Abstract
The masses of particles in a bovine milk extracellular vesicle (EV) preparation enriched for exosomes were directly determined for the first time by charge detection mass spectrometry (CDMS). In CDMS, both the mass-to-charge ratio (m/z) and z are determined simultaneously for individual particles, enabling mass determinations for particles that are far beyond the mass limit (~1.0 MDa) of conventional mass spectrometry (MS). Particle masses and charges span a wide range from m ~ 2 to ~90 MDa and z ~50 to ~1300 e (elementary charges) and are highly dependent upon the conditions used to extract and isolate the EVs. EV particles span a continuum of masses, reflecting the highly heterogeneous nature of these samples. However, evidence for unique populations of particles is obtained from correlation of the charges and masses. An analysis that uses a two-dimensional Gaussian model, provides evidence for six families of particles, four of which having masses in the range expected for exosomes. Complementary proteomics measurements and electron microscopy (EM) imaging are used to further characterize the EVs and confirm that these samples have been enriched in exosomes. The ability to characterize such extremely heterogeneous mixtures of large particles with rapid, sensitive and high-resolution MS techniques is critical to ongoing analytical efforts to separate and purify exosomes and exosome subpopulations. Direct measurement of each particle’s mass and charge is a new means of characterizing the physical and chemical properties of exosomes and other EVs.
Graphical Abstract
Introduction
Extracellular vesicles (EVs) are heterogeneous mixtures of membrane-encapsulated particles such as exosomes, apoptotic bodies, and other microvesicles that are secreted by eukaryotic cells.1–3 Currently exosomes, which are differentiated from other types of EVs based on size, biogenesis, and the type of molecular cargo they encapsulate,4 are attracting considerable attention. In addition to lipids, proteins, and other small molecules, some exosomes carry genetic material (e.g., miRNA, mRNA, and DNA5) and are associated with functional and phenotypical changes of other cells6 - within and between organisms.7, 8 These particles play central roles in cell-to-cell communication and are implicated in numerous pathological processes including inflammation,6 immunity,9, 10 tumor progression11–14 and neurodegeneration.15, 16 Given their ability to target and alter specific cells, there is growing interest in developing exosomes as therapeutics17–19 and a need to understand the structures, molecular compositions, and biological functions of these particles.
Several existing bioanalytical strategies for purifying and characterizing exosomes have allowed for fundamental progress to be made. Mixtures of EVs can be enriched for exosomes by techniques such as ultracentrifugation,20–22 size-exclusion chromatography,23,24 ultrafiltration,25 and field flow fractionation.26–28 But, these processes require large amounts of material that are often difficult to obtain and many different types of particles have similar sizes and densities (see Table 1). It is likely that unique subfractions within enriched samples exist, particularly in complex biological matrices such as blood, urine or milk. But, such subfractions remain difficult to characterize and isolate with existing analytical methods. Flow cytometry techniques are especially promising – allowing particles with targeted surface proteins to be isolated.29, 30 Electron microscopy (EM)31 and nanoparticle tracking techniques32 provide information about particle size distributions.
Table 1.
Characteristics of biological extracellular vesicles and other particlesa
| particle type | diameter (nm) | density (g·mℓ−1) | mass (MDa) |
|---|---|---|---|
| High density lipoprotein b | 5 to 15 | 1.06 to 1.21 | ~0.05 to 0.6 |
| Low density lipoprotein b | 18 to 28 | 1.03- 1.063 | ~2 to 7 |
| Very low density lipoprotein b | 30 to 80 | ~1.006 | ~8 to 80 |
| Exomeres c | ~30 to 50 | 0.93 to 1.06 | ~8 to 40 |
| Exosomes d | 30 to 120 | 1.12 to 1.21 | ~10 to 1200 |
| Microvesicles d | 50 to 1000 | 1.16 | ~50 to 4 × 106 |
| Virions d | ~30 to ~140 | 1.16 to 1.18 | ~10 to 1000 |
| Casein micelle e | 50 to 500 | 1.06 | ~1 to 1000 |
Diameters, densities, and masses are derived from literature data as indicated in the text unless otherwise noted here. In cases where we derive a physical parameter from others we have assumed that particles are spherical.
Particle diameters and densities are taken from reference 78.
The diameters and densities of microvesicles, exosomes and virions are taken from reference 76, 77.
Values for casein micelle diameters and densities are taken from reference 48.
The molecular components of lysed and digested particles can be analyzed by proteomics, glycomics, and lipidomics approaches. However, even these advanced methods provide yield only a limited understanding and there is a need for new technologies that can complement existing approaches.
In the work described below, we present the first mass spectrum of intact exosome particles. These data were recorded using charge detection mass spectrometry (CDMS).33–37 In CDMS, individual particles are reflected back and forth through an electrostatic ion trap where they pass through a sensitive charge detector. Each time a trapped particle enters and exits the detector, its charge (z) and mass-to-charge (m/z) ratio is measured. The combined measurements make it possible to directly determine the masses of large species that are far beyond the ~1 MDa mass limit of conventional mass spectrometers.38–42 In the present experiments, particles with masses from ~0.5 MDa to as high as ~90 MDa have been detected – a mass range that includes the region expected for small exosomes. Complementary proteomics and electron microscopy experiments were used to corroborate the detection of exosomes by CDMS. From the proteomics analysis we find that 71% of the proteins found in the exosome-enriched samples have been previously characterized as exosomal. Additionally, the sizes of these particles measured by electron microscopy are in range expected for exosomes. Overall, these results corroborate the detection of intact exosomal particles by mass spectrometry for the first time.
We have chosen EVs from bovine milk to demonstrate detection of exosomes by mass spectrometry because these particles are important biologically7, 43. Exosomes from bovine milk can induce physiological responses within species (when transferred from mother to calf) and as well as in humans.44 Thus, milk exosomes are potentially relevant in new types of therapeutics.45–47 Additionally, bovine milk is readily available - making it a natural starting point for analysis by CDMS. This source provides a unique mixture model for developing better physical separation and fractionation tools. While availability is an advantage, we note that raw milk is an extremely complex body fluid, containing abundant non-EV proteins, milk fat, sugars, and other components.48 Thus, characterizing exosomes from this material presents a significant analytical challenge. Exosomes from other sources are also of great interest.46, 49, 50
Experimental
All of the details associated with sample preparation and the experimental methods used for characterizing exosome samples (including CDMS, electron microscopy, proteomic confirmation of exosome enrichment, and statistical analysis of CDMS data) are provided in the Supplemental information.
Results and Discussion
Example CDMS dataset.
As described above, CDMS determines m/z and z for individual ions and m is obtained by multiplying these two values. A mass vs charge spectrum is obtained by accumulating this information from many independent measurements of single particles. Figure 1 shows the mass vs charge spectrum for 3,586 individual particles recorded for one of our bovine EV samples. The plot shows that particle masses and charges are observed as an extremely broad distribution that spans a wide range of masses and charges - from m ~ 2 to 70 MDa, and z ~ 50 to 920 e (elementary charges). As a first check to see if these values are reasonable for EVs from milk, we assume particles are spherical and estimate the expected masses of different classes of EVs from reported densities and particle diameters. A summary of these values from the literature, along with their estimated masses, is provided in Table 1. We can see from this comparison that our measured mass range is consistent with several types of species that may be present in these samples, including: relatively small low-density lipoproteins which span a range of masses, from (m ~ 4 to 120 MDa); small viruses, such as HBV capsid (m ~ 2 to 5 MDa)35, 42, 59; small exomeres (m ~4 to 120 MDa); and small exosomes (m ~10 to 1200 MDa).
Figure 1.
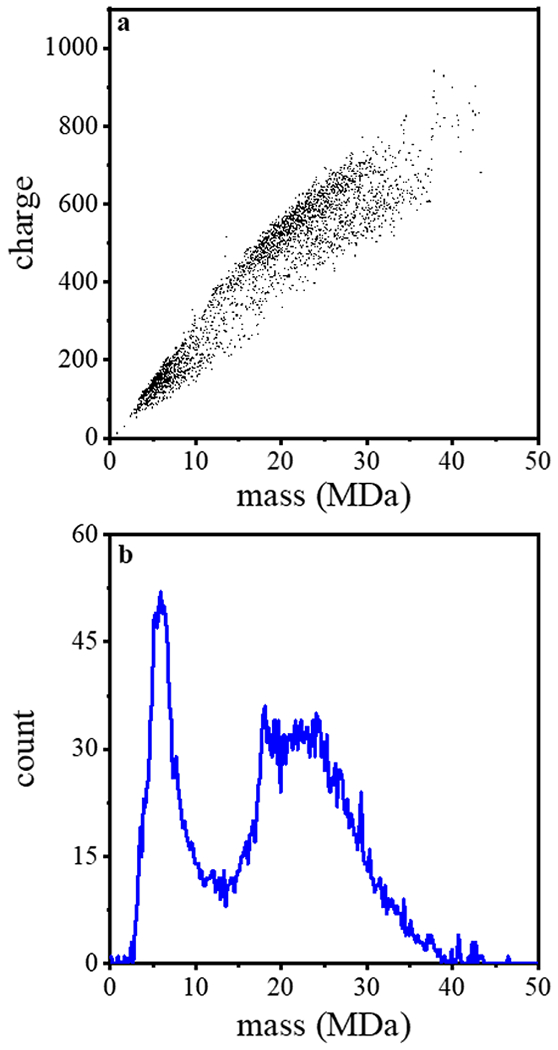
(top) The first CDMS measurement of mass verses charge for particles from an exosome-enriched bovine milk sample. In total, 3586 ions were analyzed in this measurement. (bottom) Mass spectrum generated upon integrating the ion signal across the charge dimension using 0.2 MDa bins.
While the overall mass vs charge spectrum shows only broad features, it does appear that some particles display similar characteristics and fall into mass and charge families. From visual inspection of the two-dimensional plot in Figure 1, we can see that there are at least three types of populations: small particles, having m ~2 to 10 MDa and z ~ 50 to 250 e; and two families of larger particles – one having m ~ 10 to 45 MDa, z ~ 300 to 920 e, and a less abundant second family that spans a similar mass range but having fewer charges per particle (over a range of z ~ 300 to 700 e). We note that relatively few particles exist in the region between the smaller and larger features.
Figure 1 also shows a simple mass spectrum obtained by integrating the data across the charge dimension. The most abundant species is a relatively sharp peak centered around ~6 MDa. The abundance of this peak decreases at ~7 MDa and the ion abundance reaches a minimum of ~14 MDa before a new feature corresponding to larger particles appears. This feature is broad, plateauing from ~17 to 26 MDa. At higher masses the intensity of particles decreases until ~40 MDa. Only a few sporadic particles with higher masses are observed beyond this point. The largest particle (beyond the range of masses shown in Figure 1) was observed at m = 87 MDa and z = 1269 e.
Complementary size information from electron microscopy.
In order to obtain more insight about these samples and the capabilities of CDMS, we characterized the size distributions for each sample using electron microscopy (EM). Figure 2 shows a representative EM image of the sample that was analyzed by CDMS in Figure 1.
Figure 2.
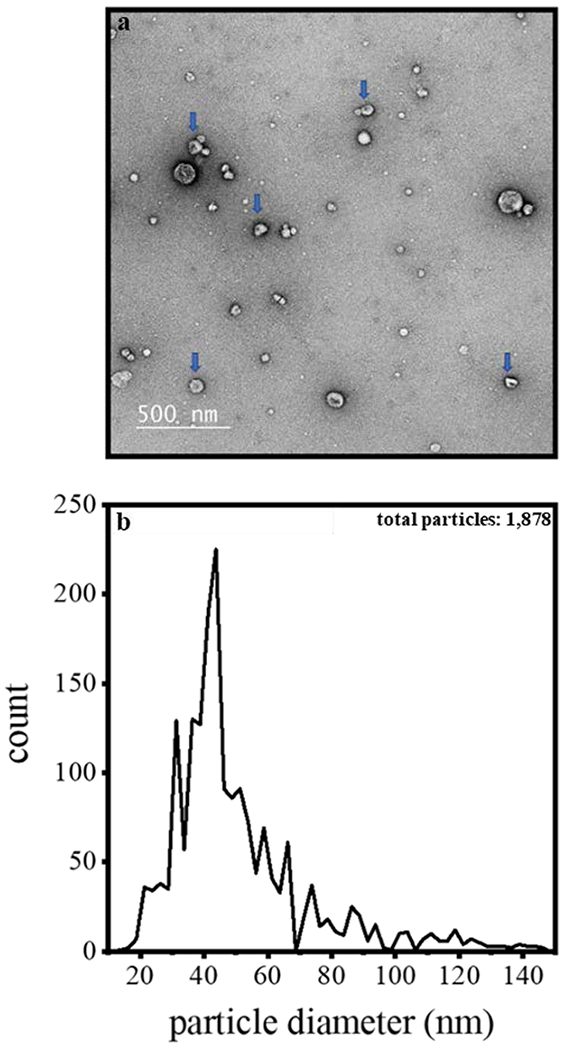
(top) Electron microscopy image of an exosome enriched sample prepared for CDMS. The blue arrows illustrate particles having diameters that are near the mean of our reported distribution. The CDMS data corresponding to this electron microscopy image for this sample are shown in Figure 1. (bottom) Size distribution (shown as diameters) determined by analyzing 1878 particles across the electron microscopy images recorded for all three of the exosome enriched milk samples reported here. Note: the particle diameter scale is binned in 2.5 nm increments; and, deformed or clearly damaged particles as well as those that clearly too small to be exosomes (below ~10 nm) were not included in this analysis.
In all of the samples that we have analyzed (with this preparation) we observed particles having diameters as small as ~10 nm to as large as ~150 nm. The arrows in Figure 2 indicate particles with diameters of ~30 to ~50 nm, consistent with small exosomes. Visual inspection shows that many smaller spherical particles are also present. These lipid-like particles that are abundant in milk are too small to be exosomes (which have a lower diameter limit of ~30 nm)65. Most particles are spherical. Careful examination of each particle shows that when the particle diameter exceeds ~30 nm, there is often visual evidence for a spherical cup-like morphology that is consistent with exosomes. We do not observe bilayer-like structure in the many smaller (~10-20 nm diameter) particles, consistent with lipid-like vesicles.
The distribution of sizes from EM analysis can be obtained from the frequency distribution shown in Figure 2. We characterized 1,878 particles across all data sets. The majority (~90%) of particles measured by EM correspond to EVs having diameters of ~20 to 60 nm, with an average diameter of ~40 nm. Overall, these values are consistent with previous exosome and EV observations that have employed similar sample preparation strategies66.
Complementary LC-MS-database search proteomics analyses corroborating the enrichment of exosomes.
Additional insight about the sample preparation and particles can be obtained by analyzing the protein content of these samples. To this end, we have carried out LC-MS-MS proteomics analyses after specific steps in the EV preparation shown in Scheme SI. This analysis allows us to determine which protein are enriched at each step of the sample workup. LC-MS analysis of the de-fatted raw milk identified 96 proteins (Supplementary Table S3). Removing cells from the sample allowed us to identify 106 proteins. Upon the acid precipitation and centrifugation, we identified 111 proteins, of which 1 are unique to this fraction. The final ultracentrifugation yielded our exosome and EV-containing pellet. From this, we identified a total of 162 proteins, of which 43 were unique. As a rough measure of enrichment, we examined those proteins uniquely identified in the exosome fraction as well as those with a 2-fold or greater relative abundance in the exosome fraction compared to the initial defatted milk. Of these proteins, 69% were annotated in both Exocarta and Vesiclepedia.67, 68
Further validating our enrichment protocol was the observation of characteristic EV marker proteins, ACTG1, Hsc70, ANXA5, CD9 and RAB1A in the EV preparation. To obtained a broader view of the EV preparation proteins, we submitted this list to DAVID (https://david.ncifcrf.gov/) for gene ontology enrichment analysis72,73, and compared these results to the bovine milk exosome proteome.67,68 Of the 130 annotated proteins in our enriched sample, ~79% are listed as being found in vesicles and ~52% are assigned to exosomes (Supplementary Table S3).
It is important to consider that other biological particles (such as those listed in Table 1) could be present in the final exosome sample that was analyzed by CDMS. Thus, we carefully monitored the presence of characteristic marker proteins from these other (non-exosomal) particles while preparing these samples. As described above, this was done by an LC-MS-database-search proteomics analysis of the raw milk sample after each step of the sample preparation (as shown in Scheme I). We note that Lutomski et al. have recorded the CDMS spectrum for HDL, LDL, and VLDL.78 The mass range associated with VLDL particles extends to as high as 70 MDa with more than 95% of lipoprotein particles having masses below ~40 MDa. Our proteomics measurements showed that after two different steps in the analysis, de-fatting and removal of cellular debris, there was evidence for several apolipoproteins, including Apolipoprotein A-IV (APOA4). However, the proteomics analysis of the final exosome sample failed to detect any of the major lipoproteins associated with these particles. Therefore, we do not believe that VLDL comprises a large fraction of the final exosome preparation.
Milk is composed primarily of caseins, lactoglobulin, albumins, lactotransferrins and immunoglobulins.48 Our intensity based MS quantification of defatted milk showed levels of these major proteins roughly in proportion to their expected amounts (Supplementary Table S4). One concern with casein is that it has been shown to be aggregate to form larger complexes or be incorporated into micelles that could precipitate along with larger particles.48 As we described above, an acid precipitation was used to deplete casein and other non-EV proteins prior to ultracentrifugation (Scheme I). From the proteomics analysis, we estimate that relative percentage of caseins is reduced by a factor of 10 in the final exosome preparation relative to the starting material. Therefore, casein micelles may comprise a small fraction of the particles detected by CDMS. While masses of casein micelles span between 1 and 1,000 MDa, the center of their distribution occurs at 100 MDa.48 We do not observe such a population of particles at 100 MDa, arguing against a significant fraction of our sample being micelles. Due to the recent discovery of exomeres and their poorly characterized proteome, we cannot assess if these particles are present since they share similar proteins with exosomes27, 74 and have similar sizes.
More insight about families of subpopulations from a Gaussian mixture model analysis of the CDMS charge verses mass datasets.
With the EM and proteomics analysis corroboration, our milk samples appear to be highly enriched in exosomes, but it is worthwhile to return to our CDMS data (Figure 1a) and analyze the charge against mass data in more detail. Specifically, we are interested in obtaining more insight about any subpopulations that may be resolved as families of particles from the CDMS data. The wide range of masses and charges that are observed in Figure 1 are consistent with the idea that these particles appear to be highly heterogeneous. Here, we develop a simple model set of subpopulations that upon summation are consistent with the complete two-dimensional CDMS dataset.
We begin by adopting the formalism of a simple, two-dimensional Gaussian mixture model (GMM) as a means of fitting the two-dimensional mass verses charge CDMS dataset. This model and statistical analysis are described in the Supplemental Information. Overall, the approach assumes that subpopulations of particles fall into families of related masses and charges, and that these distributions are normally distributed. Although we know of no biological or physical reason that requires that exosome subpopulations are Gaussian in nature, as shown below, a clustering analysis, based on the GMM assumption, results in multiple distributions of two-dimensional mass verses charge subpopulations; and, when combined, the sum of these subpopulations captures the main features of our two-dimensional CDMS data.
For simplicity the number of possible subpopulations was constrained between one and ten two-dimensional Gaussians. Except for this constraint, the analysis was unsupervised such that the algorithm determined the number of subpopulations, as well as the position, width, and abundance of each subpopulation, that when summed best fit the two-dimensional CDMS dataset.75 For the CDMS dataset shown in Figure 1a, this analysis converged on a best fit model consisting of six independent subpopulations. When applied to all of our datasets, we find similarities in position and shapes of subpopulations within different samples, suggesting that these subpopulations are conserved. From this analysis of all samples, we find evidence for eight unique subpopulations. A description of this analysis, along with the subpopulations obtained for each measurement (eight CDMS measurements of three different milk exosome samples) is provided in the Supporting Information.
The results of the GMM analysis for our first CDMS analysis of exosomes (sample 1) are shown as subpopulations in Figure 3. Table S1 provides a summary of the mean two-dimensional peak positions and associated uncertainties for this dataset. The subpopulations obtained from the GMM analysis for all eight replicate CDMS measurements measured for our three independent samples (three measurements for sample 1, three for sample 2, and two measurements for sample 3) are provided in the Supporting Information. As mentioned above, from the two-dimensional GMM analysis of all eight measurements we find evidence for eight unique subpopulations. We designate these populations from lowest to highest mass as subpopulation 1 (S1) to S8. For visual clarity, each point in the dataset is ascribed a color indicating the subpopulation to which it belongs. It should be noted that each of the points is assigned a color (and thus to a specific subpopulation) based on its highest probability of belonging to that subpopulation. This leads to artificially rigid boundaries between the families.
Figure 3.
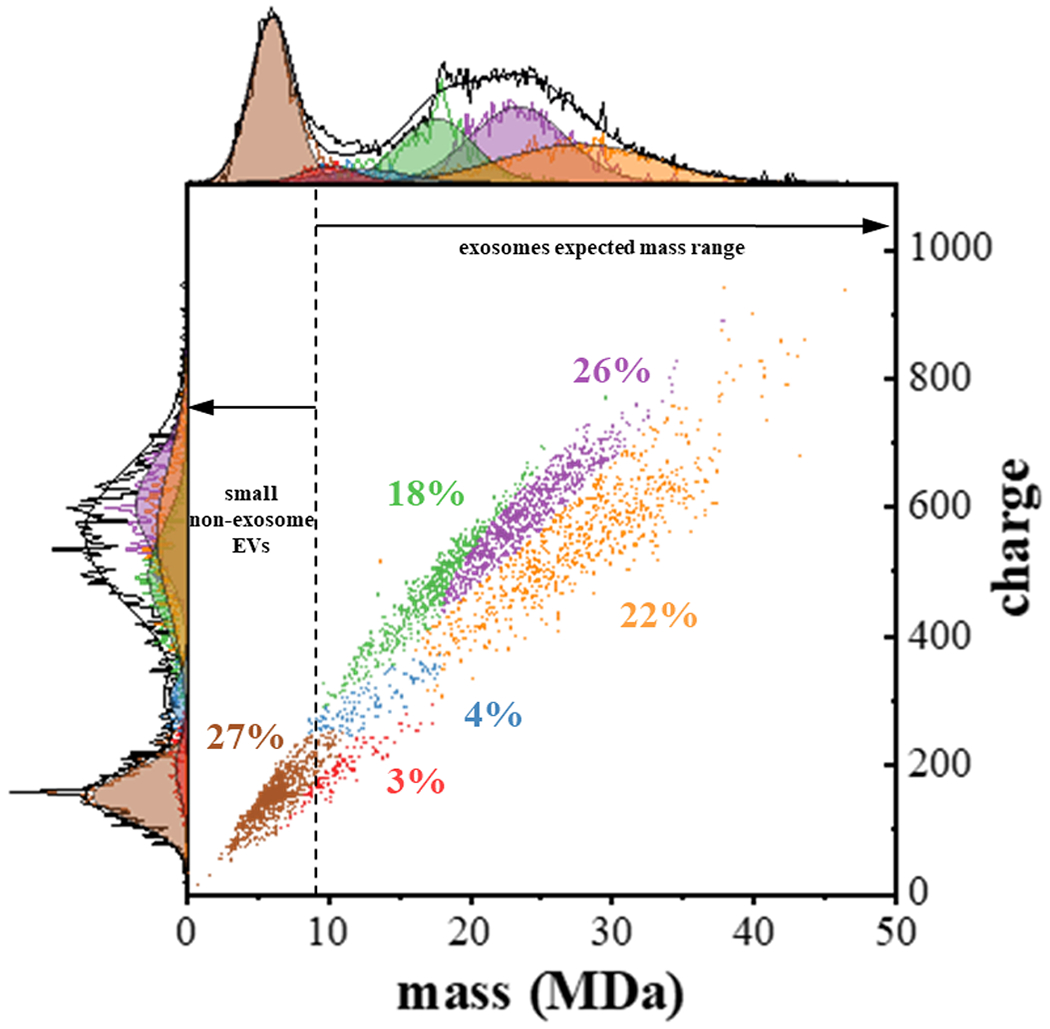
Two-dimensional mass versus charge plot showing subpopulations obtained from Gaussian fits to the experimental data for the first CDMS measurement of sample 1. See text for details. When all of the CDMS datasets are analyzed, this model finds evidence for eight subpopulations. This first measurement shows evidence for six of the eight subpopulations obtained upon analyzing all datasets [S2 (designated as brown), S3 (red), S4 (blue), S5 (green), S6 (purple) and S8 (orange)]. Each point shows the mass and charge measured for a single particle and is assigned to a subpopulation (indicated by color). Subfamily assignment is based on the highest probability of each particle belonging to a specific subfamily. Visually, this leads to boundaries that are artificially strict as in reality the subpopulations overlap. The top and left side traces show the integrated raw data for the mass and charge dimensions, respectively and corresponding sums of the Gaussian curves as black lines for these dimensions. The determined fits for each subpopulation are also shown and delineated using the same color scheme. The percentage of each subpopulation is also indicated. The dashed vertical line provides an estimate of the delineation between those particles having masses in the range that is expected for exosomes, and those particles that are too small to be exosomes.
The data in Figure 3 show that the lowest mass subpopulation that is observed in sample 1, corresponds to a relatively narrow distribution, centered at m = 5.7 ± 1.6 MDa and z = 145 ± 38 e. This is the second-lowest mass distribution that is extracted by our model. As summarized in Table S2, three GMM analyzed measurements (for two samples) show evidence for an additional well-populated (14%) narrow distribution centered at m = 3.5 ± 0.5 MDa and z = 83 ± 8 e. Finally, GMM analysis of one measurement (CDMS 2 for sample 3) uniquely found evidence for a small population (7%) of fairly massive particles centered at m = 23.7 ± 3 MDa and z = 461 ± 147 e. that was not found upon analyzing the first CDMS dataset shown in Figure 3.
The first CDMS measurement of sample 1, the lowest mass fraction that was found (i. e., the S2 population) comprises ~27% (975 out of 3586) of the total particles that were detected in this measurement. The highest mass subpopulation across all samples and measurements was observed in our first measurement of sample 1. This subpopulation (S8) is an extremely broad distribution centered at m = 27.7 ± 5.4 MDa and z = 594 ± 76 e. This subpopulation comprises ~22% of the distribution (772 out of 3586). These two subpopulations are completely resolved based on either their masses or charges. The masses, charges, and relative percentages of the four remaining subpopulations are summarized in Table S1. It is interesting to consider how these subpopulations vary in mass and charge. The S3 family, centered at m = 10 ± 2 MDa and z = 189 ± 44 e, accounts for only 3% of distribution, making it the lowest abundance subtype. This family, along with the S4 family (m = 12.5 ± 3 MDa, z = 296 ± 31 e) are both substantially more resolved based on charge compared with mass. This suggests that these families are comprised of similarly sized particles that differ substantially at the molecular level, thus influencing each particle’s charge more than its mass. We speculate that the charging differences between S3 and S4 families may reflect differences in surface proteins. The S5 (m = 17.6 ± 3 MDa, z = 488 ± 76 e) and S6 (m = 23.4 ± 3 MDa, z = 550 ± 113 e) families appear to be more resolved based on mass compared with the charge dimension, indicating that they are more similar in charging characteristics than in size. Lastly, the S8 population (m = 27.7 ± 5 MDa, z = 594 ± 76 e) is observed as extremely broad distributions in both mass and charge. Unlike other subpopulations, this species displays a greater mass with lower charge density. Many factors might contribute to such a phenomenon. For example, this family may contain surface proteins that have a lower pI and thus are not charged as extensively; or, more extensive post-translational modifications (i.e., phosphorylation or glycosylation) may introduce negatively charged moieties that effectively cancel out sites of positive charge.
Comparison of size distributions derived from CDMS data with size distributions obtained from EM measurements.
As a final assessment of the CDMS data, we compare the measured masses with the size distributions obtained from EM imaging (Figure 4) for identical samples. In this case, the sizes of particles from EM measurements are determined from an average diameter for each particle. An examination of the EM data demonstrates that particles may vary substantially in shape. One caveat to using such measurements is that dehydration during sample processing may slightly alter exosome shape.79 Nevertheless, we can still make overall comparisons between our CDMS measurements with those obtained by EM.
Figure 4.
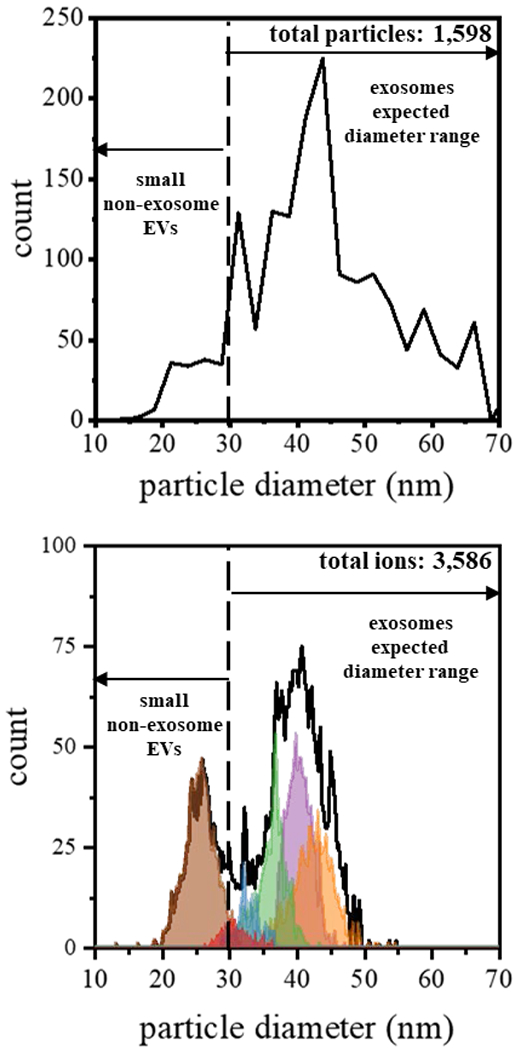
Comparison of diameters derived from CDMS measurements and EM images. (top) Plot showing the frequency distribution of electron micrograph-derived diameters (shown in Figure 2) using a bin size of 2.5 nm. (bottom) Plot of CDMS-derived diameters for each of the subpopulation (shown in Figure 3) using a bin size of 0.5 nm. Particle diameters from CDMS were determined by assuming a spherical geometry and a density of 1.15 g·cm−3.
To convert our CDMS mass measurements into diameters, we use an average exosome density of 1.15 g·cm−3,76,77 and also assume our particles are spherical. As the exosomes are desolvated during electrospray and enter the gaseous phase we assume that the contents inside the vesicle do not change, and therefore the density would be the same as in solution. Figure 4 shows a comparison of these estimated size distributions (for each of our CDMS subpopulations derived upon GMM analysis of sample 1) with the size distribution obtained upon analyzing the EM measurements (taken as the sum of all samples). This treatment of the EM data suggests that there may be favored types of particles; however, we note that these data are intrinsically noisy and a larger statistical analysis would be required to make this conclusion.
Overall, the CDMS-derived particle diameters subpopulations have a broad bimodal distribution extending from ~20 to ~50 nm. This range of particles is similar to that measured from EM, ~20 to ~70 nm. Additionally, the population maximum observed in both analyses appears just above 40 nm, demonstrating a reasonably good agreement. It does appear that CDMS may be less sensitive to the population of particles above ~50 nm observed by EM (~15% of the total particles). However, some of this disparity is definitely associated with the assumption of spherical particles used to determine diameters from EM data. Diameters for flattened species, most prominent for larger particles, are overestimated by this assumption, and this, in turn, overestimates their masses.
Total fraction of exosome particles.
From the considerations of the measured masses it is possible to estimate the fraction of particles that are exosomes. In total, across all eight of our CDMS measurements we detected 57,350 particles. Of these, the S1 (m = 3.5 ± 0.5 MDa), S2 (m = 6.0 ± 0.3 MDa), and S3 (m = 8.3 ± 1.4 MDa) subpopulations are perhaps too small to be exosomes – as m > 9.8 MDa is expected for particles larger than 30 nm (i.e., the smallest exosomes). Thus, we determine that in total 45,229 (or ~79%) particles are within the mass range expected for exosomes. This illustrates an important aspect of this analysis. To the extent that our GMM model is correct for characterizing these subpopulations, it will be possible to discern the exosome content from different sample preparations. For example, from our analysis of Sample 1, we find populations of 0%, 27%, and 3% for the S1, S2, and S3 non-exosomal subpopulations. We thus estimate that this sample is enriched to an exosome fraction of ~70%. As new sample preparation methods, aimed at purifying specific types of exosomes from different cell lines, tissues, and other body fluids continue to evolve, rapid and sensitive CDMS measurements of the physical properties of mass and charge may become an important means of assessing the efficacy of different protocols.
Conclusions
The masses of particles in three different bovine milk samples that have been enriched for exosomes have been analyzed using CDMS. In total, 57,350 particles were detected from eight CDMS measurements. A simple two-dimensional Gaussian model suggests that eight unique subpopulations of particles may be resolvable based on charge and mass. Complementary EM and proteomics analyses confirm that samples are enriched for exosomes. Particles associated with the S1, S2, and S3 families that are centered at ~ 3.5, ~5.9, and ~8.3 MDa, respectively, appear too small to be ascribed to exosomes. The remaining 45,229 (79%) particles detected by CDMS are within the mass range expected for exosomes. While CDMS measurements are at an early stage of development, this approach appears to provide a new physical basis for separating and characterizing EV particles.
Supplementary Material
Acknowledgements
This work was funded in part by a grant from the national institutes of health, NIH (R01 GM131100-01). BAB is supported by a fellowship from Indiana University Quantitative Chemical Biology Fellowship (T32GM109825 and T32GM131994).
Footnotes
Publisher's Disclaimer: Just Accepted
Publisher's Disclaimer: “Just Accepted” manuscripts have been peer-reviewed and accepted for publication. They are posted online prior to technical editing, formatting for publication and author proofing. The American Chemical Society provides “Just Accepted” as a service to the research community to expedite the dissemination of scientific material as soon as possible after acceptance. “Just Accepted” manuscripts appear in full in PDF format accompanied by an HTML abstract. “Just Accepted” manuscripts have been fully peer reviewed, but should not be considered the official version of record. They are citable by the Digital Object Identifier (DOI®). “Just Accepted” is an optional service offered to authors. Therefore, the “Just Accepted” Web site may not include all articles that will be published in the journal. After a manuscript is technically edited and formatted, it will be removed from the “Just Accepted” Web site and published as an ASAP article. Note that technical editing may introduce minor changes to the manuscript text and/or graphics which could affect content, and all legal disclaimers and ethical guidelines that apply to the journal pertain. ACS cannot be held responsible for errors or consequences arising from the use of information contained in these “Just Accepted” manuscripts.
References
- 1.Schorey JS; Cheng Y; Singh PP; Smith VL, Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO reports 2015, 16, 24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deatherage BL; Cookson BT, Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect immun 2012, 80, 1948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson DG; Ding Y; Jiang L, Unconventional protein secretion in plants: a critical assessment. Protoplasma 2016, 253, 31–43. [DOI] [PubMed] [Google Scholar]
- 4.Lotvall J; Hill AF; Hochberg F; Buzas EI; Di Vizio D; Gardiner C; Gho Y ; Kurochkin IV; Mathivanan S; Quesenberry P; Sahoo S; Tahara H; Wauben MH; Witwer KW; Thery C, Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pegtel DM; Gould SJ, Exosomes. Annu. Rev. Biochem 2019, 88, 487–514. [DOI] [PubMed] [Google Scholar]
- 6.Słomka A; Urban SK; Lukacs-Komek V; Żekanowska E; Kornek M, Large Extracellular Vesicles: Have We Found the Holy Grail of Inflammation? Front Immunol 2018, 9, 2723–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L; Hou D; Chen X; Li D; Zhu L; Zhang Y; Li J; Bian Z; Liang X; Cai X; Yin Y; Wang C; Zhang T; Zhu D; Zhang D; Xu J; Chen Q; Ba Y; Liu J; Wang Q; Chen J; Wang J; Wang M; Zhang Q; Zhang J; Zen K; Zhang C-Y, Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F; Paz HA; Sadri M; Cui J; Kachman SD; Fernando SC; Zempleni J, Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am. J. Physiol.: Gastrointest. Liver Physiol 2019, 317, G618–G624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatnagar S; Shinagawa K; Castellino FJ; Schorey JS, Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 2007, 110, 3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giri PK; Kruh NA; Dobos KM; Schorey JS, Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics 2010, 10, 3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang T; Lv H; Lv G; Li T; Wang C; Han Q; Yu L; Su B; Guo L; Huang S; Cao D; Tang L; Tang S; Wu M; Yang W; Wang H, Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun 2018, 9, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madeo M; Colbert PL; Vermeer DW; Lucido CT; Cain JT; Vichaya EG; Grossberg AJ; Muirhead D; Rickel AP; Hong Z; Zhao J; Weimer JM; Spanos WC; Lee JH; Dantzer R; Vermeer PD, Cancer exosomes induce tumor innervation. Nat. Commun 2018, 9, 4284–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X; Shi H; Yuan X; Jiang P; Qian H; Xu W, Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol Cancer 2018, 17, 146–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung BH; Weaver AM, Directed migration: Cells navigate by extracellular vesicles. J. Cell Biol 2018, 217, 2613–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maas SLN; Breakefield XO; Weaver AM, Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieu M; Martin-Jaular L; Lavieu G; Thery C, Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol 2019, 21, 9–17. [DOI] [PubMed] [Google Scholar]
- 17.Zhang ZG; Buller B; Chopp M, Exosomes — beyond stem cells for restorative therapy in stroke and neurological injury. Nat. Rev. Neurol 2019, 15, 193–203. [DOI] [PubMed] [Google Scholar]
- 18.Graner MW, 18 - Extracellular Vesicles as Vehicles of B Cell Antigen Presentation: Implications for Cancer Vaccine Therapies1 In Diagnostic and Therapeutic Applications of Exosomes in Cancer, Amiji M; Ramesh R, Eds. Academic Press: 2018; pp 325–341. [Google Scholar]
- 19.Min L; Garbutt C; Hornicek F; Duan Z, 16 - The Emerging Roles and Clinical Potential of Exosomes in Cancer: Drug Resistance In Diagnostic and Therapeutic Applications of Exosomes in Cancer, Amiji M; Ramesh R, Eds. Academic Press: 2018; pp 285–311. [Google Scholar]
- 20.Thery C; Amigorena S; Raposo G; Clayton A, Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol 2006, Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone RM; Adam M; Hammond JR; Orr L; Turbide C, Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987, 262, 9412–20. [PubMed] [Google Scholar]
- 22.Li P; Kaslan M; Lee SH; Yao J; Gao Z, Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Böing AN; van der Pol E; Grootemaat AE; Coumans FAW; Sturk A; Nieuwland R, Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles, 3, 23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gámez-Valero A; Monguió-Tortajada M; Carreras-Planella L; Franquesa M. l.; Beyer K; Borràs FE, Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep 2016, 6, 33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheruvanky A; Zhou H; Pisitkun T; Kopp JB; Knepper MA; Yuen PST; Star RA, Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol.: Renal, Fluid Electrolyte Physiol 2007, 292, F1657–F1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JS; Lee JC; Byeon SK; Rha KH; Moon MH, Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal. Chem 2017, 89, 2488–2496. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H; Freitas D; Kim HS; Fabijanic K; Li Z; Chen H; Mark MT; Molina H; Martin AB; Bojmar L; Fang J; Rampersaud S; Hoshino A; Matei I; Kenific CM; Nakajima M; Mutvei AP; Sansone P; Buehring W; Wang H; Jimenez JP; Cohen-Gould L; Paknejad N; Brendel M; Manova-Todorova K; Magalhães A; Ferreira JA; Osorio H; Silva AM; Massey A; Cubillos-Ruiz JR; Galletti G; Giannakakou P; Cuervo AM; Blenis J; Schwartz R; Brady MS; Peinado H; Bromberg J; Matsui H; Reis CA; Lyden D, Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol 2018, 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H; Lyden D, Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc 2019, 14, 1027–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pospichalova V; Svoboda J; Dave Z; Kotrbova A; Kaiser K; Klemova D; Ilkovics L; Hampl A; Crha I; Jandakova E; Minar L; Weinberger V; Bryja V, Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van der Vlist EJ; Nolte-’t Hoen ENM; Stoorvogel W; Arkesteijn GJA; Wauben MHM, Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat. Protoc 2012, 7, 1311. [DOI] [PubMed] [Google Scholar]
- 31.Colombo M; Raposo G; Thery C, Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol 2014, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- 32.Dragovic RA; Gardiner C; Brooks AS; Tannetta DS; Ferguson DJP; Hole P; Carr B; Redman CWG; Harris AL; Dobson PJ; Harrison P; Sargent IL, Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed-Nanotechnol. 2011, 7, 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Contino NC; Jarrold MF, Charge detection mass spectrometry for single ions with a limit of detection of 30 charges. Int. J. Mass Spectrom 2013, 345-347, 153–159. [Google Scholar]
- 34.Contino NC; Pierson EE; Keifer DZ; Jarrold MF, Charge detection mass spectrometry with resolved charge states. Int. J. Mass Spectrom 2013, 24, 101–8. [DOI] [PubMed] [Google Scholar]
- 35.Keifer DZ; Pierson EE; Jarrold MF, Charge detection mass spectrometry: weighing heavier things. Analyst 2017, 142, 1654–1671. [DOI] [PubMed] [Google Scholar]
- 36.Pierson EE; Contino NC; Keifer DZ; Jarrold MF, Charge Detection Mass Spectrometry for Single Ions with an Uncertainty in the Charge Measurement of 0.65 e. J. Am. Soc. Mass Spectrom 2015, 26, 1213–20. [DOI] [PubMed] [Google Scholar]
- 37.Pierson EE; Keifer DZ; Contino NC; Jarrold MF, Probing higher order multimers of pyruvate kinase with charge detection mass spectrometry. Int. J. Mass Spectrom 2013, 337, 50–56. [Google Scholar]
- 38.Mabbett SR; Zilch LW; Maze JT; Smith JW; Jarrold MF, Pulsed acceleration charge detection mass spectrometry: application to weighing electrosprayed droplets. Anal. Chem 2007, 79, 8431–9. [DOI] [PubMed] [Google Scholar]
- 39.Doussineau T; Kerleroux M; Dagany X; Clavier C; Barbaire M; Maurelli J; Antoine R; Dugourd P, Charging megadalton poly(ethylene oxide)s by electrospray ionization. A charge detection mass spectrometry study. Rapid Commun. Mass Spectrom 2011, 25, 617–23. [DOI] [PubMed] [Google Scholar]
- 40.Doussineau T; Désert A; Lambert O; Taveau J-C; Lansalot M; Dugourd P; Bourgeat-Lami E; Ravaine S; Duguet E; Antoine R, Charge Detection Mass Spectrometry for the Characterization of Mass and Surface Area of Composite Nanoparticles. J. Phys. Chem. C 2015, 119, 10844–10849. [Google Scholar]
- 41.Elliott AG; Harper CC; Lin HW; Williams ER, Mass, mobility and MS(n) measurements of single ions using charge detection mass spectrometry. Analyst 2017, 142, 2760–2769. [DOI] [PubMed] [Google Scholar]
- 42.Pierson EE; Keifer DZ; Selzer L; Lee LS; Contino NC; Wang JC; Zlotnick A; Jarrold MF, Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J. Am. Chem. Soc 2014, 136, 3536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haug A; Hostmark AT; Harstad OM, Bovine milk in human nutrition – a review. Lipids Health Dis. 2007, 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hata T; Murakami K; Nakatani H; Yamamoto Y; Matsuda T; Aoki N, Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem. Biophys. Res. Commun 2010, 396, 528–533. [DOI] [PubMed] [Google Scholar]
- 45.Agrawal AK; Aqil F; Jeyabalan J; Spencer WA; Beck J; Gachuki BW; Alhakeem SS; Oben K; Munagala R; Bondada S; Gupta RC, Milk-derived exosomes for oral delivery of paclitaxel. Nanomed-Nanotechnol. 2017, 13, 1627–1636. [DOI] [PubMed] [Google Scholar]
- 46.Akuma P; Okagu OD; Udenigwe CC, Naturally Occurring Exosome Vesicles as Potential Delivery Vehicle for Bioactive Compounds. Front. Sustain. Food Syst 2019, 3. [Google Scholar]
- 47.Arntz OJ; Pieters BCH; Oliveira MC; Broeren MGA; Bennink MB; de Vries M; van Lent PLEM; Koenders MI; van den Berg WB; van der Kraan PM; van de Loo FAJ, Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res 2015, 59, 1701–1712. [DOI] [PubMed] [Google Scholar]
- 48.McSweeney PLH and Fox PF, Advanced Dairy Chemistry. Boston, MA: Springer US, 2013. [Google Scholar]
- 49.Vashisht M; Rani P; Onteru SK; Singh D, Curcumin Encapsulated in Milk Exosomes Resists Human Digestion and Possesses Enhanced Intestinal Permeability in Vitro. Appl. Biochem. Biotechnol 2017, 183, 993–1007. [DOI] [PubMed] [Google Scholar]
- 50.Zhuang X; Xiang X; Grizzle W; Sun D; Zhang S; Axtell RC; Ju S; Mu J; Zhang L; Steinman L; Miller D; Zhang H-G, Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther 2011, 19, 1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zonneveld MI; Brisson AR; van Herwijnen MJ; Tan S; van de Lest CH; Redegeld FA; Garssen J; Wauben MH; Nolte-’t Hoen EN, Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J. Extracell. Vesicles 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somiya M; Yoshioka Y; Ochiya T, Biocompatibility of highly purified bovine milk-derived extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1440132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keifer DZ; Shinholt DL; Jarrold MF, Charge Detection Mass Spectrometry with Almost Perfect Charge Accuracy. Anal. Chem 2015, 87, 10330–7. [DOI] [PubMed] [Google Scholar]
- 54.Draper BE; Anthony SN; Jarrold MF, The FUNPET—a New Hybrid Ion Funnel-Ion Carpet Atmospheric Pressure Interface for the Simultaneous Transmission of a Broad Mass Range. J. Am. Soc. Mass Spectrom 2018, 29, 2160–2172. [DOI] [PubMed] [Google Scholar]
- 55.Shelton H Jr., C. D. H; Wuerker RF, Electrostatic Acceleration of Microparticles to Hypervelocities. J. Appl. Phys 1960, 31, 1243–1246. [Google Scholar]
- 56.Fuerstenau SD; Benner WH, Molecular weight determination of megadalton DNA electrospray ions using charge detection time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 1995, 9, 1528–38. [DOI] [PubMed] [Google Scholar]
- 57.Benner WH, A Gated Electrostatic Ion Trap To Repetitiously Measure the Charge and m/z of Large Electrospray Ions. Anal. Chem 1997, 69, 4162–4168. [Google Scholar]
- 58.Doussineau T; Mathevon C; Altamura L; Vendrely C; Dugourd P; Forge V; Antoine R, Mass Determination of Entire Amyloid Fibrils by Using Mass Spectrometry. Angew. Chem., Int. Ed 2016, 55, 2340–4. [DOI] [PubMed] [Google Scholar]
- 59.Lutomski CA; Lyktey NA; Zhao Z; Pierson EE; Zlotnick A; Jarrold MF, Hepatitis B Virus Capsid Completion Occurs through Error Correction. J. Am. Chem. Soc 2017, 139, 16932–16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keifer DZ; Motwani T; Teschke CM; Jarrold MF, Measurement of the accurate mass of a 50 MDa infectious virus. Rapid Commun. Mass Spectrom 2016, 30, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motwani T; Lokareddy RK; Dunbar CA; Cortines JR; Jarrold MF; Cingolani G; Teschke CM, A viral scaffolding protein triggers portal ring oligomerization and incorporation during procapsid assembly. Sci. Adv 2017, 3, e1700423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hogan JA; Jarrold MF, Optimized Electrostatic Linear Ion Trap for Charge Detection Mass Spectrometry. J. Am. Soc. Mass Spectrom 2018, 29, 2086–2095. [DOI] [PubMed] [Google Scholar]
- 63.Walton WH, Feret’s Statistical Diameter as a Measure of Particle Size. Nature 1948, 162, 329–330. [Google Scholar]
- 64.Schneider CA; Rasband WS; Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grigor’eva AE; Dyrkheeva NS; Bryzgunova OE; Tamkovich SN; Chelobanov BP; Ryabchikova EI, Contamination of exosome preparations, isolated from biological fluids. Biochem. (Mosc.), Suppl., Ser. B Biomed. chem 2017, 11, 265–271. [DOI] [PubMed] [Google Scholar]
- 66.Willms E; Johansson HJ; Mäger I; Lee Y; Blomberg KEM; Sadik M; Alaarg A; Smith CIE; Lehtiö J; El Andaloussi S; Wood MJA; Vader P, Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep 2016, 6, 22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keerthikumar S; Chisanga D; Ariyaratne D; Al Saffar H; Anand S; Zhao K; Samuel M; Pathan M; Jois M; Chilamkurti N; Gangoda L; Mathivanan S, ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol 2016, 428, 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalra H; Simpson RJ; Ji H; Aikawa E; Altevogt P; Askenase P; Bond VC; Borras FE; Breakefield X; Budnik V; Buzas E; Camussi G; Clayton A; Cocucci E; Falcon-Perez JM; Gabrielsson S; Gho YS; Gupta D; Harsha HC; Hendrix A; Hill AF; Inal JM; Jenster G; Kramer-Albers EM; Lim SK; Llorente A; Lotvall J; Marcilla A; Mincheva-Nilsson L; Nazarenko I; Nieuwland R; Nolte-’t Hoen EN; Pandey A; Patel T; Piper MG; Pluchino S; Prasad TS; Rajendran L; Raposo G; Record M; Reid GE; Sanchez-Madrid F; Schiffelers RM; Siljander P; Stensballe A; Stoorvogel W; Taylor D; Thery C; Valadi H; van Balkom BW; Vazquez J; Vidal M; Wauben MH; Yanez-Mo M; Zoeller M; Mathivanan S, Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinhardt TA; Lippolis JD; Nonnecke BJ; Sacco RE, Bovine milk exosome proteome. J. Proteomics 2012, 75, 1486–92. [DOI] [PubMed] [Google Scholar]
- 70.Liao Y; Alvarado R; Phinney B; Lonnerdal B, Proteomic characterization of human milk whey proteins during a twelve-month lactation period. J. Proteome. Res 2011, 10, 1746–54. [DOI] [PubMed] [Google Scholar]
- 71.Reinhardt TA; Sacco RE; Nonnecke BJ; Lippolis JD, Bovine milk proteome: quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteomics 2013, 82, 141–54. [DOI] [PubMed] [Google Scholar]
- 72.Huang da W; Sherman BT; Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 73.Huang da W; Sherman BT; Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q; Higginbotham JN; Jeppesen DK; Yang YP; Li W; McKinley ET; Graves-Deal R; Ping J; Britain CM; Dorsett KA; Hartman CL; Ford DA; Allen RM; Vickers KC; Liu Q; Franklin JL; Bellis SL; Coffey RJ, Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27, 940–954.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Redner RA; Walker HF, Mixture Densities, Maximum Likelihood and the Em Algorithm. SIAM Rev. 1984, 26, 195–239. [Google Scholar]
- 76.Kreimer S; Belov AM; Ghiran I; Murthy SK; Frank DA; Ivanov AR, Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J. Proteome. Res 2015, 14, 2367–84. [DOI] [PubMed] [Google Scholar]
- 77.Mathivanan S; Ji H; Simpson RJ, Exosomes: Extracellular organelles important in intercellular communication. J. Proteomics 2010, 73, 1907–1920. [DOI] [PubMed] [Google Scholar]
- 78.Lutomski CA; Gordon SM; Remaley AT; Jarrold MF, Resolution of Lipoprotein Subclasses by Charge Detection Mass Spectrometry. Anal. Chem 2018, 90, 6353–6356. [DOI] [PubMed] [Google Scholar]
- 79.Chopra N; Dutt Arya B; Jain N; Yadav P; Wajid S; Singh SP; Choudhury S, Biophysical Characterization and Drug Delivery Potential of Exosomes from Human Wharton’s Jelly-Derived Mesenchymal Stem Cells. ACS Omega. 2019, 4, 13143–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pierson EE; Keifer DZ; Asokan A; Jarrold MF, Resolving Adeno-Associated Viral Particle Diversity With Charge Detection Mass Spectrometry. Anal. Chem 2016, 88, 6718–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


