Abstract
Background
We performed a systematic review and meta-analysis of the Matrix metalloproteinases (MMP)-9 (C1562T), MMP-9 (R279Q), MMP-9 (P574R) and MMP-9 (R668Q) polymorphisms and risk of Coronary Artery Disease (CAD).
Methods
After a systematic literature search, pooled odds ratio (OR) and their corresponding 95% confidence interval (CI) were used to evaluate the strength of the association.
Results
We identified 40 studies with 11,792 cases and 8280 controls for C1562T, 7 case-control studies with 5525 cases and 2497 controls for R279Q, 2 studies with 1272 cases and 785 controls for P574R, and 2 studies with 1272 cases and 785 controls for R668Q. MMP-9 (C1562T) polymorphism was associated with increased risk of CAD under dominant model (OR = 1.41, P < 0.001), recessive model (OR = 1.59, P < 0.001), allelic model (OR = 1.38, P < 0.001), TT vs. CC model (OR = 1.70, P < 0.001), and CT vs. CC model (OR = 1.35, P < 0.001). Moreover, the subgroup analysis based on the continent of the study populations in this SNP indicated strong significant association in Asians but not in Europeans. Subgroup analysis was not performed in Africa, America and Oceania, due to lack of sufficient data.
Conclusions
Our meta-analysis revealed that MMP-9 (C1562T) SNP conferred a susceptibility risk for CAD in the overall analysis and Asian population. The overall analysis and subgroup analysis of the other three SNPs reject the association between MMP-9 polymorphisms and the risk of CAD. Although the results should interpret with caution because of small sample size of included studies in these three SNPs.
Keywords: Coronary artery disease, Matrix metalloproteinases, Genetic polymorphism, Meta-analysis
Background
Coronary artery disease (CAD) is a worldwide medical problem that is the leading cause of death in both developed and developing countries, especially in older people [1, 2]. Several studies have shown that the traditional risk factors, such as blood lipid, diabetes, hypertension, obesity play crucial roles in the initiation and perpetuation of CAD. However, it is nowadays accepted that genetic component has an essential role in the development of CAD [3–6]. Researches have suggested that family aggregation of CAD is not unusual, and genetic association investigations revealed that the average heritability of CAD is more than 50% [5, 7]. Epidemiological studies have found many genetic variants especially single-nucleotide polymorphism (SNP) in association with an increased risk of CAD [8]. The exact mechanism underlying the influence of polymorphism on the pathogenesis of CAD is not fully understood. Nevertheless, polymorphisms in numerous genes involved in inflammation, metabolism of lipid and glucose, blood clotting, and homocysteine may affect susceptibility to CAD [9, 10].
. This enzyme is involved in the degradation of extracellular matrix (ECM) components, such as type IV collagen, which is involved in the neovascularization, angiogenesis, inflammatory processes, and development of atherosclerosis.
MMPs are zinc containing enzymes that belong to a neutral protease family. Among the MMP family, MMP-9 is the most important enzyme of this class that is produced by the cells in the vascular wall. Moreover, inflammatory immune cells, such as neutrophils, monocytes as well as endothelial cells and vascular smooth muscle cells (VSMCs) generate MMP-9 [11]. This enzyme is involved in the degradation of extracellular matrix (ECM) components, such as type IV collagen, which is involved in the neovascularization, angiogenesis, inflammatory processes, and development of atherosclerosis [12, 13]. Several studies have shown that the levels of MMPs and their matrix-degrading activity are raised in exposed areas of atherosclerotic plaques, or following acute coronary syndrome [13, 14]. As a result, it is rational to hypothesize that genetic defects resulting in the overexpression of activated MMPs play a crucial role in the pathogenesis of coronary artery disease (CAD).
The MMP family is grouped into gelatinizes (MMP2, 9), collagenases (MMP1, 8, 13, 18), stromelysins (MMP3, 10, 11), and the membrane-type MMPs (MT-MMPs) that are coded by separate genes and have different tissue distribution and bioactive function [15].
Several studies have shown that MMP-9 family polymorphisms might be associated with the risk of CAD [16–18]. However, the results are inconsistent. For example, Mahmoodi et al. conducted a case-control study to investigate the association between -1562C > T genetic polymorphism and susceptibility to CAD. But, genotype and allele frequencies of MMP9 -1562C > T polymorphism were similar between CAD patients and controls (P > 0.05) [19]. However, Rodriguez-Perez et al. demonstrated that MMP9 (1562 C > T) allele and the CT genotype were associated with the risk of developing myocardial infraction (MI) [20]. The causes for these controversial results probably due to small sample sizes, different ethnicity, patient selection, clinical heterogeneity, low statistical power, or a combination of these factors. Therefore, we performed this meta-analysis to evaluate whether MMP-9 gene family polymorphisms play a role in CAD susceptibility.
Methods
We followed a protocol based on observational studies in epidemiology (MOOSE) guidelines [21], and results were reported based on Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline [22]. This article does not contain any studies with human participants performed by any of the authors.
Systematic search strategy
An exhaustive systematic search was conducted through electronic databases (Scopus, Medline) retrieving all potential publications considered the association between MMP-9 family gene polymorphism and susceptibility to CAD. All publications from inception to February 2020 were included (the search was updated before manuscript submission). The combination of key words and Mesh (Medical Subject Headings) terms were as follow: (“matrix metalloproteinase” [Mesh] OR “MMP” OR “gelatinase”) AND (“coronary heart disease” OR “CAD” OR “coronary syndrome” OR “ischemic heart disease” OR “vascular disease” OR “myocardial infarction” OR “MI” OR “atherosclerosis” OR “arteriosclerosis” OR “coronary stenosis” OR “coronary disease” OR “CHD” OR “angina”) AND (“single nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”). The references of review articles were cross-checked to find potential publications. Only human studies and English language publications were considered.
Inclusion and exclusion criteria
We screened retrieval publications according to following inclusion criteria: 1) observational studies (cohort or case-control design); 2) publications considered the association between MMP-9 family gene polymorphism (C1562T, R279Q, P574R and R668Q) and susceptibility to CAD; 3) publications reporting sufficient data to extract or calculate risk estimates with 95% CI; 4) publications reporting numbers or genotype frequencies in cases and healthy controls. Duplicates, book chapters, letters to editor, animal study, case reports, review articles, and studies with repetitive subjects all were excluded. The application of these criteria recognized 40, 7, 2, and 2 eligible studies for C1562T SNP, R279Q SNP, P574R SNP and R668Q SNP, respectively.
Data extraction
Two authors screened the literature and extracted data independently according to the inclusion and exclusion criteria. The following data was extracted: the first author’s name, journal and year of publication, country of origin, ethnicity, number of subjects in the case and control groups, mean or range of age, genotyping method, genotype counts in the case and control group.
Quality assessment
The quality of eligible studies was assessed by using the Newcastle-Ottawa Scale (NOS) [23]. Studies were scored based on three main components: selection, comparability, and ascertainment of outcome. This scale ranges from 1 to 9 stare and studies with scores 0–3, 4–6 or 7–9 were of low, moderate, or high-quality, respectively.
Statistical analysis
In the current meta-analysis, the strengths of association between MMP-9 family gene polymorphism and the risk of CAD was estimated via the OR and 95% CI in five genetic models: dominant model, recessive model, allelic model, homozygote contrast, and heterozygotes contrast. The potential heterogeneity was evaluated by the Q test and the I2 test [23]. According to these two test, if Q had a P value less than 0.1 and I2 exceed 50%, the random effects model (REM) was used; otherwise, the fixed effect model (FEM) was applied [24, 25]. Additionally, risk of publication bias was examined by funnel plot, Egger’s weighted regression test and Begg’s rank correlation test (P < 0.05 was regarded as statistically significant publication bias) [26, 27]. Besides, quality assessment of genotype data in case control studies was evaluated by Hardy–Weinberg equilibrium (HWE). Finally, in order to show the stability of our results, sensitivity analysis was performed. All statistical tests for this meta-analysis were performed with Stata statistical software (version 14.0; Stata Corporation, College Station, TX, USA) and SPSS (version 23.0; SPSS, Inc. Chicago, IL, USA).
Results
Study characteristics
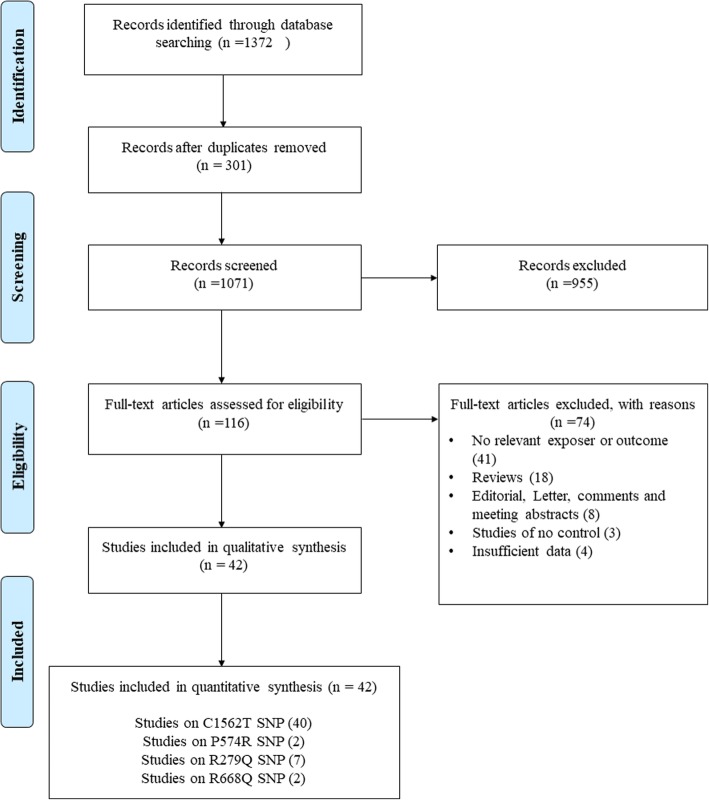
The search and screening process workflow is shown in Fig. 1. Our primary search yielded 1372 records, which 42 of them were included in quantitative analysis [16–20, 28–64]. The studies were published between 2001 to 2019 and all of them had good methodological score ranging between 5 and 8. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) as genotyping method was common between most of studies. The sample size in case and control groups of four SNPs varied between 40 to 2506 and 40 to 689 individual, respectively. The range of mean ages in case and control groups was from 33 to 94, which means studies were conducted among adults. Only one of included studies had cohort design and the other were case-control. Tables 1 and 2 summarized the characteristics and genotype frequency of the included studies.
Fig. 1.
Flow diagram of study selection process
Table 1.
Characteristics of studies included in meta-analysis of overall CAD
| Study author | Year | Country | Ethnicity | Study design | Type of CAD | Total cases/controls | Age Case / Control (Mean) |
Genotyping method | Quality score |
|---|---|---|---|---|---|---|---|---|---|
| MMP-9 (C1562T) | |||||||||
| Pollanen et al. | 2001 | Finland | European | Case-control | ACS | 109 / 167 | 33–69 / 33–69 | PCR-RFLP | 6 |
| Wang et al. | 2001 | Australia | Oceania | Case-control | Stable | 619 / 169 | 57.7 ± 0.5 / NR | PCR-RFLP | 7 |
| Cho et al. | 2002 | Korea | Asian | Case-control | Stable | 63 / 134 | NR / NR | PCR-RFLP | 5 |
| Kim et al. | 2002 | Korea | Asian | Case-control | Stable | 131 / 117 | 61.3 ± 7.9 / 59.3 ± 8.5 | PCR-RFLP | 6 |
| Jones et al. | 2002 | New Zealand | European | Case-control | Stable | 414 / 203 | 71.7 ± 7.6 / 70.8 ± 8.0 | PCR-RFLP | 8 |
| Tang et al. | 2005 | China | Asian | Case-control | ACS | 101 / 105 | NR / NR | PCR-RFLP | 5 |
| Chen et al. | 2005 | China | Asian | Case-control | ACS | 78 / 81 | NR / NR | PCR-RFLP | 5 |
| Meng et al. | 2006 | China | Asian | Case-control | Stable | 117 / 99 | NR / NR | PCR-RFLP | 5 |
| Nuzzo et al. | 2006 | Italy | European | Case-control | ACS | 49 / 123 | NR / NR | PCR-RFLP | 5 |
| Chen et al. | 2007 | China | Asian | Case-control | Stable | 150 / 70 | NR / NR | PCR-RFLP | 5 |
| Nanni et al. | 2007 | Italy | European | Case-control | ACS | 200 / 201 | 47.8 ± 6.2 / 47.0 ± 5.5 | PCR-RFLP | 7 |
| Wang et al. | 2007 | China | Asian | Case-control | ACS | 245 / 204 | NR / NR | PCR-RFLP | 8 |
| Zhang et al. | 2008 | China | Asian | Case-control | ACS | 92 / 95 | NR / NR | PCR-RFLP | 5 |
| Koh et al. | 2008 | Korea | Asian | Case-control | ACS | 206 / 173 | 61.1 ± 11.8 / 58.3 ± 11.8 | PCR-RFLP | 6 |
| Alp et al. | 2009 | Turkey | European | Case-control | Stable | 146 / 122 | 59.30 ± 9.1 / 57.30 ± 9.7 | PCR-RFLP | 6 |
| Wu et al. | 2009 | China | Asian | Case-control | ACS | 2517 / 689 | NR / 60.42 ± 9.07 | PCR-RFLP | 8 |
| Gao et al. | 2010 | China | Asian | Case-control | Stable | 96 / 78 | NR / NR | PCR-RFLP | 5 |
| Fallah et al. | 2010 | Iran | Asian | Case-control | Stable | 145 / 157 | 58.49 ± 9.12 / 55.35 ± 9.43 | PCR-RFLP | 6 |
| Yong et al. | 2010 | China | Asian | Case-control | ACS | 128 / 106 | NR / NR | PCR-RFLP | 5 |
| Ghaderian et al. | 2010 | Iran | Asian | Case-control | ACS | 400 / 200 | NR / 65.8 ± 5.9 | TaqMan | 8 |
| Zhi et al. | 2010 | China | Asian | Case-control | Stable | 762 / 555 | 67.46 ± 9.61 / 69.90 ± 11.48 | PCR-RFLP | 8 |
| Wang et al. | 2011 | China | Asian | Case-control | ACS | 384 / 451 | 55.6 ± 10.9 / 54.1 ± 10.3 | PCR-RFLP | 8 |
| Opstad et al. | 2012 | Norway | European | Case-control | Stable | 996 / 204 | 62 / NR | TaqMan | 8 |
| Han et al. | 2012 | China | Asian | Case-control | Stable | 91 / 101 | NR / NR | PCR-RFLP | 5 |
| Saracini et al. | 2012 | Italy | European | Case-control | Stable | 423 / 423 | 40–94 / 41–94 | Nano gene electronic microchip technology | 8 |
| Spurthi et al. | 2012 | India | Asian | Case-control | Stable | 100 / 100 | 56.73 ± 12.2 / 54.55 ± 14.38 | PCR-RFLP | 5 |
| Sewelam et al. | 2013 | Egypt | African | Case-control | ACS | 40 / 40 | NR / NR | PCR-RFLP | 5 |
| Wu et al. | 2013 | China | Asian | Case-control | ACS | 258 / 153 | 63.97 ± 12.32 / 63.61 ± 11.8 | PCR-RFLP | 7 |
| Xu et al. | 2013 | China | Asian | Case-control | Stable | 382 / 466 | 62 ± 12 / 62 ± 10 | PCR-RFLP | 8 |
| Rodriguez et al. | 2016 | Mexico | American | Case-control | ACS | 236 / 285 | 59 / 58 | PCR-RFLP | 8 |
| Yin et al. | 2016 | China | Asian | Case-control | Stable | 194 / 251 | 55.60 ± 10.42 / 56.21 ± 9.83 | PCR-RFLP | 7 |
| Beton et al. | 2016 | Turkey | European | Case-control | Stable | 200 / 200 | 60.2 ± 7.4 / 58.3 ± 7.7 | PCR-RFLP | 7 |
| Daraei et al. | 2016 | Iran | Asian | Case-control | ACS | 117 / 120 | 62.96 ± 12.80 / 52.55 ± 9.80 | PCR-RFLP | 6 |
| El-Aziz et al. | 2016 | Egypt | African | Case-control | ACS | 184 / 180 | 57.2 ± 10.9 / 58.8 ± 8.3 | PCR-RFLP | 7 |
| Qin et al. | 2016 | China | Asian | case-control | Stable | 261 / 261 | 58.75 ± 9.36 / 59.21 ± 10.10 | PCR-RFLP | 7 |
| Peksiene et al. | 2017 | Lithuania | European | Case-control | ACS | 518 / 645 | 61.9 ± 11.1 / 60.6 ± 11.9 | TaqMan | 8 |
| Mahmoodi et al. | 2017 | Iran | Asian | case–control | Stable | 100 / 100 | 59.4 ± 23.5 / 56.7 ± 29.5 | PCR-RFLP | 5 |
| Xu et al. | 2017 | China | Asian | Case-control | Stable | 264 / 186 | 59 ± 11.67 / 58 ± 10.72 | PCR-RFLP | 7 |
| Makrygiannis et al. | 2018 | Greece | European | Case-control | Stable | 175 / 166 | 72.7 ± 7.6 / 71.5 ± 7.1 | PCR-RFLP | 7 |
| Malkani et al. | 2019 | Iran | Asian | Case-control | Stable | 101 / 100 | 59.2 ± 10.2 / 47.3 ± 13.1 | PCR-RFLP | 5 |
| MMP-9 (R279Q) | |||||||||
| Nanni et al. | 2007 | Italy | European | Case-control | ACS | 200 / 201 | 47.8 ± 6.2 / 47.0 ± 5.5 | PCR-RFLP | 7 |
| Wu et al. | 2009 | China | Asian | Case-control | ACS | 2506 / 687 | NR / 60.42 ± 9.07 | PCR-RFLP | 8 |
| Zhi et al. | 2010 | China | Asian | Case-control | Stable | 762 / 555 | 67.46 ± 9.61 / 69.90 ± 11.48 | PCR-RFLP | 8 |
| Wang et al. | 2011 | China | Asian | Case-control | ASC | 384 / 451 | 55.6 ± 10.9 / 54.1 ± 10.3 | PCR-RFLP | 8 |
| Mishra et al. | 2012 | India | Asian | Cohort | Stable | 510 / 230 | NR/ 54.2 ± 8.5 | PCR-RFLP | 8 |
| Opstad et al. | 2012 | Norway | European | Case-control | Stable | 994 / 204 | 62 / NR | TaqMan | 8 |
| Fiotti et al. | 2017 | Italy | European | Case-control | Stable | 169 / 169 | 69–78 / 67–80 | Sequencing | 7 |
| MMP-9 (P574R) | |||||||||
| Zhi et al. | 2010 | China | Asian | Case-control | Stable | 762 / 555 | 67.46 ± 9.61 / 69.90 ± 11.48 | PCR-RFLP | 8 |
| Mishra et al. | 2012 | India | Asian | Cohort | Stable | 510 / 230 | NR / 54.2 ± 8.5 | PCR-RFLP | 8 |
| MMP-9 (R668Q) | |||||||||
| Zhi et al. | 2010 | China | Asian | Case-control | Stable | 762 / 555 | 67.46 ± 9.61 / 69.90 ± 11.48 | PCR–RFLP | 8 |
| Mishra et al. | 2012 | India | Asian | Cohort | Stable | 510 / 230 | NR / 54.2 ± 8.5 | PCR–RFLP | 8 |
NR, not reported; ACS, Acute coronary syndrome
Table 2.
Distribution of genotype and allele among CAD patients and controls
| Study author | CAD cases | Healthy control | P-HWE | MAF | ||||||||
| CC | CT | TT | C | T | CC | CT | TT | C | T | |||
| MMP-9 (C1562T) | ||||||||||||
| Pollanen et al. | 78 | 21 | 10 | 177 | 41 | 124 | 30 | 13 | 278 | 56 | 0 | 0/168 |
| Wang et al. | 479 | 128 | 12 | 1086 | 152 | 128 | 41 | 0 | 297 | 41 | 0/072 | 0/121 |
| Cho et al. | 48 | 15 | 0 | 111 | 15 | 67 | 63 | 4 | 197 | 71 | 0/016 | 0/265 |
| Kim et al. | 99 | 32 | 0 | 230 | 32 | 85 | 32 | 0 | 202 | 32 | 0/086 | 0/137 |
| Jones et al. | 257 | 145 | 12 | 659 | 169 | 145 | 57 | 1 | 347 | 59 | 0/063 | 0/145 |
| Tang et al. | 73 | 27 | 1 | 173 | 29 | 91 | 13 | 1 | 195 | 15 | 0/494 | 0/071 |
| Chen et al. | 57 | 21 | 0 | 135 | 21 | 73 | 8 | 0 | 154 | 8 | 0/640 | 0/049 |
| Meng et al. | 91 | 26 | 0 | 208 | 26 | 80 | 18 | 1 | 178 | 20 | 0/991 | 0/101 |
| Nuzzo et al. | 7 | 39 | 3 | 53 | 45 | 86 | 36 | 1 | 208 | 38 | 0/181 | 0/154 |
| Chen et al. | 97 | 48 | 5 | 242 | 58 | 61 | 6 | 3 | 128 | 12 | 0 | 0/086 |
| Nanni et al. | 136 | 62 | 2 | 334 | 66 | 135 | 63 | 3 | 333 | 69 | 0/147 | 0/172 |
| Wang et al. | 191 | 52 | 2 | 434 | 56 | 178 | 25 | 1 | 381 | 27 | 0/903 | 0/066 |
| Zhang et al. | 67 | 22 | 3 | 156 | 28 | 83 | 12 | 0 | 178 | 12 | 0/511 | 0/063 |
| Koh et al. | 151 | 52 | 3 | 354 | 58 | 142 | 31 | 0 | 315 | 31 | 0/195 | 0/090 |
| Alp et al. | 99 | 42 | 5 | 240 | 52 | 90 | 29 | 3 | 209 | 35 | 0/718 | 0/143 |
| Wu et al. | 1995 | 495 | 27 | 4485 | 549 | 545 | 143 | 1 | 1233 | 145 | 0 | 0/105 |
| Gao et al. | 49 | 38 | 9 | 136 | 56 | 59 | 18 | 1 | 136 | 20 | 0/775 | 0/128 |
| Fallah et al. | 11 | 57 | 77 | 79 | 211 | 19 | 76 | 62 | 114 | 200 | 0/558 | 0/637 |
| Yong et al. | 97 | 30 | 1 | 224 | 32 | 92 | 14 | 0 | 198 | 14 | 0/466 | 0/066 |
| Ghaderian et al. | 296 | 88 | 16 | 680 | 120 | 141 | 53 | 6 | 335 | 65 | 0/708 | 0/163 |
| Zhi et al. | 585 | 174 | 3 | 1344 | 180 | 442 | 110 | 3 | 994 | 116 | 0/164 | 0/105 |
| Wang et al. | 286 | 87 | 11 | 659 | 109 | 373 | 72 | 6 | 818 | 84 | 0/244 | 0/093 |
| Opstad et al. | 756 | 225 | 15 | 1737 | 255 | 154 | 46 | 4 | 354 | 54 | 0/794 | 0/132 |
| Han et al. | 65 | 25 | 1 | 155 | 27 | 75 | 25 | 1 | 175 | 27 | 0/489 | 0/134 |
| Saracini et al. | 313 | 98 | 12 | 724 | 122 | 307 | 101 | 15 | 715 | 131 | 0/071 | 0/155 |
| Spurthi et al. | 40 | 47 | 13 | 127 | 73 | 48 | 46 | 6 | 142 | 58 | 0/241 | 0/290 |
| Sewelam et al. | 32 | 7 | 1 | 71 | 9 | 40 | 0 | 0 | 80 | 0 | 0 | 0 |
| Wu et al. | 193 | 56 | 9 | 442 | 74 | 131 | 22 | 0 | 284 | 22 | 0/337 | 0/072 |
| Xu et al. | 268 | 109 | 5 | 645 | 119 | 361 | 103 | 2 | 825 | 107 | 0/059 | 0/115 |
| Rodriguez et al. | 210 | 26 | 0 | 446 | 26 | 271 | 14 | 0 | 556 | 14 | 0/670 | 0/025 |
| Yin et al. | 98 | 73 | 23 | 269 | 119 | 157 | 84 | 10 | 398 | 104 | 0/766 | 0/207 |
| Beton et al. | 158 | 38 | 4 | 354 | 46 | 154 | 43 | 3 | 351 | 49 | 0/999 | 0/123 |
| Daraei et al. | 66 | 50 | 1 | 182 | 52 | 79 | 38 | 3 | 196 | 44 | 0/528 | 0/183 |
| El-Aziz et al. | 125 | 52 | 7 | 302 | 66 | 141 | 36 | 3 | 318 | 42 | 0/690 | 0/117 |
| Qin et al. | 134 | 100 | 27 | 368 | 154 | 171 | 85 | 5 | 427 | 95 | 0/129 | 0/182 |
| Peksiene et al. | 340 | 156 | 22 | 836 | 200 | 431 | 185 | 29 | 1047 | 243 | 0/115 | 0/188 |
| Mahmoodi et al. | 68 | 27 | 5 | 163 | 37 | 72 | 26 | 2 | 170 | 30 | 0/844 | 0/150 |
| Xu et al. | 188 | 69 | 7 | 445 | 83 | 151 | 31 | 4 | 333 | 39 | 0/126 | 0/105 |
| Makrygiannis et al. | 133 | 40 | 2 | 306 | 44 | 133 | 31 | 2 | 297 | 35 | 0/898 | 0/105 |
| Malkani et al. | 79 | 3 | 19 | 161 | 41 | 100 | 0 | 0 | 200 | 0 | 0 | 0 |
| Study author | CAD cases | Healthy control | P-HWE | MAF | ||||||||
| AA | AG | GG | A | G | AA | AG | GG | A | G | |||
| MMP-9 (R279Q) | ||||||||||||
| Nanni et al. | 85 | 94 | 21 | 264 | 136 | 94 | 87 | 20 | 275 | 127 | 0/984 | 0/316 |
| Wu et al. | 1177 | 1102 | 227 | 3456 | 1556 | 297 | 312 | 78 | 906 | 468 | 0/772 | 0/341 |
| Zhi et al. | 398 | 296 | 68 | 1092 | 432 | 267 | 226 | 62 | 760 | 350 | 0/179 | 0/315 |
| Wang et al. | 185 | 150 | 49 | 520 | 248 | 239 | 167 | 45 | 645 | 257 | 0/052 | 0/285 |
| Mishra et al. | 114 | 253 | 143 | 481 | 539 | 53 | 103 | 74 | 209 | 251 | 0/142 | 0/546 |
| Opstad et al. | 405 | 472 | 117 | 1282 | 706 | 79 | 98 | 27 | 256 | 152 | 0/693 | 0/373 |
| Fiotti et al. | 75 | 69 | 25 | 219 | 119 | 57 | 88 | 24 | 202 | 136 | 0/282 | 0/402 |
| Study author | CAD cases | Healthy control | P-HWE | MAF | ||||||||
| PP | PR | RR | P | R | PP | PR | RR | P | R | |||
| MMP-9 (P574R) | ||||||||||||
| Zhi et al. | 406 | 296 | 60 | 1108 | 416 | 279 | 231 | 45 | 789 | 321 | 0/770 | 0/155 |
| Mishra et al. | 346 | 150 | 14 | 842 | 178 | 169 | 57 | 4 | 395 | 65 | 0/747 | 0/276 |
| Study author | CAD cases | Healthy control | P-HWE | MAF | ||||||||
| RR | RQ | R | Q | RR | RQ | R | Q | |||||
| MMP-9 (R668Q) | ||||||||||||
| Zhi et al. | 564 | 179 | 19 | 1307 | 217 | 398 | 141 | 16 | 937 | 173 | 0/416 | 0/289 |
| Mishra et al. | 191 | 286 | 33 | 668 | 352 | 113 | 107 | 10 | 333 | 127 | 0/012 | 0/141 |
P-HWE p-value for Hardy–Weinberg equilibrium, MAF minor allele frequency of control group
Meta-analysis of MMP-9 (C1562T) and risk of CAD
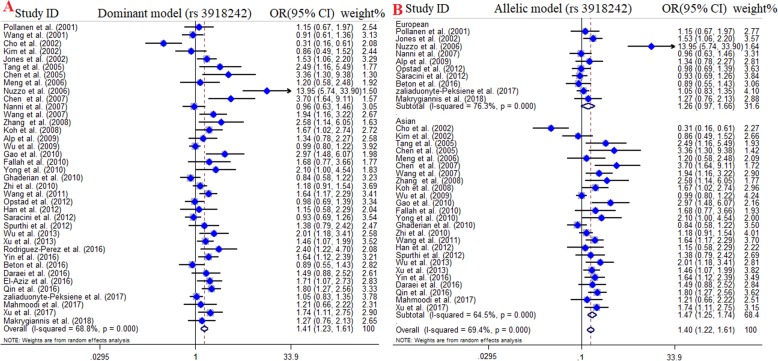
A total of 40 studies with 11,792 cases and 8280 controls were included in quantitative synthesis of the association between MMP-9 (C1562T) polymorphism and CAD susceptibility [16–20, 28–49, 51–61, 63, 64]. Among included studies, 26 studies were carried out in Asian countries, 10 studies were in European countries, 2 studies were in African countries, one study in America, and one in Oceania. Since there were only two studies for Africans and one study for American and Oceania, we excluded them from subgroup analysis. The pooled OR divulged a strong positive association between MMP-9 (C1562T) polymorphism and risk of CAD and announced this SNP as a risk factor for CAD. In details, dominant model (OR = 1.41, 95% CI = 1.23–1.61, P < 0.001), recessive model (OR = 1.59, 95% CI = 1.29–1.96, P < 0.001), allelic model (OR = 1.38, 95% CI = 1.23–1.55, P < 0.001), TT vs. CC model (OR = 1.70, 95% CI = 1.35–2.13, P < 0.001), and CT vs. CC model (OR = 1.35, 95% CI = 1.18–1.54, P < 0.001). FEM was used for recessive and homozygote compressions and REM was applied for dominant, heterozygote, and allelic models. Furthermore, the results of subgroup analysis by ethnicity remarkably showed that MMP-9 (C1562T) polymorphism increase the susceptibility of CAD in the Asian under all genotyping models; dominant model (OR = 1.47, 95% CI = 1.25–1.74, P < 0.001), recessive model (OR = 2.06, 95% CI = 1.57–2.71, P < 0.001), allelic model (OR = 1.45, 95% CI = 1.26–1.67, P < 0.001), TT vs. CC model (OR = 2.42, 95% CI = 1.77–3.32, P < 0.001), and CT vs. CC model (OR = 1.39, 95% CI = 1.19–1.64, P < 0.001) (Fig. 2). No statistically significant association was observed in Europeans.
Fig. 2.
Pooled odds OR and 95% confidence interval of individual studies and pooled data for the association between MMP-9 (C1562T) polymorphism and the risk of CAD in overall populations. a Dominat model, b Allelic model
The stratification of studies also performed based on type of CAD, including acute coronary syndrome (ACS) and stable angina. The findings demonstrated a statistically significant association between MMP9 (C1562T) polymorphism and stable angina susceptibility across all genotype model. However, the positive association between MMP-9 (C1562T) polymorphism and ACS susceptibility was observed in dominant model (OR = 1.66, 95% CI = 1.32–2.10, P < 0.001, REM), allelic model (OR = 1.57, 95% CI = 1.29–1.92, P < 0.001, REM), CT vs. CC model (OR = 1.62, 95% CI = 1.28–2.04, P < 0.001, REM), but not recessive model (OR = 1.32, 95% CI = 0.93–1.86, P = 0.12, FEM) and TT vs. CC model (OR = 1.40, 95% CI = 0.99–1.98, P = 0.06. The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
Table 3.
Main results of pooled ORs in meta-analysis of MMP9 gene polymorphisms and CAD risk
| Subgroup | Sample size | Test of association | Test of heterogeneity | Test of publication bias (Begg’s test) | Test of publication bias (Egger’s test) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic model | Case/Control | OR | 95% CI (p-value) | I2 (%) | P | Z | P | T | P | |
| MMP-9 (C1562T) | ||||||||||
| Overall | Dominant model | 11,792 / 8280 | 1.41 | 1.23–161 (≤0.001) | 68.8 | ≤0.001 | 1.48 | 0.13 | 1.62 | 0.11 |
| Recessive model | 11,792 / 8280 | 1.59 | 1.29–1.96 (≤0.001) | 18.3 | 0.19 | 1.34 | 0.17 | 1.49 | 0.14 | |
| Allelic model | 11,792 / 8280 | 1.38 | 1.23–1.55(≤0.001) | 68.7 | ≤0.001 | 1.39 | 0.16 | 1.54 | 0.13 | |
| TT vs. CC | 11,792 / 8280 | 1.70 | 1.35–2.13 (≤0.001) | 34 | 0.42 | 1.56 | 0.11 | 2.05 | 0.04 | |
| CT vs. CC | 11,792 / 8280 | 1.35 | 1.18–1.54 (≤0.001) | 65.4 | ≤0.001 | 1.48 | 0.13 | 1.62 | 0.11 | |
| Subgroup | ||||||||||
| Asian | Dominant model | 7483 / 5152 | 1.47 | 1.25–1.74 (≤0.001) | 64.5 | ≤0.001 | −0.72 | 0.47 | −0.14 | 0.88 |
| Recessive model | 7483 / 5152 | 2.06 | 1.57–2.71 (≤0.001) | 0 | 0.45 | 1.94 | 0.05 | 1.39 | 0.18 | |
| Allelic model | 7483 / 5152 | 1.45 | 1.26–1.67 (≤0.001) | 64.3 | ≤0.001 | 0.99 | 0.32 | 1.64 | 0.15 | |
| TT vs. CC | 7483 / 5152 | 2.42 | 1.77–3.32 (≤0.001) | 0 | 0.45 | 0 | 1 | −0.18 | 0.86 | |
| CT vs. CC | 7483 / 5152 | 1.39 | 1.19–1.64(≤0.001) | 60.8 | ≤0.001 | −0.25 | 0.80 | 0.15 | 0.88 | |
| European | Dominant model | 3230 / 2331 | 1.26 | 0.97–1.66 (0.08) | 76.3 | ≤0.001 | 0.78 | 0.45 | 0.84 | 0.43 |
| Recessive model | 3230 / 2331 | 1.05 | 0.75–1.47 (0.77) | 0 | 0.59 | 1.04 | 0.29 | 0.62 | 0.55 | |
| Allelic model | 3230 / 2331 | 1.22 | 0.97–1.53(0.08) | 75.5 | ≤0.001 | −1.73 | 0.08 | −0.69 | 0.51 | |
| TT vs. CC | 3230 / 2331 | 1.10 | 0.78–1.54 (0.59) | 32.1 | 0.15 | −0.21 | 0.83 | 0.2 | 0.82 | |
| CT vs. CC | 3230 / 2331 | 1.25 | 0.96–1.64 (0.09) | 74.1 | ≤0.001 | −0.25 | 0.80 | −0.68 | 0.52 | |
| ACS | Dominant model | 5862 / 4018 | 1.66 | 1.32–2.10 (≤0.001) | 76.1 | ≤0.001 | −0.25 | 0.80 | 0.89 | 0.39 |
| Recessive model | 5862 / 4018 | 1.32 | 0.93–1.86 (0.12) | 2.8 | 0.416 | −0.25 | 0.80 | −0.63 | 0.54 | |
| Allelic model | 5862 / 4018 | 1.57 | 1.29–1.92 (≤0.001) | 74.5 | ≤0.001 | −0.35 | 0.72 | −0.75 | 0.46 | |
| TT vs. CC | 5862 / 4018 | 1.40 | 0.99–1.98 (0.06) | 35.1 | 0.11 | 0.05 | 0.96 | −0.57 | 0.57 | |
| CT vs. CC | 5862 / 4018 | 1.62 | 1.28–2.04 (≤0.001) | 75 | ≤0.001 | −0.45 | 0.65 | −0.99 | 0.34 | |
| Stable | Dominant model | 5930 / 4262 | 1.26 | 1.07–1.48 (≤0.001) | 60.9 | ≤0.001 | 0.38 | 0.70 | 0.24 | 0.81 |
| Recessive model | 5930 / 4262 | 1.77 | 1.37–2.30 (≤0.001) | 23.6 | 0.18 | −0.12 | 0.90 | −0.42 | 0.68 | |
| Allelic model | 5930 / 4262 | 1.26 | 1.09–1.46 (≤0.001) | 63.9 | ≤0.001 | 0.12 | 0.90 | −0.49 | 0.63 | |
| TT vs. CC | 5930 / 4262 | 1.95 | 1.45–2.64 (≤0.001) | 31.9 | 0.10 | 1.57 | 0.11 | 14.14 | 0.04 | |
| CT vs. CC | 5930 / 4262 | 1.20 | 1.03–1.39 (0.01) | 52.2 | ≤0.001 | 0.52 | 0.60 | 0.38 | 0.76 | |
| MMP-9 (R279Q) | ||||||||||
| Overall | Dominant model | 5525 / 2497 | 0.92 | 0.83–1.02 (0.12) | 38.7 | 0.13 | 0.05 | 0.96 | −0.23 | 0.83 |
| Recessive model | 5525 / 2497 | 0.88 | 0.76–1.02 (0.08) | 0 | 0.48 | −0.18 | 0.85 | −0.21 | 0. 83 | |
| Allelic model | 5525 / 2497 | 0.93 | 0.86–1(0.05) | 38.1 | 0.13 | 0.05 | 0.96 | −0.06 | 0.95 | |
| GG vs. AA | 5525 / 2497 | 0.86 | 0.73–1.01(0.07) | 17.9 | 0.29 | 0.45 | 0.65 | 0.33 | 0.74 | |
| AG vs. AA | 5525 / 2497 | 0.94 | 0.85–1.05 (0.26) | 29.7 | 0.20 | 0.19 | 0.85 | −0.19 | 0.85 | |
| Subgroup | ||||||||||
| Asian | Dominant model | 4162/ 1923 | 0.93 | 0.83–1.04 (0.19) | 45.4 | 0.13 | −0.98 | 0.32 | −1.70 | 0.18 |
| Recessive model | 4162/ 1923 | 0.86 | 0.72–1.01 (0.06) | 36 | 0.19 | 0.56 | 0.57 | 0.37 | 0.73 | |
| Allelic model | 4162/ 1923 | 0.92 | 0.85–1 (0.06) | 59.6 | 0.06 | 0.09 | 0.92 | −0.03 | 0.97 | |
| GG vs. AA | 4162/ 1923 | 0.85 | 0.71–1.02 (0.08) | 53.7 | 0.09 | 1.16 | 0.24 | 0.92 | 0.38 | |
| AG vs. AA | 4162/ 1923 | 0.95 | 0.84–1.07 (0.41) | 15.3 | 0.31 | 1.34 | 0.18 | 1.58 | 0.15 | |
| European | Dominant model | 1363 / 574 | 0.91 | 0.74–1.13 (0.38) | 53.2 | 0.11 | 0.27 | 0.78 | 0.46 | 0.65 |
| Recessive model | 1363 / 574 | 0.96 | 0.70–1.32 (0.80) | 0 | 0.84 | 0.55 | 0.58 | 0.74 | 0.47 | |
| Allelic model | 1363 / 574 | 0.94 | 0.81–1.10 (0.45) | 10.1 | 0.32 | 0.27 | 0.78 | 0.10 | 0.92 | |
| GG vs. AA | 1363 / 574 | 0.90 | 0.64–1.26 (0.53) | 0 | 0.68 | 0 | 1 | 0.38 | 0.71 | |
| AG vs. AA | 1363 / 574 | 0.91 | 0.73–1.14 (0.39) | 58.9 | 0.08 | 0.52 | 0.60 | −0.47 | 0.72 | |
| MMP-9 (P574R) | ||||||||||
| Overall | Dominant model | 1272 / 785 | 1.05 | 0.72–1.53 (0.81) | 0.69 | 0.07 | * | * | * | * |
| Recessive model | 1272 / 785 | 1.01 | 0.69–1.49 (0.95) | 0 | 0.47 | * | * | * | * | |
| Allelic model | 1272 / 785 | 0.93 | 0.79–1.10 (0.41) | 0 | 0.41 | * | * | * | * | |
| RR vs. PP | 1272 / 785 | 0.97 | 0.55–1.44 (0.87) | 0 | 0.38 | * | * | * | * | |
| PR vs. PP | 1272 / 785 | 1.03 | 0.72–1.48 (0.87) | 63.7 | 0.09 | * | * | * | * | |
| MMP-9 (R668Q) | ||||||||||
| Overall | Dominant model | 1272 / 785 | 1.19 | 0.66–2.13 (0.56) | 88.3 | ≤0.001 | * | * | * | * |
| Recessive model | 1272 / 785 | 1.12 | 0.68–1.84 (0.64) | 21.2 | 0.26 | * | * | * | * | |
| Allelic model | 1272 / 785 | 1.11 | 0.73–1.69 (0.62) | 85.1 | 0.01 | * | * | * | * | |
| QQ vs. RR | 1272 / 785 | 1.26 | 0.55–2.89 (0.58) | 63.1 | 0.01 | * | * | * | * | |
| RQ vs. RR | 1272 / 785 | 1.18 | 0.68–2.06 (0.43) | 86.4 | ≤0.001 | * | * | * | * | |
*Begg’s and Egger’s test were not calculable
ACS acute coronary syndrome, OR odds ratio, CI confidence interval, MMP matrix metalloproteinase
Meta-analysis of MMP-9 (R279Q) and risk of CAD
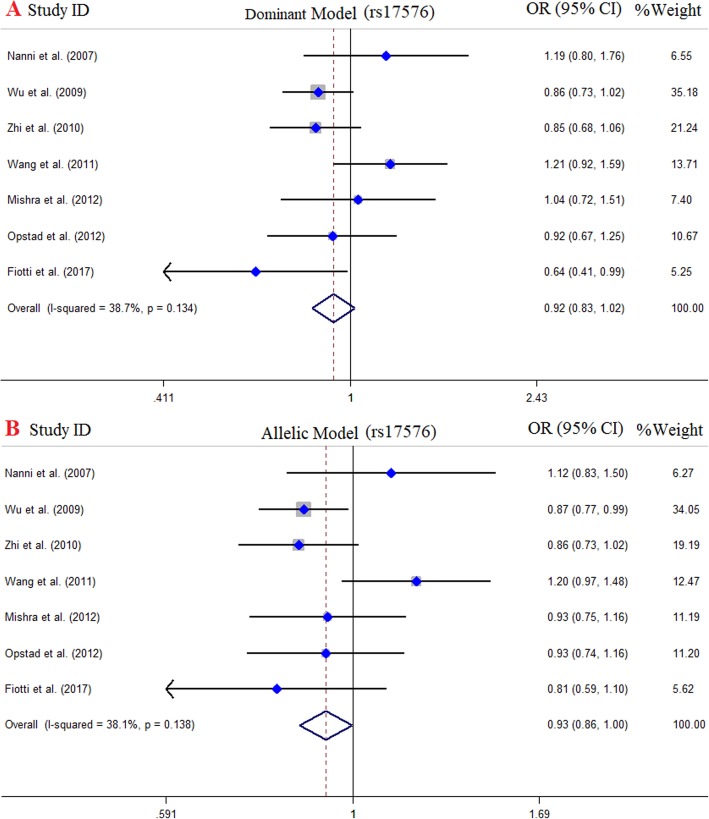
There were 7 case-control studies with 5525 cases and 2497 controls concerning MMP-9 (R279Q) polymorphism and risk of CAD [18, 38, 42, 47, 48, 50, 62]. Of those, 4 studies were performed in Asians and 3 studies were in Europeans. The pooled results indicated a negative, but not significant, association between MMP-9 (R279Q) gene polymorphism and CAD risk under all genotype models for the overall population and subgroup analysis (Fig. 3). The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
Fig. 3.
Pooled odds OR and 95% confidence interval of individual studies and pooled data for the association between MMP-9 (R279Q) polymorphism and the risk of CAD in overall populations. a Dominant model, b Allelic model
Meta-analysis of MMP-9 (P574R) and risk of CAD
For MMP-9 (P574R) SNP, two studies with 1272 case and 785 controls were included for quantitative analysis [47, 50]. Studies were carried out in China and India. The results of overall population reject any association between MMP-9 (P574R) SNP and risk of CAD across all genotype models. The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
Meta-analysis of MMP-9 (R668Q) and risk of CAD
Two studies with 1272 case and 785 controls were included for quantitative analysis for MMP-9 (R668Q) SNP [47, 50]. Studies were carried out in China and India. There was no evidence of significant association between MMP-9 (R668Q) SNP and risk of CAD under all genotype models. The results of pooled ORs, heterogeneity tests and publication bias tests in different analysis models are shown in Table 3.
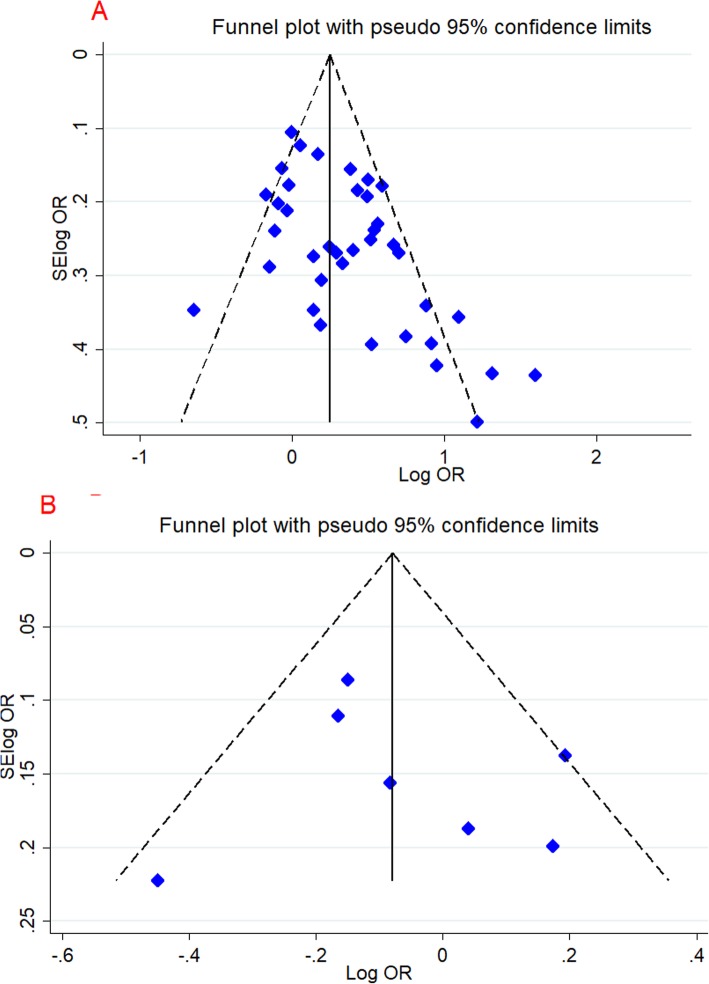
Publication bias and heterogeneity
In this study, we used Egger’s regression test, Begg’s adjusted rank correlation test and visual examination of the funnel plot (just for C1562T and R279Q) to measure publication bias (Fig. 4). Overall, no significant publication bias was detected. Besides, the estimation of heterogeneity by I2 and Q test was significant in some models (Table 3).
Fig. 4.
Begg’s funnel plot for publication bias test. Each point represents a separate study for the indicated association. a Dominant model (C1562T), b Dominant model (R279Q)
Sensitivity analysis
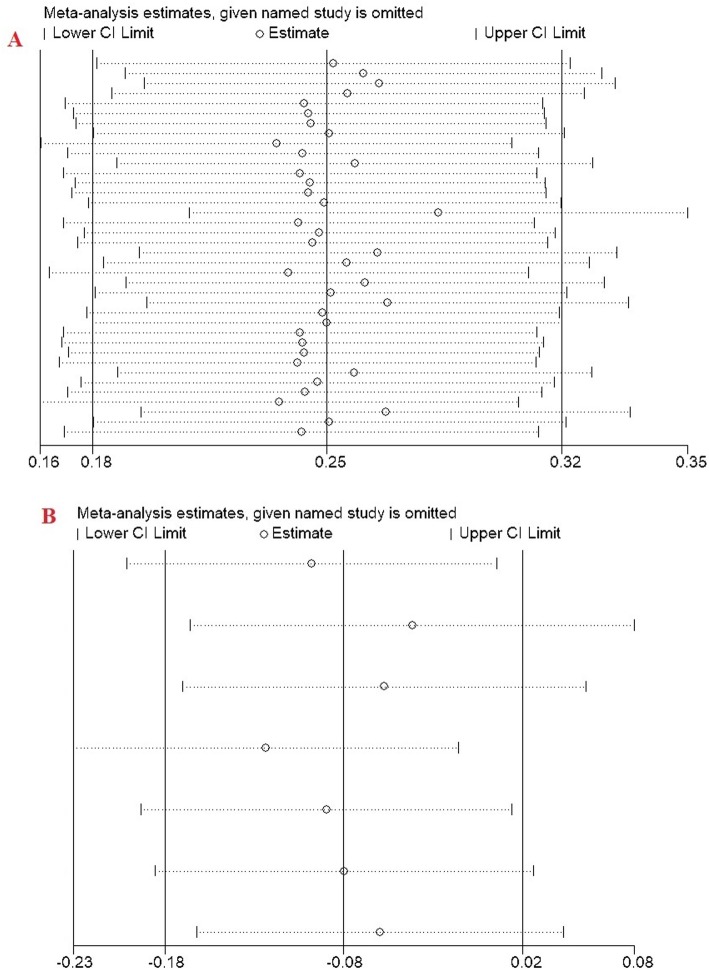
The leave-one-out method was used in the sensitivity analysis to explore the effect of individual data on the pooled ORs (just for C1562T and R279Q). The significance of ORs was not altered through omitting any single study, indicating that our results were statistically robust (Fig. 5).
Fig. 5.
Sensitivity analysis in present meta-analysis estimates the individual influence of studies on pooled results. a Dominant model (rs C1562T), b Dominant model (R279Q)
Discussion
Up until now, numerous investigations have been performed to address the association between SNPs of the MMP gene family, including MMP-9 (C1562T), MMP-9 (R279Q), MMP-9 (P574R), and MMP-9 (R668Q) polymorphisms and risk of CAD. The findings of these investigations have sometimes been in accordance with each other, but sometimes conflicting. By meeting the limitations of the individual studies, such as little statistical strength and small sample size, meta-analysis studies provide a beneficial tool to settle these limitations and confer a conclusive outcome. In order to solve this issue in respective of MMP-9 gene family polymorphisms and risk of CAD, here we carried out this meta-analysis by including the most comprehensive and up-to-date original studies worldwide to come up with valid approximation of this association. In the current meta-analysis, we included 40 studies with 11,792 cases and 8280 controls for MMP-9 (C1562T) SNP, 7 case-control studies with 5525 cases and 2497 controls for MMP-9 (R279Q) SNP, 2 studies with 1272 case and 785 controls for MMP-9 (P574R) SNP, and 2 studies with 1272 case and 785 controls for MMP-9 (R668Q) SNP. The analysis indicated that MMP-9 (C1562T) SNP was significantly associated with increased risk of CAD susceptibility in the overall analysis and Asian population particularly.
Different human and animal experiments have suggested that there is an elevated level of MMP-9 in the atherosclerotic arteries in comparison to health controls. It was also shown that MMP-9 is predominantly active in the lipid core margin of the atherosclerotic plaques, the shoulder regions of plaque, and in regions with active formation of microvessels. As a consequence, it appears that MMP-9 plays a critical role in the stability/instability of the coronary artery plaques and development of myocardial infarction during CAD [65]. In addition, researchers have observed that in mice knock-out for the MMP9 gene, migration potency of the VSMCs as well as atherosclerosis lesions were reduced in comparison to the wild-type animals [66]. According to clinical observations, upregulation of MMP-9 exhibited a correlation with instability of the atherosclerosis plaque and premature CAD development [67]. Based on the prospective studies, serum levels of MMP-9 could confer a tool to estimate the mortality risk during the cardiovascular diseases [68].
Zhang et al. in the 214 meta-analysis, by including 26 studies containing 12,776 cases and 6371 controls, indicated that MMP-9 (C1562T) polymorphism was not associated with the risk of CAD in the overall results [69]. However, they reported that MMP-9 (C1562T) SNP is involved in the decrease susceptibility to CAD in Asian population. In 2016, a meta-analysis was conducted on 10 case-control studies to assess the possible relationship between the MMP-9 (C1562T) SNP and CAD in the Chinese Han population. This study indicated that all genetic comparisons of the MMP-9 (C1562T) SNP increased the risk of CAD in the Chinese Han population [70]. In the current meta-analysis, association between SNPs of the MMP-9 gene family, including MMP-9 (C1562T), MMP-9 (R279Q), MMP-9 (P574R), and MMP-9 (R668Q) polymorphisms and risk of CAD was evaluated. Our literature search led to identification and inclusion of 40 studies with 11,792 cases and 8280 controls for MMP-9 (C1562T) SNP, 7 case-control studies with 5525 cases and 2497 controls for MMP-9 (R279Q) SNP, 2 studies with 1272 case and 785 controls for MMP-9 (P574R) SNP, and 2 studies with 1272 case and 785 controls for MMP-9 (R668Q) SNP. Therefore, this is the most comprehensive meta-analysis of MMP-9 gene family polymorphisms and risk of CAD to date (March 2020). Our analysis revealed that MMP-9 (C1562T) polymorphism increased the risk of CAD in the overall analysis under dominant (OR = 1.41), recessive (OR = 1.59), allelic (OR = 1.38), homozygous TT vs. CC (OR = 1.70), and heterozygous CT vs. CC (OR = 1.35) models. In contrast to Zhang et al. [69] study, we noticed that MMP-9 (C1562T) polymorphism increased the susceptibility of CAD risk in the Asian population under all genotyping models; dominant (OR = 1.47), recessive (OR = 2.06), allelic (OR = 1.45), homozygous TT vs. CC (OR = 2.42), and heterozygous CT vs. CC (OR = 1.39) models. However, other three polymorphisms of the MM9 gene, including MMP-9 (R279Q), MMP-9 (P574R), and MMP-9 (R668Q) polymorphisms, were not associated with CAD risk.
Regulatory mechanisms at the transcriptional level is involved in the modulation of MMP-9 expression. The MMP-9 (C1562T) SNP is harbored within the 9 bp sequence GCGCAC/TGCC (− 1567 → − 1559), which is considered as a regulatory element of the gene and confers a site for binding of molecules involved in the inhibition of transcription [71]. It was found that an alteration in the binding site structure by substitution of the MMP-9-1562 C allele with − 1562 T allele led to decreased binding potential of the proteins involved in the inhibition of transcription to the DNA sequence [30]. As a result, MMP-9 (C1562T) SNP plays a role in orchestrating the transcription activity of MMP-9 and, hence, modulate the susceptibility risk to several diseases. Therefore, we analyzed the available data to gain a wide understanding of this SNP in case of CAD. We noticed that T allele representation was increased in all models of MMP-9 (C1562T) SNP comparison in the overall analysis, including dominant model (OR = 1.41, 95% CI = 1.23–1.61, P < 0.001), recessive model (OR = 1.59, 95% CI = 1.29–1.96, P < 0.001), allelic model (OR = 1.38, 95% CI = 1.23–1.55, P < 0.001), TT vs. CC model (OR = 1.70, 95% CI = 1.35–2.13, P < 0.001), and CT vs. CC model (OR = 1.35, 95% CI = 1.18–1.54, P < 0.001), which was associated with an increased risk of CAD significantly. Upregulation of MMP-9 may be involved in the CAD development by multiple approaches, including increased proliferation and migration of VSMCs, remodeling of the injured vascular cells, and enhancing the plaque instability and rupture (that leads to the development of thrombosis), eventuating in myocardial infarction and CAD [72].
In spite of an attempt to perform the most comprehensive meta-analysis of the MMP9 gene SNPs and the risk of CAD, a number of limitations and caveats of this meta-analysis study should be taken into consideration. First, the number of studies and sample size for MMP-9 (R279Q), MMP-9 (P574R), and MMP-9 (R668Q) polymorphisms in this meta-analysis was relatively small to conclude a valid report of the association of these SNPs and CAD risk. Second, we searched for the articles published in only the English language and a number of potential studies might be omitted. Third, this meta-analysis was based on a crude analysis of the genetic polymorphisms, and the adjusting the analysis by gender, age, and other environmental factors were not implemented. Fourth, we detected some degrees of heterogeneity for the analyzed SNPs, that might stem from difference in genetic stratification and ethnicity, diversity in the environmental factors in different populations, and the detection methods.
Conclusion
Taken all the evidence into conclusion, this was the most comprehensive evaluation of the four MMP9 gene SNPs in association with CAD. We reported that MMP-9 (C1562T) SNP conferred a susceptibility risk for CAD in the overall analysis and Asian population. That notwithstanding, other three polymorphisms were not associated with disease risk, probably due to little sample size. Hence, we warrant further studies with respect to evaluation of other MMP9 gene SNPs in association with CAD. Furthermore, the role of other factors, such as age, gender, environmental contributing factors as well as other MMP9 gene variations in the analyses ahead will hopefully shed further light on the bona fide association of MMP9 gene polymorphisms and risk of CAD susceptibility.
Acknowledgements
The authors are grateful of Deputy of Research from Neyshabur University of Medical Science.
Abbreviations
- MMPs
Matrix metallo proteinases
- CAD
Coronary artery disease
- VSMCs
Vascular smooth muscle cells
- ECM
Extracellular matrix
- CI
Confidence interval
- OR
Odds ratio
- SNP
Single-nucleotide polymorphism
- PRISMA
Preferred reporting items for systematic reviews and meta-analyses
- NOS
Newcastle–Ottawa scale
- HWE
Hardy–Weinberg equilibrium
Authors’ contributions
RH and DI originated the study, acquired data. BR and SA performed statistical analysis, interpreted data, drafted the manuscript. ST revised the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data that support the conclusions of this manuscript are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Okrainec K, Banerjee DK, Eisenberg MJ. Coronary artery disease in the developing world. Am Heart J. 2004;148(1):7–15. doi: 10.1016/j.ahj.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 2.Sharma K, Gulati M. Coronary artery disease in women: a 2013 update. Glob Heart. 2013;8(2):105–112. doi: 10.1016/j.gheart.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Giannakoulas G, et al. Burden of coronary artery disease in adults with congenital heart disease and its relation to congenital and traditional heart risk factors. Am J Cardiol. 2009;103(10):1445–1450. doi: 10.1016/j.amjcard.2009.01.353. [DOI] [PubMed] [Google Scholar]
- 4.Mack M, Gopal A. Epidemiology, traditional and novel risk factors in coronary artery disease. Heart Fail Clin. 2016;12(1):1–10. doi: 10.1016/j.hfc.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Mayer B, Erdmann J, Schunkert H. Genetics and heritability of coronary artery disease and myocardial infarction. Clin Res Cardiol. 2007;96(1):1–7. doi: 10.1007/s00392-006-0447-y. [DOI] [PubMed] [Google Scholar]
- 6.Sanjadi M, et al. Atherosclerosis and autoimmunity: a growing relationship. Int J Rheum Dis. 2018;21(5):908–921. doi: 10.1111/1756-185X.13309. [DOI] [PubMed] [Google Scholar]
- 7.Evans A, et al. The genetics of coronary heart disease: the contribution of twin studies. Twin Res Hum Genet. 2003;6(5):432–441. doi: 10.1375/136905203770326439. [DOI] [PubMed] [Google Scholar]
- 8.Sayols-Baixeras S, et al. Pathogenesis of coronary artery disease: focus on genetic risk factors and identification of genetic variants. Appl Clin Genet. 2014;7:15. doi: 10.2147/TACG.S35301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onrat ST, et al. Prevalence of myocardial infarction polymorphisms in Afyonkarahisar, Western Turkey. Mol Biol Rep. 2012;39(9):9257–9264. doi: 10.1007/s11033-012-1799-1. [DOI] [PubMed] [Google Scholar]
- 10.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 11.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278(1):16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 12.Johnson JL, et al. Activation of matrix-degrading metalloproteinases by mast cell proteases in atherosclerotic plaques. Arterioscler Thromb Vasc Biol. 1998;18(11):1707–1715. doi: 10.1161/01.atv.18.11.1707. [DOI] [PubMed] [Google Scholar]
- 13.Galis ZS, et al. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1994;94(6):2493–2503. doi: 10.1172/JCI117619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayes-Genis A, et al. Pregnancy-associated plasma protein a as a marker of acute coronary syndromes. N Engl J Med. 2001;345(14):1022–1029. doi: 10.1056/NEJMoa003147. [DOI] [PubMed] [Google Scholar]
- 15.Bäck M, Ketelhuth DF, Agewall S. Matrix metalloproteinases in atherothrombosis. Prog Cardiovasc Dis. 2010;52(5):410–428. doi: 10.1016/j.pcad.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, et al. Impact of matrix metalloproteinase 9 rs3918242 genetic variant on lipid-lowering efficacy of simvastatin therapy in Chinese patients with coronary heart disease. BMC Pharmacol Toxicol. 2017;18(1):28. doi: 10.1186/s40360-017-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh YS, et al. A close relationship between functional polymorphism in the promoter region of matrix metalloproteinase-9 and acute myocardial infarction. Int J Cardiol. 2008;127(3):430–432. doi: 10.1016/j.ijcard.2007.04.107. [DOI] [PubMed] [Google Scholar]
- 18.Opstad TB, et al. Genetic variation, gene-expression and circulating levels of matrix metalloproteinase-9 in patients with stable coronary artery disease. Clin Chim Acta. 2012;413(1–2):113–120. doi: 10.1016/j.cca.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Mahmoodi K, et al. Plasma concentration, genetic variation, and gene expression levels of matrix metalloproteinase 9 in Iranian patients with coronary artery disease. J Res Med Sci. 2017;22(8):357-62. [DOI] [PMC free article] [PubMed]
- 20.Rodríguez-Pérez J, et al. rs3918242 MMP9 gene polymorphism is associated with myocardial infarction in Mexican patients. Genet Mol Res. 2016;15(1):15017776. doi: 10.4238/gmr.15017776. [DOI] [PubMed] [Google Scholar]
- 21.Stroup DF, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 22.Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 26.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;8(1):1088–101. [PubMed]
- 27.Egger M, et al. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pöllänen PJ, et al. Coronary artery complicated lesion area is related to functional polymorphism of matrix metalloproteinase 9 gene: an autopsy study. Arterioscler Thromb Vasc Biol. 2001;21(9):1446–1450. doi: 10.1161/hq0901.095545. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, et al. Polymorphism in the gelatinase B gene and the severity of coronary arterial stenosis. Clin Sci. 2001;101(1):87–92. [PubMed] [Google Scholar]
- 30.Cho H-J, et al. Functional polymorphism in the promoter region of the gelatinase B gene in relation to coronary artery disease and restenosis after percutaneous coronary intervention. J Hum Genet. 2002;47(2):88–91. doi: 10.1007/s100380200006. [DOI] [PubMed] [Google Scholar]
- 31.Kim JS, et al. The roles of stromelysin-1 and the gelatinase B gene polymorphism in stable angina. Yonsei Med J. 2002;43(4):473–481. doi: 10.3349/ymj.2002.43.4.473. [DOI] [PubMed] [Google Scholar]
- 32.Jones GT, et al. Functional matrix metalloproteinase-9 polymorphism (C-1562T) associated with abdominal aortic aneurysm. J Vasc Surg. 2003;38(6):1363–1367. doi: 10.1016/s0741-5214(03)01027-9. [DOI] [PubMed] [Google Scholar]
- 33.Chen XF, Tang L, Zhu M, Jiang JJ, Shen WF, Du YX. Matrix metalloproteinase-9 polymorphism (C1562T) and the susceptibility to myocardial infaction in Han population of China. Chin J Arterioscler. 2005;13:775–778. [Google Scholar]
- 34.Tang L, et al. Study of relations between matrix metalloproteinase-9 polymorphism (C-1562T) and acute coronary syndrome in Han population of China. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22(3):313–316. [PubMed] [Google Scholar]
- 35.Meng D, Mao Y, Chen Q. Relationship between polymorphisms of matrix metalloproteinase and coronary heart disease. Tianjin Med J. 2006;34(5):295. [Google Scholar]
- 36.Nuzzo D, et al. Role of Proinflammatory alleles in longevity and atherosclerosis: results of studies performed on− 1562C/T MMP-9 in centenarians and myocardial infarction patients from Sicily. Ann N Y Acad Sci. 2006;1089(1):496–501. doi: 10.1196/annals.1386.048. [DOI] [PubMed] [Google Scholar]
- 37.Chen B, et al. The relationship between matrix metalloproteinase-9 polymorphism (C1562T) and acute coronary syndrome. Chin J Arterioscler. 2007;15(3):209. [Google Scholar]
- 38.Nanni S, et al. Matrix metalloproteinases in premature coronary atherosclerosis: influence of inhibitors, inflammation, and genetic polymorphisms. Transl Res. 2007;149(3):137–144. doi: 10.1016/j.trsl.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Zhu T, Li Y. Relationship between matrix metalloproteinase-9 polymorphism and acute coronary syndrome. J Nanjing Med Univ. 2007;21(3):147–150. [Google Scholar]
- 40.Zhang, Y., et al., Relationship between matrix metalloproteinase-2/9 polymorphism and susceptibility to premature coronary heart disease/基质金属蛋白酶 2, 9 基因多态性与早发冠心病遗传易感性的研究. Xi'an jiao tong da xue xue bao. Yi xue ban, 2010;4: 429.
- 41.Alp E, et al. Lack of association between matrix metalloproteinase-9 and endothelial nitric oxide synthase gene polymorphisms and coronary artery disease in Turkish population. DNA Cell Biol. 2009;28(7):343–350. doi: 10.1089/dna.2009.0866. [DOI] [PubMed] [Google Scholar]
- 42.Wu N, et al. Haplotype analysis of the stromelysin-1 (MMP3) and gelatinase B (MMP9) genes in relation to coronary heart disease. Ann Hum Genet. 2009;73(4):404–410. doi: 10.1111/j.1469-1809.2009.00522.x. [DOI] [PubMed] [Google Scholar]
- 43.Fallah S, et al. Matrix metalloproteinase-9 and paraoxonase 1 Q/R192 gene polymorphisms and the risk of coronary artery stenosis in Iranian subjects. J Clin Lab Anal. 2010;24(5):305–310. doi: 10.1002/jcla.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao C, Wang Y. Research on the matrix metalloproteinase-9 gene polymorphism and risk of coronary heart disease. Mod Prev Med. 2010;37(2):328–332. [Google Scholar]
- 45.Ghaderian SMH, Najar RA, Panah AST. Genetic polymorphisms and plasma levels of matrix metalloproteinases and their relationships with developing acute myocardial infarction. Coron Artery Dis. 2010;21(6):330–335. doi: 10.1097/MCA.0b013e32833ce065. [DOI] [PubMed] [Google Scholar]
- 46.Yong F, Shi Y. The polymorphism in the MMP-9 gene in relation to coronary heart disease. J Zhejiang Univ Sch Med. 2010;16:1017–1032. [Google Scholar]
- 47.Zhi H, et al. Functional polymorphisms of matrix metallopeptidase-9 and risk of coronary artery disease in a Chinese population. Mol Biol Rep. 2010;37(1):13–20. doi: 10.1007/s11033-009-9482-x. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, et al. Interaction between MMP-9 gene polymorphisms and smoking in relation to myocardial infarction in a Uighur population. Clin Appl Thromb Hemost. 2012;18(1):72–78. doi: 10.1177/1076029611412365. [DOI] [PubMed] [Google Scholar]
- 49.Han Y, Zhang J, Su M. Correlation between the matrix metalloproteinase-9 gene single nucleotide polymorphim and coronary artery stenosis degree. Chin J Gerontol. 2012;1:5–6. [Google Scholar]
- 50.Mishra A, et al. Association of matrix metalloproteinases (MMP2, MMP7 and MMP9) genetic variants with left ventricular dysfunction in coronary artery disease patients. Clin Chim Acta. 2012;413(19–20):1668–1674. doi: 10.1016/j.cca.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 51.Saracini C, et al. Polymorphisms of genes involved in extracellular matrix remodeling and abdominal aortic aneurysm. J Vasc Surg. 2012;55(1):171–179. e2. doi: 10.1016/j.jvs.2011.07.051. [DOI] [PubMed] [Google Scholar]
- 52.Spurthi KM, et al. Influence of gelatinase B polymorphic variants and its serum levels in atherosclerosis. Genet Test Mol Biomarkers. 2012;16(8):850–854. doi: 10.1089/gtmb.2011.0299. [DOI] [PubMed] [Google Scholar]
- 53.Sewelam NI, et al. Association between the polymorphisms of matrix metalloproteinases 9 and 3 genes and risk of myocardial infarction in Egyptian patients. Egypt J Med Hum Genet. 2013;14(2):43–48.
- 54.Wu H-D, et al. Association of genetic polymorphisms in matrix Metalloproteinase-9 and coronary artery disease in the chinese han population: A case–control study. Genet Test Mol Biomarkers. 2013;17(9):707–712. doi: 10.1089/gtmb.2013.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X, Wang L, Xu C, Zhang P, Yong F, Liu H, Wang J, Shi Y. Variations in matrix metalloproteinase-1, −3, and −9 genes and the risk of acute coronary syndrome and coronary artery disease in the Chinese Han populatio. Pathophysiol Nat Hist. 2013;24:259–265. doi: 10.1097/MCA.0b013e32835ea3af. [DOI] [PubMed] [Google Scholar]
- 56.Beton O, et al. Association between MMP-3 and MMP-9 polymorphisms and coronary artery disease. Biomed Rep. 2016;5(6):709–714. doi: 10.3892/br.2016.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin L, et al. Association between matrix metalloproteinase-9 rs3918242 polymorphism and development of coronary artery disease in a Chinese population. Genet Mol Res. 2016;15(2):2–8. [DOI] [PubMed]
- 58.Yin H, et al. Polymorphism in matrix metalloproteinase-9 1562 C/T contributes to the risk of coronary artery disease. Int J Clin Exp Pathol. 2016;9(2):2277–2282. [Google Scholar]
- 59.Daraei A, et al. Influences of IL-1b-3953 C> T and MMP-9-1562C> T gene variants on myocardial infarction susceptibility in a subset of the Iranian population. Genet Test Mol Biomarkers. 2017;21(1):33–38. doi: 10.1089/gtmb.2016.0240. [DOI] [PubMed] [Google Scholar]
- 60.El-Aziz TAA, Mohamed RH. Matrix metalloproteinase-9 polymorphism and outcome after acute myocardial infarction. Int J Cardiol. 2017;227:524–528. doi: 10.1016/j.ijcard.2016.10.109. [DOI] [PubMed] [Google Scholar]
- 61.Žaliaduonytė-Pekšienė D, et al. Association of the genetic and traditional risk factors of ischaemic heart disease with STEMI and NSTEMI development. J Renin-Angiotensin-Aldosterone Syst. 2017;18(4):1470320317739987. doi: 10.1177/1470320317739987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiotti N, et al. Multiple sites of vascular dilation or aneurysmal disease and matrix metalloproteinase genetic variants in patients with abdominal aortic aneurysm. J Vasc Surg. 2018;67(6):1727–1735. doi: 10.1016/j.jvs.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 63.Makrygiannis G, et al. Risk factor assessment in a Greek cohort of patients with large abdominal aortic aneurysms. Angiology. 2019;70(1):35–40. doi: 10.1177/0003319718774474. [DOI] [PubMed] [Google Scholar]
- 64.Malkani N, et al. Matrix metalloprotease-9 polymorphism and its association with Atherosclerosis—A case-control study in Pakistani population. Population. 2019;69(1416):1416–20. [PubMed]
- 65.Speidl WS, et al. The complement component C5a is present in human coronary lesions in vivo and induces the expression of MMP-1 and MMP-9 in human macrophages in vitro. FASEB J. 2011;25(1):35–44. doi: 10.1096/fj.10-156083. [DOI] [PubMed] [Google Scholar]
- 66.Ye S. Influence of matrix metalloproteinase genotype on cardiovascular disease susceptibility and outcome. Cardiovasc Res. 2006;69(3):636–645. doi: 10.1016/j.cardiores.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 67.Jones GT, et al. Elevated plasma active matrix metalloproteinase-9 level is associated with coronary artery in-stent restenosis. Arterioscler Thromb Vasc Biol. 2006;26(7):e121–e125. doi: 10.1161/01.ATV.0000226544.93089.7f. [DOI] [PubMed] [Google Scholar]
- 68.Blankenberg S, et al. Plasma concentrations and genetic variation of matrix metalloproteinase 9 and prognosis of patients with cardiovascular disease. Circulation. 2003;107(12):1579–1585. doi: 10.1161/01.CIR.0000058700.41738.12. [DOI] [PubMed] [Google Scholar]
- 69.Zhang F-X, et al. Association between− 1562C> T polymorphism in the promoter region of matrix metalloproteinase-9 and coronary artery disease: a meta-analysis. Genet Test Mol Biomarkers. 2014;18(2):98–105. doi: 10.1089/gtmb.2013.0369. [DOI] [PubMed] [Google Scholar]
- 70.Li Y-Y, et al. Matrix metalloproteinase-9 gene-1562C> T gene polymorphism and coronary artery disease in the Chinese Han population: a meta-analysis of 5468 subjects. Front Physiol. 2016;7:212. doi: 10.3389/fphys.2016.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang B, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99(14):1788–1794. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- 72.Galis ZS, et al. Targeted disruption of the matrix metalloproteinase-9 gene impairs smooth muscle cell migration and geometrical arterial remodeling. Circ Res. 2002;91(9):852–859. doi: 10.1161/01.res.0000041036.86977.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that support the conclusions of this manuscript are included within the article.