Abstract
The development of clustered regularly interspaced short-palindromic repeat (CRISPR)-based biotechnologies has revolutionized the life sciences and introduced new therapeutic modalities with the potential to treat a wide range of diseases. Here, we describe CRISPR-based strategies to improve human health, with an emphasis on the delivery of CRISPR therapeutics directly into the human body using adeno-associated virus (AAV) vectors. We also discuss challenges facing broad deployment of CRISPR-based therapeutics and highlight areas where continued discovery and technological development can further advance these revolutionary new treatments.
Gao eTOC blurb
Wang, Zhang & Gao discuss the progress, concerns and challenges currently facing CRISPR-based therapeutics, a field that has inspired renewed but cautious interest in human genome editing.
INTRODUCTION
Numerous human diseases arise from mutations that diminish or damage gene products. Gene therapy as a strategy to treat genetic diseases was formally proposed in 1972 (Friedmann and Roblin, 1972), introducing the concept that ‘genes can be medicine’. In the ensuing decades, implementation of this medical concept was met with initial excitement, serious setbacks, resurgence of interest, and more recently, clinical successes (Dunbar et al., 2018; High and Roncarolo, 2019). Despite these successes, however, delivering a functional gene copy to replace a mutated one is not a perfect solution for many diseases. For example, an exogenous gene copy lacks many regulatory elements that are important for endogenous gene expression and function. In addition, for gain-of-function pathogenic mutations, simply supplying a wild-type copy of the gene is ineffective. These and other limitations can be theoretically addressed by directly ‘editing’ a mutated gene, thereby restoring gene function in its natural context.
Indeed, many of our cells carry out something like this thousands of times a day, using a variety of DNA repair mechanisms that guard genome integrity in response to DNA damage. Similar to DNA damage, a targeted double-stranded break (DSB) can also trigger these cellular repair mechanisms, mainly nonhomologous end-joining (NHEJ) and homology-directed repair (HDR), which can potentially introduce DNA sequence changes during repair of the DSB. HDR, which is a templated process, allows for the introduction of specific DNA changes, a phenomenon that has been leveraged to achieve insertion of new DNA sequences (Thomas and Capecchi, 1987). HDR-mediated insertion, however, requires the presence of a correct template (which is missing in most natural cases of genetic mutations) and is generally less efficient than NHEJ.
More efficient methods for gene editing arose from the observation that a targeted DSB generated by an endonuclease can dramatically stimulate HDR in eukaryotic cells (Rouet et al., 1994; Smih et al., 1995). This observation spurred a quest for programmable and efficient endonucleases (Urnov, 2018), leading to the development of meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short-palindromic repeat (CRISPR)-associated (Cas) proteins (Gaj et al., 2013; Hsu et al., 2014), the latter of which has been most widely adopted in research. Fundamental to the popularity of Cas nucleases is that they are guided by a short RNA sequence that recognizes the target DNA sequence through Watson-Crick base pairing (Brouns et al., 2008; Jinek et al., 2012), whereas the other enzymes rely on protein-DNA interactions to achieve target specificity. Following the demonstration of CRISPR-Cas9-mediated genome editing (Cong et al., 2013; Mali et al., 2013), CRISPR-based technology has rapidly advanced, benefitting greatly from a collective endeavor to characterize, improve, expand, and share the CRISPR-based molecular toolbox (Doudna, 2020; Doudna and Charpentier, 2014; Zhang, 2019). In addition to accelerating basic research, CRISPR-based technology also holds enormous potential as a therapeutic, offering an approach to permanently correct disease-causing mutations.
Deploying CRISPR-based therapeutics directly into the human body holds great promise for treating numerous diseases. Although the CRISPR-based toolbox enables diverse operations ranging from DNA and RNA editing to gene expression modulation, delivery remains a bottleneck for therapy development. Currently, adeno-associated virus (AAV) vector is the leading platform for in vivo gene therapy delivery (Wang et al., 2019). AAV is safe, capable of delivering its single-stranded DNA (ssDNA) vector genome to various tissues and cell types, and only mildly immunogenic within a wide range of dosing regimens. Although the vector genome largely remains episomal inside host cells, it is stabilized through concatemerization and circularization to mediate long-term transgene expression in post-mitotic cells, leading to durable therapeutic efficacy. The successes of AAV vectors in delivering gene therapies to disease animal models and patients propelled their adoption for in vivo delivery of CRISPR-based therapeutics.
In this review, we focus on recent advances in CRISPR-based therapeutic strategies and in vivo delivery of CRISPR machinery using AAV vectors. We also discuss the limitations of using AAV vectors to deliver CRISPR-based therapeutics. Although the lessons learned from AAV gene therapy are generally applicable to delivering CRISPR-based tools, many challenges are unique to this new class of cargoes and call for distinct solutions from both the CRISPR and AAV fields to fully unleash the power of CRISPR-based therapeutic gene editing.
HUMAN HEALTH APPLICATIONS
Editing strategies based on nuclease activity
Gene disruption by NHEJ
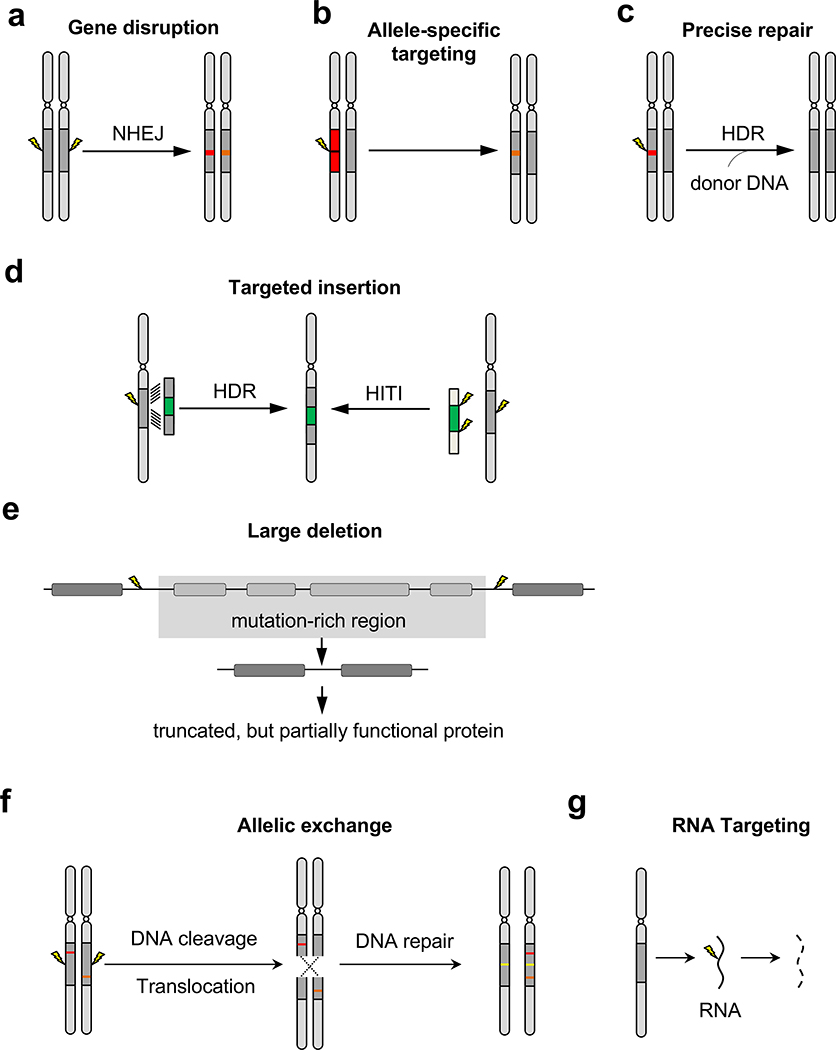
Typically, DSBs introduced with CRISPR are repaired via NHEJ, an efficient and prevalent DNA repair mechanism in human cells. NHEJ ligates two broken DNA ends together at the break site (Chapman et al., 2012). This process of targeted cleavage and repair can take place repeatedly until an insertion or deletion (indel) occurs that prevents further recognition of the target site by the nuclease. An indel mutation in a protein-coding gene can cause frameshifting or exon skipping, thereby disrupting gene function (Figure 1a). This seemingly disruptive gene editing approach has several therapeutic potentials. For example, the PCSK9 gene encodes an enzyme that binds to the cell surface receptor for low-density lipoprotein (LDLR) and triggers the lysosomal degradation of LDLR. When PCSK9 is diminished by gene editing, LDLR can return to the cell surface and continue to remove LDL, thereby lowering cholesterol levels (Ding et al., 2014; Ran et al., 2015). Similarly, loss of CCR5 - a co-receptor exploited by HIV to infect T cells - confers HIV resistance on edited T cells (Xu et al., 2019b; Xu et al., 2017). Another application of gene disruption is silencing dominant negative mutations. Through careful design of the guide RNA, the mutant allele can differentially disrupted while preserving the normal one (Bakondi et al., 2016; Gyorgy et al., 2019; Rabai et al., 2019) (Figure 1b).
Figure 1. Therapeutic editing strategies based on nuclease activity.
(a) Gene disruption introduces indel mutations (red and orange bars) into a gene (dark gray bars), silencing the gene function, (b) Targeting can be specific to the mutant allele (red bar) and spare the normal allele, (c) HDR mediates precise mutation repair in the presence of a donor template, (d) Targeted insertion of a therapeutic gene (green bar) can be achieved by either HDR or HITI. (e) Two cuts can delete a large gene fragment harboring a mutation, resulting in the production of truncated by partially functional protein, (f) Allelic exchange converts a compound heterozygous genotype to heterozygous through translation between two homologous chromosomes (dashed lines). The targeting site is designed to be intronic, so that the indel mutations caused by DNA repair (yellow bars) do not compromise gene expression, (g) RNA targeting achieves gene silencing by degrading RNA instead of DNA. NHEJ: non-homologous end joining. HDR: homology-directed repair. HITI: homology-independent targeted integration.
Predictable editing with a single cut
Although the indel mutations generated by Cas9-initiated NHEJ repair are heterogenous, the mutation spectrum is not random but reproducible and dependent on the target site and sequence context (van Overbeek et al., 2016). A data-trained machine learning model was developed to predict the types and frequencies of Cas9-mediated small indels in human and mouse cells with high accuracy (Shen et al., 2018). This model identifies human pathogenic variants that, following Cas9 cleavage, can be corrected by the predicted predominant indels. This template-free editing method was used to correct frameshift mutations and microduplication mutations in human cells (Iyer et al., 2019; Shen et al., 2018). Another class of predictable editing is targeting a splicing signal to induce exon skipping (Amoasii et al., 2017; Long et al., 2018). Compared with a 2-cut approach (see below), using a single sgRNA to destroy an exonic splicing enhancer, a splicing acceptor site, or a splicing donor site offers a simplified therapeutic design.
Precise mutation repair by HDR
Although precise correction of DNA mutations by HDR is an intuitive therapeutic approach (Figure 1c), this DNA repair mechanism is inefficient in human cells, especially in post-mitotic cells such as myofibers and neurons (Chapman et al., 2012). Furthermore, it requires the co-delivery of a repair donor template that carries homology arms matching the targeted locus, which complicates the therapy. Nevertheless, this is a powerful therapeutic strategy, especially in cases where a small number of corrected cells can improve symptoms or in cases where regenerative tissues are targeted. For example, hereditary tyrosinemia type I (HT-1) is a metabolic disease caused by mutations in the FAH gene, which is expressed in actively dividing hepatocytes. In a mouse model of HT-1, delivery of Cas9, sgRNA, and a donor template to the liver corrected a disease-causing Fah mutation in some hepatocytes, which then had a growth advantage compared to unedited cells, ultimately driving rescue of the disease phenotype (Yin et al., 2016; Yin et al., 2014).
Targeted gene insertion
Another application of HDR-based gene editing is targeted insertion of exogenous DNA sequence into the genome (Figure 1d). This is best exemplified by integrating a chimeric antigen receptor (CAR) gene into isolated T-cells; after being infused back into the patient, these armed T-cells are capable of recognizing and killing tumor cells specified by the CAR (Bailey and Maus, 2019). Using CRISPR to achieve targeted integration of the CAR by HDR can achieve concomitant disruption of the T-cell receptor a constant (TRAC) locus, resulting in engineered CAR-T cells that may be more amenable to allogeneic infusion (Eyquem et al., 2017). In addition to HDR-based approaches, homology-independent targeted integration (HITI) was developed to utilize the more robust NHEJ repair pathway for gene integration, which the authors reported achieved efficient rescue of a disease phenotype in mice (Suzuki et al., 2016). Notably, a single AAV vector design can incorporate elements allowing for both HDR-mediated gene insertion and HITI to achieve efficient genetic correction (Ohmori et al., 2017).
Large-scale DNA editing
CRISPR is inherently capable of multiplex editing, and this natural feature has been leveraged to achieve deletion of large sections of DNA. By introducing two guide RNAs targeting separate sites, fragments as large as several megabase pairs (Mbp) can be excised from the genome (Figure 1e). This approach can remove deleterious mutations while maintaining the open reading frame by deleting one or more exons. This strategy is particularly well suited for Duchene muscular dystrophy (DMD), which is caused by several different mutations, all of which can be treated with this strategy because truncated dystrophin protein with internal deletions is partially functional (England et al., 1990). In a DMD mouse model carrying a nonsense mutation in the dystrophin gene in exon 23, AAV delivery to the muscle of Cas9 and a pair of sgRNAs flanking exon 23 mediated deletion of this exon. Splicing between exon 22 and exon 24 preserved the normal reading frame, generating truncated but functional dystrophin that rescued the disease phenotype (Long et al., 2016; Nelson et al., 2016; Tabebordbar et al., 2016). Another example of this therapeutic strategy is removal of the IVS26 mutation in the CEP290 gene, which causes Leber congenital amaurosis type 10 (LCA10) (Maeder et al., 2019). This point mutation is in intron 26 and causes aberrant splicing. Using a similar AAV-based approach for eye delivery in a humanized mouse model carrying the IVS26 mutation, a pair of sgRNAs was used to guide Cas9 to excise a segment of intron 26 that contains the mutation, restoring normal splicing between exon 26 and exon 27. The editing efficiency met the targeted therapeutic threshold and was translatable to nonhuman primates (Maeder et al., 2019). Based on these preclinical results, a phase 1/2 clinical trial for the treatment of LCA10 is currently open (ClinicalTrials.gov Identifier: NCT03872479), and it is expected to be the first in vivo clinical application of CRISPR-based gene editing. At an even larger scale, recessive compound heterozygous mutations - different mutations damaging the same gene - can be corrected by allelic exchange mediated by DNA cuts at two homologous chromosomes and inter-homologue translocation (Wang et al., 2018) (Figure 1f). Although compound heterozygous mutations are prevalent in patients, application of allelic exchange in a broad range of diseases will require enhancing the efficiency of allelic exchange.
RNA targeting
Following the discovery and development of RNA-targeting Cas enzymes for use in human cells (Abudayyeh et al., 2017; Konermann et al., 2018; Shmakov et al., 2015; Smargon et al., 2017; Yan et al., 2018), a number of human health applications for these novel proteins have been pursued (Figure 1g). Compared with editing at the level of the genome, gene knock-down at the RNA level is a potentially reversible therapeutic approach. In addition, RNA targeting enzymes, such as Cas13, offer more flexible target selection than Cas9, and more specific target degradation than RNA interference (RNAi) (Abudayyeh et al., 2017). Repeated intratumoral delivery of Cas13a and guide RNA targeting a mutant KRAS transcript in the form of ribonucleoprotein (RNP) was shown to slow tumor growth in a xenograft mouse model (Zhao et al., 2018). To achieve long-lasting in vivo therapeutic effects by targeting RNA, AAV vectors are an advantageous delivery platform for Cas13-based tools because they allow for durable expression of the editing system.
Cas-effector fusion platforms
CRISPR interference and activation (CRISPRi/a)
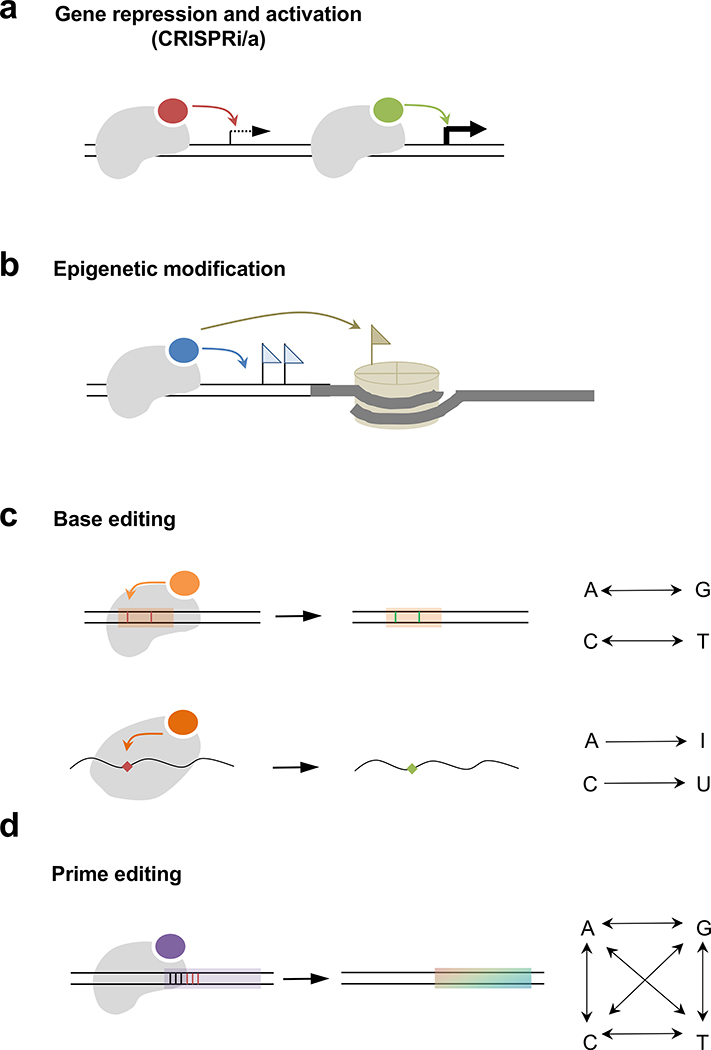
In addition to its use for gene editing, Cas9 has been repurposed as an RNA-guided DNA binding domain by introducing mutations in the RuvC and HNH nuclease domains (Bikard et al., 2013; Qi et al., 2013). In mammalian cells, robust gene repression was achieved by fusing a nuclease-deactivated Cas9 (dCas9) to a repressor domain such as the Kruppel-associated box (KRAB) domain and targeting it to the transcription start site (Gilbert et al., 2014; Gilbert et al., 2013) (Figure 2a). AAV delivery of CRISPRi targeting Pcsk9 in mouse liver effectively silenced transcription, and lowered serum PCSK9 and cholesterol levels (Thakore et al., 2018). Replacing the KRAB domain with a gene activating domain, such as VP64, converts a gene repressor to a gene activator (Cheng et al., 2013; Gilbert et al., 2013; Konermann et al., 2013; Maeder et al., 2013; Perez-Pinera et al., 2013) (Figure 2a). CRISPRa is useful for treatment of haploinsufficiency, in which a mutant allele renders the remaining normal gene copy inadequate to produce enough gene product. CRISPRa enhances transcription of the normal allele to compensate for the damaged allele, thereby restoring normal gene function. This therapeutic approach was demonstrated in a study involving two haploinsufficient mouse models of obesity caused by heterozygous mutations in the Sim1 and Mcr4 genes (Matharu et al., 2019). AAV delivery of dCas9-VP64 and a sgRNA targeting the promoter region of Sim1 or Mcr4 to hypothalamus upregulated gene expression to or beyond normal levels and reversed the obesity phenotype. In a more recent report, a similar AAV-mediated CRISPRa approach restored Scnla gene expression in the brain of a Scnla haploinsuffcient Dravet syndrome mouse model, which attenuated seizures (Colasante et al., 2019). Although CRISPRa can operate on large genes that are not amenable to viral vector delivery, this approach potentially risks toxicity associated with gene overexpression.
Figure 2. Cas-effector fusion platforms for therapeutic purposes.
(a) Cas protein (gray shape) fused a transcriptional inhibitor (red oval) or activator (green oval) can repress or boost transcription, respectively, (b) Cas protein fused with an enzyme that can modify epigenetic marks of DNA (blue flags) or histone (brown flag) is useful to change gene expression status, (c) A DNA base editor consists of a dCas or nCas protein, and an adenine or cytosine deaminase (orange oval) that converts A to G or C to T, respectively (red bars to green bars). Note that the editing window (orange shade) may contain by-stander editing targets. An RNA base editor can mediate A to I or C to U base change (red diamond to green diamond), (d) Prime editor comprises an nCas protein fused with a reverse transcriptase (purple oval). Note that the editing window (purple shade) starts from 3 base pairs upstream of the PAM (red bars) of SpCas9, and the editing outcome can be diverse (rainbow shade), including all single base changes and small indels (not shown).
Editing the epigenome
Epigenetic markers, such as DNA methylation and histone modification, are crucial to proper gene expression and have been implicated in human disease (Jaenisch and Bird, 2003). Epigenetic marks are particularly important for brain function, and their dysregulation underlies many neurological disorders (Landgrave-Gomez et al., 2015). These marks are deposited and erased by many enzymes, some of which have been fused with dCas9 to generate target-specific epigenetic writers and erasers of therapeutic value (Liu and Jaenisch, 2019) (Figure 2b). For example, fragile X syndrome (FXS) is caused by CGG trinucleotide repeat expansion in the 5’ untranslated region (UTR) of the FMR1 gene. This mutation leads to a series of epigenetic changes that silence FMR1, such as DNA hypermethylation of the CGG repeats, decreased activating histone modifications (H3K27Ac, HK4me3), and increased inhibitory histone modifications (H3K9me3). A CRISPR-based epigenetic tool comprised of dCas9 fused with the catalytic domain of a DNA demethylation enzyme TET1 (dCas9-TET1) was shown to demethylate the CGG repeats in human cells (Liu et al., 2018). This epigenetic editing converted the mutated FMR1 promoter region to an active state, leading to gene reactivation and functional restoration in various cell types including FXS neurons. Although epigenomic editing was maintained after the edited neurons were transplanted into the mouse brain (Liu et al., 2018), the in vivo therapeutic efficacy and durability of this approach remain to be evaluated.
Base editing
Point mutations represent more than half of the known pathogenic human genetic variants. Therefore, the development of programmable DNA and RNA base editors greatly expanded the CRISPR-based toolbox for therapeutic applications (Rees and Liu, 2018) (Figure 2c). Base editors have been shown to correct disease-causing point mutations in human cells, mammalian embryos, and mouse disease models (Molla and Yang, 2019). In particular, base editing can correct nonsense mutations that account for ~11% of pathogenic mutations, which has been demonstrated in mouse models of DMD (Ryu et al., 2018), phenylketonuria (Villiger et al., 2018), HT-1 (Song et al., 2020), and Niemann-Pick disease type C (Levy et al., 2020). In addition, base editing was used to silence Pcsk9 gene function by introducing a nonsense mutation into the gene (Chadwick et al., 2017) and to correct a SOD1 missense mutation in a mouse model of amyotrophic lateral sclerosis (Lim et al., 2020). Compared with HDR-based gene correction, base editing functions in both dividing and post-mitotic cells, and generally exhibits higher on-target editing efficiency and lower indel rate (Roccio et al., 2015; Yeh et al., 2018). However, by-stander editing within a window of several base pairs may cause undesired changes, which has prompted efforts to refine the editing window and increase editing precision (Thuronyi et al., 2019). RNA base editors were developed by fusing dCas13 with the deaminase domain of ADAR2 (ADARdd) (REPAIR) or an evolved cytosine deaminase (RESCUE) (Abudayyeh et al., 2019; Cox et al., 2017). REPAIR and RESCUE provide a programmable, reversible, and multiplexable approach to correct genetic mutations at the RNA level. Because the target base is defined by a mismatch between the crRNA and substrate RNA, by-stander editing does not occur. A better understanding of the interplay between RNA editing machinery and endogenous RNA metabolism pathways such as splicing and nonsense-mediated decay will facilitate its therapeutic utilization in vivo.
Prime editing
Current base editors are limited to four transition mutations (a purine to another purine, or a pyrimidine or another pyrimidine), leaving eight transversion mutations (a purine to a pyrimidine, or vice versa) and indel mutations uncorrectable by base editing. Recently, prime editing was developed to expand the scope of donor-free precise DNA editing (Anzalone et al., 2019) (Figure 2d). The prime editor (PE) consists of Cas9 nickase mutant (nCas9) fused with an engineered reverse transcriptase. PE is guided by a prime editing guide RNA (pegRNA) that serves three functions. First, pegRNA dictates target DNA specificity. Second, following nCas9 nicking at the target site, pegRNA hybridizes with the single-stranded DNA to initiate reverse transcription. Third, pegRNA encodes the desired editing information that can be reverse transcribed and incorporated into the target site. Overall, prime editing is a versatile genome editing tool that can correct not only all 12 types of point mutations, but also small indel mutations. The therapeutic potential of prime editing was demonstrated in the initial report, where a diverse range of disease-causing mutations were corrected in various cell types (Anzalone et al., 2019). Future work will likely improve prime editing efficiency and precision, and assess genome-wide off-target editing.
DELIVERY USING AAV VECTORS
Ex vivo and in vivo approaches
The delivery of genome editing therapeutics can be broadly categorized into ex vivo and in vivo approaches. Both approaches have been extensively practiced in the broader gene and cell therapy field and have achieved clinical successes (Dunbar et al., 2018; High and Roncarolo, 2019). The lessons on gene delivery are broadly translatable to delivery of CRISPR-based therapeutics. In ex vivo delivery, genome editing reagents are introduced into isolated human cells to achieve the desired genetic modification. After expansion, the genetically modified cells are infused into patients to confer a therapeutic effect. In vivo delivery aims to introduce genome editing reagents into patients systemically or locally to directly manipulate cells in the body.
Ex vivo delivery has several advantages. First, multiple robust methods have been established to introduce nucleic acid and protein into cell culture, such as transduction by lentiviral vectors, transfection with DNA or RNA, or electroporation with ribonucleoproteins (RNPs). Furthermore, infection by AAV vectors with a ssDNA vector genome can provide donor template for HDR-mediated strategies (Bak et al., 2017). Second, before the edited cells are infused into patients, they can be scrutinized to ensure gene editing efficiency and accuracy. Third, delivery of CRISPR-based genome editing reagents into cell culture is not limited by host immune responses. For these reasons, ex vivo cell therapy has rapidly advanced to a number of clinical trials involving various cell types and targeting multiple diseases (Table 1), such as disrupting CCR5 in T-cells for HIV infection, engineering immune cells to combat cancer, and editing the BCL11A gene in hematopoietic stem cells for treating β-hemoglobinopathies (Porteus, 2019). However, the human cell types amenable to isolation and ex vivo manipulation are limited, and most cell types, especially highly differentiated and post-mitotic cells, are only functional and manipulatable in vivo. Therefore, in vivo genome editing arguably has the potential to modify more cell types relevant to a broader range of human diseases.
Table 1.
Ongoing interventional clinical trials involving CRISPR-based gene editing.
| Condition | Editing strategy | Target gene | Delivery mode | Phase | Sponsor | ClinicalTrials.gov identifier |
|---|---|---|---|---|---|---|
| Beta-thalassemia | ND | ND | ex vivo (HSCs) | NA | Shanghai Bioray Laboratory Inc. | NCT04211480 |
| ND | ND | ex vivo (HSCs) | Phase 1/2 | Shanghai Bioray Laboratory Inc. | NCT04205435 | |
| Gene knock-out | BCL11A | ex vivo (CD34+ HSPCs) | Phase 1/2 | Vertex Pharmaceuticals Inc. | NCT03655678 | |
| Sickle cell disease | Gene knock-out | BCL11A | ex vivo (CD34+ HSPCs) | Phase 1/2 | Vertex Pharmaceuticals Inc. | NCT03745287 |
| Advanced stage EBV-associated malignancies | Gene knock-out | PD-1 | ex vivo (CTL) | Phase 1/2 | The Affiliated Nanjing Drum Tower Hospital of Nanjing University Medical School | NCT03044743 |
| B cell malignancy | Gene knock-out & insertion of CAR using lentiviral vector | TCR, B2M | ex vivo (T cells) | Phase 1/2 | Chinese PLA General Hospital | NCT03166878 |
| ND | ND | ex vivo (T cells) | Phase 1/2 | Chinese PLA General Hospital | NCT03398967 | |
| Gene knock-out and insertion | TRAC, B2M, CAR | ex vivo (T cells) | Phase 1/2 | CRISPR Therapeutics | NCT04035434 | |
| Gene knock-out & insertion of CAR using lentiviral vector | HPK1 | ex vivo (T cells) | Phase 1 | Xijing Hospital | NCT04037566 | |
| Esophageal cancer | Gene knock-out | PD-1 | ex vivo (T cells) | NA | Hangzhou Cancer Hospital | NCT03081715 |
| Metastatic non-small cell lung cancer | Gene knock-out | PD-1 | ex vivo (T cells) | Phase 1 | Sichuan University | NCT02793856 |
| Multiple myeloma, Melanoma, Synovial sarcoma, Myxoid/round Cell Liposarcoma | Gene knock-out & insertion of NYESO-1 using lentiviral vector | TCR, PD-1 | ex vivo (T cells) | Phase 1 | University of Pennsylvania | NCT03399448 |
| Multiple solid tumors | Gene knock-out & insertion of CAR using unspecified method | TCR, PD-1 | ex vivo (T cells) | Phase 1 | Chinese PLA General Hospital | NCT03545815 |
| Gene knock-out & insertion of CAR using unspecified method | PD-1 | ex vivo (T cells) | Phase 1 | Chinese PLA General Hospital | NCT03747965 | |
| Multiple myeloma | Gene knock-out and insertion of CAR | TRAC, B2M, CAR | ex vivo (T cells) | Phase 1 | CRISPR Therapeutics | NCT04244656 |
| T cell malignancy | Gene knock-out & insertion of CAR using unspecified method | CD7 | ex vivo (T cells) | Phase 1 | Baylor College of Medicine | NCT03690011 |
| HIV infection | Gene knock-out | CCR5 | ex vivo (CD34+ HSPCs) | NA | Affiliated Hospital to Academy of Military Medical Sciences | NCT03164135 |
| HPV-related cervical intraepithelial neoplasia I | Gene knock-out | HPV E6/E7 | in vivo (plasmid in gel) | Phase 1 | First Affiliated Hospital, Sun Yat-Sen University | NCT03057912 |
| LCA10 | Gene correction | CEP290 | in vivo (rAAV) | Phase 1/2 | Allergan | NCT03872479 |
EBV: Epstein-Barr virus; HIV: human immunodeficiency virus; HPV: human papillomavirus; LCA10: Leber's congenital amaurosis 10; ND: not disclosed; CAR: chimeric antigen receptor; HSC: hematopoietic stem cell; HSPC: hematopoietic stem and progenitor cell; CTL: cytotoxic T lymphocyte; rAAV: recombinant adeno-associated virus; NA: not applicable. Data are from www.clinicaltrials.gov (accessed on February 6, 2020).
Key to the success of in vivo genome editing is safe and effective delivery of genome editing reagents to the target tissue and cell types. AAV vectors have been widely used for in vivo gene therapy delivery (Wang et al., 2019). Using AAV vectors for in vivo delivery of CRISPR-based genome editing therapeutics has been reported in numerous studies involving disease models and wild-type animals (Lau and Suh, 2017). Along with these continuing successes in proof-of-concept animal studies, the first human application has gained regulatory approval. A phase 1/2 clinical trial employing AAV-CRISPR delivery directly to the eye to correct a CEP290 mutation is currently open for LCA10 patient enrollment (ClinicalTrials.gov Identifier: NCT03872479). Although the success of AAV vectors in gene therapy has paved the way for delivering genome editing therapeutics, the nature of CRISPR-based reagents presents unique challenges. Here, we focus on two issues associated with using AAV vectors for CRISPR delivery, namely the packaging size limit of AAV vectors and undesired editing outcomes.
Fitting big CRISPR into small AAV
Smaller Cas proteins
AAV is a small non-enveloped virus with a diameter of ~25 nm. The icosahedral protein capsid packs a ssDNA genome of ~4.7 kb flanked by two inverted terminal repeats (ITRs). Most of the viral genome sequence can be replaced with a transgene cassette generating recombinant AAV (rAAV), and only the two ITRs totaling ~0.3 kb are required during rAAV production. Despite decades of research, the vector genome seems to be limited to ~5.0 kb (Dong et al., 1996), leaving a maximal length of ~4.7 kb for the transgene cassette. Cas9 from Streptococcus pyogenes (SpyCas9), which has a permissive PAM and is widely used, is 4.2 kb, and therefore necessitates the use of short gene regulatory elements including promoter and polyadenylation signal totaling less than 0.5 kb. This excludes some promoters that have desired expression strength and specificity. Furthermore, other genome editing components must be carried on a separate AAV vector, such as sgRNA expression cassette(s) and donor template for HDR or gene insertion. A dual-vector delivery scheme can achieve genome editing only when both vectors are taken up by the same cell, potentially limiting editing efficiency. Nevertheless, this approach is being pursued for many applications, some of which are discussed below.
An early approach to overcome this challenge was to identify smaller orthologs of Cas9 (Ran et al., 2015). Additional smaller Cas proteins have since been discovered or engineered and experimentally validated as effective genome editing tools in mammalian cells (Kim et al., 2017; Konermann et al., 2018; Liu et al., 2019; Strecker et al., 2019; Teng et al., 2018) (Table 2). These Cas proteins are more compatible with AAV delivery, enabling additional vector design options such as expanded promoter choices and a streamlined delivery scheme. For example, Cas9 from Staphylococcus aureus (SauCas9) has a gene size of 3.2 kb, allowing a single AAV vector to express SauCas9 together with one or two sgRNAs (Ran et al., 2015). Recently, an “all-in-one” AAV8 vector was reported to not only express SauCas9 and two sgRNAs, but also carry a self-linearizing repair template. Together, this vector was shown to correct an Fah mutation in mice to treat HT-1 (Krooss et al., 2020).
Table 2.
Gene sizes of selected CRISPR-based tools in comparison with AAV vector genome size.
| Tool | Function | Gene size (kb) | Ref. | |
| Within AAV packaging limit | SpCas9 | Nuclease | 4.2 | Cong et al., 2013 |
| SaCas9 | Nuclease | 3.2 | Ran et al., 2015 | |
| PspCas13b | Ribonuclease | 3.4 | Cox et al., 2017 | |
| RfxCas13d | Ribonuclease | 2.9 | Konermann et al., 2018 | |
| dPspCas13b(tr)-ADARdd | RNA editing | 4.1 | Cox et al., 2017 | |
| dSaCas9-KRAB | CRISPRi | 3.6 | Thakore et al., 2018 | |
| FnCas12a | Nuclease | 3.9 | Zetsche et al., 2015a | |
| BhCas12b | Nuclease | 3.3 | Strecker et al., 2019 | |
| Beyond AAV packaging limit | BE4 | Cytosine base editing | 5.6 | Komor et al., 2017 |
| ABE7.10 | Adenine base editing | 5.3 | Gaudelli et al., 2017 | |
| dSpCas9-TET1 | Editing DNA methylation | 6.4 | Liu et al., 2016 | |
| PE2 | Prime editing | 6.4 | Anzalone et al., 2019 | |
| AAV vector component | Function | Size (kb) | ||
| ITR × 2 | Vector genome rescue and replication during packaging; Packaging signal | 0.3 total | ||
| Transgene cassette | Transgene expression | < 4.7 | ||
Generating full-length protein using dual AAV vectors
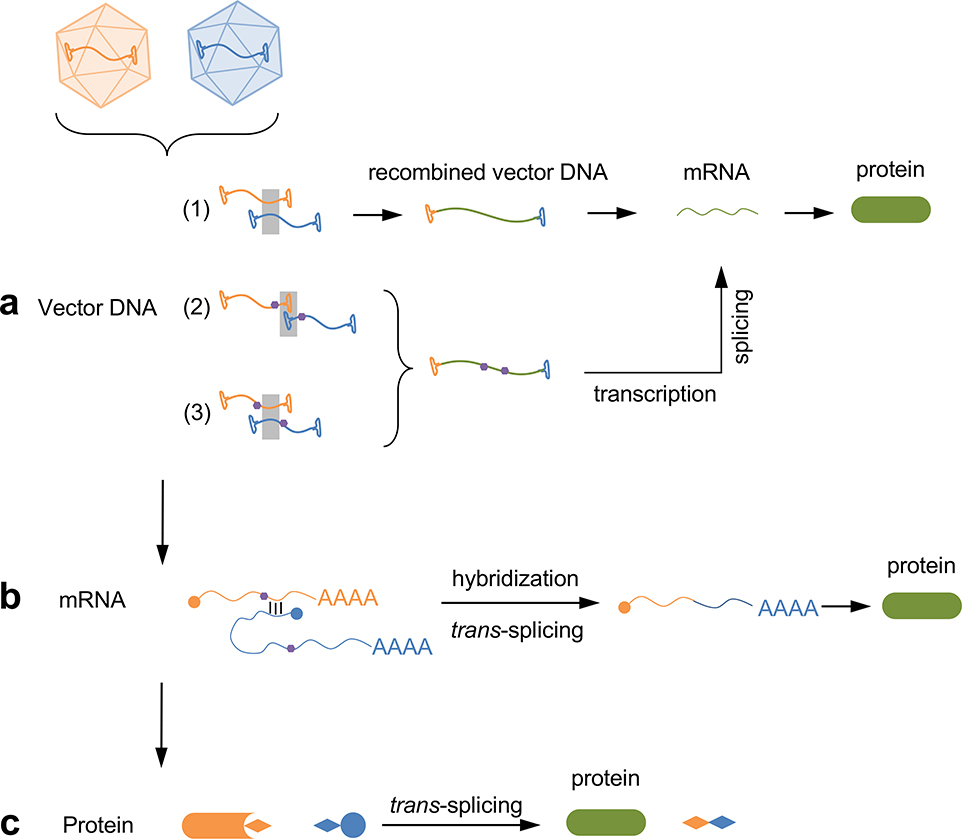
The above-mentioned Cas-effector fusion platforms, especially base editors and prime editors, require delivering even larger cargoes that exceed the packaging limit of AAV (Table 2). In general, delivering over-sized transgenes has been a longstanding hurdle in the AAV gene therapy field, and several approaches have been developed to address this challenge (Patel et al., 2019; Tornabene and Trapani, 2020). The principle is to split the large transgene into two or more segments, each packaged into an individual AAV vector. Delivering all AAV vectors to the same cell leads to the reconstitution of the full-length gene product (Figure 3). This can be achieved at any level along the flow of genetic information. At the DNA level, AAV ITR, a partial transgene sequence, or an optimized recombinogenic sequence present in both vector genomes can promote inter-vector DNA recombination (Duan et al., 2000; Ghosh et al., 2008; Lai et al., 2005; Nakai et al., 2000; Sun et al., 2000) (Figure 3a). The recombined vector genome undergoes transgene expression, during which purposefully designed splicing signals mediate pre-mRNA splicing to remove the overlapping sequence, generating a mature transcript that is translated into the desired protein. This strategy was employed to deliver a base editor using dual AAV vectors into a DMD mouse model (Ryu et al., 2018). At the RNA level, two transcripts derived from separate AAV vector genomes can partially hybridize to promote trans-splicing mediated by the splicing donor and acceptor present in the two transcripts, respectively (Pergolizzi et al., 2003; Song et al., 2009) (Figure 3b). The fully spliced mature transcript leads to full-length protein production.
Figure 3. Reconstitution of full-length protein expression from two AAV vectors.
(a) The two AAV vector genomes can undergo recombination mediated by an overlapping sequence (1), ITR (2), or a recombinogenic sequence (3) to form a recombined vector DNA that encodes the full-length protein. The transcript derived from the recombined vector DNA may undergo splicing by engineered splicing signals (purple pentagons), (b) The two mRNAs transcribed from individual vector genomes can undergo trans-splicing to form a chimeric mRNA encoding the full-length protein, which is promoted by the hybridization domains (short black bars), (c) The two half proteins expressed from the two AAV vectors can undergo protein trans-splicing mediated by split inteins (diamonds) to form a full-length protein.
Reconstitution at the protein level by split inteins has been increasingly investigated as another approach to express large proteins using AAV vectors (Chew et al., 2016; Fine et al., 2015; Li et al., 2008; Truong et al., 2015; Villiger et al., 2018). An intein is a protein segment that can excise itself and ligate the nearby protein segments with a peptide bond, similar to an intron in pre-mRNA splicing (Shah and Muir, 2014). Split inteins are a pair of natural polypeptides that, when present at the termini of two other proteins, mediate protein trans-splicing (PTS) (Aranko et al., 2014) (Figure 3c). In contrast to pre-mRNA splicing that requires complex cellular machineries and has a certain promiscuity, split inteins are self-sufficient to mediate precise PTS. Therefore, a split intern-mediated protein reconstitution strategy is potentially robust and precise in various cell types, which is an important feature for therapeutics. In a recent report, base editors were systematically studied for split intein reconstitution and AAV vector delivery to mice (Levy et al., 2020). The optimized base editors achieved genome editing in multiple tissues including the brain, liver, retina, heart, and skeletal muscle, with editing efficiencies ranging from 9% - 59%. As expected, the choices of split intein, split site, and base editor design are important parameters. In addition, AAV capsid, dose, and route of administration also have a profound impact on the final editing efficiency, highlighting the synergy between CRISPR-based reagents and AAV-mediated delivery to fully unleash the therapeutic potential. Notably, base editing constructs delivered by intein-mediated PTS was more efficient than reconstitution at the AAV vector DNA level in a side-by-side comparison, likely due to the multiple cellular processes in the latter approach that together compromise efficiency (Levy et al., 2020).
Control of undesired genome editing outcomes
Inducible expression
Although AAV delivery can mediate durable transgene expression in post-mitotic cells, for many genome editing applications, this is not necessary because the mutation is permanently corrected after being edited. Furthermore, long-term expression of an active genome editing system poses a safety concern, because it has been shown to increase off-target cleavage (Zuris et al., 2015). Therefore, transient expression of CRISPR-based genome editing machineries is preferred in many therapeutic settings. To achieve this, non-viral delivery methods can be used to deliver mRNA or proteins, which will eventually be degraded in cells (We refer the reader to recent reviews for details about progress in this area (Chen et al., 2019; Wan et al., 2019; Xu et al., 2019a)). While these methods hold great potential for in vivo applications such as those targeting the liver (Finn et al., 2018; Yin et al., 2016), their delivery efficiency and range of targetable tissues and cell types beyond liver are currently limited compared to AAV vectors. A straightforward way to make AAV vectors compatible with transient delivery is to incorporate a molecular apparatus for inducible expression. Many approaches for this purpose have been reported to function in cell culture, such as using a drug- inducible promoter for gene expression, protein activation by light, drug-inducible assembly or reconstitution from a split construct, and protein inactivation by a drug-inducible destabilizing domain (Davis et al., 2015; Dow et al., 2015; Kumar et al., 2018; Nguyen et al., 2016; Nihongaki et al., 2015; Rose et al., 2017; Tak et al., 2017; Truong et al., 2015; Wright et al., 2015; Zetsche et al., 2015). The translational value of these methods for in vivo applications remains to be investigated. Ideally, an AAV-compatible, inducible genome editing system would be a molecular circuit that comprises a simple transgene design to fit in AAV vectors and a safe trigger for activation or inactivation. Such a system was explored (Ye et al., 1999), but remains to be translated to clinical applications.
Self-cleavage AAV-CRISPR
An interesting idea to achieve transient function of AAV-CRISPR is to engineer an intrinsic destabilization mechanism into the vector itself by taking advantage of the DNA cleavage activity of the delivered CRISPR machinery. Along with the desired therapeutic genome editing event, DNA cleavage of the vector genome occurs simultaneously (Li et al., 2019a; Li et al., 2019b). This intriguing approach has been shown to limit SauCas9 expression in mice without significantly compromising on-target editing. However, the reduction in SauCas9 protein levels was not complete, leaving some possibility of off-target cleavage by the remaining vector expression (Li et al., 2019a). Moreover, the cleaved AAV vector DNA was found to integrate into the on-target genomic site (Li et al., 2019a), and the potential impact of this on-target but undesired editing on safety remains to be evaluated.
Spatial control of genome editing
In addition to temporal control, avoiding genome editing in unintended tissues and cell types will also improve the clinical safety profile of CRISPR-based therapeutics. Although different AAV capsids provide a spectrum of tissue tropism, they often differ only in the tropism strength but not absolute specificity. Tissue-specific promoters are widely used in AAV vector design for spatial control of transgene expression. As mentioned earlier, the large gene sizes of Cas proteins require compact promoters that usually do not exhibit high specificity. A potentially viable approach to exclude Cas protein expression in certain tissue types is to include binding sites for an endogenous microRNA (miRNA) in the 3’ UTR of the Cas protein gene cassette (Brown et al., 2006; Geisler et al., 2011; Qiao et al., 2011; Xiao et al., 2019; Xie et al., 2011). Delivery to tissues that highly express the miRNA will silence Cas protein expression, whereas other tissues lacking the miRNA will express the Cas protein for genome editing. In addition, silencing Cas protein expression in antigen presenting cells may help mitigate immune responses against Cas9 (Xiao et al., 2019). Utilizing this detargeting concept and combining it with natural inhibitors of Cas proteins, known as anti-CRISPRs (Acr), can lead to cell- and tissue-specific targeted genome editing (Hoffmann et al., 2019; Lee et al., 2019). In this elegant system, Cas and Acr proteins are co-delivered by AAV vectors. Acr protein expression is subjected to tissue-specific miRNA-mediated suppression, which in turn allows for Cas protein expression and genome editing in that tissue.
AAV vector DNA integration at on-target DSBs
It was shown that AAV vector DNA can integrate at pre-existing chromosomal breaks induced by the endonuclease I-SceI or irradiation in human cells (Miller et al., 2004). Similarly, when AAV was used to deliver CRISPR to generate DSBs in model animals in vivo, AAV vector DNA integration at the on-target cleavage site occurred with frequencies approaching or exceeding that of indels in some studies (Hanlon et al., 2019; Jarrett et al., 2017; Maeder et al., 2019; McCullough et al., 2019; Nelson et al., 2019; Yoon et al., 2018). In general, such integration events are undesired, and their biological consequences remain to be evaluated in the context of specific applications, where parameters including the AAV vector design, on-target and off-target genomic loci, and editing goals will all need to be taken into consideration. Furthermore, it is worth exploring whether AAV vector integration is a therapeutically relevant editing outcome in certain applications, such as to mediate permanent gene insertion in dividing cells (Wang et al., 2020). It has been suggested that AAV ITRs facilitate vector integration at DSBs (Miller et al., 2004), and the ITR has been found in most integrated sequences (Jarrett et al., 2017). Therefore, a better understanding of the mechanistic role of ITR in mediating AAV vector integration at DSBs may provide not only insight into this genome editing outcome, but also clues to harness this phenomenon for meaningful applications.
CHALLENGES AND FUTURE PERSPECTIVES
The advancement of AAV-CRISPR therapeutics faces the same set of obstacles facing the development of AAV-based gene therapy in general, such as pre-existing immunity against AAV capsids and vector-induced immune responses, delivery efficiency and specificity, and AAV vector manufacturing. These topics were discussed in depth in recent reviews (Colella et al., 2018; Domenger and Grimm, 2019; Rabinowitz et al., 2019; Verdera et al., 2020; Wang et al., 2019). In addition, clinical application of AAV-CRISPR presents unique challenges that require distinct solutions. For example, pre-existing humoral and cellular immunity against commonly used Cas9 orthologs has been reported in general human populations due to widespread infections of the bacteria from which these proteins are derived (Charlesworth et al., 2019; Simhadri et al., 2018; Wagner et al., 2019). It remains to be evaluated whether delivering vectored Cas9 proteins directly into the human body in the presence of pre-existing anti-Cas9 immunity will compromise safety or therapeutic efficacy (Crudele and Chamberlain, 2018). Potential routes to addressing the issue of immunogenicity includes use of immune-orthogonal Cas9 orthologs (Moreno et al., 2019) and further exploration of the natural diversity of CRISPR systems to identify new enzymes that may be less immunogenic in human cells. Furthermore, various protein engineering approaches can improve clinically relevant features, such as mapping and editing epitopes for a better immunological profile.
Off-target editing needs to be carefully assessed for any genome editing therapy. A number of methods to characterize off-targets of CRISPR-based reagents have been developed (Akcakaya et al., 2018; Cameron et al., 2017; Crosetto et al., 2013; Kim et al., 2015; Tsai et al., 2017; Tsai et al., 2015; Wienert et al., 2019; Yan et al., 2017; Zuo et al., 2019). Although these methods provide sensitive measurement of off-targets at nucleic acid sequence level, the potential biological consequences stemming from any off-target event largely have to be evaluated empirically. Because off-targets heavily depend on the species-specific genomic sequence and the in vivo delivery method, using animal models that are humanized at the cellular or tissue level can provide valuable pre-clinical information. To improve specificity, various protein engineering approaches have yielded high-fidelity versions of Cas and Cas-effector fusion proteins (Casini et al., 2018; Chen et al., 2017; Hu et al., 2018; Kleinstiver et al., 2016; Kleinstiver et al., 2019; Lee et al., 2018; Slaymaker et al., 2015; Vakulskas et al., 2018). The other components of CRISPR-based therapeutics, such as sgRNA and donor template, have also been subjected to modifications to enhance editing precision and efficiency (Cromwell et al., 2018; Fu et al., 2014; Kocak et al., 2019; Mir et al., 2018; Yin et al., 2018).
In addition to off-target editing, on-target editing can also lead to undesired editing events, such as large deletions extending to nearby genes (Adikusuma et al., 2018; Kosicki et al., 2018), chromosome rearrangements including inversions and translocations (Frock et al., 2015; Maeder et al., 2019), and AAV vector genome integration (Hanlon et al., 2019; Nelson et al., 2019). Profiling these events usually requires a set of techniques different from the ones used to monitor off-target cleavage (Frock et al., 2015). It is worth emphasizing that a therapeutic genome editing strategy synergizes the action of the editor and multiple cellular processes triggered by the editor, such as DNA repair pathways in the case of utilizing the nuclease activity of Cas9. Therefore, a better understanding of these relevant cellular processes will provide valuable insights into genome editing outcomes and safety (Yeh et al., 2019).
Disease animal models are critical in translational research and therapy development. Because in vivo delivery is a major bottleneck in AAV-CRISPR therapeutics, directly testing in disease animal models is the gold standard to evaluate therapeutic efficacy. For AAV gene replacement therapy that aims to deliver a functional gene copy to correct recessive diseases, animal models created by various gene knock-out strategies are usually sufficient. In contrast, many genome editing strategies, such as base editing and prime editing, often target a particular mutation. Therefore, animal models harboring deliberately engineered mutations are required to provide a suitable testing platform. For example, several DMD mouse models with representative dystrophin gene mutations were purposefully generated to demonstrate the effectiveness of various genome editing strategies (Amoasii et al., 2017). Although the advent of CRISPR-based genome editing technology has greatly facilitated the generation of genetically modified animals, the lack of suitable animals with patient-relevant mutations remains a major challenge. Developing easy, robust, and rapid animal modeling approaches will likely expedite the development of precise genome editing therapeutics.
CONCLUSIONS
CRISPR and AAV are just two examples illustrating how natural systems can be harnessed for revolutionary biotechnologies, highlighting the exciting opportunities that can emerge from a deeper understanding of and appreciation for natural diversity. However, adaptation of microbial machineries to meet human needs is not guaranteed to succeed. This adaptation process involves multidisciplinary expertise spanning various research fields such as microbiology, biochemistry, molecular biology, protein engineering, structural biology, and bioinformatics. For CRISPR-based technology, the simplicity of its design and utilization should not disguise the importance of openly sharing valuable resources ranging from plasmids and sgRNA libraries to transgenic animals and bioinformatic tools, which has encouraged cross-validation and speedy adoption of the technology for novel applications and will continue to foster rapid scientific and technological advances.
In general, delivery is among the most urgent obstacles hindering in vivo gene therapy, including genome editing therapeutic approaches. AAV vectors are currently the most effective gene delivery vehicle, and the only delivery vehicle approved to introduce CRISPR-based genome editing therapeutics directly into the human body. It will be exciting to follow the on-going AAV-CRISPR clinical trial and glean new insight into the strengths and weaknesses of this approach that can then inform further development and refinement of the system. Such refinements are expected to come both from the efforts to modify CRISPR-based reagents to fit the intrinsic features of AAV vectors and from ongoing research in AAV biology and vectorology. Merging the findings from these two fields will yield a broader range of delivery platforms that can better serve emerging molecular therapeutics.
ACKNOWLEGEMENTS
D.W. is funded by the Pitt Hopkins Research Foundation, Grace Science Foundation, Believe In A Cure. F.Z. is supported by NIH grants (1R01-HG009761, 1R01-MH110049, 1DP1-HL141201, and 5R-M1HG006193-09); the Howard Hughes Medical Institute; the Harold G. and Leila Mathers, and Edward Mallinckrodt, Jr. Foundations; the Poitras Center for Psychiatric Disorders Research; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and by the Phillips family and J. and P. Poitras. G.G. is funded by the NIH, Cystic Fibrosis Foundation, Michelson Found Animals Foundation, and industrial partners under sponsored research agreements.
Footnotes
DECLARATION OF INTERESTS
D.W. is an inventor on issued patents and patent applications relating to AAV- and CRISPR- based technologies. F.Z. is an inventor on issued patents and patent applications relating to CRISPR- based technologies. F.Z. is a co-founder and scientific advisor of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences. G.G. is an inventor on issued patents and patent applications relating to AAV- and CRISPR-based technologies. G.G. is a co-founder of Voyager Therapeutics and Aspa Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Franklin B, Koob J, Kellner MJ, Ladha A, Joung J, Kirchgatterer P, Cox DBT, and Zhang F (2019). A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, and Thomas PQ (2018). Large deletions induced by Cas9 cleavage. Nature 560, E8–E9. [DOI] [PubMed] [Google Scholar]
- Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, Fellows MD, Porritt MJ, Firth MA, Carreras A, et al. (2018). In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoasii L, Long C, Li H, Mireault AA, Shelton JM, Sanchez-Ortiz E, McAnally JR, Bhattacharyya S, Schmidt F, Grimm D, et al. (2017). Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Science translational medicine 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranko AS, Wlodawer A, and Iwai H (2014). Nature’s recipe for splitting inteins. Protein engineering, design & selection : PEDS 27, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SR, and Maus MV (2019). Gene editing for immune cell therapies. Nature biotechnology 37, 1425–1434. [DOI] [PubMed] [Google Scholar]
- Bak RO, Dever DP, Reinisch A, Cruz Hernandez D, Majeti R, and Porteus MH (2017). Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AaV6. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, Levy R, Akhtar AA, Breunig JJ, Svendsen CN, et al. (2016). In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Molecular therapy : the journal of the American Society of Gene Therapy 24, 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, and Marraffini LA (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic acids research 41, 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, and van der Oost J (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L, and Naldini L (2006). Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nature medicine 12, 585–591. [DOI] [PubMed] [Google Scholar]
- Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, et al. (2017). Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nature methods 14, 600–606. [DOI] [PubMed] [Google Scholar]
- Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, Lorenzin F, Prandi D, Romanel A, Demichelis F, et al. (2018). A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nature biotechnology 36, 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick AC, Wang X, and Musunuru K (2017). In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arteriosclerosis, thrombosis, and vascular biology 37, 1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, and Boulton SJ (2012). Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47, 497–510. [DOI] [PubMed] [Google Scholar]
- Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, et al. (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 25, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Alphonse M, and Liu Q (2019). Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip Rev Nanomed Nanobiotechnol, e1609. [DOI] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, and Doudna JA (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, and Jaenisch R (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, and Church GM (2016). A multifunctional AAV-CRISPR-Cas9 and its host response. Nature methods 13, 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G, Lignani G, Brusco S, Di Berardino C, Carpenter J, Giannelli S, Valassina N, Bido S, Ricci R, Castoldi V, et al. (2019). dCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Molecular therapy : the journal of the American Society of Gene Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella P, Ronzitti G, and Mingozzi F (2018). Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol Ther Methods Clin Dev 8, 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, and Zhang F (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell CR, Sung K, Park J, Krysler AR, Jovel J, Kim SK, and Hubbard BP (2018). Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat Commun 9, 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. (2013). Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nature methods 10, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crudele JM, and Chamberlain JS (2018). Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat Commun 9, 3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Pattanayak V, Thompson DB, Zuris JA, and Liu DR (2015). Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nature chemical biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, and Musunuru K (2014). Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res 115, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenger C, and Grimm D (2019). Next-generation AAV vectors-do not judge a virus (only) by its cover. Human molecular genetics 28, R3–R14. [DOI] [PubMed] [Google Scholar]
- Dong JY, Fan PD, and Frizzell RA (1996). Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Human gene therapy 7, 2101–2112. [DOI] [PubMed] [Google Scholar]
- Doudna JA (2020). The promise and challenge of therapeutic genome editing. Nature 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, and Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- Dow LE, Fisher J, O’Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, and Lowe SW (2015). Inducible in vivo genome editing with CRISPR-Cas9. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Yue Y, Yan Z, and Engelhardt JF (2000). A new dual-vector approach to enhance recombinant adeno-associated virus-mediated gene expression through intermolecular cis activation. Nature medicine 6, 595–598. [DOI] [PubMed] [Google Scholar]
- Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, and Sadelain M (2018). Gene therapy comes of age. Science 359. [DOI] [PubMed] [Google Scholar]
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, and Davies KE (1990). Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature 343, 180–182. [DOI] [PubMed] [Google Scholar]
- Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, and Sadelain M (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine EJ, Appleton CM, White DE, Brown MT, Deshmukh H, Kemp ML, and Bao G (2015). Trans-spliced Cas9 allows cleavage of HBB and CCR5 genes in human cells using compact expression cassettes. Scientific reports 5, 10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, et al. (2018). A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep 22, 2227–2235. [DOI] [PubMed] [Google Scholar]
- Friedmann T, and Roblin R (1972). Gene therapy for human genetic disease? Science 175, 949–955. [DOI] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, and Alt FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, and Joung JK (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology 32, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, and Barbas CF 3rd (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler A, Jungmann A, Kurreck J, Poller W, Katus HA, Vetter R, Fechner H, and Muller OJ (2011). microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene therapy 18, 199–209. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Yue Y, Lai Y, and Duan D (2008). A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Molecular therapy : the journal of the American Society of Gene Therapy 16, 124–130. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy B, Nist-Lund C, Pan B, Asai Y, Karavitaki KD, Kleinstiver BP, Garcia SP, Zaborowski MP, Solanes P, Spataro S, et al. (2019). Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nature medicine 25, 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE, et al. (2019). High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 10, 4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High KA, and Roncarolo MG (2019). Gene Therapy. The New England journal of medicine 381, 455–464. [DOI] [PubMed] [Google Scholar]
- Hoffmann MD, Aschenbrenner S, Grosse S, Rapti K, Domenger C, Fakhiri J, Mastel M, Borner K, Eils R, Grimm D, et al. (2019). Cell-specific CRISPR-Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic acids research 47, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, and Zhang F (2014). Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, et al. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Suresh S, Guo D, Daman K, Chen JCJ, Liu P, Zieger M, Luk K, Roscoe BP, Mueller C, et al. (2019). Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, and Bird A (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics 33 Suppl, 245–254. [DOI] [PubMed] [Google Scholar]
- Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta R, Zhang M, Rodriguez PJ, Lee CS, Gillard BK, Bissig KD, et al. (2017). Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Scientific reports 7, 44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, and Kim JS (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nature methods 12, 237–243, 231 p following 243. [DOI] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, et al. (2017). In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8, 14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, and Joung JK (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, et al. (2019). Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocak DD, Josephs EA, Bhandarkar V, Adkar SS, Kwon JB, and Gersbach CA (2019). Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nature biotechnology 37, 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, and Zhang F (2013). Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, and Hsu PD (2018). Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173, 665–676 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, and Bradley A (2018). Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krooss SA, Dai Z, Schmidt F, Rovai A, Fakhiri J, Dhingra A, Yuan Q, Yang T, Balakrishnan A, Steinbruck L, et al. (2020). Ex Vivo/In vivo Gene Editing in Hepatocytes Using “All-in-One” CRISPR-Adeno-Associated Virus Vectors with a Self-Linearizing Repair Template. iScience 23, 100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Stanford W, de Solis C, Aradhana, Abraham ND, Dao TJ, Thaseen S, Sairavi A, Gonzalez CU, and Ploski JE (2018). The Development of an AAV-Based CRISPR SaCas9 Genome Editing System That Can Be Delivered to Neurons in vivo and Regulated via Doxycycline and Cre-Recombinase. Front Mol Neurosci 11, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Yue Y, Liu M, Ghosh A, Engelhardt JF, Chamberlain JS, and Duan D (2005). Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nature biotechnology 23, 1435–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgrave-Gomez J, Mercado-Gomez O, and Guevara-Guzman R (2015). Epigenetic mechanisms in neurological and neurodegenerative diseases. Frontiers in cellular neuroscience 9, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CH, and Suh Y (2017). In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Res 6, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mou H, Ibraheim R, Liang SQ, Liu P, Xue W, and Sontheimer EJ (2019). Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. Rna 25, 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Jung I, Kim D, Kim S, et al.Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 9, 3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JM, Yeh WH, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, and Liu DR (2020). Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng 4, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Lee CM, Hurley AE, Jarrett KE, De Giorgi M, Lu W, Balderrama KS, Doerfler AM, Deshmukh H, Ray A, et al. (2019a). A Self-Deleting AAV-CRISPR System for In Vivo Genome Editing. Mol Ther Methods Clin Dev 12, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Hung SSC, Mohd Khalid MKN, Wang JH, Chrysostomou V, Wong VHY, Singh V, Wing K, Tu L, Bender JA, et al. (2019b). Utility of Self-Destructing CRISPR/Cas Constructs for Targeted Gene Editing in the Retina. Human gene therapy 30, 1349–1360. [DOI] [PubMed] [Google Scholar]
- Li J, Sun W, Wang B, Xiao X, and Liu XQ (2008). Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Human gene therapy 19, 958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CKW, Gapinske M, Brooks AK, Woods WS, Powell JE, Zeballos CM, Winter J, Perez-Pinera P, and Gaj T (2020). Treatment of a Mouse Model of ALS by In Vivo Base Editing. Molecular therapy : the journal of the American Society of Gene Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, Chuck J, Tan D, Knott GJ, Harrington LB, et al. (2019). CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, and Jaenisch R (2019). Editing the Epigenome to Tackle Brain Disorders. Trends Neurosci 42, 861–870. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, et al. (2018). Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992 e976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, and Olson EN (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H, Zhang Y, Min YL, Shelton JM, Mammen PPA, et al. (2018). Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 4, eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, and Joung JK (2013). CRISPR RNA-guided activation of endogenous human genes. Nature methods 10, 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, et al. (2019). Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nature medicine 25, 229–233. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A, Hardin A, Eckalbar WL, Vaisse C, and Ahituv N (2019). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough KT, Boye SL, Fajardo D, Calabro K, Peterson JJ, Strang CE, Chakraborty D, Gloskowski S, Haskett S, Samuelsson S, et al. (2019). Somatic Gene Editing of GUCY2D by AAV-CRISPR/Cas9 Alters Retinal Structure and Function in Mouse and Macaque. Human gene therapy 30, 571–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Petek LM, and Russell DW (2004). Adeno-associated virus vectors integrate at chromosome breakage sites. Nature genetics 36, 767–773. [DOI] [PubMed] [Google Scholar]
- Mir A, Alterman JF, Hassler MR, Debacker AJ, Hudgens E, Echeverria D, Brodsky MH, Khvorova A, Watts JK, and Sontheimer EJ (2018). Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat Commun 9, 2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla KA, and Yang Y (2019). CRISPR/Cas-Mediated Base Editing: Technical Considerations and Practical Applications. Trends in biotechnology 37, 1121–1142. [DOI] [PubMed] [Google Scholar]
- Moreno AM, Palmer N, Aleman F, Chen G, Pla A, Jiang N, Leong Chew W, Law M, and Mali P (2019). Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat Biomed Eng 3, 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Storm TA, and Kay MA (2000). Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nature biotechnology 18, 527–532. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, Robinson-Hamm JN, Bulaklak K, Castellanos Rivera RM, Collier JH, et al. (2019). Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nature medicine 25, 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DP, Miyaoka Y, Gilbert LA, Mayerl SJ, Lee BH, Weissman JS, Conklin BR, and Wells JA (2016). Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat Commun 7, 12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Kawano F, Nakajima T, and Sato M (2015). Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nature biotechnology 33, 755–760. [DOI] [PubMed] [Google Scholar]
- Ohmori T, Nagao Y, Mizukami H, Sakata A, Muramatsu SI, Ozawa K, Tominaga SI, Hanazono Y, Nishimura S, Nureki O, et al. (2017). CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Scientific reports 7, 4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Zhao J, Duan D, and Lai Y (2019). Design of AAV Vectors for Delivery of Large or Multiple Transgenes. Methods Mol Biol 1950, 19–33. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, and Gersbach CA (2013). Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nature methods 10, 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergolizzi RG, Ropper AE, Dragos R, Reid AC, Nakayama K, Tan Y, Ehteshami JR, Coleman SH, Silver RB, Hackett NR, et al. (2003). In vivo trans-splicing of 5’ and 3’ segments of pre-mRNA directed by corresponding DNA sequences delivered by gene transfer. Molecular therapy : the journal of the American Society of Gene Therapy 8, 999–1008. [DOI] [PubMed] [Google Scholar]
- Porteus MH (2019). A New Class of Medicines through DNA Editing. The New England journal of medicine 380, 947–959. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, and Lim WA (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao C, Yuan Z, Li J, He B, Zheng H, Mayer C, Li J, and Xiao X (2011). Liver-specific microRNA-122 target sequences incorporated in AAV vectors efficiently inhibits transgene expression in the liver. Gene therapy 18, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabai A, Reisser L, Reina-San-Martin B, Mamchaoui K, Cowling BS, Nicot AS, and Laporte J Allele-Specific CRISPR/Cas9 Correction of a Heterozygous DNM2 Mutation Rescues Centronuclear Myopathy Cell Phenotypes. Molecular therapy Nucleic acids 16, 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J, Chan YK, and Samulski RJ (2019). Adeno-associated Virus (AAV) versus Immune Response. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, and Liu DR (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nature reviews Genetics 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccio M, Hahnewald S, Perny M, and Senn P (2015). Cell cycle reactivation of cochlear progenitor cells in neonatal FUCCI mice by a GSK3 small molecule inhibitor. Scientific reports 5, 17886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JC, Stephany JJ, Valente WJ, Trevillian BM, Dang HV, Bielas JH, Maly DJ, and Fowler DM (2017). Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nat Methods 14, 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, and Jasin M (1994). Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 91, 6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, et al. (2018). Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nature biotechnology 36, 536–539. [DOI] [PubMed] [Google Scholar]
- Shah NH, and Muir TW (2014). Inteins: Nature’s Gift to Protein Chemists. Chemical science 5, 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, and Sherwood RI (2018). Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. (2015). Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 60, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, and Sauna ZE (2018). Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol Ther Methods Clin Dev 10, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, and Zhang F (2015). Rationally engineered Cas9 nucleases with improved specificity. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, et al. (2017). Cas13b Is a Type VI-B CRISPR- Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell 65, 618–630 e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smih F, Rouet P, Romanienko PJ, and Jasin M (1995). Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic acids research 23, 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CQ, Jiang T, Richter M, Rhym LH, Koblan LW, Zafra MP, Schatoff EM, Doman JL, Cao Y, Dow LE, et al. (2020). Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng 4, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Lou HH, Boyer JL, Limberis MP, Vandenberghe LH, Hackett NR, Leopold PL, Wilson JM, and Crystal RG (2009). Functional cystic fibrosis transmembrane conductance regulator expression in cystic fibrosis airway epithelial cells by AAV6.2-mediated segmental trans-splicing. Human gene therapy 20, 267–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, Makarova KS, Koonin EV, and Zhang F (2019). Engineering of CRISPR-Cas12b for human genome editing. Nat Commun 10, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Li J, and Xiao X (2000). Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nature medicine 6, 599–602. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology- independent targeted integration. Nature 540, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. (2016). In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YE, Kleinstiver BP, Nunez JK, Hsu JY, Horng JE, Gong J, Weissman JS, and Joung JK (2017). Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nature methods 14, 1163–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, Li T, Li J, Zhou Q, and Li W (2018). Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, Kwon JB, Nelson CE, Rouse DC, Gemberling MP, Oliver ML, and Gersbach CA (2018). RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat Commun 9, 1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, and Capecchi MR (1987). Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503–512. [DOI] [PubMed] [Google Scholar]
- Thuronyi BW, Koblan LW, Levy JM, Yeh WH, Zheng C, Newby GA, Wilson C, Bhaumik M, Shubina-Oleinik O, Holt JR, et al. (2019). Continuous evolution of base editors with expanded target compatibility and improved activity. Nature biotechnology 37, 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornabene P, and Trapani I (2020). Can Adeno-Associated Viral Vectors Deliver Effectively Large Genes? Human gene therapy 31, 47–56. [DOI] [PubMed] [Google Scholar]
- Truong DJ, Kuhner K, Kuhn R, Werfel S, Engelhardt S, Wurst W, and Ortiz O (2015). Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic acids research 43, 6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, and Joung JK (2017). CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nature methods 14, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD (2018). Genome Editing B.C. (Before CRISPR): Lasting Lessons from the “Old Testament”. CRISPR J 1, 34–46. [DOI] [PubMed] [Google Scholar]
- Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, Bode NM, McNeill MS, Yan S, Camarena J, et al. (2018). A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nature medicine 24, 1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, Capurso D, Carter MM, Thompson MS, Frias E, Russ C, Reece-Hoyes JS, Nye C, Gradia S, Vidal B, et al. (2016). DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol Cell. [DOI] [PubMed] [Google Scholar]
- Verdera HC, Kuranda K, and Mingozzi F (2020). AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Molecular therapy : the journal of the American Society of Gene Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, Fingerhut R, Haberle J, Matos J, Robinson MD, et al. (2018). Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nature medicine 24, 1519–1525. [DOI] [PubMed] [Google Scholar]
- Wagner DL, Amini L, Wendering DJ, Burkhardt LM, Akyuz L, Reinke P, Volk HD, and Schmueck-Henneresse M (2019). High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nature medicine 25, 242–248. [DOI] [PubMed] [Google Scholar]
- Wan T, Niu D, Wu C, Xu F, Church G, and Ping Y (2019). Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Materials Today 26, 27. [Google Scholar]
- Wang D, Li J, Song CQ, Tran K, Mou H, Wu PH, Tai PWL, Mendonca CA, Ren L, Wang BY, et al. (2018). Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nature biotechnology 36, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tai PWL, and Gao G (2019). Adeno-associated virus vector as a platform for gene therapy delivery. Nature reviews Drug discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang Y, Breton C, Bell P, Li M, Zhang J, Che Y, Saveliev A, He Z, White J, et al. A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency. Sci Adv 6, eaax5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, Vu JT, Kazane KR, Watry HL, et al. (2019). Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, and Doudna JA (2015). Rational design of a split-Cas9 enzyme complex. Proceedings of the National Academy of Sciences of the United States of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Muhuri M, Li S, Qin W, Xu G, Luo L, Li J, Letizia AJ, Wang SK, Chan YK, et al. (2019). Circumventing cellular immunity by miR142-mediated regulation sufficiently supports rAAV-delivered OVA expression without activating humoral immunity. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Xie Q, Zhang H, Ameres SL, Hung JH, Su Q, He R, Mu X, Seher Ahmed S, Park S, et al. (2011). MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Molecular therapy : the journal of the American Society of Gene Therapy 19, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CF, Chen GJ, Luo YL, Zhang Y, Zhao G, Lu ZD, Czarna A, Gu Z, and Wang J (2019a). Rational designs of in vivo CRISPR-Cas delivery systems. Advanced drug delivery reviews. [DOI] [PubMed] [Google Scholar]
- Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, Wang L, Liu T, Wang X, Zhang B, et al. (2019b). CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. The New England journal of medicine 381, 1240–1247. [DOI] [PubMed] [Google Scholar]
- Xu L, Yang H, Gao Y, Chen Z, Xie L, Liu Y, Liu Y, Wang X, Li H, Lai W, et al. (2017). CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Molecular therapy : the journal of the American Society of Gene Therapy 25, 17821789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, and Scott DA (2018). Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol Cell 70, 327–339 e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, et al. (2017). BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8, 15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Rivera VM, Zoltick P, Cerasoli F Jr., Schnell MA, Gao G, Hughes JV, Gilman M, and Wilson JM (1999). Regulated delivery of therapeutic proteins after in vivo somatic cell gene transfer. Science 283, 88–91. [DOI] [PubMed] [Google Scholar]
- Yeh CD, Richardson CD, and Corn JE (2019). Advances in genome editing through control of DNA repair pathways. Nat Cell Biol 21, 1468–1478. [DOI] [PubMed] [Google Scholar]
- Yeh WH, Chiang H, Rees HA, Edge ASB, and Liu DR (2018). In vivo base editing of post-mitotic sensory cells. Nat Commun 9, 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, et al. (2016). Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nature biotechnology 34, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Suresh S, Kwan SY, Wu Q, Walsh S, Ding J, Bogorad RL, Zhu LJ, Wolfe SA, et al. (2018). Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nature chemical biology 14, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, and Anderson DG (2014). Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature biotechnology 32, 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Wang D, Tai PWL, Riley J, Gao G, and Rivera-Perez JA (2018). Streamlined ex vivo and in vivo genome editing in mouse embryos using recombinant adeno-associated viruses. Nat Commun 9, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, and Zhang F (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature biotechnology 33, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F (2019). Development of CRISPR-Cas systems for genome editing and beyond. Quarterly Reviews of Biophysics 51, 31. [Google Scholar]
- Zhao X, Liu L, Lang J, Cheng K, Wang Y, Li X, Shi J, Wang Y, and Nie G (2018). A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment. Cancer letters 431, 171–181. [DOI] [PubMed] [Google Scholar]
- Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, and Yang H (2019). Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, and Liu DR (2015). Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature biotechnology 33, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]