Abstract
Mechanisms of activation, signaling, and trafficking of adhesion G protein–coupled receptors (aGPCRs) have remained largely unknown. Several aGPCRs, including GPR56/ADGRG1 and GPR64/ADGRG2, show increased activity in the absence of their N-terminal fragment (NTF). This constitutive signaling is plausibly caused by the binding of extracellular N-terminal 15–25 amino acid–long tethered agonist to extracellular domains of the cognate aGPCRs. To test the role of NTF and tethered agonist in GPR64 signaling and endocytosis, we generated mutants that lack either NTF alone (ΔNTF) or NTF and tethered agonist (P622). We discover that unlike full-length GPR64, ΔNTF and P622 mutants interact with β-arrestin1 and β-arrestins2 and are constitutively internalized in steady states. However, only ΔNTF shows exaggerated basal activation of the Gαs–cAMP–CRE signaling cascade. Neither ΔNTF nor P622 shows constitutive activation of the Gα13–SRE pathway, but both mutants respond to exogenously added agonistic peptide via CRE and SRE. GPCR kinases and dynamin mediate the constitutive internalization of ΔNTF and P622 to early endosomes, where ΔNTF constantly induces CRE. These data suggest that NTF not only shields the tethered agonist to prevent G protein signaling but also confers a conformation that inhibits the interaction with β-arrestins and the consequent endocytosis and sustained signaling from endosomes.
Keywords: adhesion GPCR, G protein, endocytosis, GPCR kinase
Introduction
G protein–coupled receptors (GPCRs) have been the subject of basic and translational research for decades and are currently targeted by about 35% of marketed drugs.1 Although adhesion GPCRs (aGPCRs) form the second largest family of GPCR superfamily, our knowledge of their pharmacology and physiological roles is limited.2 This is mainly due to their unusual structural elements that preclude an in-depth investigation of their mechanisms of activation, signaling, and trafficking. aGPCRs have a large N-terminal fragment (NTF) that, in many aGPCRs, is cleaved from the rest of receptor at a GPCR proteolysis site (GPS). The GPS is located 15–25 amino acids before the first transmembrane domain (TM) and is generally composed of an HLT/S sequence. The cleaved NTF remains associated with the remaining receptor via noncovalent interactions to form a two-segmented structure.3
A majority of aGPCRs are still orphan and several studies have revealed their pivotal roles in neurological processes,2,4–6 cancer,7–9 inflammation,5,10,11 and the endocrine system.2,10,12,13 Recent reports on GPR126/ADGRG6,14 GPR133/ADGRD1,14 GPR56/ADGRG1,15,16 and GPR64/ADGRG212,17 point to the agonistic properties of the sequence between the GPS and the first TM of these aGPCRs. However, the mechanisms by which these tethered agonists regulate G protein and β-arrestin signaling and trafficking of aGPCRs are largely unknown. A recent study by the Hall laboratory showed that while the tethered agonist was not required for BAI1/ADGRB1 signaling, GPR56 without tethered agonist showed reduced signaling of certain pathways.18 However, the physical interaction of BAI1 and GPR56 with β-arrestin2 was not dependent on their tethered agonist. The mechanisms by which aGPCRs interact with β-arrestins in the absence of tethered agonist, and whether such interactions impact the trafficking and signaling of aGPCRs are currently unexplored. Recent discoveries on the sustained signaling of GPCRs from compartments other than plasma membrane, such as endosomes19–21 and Golgi apparatus,22 emphasize the importance of exploring the underlying mechanisms of receptor trafficking in the field of aGPCRs.
We have previously shown that a tethered agonist activates GPR64 and that the activated GPR64 crosstalks with the calcium-sensing receptor in human parathyroid adenoma cells.12 To understand the intricate role of the NTF and tethered agonist in regulation of signaling and trafficking of aGPCRs, we set out to compare the pharmacology of full-length (FL) GPR64 with its NTF-truncated (ΔNTF) and NTF/tethered agonist–truncated (P622) mutants. Our findings suggest that NTF not only inhibits G protein signalling of GPR64, but may also induce a conformation that inhibits the interaction of receptor with β-arrestins. Moreover, we found that GPCR kinase–mediated internalization of GPR64 mutants to endosomal compartments is independent from the receptor basal activation state.
Materials and methods
Cell culture
AD-293 (HEK) cells were purchased from Agilent Technologies (Santa Clara, CA #240085). HEK293 (parental WT) cells were engineered via CRISPR/Cas9 technology to knock out either Gαq/11 (ΔGNAQ/11) or Gα12 and Gα13 (ΔGNA12/13) G proteins.23 Cells were cultured in DMEM media (Sigma, St. Louis, MO #D6429) supplemented with 10% FBS (Sigma #12303C), 100 U/mL penicillin, and 100 mg/mL streptomycin (Thermo Fisher Scientific, Frederick, MD #15140–122). Cells were transiently transfected with plasmids in OptiMEM (Thermo Fisher Scientific #31985070) with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific #11668019) following the manufacturer’s instructions. In all of the following assays, cells were serum starved in DMEM (Thermo Fisher Scientific #21068028) supplemented with glutamine and 1.25 mM Ca2+ overnight and stimulation was conducted in this starvation medium.
Plasmids
Construction of pcDNA3.1 plasmids expressing human FL GPR64 and its NTF-truncated mutant (ΔNTF) was previously described.12 To construct a panel of mutants lacking amino acids from the C-terminus of the GPS site, we used the Q5 Site-Directed Mutagenesis Kit (New England BioLabs, Ipswich, MA #E0552S). FL and mutant receptors are N-terminally tagged with 3HA and C-terminally tagged with V5 (named here as FL, ΔNTF, S608, F609, and so on). In some experiments, we used plasmids expressing FL, ΔNTF, and P622 mutants that lack any N-terminal tag but have the C-terminal V5 tag (named here as FL-V5, ΔNTF-V5, and P622-V5). Also, we used ΔNTF-V5 to introduce single mutations in the first five amino acids of the tethered agonist to alanine (named here as T607A-V5, S608A-V5, F609A-V5, G610A-V5, and V611A-V5). All of the GPR64 constructs have a signal sequence from influenza hemagglutinin, as described previously.12 A list of primers is provided in Table S1 (online only). C-terminally FLAG-tagged β-arrestin1 and β-arrestin2 were purchased from Addgene. pCRE-Luc, pSRE-Luc, and pNFAT-Luc plasmids were kindly provided by Evi Kostenis (University of Bonn, Bonn, Germany) and Silvio Gutkind (University of California, San Diego, CA).
siRNA knockdown
MISSION® siRNA Universal Negative Control (#SIC001) and gene-specific siRNAs were ordered from Sigma-Aldrich and were transfected into HEK cells with Lipofectamine 2000 transfection reagent following the manufacturer’s instructions. Expression was knocked down 72 h before the assays and was confirmed by western blotting. A list of siRNAs is provided in Table S2 (online only).
Antibodies
Antibodies were purchased from the following sources: Cell Signaling Technologies (Beverly, MA): rabbit anti-HA (#3724), mouse anti-FLAG (#8146), rabbit anti-GRK2 (#3982), rabbit anti-GRK3 (#80362), rabbit anti-GRK6 (#5878), rabbit anti-β-arrestin1 (#30036), rabbit anti-β-arrestin2 (#3857), and rabbit anti-V5 (#13202); Biolegend (San Diego, CA): mouse anti-HA (#901513); Thermo Fisher Scientific: mouse anti-V5 (#R96025); Abcam (Cambridge, MA): rabbit anti-GRK4 (#ab182635); Sigma: rabbit anti-β-actin (#A2066); RnD Systems (Minneapolis, MN): goat anti-GRK5 (#AF4539).
Chemicals
P-15 peptide was synthesized by GenScript® (Piscataway, NJ) as described previously.12 Dyngo® 4a was purchased from Abcam (#ab120689).
On-cell ELISA
HEK cells were seeded in clear-bottom 96-well plates at 20,000 cells per well and were transfected with 200 ng plasmids. Surface expression of HA-tagged receptors was determined as previously described.12 To quantify the total expression, cells were permeabilized and were stained with anti-V5 tag antibody. The absorbance at 450 nm was measured in a FLEXStation III microplate reader (Molecular Devices, Sunnyvale, CA).
Reporter gene assays
Luciferase reporter assay was performed as described previously,12,24,25 with some modifications. HEK cells were seeded in 6-well plates at 100,000 cells per well and were transfected with 1 μg of GPR64 plasmids and 1 μg of pCRE-Luc, pSRE-Luc, or pNFAT-Luc reporter plasmids. Twenty-four hours later, cells were detached by phosphate-buffered saline (PBS) and were seeded in white 384-well clear-bottom plates (20,000 cells/well) for 48 hours. Serum-starved cells were then stimulated with different concentrations of P-15 or vehicle for 5 h at 37 °C. Using Steadylite reagents (PerkinELmer, Hopkinton, MA, #6066756), the luminescence was measured in a FLEXStation III device. In some experiments, cells were transfected with 500 pmole siRNA 24 h before transfection with plasmids.
cAMP production assay
Cells were transfected with 1 μg of plasmids in 6-well plates for 24 hours. Cells were detached with PBS and seeded in white 384-well half-area plates (10,000 cells/well) for 48 hours. For basal cAMP measurements, cells were incubated with 0.5 mM IBMX (Sigma #I5879) in starvation media overnight. Other cells were left untreated overnight in starvation media and then were stimulated with increasing concentrations of P-15 for 1 h in the presence of 0.5 mM IBMX. cAMP production was measured by a homogenous time-resolved fluorescence assay with a cAMP Dynamic 2 kit (Cisbio, Bedford, MA #62AM4PEC) in the FLEXStation III device.
Western blotting
Cells were lysed in RIPA buffer (Sigma #20–188) supplemented with protease and phosphatase inhibitors. Lysates were boiled in sample buffer (Thermo Fisher Scientific #NP0007) supplemented with DTT (Thermo Fisher Scientific #NP0009), and 2.5 μg total protein was separated on 4–12% Bis-Tris gels and transferred to polyvinylidene difluoride (PVDF) membranes. Following the treatment with blocking solution (5% or 10% nonfat dry milk in TBS buffer + 0.1% Tween 20), membranes were incubated with the following primary antibodies overnight at 4 °C: anti-HA (1:2000), anti-V5 (1:2000), anti-GRK2 (1:500), anti-GRK3 (1:500), anti-GRK4 (1:500), anti-GRK5 (1:500), anti-GRK6 (1:500), anti-β-arrestin1 (1:500), anti-β-arrestin2 (1:500), and anti-β-actin (1:1000). Membranes were then incubated with HRP-conjugated anti-mouse, anti-rabbit, or anti-goat antibodies (1:5000) for 2 h at room temperature (RT). Detection was performed using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific #32209).
Cell surface protein biotinylation
Cells were seeded in 6-cm dishes at 300,000 cells per dish and were transfected with 5 μg of plasmids. Expression of receptors on the cell surface was assessed by following the instructions of the Cell Surface Protein Isolation Kit (Thermo Fisher Scientific #89881). In brief, cells were washed with cold PBS and then incubated with EZ-Link Sulfo-NHS-SS-biotin for 30 min at 4 °C. Cells were then washed with quenching solution in PBS and were lysed in RIPA lysis buffer for 1 h on ice with frequent vortexing. The soluble lysate, containing 100 μg of total protein, was incubated with NeutrAvidin agarose beads with agitation for 1 h at RT. Beads were washed three times with washing buffer supplemented with protease inhibitors. The beads were then incubated with Pierce™ Lane Marker Sample Buffer (Thermo Fisher Scientific #39001) supplemented with 50 mM DTT with agitation for 1 h at RT to elute the biotinylated surface proteins. Total (input) and pulled-down surface proteins were detected by western blotting as described above.
β-Arrestin binding assay
Cells were seeded in 6-well plates at 100,000 cells per well and transfected with 1 μg of FLAG-tagged β-arrestin1 or β-arrestin2 along with 1 μg of GPR64 plasmids. After overnight starvation, cells were lysed in lysis buffer from the FLAG® immunoprecipitation kit (Sigma; #FLAGIPT1) supplemented with 1 mM MgCl2 and protease inhibitors. Cell lysates, containing 200 μg of total protein, were agitated with 20 μL anti-FLAG M2–agarose affinity gel overnight at 4 °C. After washing the gels, proteins were eluted by using FLAG peptide. Eluted protein (IP) and cell lysates (Input) were subjected to western blotting to detect V5, FLAG, and β-actin, as described above. Densitometry analysis of images by ImageJ was used to quantify the specific binding of β-arrestins to receptors and is defined as immunoprecipitated V5-tagged receptor per total FLAG-tagged β-arrestins.
Immunofluorescence staining
HEK cells were seeded on glass coverslips and were transiently transfected with FL, ΔNTF, or P622, with or without FLAG-β-arrestin1 or FLAG-β-arrestin2 plasmids. In some experiments, cells were incubated overnight with either CellLight™ Early Endosomes Rab5-GFP, BacMam 2.0 virus (Thermo Fisher Scientific #C10586), or CellLight™ Golgi N-acetylgalactosaminyltransferase-GFP, BacMam 2.0 virus (Thermo Fisher Scientific #C10592). In other experiments, cells were transfected with siRNA to knock down β-arrestin1, β-arrestin2, or GRK4 before receptor transfection. To prevent dynamin-mediated internalization of receptors, we preincubated cells with endosomal marker virus before receptor transfection and Dyngo treatment. Cells were fixed in 4% paraformaldehyde for 15 min and were washed with PBS several times. The antibodies and protocols (permeabilized or nonpermeabilized) used for each experiment are provided in corresponding figure legends. All cells were mounted in ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific; #P36971) for nuclear counterstaining. Confocal microscopy was conducted using a Plan Fluor 40× oil objective (1.3 NA) on a Nikon CSU-W1 spinning disk Ti2 microscope equipped with a sCMOS ORCAFlash4.0 Hamamatsu camera, and images were analyzed with Nikon NIS-Elements Research software.
Ab feeding assay
Cells were plated onto glass coverslips and were transfected with FL, ΔNTF, or P622 plasmids. Cells were incubated with Cell-Light™ Early Endosomes Rab5-GFP, BacMam 2.0 virus in serum starvation media overnight. Living cells were fed with mouse anti-HA antibody (Biolegend) for 30 min at 10 °C to label surface receptors in the starvation media. Subsequently, cells were washed with PBS (+Ca2+) and then were either fixed in 4% paraformaldehyde (0 h) or left in fresh starvation media at 37 °C for 6 h and then were fixed. Subsequently, cells were permeabilized with 0.3% Triton X-100, and HA antibody-bound receptors were labeled with Alexa Fluor 594–conjugated goat anti-mouse antibody. Nuclear staining and confocal imaging were performed as described above.
Cell viability assay
HEK cells were seeded at 20,000 cells per well in 96-well plates and were transfected with pcDAN3.1, FL, ΔNTF, or P622 plasmids. WT, ΔGNAQ/11, and ΔGNA12/13 HEK293 cells were seeded in the same manner. After an overnight serum starvation, the cell viability reagent WST-1 (Sigma #5015944001) was added at a 1:10 dilution and cells were kept at 37 °C and 5% CO2 for 2 hours. The absorbance was measured at 440 nm and data were analyzed following the manufacturer’s instructions.
Statistical analysis
Statistical analyses were conducted using two-tailed Student’s t-test for comparisons between two groups, one-way ANOVA with Dunnett’s test, or multiple t-test with Holm–Sidak test for multiple groups in GraphPad Prism 7.0 software (GraphPad, San Diego, CA); P < 0.05 was considered significant.
Results
Intact endogenous tethered agonist is required for constitutive activity of GPR64
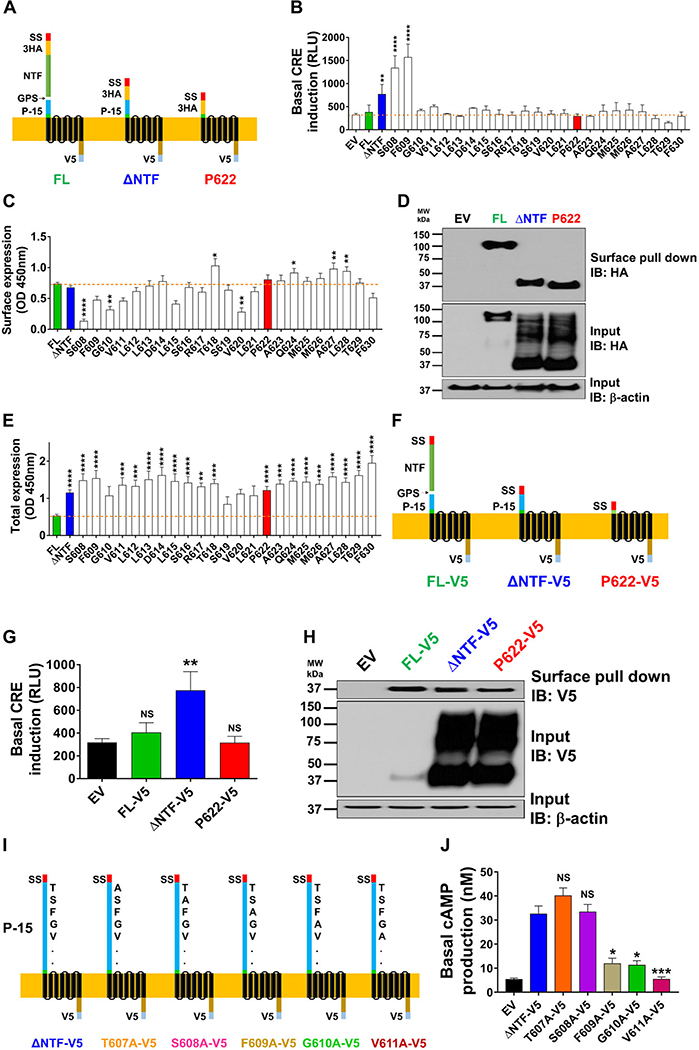
We and others have previously shown that a 15 amino acid tethered agonist (P-15) starting right after the hydrolyzed bond in the GPS site (Fig. 1A, left) activates GPR64.12,17 To test whether extracellular N-terminal residues of NTF-deficient GPR64 (ΔNTF; Fig. 1A, middle) are required for its constitutive activity, we employed different approaches. We deleted the amino acids of this tethered agonist one at a time. These mutants (from S608 to F630) have a complete 7TM region and C-terminus, are tagged N-terminally with 3HA and C-terminally with V5 tags but start at different residues denoted by amino acid initial and number (Fig. 1A and B). Consistent with our previous report,12 FL receptor only showed limited activation of CRE downstream of Gαs–cAMP pathway compared with the control plasmid (EV), and ΔNTF showed significantly higher activity compared with FL (Fig. 1B). Basal activity remained high upon deletion of two additional residues after the GPS cleavage site (in mutants S608 and F609) (Fig. 1B). However, starting from mutant G610, the constitutive activity was blunted and it remained comparable to EV until mutant F630 (starting with the second residue in the TM1 based on several domain prediction bioinformatics methods).
Figure 1.
The NTF of GPR64 inhibits its basal signaling. (A) Schemes of full-length (FL) human GPR64, its NTF-truncated mutant (ΔNTF), and a mutant that lacks both the NTF and P-15 tethered agonist (P622) are shown. SS, signal sequence; 3HA, 3 repeats of N-terminal HA tags; NTF, N-terminal fragment; GPS, GPCR-proteolysis site; P-15, 15-residue tethered agonist; V5, C-terminal tag. (B) HEK cells were transiently transfected with control (EV), FL, ΔNTF, P622, or other GPR64 mutant plasmids (lacking NTF and a varied number of residues from the N-terminus), along with pCRE-Luc plasmid. Basal induction of CRE was measured after an overnight serum starvation in a luminescence-based assay. Data are shown in relative light units (RLUs) recorded in a luminometer and are presented as mean ±SEM from a representative experiment out of three individual experiments performed in quadruplicate. ∗∗P < 0.01, ∗∗∗∗P < 0.0001. Data were compared with EV with one-way ANOVA with Dunnett’s test. (C) Cells were transfected as in B but without pCRE-Luc plasmid and were starved overnight before fixation. The expression of N-terminally HA-tagged receptors on the cell surface was measured by ELISA at OD 450 nm. Specific OD recordings (OD values of each plasmid minus that of EV) are presented as mean ± SEM from four individual experiments performed in quadruplicate. Nonspecific OD value for EV was 0.26 ± 0.02. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. Data were compared with FL with one-way ANOVA with Dunnett’s test. (D) Cells were transfected with EV, FL, ΔNTF, or P622, and after an overnight serum starvation the cell surface proteins were biotinylated as described in the Methods section. Equal amounts of protein were incubated with NeutrAvidin beads and both total and pulled-down surface receptors were detected by western blotting. Representative blots from four individual experiments are shown. (E) Cells were transfected as in C and were starved overnight before fixation and permeabilization. The expression of C-terminally V5-tagged receptors, as a surrogate for total expression, was measured by ELISA. Specific OD recordings are presented as mean ± SEM from four individual experiments performed in quadruplicate. Nonspecific OD value for EV was 0.45 ± 0.04. ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Data were compared with FL with one-way ANOVA with Dunnett’s test. (F) Schemes of receptors that lack the N-terminal tag but are tagged C-terminally with V5 (FL-V5, ΔNTF-V5, and P622-V5) are shown. (G) HEK cells were transiently transfected with EV, FL-V5, ΔNTF-V5, or P622-V5 plasmids along with pCRE-Luc plasmid. Basal activation of CRE was measured after an overnight serum starvation. Data are shown in RLUs and are presented as mean ± SEM from a representative experiment out of three individual experiments performed in duplicate. Data were compared with EV with a two-tailed Student’s t-test. ∗∗P < 0.01. (H) Cells were transfected with EV, FL-V5, ΔNTF-V5, or P622-V5 and after an overnight serum starvation the cell surface proteins were biotinylated and detected by western blotting as in D. Representative blots from three individual experiments are shown. (I) Schemes of ΔNTF-V5 and its mutants (T607A-V5, S608A-V5, F609A-V5, G610A-V5, and V611A-V5) in which one amino acid has been substituted with alanine are shown. (J) Basal cAMP production in cells transfected with EV, ΔNTF-V5, or its alanine-substituted mutants was measured after an overnight incubation with 0.5 mM IBMX in starvation media. Concentration of cAMP in nanomolar is presented as mean ± SEM from a representative experiment out of three individual experiments performed in duplicate. ∗P < 0.05, ∗∗∗P < 0.001. Data were compared with ΔNTF-V5 with one-way ANOVA with Dunnett’s test.
To test whether the higher basal activity of mutants was due to changed surface expression of receptors, we used two approaches. First, we detected HA-tag by ELISA and found that mutants have comparable (e.g., ΔNTF and P622), lower (e.g., S608), or higher (e.g., T618) surface expression compared with FL (Fig. 1C). Second, we used a surface protein biotinylation assay and found that while the total expression of ΔNTF and P622 (lacking NTF and complete P-15 sequence, Fig. 1A, right panel) mutants was higher than FL, their surface expression was essentially similar (Fig. 1D). We have previously shown that transfection of HEK cells with the same dose of FL or ΔNTF plasmids results in higher total expression of ΔNTF mutant due to polyubiquitination.12 Using ELISA assay after cell permeabilization, we measured the total expression of receptors and found that most of the mutants show elevated total expression compared with FL (Fig. 1E). To rule out the possible effect of the HA-tag on receptor signaling and expression, we generated FL, ΔNTF, and P622 mutants that lack any N-terminal tag (FL-V5, ΔNTF-V5, P622-V5; Fig. 1F). These receptors showed similar patterns of basal activity (Fig. 1G) and surface and total expression (Fig. 1H) compared with HA-tagged receptors. To find out how the total expression level affects receptor signaling, we transfected HEK cells with several doses of FL and a constant dose of either ΔNTF or P622 plasmids. Increasing the total expression of FL to reach or even exceed that of ΔNTF and P622 (Fig. S1A, online only) did not lead to similar cAMP production in response to agonistic peptide P-15 (Fig. S1B, online only). This suggests that the total expression of FL per se does not correlate with the signaling output of the receptor. In addition to deletion of N-terminal residues (Fig. 1B), we substituted the first five amino acids of the P-15 with alanine in the ΔNTF-V5 mutant (Fig. 1I). Overnight accumulation of cAMP revealed that the constitutive signaling is significantly reduced in cells that express F609A-V5, G610A-V5, and V611A-V5 compared with ΔNTF-V5, T607A-V5, and S608A-V5 (Fig. 1J). The CRE assay showed a similar pattern of basal activity among the alanine-mutated receptors (Fig. S2A, online only). These mutants showed comparable response to P-15 stimulation (Fig. S2A, online only) and similar total and surface expression (Fig. S2B, online only). Taken together, these findings signify the crucial role of the tethered agonist in self-activation of GPR64 and suggest an inhibitory role for NTF in GPR64 activation.
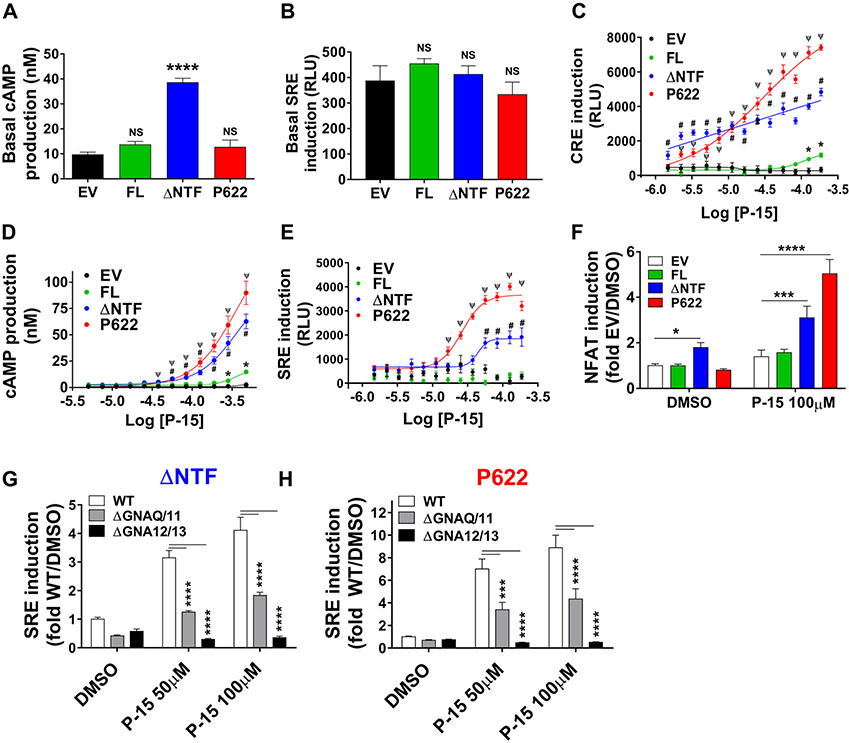
Intact endogenous tethered agonist selectively triggers GPR64 constitutive signaling pathways
Based on previous reports by our laboratory and others,17 P-15 is the most potent activator of GPR64. Therefore, we set out to investigate the impact of NTF and endogenous P-15 on GPR64 signaling by comparing the pharmacology of P622 with FL and ΔNTF. Whereas FL and P622 mutant showed modest but insignificant basal activity compared with EV (Fig. 2A), ΔNTF-expressing cells showed significantly higher basal cAMP production downstream of the Gαs pathway.12,17 However, the SRE luciferase activity, downstream of Gα13 and Gαq pathways,26 was unchanged in cells expressing any of the GPR64 plasmids compared with EV (Fig. 2B). These data confirm the agonistic property of endogenous P-15 and indicate differential activity toward distinct G protein pathways. To see how GPR64 and its mutants respond to exogenous synthetic P-15 (agonistic peptide), we incubated cells with increasing concentrations of P-15 for 5 or 1 h in luciferase and cAMP assays, respectively. The P622 mutant showed the highest maximal effect in CRE (14-fold), cAMP (83-fold), and SRE (6.2-fold) assays (Fig. 2 C–E). ΔNTF showed lower levels of induction of CRE (fourfold) and SRE (threefold) and production of cAMP (58-fold). However, FL showed marginal CRE (twofold), SRE (1.2-fold), and cAMP (13.6-fold) responses at the highest concentration of P-15. Together, these data show the exaggerated response of the mutant without endogenous tethered agonist, P622, to agonistic peptide P-15.
Figure 2.
Multiple signaling pathways are activated by endogenous tethered agonist and synthetic agonistic peptide. (A) HEK cells were transiently transfected with EV, FL, ΔNTF, or P622 plasmids and were serum starved overnight in the presence of IBMX (0.5 mM). The basal concentration of cAMP in nanomolar is presented as mean ± SEM from a representative experiment out of six individual experiments performed in quadruplicate. ∗∗∗∗P < 0.0001, NS, not significant. Data were compared with EV with a two-tailed Student’s t-test. (B) Cells were transiently transfected with EV, FL, ΔNTF, or P622 plasmids along with pSRE-Luc plasmid. Basal induction of SRE was measured after an overnight serum starvation in a luminescence-based assay. Data are shown in RLUs and are presented as mean ± SEM from a representative experiment out of four individual experiments performed in quadruplicate. NS, not significant. Data were compared with EV with a two-tailed Student’s t-test. (C) Cells were transfected with EV, FL, ΔNTF, or P622 plasmids along with pCRE-Luc plasmid and after an overnight serum starvation, cells were stimulated with increasing concentrations of synthetic agonistic peptide P-15 for 5 h at 37 °C. Luciferase assay was performed as above. Data are shown in RLUs and are presented as mean ± SEM from a representative experiment out of four individual experiments performed in triplicate. Data were compared with EV with multiple t-test with Holm–Sidak test. ∗P < 0.05 (for FL), #P < 0.0001 (for ΔNTF), and ΨP < 0.0001 (for P622). (D) Cells were transfected as in A and after an overnight serum starvation without IBMX, cells were stimulated with increasing concentrations of agonistic peptide P-15 for 1 h at 37 °C in the presence of IBMX (0.5 mM). cAMP production was measured as above. The concentration of cAMP in nanomolar is presented as mean ± SEM from a representative experiment out of five individual experiments performed in quadruplicate. Data were compared with EV with multiple t-test with Holm–Sidak test. ∗P < 0.05 (for FL), #P < 0.001 (for ΔNTF), and ΨP < 0.001 (for P622). (E) Cells were transfected as in B and after an overnight serum starvation, cells were stimulated with increasing concentrations of P-15 for 5 h at 37 °C. Luciferase assay was performed as above. Data are shown in RLUs and are presented as mean ± SEM from a representative experiment out of four individual experiments performed in triplicate. Data were compared with EV with multiple t-test with Holm–Sidak test. #P < 0.001 (for ΔNTF) and ΨP < 0.001 (for P622). (F) Cells were transiently transfected with EV or GPR64-expressing plasmids along with pNFAT-Luc reporter plasmid. After an overnight serum starvation, basal and P-15–stimulated induction of NFAT was measured. Data were normalized to that of EV (treated with DMSO; RLU value: 112.8 ± 20.4) and are presented as mean ± SEM from three individual experiments performed in triplicate. Data were compared with EV with a two-tailed Student’s t-test. ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (G, H) Parental HEK293 cells (WT), Gαq/11 KO cells (ΔGNAQ/11), or Gα12/Gα13 KO cells (ΔGNA12/13) were transiently transfected with either ΔNTF (G) or P622 (H) mutant in combination with pSRE-Luc reporter plasmid. After an overnight serum starvation, basal and P-15–stimulated induction of SRE was measured as in B. Data were normalized to that of WT cells treated with DMSO (RLU value: 785.3 ± 115.8) and are presented as mean ± SEM from a representative experiment from three individual experiments performed in triplicate. Data were compared with WT with two-tailed Student’s t-test. ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
GPR64 couples to Gαq and Gα13 G proteins
Previous studies suggested the coupling of GPR64 and its mutants to Gα proteins other than Gαs.17,26 We assessed the activation of NFAT, downstream of calcium signaling, and found that only ΔNTF shows basal activation of this transcription factor (Fig. 2F), but both ΔNTF and P622 induce NFAT in response to agonistic peptide P-15. To test the role of the G proteins Gαq and Gα13 in GPR64 signaling, we used HEK293 cells that are knocked-out for either Gαq and Gα11 (ΔGNAQ/11) or Gα12 and Gα13 (ΔGNA12/13).27 Consistent with the foregoing results derived from HEK cells, FL or mutant GPR64 receptors do not induce SRE in basal condition in WT, ΔGNAQ/11, or ΔGNA12/13 HEK293 cells (Fig. S3A, online only). We found that while P-15–induced SRE activation is completely blunted in ΔGNA12/13 cells, the ΔGNAQ/11 cells show a modest but significant impact on SRE induction (Fig. 2G and H). G protein KO cells showed similar viability compared with WT cells (Fig. S3B, online only), discounting the effect of cell viability on their signaling potential. These data demonstrate multiple signaling pathways triggered by GPR64.
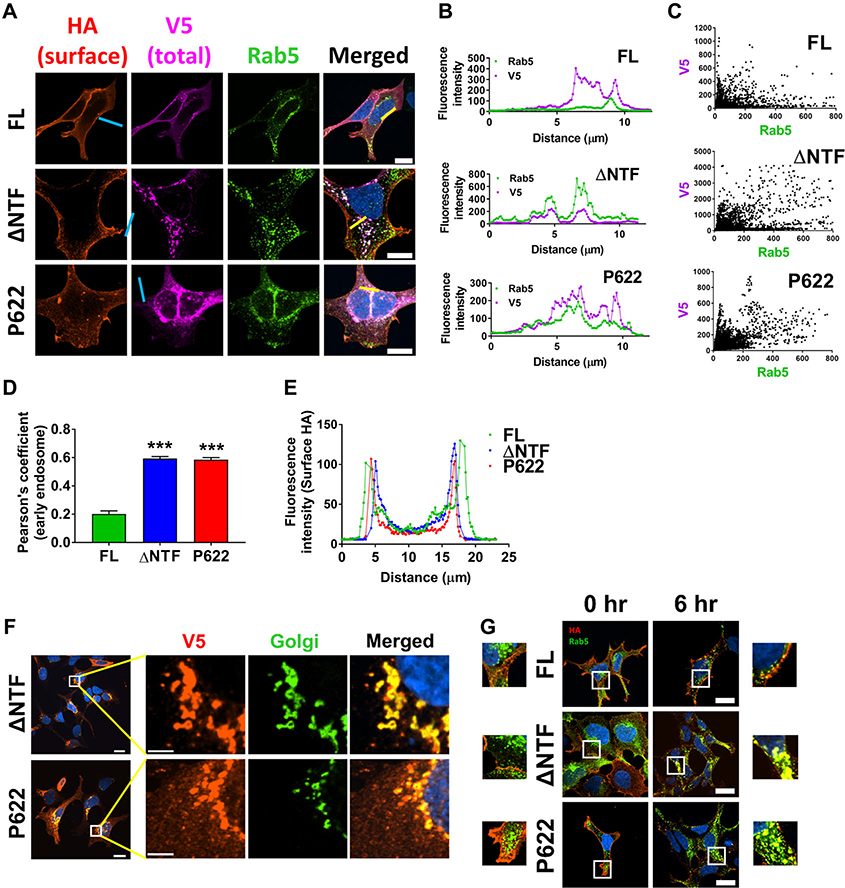
NTF inhibits constitutive trafficking of GPR64
Using confocal microscopy, we found that there is a significant colocalization of ΔNTF and P622 mutants with an early endosomal marker, Rab5 GTPase (Fig. 3A). We used quantitative analysis of confocal images from a set of cells (Fig. 3B–D) and determined that the colocalization with early endosomes is significantly higher in cells expressing ΔNTF and P622 compared with FL. However, such intracellular localization did not affect the expression of receptors on the cell surface quantified by confocal imaging (Fig. 3E), consistent with the results of the ELISA (Fig. 1C) and surface biotinylation (Fig. 1D) assays. To ascertain that localization of receptors intracellularly is not due to impaired transport from the endoplasmic reticulum (ER) to the cell surface, we used two approaches. First, we examined the colocalization of receptors with a Golgi apparatus marker and found that some of the ΔNTF and P622 receptors are present in Golgi-derived vesicles (Fig. 3F), suggesting proper transport of receptors from the ER. Second, we fed live cells with anti-HA antibody to recognize the cell surface receptors and then incubated cells for an additional 6 h at 37 °C. Staining of anti-HA antibody–bound receptors showed that unlike FL, both ΔNTF and P622 mutants constitutively internalize to early endosomes (Fig. 3G). Taken together, these data suggest sustained internalization of GPR64 in the absence of the NTF.
Figure 3.
Basally active and inactive mutants of GPR64 localize in early endosomes. (A) Cells were transfected with FL, ΔNTF, or P622 and were incubated with Cell-Light early endosome Rab5-GFP–expressing viruses in serum starvation media overnight. The cell surface N-terminal HA-tagged receptors were initially incubated with mouse IgG1 anti-HA antibody (1:1000) and labeled with anti-IgG1 secondary antibody conjugated to Alexa Fluor 594 fluorophore (1:500) in nonpermeabilizing condition. The V5-tagged receptors were recognized by mouse IgG2a anti-V5 antibody (1:1000) and were labelled with anti-IgG2a secondary antibody conjugated to Alexa Fluor 647 fluorophore (1:500) in permeabilizing condition and DAPI was used for nuclear staining. Magnification 400×, scale bar: 20 μm. (B) Histograms of the fluorescence intensity of two channels (representing V5 and Rab5) over the length of yellow lines (in A) are shown. (C) Fluorescence intensity was measured in Nikon NIS Elements software for 15–20 cells expressing each plasmid, as in B. Data are presented to show the colocalization of V5 and Rab5, and the Pearson correlation method was used to determine the significance of such colocalization. (D) Comparison of Pearson’s coefficients derived from C reveals significant colocalization of ΔNTF and P622 with early endosomes. Data are presented as mean ± SEM from three individual experiments and are compared with FL with a two-tailed Student’s t-test. ∗∗∗P < 0.001. (E) Histograms derived from the line scanning (blue lines in A) show similar fluorescence intensity representing surface HA tags. (F) HEK cells were seeded on coverslips and were transiently transfected with either ΔNTF or P622 plasmids. After an overnight serum starvation, the expression of C-terminally V5-tagged receptors was determined by a specific primary antibody, followed by an Alexa Fluor 594-conjugated secondary antibody (red) in permeabilizing condition. Golgi apparatus was labeled with N-acetylgalactosaminyltransferase-GFP marker (green) and DAPI was used for nuclear staining. Representative images show the colocalization of receptors with Golgi apparatus. Magnification 400×, scale bar: 20 μm. Insets (white boxes) are shown at higher magnification on the right (scale bar: 5 μm). (G) HEK cells were transfected with FL, ΔNTF, or P622 plasmids and were incubated with Cell-Light early endosome Rab5-GFP viruses overnight. Live cells were fed with mouse IgG1 anti-HA antibody (1:200) for 30 min at 10 °C. Cells were then washed and were either fixed or left untreated for 6 h in starvation media at 37 °C. HA-tagged receptors were then labeled with anti-IgG1 secondary antibody conjugated to Alexa Fluor 594 fluorophore (1:500) in permeabilizing condition and DAPI was used for nuclear staining. Magnification 400×, scale bar: 20 μm. Insets (white boxes) are shown at higher magnification either on the left side (for 0 h) or on the right side (for 6 h). Representative images form three independent experiments are shown.
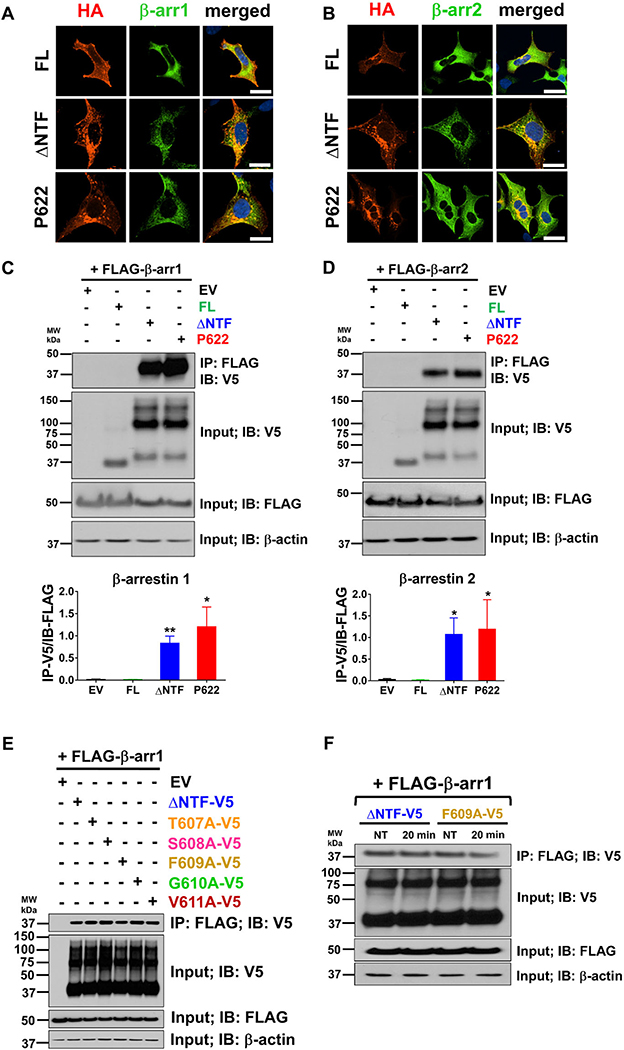
β-Arrestins regulate trafficking and signaling of GPR64
One of the major pathways of GPCR endocytosis is regulated by β-arrestins, scaffolding proteins that are recruited to activate GPCRs.28 We observed that β-arrestins1 and β-arrestin2 colocalize with ΔNTF and P622 in intracellular compartments but show negligible proximity with FL in the basal condition (Fig. 4A and B). To confirm that such colocalization represents physical interaction, we used coimmunoprecipitation assays. We found that β-arrestin1 (Fig. 4C, upper and lower panels) and −2 (Fig. 4D, upper and lower panels) physically interact with both ΔNTF and P622 in steady state. We did not detect physical interaction between β-arrestins and FL (Fig. 4C and D) in our coimmunoprecipitation assays. To rule out an effect of the N-terminal HA tag and to assess the role of the first five amino acids of the GPR64 tethered agonist, we examined whether ΔNTF-V5 (Fig. 1F) and its single-residue mutants (Fig. 1I) interact with β-arrestin1. We found that β-arrestin1 interacts with ΔNTF-V5, T607A-V5, S608-V5, F609A-V5, G610A-V5, and V611A-V5 in steady state (Fig. 4E). We stimulated the cells expressing either ΔNTF-V5 or F609A-V5 with P-15 but did not observe a change in their physical interaction with β-arrestin1 (Fig. 4F). These findings suggest that NTF not only inhibits the interaction of tethered agonist with receptor, but may also suppress β-arrestin recruitment. Furthermore, tethered agonist, its composition, and agonistic peptide P-15 do not modulate the interaction of GPR64 with β-arrestins.
Figure 4.
GPR64 mutants interact with β-arrestin1 and β-arrestin2 constitutively. HEK cells were transfected with GPR64-expressing plasmids along with either FLAG-tagged β-arrestin1 (A) or β-arrestin2 (B) plasmids. Basal localization of receptors and β-arrestins was assessed by immunofluorescence staining after an overnight serum starvation. Cells were fixed and permeabilized and HA-tagged receptors and FLAG-tagged β-arrestins were recognized by rabbit anti-HA (1:1000) and mouse IgG1 anti-FLAG (1:1000) antibodies and were then labeled with secondary antibodies conjugated to Alexa Fluor 594 and Alexa Fluor 488 fluorophores (1:500), respectively, and DAPI was used for nuclear staining. Magnification 400×, scale bar: 20 μm. (C, D) Cells were transfected as above, and cell lysates were incubated with anti-FLAG antibody–bound agarose resins. The coprecipitating proteins were eluted and along with total lysate were subjected to immunoblotting with V5 antibody. Total lysate was also subjected to immunoblotting with anti-FLAG and anti-β-actin antibodies as controls. Representative blots from four independent experiments are shown. WB band intensity was analyzed by ImageJ and specific interaction of β-arrestins and receptors was determined as the ratio of V5-tagged receptors in the eluate to the FLAG-tagged β-arrestins in the total lysate. Data are presented as mean ± SEM from four individual experiments. ∗P < 0.05, ∗∗P < 0.01. Data were compared with FL with a two-tailed Student’s t-test. (E) Cells were transfected with plasmids described in Figure 1I along with FLAG-tagged β-arrestin1 and the coimmunoprecipitation assay was performed as above. Representative blots from three independent experiments are shown. (F) Cells were transfected with either ΔNTF-V5 or F609A-V5 plasmids along with FLAG-tagged β-arrestin1. β-Arrestin recruitment at basal condition and 20 min post-stimulation with 100 μM P-15 was assessed as above. Representative blots from three independent experiments are shown.
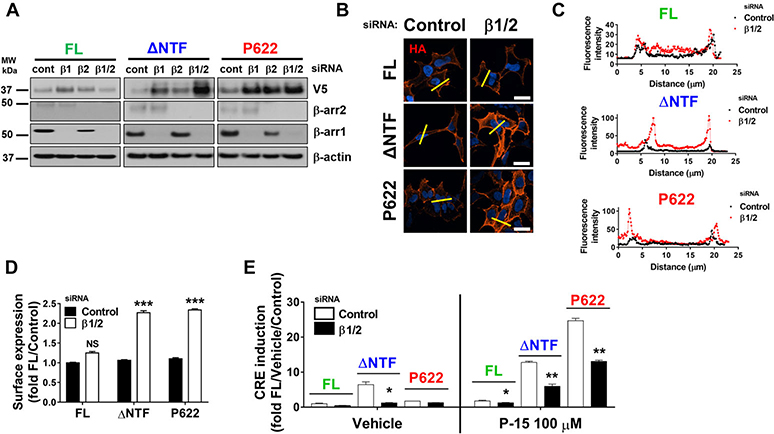
To test whether β-arrestins drive receptor internalization, we knocked down their expression by siRNA. We found that downregulation of β-arrestins increases the total expression level of ΔNTF and P622 profoundly (Fig. 5A). This was accompanied by a significant increase in surface expression of mutants and a modest increase in surface expression of FL (Fig. 5B–D). Although downregulation of β-arrestins increased total and surface levels of mutants, it reduced the basal or P-15–induced CRE induction in ΔNTF-expressing cells (Fig. 5E). P622-expressing cells showed unchanged basal and lowered P-15–induced CRE activation after β-arrestins downregulation (Fig. 5E). Interestingly, the modest response of FL to P-15 was also reduced after β-arrestin mRNA knockdown (Fig. 5E). These data indicate that GPR64 turnover and cell surface localization are in part regulated by β-arrestins, and β-arrestins potentiate GPR64 signaling.
Figure 5.
β-Arrestins regulate GPR64 trafficking and signaling. HEK cells were transfected with control siRNA (cont) or siRNAs specific for β-arrestin1 (β1) and β-arrestin2 (β2) alone or in combination (β2/2). Cells were then transiently transfected with FL, ΔNTF, or P622 plasmids. (A) Expression of receptors (V5 tag) was assessed by western blotting. Lysates were also subjected to immunoblotting with antibodies against endogenous β-arrestin1 and −2 and β-actin as appropriate controls. Representative blots from three independent experiments are shown. (B) Cells transfected as above were serum starved overnight, fixed, and the HA-tagged receptors on the cell surface were labeled with mouse anti-HA antibody and Alexa Fluor 594–conjugated anti-mouse antibody in nonpermeabilizing condition. DAPI was used for nuclear staining. Magnification 400×, scale bar: 20 μm. (C) Histograms of the fluorescence intensity of surface HA tags over the length of yellow lines (in B) are shown. (D) Cells were transfected as in B and the expression of N-terminally HA-tagged receptors on the cell surface was measured by ELISA at OD 450 nm. Data were normalized to that of cells transfected with control siRNA and FL plasmid (OD value: 0.56 ± 0.01) and are presented as mean ± SEM from three individual experiments performed in triplicate. ∗∗∗P < 0.001, NS, not significant. Data were compared with control siRNA with a two-tailed Student’s t-test. (E) Cells were transfected with either control siRNA or a combination of β-arrestin1 and −2 (β1/2) siRNAs followed by FL, ΔNTF, or P622 plasmids in combination with pCRE-Luc plasmid. CRE induction after 5 h incubation with either DMSO (vehicle) or 100 μM P-15 was measured. Data were normalized to the response induced by vehicle in cells transfected with FL and control siRNA (RLU value: 380.8 ± 153.2) and are presented as mean ± SEM from four individual experiments performed in quadruplicate. ∗P < 0.05, ∗∗P < 0.01. Data were compared with control siRNA with a two-tailed Student’s t-test.
Cellular localization and signaling of GPR64 are regulated by several GRKs
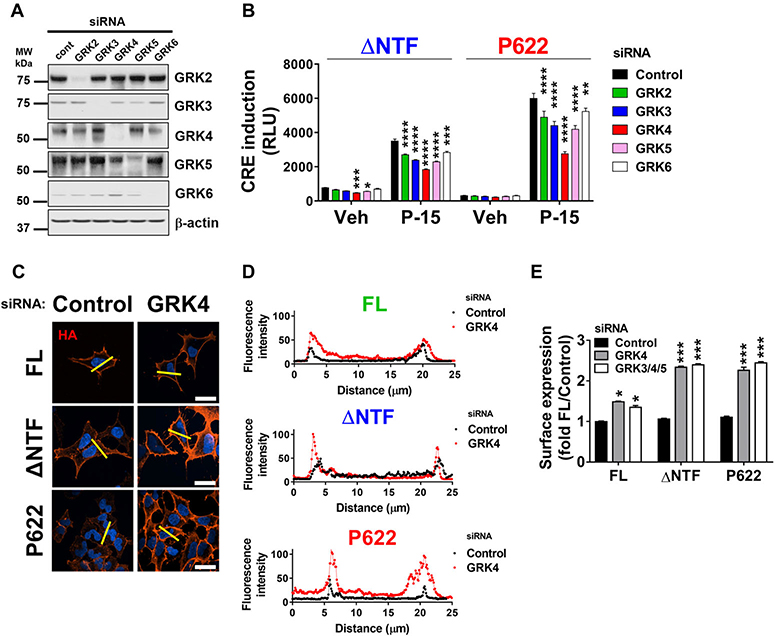
As an essential component of GPCR endocytosis, GPCR kinases (GRKs) are recruited to active GPCRs before β-arrestins. GRKs phosphorylate serine/threonine residues in the C-terminus of GPCRs, which, in turn, act as interaction spots for β-arrestins. We knocked down members of the nonvisual GRK family (GRK2–6) (Fig. 6A; Fig. S4, online only) and found that CRE induction by ΔNTF is reduced in cells with downregulated GRK4 and GRK5 (Fig. 6B). In addition, CRE induction by ΔNTF and P622 in response to P-15 was dependent on GRK2–6 activity. Among GRKs studied here, GRK4 was the most efficient regulator of signaling for both mutants. Downregulation of GRK4 alone (Fig. 6C and D) or in combination with GRK3 and GRK5 (Fig. 6E) increased the surface expression of FL, ΔNTF, and P622, albeit the greatest effect was seen for the mutants. This reduced the residence of mutants in early endosomal compartments (Fig. S5, online only). Thus, our data show that endocytosis of GPR64 is greatly dependent on the function of GRKs, and the internalized receptors may exhibit elevated signaling compared with the cell surface receptors.
Figure 6.
GPCR kinases regulate GPR64 trafficking and signaling. HEK cells were transfected with control or GRK-specific siRNAs. (A) Cleared lysates were used for immunoblotting with anti-GRK-specific antibodies or anti-β-actin antibody as a loading control. Representative blots from three independent experiments are shown. (B) Cells were first transfected with siRNAs as above and then transiently transfected with ΔNTF or P622 plasmids in combination with pCRE-Luc plasmid. After an overnight serum starvation, cells were stimulated with either DMSO (vehicle) or 100 μM P-15 for 5 h and the CRE induction was measured. Data are shown in RLUs and are presented as mean ± SEM from a representative experiment out of three individual experiments performed in triplicate. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Data were compared with control siRNA with one-way ANOVA with Dunnett’s test. (C) Cells were transfected with control or GRK4-specific siRNA followed by GPR64 plasmids. After an overnight serum starvation, cells were fixed and the HA-tagged receptors on the cell surface were labeled with mouse anti-HA antibody and Alexa Fluor 594–conjugated anti-mouse antibody in nonpermeabilizing condition. DAPI was used for nuclear staining. Magnification 400×, scale bar: 20 μm. (D) Histograms of the fluorescence intensity of surface HA-tagged receptors over the length of yellow lines (in C) are shown. (E) Cells were transfected with either control, GRK4-specific, or a combination of GRK3-specific, GRK4-specific, and GRK5-specific siRNAs followed by GPR64-expressing plasmids. The expression of HA-tagged receptors at the cell surface was measured by ELISA at OD 450 nm. Data were normalized to that of cells transfected with control siRNA and FL plasmid (OD value: 0.49 ± 0.01) and are presented as mean ± SEM from three individual experiments performed in triplicate. ∗P < 0.05, ∗∗∗P < 0.001. Data were compared with control siRNA with one-way ANOVA with Dunnett’s test.
Dynamin-dependent internalization of GPR64 elevates receptor signaling
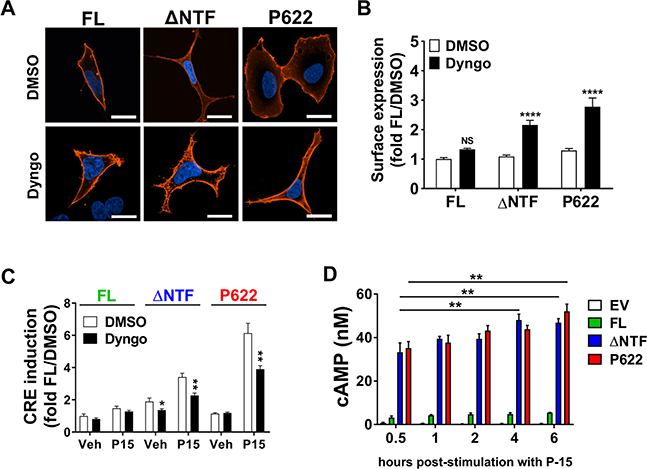
Dynamin GTPase plays a major role in endocytosis of GPCRs by scissoring vesicles at clathrin-coated pits.29 A selective inhibitor of dynamin, Dyngo, increased the surface expression of ΔNTF and P622, but showed no significant effect on surface expression of FL (Fig. 7A and B). Confocal microscopy showed that Dyngo prevents the constitutive internalization and targeting of ΔNTF to early endosomes (Fig. S6, online only). Reduced internalization was accompanied by suppression of the basal and P-15–triggered CRE induction by both mutants (Fig. 7C). We did not detect either receptor- or Dyngo-mediated reduction in cell viability, discounting a cytotoxic effect of Dyngo (Fig. S7, online only). Finally, we measured the receptor activity at different time points after the removal of the P-15 stimulant. ΔNTF and P622 showed sustained elevation of cAMP production 4–6 h after a 30-min stimulation with P-15 (Fig. 7D). Altogether, these data suggest that dynamin is an important regulator of GPR64 internalization, which potentiates receptor signaling from intracellular compartments.
Figure 7.
Dynamin controls trafficking and signaling of GPR64. (A) HEK cells were seeded on coverslips and were transiently transfected with FL, ΔNTF, or P622 plasmids. Cells were then incubated with vehicle (DMSO) or 20 μM Dyngo (dynamin inhibitor) in starvation media overnight. Surface expression of HA-tagged receptors was determined by a specific primary antibody, which was then labeled with Alexa Fluor 594 fluorophore in nonpermeabilizing condition. DAPI was used for nuclear staining. Magnification 400×, scale bar: 20 μm. (B) Cells were seeded in 96-well plates and were transfected with FL, ΔNTF, or P622 plasmids. An overnight incubation with either DMSO (vehicle) or 20 μM Dyngo was followed by an ELISA assay to measure receptor expression on the cell surface. Optical density (OD) at 450 nm was measured and the OD of all cells was normalized to that of FL-expressing cells that were kept with DMSO as vehicle control (OD value: 0.41 ± 0.02). Data are presented as mean ± SEM from three individual experiments performed in triplicate. ∗∗∗∗P < 0.0001. NS, not significant. Data were compared with DMSO with a two-tailed Student’s t-test. (C) Cells were transfected with FL, ΔNTF, or P622 plasmids in combination with pCRE-Luc plasmid. After a 1-h pretreatment with DMSO or 20 μM Dyngo, cells were stimulated with DMSO (Veh) or 100 μM P-15 for 5 hours. The CRE induction was measured in a luminescence assay. Data were normalized to response induced by vehicle in cells transfected with FL and pretreated with DMSO (RLU value: 536.8 ± 53.39) and are presented as mean ± SEM from three individual experiments performed in triplicate. ∗P < 0.05, ∗∗P < 0.01. Data were compared with DMSO pretreatment with two-tailed Student’s t-test. (D) Cells were transiently transfected with EV, FL, ΔNTF, or P622 plasmids and cells were stimulated with 100 μM P-15 for 30 min and then media was removed and fresh starvation media without P-15 but with IBMX was added. cAMP production at different time points after P-15 removal was measured. The concentration of cAMP in nanomolar is presented as mean ± SEM from a representative experiment out of three individual experiments performed in duplicate. ∗∗P < 0.01. Data were compared with a 30-min time point with a two-tailed Student’s t-test.
Discussion
Several groups have reported the constitutive activity of NTF-deficient aGPCRs.14,15,30 An extracellular short peptide at the N-terminus of a truncated aGPCR is regarded as the agonist that triggers such basal activity.14,16 Our study suggests that the NTF inhibits GPR64 signaling and β-arrestin recruitment and that the tethered agonist activates the receptor. In addition, we provide the evidence that β-arrestins and GRKs mediate the constitutive trafficking of GPR64 to endosomal compartments, which potentiates its sustained signaling.
The comprehensive mutational analysis on GPR64 tethered agonist, either by deletion or alanine scanning, revealed the residues that have the highest impact on receptor signaling. The ΔNTF mutant starts from Thr607 and shows elevated cAMP production and CRE induction compared with FL GPR64. It was previously reported that the threonine residue of the GPS site has an inhibitory function in GPR126 signaling.14 In our study, however, we found that while deletion of Thr607, in the GPR64 mutant starting with Ser608, increased the basal activity significantly, mutation of Thr607 to alanine only led to a modest but insignificant increase in activity. Deletion of Ser608, in the GPR64 mutant starting with Phe609, did not change the receptor activity compared with the receptor starting with Ser608. However, deletion of Phe609 and preceding residues in the mutant starting from Gly610 completely blunted the basal activity of GPR64 and further deletions of residues did not restore the basal activity. To complement this approach, we mutated the first five residues of ΔNTF-V5 to alanine, one at a time (Fig. 1I). Basal cAMP production was not altered in cells expressing mutants T607A-V5 or S608A-V5 compared with ΔNTF-V5. However, mutants F609A-V5, G610A-V5, and V611A-V5 showed significantly reduced basal cAMP production. These two approaches suggest important roles for the highly conserved Phe609 residue,16 Gly610, and Val611 in self-activation of GPR64.
The basal CRE induction and lack of basal SRE induction by the ΔNTF mutant suggest that GPR64 may engage different G proteins. Lack of P-15 in the mutant starting with Pro622 (P622) provided an opportunity to suppress basal activation of both CRE and SRE and to test the effect of P-15 agonistic peptide on these Gαs, Gαq, and Gα13 pathway readouts. In response to the P-15, the P622 mutant showed the largest fold activation compared with FL and ΔNTF. This may be due to two reasons; first, the occupation of the P-15 binding pocket by the endogenous P-15 in ΔNTF mutant can reduce the access of agonistic peptide P-15, and second, the constitutive activity of ΔNTF may desensitize the components of these signaling cascades so that their response to synthetic P-15 is not at maximal level. In all three assays used (CRE, SRE, and cAMP), the FL receptor only showed limited response at the very high concentrations of P-15, suggesting that NTF not only masks the endogenous tethered agonist but also may shield the P-15 binding pocket in the conditions and cells used in these experiments. Future discovery of the natural ligand(s) of GPR64 can reveal the full potential of the FL.
FL GPR64 was mainly localized to plasma membrane and did not show strong interactions with β-arrestins in the basal condition. However, ΔNTF and P622 mutants constitutively interacted with β-arrestins and were located in early endosomes. We show that GRK4, β-arrestins, and dynamin play major roles in constitutive internalization of ΔNTF and P622 mutants. Interaction of P622 mutant with β-arrestins was unexpected because for most GPCRs, G protein activation precedes the phosphorylation by GRKs and the recruitment of β-arrestins to the activated receptor.31,32 We did not observe P622-mediated constitutive activation of adenylyl cyclase, CRE, or SRE, readouts used to probe G protein activation. Such pharmacology of GPR64 is consistent with the recent report on the G protein–independent recruitment of β-arrestins to agonist-activated D prostanoid receptor-2 (Gαi-coupled), GPR17 (Gαi/q-coupled), and free fatty acid receptor-2 (FFA2, Gαi/q/12-coupled), and the consequent internalization of these receptors.33
Previous studies have shown that β2-adrenergic,34 M2 muscarinic,34 and dopamine D235 receptors interact with certain members of the GRK family in the absence of their cognate agonists. The underlying cause of GRK and β-arrestin recruitment to P622 needs further investigation. Our findings reveal that although the tethered agonist initiates G protein signaling, it may be dispensable for the interaction with GRKs and β-arrestins. In addition, our data suggest that NTF not only inhibits G protein signaling by FL, but may also confer a certain conformation that prevents interaction of GRK4 and β-arrestins with GPR64. Recent structural studies have revealed that GPCRs go through multiple distinct inactive and intermediate states before adopting a fully active ligand-bound state.36–38 This transition is manifested by relative movements of the TM domains, leading to the formation of an opening in the intracellular side of receptor for G protein binding.39,40 Each of these inactive states can potentially have differential signaling effects. Whether NTF directly induces a conformational state in TM domains that is distinct between FL and P622 and whether these conformational changes lead to the constitutive interaction of P622 with β-arrestins is unclear.
Among nonvisual GRKs (GRK2–6), GRK4 showed the most prominent effect on both signaling and trafficking of GPR64. The basis for such a biased effect is not currently understood, but Li et al. have previously shown a GRK selectivity phenomenon for various GPCRs.34 The restricted expression of GRK4 compared with other GRKs34,41 and its coexpression with GPR64 in the epididymis (The Human Protein Atlas) may suggest GRK4 as a potential regulator of GPR64.
Our findings are in line with some of the reported pharmacology of GPR56. For instance, GPR56 mutants that lack either NTF alone or NTF and tethered agonist showed increased total expression but similar surface expression compared with the FL GPR56.16,18 Like P622 mutant of GPR64, NTF/tethered agonist–truncated GPR56 constitutively interacts with β-arrestin2.18 However, this GPR56 mutant actively engages G protein signaling. The reason behind G protein signaling by GPR56 in the absence of tethered agonist is not well understood. Structurally, a common feature of GPR56 and GPR64 lies in the clusters of serine/threonine residues in their C-terminus. Such clusters were shown to be crucial for formation of megaplexes between some of the class B GPCRs (i.e., PTHR and V2R) and β-arrestins and their sustained signaling from endosomal compartments.21 The latter study provided compelling evidence for the role of β-arrestins in promoting rather than desensitizing GPCR signaling.
These features of β-arrestins have been since reported for other GPCRs in different cell and animal models. For instance, the persistent hyperexcitability of sensory neurons in patients with irritable bowel syndrome is caused by β-arrestin–mediated internalization and sustained MAPK signaling of protease-activated receptor 2.42 Also, the internalization of calcitonin receptor-like receptor to early endosomes via coordinated function of β-arrestins, clathrin, and dynamin was shown to mediate protein kinase C (PKC) and ERK1/2 activation in response to calcitonin gene–related protein (CGRP).43 Inhibitors of dynamin and clathrin abolished the CGRP-induced PKC activation in HEK293 cells and the sustained excitation in rat spinal cord slices. These studies prove the pathophysiological implications of GPCR interaction with β-arrestins and the consequent signaling from intracellular compartments. Interestingly, we show that preventing the constitutive internalization of ΔNTF, by either downregulation of GRK4 and β-arrestins or inhibition of dynamin, ablates GPR64 signaling. In line with these findings, we observed the sustained production of cAMP by truncated GPR64 even after the removal of the agonistic peptide P-15. This, for the first time, suggests that an aGPCR is more active while in intracellular compartments rather than when it is accessible on the surface.
We have previously shown that unlike the FL, the ΔNTF mutant is highly ubiquitinated in basal conditions.12 The effect of β-arrestins on turnover of ΔNTF and P622 mutants (Fig. 5A) suggests a dual role for β-arrestins. On the one hand, they internalize receptors to endosomal compartments for further signaling, and on the other hand, they may mediate their degradation. A recent study showed that β-arrestin1 plays a crucial role in male fertility via its interaction with GPR64 in the efferent ductules, one of the few tissues where GPR64 is expressed.44 This physiological function of β-arrestin and GPR64 emphasizes the importance of understanding the mechanisms of ubiquitination of GPR64 and the impact of such posttranslational modifications on temporal and spatial receptor signaling.
Our study has some limitations. The reason for differential impact of GRKs and β-arrestins on the surface expression of FL GPR64 is unclear. Also, we did not compare the effect of GRKs and β-arrestins on the signaling of the plasma membrane–localized versus endosome-localized receptors. Such direct assessments require a bystander BRET assay45 that quantifies the activity of receptors based on the energy transfer specifically in the plasma membrane or early endosomes. Interaction of β-arrestins with GPR64 and receptor endocytosis in G protein KO cells remains to be investigated to determine whether such events are preceded by G protein activation. β-Arrestin–mediated ERK1/2 activation46 downstream of GPR64 and recruitment of E3 ligase(s)47 for GPR64 ubiquitination are also unknown.
To summarize, our study shows that the NTF and tethered agonist play significant roles in GPR64 signaling and trafficking by regulating the recruitment of GRKs and β-arrestins. The role of GRKs and β-arrestins in potentiating GPR64 signaling via its internalization is novel in the field of aGPCRs. Further studies of other aGPCRs in cell lines and primary cells can reveal how universal these features are in the aGPCR family.
Supplementary Material
Acknowledgments
We acknowledge the assistance from the staff of the Confocal Microscopy Core at the University of Maryland School of Medicine, Center for Innovative Biomedical Resources, Baltimore, Maryland. We are grateful to Dr. John Olson for the infrastructure support. This work was supported by the American Cancer Society Institutional Research Grant and Research Starter Grant in Translational Medicine and Therapeutics from the PhRMA Foundation to N.B. A.I. was supported by the PRIME JP18gm5910013h and the LEAP JP18gm0010004h from the Japan Agency for Medical Research and Development (AMED). S.C.T.-L was supported by the Meyerhoff Fellows Program.
Footnotes
Competing interests
The authors declare no competing interests.
Supporting information
Additional supporting information may be found in the online version of this article.
References
- 1.Hauser AS, Attwood MM, Rask-Andersen M, et al. 2017. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov 16: 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamann J et al. 2015. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev 67: 338–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arac D et al. 2012. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31: 1364–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson JR et al. 2013. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J. Biol. Chem 288: 22248–22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paavola KJ & Hall RA. 2012. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol 82: 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monk KR et al. 2015. Adhesion G protein-coupled receptors: from in vitro pharmacology to in vivo mechanisms. Mol. Pharmacol 88: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aust G, Zhu D, Van Meir EG & Xu L. 2016. Adhesion GPCRs in tumorigenesis. Handb. Exp. Pharmacol 234: 369–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z et al. 2017. Expression of orphan GPR56 correlates with tumor progression in human epithelial ovarian cancer. Neoplasma 64: 32–39. [DOI] [PubMed] [Google Scholar]
- 9.Luo R, Jin Z, Deng Y, et al. 2012. Disease-associated mutations prevent GPR56–collagen III interaction. PLoS One 7: e29818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veninga H et al. 2011. CD97 antibody depletes granulocytes in mice under conditions of acute inflammation via a Fc receptor-dependent mechanism. J. Leukoc. Biol 89: 413–421. [DOI] [PubMed] [Google Scholar]
- 11.Krishnan A, Nijmeijer S, de Graaf C & Schioth HB. 2016. Classification, nomenclature, and structural aspects of adhesion GPCRs. Handb. Exp. Pharmacol 234: 15–41. [DOI] [PubMed] [Google Scholar]
- 12.Balenga N et al. 2017. Orphan adhesion GPCR GPR64/ADGRG2 is overexpressed in parathyroid tumors and attenuates calcium-sensing receptor-mediated signaling. J. Bone Miner. Res 32: 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amisten S et al. 2017. A comparative analysis of human and mouse islet G-protein coupled receptor expression. Sci. Rep 7: 46600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebscher I et al. 2014. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 9: 2018–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paavola KJ, Stephenson JR, Ritter SL, et al. 2011. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J. Biol. Chem 286: 28914–28921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoveken HM, Hajduczok AG, Xu L & Tall GG. 2015. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl. Acad. Sci. USA 112: 6194–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demberg LM, Rothemund S, Schoneberg T & Liebscher I. 2015. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem. Biophys. Res. Commun 464: 743–747. [DOI] [PubMed] [Google Scholar]
- 18.Kishore A, Purcell RH, Nassiri-Toosi Z & Hall RA. 2016. Stalk-dependent and stalk-independent signaling by the Adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J. Biol. Chem 291: 3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calebiro D et al. 2009. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 7: e1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrandon S et al. 2009. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat. Chem. Biol 5: 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomsen AR et al. 2016. GPCR-G protein–β-arrestin super-complex mediates sustained G protein signaling. Cell 166: 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irannejad R et al. 2017. Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol 13: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrage R, Schmitz AL, Gaffal E, et al. 2015. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun 6: 10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balenga NA, Aflaki E, Kargl J, et al. 2011. GPR55 regulates cannabinoid 2 receptor-mediated responses in human neutrophils. Cell Res. 21: 1452–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henstringe CM, Balenga NA, Ford LA, et al. 2009. The GPR55 ligand L-alpha-lysophosphatidylinositol promotes RhoA-dependent Ca2+ signaling and NFAT activation. FASEB J. 23: 813–193. [DOI] [PubMed] [Google Scholar]
- 26.Peeters MC et al. 2015. The adhesion G protein-coupled receptor G2 (ADGRG2/GPR64) constitutively activates SRE and NFkappaB and is involved in cell adhesion and migration. Cell Signal. 27: 2579–2588. [DOI] [PubMed] [Google Scholar]
- 27.Alvarez-Curto E et al. 2016. Targeted elimination of G proteins and arrestins defines their specific contributions to both intensity and duration of G protein-coupled receptor signaling. J. Biol. Chem 291: 27147–27159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenoy SK & Lefkowitz RJ. 2011. β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol. Sci 32: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfe BL & Trejo J. 2007. Clathrin-dependent mechanisms of G protein-coupled receptor endocytosis. Traffic 8: 462–470. [DOI] [PubMed] [Google Scholar]
- 30.Bohnekamp J & Schoneberg T. 2011. Cell adhesion receptor GPR133 couples to Gs protein. J.Biol.Chem 286: 41912–41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Premont RT, Inglese J & Lefkowitz RJ. 1995. Protein kinases that phosphorylate activated G protein-coupled receptors. FASEB J. 9: 175–182. [DOI] [PubMed] [Google Scholar]
- 32.Pitcher JA et al. 1992. Role of beta gamma subunits of G proteins in targeting the beta-adrenergic receptor kinase to membrane-bound receptors. Science 257: 1264–1267. [DOI] [PubMed] [Google Scholar]
- 33.Grundmann M et al. 2018. Lack of beta-arrestin signaling in the absence of active G proteins. Nat. Commun 9: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L et al. 2015. G protein-coupled receptor kinases of the GRK4 protein subfamily phosphorylate inactive G protein-coupled receptors (GPCRs). J. Biol. Chem 290: 10775–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pack TF, Orlen MI, Ray C, et al. 2018. The dopamine D2 receptor can directly recruit and activate GRK2 without G protein activation. J. Biol. Chem 293: 6161–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wootten D, Christopoulos A, Marti-Solano M, et al. 2018. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol 19: 638–653. [DOI] [PubMed] [Google Scholar]
- 37.Nygaard R et al. 2013. The dynamic process of β(2)-adrenergic receptor activation. Cell 152: 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staus DP et al. 2016. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latorraca NR, Venkatakrishnan AJ & Dror RO. 2017. GPCR dynamics: structures in motion. Chem. Rev 117: 139–155. [DOI] [PubMed] [Google Scholar]
- 40.Dror RO et al. 2011. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA 108: 18684–18689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gurevich EV, Tesmer JJ, Mushegian A & Gurevich VV. 2012. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol. Ther 133: 40–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jimenez-Vargas NN et al. 2018. Protease-activated receptor-2 in endosomes signals persistent pain of irritable bowel syndrome. Proc. Natl. Acad. Sci. USA 115: E7438–E7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarwood RE et al. 2017. Endosomal signaling of the receptor for calcitonin gene-related peptide mediates pain transmission. Proc. Natl. Acad. Sci. USA 114: 12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang DL et al. 2018. Gq activity- and β-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. elife 7 10.7554/eLife.33432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan Q et al. 2018. Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J. Biol. Chem 293: 7466–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shenoy SK et al. 2006. Beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J. Biol. Chem 281: 1261–1273. [DOI] [PubMed] [Google Scholar]
- 47.Shenoy SK et al. 2008. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J. Biol. Chem 283: 22166–22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.