Abstract
Systemic sclerosis (SSc) is a life-threatening connective tissue disorder of unknown etiology characterized by widespread vascular injury and dysfunction, impaired angiogenesis, immune dysregulation and progressive fibrosis of the skin and internal organs. Over the past few years, a new trend of investigations is increasingly reporting aberrant epigenetic modifications in genes related to the pathogenesis of SSc, suggesting that, besides genetics, epigenetics may play a pivotal role in disease development and clinical manifestations. Like many other autoimmune diseases, SSc presents a striking female predominance, and even if the reason for this gender imbalance has yet to be completely understood, it appears that the X chromosome, which contains many gender and immune-related genes, could play a role in such gender-biased prevalence. Besides a short summary of the genetic background of SSc, in this review we provide a comprehensive overview of the most recent insights into the epigenetic modifications which underlie the pathophysiology of SSc. A particular focus is given to genetic variations in genes located on the X chromosome as well as to the main X-linked epigenetic modifications that can influence SSc susceptibility and clinical phenotype. On the basis of the most recent advances, there is realistic hope that integrating epigenetic data with genomic, transcriptomic, proteomic and metabolomic analyses may provide in the future a better picture of their functional implications in SSc, paving the right way for a better understanding of disease pathogenesis and the development of innovative therapeutic approaches.
Keywords: epigenetics, gender dimorphism, genetics, lncRNA, miRNA, scleroderma, systemic sclerosis, X chromosome
Introduction
Systemic sclerosis (SSc, scleroderma) is a multisystem connective tissue disorder characterized by immune system activation/dysregulation, vasculopathy and progressive fibrosis of the skin and internal organs.1 Although the pathogenesis of SSc is extremely heterogeneous and complex, it appears that disease onset and progression may be orchestrated by the interplay between genetics and specific environmental agents.1 Indeed, the exposition to chemical compounds (i.e. silica or organic solvents) or infectious agents has long been associated with increased disease susceptibility.2 More recently, a new trend of investigations is increasingly reporting aberrant epigenetic modifications in genes related to the pathogenesis of SSc, suggesting that both genetics and epigenetics may play pivotal roles in SSc development.2
Epigenetics refers to the study of stable and mitotically heritable modifications in both gene expression and function that do not involve changes in the DNA sequence. Epigenetic modifications are deeply influenced by environmental factors and may contribute to the breakdown of immune tolerance and the development of SSc in individuals with a particular genetic background.2,3 In particular, several studies have focused on genetic and epigenetic differences in an attempt to explain the female prevalence of SSc.2,3 Indeed, like many other autoimmune diseases, SSc presents a striking female predominance, with a sex ratio (F/M) ranging from 5:1 to 12:1.4,5 However, the reason for this gender imbalance has yet to be completely understood. Moreover, SSc female preponderance and multiple clinical features of the disease vary in different geographical areas and ethnicities6 and depend on the age, as suggested by the higher F/M incidence ratio of SSc during the childbearing years with respect to the postmenopausal years.7 It is also noteworthy that female preponderance merely reflects a different disease incidence between the sexes, but not a different disease severity. Indeed, male SSc patients usually have a more severe prognosis compared to female patients.8
Besides a short summary of the genetic background of SSc, in this review we will provide a comprehensive overview of the most recent discoveries regarding the epigenetic modifications which may offer insights into the pathophysiology of SSc. A particular focus will be given to the genetic and epigenetic modifications responsible for alterations in X chromosome-related gene expression that may underlie SSc gender dimorphism.
Genetic background of SSc
Although SSc is not inherited in a Mendelian fashion, a large body of evidence indicates that multiple gene variants may influence both disease susceptibility and differences in clinical expression and progression.9 The overall genetic burden is modest (i.e. only 2.6% of patients’ siblings develop SSc),10 but the incidence of SSc is higher in individuals with a family history than in the general population.11 Indeed, a positive family history for SSc significantly increases the relative risk by 15–19-fold in siblings and by 13–15-fold in first-degree relatives.12 The involvement of genetics in SSc pathogenesis is further supported by a racial difference in disease prevalence and clinical manifestations, as testified by the evidence that some ethnic groups or subpopulations have an increased SSc prevalence compared to the general population.12
To date, the most susceptible locus for SSc is the major histocompatibility complex (MHC), a genetic region of chromosome 6 with high gene density and long-range linkage disequilibrium patterns. In humans, the MHC is one of the most polymorphic regions of the genome and its gene products are called human leukocyte antigen (HLA) complex. Some HLA variants have been associated with SSc (HLA-DRB1*01, HLA-DRB1*11, HLA-A*30, and HLA-A*32), while others (HLA-DRB1*07, HLA-B*57, and HLA-Cw*14) are protective against the disease. The HLA-class I complexes HLA-A, B, C, and G and HLA-class II complexes HLA-DP, DQ, and DR have been reported to increase the risk of developing SSc.12,13 As far as non-HLA genes are concerned, several candidate genes have been implicated in SSc susceptibility. However, they all appear to be shared by other autoimmune diseases and do not explain the clinical heterogeneity of SSc.9,13,14
Recently, whole-exome sequencing (WES) studies in SSc patients have identified variants in ATP8B4, a gene encoding a phospholipid transporter, as a novel locus for SSc susceptibility and progression in European Americans but not in African Americans.15,16 WES has also been specifically performed in patients with diffuse cutaneous SSc (dcSSc), reporting a significant association of the extracellular matrix-related pathway with enrichment of variants within the COL4A3, COL4A4, COL5A2, COL13A1, and COL22A1 genes.17 Collectively, it is clear that modifications in DNA sequence alone cannot explain SSc heterogeneity, as further indicated by the evidence that monozygotic twins, even if sharing identical DNA sequences, present low concordance rates for the disease and may display different clinical phenotypes.2,11 Apart from inheritance, in the development of SSc a major role could therefore be played by epigenetic modifications.14,18,19
Epigenetics of SSc
As already mentioned, genetic abnormalities and the concomitant influence of environmental agents cannot fully explain SSc heterogeneity. In this context, epigenetic modifications that are able to modulate gene expression without altering the DNA sequence are regarded as a unique crossroad between genetics and environmental factors.2 Epigenetic mechanisms include DNA methylation, histone modifications, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs).
DNA methylation
DNA methylation is the most widely investigated epigenetic mechanism. The process is catalyzed by specific enzymes called DNA methyltransferases (DNMTs) and consists of the transfer of a methyl group from S-adenyl methionine to the pyrimidine C5 position of cytosine residues, forming 5-methylcytosine (5-mC). This usually occurs on CpG sites, which are sequences characterized by a cytosine preceding a guanine nucleotide.9,20 DNMTs are classified into ‘maintenance DNMTs’ (DNMT1, DNMT2), which are involved in maintaining the existing pattern of DNA methylation during cell replication, and ‘de novo DNMTs’ (DNMT3a, DNMT3b and DNMT3L), which control methylation during embryonic development.9,20 If the promoter region of a gene is sufficiently methylated, the transcription of that gene will be inhibited due to the reduced capability of transcription factors to bind to the gene promoter. On the contrary, a low methylation of the promoter activates DNA transcription.9,20 The active demethylation of DNA, which is linked to transcriptional activation and gene expression, consists of the removal of the methyl group, with the conversion of 5-mC to 5-hydroxymethylcytosine (5-hmC). This conversion is an oxidation reaction catalyzed by the ten eleven translocation (TET) family of enzymes.21
The DNA methylation state has been extensively studied in a variety of autoimmune diseases including systemic lupus erythematosus, rheumatoid arthritis, multiple sclerosis and Sjögren’s syndrome.22,23 As far as SSc is concerned, abnormalities in DNA methylation have been mainly reported in autosomal genes of fibroblasts, immune cells and endothelial cells.24
Fibroblasts
SSc is characterized by persistently activated fibroblasts responsible for an excessive production of collagen and other extracellular matrix components. As reported in a genome-wide DNA methylation study, the pathological phenotype of SSc fibroblasts seems to be determined by an altered global hypomethylation state.25 In this large-scale analysis, fibroblasts from the dcSSc and the limited cutaneous SSc (lcSSc) subsets revealed different and characteristic methylation patterns, with 916 CpG hypomethylated sites in lcSSc fibroblasts as compared with 1653 CpG hypomethylated sites in dcSSc fibroblasts. In particular, an abnormal DNA methylation profile was detected in several genes involved in fibrosis-related pathways (i.e. transforming growth factor-β (TGF-β) and Wnt/β-catenin signaling pathways), highlighting the potential role of DNA methylation changes in SSc pathogenesis.25
Conversely, increased promoter methylation and consequent downregulation of friend leukemia integration 1 (Fli1) transcription factor resulting in enhanced type I collagen gene expression have been reported in SSc fibroblasts.26 Fli1 acts as an important suppressor of type I collagen gene transcription and has been found to be constitutively downregulated in cultured dermal fibroblasts from clinically involved SSc skin.27,28 Of note, treatment of SSc fibroblasts with DNA methyltransferase inhibitor 2-deoxy-5-azaC (5-aza) could reverse Fli1 downregulation and normalize type I collagen expression.26 In another study, altered DNA methylation at Krüppel-like factor 5 (KLF5) gene promoter, which encodes a transcription factor that works synergistically with Fli1, contributed to impaired KLF5-Fli1 activity in SSc fibroblasts.29
In addition, fibroblasts from SSc patients were found to exhibit a hypermethylation profile of the promoters of genes encoding for the endogenous Wnt antagonists dickkopf-related protein 1 (DKK1) and secreted-frizzled protein 1 (SFRP1), with a consequent pathological activation of canonical Wnt/β-catenin signaling and a Smad-dependent fibrotic response.30,31 Of note, inhibition of DNA methylation with 5-aza completely restored DKK1 and SFRP1 expression and reduced the activation of canonical Wnt/β-catenin signaling, effectively inhibiting the fibrotic process either in vitro in cultured SSc fibroblasts or in vivo in the bleomycin-induced scleroderma mouse model.31
A recent DNA methylation analysis in SSc fibroblasts showed a significant hypermethylation in the promoter of poly(ADP-ribose)polymerase-1 (PARP1), a NAD+-dependent DNA repair enzyme, whose reduced expression was found to contribute to persistent fibroblast activation and progression of fibrosis.32 Remarkably, treatment of SSc fibroblasts with 5-aza gradually increased PARP1 expression, strengthening the notion that hypermethylation of PARP1 promoter is responsible for its downregulation in SSc.32
As far as active DNA demethylation is concerned, its involvement in the pathogenesis of SSc is supported by the evidence that, among TET family members, TET1 expression was found to be specifically upregulated under hypoxic conditions via hypoxia-inducible factor (HIF)-1α-independent pathways in SSc fibroblasts, but not in normal fibroblasts. Interestingly, such aberrant TET1 expression in SSc fibroblasts was accompanied by global DNA hypomethylation.33
Immune cells
Similar to dermal fibroblasts, a global hypomethylation pattern was first reported in SSc CD4+ T cells, presumably due to a reduced expression of DNMT1.34 A subsequent genome-wide DNA methylation analysis in both CD4+ and CD8+ T cells revealed a predominant hypomethylation profile in type I interferon (IFN)-related genes such as IFI44L, IFITM1, MX1 and PARP9 associated with an increase in circulating levels of type I IFN, suggesting that hypomethylation and consequent upregulation of the IFN signaling pathway might be critical in SSc pathogenesis.35 Widespread hypomethylation status of CpG sites located at genes involved in type I IFN signaling in CD4+ T cells from SSc and other autoimmune diseases was confirmed in a very recent genome-wide DNA methylation study.36 Moreover, in SSc peripheral blood mononuclear cells (PBMCs), the analysis of the methylation status of 16 CpG sites at the promoter region of IRF7, a gene encoding a type I IFN transcription factor associated with the production of autoantibodies in SSc,37 revealed a significant hypomethylation profile in comparison to healthy cells.38
Another gene whose promoter region was found to be hypomethylated in SSc CD4+ T cells is ITGAL, which encodes CD11a, a cell surface antigen involved in T cell stimulation.39 In particular, the extent of ITGAL hypomethylation was found to be inversely correlated with the scleroderma disease activity index determined by skin thickness, patient-reported worsening of symptoms, digital necrosis, and inflammation markers.39
The unique type I IFN signature found in SSc stimulated further studies focused on SSc plasmacytoid dendritic cells (pDCs), that is specialized antigen-presenting cells capable of immediately producing a massive amount of type I IFN upon activation.40 Interestingly, a higher methylation status of the runt-related transcription factor 3 (RUNX3) gene was correlated with lower RUNX3 expression in SSc patients. RUNX3 transcription factor is known to contribute to differentiation and regulation of the dendritic cell lineage, and its downregulation was found to impair pDC functionality in mouse models. Notably, specific pDC RUNX3 ablation resulted in an increase in the severity of bleomycin-induced skin inflammation and fibrosis with respect to wild type mice. Thus, it has been proposed that RUNX3 might play an important role in regulating pDC function and the fibrotic process in SSc pathogenesis.40
Finally, although peripheral blood cells of patients with SSc display an increased expression of several selectin and integrin genes such as ITGB2, encoding integrin β2 protein, no significant differences between SSc patients and healthy individuals were observed in the methylation status of the ITGB2 promoter, suggesting that its upregulation is probably due to other yet unknown mechanisms.41
Endothelial cells
At variance with fibroblasts and immune cells, notions regarding an altered DNA methylation in endothelial cells from patients with SSc are limited to a single study reporting a reduced expression of the bone morphogenic protein receptor II (BMPRII) and consequent decrease in cell survival and apoptosis resistance in SSc microvascular endothelial cells (MVECs) compared with healthy cells.42 Indeed, through BMPRII, bone morphogenic proteins coordinate cell proliferation, differentiation and survival. In this study, the downregulation of BMPRII expression was attributable to heavily methylated CpG sites in the BMPRII gene promoter region, as demonstrated by the evidence that treatment of SSc MVECs with the DNA methyltransferase inhibitor 5-aza could normalize BMPRII expression levels and restore cell apoptotic response to levels comparable to healthy cells.42
The most relevant SSc-related global and gene-specific DNA methylation modifications are listed in Table 1.
Table 1.
Summary of epigenetic modifications in SSc.
| Epigenetic process | Modification | Cell type | Effect | References |
|---|---|---|---|---|
| Global DNA methylation | ||||
| ↓ | Fibroblasts | Profibrotic | Altorok et al.25 | |
| ↓ | CD4+ T cells | Supposed increased expression of autoimmune-related genes in lymphocytes | Lei et al.34 | |
| Skewed X chromosome inactivation | PBMCs | Immunosenescence, autoantibody production | Kanaan et al.43 | |
| Ozbalkan et al.44 | ||||
| Gene-specific DNA methylation | ||||
| Fli1 | ↑ | Fibroblasts | Profibrotic | Wang et al.26 |
| KLF5 | ↑ | Fibroblasts | Profibrotic | Noda et al.29 |
| DKK1, SFRP1 | ↑ | Fibroblasts | Profibrotic | Dees et al.31 |
| PARP1 | ↑ | Fibroblasts | Profibrotic | Zhang et al.32 |
| IFN-related genes (IFI44L, IFITM1, MX1, PARP9) | ↓ | CD4+ and CD8+ T cells | Increased IFN production | Ding et al.35 |
| Chen et al.36 | ||||
| IRF7 | ↓ | PBMCs | N.D. | Rezaei et al.38 |
| ITGAL | ↓ | CD4+ T cells | Increased proliferation of CD4+ T cells, IgG overproduction by B cells, and excessive collagen synthesis by fibroblasts | Wang et al.39 |
| RUNX3 | ↑ | pDCs | Profibrotic | Affandi et al.40 |
| BMPR2 | ↑ | MVECs | Proapoptotic | Wang and Kahaleh42 |
| CD40L (X chromosome) | ↓ | CD4+ T cells | Altered immune response | Lian et al.45 |
| ARX, HSFX1, IL1RAPL2 (X chromosome) | ↑ | PBMCs | Disease susceptibility | Selmi et al.46 |
| ZBED1, ZNF41, PGMRC1 (X chromosome) | ↓ | PBMCs | Disease susceptibility | Selmi et al.46 |
| FOXP3 (X chromosome) | ↑ | CD4+ T cells | Treg reduction | Wang et al.47 |
| Global histone acetylation | ||||
| ↓ HDAC2, HDAC7 H4 hyperacetylation |
B cells | B cell dysfunction | Wang et al.48 | |
| altered H3K4me3, H3K27ac |
Monocytes | Altered phenotype | Van der Kroef et al.49 | |
| ↑ HDAC5 | Endothelial cells | Impaired angiogenesis | Tsou et al.50 | |
| ↓ SIRT1, 3, 7 | Fibroblasts | Profibrotic | Chu et al.51 | |
| Wyman et al.52 | ||||
| Sosulski et al.53 | ||||
| ↑ p300 | Fibroblasts | Profibrotic | Ghosh et al.54 | |
| Gene-specific histone acetylation | ||||
| KLF5 | H3, H4 hypoacetylation | Fibroblasts | Profibrotic | Noda et al.29 |
| FLI1 | H3, H4 hypoacetylation | Fibroblasts | Profibrotic | Noda et al.29 |
| Wang et al.55 | ||||
| Global histone methylation | ||||
| ↑ H3K27me3 ↑ EZH2 |
Fibroblasts, endothelial cells | Profibrotic, impaired angiogenesis | Xiao et al.56 | |
| Tsou et al.57 | ||||
| ↑ H3K27me3 ↑ EZH2 |
Fibroblasts | Antifibrotic | Krämer et al.58 | |
| ↓ H3K27me3 ↑ JMJD3 |
CD4+ T cells | N.D. | Wang et al.59 | |
| H3K9 hypomethylation ↓ SUV39H2 ↑ JHDM2 |
B cells | N.D. | Wang et al.48 | |
| Gene-specific histone methylation | ||||
| FOSL2 | ↓ H3K27me3 ↑ JMJD3 = UTX |
Fibroblasts | Profibrotic | Bergmann et al.60 |
PBMC, peripheral blood mononuclear cell; pDC, plasmacytoid dendritic cell: MVEC, microvascular endothelial cell; N.D., not determined; SSc, systemic sclerosis.
Histone post-translational modifications
While DNA is primarily methylated, histone proteins can undergo a wide array of post-translational modifications, representing the target of another major epigenetic regulatory mechanism. Histones are conserved nuclear proteins forming the core center of nucleosomes, the basic subunits of eukaryotic chromatin consisting of 146 base pairs of DNA wrapped around an octamer of two pairs of four core histones (H2A, H2B, H3, and H4). Histone post-translational modifications include lysine acetylation, lysine and arginine methylation, serine and threonine phosphorylation, lysine ubiquitination, sumoylation, citrullination, ADP-ribosylation, and proline isomerization. These modifications serve as signals and regulate the transcription process. Histone modifications can affect the structure of nucleosomes in two ways: (a) the alteration in the electrostatic charge of the histone results in a conformational change and (b) protein-binding is altered by the modification.61 Of note, there is a complex interplay between histone modifications and DNA methylation. Emerging evidence, indeed, indicates that the modification state of DNA can affect the methylation pattern of accompanying histones, while the histone lysine methylation state of chromatin can in turn influence modification of the DNA itself. In particular, it has been reported that DNA methylation, which results in chromatin compaction, elicits additional effects including histone deacetylation and methylation. In fact, biochemical works studying methyl-CpG binding proteins revealed that these proteins not only specifically associate with methylated CpG sites, but also bind to a multitude of different chromatin modifying enzymes, including histone deacetylases and histone lysine methyltransferases.62 On the other hand, it has also been shown that DNA methylation by de novo methyltransferases DNMT3a/b is dependent on pre-existing histone methylation.62
Aberrant gene expression resulting from histone post-translational modifications is known to be involved in SSc pathogenesis. In particular, histone acetylation and methylation are the most commonly described epigenetic modifications in SSc.
Histone acetylation/deacetylation
Acetylation is the most common histone modification and is tightly controlled by the balance of two antagonizing enzymes, namely histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs catalyze the addition of an acetyl group from acetyl coenzyme A to the lysine side chains of histones, neutralizing their positive charge and weakening their interaction with DNA. By leading to a more open chromatin conformation, histone acetylation enables the binding of the transcriptional machinery, and is thus generally followed by transcriptional activation. In contrast, HDACs remove acetyl groups from target histones, restoring their positive charge and leading to a closed chromatin structure. Histone deacetylation is therefore involved in the suppression of gene expression.61
Over the past few years, several studies highlighted the potential effect of chromatin deacetylation in SSc pathogenesis. In 2014, Noda and coworkers reported a significant lower expression of KLF5 in SSc dermal fibroblasts compared with healthy fibroblasts, and attributed this downregulation to a significant suppression of the acetylated forms of histones H3 and H4 in the KLF5 gene promoter.29 Indeed, the recovery of KLF5 expression levels was obtained after treating SSc fibroblasts with the HDAC inhibitor trichostatin A.29 A similar epigenetic repression was also observed for the promoter of FLI1 gene.29,55 As KLF5 and Fli1 synergistically inhibit the expression of connective tissue growth factor (CTGF), which is known to be a regulator of tissue remodeling and fibrosis, their epigenetic repression in fibroblasts from SSc patients might contribute to the increased CTGF expression and the consequent profibrotic phenotype observed in these cells.29
Recent data support the notion that histone modifications are associated with B cell development, activation, differentiation, apoptosis, and autoantibody production.48 In this context, a study aimed at clarifying the mechanisms underlying B cell activation in SSc reported a significant decrease in the expression of the histone deacetylases HDAC2 and HDAC7 and concomitant global histone H4 hyperacetylation in B cells from SSc patients. In that study, the degree of histone H4 acetylation positively correlated with disease activity and the expression of HDAC2 protein negatively correlated with skin thickness, suggesting that a dysregulation of histone acetylation might contribute to B cell dysfunction in the pathogenesis of SSc.48
Besides fibroblasts and B cells, histone modifications at a genome-wide level have been implicated in the altered phenotype of SSc monocytes.49
An indirect role of histone deacetylation in the fibrotic process of SSc has been suggested based on the evidence that treatment of SSc skin fibroblasts with trichostatin A suppressed TGF-β-induced mRNA expression of type I collagen and fibronectin and prevented dermal deposition of extracellular matrix in experimental scleroderma.63 The contribution of histone acetylation to SSc pathogenesis is further supported by aberrant cell-specific expression patterns of the histone-modifying enzymes regulating this epigenetic process. Indeed, endothelial cells isolated from patients with SSc were found to display an increased expression of HDAC5, a histone deacetylase regulating several genes involved in angiogenesis, suggesting that HDAC5 may contribute to the disrupted angiogenic process in SSc.50 Moreover, a decrease in the levels of sirtuins SIRT1, SIRT3 and SIRT7, which are class III HDACs regulating TGF-β signaling in fibroblasts, was found in dermal and pulmonary SSc fibroblasts, indicating a possible role of these proteins in the pathogenesis of SSc-related multiorgan fibrosis.51–53 Indeed, both SIRT1 and SIRT3 activation with resveratrol or hexafluoro, respectively, was shown to ameliorate fibrosis either in vitro or in vivo.51,64
Finally, a high expression of the p300 HAT, whose availability guides the fibrotic process, has been reported in SSc skin biopsies and in TGF-β-treated fibroblasts. Increased p300 accumulation was associated with histone hyperacetylation, whereas p300 depletion or selective pharmacological blockade of its acetyltransferase activity attenuated TGF-β-induced responses in fibroblasts. Thus, it has been suggested that targeted disruption of p300-mediated histone acetylation might represent an additional strategy against fibrotic diseases.54 In a very recent study, chromatin accessibility and transcriptome profiling coupled with targeted epigenetic editing revealed the constitutive activation of a previously unannotated TGFB2 gene enhancer maintained through epigenetic memory in fibroblasts isolated from clinically affected SSc skin. In particular, it could be demonstrated that elevated acetylation of histone H3 on lysine 27 (H3K27ac) and occupancy by p300 were the main epigenetic modifications responsible for the activation of the TGFB2 enhancer and the consequent maintenance of a profibrotic state in SSc fibroblasts.65
A summary of SSc-related global and gene-specific histone acetylation modifications is reported in Table 1.
Histone methylation/demethylation
Besides acetylation, histones can be methylated by histone methyltransferases (HMTs), which transfer up to three methyl groups from S-adenosyl-l-methionine to the lysine or arginine residues of the histone tail, as well as demethylated by histone demethylases (HDMs). Unlike DNA methylation, histone methylation can result in either increased or suppressed transcription of the nearby gene, typically depending on the specific methylated residue. In general, methylation of histone 3 on lysine 4 (H3K4) is associated with active chromatin and gene expression, whereas methylation of histone 3 on lysine 9 (H3K9) and trimethylation of histone H3 on lysine 27 (H3K27me3) are associated with condensed heterochromatin and transcriptional repression. In particular, H3K27me3, mediated by the two histone-lysine N-methyltransferases enhancer of zeste homolog 1 and 2 (EZH1 and EZH2) and the two histone demethylases jumonji domain-containing protein 3 (JMJD3) and UTX, represents a particular histone modification which seems to play a central role in SSc.60 Indeed, reduced H3K27me3 levels at the promoter of FOSL2, a gene encoding the profibrotic transcription factor fos-related antigen 2 (FRA2) whose expression is increased in SSc fibroblasts, have been detected in either SSc or TGF-β-treated fibroblasts compared to control cells.60 Such a decrease inversely correlated with an increase in JMJD3 expression, whereas the expression of UTX remained unchanged.60 Of note, the blockade of JMJD3 with the inhibitor GSKJ4 increased H3K27me3 levels, significantly limiting the aberrant activation of SSc fibroblasts and exerting antifibrotic effects in both bleomycin and topoisomerase-I mice models of dermal fibrosis.60 In addition, EZH2 blockade with its inhibitor 3-deazaneplanocin A (DZNep) was found to ameliorate lung fibrosis in a scleroderma mouse model and to prevent fibrosis and restore normal angiogenesis in SSc fibroblasts and endothelial cells, respectively.56,57 These data further support the importance of H3K27me3 in SSc and provide evidence that EZH2 might play a critical role in SSc-related fibrosis and vasculopathy. The role of EZH2 in SSc had also been examined in a previous study, but with conflicting results.58 Indeed, Krämer et al. showed that treatment with DZNep exacerbated fibrosis in SSc dermal fibroblasts and in the bleomycin mouse model of scleroderma. However, as neither EZH2 nor H3K27me3 levels were determined after DZNep treatment, it is uncertain whether EZH2 was effectively inhibited in SSc fibroblasts and, as far the mouse model is concerned, it is likely the dosing regimen used might have been insufficient to achieve EZH2 inhibition.58 Besides SSc fibroblasts, a reduced H3K27me3 pattern inversely correlated with JMJD3 levels was also reported in SSc CD4+ T cells, reinforcing the notion that JMJD3 may represent the specific histone demethylase responsible for H3K27me3 changes in SSc.59
Finally, as far as B cells are concerned, a global histone H3K9 hypomethylation was observed in B cells isolated from SSc patients compared with controls.48 In particular, the decrease in total H3K9 methylation was attributable to the concomitant reduction in the methyltransferase SUV39H2 and induction of the histone demethylase JHDM2. In addition, decreased H3K9 methylation was positively correlated with the degree of SUV39H2 protein reduction but did not correlate with skin thickness and disease activity.48
The most important SSc-related global and gene-specific histone methylation modifications are included in Table 1.
Non-coding RNAs
LncRNAs
LncRNAs comprise a large class of transcribed RNA molecules (more than 200 nucleotides long) that are not translated into proteins but are able to regulate gene expression both at the transcriptional and post-transcriptional levels.66 LncRNAs have been reported to be critically involved in several biological and immunological processes, including different pathways related to innate immunity.66 However, their role in SSc pathogenesis remains poorly understood. With the use of next-generation sequencing, an in-depth transcriptomic analysis of deregulated lncRNAs in skin tissue from SSc patients identified 676 lncRNAs differentially expressed between patients and healthy individuals.67 Interestingly, 257 lncRNAs out of the 676 identified were classified as antisense lncRNAs, which are supposed to function as co-regulators of their sense genes. Among them, the top three deregulated antisense lncRNAs were CTBP1-AS2, OTUD6B-AS1 and AGAP2-AS1, whose expression was strongly correlated with the expression of their paired sense genes.67 A strong deregulation of OTUD6B-AS1 in SSc skin biopsies was also reported in a recent RNA sequencing study.68 In particular, OTUD6B-AS1 was found to be significantly downregulated either in SSc fibroblasts or in healthy dermis after treatment with platelet-derived growth factor. Moreover, when healthy fibroblasts were silenced for OTUD6B-AS1 in order to mimic the downregulation seen in SSc patients, they showed increased Cyclin D1 expression, reduced proliferation and suppressed apoptosis. Collectively, these data suggest that OTUD6B-AS1 regulates fibroblast proliferation and apoptosis via cyclin D1 expression, shedding light on a possible novel apoptosis resistance mechanism that might be relevant for SSc pathogenesis.68 In another recent study, a total of 542,500 transcripts were profiled in PBMCs from SSc patients and healthy donors, and only the heterogeneous nuclear ribonucleoprotein U processed transcript (ncRNA00201) lncRNAs was found to be significantly downregulated in SSc patients.69 The authors also observed that ncRNA00201 alone may control different biological pathways closely related to the three main features of SSc, namely immune/inflammatory response, vasculopathy and fibrosis, thus providing new insights into disease pathogenesis and opening new avenues for the design of therapeutic strategies.69 In addition, by RNA sequencing, a group of lncRNAs related to the IFN and anti-viral response were shown to be modulated in a type I IFN-dependent manner in human monocytes in response to TLR4 activation.70 Among these lncRNAs, the negative regulator of the IFN response (NRIR) was found to be significantly upregulated in SSc monocytes and to affect the expression of IFN-stimulated genes. Thus, dysregulation of NRIR in SSc monocytes might partly contribute to the aberrant IFN response present in SSc patients.70 Finally, another lncRNA recently reported to be implicated in SSc pathogenesis is HOX transcript antisense RNA (HOTAIR) that was found to induce profibrotic activation and myofibroblastic transformation of dermal fibroblasts in vitro by driving the specific methylation profile of the histone methyltransferase EZH2, which in turn increases Notch transcription through the methylation and consequent repression of the negative regulator of Notch expression miRNA-34a.71
Table 2 includes a list of the most relevant lncRNAs involved in SSc pathogenesis.
Table 2.
LncRNAs implicated in SSc.
| LncRNAs | Modification | Cell type/tissue | Effect | References |
|---|---|---|---|---|
| CTBP1-AS2, AGAP2-AS1 | ↑ | Skin | Differentially expressed between SSc patients and healthy controls | Messemaker et al.67 |
| OTUD6B-AS1 | ↓ | Fibroblasts, skin | Differentially expressed between SSc patients and healthy controls; antiapoptotic | Messemaker et al.67 |
| Takata et al.68 | ||||
| ncRNA00201 | ↓ | PBMCs | Regulates genes and pathways involved in vasculopathy, fibrosis and autoimmunity | Dolcino et al.69 |
| NRIR | ↑ | Monocytes | Aberrant IFN response | Mariotti et al.70 |
| TSIX (X chromosome) | ↑ | Fibroblasts, serum | Profibrotic | Wang et al.72 |
LncRNA, long non-coding RNA; NRIR, negative regulator of the IFN response; PBMC, peripheral blood mononuclear cell; SSc, systemic sclerosis.
MiRNAs
MiRNAs are short (20–25 nucleotides) single-stranded non-coding RNAs that function as post-transcriptional regulators of gene expression, leading to translational suppression by binding to the 3′ untranslated region (UTR) of specific mRNAs. MiRNAs are expressed in a tissue and cell type-specific manner and show a close interplay with other epigenetics mechanisms, such as DNA methylation and histone modifications.20,61,66,73 MiRNAs regulate a variety of biological processes such as cell growth, differentiation, and immune functions and have been extensively implicated in SSc, where they seem to be mainly implicated in the fibrotic process. In this context, one of the most studied miRNAs is miR-29a, which is known to bind the 3′ UTR of the COL1A1 gene and to exhert potent antifibrotic effects.74 MiR-29a was found to be significantly downregulated in SSc fibroblasts, SSc skin biopsies and skin samples from bleomycin-treated mice, while its induced overexpression in SSc fibroblasts decreased type I and type III collagen synthesis, supporting the notion that miR-29a may play an important role in SSc-related fibrosis.74,75 The antifibrotic effect of miR-29a is further sustained by the evidence that this miRNA reduces the expression of TGF-β-activated kinase 1 binding protein 1 (TAB1), a protein involved in the downregulation of tissue inhibitor of metalloproteinase 1 (TIMP1) expression and collagen degradation.76 Another antifibrotic miRNA able to regulate type I collagen expression is miR-196a, which was found to be significantly reduced both in dermal fibroblasts and the skin of SSc patients.77,78 Several members of the let-7 family of miRNAs have also been shown to be involved in SSc dysregulated fibrosis. Among these, miR-let-7d and miR-let-7a were reported to be significantly downregulated in skin biopsies from SSc patients.79,80 MiR-let-7d levels were negatively correlated with patient pulmonary arterial pressure, which suggests a potential role for this miRNA in the regulation of SSc-related pulmonary hypertension,79 while miR-let-7a downregulation, also found in SSc serum and fibroblasts, was implicated in the abnormally increased expression of type I collagen.80 In addition, the intraperitoneal administration of miR-let-7a to mice with bleomycin-induced scleroderma improved the cutaneous fibrotic process, suggesting a possible use of miR-let-7a analogs as antifibrotic drugs.80 Collagen production was found to be induced in SSc fibroblasts also by the suppression of miR-135b and the concomitant activation of STAT6-dependent interleukin-13 (IL-13) signaling.81 In addition, reduced levels of miR-135b were found in SSc serum and monocytes and the expression of this miRNA was proposed to be regulated by methylation events.81 Another miRNA that is downregulated in SSc fibroblasts and mediates antifibrotic effects is miR-132. By using a specific 3′ UTR luciferase assay, it was demonstrated that miR-132 directly downregulates methyl cap binding protein 2 (MeCP2), a transcriptional regulator which positively modulates the expression of extracellular matrix through epigenetic repression of the Wnt antagonist sFRP-1 with consequent enhanced Wnt signaling and fibrosis.82 However, the role of MeCP2 in SSc-related fibrosis remains controversial, because another study reported an antifibrotic effect of this transcriptional factor in dermal fibroblasts, with overexpression of MeCP2 in early dcSSc fibroblasts proposed as a possible defence mechanism to counteract the profibrotic nature of the disease in its early stages.83 Using miRNA array analysis, a recent study indicated that miR-202-3p was increased in SSc skin and demonstrated that miR-202-3p upregulation contributed to the suppression of matrix metalloproteinase 1 (MMP1) and a consequent increase in collagen deposition.84 Thus, miR-202-3p appears to function as a novel profibrotic miRNA in SSc. Another miRNA which has been found to be overexpressed in the serum, affected skin and explanted fibroblasts from SSc patients is miR-155.85–87 Moreover, it has recently been demonstrated that in SSc fibroblasts the activation of the NOD, LRR and pyrin domain-containing 3 (NLRP3) inflammasome drives miR-155 expression via IL-1 autocrine signaling, that further enhances IL-1 transcription leading to increased collagen production and consequent fibrosis.87 Besides collagen, miRNAs are also thought to contribute to the regulation of other SSc-related molecules or cytokines. For instance miR-150, which is the direct regulator of integrin-β3, a key molecule that is known to be overexpressed in SSc dermal fibroblasts and to activate TGF-β signaling, was found to be constitutively downregulated in SSc fibroblasts.88 The induced overexpression of this miRNA in SSc fibroblasts downregulated integrin-β3, phosphorylated Smad3 and type I collagen expression, while miR-150 knockdown in healthy fibroblasts exerted an opposite effect.88
Aside from exclusively regulating collagen production and fibroblast functionality, several miRNAs such as miR-21, miR-145, miR-193b and miR-130b have been implicated in both SSc vasculopathy and the associated fibrotic alterations. MiR-21 and miR-145 are two miRNAs closely related to the TGF-β signaling Smad pathways, as the profibrotic miR-21 downregulates the expression of Smad7 and is a promoter of endothelial-to-mesenchymal transition, while the antifibrotic miR-145 inhibits Smad3.73 MiR-21 was found to regulate apoptosis in SSc fibroblasts89 and to be overexpressed in skin biopsies, fibroblasts and serum from SSc patients as well as in healthy MVECs treated with SSc serum, with inhibitors of miR-21 attenuating the TGF-β-driven upregulation of profibrotic markers.89–92 Similarly, miR-146b, miR-130b, miR-31 and miR-34a were found to be upregulated in skin tissues and fibroblasts of SSc patients and in healthy MVECs stimulated with SSc serum.92,93 In contrast, the antifibrotic miR-145 was found to be reduced both in SSc skin and fibroblasts and in SSc serum-treated MVECs.92 Although these miRNAs are known to regulate several mechanisms involved in the pathogenesis of SSc such as toll-like receptors (TLRs), TGF-β and Wnt signaling pathways, further studies are needed to clarify their role fully in both the fibrotic and vascular aspects of the disease. Another miRNA which is thought to connect SSc vasculopathy and fibroproliferation is miR-193b, whose concentrations have been found to be decreased in fibroblasts and skin from SSc patients, resulting in a concurrent upregulation of urokinase-type plasminogen activator (uPA).94 As an increase in uPA levels is known to inhibit apoptosis and stimulate vascular smooth muscle cell proliferation, miR-193b downregulation might contribute to proliferative vasculopathy in SSc.94
As far as immune cells are concerned, a recent study has revealed a significant overexpression of miR-618 in SSc pDCs.95 In this work, it was suggested that miR-618 upregulation leads to the downregulation of IFN regulator factor 8, a crucial transcription factor for dendritic cell development. Indeed, miR-618 overexpression inhibited pDC differentiation and activation and led to increased production of IFNα upon TLR9 stimulation.95 In addition, an upregulation of miRNA-5196 in peripheral blood monocytes from patients with SSc was recently reported.96 As this miRNA exerts antifibrotic effects by inhibiting the expression of TIMP1 and FRA2, two proteins playing an important role in SSc development, the authors proposed that its overexpression might serve as a compensatory mechanism to reverse the profibrotic phenotype in SSc monocytes.96
MiRNAs can be detected not only inside the cells, but also in several body fluids, including serum, plasma and saliva.9 Although it has been supposed that there is an immediate degradation of circulating miRNAs by RNases, once they are secreted into extracellular spaces they can be stabilized in at least four ways: (a) protected in shedding vesicles; (b) covered under membranous microvescicles called exosomes; (c) surrounded by apoptotic cells; and (d) complexed with proteins or lipoproteins.9 In order to identify novel blood-based biomarkers, several studies have analyzed a broad range of circulating miRNAs in SSc. In 2015, the analysis of 45 circulating miRNAs in plasma from SSc patients and healthy individuals identified 21 miRNAs differentially expressed between the two groups. In particular, the different miRNA profile comprised miRNAs belonging to the miRNA-17~92 cluster, miR-16, miR-223, and miR-638.97 In the same year, miR-223, miR-181b, miR-342-3p and miR-184 were found to be differentially expressed between the limited and diffuse cutaneous SSc subsets, while miR-409, miR-184, miR-92a, miR-29a and miR-101 were reported to correlate with the disease autoantibody profiles.98 In a subsequent work, the screening of 758 serum miRNAs identified 30 miRNAs that were significantly increased in patients with SSc with respect to controls. Among these, miR-483-5p was elevated in patients with early stage SSc.99 In addition, an increase in six profibrotic miRNAs, including the previously mentioned miR-483-5p, and a reduction in ten antifibrotic miRNAs were found in serum exosomes isolated from SSc patients compared to healthy controls. Interestingly, exosomes isolated from patients with SSc were able to stimulate normal human dermal fibroblasts to express profibrotic genes, suggesting that exosomal miRNAs may contribute to spread fibrotic signals to distant sites not yet affected.99,100 Finally, two recent studies based on bioinformatics analysis were performed to identify miRNAs that are differentially expressed in SSc and might potentially contribute to disease pathogenesis. In the first study, the authors reported in SSc patients with interstitial lung disease (ILD) a panel of differentially expressed miRNAs involved in pathways related to inflammation and fibroblast regulation, providing novel insights into the molecular mechanisms underlying the pathogenesis of SSc-ILD.101 In the second study, miR-4484 was identified as having the highest (18-fold) upregulation in SSc patients compared to healthy subjects. Moreover, bioinformatics analysis of miR-4484 target genes showed that this miRNA is potentially involved in the TGF-β signaling pathway, extracellular matrix-receptor interaction and MMP expression, and might therefore contribute to pathological fibrosis in SSc.102
Table 3 summarizes the main miRNAs that have been implicated in SSc.
Table 3.
MiRNAs implicated in SSc.
| miRNAs (chromosome) |
Expression | Cell type/tissue | Target gene | Effect | References |
|---|---|---|---|---|---|
|
miR-29a
(chromosome 7) |
↓ | Fibroblasts Skin Skin of bleomycin-treated mice |
COL1A1
COL3A1 TAB1 |
Antifibrotic | Maurer et al.74 |
| Jafarinejad-Farsangi et al.75 | |||||
| Ciechomska et al.76 | |||||
|
miR-196a
(chromosome 12) |
↓ | Fibroblasts Skin |
COL1A1
COL1A2 |
Antifibrotic | Makino et al.77 |
| Honda et al.78 | |||||
|
miR-let-7d
(chromosome 9) |
↓ | Skin | N.D. | N.D. | Izumiya et al.79 |
|
miR-let-7a
(chromosome 9) |
↓ | Fibroblasts Skin Serum |
COL1A1
COL1A2 |
Antifibrotic | Makino et al.80 |
|
miR-135b
(chromosome 1) |
↓ | Fibroblasts Serum Monocytes |
STAT6 | Antifibrotic | O’Reilly et al.81 |
|
miR-132
(chromosome 17) |
↓ | Fibroblasts | MeCP2 | Antifibrotic | Henderson et al.82 |
|
miR-202-3p
(chromosome 10) |
↑ | Skin | MMP1 | Profibrotic | Zhou et al.84 |
|
miR-155
(chromosome 21) |
↑ | Fibroblasts Skin Serum |
CSNK1A1
SHIP1 |
Profibrotic | Yan et al.85 |
| Dolcino et al.86 | |||||
| Artlett et al.87 | |||||
|
miR-150
(chromosome 19) |
↓ | Fibroblasts | ITGB3 | Antifibrotic | Honda et al.88 |
|
miR-21
(chromosome 17) |
↑ | Fibroblasts Skin Serum SSc serum-treated MVECs |
SMAD7 | Profibrotic Antiapoptotic |
Henry et al.73 |
| Jafarinejad-Farsangi et al.89 | |||||
|
miR-145
(chromosome 5) |
↓ | Fibroblasts Skin SSc serum-treated MVECs |
SMAD3 | Antifibrotic | Henry et al.73 |
| Zhou et al.92 | |||||
|
miR-146b
(chromosome 10) miR-130b (chromosome 22) miR-31 (chromosome 9) miR-34a (chromosome 1) |
↑ | Fibroblasts Skin SSc serum-treated MVECs |
N.D. | N.D. | Zhou et al.92 |
| Lou et al.93 | |||||
|
miR-193b
(chromosome 16) |
↓ | Fibroblasts Skin |
PLAU | Regulation of uPA expression | Iwamoto et al.94 |
|
miR-618
(chromosome 12) |
↑ | pDCs | IRF8 | Inhibition of pDCs differentiation | Rossato et al.95 |
|
miR-5196
(chromosome 19) |
↑ | Monocytes |
TIMP1
FRA2 |
Antifibrotic | Steen et al.97 |
|
miR-17~92
(autosomal) miR-16 (chromosome 13) miR-223 (X chromosome) miR-638 (chromosome 19) |
Differentially expressed between SSc patients and healthy controls | Plasma | N.D. | N.D. | Steen et al.97 |
|
miR-223
(X chromosome) miR-181b (chromosome 1) miR-342 (chromosome 14) miR-184 (chromosome 15) |
Differentially expressed between lcSSc and dcSSc | Plasma | N.D. | N.D. | Wuttge et al.98 |
|
miR-409-3p
(chromosome 14) miR-184 (chromosome 15) miR-92a (X chromosome) miR-29a (chromosome 7) miR-101 (chromosome 1) |
Correlating with the disease autoantibody profiles | Plasma | N.D. | N.D. | Wuttge et al.98 |
|
miR-483-5p
(chromosome 11) |
↑ | Serum | N.D. | Profibrotic | Chouri et al.99 |
|
miR-let-7a-5p
(chromosome 9) miR-26b-5p (chromosome 2) miR-29b-3p (chromosome 7) miR-129-5p (chromosome 7) miR-133a-3p (chromosome 18) miR-140-5p (chromosome 16) miR-145-5p (chromosome 5) miR-146a-5p (chromosome 5) miR-196a-5p (chromosome 12) miR-200a-3p (chromosome 1) miR-223-3p (X chromosome) |
↓ | Serum exosomes | N.D. | Antifibrotic | Chouri et al.99 |
| Wermuth et al.100 | |||||
|
miR-let-7g-5p
(chromosome 3) miR-17-5p (chromosome 13) miR-21-5p (chromosome 17) miR-23b-5p (chromosome 9) miR-29a-3p (chromosome 7) miR-150-5p (chromosome 19) miR-155-5p (chromosome 21) miR-215-5p (chromosome 1) miR-503-5p (X chromosome) |
↑ | Serum exosomes | N.D. | Profibrotic | Chouri et al.99 |
| Wermuth et al.100 | |||||
|
miR-4484
(chromosome 10) |
↑ | Serum | MMP21 | Profibrotic | Rusek et al.102 |
|
miR-92a
(X chromosome) |
↑ | Fibroblasts Serum TGF-β-treated healthy fibroblasts |
MMP1 | Profibrotic | Sing et al.103 |
|
miR-542-3p
(X chromosome) |
↓ | Fibroblasts | BIRC5 | Proapoptotic | Vahidi Manesh et al.104 |
pDC, plasmacytoid dendritic cell: MVEC, microvascular endothelial cell; N.D., not determined; SSc, systemic sclerosis; TGF-β, transforming growth factor beta.
X-linked genetic and epigenetic modifications in SSc
As already mentioned, SSc presents a striking female predominance and the X chromosome, which contains many gender and immune-related genes, may play a role in this sex-biased prevalence.105 Thus, in the second part of this review we will provide an overview of genetic variations in genes located on the X chromosome and the main X-linked epigenetic modifications that can influence SSc susceptibility and clinical phenotype.
X-linked single nucleotide polymorphisms
A possible association between single nucleotide polymorphisms (SNPs) in IL-13 receptor subunit α1 (IL13RA1) and α2 (IL13RA2) genes and SSc was investigated in a Caucasian population comprising 97 women affected by SSc and 109 sex-matched healthy controls.106 IL13RA1 and IL13RA2 genes are located on the X chromosome and their products are receptors for IL-13, a cytokine playing an important role in normal tissue repair and Th2-mediated pathological fibrosis. In this study, IL13RA2 rs638376G allele frequency was higher in SSc patients and in the subgroup with dcSSc than in controls, while the IL13RA2 rs5946040G allele was more common only in patients with dcSSc with respect to controls.106
IL-1 receptor-associated kinase 1 (IRAK1, encoded by the IRAK1 gene) is a serine/threonine protein kinase able to regulate NF-κB activity and known to be involved in the TLR pathway.107 An association between some IRAK1 SNPs and dcSSc as well as anti-topo-I positive SSc patients was found in a discovery cohort including 849 SSc female patients and 625 sex-matched controls.107 The replication cohort confirmed a strong association between the IRAK1 rs1059702 TT risk genotype and both the dcSSc and anti-topo I-positive SSc subsets.107 However, in a subsequent study by Carmona et al., the IRAK1 SNP rs1059702 was found to be associated only with the susceptibility to SSc-related lung fibrosis.108 In the same study, the rs17345 SNP of MECP2, a gene encoding the protein MeCP2, which participates in epigenetic mechanisms binding to methylated DNA, was found to be associated with the dcSSc subtype in women of Caucasian ancestry.108 However, as IRAK1 and MECP2 genes are in moderate linkage disequilibrium, the real contribution of these genes to the susceptibility to SSc remains to be fully clarified.107,108
The X chromosome-located FOXP3 gene is primarily expressed in regulatory T cells (Treg) and encodes forkhead box P3 (FOXP3), a transcription factor that regulates T cell activation. Polymorphisms in this gene have been reported to alter FOXP3, causing Treg dysfunction and the consequent development of autoimmune diseases.109,110 A case control study evaluating the possible influence of FOXP3 SNPs rs3761548 and rs2280883 in SSc susceptibility was performed in a population of 228 Italian SSc patients (206 women and 22 men) and 239 healthy subjects. The authors found that the rs2280883 genetic variant was associated with the presence of anti-centromere antibodies and the lcSSc subset only in female patients.109 In a second study, the same authors investigated the association of FOXP3 rs2294020, inducible T cell co-stimulatory (ICOS) rs6726035 and ICOS ligand (ICOSL) rs378299 SNPs with either SSc susceptibility or the progression towards the disease in an Italian population. Although no significant associations were found between these SNPs and SSc susceptibility, the occurrence of FOXP3 rs2294020 in female patients was found to be correlated with a decreased time to progression from early to definite SSc.110 Even if these findings need to be replicated in a larger cohort and other populations, rs2294020 may be considered a disease-modifying gene variant rather than a disease-susceptibility SNP in SSc.110 In a recent study, Vreca et al.111 performed an analysis of both gene variants and mRNA expression levels of IRAK1 and its regulator miR-146a in SSc patients compared with healthy controls. Although neither IRAK1 rs3027898 nor miR-146a rs2910164 variants were directly associated with SSc susceptibility, miR-146a rs2910164 genotype and allele distribution correlated with the presence of lung fibrosis.111 As far as gene expression is concerned, both IRAK1 and miR-146a were found to be downregulated in PBMCs from SSc patients, with a strong negative correlation between IRAK1 and miR-146a expression levels when SSc patients were stratified by gender. Indeed, a significant increase in miR-146a expression in male SSc patients was accompanied by decreased IRAK1 mRNA levels, suggesting a direct regulation of IRAK1 gene expression by this specific miRNA.111
Finally, a case–control study involving a total of 461 individuals of Italian Caucasian origin (228 SSc patients and 233 healthy control subjects) was performed in order to evaluate a possible association between the rs4898 of the TIMP1 gene and SSc susceptibility and digital ulcers.112 Indeed, TIMP1, which is an inhibitor of MMPs, was found to be increased in SSc serum and excessively produced by dermal fibroblasts isolated from SSc patients. In that study, TIMP1 rs4898 did not show any association with SSc in male subjects, while women with C/C or T/C genotypes appeared to be less prone to the development of digital ulcers, suggesting that this SNP may play a protective role in the susceptibility to SSc in women, particularly to digital ulcer formation.112
X-linked epigenetic modifications in SSc
X-linked DNA methylation
DNA methylation is known to play a central role in X-chromosome inactivation in women, a mechanism of dosage compensation evolved to balance the levels of X-linked gene products between genders. Indeed, as women have two X chromosomes whereas men have only one, in the early stages of female embryogenesis one of the two X chromosomes is randomly silenced, leading to the formation of the heterochromatic Barr body. As a result, both men and women have only one active X chromosome (Xa).113 Several studies have shown that DNA methylation of the inactive X (Xi) is crucial for the maintenance of its inactive state. In particular, it has been demonstrated that the CpG islands have a tendency to be methylated on Xi and unmethylated on Xa.114 As the process of inactivation involves randomly one of the two X chromosomes and is irreversible during the lifetime of the cell and its offspring, each tissue of an adult woman is composed of cells expressing either the maternal or the paternal X chromosome, a phenomenon called female mosaicism.5 Not all the genes on Xi are silenced, as those located in pseudoautosomal regions, that is regions of the X chromosome homologous to the Y one, escape X-inactivation. Besides pseudoautosomal region genes, other genes may escape silencing.115
As several autoimmune diseases are known to have a striking female preponderance, numerous studies evaluated the involvement of different methylation profiles in X-linked genes in these pathologies.43,116,117 As far as SSc is concerned, a few studies hypothesized that the female gender bias of the disease could be explained by either the reactivation of genes that are typically silenced in the Xi chromosomes or the inhibition of genes normally expressed in the Xa chromosomes of female patients. In particular, the CD40L promoter region on Xi was found to be demethylated in CD4+ T cells from SSc female patients, resulting in female-specific CD40 ligand (CD40L/CD154/TRAP) overexpression.45 Of note, the interaction between CD40 ligand and its receptor CD40 plays a pivotal role in various autoimmune diseases including SSc.45 In fact, the combination of the CD40 ligand (on T cells) and CD40 (on B cells) acts as a signal to initiate immune responses, including B cell activation and differentiation, and the production of pathogenic autoantibodies.45 In another study, genome-wide methylation profiles analyzed in PBMCs from monozygotic twins discordant and concordant for SSc demonstrated consistent differences between the investigated twins only in genes located on the X chromosome.46 In particular, the biostatistical analysis identified 18 hypermethylated and 25 hypomethylated genes that included transcription factors (ARX, HSFX1, ZBED1, ZNF41) and surface antigens (IL1RAPL2, PGRMC1) involved in cell proliferation, apoptosis, inflammation and oxidative stress. Therefore, it has been suggested that the X chromosome genes, with different methylation profiles in monozygotic twin pairs, may be candidates for SSc susceptibility.46 Finally, it has been observed that a reduced expression of FOXP3, a key transcription factor that regulates Treg generation, was due to hypermethylation of the FOXP3 promoter region in CD4+ T cells of SSc patients.47 This hypermethylation status contributed to a reduced number of Tregs and was suggested to influence disease severity.47
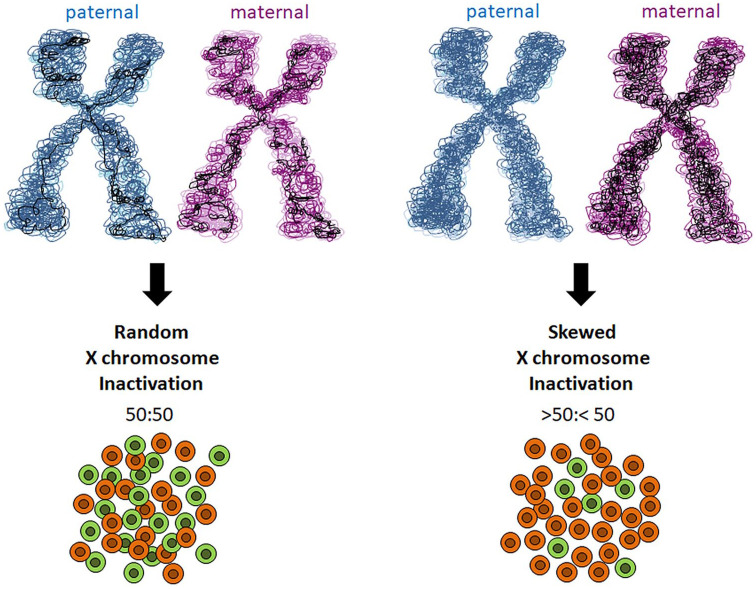
X-chromosome inactivation by DNA methylation is random, with an equal probability for the maternally or paternally derived X chromosome to be inactivated. This results in a mosaic distribution of cells, approximately half with the paternally derived Xi chromosome and half with the maternally derived Xi chromosome. However, when the inactivation of one X chromosome is favored over the other, leading to an uneven number of cells with each chromosome inactivated, skewed (non-random) X chromosome inactivation occurs (i.e. silencing of the same X chromosome in most cells of a specific tissue) (Figure 1). A skewed result is defined as one allele being inactivated at >75%, while extreme skewing represents an inactivation of >90%. Skewed X chromosome inactivation, although infrequent, may take place in normal women, and can be a primary event occurring in embryonic stem cells (primary non-random inactivation), or can be acquired with age. However, an extremely skewed pattern has the potential to unmask unfavorable X-linked alleles carrying mutations, thus leading to disease onset. Indeed, extreme X chromosome inactivation skewing (or loss of mosaicism) is often associated with a variety of diagnoses, including premature ovarian failure, recurrent spontaneous abortion, some cancers and several autoimmune disorders including SSc.118 In this context, skewed X chromosome inactivation has been observed in PBMCs of patients with SSc.43,44 Interestingly, a significant proportion of women with SSc have been found to manifest higher frequencies of PBMCs with X monosomy than healthy women over 50 years of age.119 Both extreme X chromosome inactivation skewing and X monosomy may contribute to haploinsufficienty of X-linked genes, a condition that in peripheral lymphocytes has been proposed to be responsible for immunosenescence and autoantibody production. However, in SSc, skewed X chromosome inactivation is not sufficient to explain the onset of the disease and should be considered as a possible cofactor in its pathogenic cascade. Indeed other factors, including genomic predisposition and environmental factors, are supposed to contribute to the disease phenotype.
Figure 1.
Schematic representation of skewed X chromosome inactivation. X chromosome inactivation occurs randomly, with an equal probability for the maternally or paternally derived X chromosome to be inactivated, resulting in a mosaic distribution of cells (50:50). Skewed X chromosome inactivation occurs when the inactivation of one X chromosome is favored over the other, leading to an uneven number of cells with each chromosome inactivated (>50:<50).
A summary of the principal X-linked DNA modifications described in SSc is shown in Table 1.
X-linked non-coding RNAs
Increasing evidence supports a dysregulated expression pattern of X-linked non-coding RNAs in autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis and Sjögren’s syndrome.120,121 As far as SSc is concerned, the lncRNA TSIX was found to be upregulated in dermal SSc fibroblasts, where it stabilizes type I collagen mRNA thus fostering collagen deposition. Moreover, TSIX levels were increased also in the serum of SSc patients, suggesting that this lncRNA might serve as a diagnostic disease biomarker (Table 2).72
Among non-coding RNAs, about 10% of miRNAs are located on X chromosome and may escape inactivation or be subjected to skewed X inactivation. As the X chromosome is known to contain the largest number of immune-related genes of the whole human genome, a dysregulation of X-linked miRNAs may influence immune response in women.122 As already discussed, many miRNAs have been found to be dysregulated both in the circulation and skin of SSc patients and to be primarily involved in the modulation of fibrosis.20–24 Among these, only few dysregulated X-linked miRNAs have been observed in SSc (Table 3).87,97,103 In particular, two studies reported elevated miR-92a levels in SSc serum87,103 and dermal fibroblasts,103 as well as in normal fibroblasts cultured in the presence of TGF-β.103 Furthermore, the induction of miR-92a overexpression in normal fibroblasts resulted in MMP-1 downregulation, suggesting that higher miR-92a levels might contribute to increased collagen deposition in SSc.103 However, a subsequent study evaluating plasma miRNA profiles reported lower levels of the miRNA-17~92 cluster in SSc patients with respect to healthy individuals.97 Finally, a significant downregulation in dermal SSc fibroblasts was reported for miR-542-3p. In particular, decreased miR-542-3p led to increased expression of survivin, a protein that belongs to the inhibitor of apoptosis (IAP) protein family. Thus, miR-542-3p downregulation might contribute to foster apoptosis resistance in SSc fibroblasts.104 Other miRNAs located on the X chromosome, such as miR-106a, miR-223, miR-221 and miR-503-5p, were found to be dysregulated in the circulation of SSc patients, although their functional significance in disease pathogenesis remains to be fully clarified.97,100
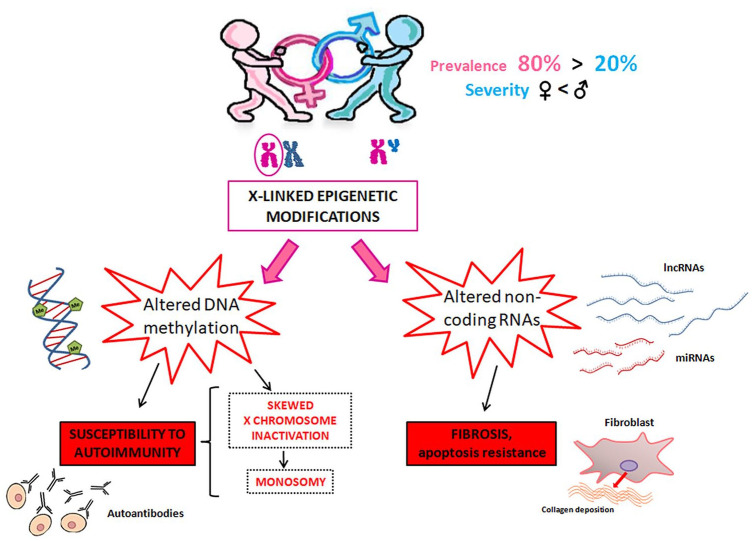
A schematic representation of the most important X-linked epigenetic mechanisms implicated in SSc pathogenesis is summarized in Figure 2.
Figure 2.
Schematic representation of the main X-linked epigenetic modifications in systemic sclerosis (SSc). SSc is characterized by a striking female predominance which is not reflected by a greater disease severity. Indeed, male SSc patients usually have a more severe prognosis compared to women. The causes of this gender imbalance have yet to be completely understood, but it appears that the X chromosome, which is known to contain the largest number of immune-related genes of the whole human genome, may play an important role in this sex-biased prevalence. X-linked epigenetic modifications reported to be altered and implicated in SSc pathogenesis are DNA methylation and non-coding RNAs. When altered, DNA methylation, which is known to play a central role in the X-chromosome inactivation in women, has the potential to reactivate genes typically silenced in the inactivated chromosome or inhibit genes normally expressed in the activated chromosome, thus fostering autoimmunity susceptibility and leading to SSc onset. In addition, a dysregulated expression pattern of X-linked non-coding RNAs (lncRNAs and miRNAs) has been shown to influence both the fibrotic and the apoptotic processes.
Conclusion
Over the past few years, growing evidence has suggested that SSc pathophysiology is the result of a complex interplay between genetic predisposition, environmental factors and epigenetics. Genetic variants thus far associated with SSc, although identified in studies performed on a large scale, are not sufficient to explain the multiple altered biological processes underlying this autoimmune disorder. Moreover, it has become clear that exposure to specific environmental agents participates in modulating both the epigenome (i.e. the set of chemical modifications of chromatin, alterations in chromatin constituents, and changes in the spatial chromatin organization that regulate gene expression) and the genetic disease predisposition. As outlined in this review, epigenetic modifications may have a pivotal contribution in the pathogenesis of SSc, mainly in the context of the disease-related fibrotic phenotype. Unlike genetic mutations, epigenetics consists of reversible changes amenable to modifications in dividing cells. Thus, a deeper exploration of the complex epigenetic mechanisms underlying disease onset and development are extremely important to favor the development of novel promising therapeutic strategies for the treatment of SSc. Furthermore, epigenetic marks are emerging as attractive biomarkers for disease diagnosis, monitoring and even prediction and assessment of the therapeutic response. It is also well known that SSc affects mainly the female gender while men are usually exposed to a more aggressive disease. Therefore, the contribution of the X chromosome to the development of SSc still remains to be unveiled. In fact, its role in the onset and rapid evolution of the disease in men is a matter of future investigation. Thus, further in-depth studies of X chromosome-linked epigenetic modifications are needed to provide novel insights into SSc female predominance and possibly to identify new promising gender-specific therapeutic targets.
On the basis of the most recent advances, there is realistic hope that integrating epigenetic data with genomic, transcriptomic, proteomic and metabolomic analyses may provide in the future a better picture of the functional implications in SSc, paving the right way for a better understanding of disease pathogenesis and the development of innovative therapeutic approaches.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Bianca Saveria Fioretto  https://orcid.org/0000-0001-9275-7593
https://orcid.org/0000-0001-9275-7593
Mirko Manetti  https://orcid.org/0000-0003-3956-8480
https://orcid.org/0000-0003-3956-8480
Contributor Information
Bianca Saveria Fioretto, Department of Experimental and Clinical Medicine, Division of Rheumatology, University of Florence, Viale Pieraccini 6, Florence, 50139, Italy.
Irene Rosa, Department of Experimental and Clinical Medicine, Division of Rheumatology, University of Florence and Scleroderma Unit, Azienda Ospedaliero-Universitaria Careggi (AOUC),Florence, Italy Department of Experimental and Clinical Medicine, Section of Anatomy and Histology, University of Florence, Florence, Italy.
Eloisa Romano, Department of Experimental and Clinical Medicine, Division of Rheumatology, University of Florence and Scleroderma Unit, Azienda Ospedaliero-Universitaria Careggi (AOUC), Florence, Italy.
Yukai Wang, Department of Rheumatology and Immunology, Shantou Central Hospital, Shantou, China.
Serena Guiducci, Department of Experimental and Clinical Medicine, Division of Rheumatology, University of Florence and Scleroderma Unit, Azienda Ospedaliero-Universitaria Careggi (AOUC), Florence, Italy.
Guohong Zhang, Department of Pathology, Shantou University Medical College, Shantou, China.
Mirko Manetti, Department of Experimental and Clinical Medicine, Section of Anatomy and Histology, University of Florence, Florence, Italy.
Marco Matucci-Cerinic, Department of Experimental and Clinical Medicine, Division of Rheumatology, University of Florence and Scleroderma Unit, Azienda Ospedaliero-Universitaria Careggi (AOUC), Florence, Italy.
References
- 1. Varga J, Trojanowska M, Kuwana M. Pathogenesis of systemic sclerosis: recent insights of molecular and cellular mechanisms and therapeutic opportunities. J Scleroderma Relat Disord 2017; 2: 137–152. [Google Scholar]
- 2. Aslani S, Sobhani S, Gharibdoost F, et al. Epigenetics and pathogenesis of systemic sclerosis; the ins and outs. Hum Immunol 2018; 79: 178–187. [DOI] [PubMed] [Google Scholar]
- 3. Salazar G, Mayes MD. Genetics, epigenetics, and genomics of systemic sclerosis. Rheum Dis Clin North Am 2015; 41: 345–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peoples C, Medsger TA, Jr, Lucas M, et al. Gender differences in systemic sclerosis: relationship to clinical features, serologic status and outcomes. J Scleroderma Relat Disord 2016; 1: 177–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Invernizzi P, Pasini S, Selmi C, et al. Female predominance and X chromosome defects in autoimmune diseases. J Autoimmun 2009; 33: 12–16. [DOI] [PubMed] [Google Scholar]
- 6. Nietert PJ, Mitchell HC, Bolster MB, et al. Racial variation in clinical and immunological manifestations of systemic sclerosis. J Rheumatol 2006; 33: 263–268. [PubMed] [Google Scholar]
- 7. Steen VD, Oddis CV, Conte CG, et al. Incidence of systemic sclerosis in Allegheny County, Pennsylvania. A twenty-year study of hospital-diagnosed cases, 1963–1982. Arthritis Rheum 1997; 40: 441–445. [DOI] [PubMed] [Google Scholar]
- 8. Hoffmann-Vold AM, Molberg Ø, Midtvedt Ø, et al. Survival and causes of death in an unselected and complete cohort of Norwegian patients with systemic sclerosis. J Rheumatol 2013; 40: 1127–1133. [DOI] [PubMed] [Google Scholar]
- 9. Makino T, Jinnin M. Genetic and epigenetic abnormalities in systemic sclerosis. J Dermatol 2016; 43: 10–18. [DOI] [PubMed] [Google Scholar]
- 10. Korman BD, Criswell LA. Recent advances in the genetics of systemic sclerosis: toward biological and clinical significance. Curr Rheumatol Rep 2015; 17: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feghali-Bostwick C, Medsger TA, Jr, Wright TM. Analysis of systemic sclerosis in twins reveals low concordance for disease and high concordance for the presence of antinuclear antibodies. Arthritis Rheum 2003; 48: 1956–1963. [DOI] [PubMed] [Google Scholar]
- 12. Luo Y, Wang Y, Wang Q, et al. Systemic sclerosis: genetics and epigenetics. J Autoimmun 2013; 41: 161–167. [DOI] [PubMed] [Google Scholar]
- 13. Romano E, Manetti M, Guiducci S, et al. The genetics of systemic sclerosis: an update. Clin Exp Rheumatol 2011; 29: S75–S86. [PubMed] [Google Scholar]
- 14. Murdaca G, Contatore M, Gulli R, et al. Genetic factors and systemic sclerosis. Autoimmun Rev 2016; 15: 427–432. [DOI] [PubMed] [Google Scholar]
- 15. Gao L, Emond MJ, Louie T, et al. Identification of rare variants in ATP8B4 as a risk factor for systemic sclerosis by whole-exome sequencing. Arthritis Rheumatol 2016; 68: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gourh P, Remmers EF, Boyden SE, et al. Brief report: whole-exome sequencing to identify rare variants and gene networks that increase susceptibility to scleroderma in African Americans. Arthritis Rheumatol 2018; 70: 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mak AC, Tang PL, Cleveland C, et al. Brief report: whole-exome sequencing for identification of potential causal variants for diffuse cutaneous systemic sclerosis. Arthritis Rheumatol 2016; 68: 2257–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramos PS. Epigenetics of scleroderma: integrating genetic, ethnic, age, and environmental effects. J Scleroderma Relal Disord 2019; 4: 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiang Z, Yang Y, Chang C, et al. The epigenetic mechanism for discordance of autoimmunity in monozygotic twins. Autoimmunity 2017; 83: 43–50. [DOI] [PubMed] [Google Scholar]
- 20. Henderson J, Distler J, O’Reilly S. The role of epigenetic modifications in systemic sclerosis: a druggable target. Trends Mol Med 2019; 25: 395–411. [DOI] [PubMed] [Google Scholar]
- 21. Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017; 18: 517–534. [DOI] [PubMed] [Google Scholar]
- 22. Meda F, Folci M, Baccarelli A, et al. The epigenetics of autoimmunity. Cell Mol Immunol 2011; 8: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walczyk M, Paradowska-Gorycka A, Olesinska M. Epigenetics: the future direction in systemic sclerosis. Scand J Immunol 2017; 86: 427–435. [DOI] [PubMed] [Google Scholar]
- 24. Altorok N, Almeshal N, Wang Y, et al. Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatology (Oxford) 2015; 54: 1759–1770. [DOI] [PubMed] [Google Scholar]
- 25. Altorok N, Tsou PS, Coit P, et al. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis 2015; 74: 1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum 2006; 54: 2271–2279. [DOI] [PubMed] [Google Scholar]
- 27. Asano Y, Bujor AM, Trojanowska M. The impact of Fli1 deficiency on the pathogenesis of systemic sclerosis. J Dermatol Sci 2010; 59: 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manetti M. Fli1 deficiency and beyond: a unique pathway linking peripheral vasculopathy and dermal fibrosis in systemic sclerosis. Exp Dermatol 2015; 24: 256–257. [DOI] [PubMed] [Google Scholar]
- 29. Noda S, Asano Y, Nishimura S, et al. Simultaneous downregulation of KLF5 and Fli1 is a key feature underlying systemic sclerosis. Nat Commun 2014; 5: 5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei J, Fang F, Lam AP, et al. Wnt/β-catenin signaling is hyperactivated in systemic sclerosis and induces Smad-dependent fibrotic responses in mesenchymal cells. Arthritis Rheum 2012; 64: 2734–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dees C, Schlottmann I, Funke R, et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis 2014; 73: 1232–1239. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Pötter S, Chen CW, et al. Poly(ADP-ribose)polymerase-1 regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis 2018; 77: 744–751. [DOI] [PubMed] [Google Scholar]
- 33. Hattori M, Yokoyama Y, Hattori T, et al. Global DNA hypomethylation and hypoxia-induced expression of the ten eleven translocation (TET) family, TET1, in scleroderma fibroblasts. Exp Dermatol 2015; 24: 841–846. [DOI] [PubMed] [Google Scholar]
- 34. Lei W, Luo Y, Lei W, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol 2009; 38: 369–374. [DOI] [PubMed] [Google Scholar]
- 35. Ding W, Pu W, Wang L, et al. Genome-wide DNA methylation analysis in systemic sclerosis reveals hypomethylation of IFN-associated genes in CD4(+) and CD8(+) T cells. J Invest Dermatol 2018; 138: 1069–1077. [DOI] [PubMed] [Google Scholar]
- 36. Chen S, Pu W, Guo S, et al. Genome-wide DNA methylation profiles reveal common epigenetic patterns of interferon-related genes in multiple autoimmune diseases. Front Genet 2019; 10: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Angiolilli C, Marut W, Van der Kroef M, et al. New insights into the genetics and epigenetics of systemic sclerosis. Nat Rev Rheumatol 2018; 14: 657–673. [DOI] [PubMed] [Google Scholar]
- 38. Rezaei R, Mahmoudi M, Gharibdoost F, et al. IRF7 gene expression profile and methylation of its promoter region in patients with systemic sclerosis. Int J Rheum Dis 2017; 20: 1551–1561. [DOI] [PubMed] [Google Scholar]
- 39. Wang Y, Shu Y, Xiao Y, et al. Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenet 2014; 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Affandi AJ, Carvalheiro T, Ottria A, et al. Low RUNX3 expression alters dendritic cell function in patients with systemic sclerosis and contributes to enhanced fibrosis. Ann Rheum Dis 2019; 78: 1249–1259. [DOI] [PubMed] [Google Scholar]
- 41. Dashti N, Mahmoudi M, Gharibdoost F, et al. Evaluation of ITGB2 (CD18) and SELL (CD62L) genes expression and methylation of ITGB2 promoter region in patients with systemic sclerosis. Rheumatol Int 2018; 38: 489–498. [DOI] [PubMed] [Google Scholar]
- 42. Wang Y, Kahaleh B. Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med 2013; 17: 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kanaan SB, Onat OE, Balandraud N, et al. Evaluation of X chromosome inactivation with respect to HLA genetic susceptibility in rheumatoid arthritis and systemic sclerosis. PLoS One 2016; 11: e0158550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ozbalkan Z, Bagişlar S, Kiraz S, et al. Skewed X chromosome inactivation in blood cells of women with scleroderma. Arthritis Rheum 2005; 52: 1564–1570. [DOI] [PubMed] [Google Scholar]
- 45. Lian X, Xiao R, Hu X, et al. DNA demethylation of CD40L in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum 2012; 64: 2338–2345. [DOI] [PubMed] [Google Scholar]
- 46. Selmi C, Feghali-Bostwick CA, Lleo A, et al. X chromosome gene methylation in peripheral lymphocytes from monozygotic twins discordant for scleroderma. Clin Exp Immunol 2012; 169: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang YY, Wang Q, Sun XH, et al. DNA hypermethylation of the forkhead box protein 3 (FOXP3) promoter in CD4+ T cells of patients with systemic sclerosis. Br J Dermatol 2014; 171: 39–47. [DOI] [PubMed] [Google Scholar]
- 48. Wang Y, Yang Y, Luo Y, et al. Aberrant histone modification in peripheral blood B cells from patients with systemic sclerosis. Clin Immunol 2013; 149: 46–54. [DOI] [PubMed] [Google Scholar]
- 49. Van der Kroef M, Castellucci M, Mokry M, et al. Histone modifications underlie monocyte dysregulation in patients with systemic sclerosis, underlining the treatment potential of epigenetic targeting. Ann Rheum Dis 2019; 78: 529–538. [DOI] [PubMed] [Google Scholar]
- 50. Tsou PS, Wren JD, Amin MA, et al. Histone deacetylase 5 is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis Rheumatol 2016; 68: 2975–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chu H, Jiang S, Liu Q, et al. Sirtuin 1 protects against systemic sclerosis-related pulmonary fibrosis by decreasing proinflammatory and profibrotic processes. Am J Respir Cell Mol Biol 2018; 58: 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wyman AE, Noor Z, Fishelevich R, et al. Sirtuin 7 is decreased in pulmonary fibrosis and regulates the fibrotic phenotype of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol 2017; 312: L945–L958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sosulski ML, Gongora R, Feghali-Bostwick C, et al. Sirtuin 3 deregulation promotes pulmonary fibrosis. J Gerontol A Biol Sci Med Sci 2017; 72: 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ghosh AK, Bhattacharyya S, Lafyatis R, et al. p300 is elevated in systemic sclerosis and its expression is positively regulated by TGF-β: epigenetic feed-forward amplification of fibrosis. J Invest Dermatol 2013; 133: 1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum 2006; 54: 2271–2279. [DOI] [PubMed] [Google Scholar]
- 56. Xiao X, Senavirathna LK, Gou X, et al. EZH2 enhances the differentiation of fibroblasts into myofibroblasts in idiopathic pulmonary fibrosis. Physiol Rep 2016; 4: e12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsou PS, Campbell P, Amin MA, et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc Natl Acad Sci USA 2019; 116: 3695–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krämer M, Dees C, Huang J, et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis 2013; 72: 614–620. [DOI] [PubMed] [Google Scholar]
- 59. Wang Q, Xiao Y, Shi Y, et al. Overexpression of JMJD3 may contribute to demethylation of H3K27me3 in CD4+ T cells from patients with systemic sclerosis. Clin Immunol 2015; 161: 396–399. [DOI] [PubMed] [Google Scholar]
- 60. Bergmann C, Brandt A, Merlevede B, et al. The histone demethylase Jumonji domain-containing protein 3 (JMJD3) regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis 2018; 77: 150–158. [DOI] [PubMed] [Google Scholar]
- 61. Deng Q, Luo Y, Chang C, et al. The emerging epigenetic role of CD8+ T cells in autoimmune diseases: a systematic review. Front Immunol 2019; 10: 856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta 2014; 1839: 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huber LC, Distler JH, Moritz F, et al. Trichostatin A prevents the accumulation of extracellular matrix in a mouse model of bleomycin-induced skin fibrosis. Arthritis Rheum 2007; 56: 2755–2764. [DOI] [PubMed] [Google Scholar]
- 64. Akamata K, Wei J, Bhattacharyya M, et al. SIRT3 is attenuated in systemic sclerosis skin and lungs, and its pharmacologic activation mitigates organ fibrosis. Oncotarget 2016; 7: 69321–69336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shin JY, Beckett JD, Bagirzadeh R, et al. Epigenetic activation and memory at a TGFB2 enhancer in systemic sclerosis. Sci Transl Med 2019; 11: eaaw0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mazzone R, Zwergel C, Artico M, et al. The emerging role of epigenetics in human autoimmune disorders. Clin Epigenetics 2019; 11: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Messemaker TC, Chadli L, Cai G, et al. Antisense long non-coding RNAs are deregulated in skin tissue of patients with systemic sclerosis. J Invest Dermatol 2018; 138: 826–835. [DOI] [PubMed] [Google Scholar]
- 68. Takata M, Pachera E, Frank-Bertoncelj M, et al. OTUD6B-AS1 might be a novel regulator of apoptosis in systemic sclerosis. Front Immunol 2019; 10: 1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dolcino M, Tinazzi E, Puccetti A, et al. In systemic sclerosis, a unique long non coding RNA regulates genes and pathways involved in the three main features of the disease (vasculopathy, fibrosis and autoimmunity) and in carcinogenesis. J Clin Med 2019; 8: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mariotti B, Servaas NH, Rossato M, et al. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front Immunol 2019; 10: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wasson CW, Abignano G, Hermes H, et al. Long non-coding RNA HOTAIR drives EZH2-dependent myofibroblast activation in systemic sclerosis through miRNA 34a-dependent activation of NOTCH. Ann Rheum Dis 2020; 79: 507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Z, Jinnin M, Nakamura K, et al. Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp Dermatol 2016; 25: 131–136. [DOI] [PubMed] [Google Scholar]
- 73. Henry TW, Mendoza FA, Jimenez SA. Role of microRNA in the pathogenesis of systemic sclerosis tissue fibrosis and vasculopathy. Autoimmun Rev 2019; 18: 102396. [DOI] [PubMed] [Google Scholar]
- 74. Maurer B, Stanczyk J, Jüngel A, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 2010; 62: 1733–1743. [DOI] [PubMed] [Google Scholar]
- 75. Jafarinejad-Farsangi S, Gharibdoost F, Farazmand A, et al. MicroRNA-21 and microRNA-29a modulate the expression of collagen in dermal fibroblasts of patients with systemic sclerosis. Autoimmunity 2019; 52: 108–116. [DOI] [PubMed] [Google Scholar]
- 76. Ciechomska M, O’Reilly S, Suwara M, et al. MiR-29a reduces TIMP-1 production by dermal fibroblasts via targeting TGF-β activated kinase 1 binding protein 1, implications for systemic sclerosis. PLoS One 2014; 9: e115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Makino T, Jinnin M, Etoh M, et al. Down-regulation of microRNA-196a in the sera and involved skin of localized scleroderma patients. Eur J Dermatol 2014; 24: 470–476. [DOI] [PubMed] [Google Scholar]
- 78. Honda N, Jinnin M, Kajihara I, et al. TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol 2012; 188: 3323–3331. [DOI] [PubMed] [Google Scholar]
- 79. Izumiya Y, Jinnin M, Kimura Y, et al. Expression of Let-7 family microRNAs in skin correlates negatively with severity of pulmonary hypertension in patients with systemic scleroderma. J Cardiol Heart Vasc 2015; 8: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Makino K, Jinnin M, Hirano A, et al. The downregulation of miR-let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol 2013; 190: 3905–3915. [DOI] [PubMed] [Google Scholar]
- 81. O’Reilly S, Ciechomska M, Fullard N, et al. IL-13 mediates collagen deposition via STAT6 and microRNA-135b: a role for epigenetics. Sci Rep 2016; 6: 25066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Henderson J, Brown M, Horsburgh S, et al. Methyl cap binding protein 2: a key epigenetic protein in systemic sclerosis. Rheumatology 2019; 58: 527–535. [DOI] [PubMed] [Google Scholar]
- 83. He Y, Tsou PS, Khanna D, et al. Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann Rheum Dis 2018; 77: 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhou B, Zhu H, Luo H, et al. MicroRNA-202-3p regulates scleroderma fibrosis by targeting matrix metalloproteinase 1. Biomed Pharmacother 2017; 87: 412–418. [DOI] [PubMed] [Google Scholar]
- 85. Yan Q, Chen J, Li W, et al. Targeting miR-155 to treat experimental scleroderma. Sci Rep 2016; 6: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dolcino M, Pelosi A, Fiore PF, et al. Gene profiling in patients with systemic sclerosis reveals the presence of oncogenic gene signatures. Front Immunol 2018; 9: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Artlett CM, Sassi-Gaha S, Hope JL, et al. Mir-155 is overexpressed in systemic sclerosis fibroblasts and is required for NLRP3 inflammasome-mediated collagen synthesis during fibrosis. Arthritis Res Ther 2017; 19: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Honda N, Jinnin M, Kira-Etoh T, et al. MiR-150 down-regulation contributes to the constitutive type I collagen overexpression in scleroderma dermal fibroblasts via the induction of integrin β3. Am J Pathol 2013; 182: 206–216. [DOI] [PubMed] [Google Scholar]
- 89. Jafarinejad-Farsangi S, Farazmand A, Gharibdoost F, et al. Inhibition of microRNA-21 induces apoptosis in dermal fibroblasts of patients with systemic sclerosis. Int J Dermatol 2016; 55: 1259–1267. [DOI] [PubMed] [Google Scholar]
- 90. Zhu H, Li Y, Qu S, et al. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol 2012; 32: 514–522. [DOI] [PubMed] [Google Scholar]
- 91. Zhu H, Luo H, Li Y, et al. MicroRNA-21 in scleroderma fibrosis and its function in TGF-β-regulated fibrosis-related genes expression. J Clin Immunol 2013; 33: 1100–1109. [DOI] [PubMed] [Google Scholar]
- 92. Zhou B, Zuo XX, Li YS, et al. Integration of microRNA and mRNA expression profiles in the skin of systemic sclerosis patients. Sci Rep 2017; 7: 42899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Luo H, Zhu H, Zhou B, et al. MicroRNA-130b regulates scleroderma fibrosis by targeting peroxisome proliferator-activated receptor γ. Mod Rheumatol 2015; 25: 595–602. [DOI] [PubMed] [Google Scholar]
- 94. Iwamoto N, Vettori S, Maurer B, et al. Downregulation of miR-193b in systemic sclerosis regulates the proliferative vasculopathy by urokinase-type plasminogen activator expression. Ann Rheum Dis 2016; 75: 303–310. [DOI] [PubMed] [Google Scholar]
- 95. Rossato M, Affandi AJ, Thordardottir S, et al. Association of microRNA-618 expression with altered frequency and activation of plasmacytoid dendritic cells in patients with systemic sclerosis. Arthritis Rheumatol 2017; 69: 1891–1902. [DOI] [PubMed] [Google Scholar]
- 96. Ciechomska M, Zarecki P, Merdas M, et al. The role of microRNA-5196 in the pathogenesis of systemic sclerosis. Eur J Clin Invest 2017; 47: 555–564. [DOI] [PubMed] [Google Scholar]
- 97. Steen SO, Iversen LV, Carlsen AL, et al. The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J Rheumatol 2015; 42: 214–221. [DOI] [PubMed] [Google Scholar]
- 98. Wuttge DM, Carlsen AL, Teku G, et al. Specific autoantibody profiles and disease subgroups correlate with circulating micro-RNA in systemic sclerosis. Rheumatology (Oxford) 2015; 54: 2100–2107. [DOI] [PubMed] [Google Scholar]
- 99. Chouri E, Servaas NH, Bekker CPJ, et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J Autoimmun 2018; 89: 162–170. [DOI] [PubMed] [Google Scholar]
- 100. Wermuth PJ, Piera-Velazquez S, Jimenez SA. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin Exp Rheumatol 2017; 106: 21–30. [PMC free article] [PubMed] [Google Scholar]
- 101. He Y, Liu H, Wang S, et al. In silico detection and characterization of microRNAs and their target genes in microRNA microarray datasets from patients with systemic sclerosis-interstitial lung disease. DNA Cell Biol 2019; 38: 933–944. [DOI] [PubMed] [Google Scholar]
- 102. Rusek M, Michalska-Jakubus M, Kowal M, et al. A novel miRNA-4484 is up-regulated on microarray and associated with increased MMP-21 expression in serum of systemic sclerosis patients. Sci Rep 2019; 9: 14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sing T, Jinnin M, Yamane K, et al. MicroRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford) 2012; 51: 1550–1556. [DOI] [PubMed] [Google Scholar]
- 104. Vahidi Manesh P, Farazmand A, Gharibdoost F, et al. Downregulation of miR-542-3p contributes to apoptosis resistance in dermal fibroblasts from systemic sclerosis patients via survivin overexpression. Iran J Allergy Asthma Immunol 2019; 18: 173–181. [PubMed] [Google Scholar]
- 105. D’Amico F, Skarmoutsou E, Mazzarino MC. The sex bias in systemic sclerosis: on the possible mechanisms underlying the female disease preponderance. Clin Rev Allergy Immunol 2014; 47: 334–343. [DOI] [PubMed] [Google Scholar]
- 106. Granel B, Allanore Y, Chevillard C, et al. IL13RA2 gene polymorphisms are associated with systemic sclerosis. J Rheumatol 2006; 33: 2015–2019. [PubMed] [Google Scholar]
- 107. Dieudé P, Bouaziz M, Guedj M, et al. Evidence of the contribution of the X chromosome to systemic sclerosis susceptibility: association with the functional IRAK1 196Phe/532Ser haplotype. Arthritis Rheum 2011; 63: 3979–3987. [DOI] [PubMed] [Google Scholar]
- 108. Carmona FD, Cénit MC, Diaz-Gallo LM, et al. New insight on the Xq28 association with systemic sclerosis. Ann Rheum Dis 2013; 72: 2032–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. D’Amico F, Skarmoutsou E, Marchini M, et al. Genetic polymorphisms of FOXP3 in Italian patients with systemic sclerosis. Immunol Lett 2013; 152: 109–113. [DOI] [PubMed] [Google Scholar]
- 110. D’Amico F, Fiorito G, Skarmoutsou E, et al. FOXP3, ICOS and ICOSL gene polymorphisms in systemic sclerosis: FOXP3 rs2294020 is associated with disease progression in a female Italian population. Immunobiology 2018; 223: 112–117. [DOI] [PubMed] [Google Scholar]
- 111. Vreca M, Andjelkovic M, Tosic N, et al. Impact of alterations in X-linked IRAK1gene and miR-146a on susceptibility and clinical manifestations in patients with systemic sclerosis. Immunol Lett 2018; 204: 1–8. [DOI] [PubMed] [Google Scholar]
- 112. Skarmoutsou E, D’Amico F, Marchini M, et al. Association of TIMP-1 +372 SNP with digital ulcer manifestation in female systemic sclerosis patients. Hum Immunol 2012; 73, 950–953. [DOI] [PubMed] [Google Scholar]
- 113. Lambert NC. Nonendocrine mechanisms of sex bias in rheumatic diseases. Nat Rev Rheumatol 2019; 15: 673–686. [DOI] [PubMed] [Google Scholar]
- 114. Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science 2007; 315: 1141–1143. [DOI] [PubMed] [Google Scholar]
- 115. Selmi C. The X in sex: how autoimmune diseases revolve around sex chromosomes. Best Pract Res Clin Rheumatol 2008; 22: 913–922. [DOI] [PubMed] [Google Scholar]
- 116. Mougeot JL, Noll BD, Bahrani Mougeot FK. Sjögren’s syndrome X-chromosome dose effect: an epigenetic perspective. Oral Dis 2019; 25: 372–384. [DOI] [PubMed] [Google Scholar]
- 117. Long H, Yin H, Wang L, et al. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J Autoimmun 2016; 74: 118–138. [DOI] [PubMed] [Google Scholar]
- 118. Minks J, Robinson WP, Brown CJ. A skewed view of X chromosome inactivation. J Clin Invest 2008; 118: 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Svyryd Y, Hernández-Molina G, Vargas F, et al. X chromosome monosomy in primary and overlapping autoimmune diseases. Autoimmun Rev 2012; 11: 301–304. [DOI] [PubMed] [Google Scholar]
- 120. Long H, Wang X, Chen Y, et al. Dysregulation of microRNAs in autoimmune diseases: pathogenesis, biomarkers and potential therapeutic targets. Cancer Lett 2018; 428: 90–103. [DOI] [PubMed] [Google Scholar]
- 121. Soltanzadeh-Yamchi M, Shahbazi M, Aslani S, et al. MicroRNA signature of regulatory T cells in health and autoimmunity. Biomed Pharmacother 2018; 100: 316–323. [DOI] [PubMed] [Google Scholar]
- 122. Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays 2011; 33: 791–802. [DOI] [PubMed] [Google Scholar]