Abstract

Protein–protein interactions (PPIs) play an important role in numerous biological processes such as cell-cycle regulation and multiple diseases. The family of 14-3-3 proteins is an attractive target as they serve as binding partner to various proteins and are therefore capable of regulating their biological activities. Discovering small-molecule modulators, in particular stabilizers, of such complexes via traditional screening approaches is a challenging task. Herein, we pioneered the first application of dynamic combinatorial chemistry (DCC) to a PPI target, to find modulators of 14-3-3 proteins. Evaluation of the amplified hits from the DCC experiments for their binding affinity via surface plasmon resonance (SPR), revealed that the low-micromolar (KD 15–16 μM) acylhydrazones are 14-3-3/synaptopodin PPI stabilizers. Thus, DCC appears to be ideally suited for the discovery of not only modulators but even the more elusive stabilizers of notoriously challenging PPIs.
Keywords: Protein−protein interactions, DCC, hit-identification strategy, small-molecule stabilizers
The family of 14-3-3 proteins is present in all eukaryotic cell types, and its members are involved in a myriad of processes in the human body.1 They play significant roles, ranging from signal transduction, to regulation of metabolism, to cell-death, and they are correlated to diseases such as Alzheimer’s and Noonan syndrome.2,3 This is attributed to the ability of 14-3-3 to establish protein–protein interactions (PPIs) with more than 500 protein partners.4 There are seven known human isoforms of 14-3-3: beta (β), epsilon (ε), eta (η), gamma (γ), tau (τ), sigma (σ), and zeta (ζ). The binding partners feature three conserved binding motifs for the binding groove in 14-3-3: RSXpSXP (mode 1), RXXXpSXP (mode 2), and pS/TX-COOH (mode 3), where pS/T denotes a phosphoserine or threonine residue.5−7 In this study, we used the actin binding protein synaptopodin as a binding partner of 14-3-3 using mode 2 for interaction. Synaptopodin maintains the actin cytoskeleton and urine-filtering capabilities of podocytes in the kidney glomerulus. It is regulated through phosphorylation and subsequent 14-3-3 binding.8 14-3-3 proteins are potential drug targets and an increasing number of chemical classes that modulate 14-3-3 PPIs have been reported, as recently reviewed.9,10 Modulators of 14-3-3 PPIs can be inhibitors, mostly small synthetic molecules, and stabilizers, which include bigger scaffolds, e.g., pyrrolidone1 and (semi-) natural products, e.g., fusicoccin-A.9,11−14 Dynamic combinatorial chemistry (DCC) has become an established technique for hit identification. Briefly, it allows a target-based amplification of the best binder(s) from a pool of reacting building blocks and the corresponding products existing under thermodynamic equilibrium. The types of reversible linkages that can be applied in DCC, reaction conditions, and analysis of the dynamic combinatorial library (DCL) have been comprehensively reviewed before.15−17 In the present work, we set out to demonstrate the full potential of DCC by pioneering its application to the hitherto unexplored class of PPI target, thus we exploited the power of DCC to identify new PPI modulators targeting the 14-3-3(ζ) isoform. For the DCC experiments, we used acylhydrazone formation from the corresponding hydrazide and aldehyde as a reversible reaction. The acylhydrazone linkage can take part in binding with the desired target, as it offers H-bond donor and -acceptor sites. The acylhydrazone formation is sufficiently reversible in acidic media, but also stable against hydrolysis.18 In basic media, the reaction is very slow, therefore high pH values are used to freeze the equilibrium prior to DCL analysis. Bhat et al. showed that the use of aniline can accelerate the formation of the equilibrium to only 6 h while applying a closer to physiological pH of 6.2.19,20 As a prerequisite for acylhydrazone-based DCC, we systematically monitored the stability of our target protein 14-3-3(ζ) under acidic conditions using different buffers and pH values.25 We found that in MES buffer at pH 6.5, 14-3-3(ζ) is stable at room temperature up to 7 days. Therefore, we selected these conditions in the presence of 10 mM aniline as a nucleophilic catalyst for the DCC experiments. In order to confirm that the protein is folded correctly, we recorded the circular dichroism (CD) of 14-3-3. The obtained CD-spectrum matches that reported in the literature (Supporting Information, Figure S4).21
Results and Discussion
The initial design of the DCL and choice of building blocks were inspired by compound 1, a small-molecule inhibitor of 14-3-3 discovered by virtual screening.22 We envisioned the acylhydrazone linkage between the two aromatic rings, resulting in compound 2 (Figure 1). This modification should maintain the length of the linker between the two aromatic moieties of 1 owing to the restricted flexibility of the acylhydrazone group. Accordingly, compound 2 was synthesized in three consecutive steps starting with a condensation reaction between the commercially available 2-iodobenzhydrazide and 2,3-dichlorobenzaldehyde to form the acylhydrazone 3 (Scheme 1). A palladium-catalyzed coupling between the aryl iodide 3 and triethyl phosphite followed, to afford the phosphate ester 4. This was achieved by first formation of a palladium phosphonate complex at 150 °C, followed by the addition of the acylhydrazone 3 at 100 °C. It was important to lower the temperature before adding the acylhydrazone 3 in order to prevent its decomposition. Deprotection of 4 was achieved by TMSBr, resulting in the target compound 2. It is noteworthy that trials to prepare the hydrazide building block with o-phosphonic acid moiety, required for DCC, using the above-mentioned coupling conditions were unsuccessful, and only an intramolecular N-arylation product was obtained. We therefore had to use compound 2 in our DCC experiments for in situ generation of the corresponding hydrazide and aldehyde.
Figure 1.
Design of DCL for 14-3-3 PPIs modulation based on the known small-molecule inhibitor 1.
Scheme 1. Synthetic Route toward Compound 2.
We evaluated compound 2 for its biochemical properties via surface plasmon resonance (SPR) and fluorescence polarization (FP) assay. For the SPR study, we first determined the binding affinity (KD) of a synaptopodin fragment containing the binding motif to 14-3-3(ζ). The 29-amino acid peptide showed a clear binding response with the same KD value of 1.38 μM obtained from either the Langmuir isotherm or the kinetic curves (Figure S7). This showed us that the immobilized protein is still in the native folded state and can engage in PPIs. Next, we assessed compound 2, which showed low millimolar affinity (KD value 1.01 mM) toward 14-3-3(ζ) (Figure 2). In the FP assay, compound 2 was titrated to fixed concentrations of 14-3-3(ζ) and a fluorescently labeled 29-mer peptide fragment of synaptopodin. A known 14-3-3 PPI stabilizing molecule, fusicoccin-A, was used as a positive control resulting in a dose-dependent increase in FP signal (Figure S5). In agreement with our design approach, titrating compound 2 to the protein–peptide complex decreases the signal indicating an inhibitory effect with an EC50 value of 120 μM (Figure S5).
Figure 2.
SPR binding assay of 2: (a) Overlay of sensorgrams (black) of 2 at concentrations of 1.6–1000.0 μM running over an immobilized 14-3-3(ζ). Global fitting of the association and dissociation curves (red), (b) Langmuir binding isotherm (KD = 1.01 ± 0.03 mM). Data obtained from single experiment.
Encouraged by these results we designed a DCL based on compound 2 (Figure 1 and Scheme S1). Besides the o-phenylphosphonic acid moiety, we used diverse aryl-, heteroaryl-, alicylic-, and alkylhydrazides to explore other scaffolds for potential binding to 14-3-3(ζ). As an aldehyde component, four building blocks with similar reactivity were selected including haloaryl- and heteroarylaldehydes. However, LC-MS analysis of the DCC experiment did not reveal all of the possible acylhydrazones bearing o-phosphonic or o-sulfonic acid moieties (Figure S1). We therefore modified the DCL to include three aldehydes A1–A3 and six hydrazides H1–H6 and exclude the acidic motifs as shown in Scheme 2. Consequently, we ran three DCC experiments: (a) a library in which building blocks were present in combination with the 14-3-3(ζ)/unlabeled synaptopodin complex as a PPI model; (b) a library containing the building blocks and 14-3-3 protein; and (c) a “blank” in which only building blocks were present (Scheme 2).
Scheme 2. Dynamic Combinatorial Library (DCL) of Acylhydrazones with Aldehydes (100 μM Each), Hydrazides (300 μM Each), DMSO (5%), Aniline (10 mM), and (a) 14-3-3(ζ) (10 μM) and Synaptopodin (10 μM); (b) Control with 14-3-3(ζ) (10 μM); and (c) Control without Protein or Synaptopodin.
LC-MS analysis of the DCLs that were allowed to equilibrate for 6 h resulted in the chromatograms shown in Figure 3. The most obvious differences are the two peaks at retention times of 13.5 and 13.9 min, corresponding to the compounds A1H3 and A2H3, respectively. In the presence of 14-3-3(ζ) only and in the PPI-complex (14-3-3(ζ)/synaptopodin), these two acylhydrazones show a significant amplification (about 148%) compared to the DCL in the absence of protein. Table 1 shows the ratios of the relative areas of each peak compared to the blank DCL. These two hits were then synthesized to confirm the identity of peaks (Figure S3) and for biochemical characterization.
Figure 3.
UV-chromatograms at 290 nm of DCLs in duplicate, blank (B), in the presence of protein (P), and in the presence of protein plus synaptopodin (PS) after 6 h. Compounds A1H3 and A2H3 are amplified in P as well as PS compared to B.
Table 1. Amplification Factors of the Products Formed in the DCC Experiments Analyzed via the Relative Areas of Peaks in the UV-Chromatogramsa.
| Compd | tR (min)b | % Amplification in the presence of 14-3-3c | % Amplification in the presence of 14-3-3–synaptopodin complexd |
|---|---|---|---|
| A3H2 | 4.76 | –59.0 | –44.0 |
| A3H4 | 5.66 | –33.9 | –35.9 |
| A3H6 | 6.62 | –25.7 | –16.4 |
| A3H1 | 7.93 | –33.1 | –18.1 |
| A1H2, A3H5 | 8.21 | –32.3 | –35.1 |
| A2H2, A1H4 | 9.09 | –28.0 | –26.0 |
| A2H4 | 9.73 | –36.7 | –32.9 |
| A1H1, A1H6 | 10.66 | –20.4 | –23.1 |
| A3H3 | 10.79 | –1.3 | –15.0 |
| A2H6 | 11.29 | –22.5 | –23.6 |
| A2H1 | 11.49 | 32.2 | 29.5 |
| A1H5 | 11.91 | –12.2 | –16.0 |
| A2H5 | 12.49 | 12.8 | 14.6 |
| A1H3 | 13.46 | 148.0 | 148.4 |
| A2H3 | 13.91 | 148.5 | 153.6 |
Values represent the average of two independent experiments.
Retention time.
Calculated
as 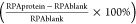 .
.
Calculated as 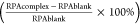 .
.
Synthesis was accomplished through the reaction of the hydrazide H3 with a stoichiometric amount of the appropriate aldehyde at reflux overnight to afford the corresponding acylhydrazones in good to quantitative yield (Scheme 3).
Scheme 3. Synthetic Route toward Compounds A1H3 and A2H3.
We used SPR binding assays to analyze binding events (affinity and binding kinetics) of the synthesized DCC hits to the 14-3-3(ζ) protein. The acylhydrazones A1H3 and A2H3 showed low micromolar affinity to 14-3-3(ζ) (KD values 16 and 15 μM, respectively). Interestingly, compound A2H3 exhibited different on- and off-rates with a longer residence time compared to A1H3 although with equal binding affinities (Table 2, Figure S8 and S9).
Table 2. Kinetic Parameters of Hit Compounds A1H3 and A2H3 to 14-3-3(ζ)a.
| Compd | Rmax (RU) | kon (M–1s–1) | koff (s–1) | t1/2 (s) | KD (μM) |
|---|---|---|---|---|---|
| A1H3 | 4.6 ± 0.2 | 2.6 ± 0.2 × 103 | 4.1 ± 0.1 × 10–2 | 17 | 16 ± 1 |
| A2H3 | 4.5 ± 0.2 | 5.8 ± 0.5 × 102 | 8.7 ± 0.4 × 10–3 | 80 | 15 ± 1 |
Rmax: maximum analyte binding capacity; kon: association rate constant; koff: dissociation rate constant; t1/2: dissociative half-life; KD: equilibrium dissociation constant.
Intrigued by these findings, we next investigated the mechanism of action and binding site of the new 14-3-3 PPI modulators (compounds 2, A1H3, and A2H3) by SPR competition assays using synaptopodin as a reference, which occupies the 14-3-3 main binding pocket. Modulators that inhibit 14-3-3 PPIs bind in the phosphorylation binding pocket, whereas stabilizers bind allosterically to the binding pocket or at the interface between 14-3-3 and its protein partners.23 To check whether the compounds bind to the active site or elsewhere, we injected compound 2 (1000 μM), synaptopodin (1 μM), and a mixture of both at the same concentration in sequence over immobilized 14-3-3(ζ). The obtained response unit (RU) value of the mixture was compared to the theoretical sum of the RU values for the individual compounds. If 2 bound to the main binding pocket of 14-3-3, it would compete with synaptopodin and the response of the mixture should be less than the sum of RU values determined for the single compounds. On the other hand, if 2 bound allosterically to the active site, no competition would occur and the response of the mixture should be equal to the sum of RU values of the individual compounds. We found that the RU value of 2 in combination with synaptopodin was less than the sum of the individual responses (Figure 4 and S10). This suggests that compound 2 competes with synaptopodin for the same binding pocket. No compound, however, could completely displace the other at the tested concentrations. To verify this, we recalculated the sum of responses considering a fractional occupancy (FO) of 2 and synaptopodin according to Perspicace et al.24 Indeed, the experimental RU value of the mixture was equal to the new estimated one (Figure 4), indicating that the compounds compete for the same binding site. Using different concentrations of 2 and synaptopodin (1000 μM vs 25 μM; and 200 μM vs 1 μM) afforded the same results (Figure S11 and S12). In line with the FP-assay data, these results clearly indicate that compound 2 binds to the active site of 14-3-3, leading to disruption of PPIs.
Figure 4.

SPR responses of compounds 2, A1H3, and A2H3 in the competition assays, using synaptopodin as a reference compound binding to the active site of 14-3-3.
Using the same approach, we injected the DCC hit compounds A1H3 (25 μM) and A2H3 (25 μM) alone and as a mixture with synaptopodin (1 μM) over immobilized 14-3-3, and analyzed their binding responses (Figure 5). Interestingly, the RU values of the mixtures containing A1H3 or A2H3 with synaptopodin were equal to or more than the sum of the individual responses (Figure 4 and 5). This indicates that these two compounds bind to 14-3-3 in a different pocket than that of synaptopodin. Moreover, the increased binding response of the mixture compared to the calculated sum indicates a stabilizing effect of the acylhydrazones to the complex of synaptopodin with 14-3-3(ζ). This confirms our findings from the DCC experiments, where the same amplification factors for these hits were obtained in the presence of 14-3-3(ζ) alone as well as the 14-3-3(ζ)/synaptopodin complex (Table 1). Therefore, their binding site could be allosteric to the main binding pocket, e.g. at the interface of the 14-3-3(ζ)/synaptopodin complex.
Figure 5.
SPR competition assays: (a) Sensorgram overlay of A1H3 (25 μM, blue), synaptopodin (1 μM, red), and a A1H3–synaptopodin mixture (black) showing an additive effect, indicating a noncompetitive binding; (b) sensorgrams of A2H3 (25 μM, blue), synaptopodin (1 μM, red) and a A2H3–synaptopodin mixture (black) showing a synergistic effect, indicating a noncompetitive binding and a stabilizing effect.
Conclusions
We set out to develop novel PPI modulators targeting the versatile 14-3-3 protein family. First, we pursued a ligand-based design of the acylhydrazone 2, which turned out to be an inhibitor of 14-3-3 PPIs. Next, we applied a DCC approach, using 14-3-3(ζ) in complex with synaptopodin as a PPI model, resulting in the discovery of two modulators A1H3 and A2H3. No significant change was observed in the DCL composition in the presence of only 14-3-3 compared to the 14-3-3(ζ)/synaptopodin complex, indicating that the hit compounds bind independently to a different site than the main binding groove of 14-3-3 involved in PPIs. Finally, we determined the binding affinities and kinetics of the hits via SPR and investigated their binding site on 14-3-3. Results of the SPR competition assays support our initial findings from the DCC experiments, suggesting that these small molecules stabilize the 14-3-3(ζ)/synaptopodin complex either directly or allosterically.
Glossary
Abbreviations
- PPIs
protein–protein interactions
- DCC
dynamic combinatorial chemistry
- DCL
dynamic combinatorial library
- SPR
surface plasmon resonance
- RU
response unit
- FO
fractional occupancy
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.9b00541.
Synthesis and characterization of the described compounds, biochemical assays, protein expression, purification details, and NMR spectra (PDF)
Author Present Address
∥ Sebastian Andrei, Department of Chemistry, Imperial College London, London, UK.
Author Contributions
A. M. Hartman was involved in the design of the project, performing the DCC experiments, synthesis of compounds, binding study via SPR, and writing of the manuscript. W. A. M. Elgaher was involved in the design and analysis of the SPR study and editing the manuscript. N. Hertrich was involved in the DCC experiments and the synthesis of the compounds. S. A. Andrei was involved in the expression and purification of 14-3-3, the synthesis of the synaptopodin fragment, and the FP assay. C. Ottmann and A. K. H. Hirsch were involved in the design of the project, supervision of the project, and editing the manuscript.
Funding from The Netherlands Organization for Scientific Research (NWO) (VIDI grant: 723.014.008 (A.K.H.H.) and ECHO grant 717.014.001 (C.O.)) is gratefully acknowledged. A.K.H.H. is grateful for financial support from the European Research Council, ERC starting grant 2017 (NovAnI) 757913 and from the Helmholtz Association’s Initiative and Networking Fund.
The authors declare no competing financial interest.
Supplementary Material
References
- Paul G.; van Heusden H. 14–3-3 Proteins: Regulators of Numerous Eukaryotic Proteins. IUBMB Life 2005, 57 (9), 623–629. 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- Molzan M.; Schumacher B.; Ottmann C.; Baljuls A.; Polzien L.; Weyand M.; Thiel P.; Rose R.; Rose M.; Kuhenne P.; et al. Impaired Binding of 14–3-3 to C-RAF in Noonan Syndrome Suggests New Approaches in Diseases with Increased Ras Signaling. Mol. Cell. Biol. 2010, 30, 4698–4711. 10.1128/MCB.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milroy L. G.; Bartel M.; Henen M. A.; Leysen S.; Adriaans J. M. C.; Brunsveld L.; Landrieu I.; Ottmann C. Stabilizer-Guided Inhibition of Protein-Protein Interactions. Angew. Chem., Int. Ed. 2015, 54, 15720–15724. 10.1002/anie.201507976. [DOI] [PubMed] [Google Scholar]
- Ottmann C. Small-Molecule Modulators of 14–3-3 Protein-Protein Interactions. Bioorg. Med. Chem. 2013, 21, 4058–4062. 10.1016/j.bmc.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Tzivion G.; Avruch J. 14–3-3 Proteins: Active Cofactors in Cellular Regulation by Serine/Threonine Phosphorylation. J. Biol. Chem. 2002, 277, 3061–3064. 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- Coblitz B.; Wu M.; Shikano S.; Li M. C-Terminal Binding: An Expanded Repertoire and Function of 14–3-3 Proteins. FEBS Lett. 2006, 580 (6), 1531–1535. 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Ganguly S.; Weller J. L.; Ho A.; Chemineau P.; Malpaux B.; Klein D. C. Melatonin Synthesis: 14–3-3-Dependent Activation and Inhibition of Arylalkylamine N-Acetyltransferase Mediated by Phosphoserine-205. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (4), 1222–1227. 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul C.; Donnelly M.; Merscher-Gomez S.; Chang Y. H.; Franz S.; Delfgaauw J.; Chang J.-M.; Choi H. Y.; Campbell K. N.; Kim K.; Reiser J.; Mundel P. The Actin Cytoskeleton of Kidney Podocytes is a Direct Target of the Antiproteinuric Effect of Cyclosporine A. Nat. Med. 2008, 14, 931–938. 10.1038/nm.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A. M.; Hirsch A. K. H. Molecular Insight into Specific 14–3-3 Modulators: Inhibitors and Stabilisers of Protein–Protein Interactions of 14–3-3. Eur. J. Med. Chem. 2017, 136, 573–584. 10.1016/j.ejmech.2017.04.058. [DOI] [PubMed] [Google Scholar]
- Stevers L. M.; Sijbesma E.; Botta M.; MacKintosh C.; Obsil T.; Landrieu I.; Cau Y.; Wilson A. J.; Karawajczyk A.; Eickhoff J.; et al. Modulators of 14–3-3 Protein–Protein Interactions. J. Med. Chem. 2018, 61 (9), 3755–3778. 10.1021/acs.jmedchem.7b00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi V.; Mancini M.; Manetti F.; Petta S.; Santucci M. A.; Botta M. Identification of the First Non-Peptidic Small Molecule Inhibitor of the c-Abl/14–3-3 Protein-Protein Interactions Able to Drive Sensitive and Imatinib-Resistant Leukemia Cells to Apoptosis. Bioorg. Med. Chem. Lett. 2010, 20, 6133–6137. 10.1016/j.bmcl.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Du Y.; Horton J. R.; Upadhyay a. K.; Lou B.; Bai Y.; Zhang X.; Du L.; Li M.; Wang B.; et al. Discovery and Structural Characterization of a Small Molecule 14–3-3 Protein-Protein Interaction Inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16212–16216. 10.1073/pnas.1100012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevers L. M.; Lam C. V.; Leysen S. F. R.; Meijer F. A.; van Scheppingen D. S.; de Vries R. M. J. M.; Carlile G. W.; Milroy L. G.; Thomas D. Y.; Brunsveld L.; et al. Characterization and Small-Molecule Stabilization of the Multisite Tandem Binding between 14–3-3 and the R Domain of CFTR. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E1152–E1161. 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C.; Higuchi Y.; Koschinsky K.; Bartel M.; Schumacher B.; Thiel P.; Nitta H.; Preisig-Müller R.; Schlichthörl G.; Renigunta V.; et al. A Semisynthetic Fusicoccane Stabilizes a Protein-Protein Interaction and Enhances the Expression of K+ Channels at the Cell Surface. Chem. Biol. 2013, 20, 583–593. 10.1016/j.chembiol.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Mondal M.; Hirsch A. K. H. Dynamic Combinatorial Chemistry: A Tool to Facilitate the Identification of Inhibitors for Protein Targets. Chem. Soc. Rev. 2015, 44, 2455–2488. 10.1039/C4CS00493K. [DOI] [PubMed] [Google Scholar]
- Van der Vlag R.; Hirsch A. K. H.. 5. 20 Analytical Methods in Protein-Templated Dynamic Combinatorial Chemistry. In Comprehensive Supramolecular Chemistry 2; Elsevier, 2017; Vol. 5, pp 487–509. [Google Scholar]
- Frei P.; Hevey R.; Ernst B. Dynamic Combinatorial Chemistry: A New Methodology Comes of Age. Chem. - Eur. J. 2019, 25, 60–73. 10.1002/chem.201803365. [DOI] [PubMed] [Google Scholar]
- Mondal M.; Radeva N.; Köster H.; Park A.; Potamitis C.; Zervou M.; Klebe G.; Hirsch A. K. H. Structure-Based Design of Inhibitors of the Aspartic Protease Endothiapepsin by Exploiting Dynamic Combinatorial Chemistry. Angew. Chem., Int. Ed. 2014, 53, 3259–3263. 10.1002/anie.201309682. [DOI] [PubMed] [Google Scholar]
- Bhat V. T.; Caniard A. M.; Luksch T.; Brenk R.; Campopiano D. J.; Greaney M. F. Nucleophilic Catalysis of Acylhydrazone Equilibration for Protein-Directed Dynamic Covalent Chemistry. Nat. Chem. 2010, 2, 490–497. 10.1038/nchem.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisalli P.; Kool E. T. Water-Soluble Organocatalysts for Hydrazone and Oxime Formation. J. Org. Chem. 2013, 78, 1184–1189. 10.1021/jo302746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A. M.; Gierse R. M.; Hirsch A. K. H. Protein-Templated Dynamic Combinatorial Chemistry: Brief Overview and Experimental Protocol. Eur. J. Org. Chem. 2019, 2019, 3581–3590. 10.1002/ejoc.201900327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.; Ratha B. N.; Gayen N.; Mroue K. H.; Kar R. K.; Mandal A. K.; Bhunia A. Biophysical Characterization of Essential Phosphorylation at the Flexible C-Terminal Region of C-Raf with 14–3-3 ζ Protein. PLoS One 2015, 10, 1–21. 10.1371/journal.pone.0135976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel P.; Röglin L.; Meissner N.; Hennig S.; Kohlbacher O.; Ottmann C. Virtual Screening and Experimental Validation Reveal Novel Small-Molecule Inhibitors of 14–3-3 Protein-Protein Interactions. Chem. Commun. (Cambridge, U. K.) 2013, 49, 8468–8470. 10.1039/c3cc44612c. [DOI] [PubMed] [Google Scholar]
- Rose R.; Erdmann S.; Bovens S.; Wolf A.; Rose M.; Hennig S.; Waldmann H.; Ottmann C. Identification and Structure of Small-Molecule Stabilizers of 14–3-3 Protein-Protein Interactions. Angew. Chem., Int. Ed. 2010, 49, 4129–4132. 10.1002/anie.200907203. [DOI] [PubMed] [Google Scholar]
- Perspicace S.; Banner D.; Benz J.; Müller F.; Schlatter D.; Huber W. Fragment-Based Screening Using Surface Plasmon Resonance Technology. J. Biomol. Screening 2009, 14 (4), 337–349. 10.1177/1087057109332595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









