To the editor: We introduce a web-enabled mass spectrometry (MS) search engine named MASST (Mass Spectrometry Search Tool) (https://proteosafe-extensions.ucsd.edu/masst/). By enabling searches of all small-molecule tandem MS data in public metabolomics repositories, we posit that MASST will unlock these resources for clinical, environmental and natural product applications.
Introduced in 1990, a tool for discovering related protein or gene sequences, named Basic Local Alignment Search Tool (BLAST) enabled researchers to query entire public sequence data repositories through a web interface (WebBLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi).1 WebBLAST is one of the most widely cited and used bioinformatics tools because it permits any researcher to answer simple questions such as ‘is a protein or DNA sequence common or rare? In the early days of public gene and protein databases, metadata, which includes descriptions of sample, population or technical details was limited and no deposition standards existed, but short read archive and European nucleotide archive that includes experimental details for sequencing, instrumental details, and sample description such as source of sample. The current status of much of the mass spectrometry data in the public domain is reminiscent of the DNA databanks of the 1990s. In order to increase usage and unlock the potential of openly available MS resources, we set out to build an infrastructure to enable ‘WebBLAST for MS’.
Algorithms developed for mass spectrometry data, including molecular networking2 and fragmentation trees3, enable similarity searches against reference libraries of known molecules, while powerful metabolomics analysis software infrastructures, such as MS-DIAL4, MetaboAnalyst5, XCMS Online6, HMDB7 focus on annotation of MS/MS spectra, or finding statistical relationships between molecular features. However, none of the existing tools enable searching a single MS/MS spectrum for identical or analogous MS/MS spectra against public data in repositories, including unknown molecules. Finding specific MS/MS spectra of interest, including unannotated spectra or structural analogs, in public metabolomics’ and natural product’s mass spectrometry data repositories, is not possible. Deposition of untargeted mass spectrometry data in the public domain is experiencing rapid growth. In March, 2017 there were 910 metabolomics datasets available8 but in January 2019 there were more than 2,000 downloadable metabolomics datasets (about half of these datasets contain MS/MS data).9 Despite the availability of metabolomics and natural products data, including environmental and clinical mass spectrometry datasets, public small molecule mass spectrometry data is hardly reused.10 Now that there is a huge amount of small molecule untargeted mass spectrometry datasets publicly available (~1,100 untargeted datasets and ~110,000,000 spectra in ~150,000 files as of Dec 11, 2018) we felt that the time was right to develop MASST, to enable reuse of these mass spectrometry data.
MASST comprises a web-based system to search the public data repository part of the GNPS/MassIVE knowledge base11 and an analysis infrastructure for a single MS/MS spectrum. The developments required for MASST searches included converting deposited public data to a uniform open format12 (irrespective of instrument type and original data format), the ability to trace the file from which each MS/MS spectrum originated, and a reporting system that shows all identical or similar MS/MS spectra found in public data along with their associated metadata. Reasons why MASST development is possible include increased adoption of universal, non-vendor specific MS data formats, which means that multiple publicly available datasets have been converted to the same data format13, and a recently developed ability to connect all public data in GNPS/MassIVE and connect each MS/MS spectrum to its metadata entries had not been developed yet.
A MASST report also includes matches to any reference spectra in public MS/MS spectral libraries, if the matches are within the user-specified search parameters. Libraries include GNPS user contributed spectra11, GNPS libraries11, all three MassBanks14,15,16, ReSpect17, MIADB/Beniddir18, Sumner/Bruker, CASMI19, PNNL lipids20, Sirenas/Gates, EMBL MCF and several other libraries that can be found here: https://gnps.ucsd.edu/ProteoSAFe/libraries.jsp. Visualization of the MASST matches uses a mirror view (Figure 1c).
Figure 1. MASST search, reporting and match visualization.
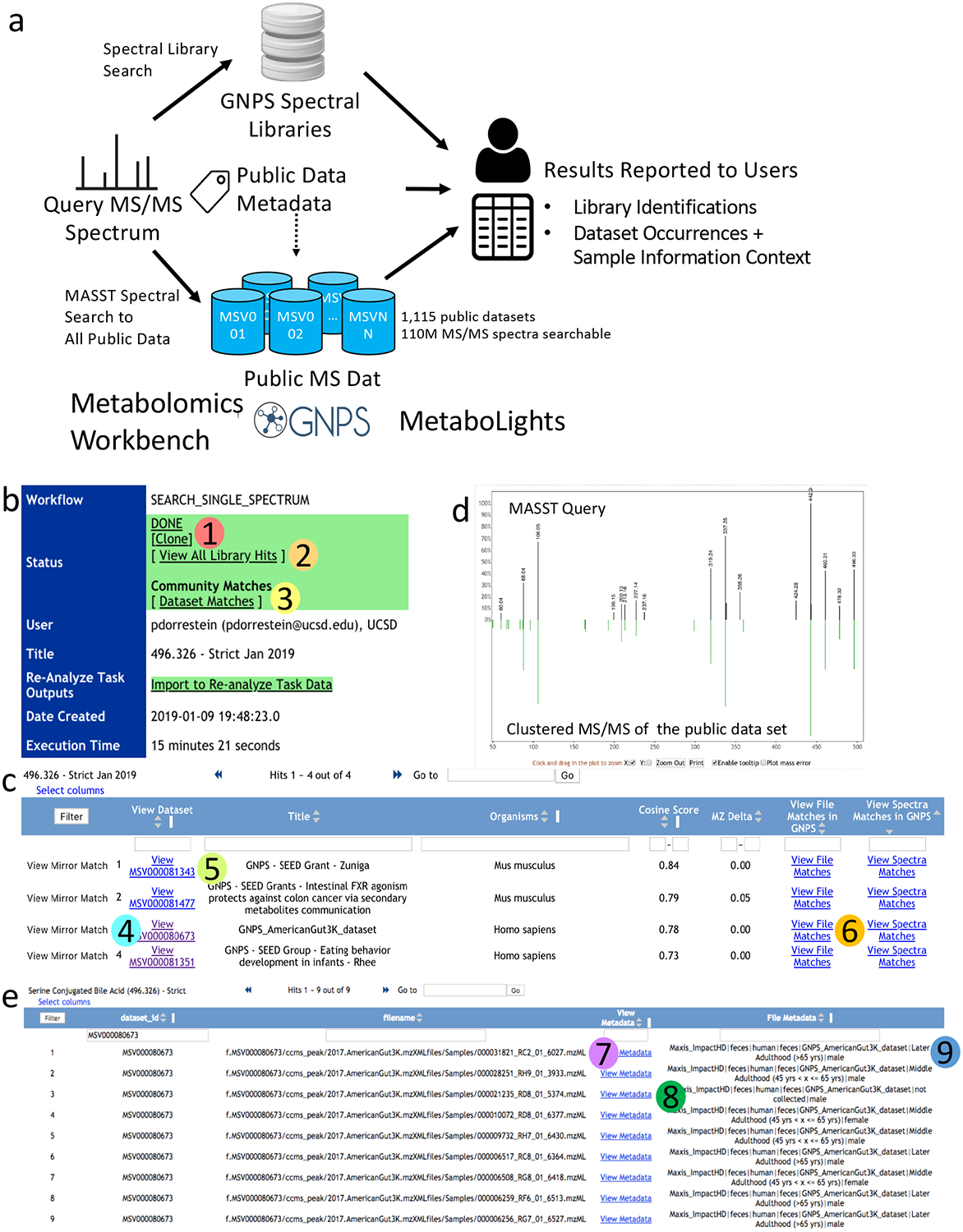
a. Overview of MASST query procedure. MASST queries MS/MS spectra against all public metabolomics data, including spectra deposited in GNPS, Metabolomics Workbench and Metabolights. Combining these matches with sample information provides users with a report containing MS/MS compound annotation and MS/MS sample information (metadata). Once a MASST search is completed at https://proteosafe-extensions.ucsd.edu/masst/, the results can be found in the user’s job tab or using a link provided over email. b. The opening page is shown. There are two options (2 and 3) for inspecting the data and additional options for cloning a job (1). Clicking (2) will reveal all MS/MS spectral matches within the user defined settings. There can be none, one, or more than one match for a given input spectrum. c. Clicking (3) will reveal all data sets that contain an MS/MS spectrum that has a match to the input spectrum and any associated metadata. d. clicking on “View Mirror Match” (4) shows the mirror match between the input spectrum and the merged MS/MS spectrum enabling manual inspection of a match; “View MSV0000…..” (5) brings the user to the data set: all uploaded information associated with this data set can be found or is linked in this location. (6) Opens up the file information window and tabulated metadata. (7) shows the files where MS/MS matches are found and (8) Links-out to full sample information for the file. (9) Displays the abbreviated (and filterable) sample information associated with the files. If no sample information has been uploaded with the original data, then this field will be blank. The MASST_GNPS job link for this search to enable the reader to navigate the same results can be found here. https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=bac3d3788e704af59e4a15a5146e4d6b
MASST can search against various repositories including GNPS/MassIVE11, Metabolomics Workbench21, Metabolights22 or the non-redundant (nr) MS/MS library of all unique MS/MS spectra from all three repositories combined. MASST searching using multiple repositories was enabled by converting data uploaded to the Metabolomics Workbench and MetaboLights repositories to the same open mass spectrometry format in the GNPS/MassIVE data storage environment. Instructions on how to upload to GNPS/MassIVE can be found here https://ccms-ucsd.github.io/GNPSDocumentation/datasets/. All public data in GNPS/MassIVE becomes MASST searchable. MASST searches output results according to a user-defined search parameters. The report returns the origin of the matched MS/MS spectrum with respect to the dataset and file information, and any metadata associated with the file (Figure 1 a–e). Further, datasets and files can be tagged with sample or spectral information by the community of MASST users, and this information then becomes part of the metadata reported back in future MASST searches. We have also curated approximately 34,000 additional mass spectrometry files with ~340,000 tags, mostly from human-associated samples, but also from microbes, food and indoor and outdoor environments, to provide a good foundation for MASST searches.
Metadata can be associated with MS/MS spectra in the GNPS/MassIVE upload portal at the dataset level, file level or single annotated spectrum level. Examples of metadata include instrument type, phylogeny (according to NCBI taxonomy) and keywords at the dataset level, phylogeny, sample type, age, sex, body site (defined using the Uberon anatomy ontology23), and disease24 at the file level, and source, biological activity, and structural class information at the single annotated spectrum level. In addition, GNPS/MassIVE is compatible with metadata formats from other software tools, e.g. QIIME2 and Qiita, which are used to analyze microbiome data and have a controlled vocabulary that can be imported.25,26 Further, any sample information uploaded to GNPS/MassIVE from another repository, e.g. from Metabolights and Metabolomics workbench is also included in the MASST report.
At present there is only limited metadata at the dataset and file level, but the metadata in the public domain can provide insights into the types of MS/MS signals being analysed (Box 1 contains examples of usage). Although the amount and quality of metadata is increasing27, datasets do not always have detailed metadata. To allay this problem, re-annotation of metadata as knowledge increases, while retaining provenance of all changes, is possible in GNPS.11 If insufficient metadata is available for interpretation of a public dataset search results, the original depositors of the public data can be contacted. We expect this feature in MASST to foster collaborations worldwide.
Box 1. Ten example applications and questions that can be addressed with MASST.
1). Are specific molecular features detected via mass spectrometry in one clinical cohort also observed elsewhere?
Human studies of disease vs healthy cohorts are confounded by different exercise, diet, medications in the diseased cohort vs healthy cohort. MASST-ing an MS/MS feature found to be differentiated with non-alcoholic fatty acid liver disease (NAFLD) in people revealed the same MS/MS could be found in other liver disease studies. The video describing this MASST search can be found here https://youtu.be/sHHlVTCoQJY. An expanded description of the MASST and discovery of the new bile acid can be found in the supplementary note.
2). Can findings about a molecule identified in model organism studies be translated to humans?
One major application expected will be the translation of molecular information from animal models to humans. A MASST-ing the MS/MS of a mass spectrometry feature that was differentiated in a mouse model infected with lymphocytic choriomeningitis virus Armstrong resulted in the discovery that a new molecule, cholylserine, is also found in human studies. The details of this MASST job can be found here https://youtu.be/SExVUrD56-s, and supplementary note.
3). Can MASST be used to reveal the presence and distribution of environmental toxins?
In this example it was revealed that domoic acid, the neurotoxin poison that became famous through the novel “The Birds” by Daphne du Maurier and a film from Alfred Hitchcock as it caused seagulls to attack humans, was found in seven different public datasets, including San Diego, Narragansett Bay and Hawaii. A description of the results of this MASST analysis job can be found here https://youtu.be/vm6UkYwDGn4 and supplementary note.
4). In what datasets can we find a published MS/MS spectrum?
Here a published MS/MS spectrum was searched. A description of MASST using the MS/MS of 3-hydroxyhexadecanoyl glycine and 3-hydroxypentadecanoyl lysine, both N-acyl lipids, suggests that these molecules have a very wide ecological distribution can be found here https://youtu.be/8W2BCxtszIA and supplementary note.
5). Are specific natural products observed in cultured microbes also observed in non-laboratory settings?
An example using orfamides revealed four datasets that contained this molecular ion including field-collected Trachymyrmex septentrionalis fungus gardens. More detail can be found in this video https://youtu.be/4Zb5gZIabBU and supporting note.
6). Where do we find agricultural fungicides in the environment? Is there evidence that people may be in contact with these fungicides?
A MASST search with the MS/MS spectrum of azoxystrobin, a fungicide can be found here https://youtu.be/hGemmjdeOY0 and supporting note.
7). Are known toxins from food found in/on people?
A MASST search with the MS/MS spectrum of the mycotoxin roquefortine C it was found in human stool (infants and adults). This MASST search is described in more detail in this video https://youtu.be/04RSsOY0oGM and supporting note.
8). Can we use approximate matches to a natural product to find datasets that may contain analogs?
A for staurosporine derivatives among the public datasets with MASST took less than 15 min, and suggests that there are still yet to be discovered reservoirs of unique staurosporine derivatives as shown in this video https://youtu.be/04RSsOY0oGM and supporting note.
9). Can MASST be used to track sunscreens in human and environmental samples?
A MASST search of the MS/MS spectra of two active ingredients of sunscreen - avobenzone and octocrylene – reveals, as expected, their presence in many human skin datasets, personal objects, meat for human consumption, corals, and even in coral reef in remote areas such as Moorea. This MASST analysis job is described here https://youtu.be/Sjv00dpMSQ8 and supporting note.
10). Can we find evidence of opioids exposure in public data?
By searching the MS/MS of methadone and cocaine using MASST, we found a matching MS/MS spectrum in five datasets. A description with this MASST analysis job can be found here https://youtu.be/9hTsXJ6l1Is and supporting note.
MASST can be accessed at https://proteosafe-extensions.ucsd.edu/masst/,mas by copying/pasting the MS/MS spectrum peak list reported as m/z and intensity separated by a space for each fragment ion (aka product ion) that can also be extracted from the open mass spectrometry formats, such as .mzML, .mzXML, .MGF. Finally, MASST can be accessed as part of a GNPS data analysis. Manual entry at https://proteosafe-extensions.ucsd.edu/masst/provides researchers with the ability to enter data from theoretical spectra, or spectra from published papers or supporting information, without needing access to the original experimental data. In GNPS users can launch a MASST search using links provided in classical and feature based molecular networking output created within the GNPS infrastructure11, which automatically redirects to the MASST search page with prepopulated spectral data by clicking a simple MASST spectrum button. The MS/MS spectrum provided via the MASST website or as a link-out from a GNPS search is then searched against all public data with user defined parameters of minimum number of ions to match, precursor (parent) and product (fragment) ion tolerances, and analog similarity searches based on non-identical precursor masses.2 An instruction video for running MASST jobs is available https://youtu.be/4yBKomKzEKU. MASST searches retrieve all associated sample information (dataset and files) that match the MS/MS input spectrum query. A typical search takes about 10–20 min. Multiple searches queries are placed in a queue for parallel execution as resources become available.
To promote data analysis reproducibility, the results of every job are stored in each user’s space and can be found under the “Jobs” tab accessible through the banner in the GNPS browser (http://gnps.ucsd.edu). Only MASST jobs run while logged in to GNPS will be retained. Search parameters are also retained with each job and constitute a provenance record that can be provided as hyperlinks to share with others, e.g. collaborators, or in publications. These jobs can be shared, cloned and rerun with or without alterations of the input parameters (examples of links to jobs are shown in Box1). This feature could enable new matches to be made when relevant public data are uploaded. The matches of MS/MS spectra among datasets are the equivalent to level two (putative annotation based on spectral library similarity) or three (putatively characterized compound class based on spectral similarity to known compounds of a chemical class) according to the 2007 metabolomics standards initiative28. Similar to short sequence reads, MASST searches will not distinguish chemicals that have nearly identical fragmentation patterns, such as isomeric compounds, which would require an authentic standard and the use of an orthogonal property (such as the retention time). In cases when a MASST search returns no matches, its possible that either there is no matching data or that MS/MS matches are possible but fall outside the specified search parameters. MASST should be used with these caveats in mind.
MASST, like WebBLAST, will likely find broad application. Uses of MASST might include translation of in vitro or in vivo data from model organisms to humans, or broad ecological questions. Box 1 contains ten example uses to highlight the types of discoveries possible with access, via MASST, to the entire body of public MS/MS data. These examples are illustrative, and we expect the user community to find multiple, innovative ways to use MASST.
Supplementary Material
Acknowledgements:
Conversion of data from different repositories was supported by R03 CA211211 on reuse of metabolomics data. The development of a user-friendly interface was in part supported by Gordon and Betty Moore Foundation through Grant GBMF7622. The UC San Diego Center for Microbiome Innovation supported the campus wide SEED grant awards for data collection that enabled the development of much of this infrastructure. AKJ thanks the American Society for Mass Spectrometry for the 2018 Postdoctoral Career Development Award. We further acknowledge Claire O’Donovan and Kenneth Haug for help with navigating the MetaboLights data repository. JVDH was supported by a ASDI eScience grant (ASDI.2017.030) from the Netherlands eScience Center (NLeSC). EIZ and LLB were supported by NIH grants AI081923 and AI113923. AMCR, KEK, SPP, JLK, MJB, and PCD were supported by NSF grant IOS-1656475. AB was supported by National Institute of Justice Award 2015-DN-BX-K047. FV was supported by the Department of Navy, Office of Naval Research Multidisciplinary University Research Initiative (MURI) Award, Award number N00014-15-1-2809. DP was supported by the German Research Foundation (DFG) with Grant PE 2600/1. Additional support for data acquisition and data storage was provided by P41 GM103484 Center for Computational Mass Spectrometry, Instrument support though NIH S10RR029121, RL is supported by NIH grants R01DK106419, 5P42ES010337, and 5UL1TR001442, NIH K01DK116917 to J.D.W. The development of the web interface and harmonization with Qiita was in part supported by the Sloan Foundation.
Footnotes
Data availability: All data used for testing and validating MASST is deposited in GNPS/MassIVE. MASST is a web-based application that is embedded in GNPS, which is a community service in which all public data is public. We are not able to provide server installation, software engineers or administrator support for individual installations of MASST. The MASST platform is built as a workflow on top of the web repository workflow platform ProteoSAFe (https://github.com/CCMS-UCSD/ProteoSAFe). Each step of the MASST query is written in Python. Web rendering of the results is displayed by ProteoSAFe in the browser.
Code availability: For those who wish to build out MASST and recruit their own programmers, software engineers and system administrators we have deposited the code at github https://github.com/CCMS-UCSD/GNPS_Workflows/tree/master/search_single_spectrum. The standalone MASST query interface is written in Python and Flask with a web front end written in HTML and javascript. It is open source: https://github.com/mwang87/GNPS_MASST and released under an LGPL-3 license.”
Competing interest: Mingxun Wang is the founder of Ometa and consults for Sirenas and Pieter C. Dorrestein is on the scientific advisory board of Sirenas.
References:
- 1.Altschul SF et al. J Mol Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Watrous J et al. Proc Natl Acad Sci U S A. 109, 1743–52 (2012).22232671 [Google Scholar]
- 3.Rasche F Anal Chem. 83, 1243–51 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Lai Z et al. Nat Methods 15, 53–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong J et al. Nucleic Acids Res. 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tautenhahn R et al. Anal Chem. 84, 5035–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishart DS et al. Nucleic Acids Res. 46, D608–D617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aksenov AA et al. Nat. Rev. Chem 1, 0054 (2017) [Google Scholar]
- 9.Perez-Riverol Y et al. Nat Biotechnol. 35, 406–409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocca-Serra P et al. Metabolomics 12, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M et al. Nat Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchner M et al. J Proteome Res. 9, 2762–3 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Kessner D et al. Bioinformatics 24, 2534–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horai H et al. J Mass Spectrom. 45, 703–14 (2010). [DOI] [PubMed] [Google Scholar]
- 15. https://massbank.eu/MassBank/
- 16. http://mona.fiehnlab.ucdavis.edu/
- 17.Sawada Y et al. Phytochemistry 82, 38–45 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Otogo N’Nang E et al. Org Lett. 20, 6596–6600 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Schymanski EL et al. Metabolites. 3, 517–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyle JE et al. 33, 1744–1746 (2017). [Google Scholar]
- 21.Haug K et al. Nucleic Acids Res. 41, D781–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sud M et al. Nucleic Acids Res. 44, D463–70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mungall CJ et al. Genome Biol. 13, R5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schriml LM et al. Nucleic Acids Res. 47, D955–D962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolyen E et al. Nat Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez A et al. Nat Methods 15, 796–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarmusch AK. et al. Preprint at https://www.biorxiv.org/content/10.1101/750471v1.
- 28.Sumner LW et al. Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


