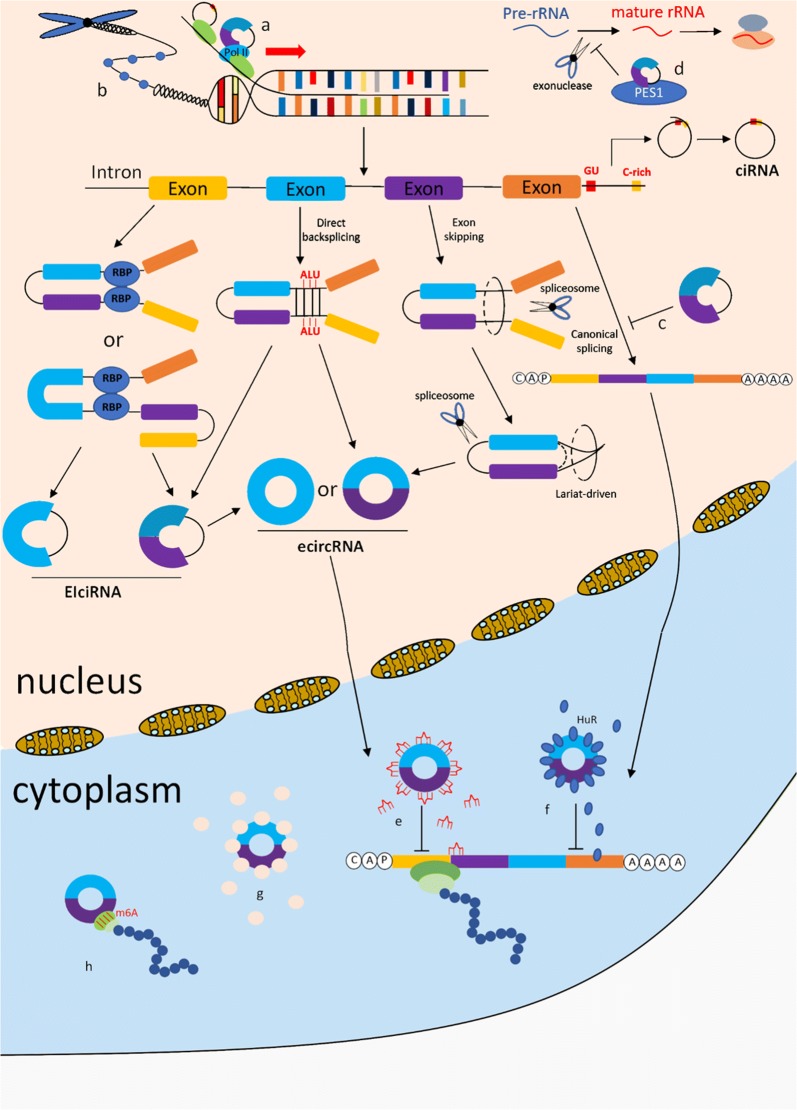
The summary of biogenesis and function characteristics of circRNAs shows in Fig. 3. In recent years, numerous of circRNAs have been found and investigated. circRNAs affect the expression of related genes by acting as miRNA sponges. Additionally, circRNAs are abundant and stable in cells, blood, tissues, exosomes and saliva, providing a new way of understanding the occurrence and development of diseases.
Fig. 3.
The whole view of the biogenesis and function of circRNAs. The biogenesis of three kinds of circRNA (EIciRNA, ecircRNA and ciRNA) and eight kinds of function, including circRNA-miRNA-mRNA axis, were presented in the picture. a RNAP II elongation. b Histone methylation. c Alternative splicing. d RNA maturation. e miRNA sponge. f Translation splicing. g Protein sponge. h Protein translation (including m6A-driven)
Despite the discovery of thousands of circRNAs, our understanding of circRNAs is still limited, especially in terms of their biogenesis and functions. Moreover, although the circRNAs are known as a new kinds of potential cancer biological markers and therapeutic targets, there still exist many problems in the clinical application of circRNAs, such as high medical expense, complex monitoring approach, and so on. Therefore, we need universal study to shed light on the development and clinical application of circRNAs so that it may provide new ideas for early diagnosis and treatment of lung cancer.
Acknowledgements
Not applicable.
Abbreviations
- ncRNA
Non-coding RNA
- miRNA
MicroRNA
- lncRNA
Long non-coding RNA
- circRNA
CIRCULAR RNA
- RNA-seq
RNA sequencing
- mRNA
Message RNA
- pre-mRNA
Precursor mRNA
- ecircRNA
Exonic circRNA
- EIciRNA
Exonic-intronic circRNA
- ciRNA
intronic circRNA
- tricRNA
tRNA intronic circRNA
- circrRNA
CircRNA rRNA
- CS
Constitutive splicing
- AS
Alternative splicing
- 3′ ss
3′ upstream splice site
- 5′ ss
5′ downstream splice site
- RBP
RNA binding protein
- MBL
Muscleblind
- QKI
Quaking
- FUS
Fused-in sarcoma
- HCC
Hepatocellular carcinoma
- EMT
Epithelial-to-mesenchymal transition
- ORF
Open reading frame
- m6A
N6-methyladenosine
- PES1
Pescadillo homologue 1
- ceRNA
Competing endogenous RNA
- MRE
miRNA response element
- ESCC
Esophageal squamous cell carcinoma
- TNBC
Triple negative breast cancer
- MMP
Matrix metalloproteinase
- SIRT1
Sirtuin 1
- TME
Tumor microenvironment
- PD-1/PD-L1
Programmed death 1/programmed death-ligand 1
- NK
Natural killer
- GEC
Glioma associated endothelial cell
Authors’ contributions
ZZL and CG were mainly responsible for data collection and drafted the manuscript. MMZ took part in the design of the paper and provided the direction through writing. MMZ, PM and TTZ helped revise the manuscript. All authors read and approved the final manuscript.
Funding
This review is supported the Natural Science Foundation of Guangdong province (Grant Number 2017A030313525).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhuo-Zheng Liang and Cheng Guo contributed equally to this work
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sui H, Ma N, Wang Y, Li H, Liu X, Su Y, Yang J. Anti-PD-1/PD-L1 therapy for non-small-cell lung cancer: toward personalized medicine and combination strategies. J Immunol Res. 2018;2018:10–1155. doi: 10.1155/2018/6984948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ai X, Guo X, Wang J, Stancu AL, Joslin P, Zhang D, Zhu S. Targeted therapies for advanced non-small cell lung cancer. Oncotarget. 2018;9(101):37589–37607. doi: 10.18632/oncotarget.26428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Zhang X, Wang YZ, Tian H, Xu S. Research progress of circular RNAs in lung cancer. Cancer Biol Ther. 2019;20(2):123–129. doi: 10.1080/15384047.2018.1523848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang J, Yang L, Chen L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 8.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 10.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7(2):e30710–e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102(4):837. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Kearse MG, Huang C. The nuclear export of circular RNAs is primarily defined by their length. RNA Biol. 2018 doi: 10.1080/15476286.2018.1557498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18(1):10–1186. doi: 10.1186/s12885-018-4213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang X, Wang H, Lu X, Chen L, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16(4):10–1186. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 19.Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2016;14(8):978–984. doi: 10.1080/15476286.2017.1317911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2016;14(8):992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L, Zhu Y, Dai D, Wang X, Jin H. Epigenetic regulation of alternative splicing. Am J Cancer Res. 2018;8(12):2346–2358. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Zhu M, Pan J, Chen C, Xia S, Song Y. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20(3):10–1186. doi: 10.1186/s12931-018-0962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang F, Zhou Q, Zhu W, Song YY, Zhan BT, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6(5):563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R, Peruzzi G, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8(14741):10–1038. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta SK, Garg A, Bär C, Chatterjee S, Foinquinos A, Milting H, Streckfuß-Bömeke K, Fiedler J, Thum T. QUAKING inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. 2018;122(2):246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67(2):214–227. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Aktaş T, Ilık IA, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 31.Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhao SG, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar L, Haque R, Baghel T, Nazir A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol. 2017;54(9):7224–7234. doi: 10.1007/s12035-016-0213-8. [DOI] [PubMed] [Google Scholar]
- 33.Yu J, Xu Q, Wang Z, Yang Y, Zhang L, Ma J, Sun S, Yang F, Zhou W. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Daniel C, Behm M, Öhman M. The role of Alu elements in the cis-regulation of RNA processing. Cell Mol Life Sci. 2015;72(21):4063–4076. doi: 10.1007/s00018-015-1990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Gene Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Ruocco F, Basso V, Rivoire M, Mehlen P, Ambati J, De Falco S, Tarallo V. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene. 2018;37(5):627–637. doi: 10.1038/onc.2017.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- 39.Bolisetty MT, Graveley BR. Circuitous route to transcription regulation. Mol Cell. 2013;51(6):705–706. doi: 10.1016/j.molcel.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Tian F, Wang Y, Xiao Z, Zhu X. Circular RNA CircHIPK3 promotes NCI-H1299 and NCI-H2170 cell proliferation through miR-379 and its target IGF1. Zhongguo Fei Ai Za Zhi. 2017;20(7):459–467. doi: 10.3779/j.issn.1009-3419.2017.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, Xu W, Zhang E, Wang J, Fang T, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78(11):2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 43.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation byCircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 45.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7(17429):10–1038. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du WW, Yang W, Chen Y, Wu Z, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 49.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001210–e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 51.Lei K, Bai H, Wei Z, Xie C, Wang J, Li J, Chen Q. The mechanism and function of circular RNAs in human diseases. Exp Cell Res. 2018;368(2):147–158. doi: 10.1016/j.yexcr.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG, Wang WF, Fang MH. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9(8):810–838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sang M, Meng L, Liu S, Ding P, Chang S, Ju Y, Liu F, Gu L, Lian Y, Geng C. Circular RNA ciRS-7 maintains metastatic phenotypes as a ceRNA of miR-1299 to target MMPs. Mol Cancer Res. 2018;16(11):1665–1675. doi: 10.1158/1541-7786.MCR-18-0284. [DOI] [PubMed] [Google Scholar]
- 54.Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. J Cell Sci. 2012;125(Pt 23):5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, Li X, Sun Z. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi: 10.1186/s12943-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 58.Shi MH, Xing YF, Zhang ZL, Huang JA, Chen YJ. Effect of soluble PD-L1 released by lung cancer cells in regulating the function of T lymphocytes. Zhonghua Zhong Liu Za Zhi. 2013;35(2):85–88. doi: 10.3760/cma.j.issn.0253-3766.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 59.Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. 2007;121(1):1–14. doi: 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ou ZL, Luo Z, Wei W, Liang S, Gao TL, Lu YB. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol. 2019;16(11):1592–1603. doi: 10.1080/15476286.2019.1649585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 62.He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, Ma J, Shao L, Wang D, Shen S, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J Exp Clin Cancer Res. 2019;38(1):65. doi: 10.1186/s13046-019-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, Cai H, Ma J, Liu L, Wang P, et al. circ-SHKBP1 Regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids. 2018;10:331–348. doi: 10.1016/j.omtn.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang Y, Lin D, Taniguchi CM. Hypoxia inducible factor (HIF) in the tumor microenvironment: friend or foe? Sci China Life Sci. 2017;60(10):1114–1124. doi: 10.1007/s11427-017-9178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015;117(10):884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 66.Ren S, Liu J, Feng Y, Li Z, He L, Li L, Cao X, Wang Z, Zhang Y. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J Exp Clin Cancer Res. 2019;38(1):388. doi: 10.1186/s13046-019-1398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su H, Zou D, Sun Y, Dai Y. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell Mol Biol Lett. 2019;24:24. doi: 10.1186/s11658-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):10–1186. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, Han S, Wu G. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 70.Sun C, Li S, Li D. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7(24):35960–35978. doi: 10.18632/oncotarget.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22(6):3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther. 2018;11:3979–3987. doi: 10.2147/OTT.S158316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. CircFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38(4):10–1042. doi: 10.1042/BSR20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao N, Wang FX, Wang G, Zhao QS. Targeting the HMGA2 oncogene by miR-498 inhibits non-small cell lung cancer biological behaviors. Eur Rev Med Pharmacol Sci. 2018;22(6):1693–1699. doi: 10.26355/eurrev_201803_14582. [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Zhu X, Zhang H, Wei S, Chen Y, Chen Y, Wang F, Fan X, Han S, Wu G. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun. 2018;496(4):1069–1075. doi: 10.1016/j.bbrc.2018.01.126. [DOI] [PubMed] [Google Scholar]
- 76.Wu S, Li H, Lu C, Zhang F, Wang H, Lu X, Zhang G. Aberrant expression of hsa-circ-0025036 in lung adenocarcinoma and its potential roles in regulating cell proliferation and apoptosis. Biol Chem. 2018;399(12):1457–1467. doi: 10.1515/hsz-2018-0303. [DOI] [PubMed] [Google Scholar]
- 77.Shi Q, Zhou Z, Ye N, Chen Q, Zheng X, Fang M. MiR-181a inhibits non-small cell lung cancer cell proliferation by targeting CDK1. Cancer Biomark. 2017;20(4):539–546. doi: 10.3233/CBM-170350. [DOI] [PubMed] [Google Scholar]
- 78.Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, Di QG. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–674. doi: 10.1007/s00432-017-2576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang P, Ye B, Yang Y, Shi J, Zhao H. MicroRNA-181 functions as a tumor suppressor in non-small cell lung cancer (NSCLC) by targeting Bcl-2. Tumor Biol. 2015;36(5):3381–3387. doi: 10.1007/s13277-014-2972-z. [DOI] [PubMed] [Google Scholar]
- 80.Ma X, Yang X, Bao W, Li S, Liang S, Sun Y, Zhao Y, Wang J, Zhao C. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498(4):1009–1015. doi: 10.1016/j.bbrc.2018.03.105. [DOI] [PubMed] [Google Scholar]
- 81.Li S, Sun X, Miao S, Lu T, Wang Y, Liu J, Jiao W. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer. 2018;9(8):924–930. doi: 10.1111/1759-7714.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, Cui J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–2435. doi: 10.1016/j.bbrc.2018.06.172. [DOI] [PubMed] [Google Scholar]
- 83.Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X, Guan G, Dong X. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun. 2018;503(3):2089–2094. doi: 10.1016/j.bbrc.2018.07.164. [DOI] [PubMed] [Google Scholar]
- 84.Yu W, Jiang H, Zhang H, Li J. Hsa_circ_0003998 promotes cell proliferation and invasion by targeting miR-326 in non-small cell lung cancer. Onco Targets Ther. 2018;11:5569–5577. doi: 10.2147/OTT.S174750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8(8):1387–1402. [PMC free article] [PubMed] [Google Scholar]
- 86.Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi: 10.1016/j.bbrc.2018.10.087. [DOI] [PubMed] [Google Scholar]
- 87.Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019;120(3):3765–3772. doi: 10.1002/jcb.27657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen J, Xu S, Chen S, Zong Z, Han X, Zhao Y, Shang H. CircPUM1 promotes the malignant behavior of lung adenocarcinoma by regulating miR-326. Biochem Biophys Res Commun. 2019;508(3):844–849. doi: 10.1016/j.bbrc.2018.11.176. [DOI] [PubMed] [Google Scholar]
- 89.Qiu BQ, Zhang PF, Xiong D, Xu JJ, Long X, Zhu SQ, Ye XD, Wu Y, Pei X, Zhang XM, et al. CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non-small cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. J Cell Physiol. 2018;234(7):11256–11264. doi: 10.1002/jcp.27783. [DOI] [PubMed] [Google Scholar]
- 90.Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi: 10.1016/j.bbrc.2018.04.172. [DOI] [PubMed] [Google Scholar]
- 91.Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan H, Niu Y, Liu F, Gu H, Fan X, et al. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol Biochem. 2018;51(5):2324–2340. doi: 10.1159/000495876. [DOI] [PubMed] [Google Scholar]
- 92.Qin S, Zhao Y, Lim G, Lin H, Zhang X, Zhang X. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed Pharmacother. 2018;111:244–250. doi: 10.1016/j.biopha.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 93.Yao Y, Hua Q, Zhou Y, Shen H. CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2019;111:1367–1375. doi: 10.1016/j.biopha.2018.12.120. [DOI] [PubMed] [Google Scholar]
- 94.Li W, Jiang W, Liu T, Lv J, Guan J. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem Biophys Res Commun. 2019;510(3):467–471. doi: 10.1016/j.bbrc.2019.01.134. [DOI] [PubMed] [Google Scholar]
- 95.Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, Dai J, Zhang S, Yang Q, Yi Y, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):10–13. doi: 10.1186/s12943-019-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cui J, Li W, Liu G, Chen X, Gao X, Lu H, Lin D. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47(1):810–821. doi: 10.1080/21691401.2019.1575847. [DOI] [PubMed] [Google Scholar]
- 97.Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593–3p/CCND2 and miR-653–5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;510(3):462–466. doi: 10.1016/j.bbrc.2019.01.131. [DOI] [PubMed] [Google Scholar]
- 98.An J, Shi H, Zhang N, Song S. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019 doi: 10.1016/j.bbrc.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 99.Zhou Y, Zheng X, Xu B, Chen L, Wang Q, Deng H, Jiang J. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun. 2019;508(2):527–535. doi: 10.1016/j.bbrc.2018.11.157. [DOI] [PubMed] [Google Scholar]
- 100.Wang T, Wang X, Du Q, Wu N, Liu X, Chen Y, Wang X. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513(4):904–911. doi: 10.1016/j.bbrc.2019.04.108. [DOI] [PubMed] [Google Scholar]
- 101.Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother. 2019;112:10–1016. doi: 10.1016/j.biopha.2019.108743. [DOI] [PubMed] [Google Scholar]
- 102.Liu G, Shi H, Deng L, Zheng H, Kong W, Wen X, Bi H. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513(1):207–212. doi: 10.1016/j.bbrc.2019.03.213. [DOI] [PubMed] [Google Scholar]
- 103.Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, Pan X, Zhang X, Zhou L, Yu D, et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res. 2019;25(4):1302–1317. doi: 10.1158/1078-0432.CCR-18-1447. [DOI] [PubMed] [Google Scholar]
- 104.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin Pathway. Biomed Res Int. 2016;2016:10–1155. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang L, Wang J, Fan Y, Yu K, Jiao B, Su X. Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respir Res. 2018;19(115):10–1186. doi: 10.1186/s12931-018-0819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17(140):10–1186. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080–2090. doi: 10.1080/15384101.2018.1515553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503(3):1503–1509. doi: 10.1016/j.bbrc.2018.07.070. [DOI] [PubMed] [Google Scholar]
- 109.Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z, Wang Z, Yang C, Jiang Y. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA 0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39(8):981–992. doi: 10.1093/carcin/bgy061. [DOI] [PubMed] [Google Scholar]
- 110.Zhang Y, Zhao H, Zhang L. Identification of the tumorsuppressive function of circular RNA FOXO3 in nonsmall cell lung cancer through sponging miR155. Mol Med Rep. 2018;17(6):7692–7700. doi: 10.3892/mmr.2018.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang L, Liu S, Mao Y, Xu J, Yang S, Shen H, Xu W, Fan W, Wang J. CircRNF13 regulates the invasion and metastasis in lung adenocarcinoma by targeting miR-93-5p. Gene. 2018 doi: 10.1016/j.gene.2018.04.069. [DOI] [PubMed] [Google Scholar]
- 112.Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783–3p/DKK1 axis and inactivating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2019;508(1):37–45. doi: 10.1016/j.bbrc.2018.11.079. [DOI] [PubMed] [Google Scholar]
- 113.Yu C, Tian F, Liu J, Su M, Wu M, Zhu X, Qian W. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway. Cell Proliferat. 2019 doi: 10.1111/cpr.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.World Health statistics 2018 monitoring health for the SDGs. WHO. 2018.
- 115.Fennell DA, Summers Y, Cadranel J, Benepal T, Christoph DC, Lal R, Das M, Maxwell F, Visseren-Grul C, Ferry D. Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat Rev. 2016;44:42–50. doi: 10.1016/j.ctrv.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 116.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7(9):493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. OncoTargets Ther. 2018;11:3979–3987. doi: 10.2147/OTT.S158316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee BB, Kim Y, Kim D, Cho EY, Han J, Kim HK, Shim YM, Kim D. Metformin and tenovin-6 synergistically induces apoptosis through LKB1-independent SIRT1 down-regulation in non-small cell lung cancer cells. J Cell Mol Med. 2019;23(4):2872–2889. doi: 10.1111/jcmm.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ding L, Yao W, Lu J, Gong J, Zhang X. Upregulation of circ_001569 predicts poor prognosis and promotes cell proliferation in non-small cell lung cancer by regulating the Wnt/beta-catenin pathway. Oncol Lett. 2018;16(1):453–458. doi: 10.3892/ol.2018.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW, Wen J. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.