Abstract
Organisms from all kingdoms of life have evolved a vast array of peptidic natural products to defend against microbes. These are known collectively as antimicrobial peptides (AMPs) or host defense peptides, reflecting their abilities to not only directly kill microbes, but also to modulate host immune responses. Despite decades of investigation, AMPs have yet to live up to their promise as lead therapeutics, a reality that reflects, in part, our incomplete understanding of these diverse agents in their various physiological contexts. Toward improving our understanding of AMP biology and the ways in which this can be best leveraged for therapeutic development, we are interested in large-scale comparisons of the antimicrobial and immunological activities of human AMPs, an undertaking that requires an efficient workflow for AMP synthesis and subsequent characterization. We describe here the application of flow chemistry and reverse phase flash chromatography to the generation of 43 AMPs, approaches that, when combined, significantly expedite synthesis and purification, potentially facilitating more systematic approaches to downstream testing and engineering.
INTRODUCTION
Antimicrobial peptides (AMPs) are miniproteins made by diverse organisms to defend against microbes. These natural compounds have been studied in depth since the 1980s with the goal of using them in future generations of antibiotics. While certain agents based on the cyclic natural products of non-ribosomal peptide synthetases such as daptomycin have entered clinical use(1), no ribosomally synthesized AMP has had such success to date despite extensive effort and a number of promising results(2–4).
Multiple barriers have thwarted attempts to adapt AMPs for clinical use, including instability, toxicity, and limited potency. Despite these drawbacks, we posit that the failure of AMPs to date as therapeutic agents reflects not an intrinsic shortcoming of AMPs themselves, but rather our own tendency to think of these molecules as drugs rather than as peptides and miniproteins that evolve to confer upon their hosts a selective advantage, a distinction that may impact both the study and the clinical application of AMPs(5,6).
A further limitation in the field is the diversity of materials and methods employed in the study of AMPs, which makes an appreciation of the relative biological properties of each variant difficult. It is also generally true that, despite the thousands of papers written to date about AMPs, large gaps remain in our ability to answer even basic questions. For example, the activity of most AMPs against a number of organisms commonly encountered in clinical practice is unknown, as is the activity of these AMPs against appreciable numbers of clinical isolates of any single species. There is further a persistent emphasis in the literature on the membranolytic effects of certain AMPs despite the fact that these peptides are endowed with a variety of other functions, some of which might prove therapeutically useful, not the least of which is the ability of AMPs to recruit protective immune responses(7–9).
Our goal is to systematically address these knowledge gaps with the expectation that, in doing so, we will gain the insight necessary to engineer and deploy AMPs for optimal effect in specific contexts. Such a systematic approach is complicated, however, by the cost either in time or in treasure of obtaining sufficient quantities of AMP for testing all the conditions one might want to study. A brief survey of commercially available LL-37, for example, reveals a cost range of 179–1,900 USD per mg of material. For more synthetically difficult targets such as hBD- 3, often made by recombinant techniques, this rises to 2,220–6,580 USD per mg. We estimate that at least 6 mg for larger AMPs such as LL-37 may be required for initial testing against selected species under a range of microbiological conditions representative of the physiology underlying conditions of infection, with additional material required for expanded species testing among selected AMPs and more still to assay immunological functions. There is further literature precedent to suggest that performing synthesis and quality control in-house may improve the ability to draw meaningful biological conclusions(10), and our own experience has been that commercial synthesis fails to yield the desired product in more than 40% of cases(11).
Here we describe the application of automated flow chemistry developed in our group(12) as well as reverse phase flash chromatography to the efficient synthesis and purification of synthetic AMPs for biological studies, our goal being the generation of a library containing all human-derived AMPs described in the APD3 antimicrobial peptide database(13) for subsequent systematic testing of antimicrobial and immunological functions.
RESULTS AND DISCUSSION
Overview
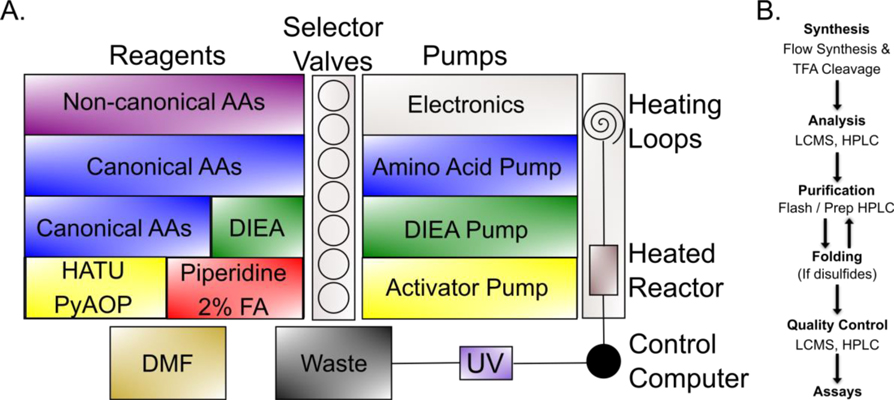
Synthesis was completed primarily on a 3rd generation automated fast-flow peptide synthesizer (AFPS) based on the original instrument described in 2017(12), which in turn follows prior work on the implementation of manual flow peptide synthesis from our group(14). A schematic of a typical AFPS instrument is shown in Figure 1. AFPS instruments permit automated solid phase fluorenylmethyloxycarbonyl (Fmoc) chemistry at 90 °C under flow for improved speed and fidelity. Under rapid synthesis conditions, which were used for most of the AMPs described here as specified in the legends to Supplementary Figures 1–43, coupling of a single amino acid can be completed in as few as 40 seconds, though more difficult amino acid couplings require more time. The average coupling time per amino acid across all peptides described here was approximately 80 seconds. A subset of the peptides were synthesized under modified conditions optimized for the synthesis of long peptides; average synthesis time for these peptides was approximately 150 seconds per amino acid. Fmoc deprotection is monitored throughout to verify expected progression of synthesis and to assist in the identification of problematic couplings; the B and C panels of Supplementary Figures 1–43 show the UV traces and integrals thereof for each AMP reported.
Figure 1:
A. Schematic of the flow synthesizers used in this study. Three pumps on the right dedicated to amino acids (blue), base (green), and activators or deprotection reagents (yellow - depending on cycle step) control flow of selected reagents (amino acids in blue / purple, base in green, activators in yellow, deprotection solution in red) and solvent (brown) on the left through the selector valves in the center and into to the heating loops and heated reactor on the right, eventually passing through an ultraviolet (UV) detector and on to the central waste bin. All functions are computer-controlled. See also Experimental. B. Workflow for peptide synthesis and characterization. In brief, flow-synthesized peptides are treated with strong acid to effect sidechain deprotection and release of the linear polypeptide from resin followed by trituration, lyophilization, and characterization by liquid chromatography-mass spectrometry (LCMS) and high performance liquid chromatography (HPLC). Crude peptides are purified by reverse phase flash chromatography (RPFC) or preparative HPLC and folded if required. Quality control LCMS and HPLC are then used to characterize purified peptides ahead of assays.
Synthesis yielded the desired AMP as the major product in most cases, with an average crude purity of 65% across all peptides and an average crude yield of 77 mg, which varied with the AMP and with the amount of resin cleaved - typically in the range of 50–75% of 100–200 mg of 4-(4-hydroxymethyl-3-methoxyphenoxy)butyric acid (HMPB) resin at approximately 0.44 mmol/g loading, with the remainder retained for later modification or workup as needed. This paper describes the synthesis of all linear human AMPs annotated in the APD3 antimicrobial peptide database(13). For ease of presentation, we have divided these into four groups, with the specifics of each synthesis summarized in Tables 1–4 for each class - Cathelicidins, Histatins, Neuropeptides, and Miscellaneous peptides. Of note, Cathelicidins derive from a single gene and Histatins derive from two genes in humans, each of which results in multiple antimicrobial fragments through proteolytic cleavage with substantial sequence overlap (Supplementary Figure 44A-B). In contrast, the groupings of “Neuropeptides” (peptides that localize in some way to nervous system tissues, as annotated in the APD3) and Miscellaneous peptides are arbitrary and without similar genetic basis. Cystine-containing human AMPs such as a and p defensins will be described in a separate manuscript to follow.
Table 1:
Cathelicidin Yield and PurityA
| AMP | Length (AA) | Avg. MW (Da) | Crude Yield (mg) | Crude Purity % (RTE) | Crude Loaded (mg) | Pure Yield (mg) | Recovery % | Purity % (RTE) |
|---|---|---|---|---|---|---|---|---|
| KR-20D | 20 | 2468.9 | 154 | 90 (29.7) | 100 | 22 | 24 | 99% (29.6) |
| LL-23B | 23 | 2823.4 | 81 | 65 (27.4) | 60 | 33 | 85 | 92 (27.0) |
| KS-27B | 27 | 3327.0 | 89 | 78 (39.2) | 60 | 46 | 98 | 96 (38.8) |
| LL-29B | 29 | 3596.3 | 90 | 67 (38.6) | 68 | 30 | 66 | 92 (38.8) |
| KS-30B,C | 30 | 3644.3 | 34 | 83 (38.6) | 30 | 19 | 76 | 98 (38.0) |
| RK-31B | 31 | 3715.4 | 106 | 83 (39.1) | 57 | 32 | 68 | 78 (38.9) |
| LL-37B,C | 37 | 4493.3 | 61 | 77 (42.9) | 51 | 32 | 81 | 95 (43.2) |
| ALL-38B | 38 | 4564.4 | 151 | 67 (43.9) | 69 | 15 | 32 | 71 (44.3) |
| TLN-58D | 58 | 6861.9 | 162 | 74 (44.7) | 98 | 19 | 26 | 98 (44.6) |
| Avg. | 32.6 | 3943.9 | 103 | 76 | 66 | 28 | 62 | 91 |
Characterization in Supplementary Figures 1–9. All synthesized on a 3rd Generation flow synthesizer set for speed and purified by RPFC Method 2 unless otherwise specified.
Purified by RPFC Method 1.
Synthesized on a 3rd Generation flow synthesizer set for length.
Synthesized on a 4th Generation flow synthesizer set for length.
RT = retention time in minutes.
Table 4:
Miscellaneous Linear AMP Yield and PurityA
| AMP | Length (AA) | Avg. MW (Da) | Crude Yield (mg) | Crude Purity % (RTE) | Crude Loaded (mg) | Pure Yield (mg) | Recovery % | Purity % (RTE) |
|---|---|---|---|---|---|---|---|---|
| Buforin I | 39 | 4250.9 | 102 | 28 (22.2) | 102 | 6.0 | 21 | >99 (20.9) |
| CalciterminB | 15 | 1688.9 | 57 | 82 (14.9) | 48 | 13 | 33 | 75 (15.4) |
| β-Casein 197 | 17 | 2005.3 | 39 | 53 (28.5) | 39 | 14 | 68 | 98 (28.1) |
| Dermcidin | 47 | 4705.4 | 126 | 32 (38.1) | 126 | 11 | 27 | 88 (37.3) |
| GHH20 | 21 | 2417.5 | 80 | 34 (15.3) | 80 | 8.5 | 31 | 55 (15.2) |
| hGAPDHB | 31 | 3186.7 | 94 | 61 (28.1) | 52 | 35 | >99% | 80 (28.1) |
| KDAMP 19-mer | 19 | 1767.0 | 57 | 37 (18.9) | 57 | 17 | 81 | 85 (18.6) |
| PDC213 | 14 | 1471.7 | 31 | 70 (20.9) | 31 | 12 | 55 | >99 (20.8) |
| Salusin β | 20 | 2342.8 | 17 | 62 (38.4) | 17 | 8.0 | 76 | 88 (38.5) |
| Salvic | 46 | 5258.1 | 21 | 45 (54.4) | 21 | n/a | n/a | n/a |
| SgI-29 | 29 | 3377.7 | 104 | 67 (11.5) | 104 | 26 | 37 | 98 (11.4) |
| SgII Peptide AB | 29 | 3309.7 | 95 | 65 (15.0) | 52 | 6.1 | 18 | 79 (15.3) |
| Cathepsin G (1–5)C | 5 | 514.6 | 39 | 90 (14.3) | n/a | n/a | n/a | n/a |
| Ubiquicidin | 59 | 6647.8 | 117 | 61 (19.7) | 117 | 17 | 24 | 98 (9.1)D |
| Avg. | 27.9 | 3067.4 | 70 | 56 | 65 | 14 | 47 | 87 |
Characterization in Supplementary Figures 30–43. All synthesized on a 3rd Generation flow synthesizer set for speed and purified by RPFC Method 2 unless otherwise specified.
Purified by RPFC Method 1.
Synthesized on a 4th Generation flow synthesizer set for length.
Derived from HPLC Method 3 (30-minutes); crude characterized with HPLC Method 1 (60-minutes).
RT = retention time in minutes.
Cathelicidins - Synthesis, Characterization, and Purification
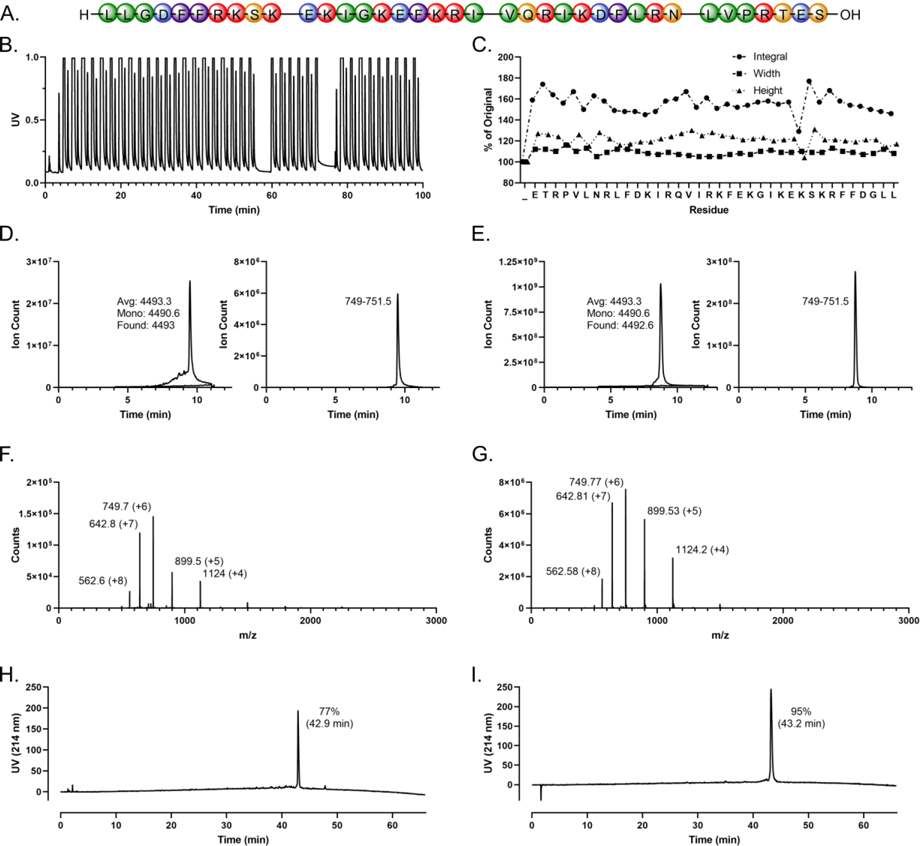
Data collected in the course of each synthesis are presented in Supplementary Figures 1–43; for reference within the main text, Figure 2 reproduces the data from Supplementary Figure 7 on LL-37 synthesis.
Figure 2:
Automated flow synthesis of antimicrobial peptide LL-37 (reproduced here from Supplementary Figure 7). A. LL-37 sequence (red = cationic, blue = anionic, orange = polar, green = nonpolar, purple = aromatic). B. Synthesizer UV trace showing resolved Fmoc deprotection peaks alternating with saturated amino acid coupling peaks. Synthesizer settings are specified for each peptide; here, 3rd Generation synthesizer - optimized for Length. Spaces in the x-axis represent optional, user-initiated pauses. C. Fmoc deprotection integrals and peak width and height expressed as percentages relative to the first cycle. D. Left panel: total ion chromatogram (TIC) of crude AMP overlaid on Blank run with the predicted average and monoisotopic masses as well as the observed mass as calculated from the most abundant ion. Right panel: extracted ion chromatogram (EIC) of crude AMP for the specified m/z range. The LCMS method is specified for each AMP; here, LCMS Method 5. E. TIC and EIC of purified AMP, LCMS Method 3. F-G. Mass spectra associated with the dominant peaks of D and E, respectively. The charge states of the labeled ions are indicated in parentheses. H-I. Analytical high performance liquid chromatography (HPLC) traces of crude and purified peptide, respectively, with the integrated percentage of the dominant peak (retention time in parentheses). The HPLC method is specified for each AMP; here, HPLC Method 1.
Among the linear human AMPs, cathelicidins, including the canonical AMP LL-37 and eight previously described related cleavage fragments thereof were synthesized with the highest average group purity among the AMP classes studied here at 76% (Table 1). With conditions optimized for the synthesis of longer peptides (see Experimental), crude purity reached up to 90% for shorter peptides (Supplementary Figure 1, a 20-mer) and 74% for the longest sequence in this group (Supplementary Figure 9, a 58-mer). No problematic sequences were noted among cathelicidins.
Although we initially utilized standard, preparative reverse phase HPLC for AMP purification, we found that, given the general ease of synthesis using our AFPS synthesizers, purification by this method became a rate-limiting step in our workflow. This was further complicated by the potential loss of material during the filtration step prior to loading due to poor solubility of many crude AMP preparations in water (data not shown). We hypothesized, however, that when starting from a high crude purity, as was typical of the cathelicidins in particular, these barriers might be overcome by using reverse phase flash chromatography (RPFC) in place of HPLC, which in addition to being a faster purification method permits sample loading as a suspension.
Initial attempts at using RPFC for AMP purification demonstrated the need for further optimization to ensure consistent results with this methodology. Although some AMPs with a high crude purity such as certain cathelicidins (Supplementary Figures 2–5, 7) could be purified with a generic A - water:B - acetonitrile (each with 0.1% trifluoroacetic acid (TFA)) gradient of 1–91% B over 20 column volumes (CV) on Biotage RPFC columns (see Experimental), this approach yielded little or no enrichment of the desired product with other crude materials, which was true to some extent among cathelicidins (Supplementary Figures 6, 8), and which was clearly the case for a broader range of AMPs (Supplementary Figures 24, 29, 31, 35, 41, and others not reported here). Overall, average crude purity for peptides purified once with RPFC using the above gradient or a minor variation thereof was 70%, while their post-RFPC purity was 82% - 74% crude and 89% pure among cathelicidins, 63% crude and 71% pure among others.
To optimize RPFC for AMP purification, we transitioned to a strategy in which we utilized a shallow gradient of 20% B over 30 CV centered on the estimated % B at elution derived from analytical HPLC. Although retention using this approach as a predictor was often slightly longer than anticipated, as might be expected given the more hydrophobic character of the RPFC C18 columns compared with the analytical HPLC C4 column used for most AMPs, this approach resulted in an average final purity of 91% (over an average 62% crude purity). Recovery of desired peptide from the amount theoretically contained in the crude material was also robust, averaging 62% among cathelicidins. At less than 30 minutes per purification and approximately 0.8 L of solvent on the 10 g column most commonly used (<45 minutes with around 2 L of solvent on a 25 g column), this remained a substantial improvement over our typical preparative HPLC methods, which require approximately 90 minutes and more than 1.6 L of solvent per purification inclusive of equilibration and loading steps on a 21.2 mm internal diameter column. Overall purity of the isolated products among cathelicidins was 91% with an average purified yield of 28 mg (Table 1).
Histatins - Synthesis, Characterization, and Purification
Histatin synthesis on our flow synthesizers resulted in a slightly lower crude purity, 63% on average, than was obtained with the cathelicidins (Table 2). Although only one of these, Histatin 5, was synthesized with methods optimized for length, the crude purity of this AMP was somewhat higher than those of others in this group at 85%. Although no specific problematic sequences were noted among the histatins, sequential histidines did tend to produce a decrease in Fmoc deprotection integrals among the longer histatins (Panel C of Supplementary Figures 10–13, 15, less prominent or absent in Supplementary Figures 14, 16–18). It may be possible to further optimize synthesis by considering both the preceding amino acid and the amino acid being coupled, though we have not yet performed any investigations along these lines.
Table 2:
Histatin Yield and PurityA
| AMP | Length (AA) | Avg. MW (Da) | Crude Yield (mg) | Crude Purity % (RTC) | Crude Loaded (mg) | Pure Yield (mg) | Recovery % | Purity % (RTC) |
|---|---|---|---|---|---|---|---|---|
| Histatin 1 | 38 | 4928.1 | 62 | 42 (28.0) | 62 | 22 | 84 | 92 (28.0) |
| Histatin 2 | 27 | 3444.7 | 100 | 51 (28.7) | 21 | 14 | >99% | 87 (28.8) |
| Histatin 3 | 32 | 4062.4 | 43 | 54 (21.5) | 42 | 12 | 53 | 79 (21.6) |
| Histatin 4 | 21 | 2745.0 | 61 | 59 (21.4) | 61 | 17 | 47 | 77 (21.4) |
| Histatin 5B | 24 | 3036.3 | 124 | 85 (19.2) | 22 | 23 | >99% | 99 (18.6) |
| Histatin 6 | 25 | 3192.5 | 65 | 69 (18.3) | 64 | 20 | 45 | 97 (18.3) |
| Histatin 7 | 13 | 1718.9 | 58 | 74 (17.9) | 23 | 13 | 76 | 89 (17.4) |
| Histatin 8 | 12 | 1562.7 | 49 | 71 (17.8) | 39 | 21 | 76 | 92 (17.1) |
| Histatin 9 | 14 | 1875.1 | 77 | 65 (17.3) | 76 | 13 | 26 | 75 (16.7) |
| Avg. | 22.9 | 2951.7 | 71 | 63 | 46 | 17 | 67 | 87 |
Characterization in Supplementary Figures 10–18. All synthesized on a 3rd Generation flow synthesizer set for speed and purified by RPFC Method 2 unless otherwise specified.
Synthesized on a 3rd Generation flow synthesizer set for length.
RT = retention time in minutes.
Histatin 1 in this series required the addition of a known post-translational modification, phosphorylation at Ser2. This was introduced by batch coupling the modified amino acid following flow synthesis of the bulk of the peptide. We have not yet tested whether phosphorylated amino acids or amino acids carrying other modifications found in vivo may be incorporated directly using our flow synthesizers. The main benefit of batch synthesis in this scenario is the ability to minimize the amount of material used when multiple couplings of a given modified amino acid are not needed.
Histatin analysis was carried out on Luna C18 columns for both LCMS and analytical HPLC, as retention of the shorter members of this group (Histatins 5–6 and particularly 7–9) was best achieved on this column (see Experimental). Of note, the lower end of the mass spectra obtained for some purified histatins contained a number of low molecular weight ions compared with those from the corresponding crude peptides (Supplementary Figures 14–18 panels F-G), which was attributed to higher energy ESI and associated fragmentation during analysis of some histatins with greater charge-to-length ratios (54–64% positively charged residues among Histatins 5–9 versus a range of 30–47% in Histatins 1–4)1. Despite this, the predicted ions were evident for each histatin, while analytical HPLC suggested a single, major product (Supplementary Figures 14–18, panel I).
Despite use of the optimized RFPC Method 2 that generally resulted in more reliable purification among all peptides as above, final histatin purity was slightly lower than that seen among the cathelicidins at 87%, with an average recovery of 67% and an average pure yield of 17 mg (Table 2). We anticipate that further optimization of our RPFC methods may allow for further improvements among histatins and other exceptionally polar or otherwise difficult-to- purify AMPs.
Neuropeptides - Synthesis, Characterization, and Purification
With the exception of the two p-amyloid derivatives described here (Supplementary Figure 44C), there are no sequence relationships among neuropeptides, which are instead defined by a common localization to neural tissues as annotated in the APD3. Syntheses are thus generally unique to each peptide. Group crude purity was 70% by HPLC (Table 3), though this is somewhat skewed by neurotensin and cathepsin G as discussed below, with an average recovery of 40% for an average yield of 15 mg with an average purity of 88% (97% if considering only the six neuropeptides purified by RPFC Method 2, see Table 3).
Table 3:
Neuropeptide Yield and PurityA
| AMP | Length (AA) | Avg. MW (Da) | Crude Yield (mg) | Crude Purity % (RTE) | Crude Loaded (mg) | Pure Yield (mg) | Recovery % | Purity % (RTE) |
|---|---|---|---|---|---|---|---|---|
| Alarin | 25 | 2894.3 | 97 | 54 (19.1) | 96 | 10 | 19 | >99% (19.0) |
| Amyloid β 1–40C | 40 | 4329.8 | 32 | 65 (31.9) | 22 | 2.9 | 20 | 96 (32.0) |
| Amyloid β 1–42C | 42 | 4514.1 | 80 | 65 (33.8) | 21 | 5.4 | 40 | 77 (34.0) |
| Bradykinin | 9 | 1060.2 | 17 | 91 (17.6) | n/a | n/a | n/a | n/a |
| Catestatin | 21 | 2326.7 | 81 | 74 (24.5) | 81 | 16 | 27 | 96 (23.7) |
| CGA-N46B | 46 | 5363.1 | 117 | 51 (34.6) | 51 | 13 | 50 | 53 (35.3) |
| α MSHD | 13 | 1623.8 | 73 | 88 (21.2) | 73 | 28 | 44 | >99% (21.2) |
| Neurotensin | 13 | 1671.9 | 45 | 90 (22.1) | 45 | 13 | 32 | 95 (22.0) |
| Neuropeptide Y | 36 | 4271.7 | 66 | 48 (35.1) | 65 | 19 | 61 | 96 (35.1) |
| Substance PD | 11 | 1348.6 | 71 | 90 (24.0) | 70 | 15 | 24 | >99% (24.1) |
| Vasoactive Intestinal PeptideB | 28 | 3326.8 | 79 | 56 (27.0) | 54 | 25 | 83 | 69 (27.6) |
| Avg. | 25.8 | 2975.5 | 69 | 70 | 58 | 15 | 40 | 88 |
Characterization in Supplementary Figures 19–29. All synthesized on a 3rd Generation flow synthesizer set for speed and purified by RPFC Method 2 unless otherwise specified.
Purified by RPFC Method 1.
Purified by RP-HPLC Method 1.
Synthesized on a 4th Generation flow synthesizer set for length.
RT = retention time in minutes.
Neurotensin in this series involved batch addition of unprotected pyroglutamate to the N- terminus of the flow-synthesized core peptide to reflect a known post-translational modification. This resulted in apparent dipeptide addition of the unprotected amino acid in roughly equimolar quantities with the desired product, which proved inseparable from its larger counterpart (Supplementary Figure 26, wherein the left shoulders of the D and E panel TIC peaks contain the unintended product). The other post-translational modification included among the neuropeptides, C-terminal amidation of Neuropeptide Y (Supplementary Figure 27), was introduced via synthesis on Rink Amide, whereas the remainder of the peptides described here contain C-terminal acids (see Experimental).
As discussed above for cathelicidins, purification of neuropeptides using the generic gradient in RPFC Method 1 resulted in suboptimal separation and final purities of 53% and 69% in the two instances where this was attempted - for CGA-N46 and Vasoactive Intestinal Peptide (Table 3, Supplementary Figures 24I and 29I). While most of the impurities evident by LCMS in the AMPs described here are smaller fragments more consistent with truncations, degradation products, or column contaminants, the prominent co-eluting shoulder in the TIC for CGA-N46 contains a mass shift of −200 Da, likely representing a compound deletion of two amino acids (Supplementary Figure 24E); five such combinations are possible in this sequence (Supplementary Figure 24C). The most prominent co-eluting shoulder for Vasoactive Intestinal Peptide in Supplementary Figure 29E contains a mass shift of −18, which could represent any of several alterations, though aspartimide formation at the C-terminal Asn24-Ser25 appears the most likely.
Purification of p-amyloid 1–42 (Supplementary Figures 20–21) by RPFC Method 2 was unsuccessful due to apparent aggregation of these peptides on the column (data not shown). Prior descriptions of p-amyloid purification have emphasized the importance of column heating(15), a feature not available with our RPFC columns. We therefore returned to preparative HPLC for purification of p-amyloid 1–40 and 1–42 with initial solubilization in DMSO followed by dilution in deionized water prior to loading with column heating to 60 °C for the duration of separation, which resulted in modest recovery (20 and 40%, respectively) and purity (96 and 77%, respectively).
Miscellaneous - Synthesis, Characterization, and Purification
Like neuropeptides, there are no sequence relationships among the Miscellaneous AMPs described here with the exception of the two AMPs derived from Semenogelins I and II, respectively (Table 4, Supplementary Figures 40, 41, 44). Reflecting their diversity, this group was also the most difficult to work with as a whole, including the only peptide in this dataset wherein the desired product was not the major crude product (SgI-29, Supplementary Figure 40) and the only two peptides in this dataset for which purification was either unsuccessful (Salvic, Supplementary Figure 39) or deferred (Cathepsin G 1–5, Supplementary Figure 42). Average crude purity in this group was 56% (Table 4), with several especially impure crude products such as Buforin I (28%, Supplementary Figure 30), Dermcidin (32%, Supplementary Figure 33), KDAMP 19-mer (37%, Supplementary Figure 36), and Salvic (45%, Supplementary Figure 39). As in the other AMPs described here, impurities were generally not identifiable as minor alterations of the core peptide (e.g., aspartimide formation, deamidation,etc.), but rather tended to be either amino acid deletions or smaller fragments more consistent with truncations, degradation products, or column contaminants.
Another peptide with poor crude yield in this series was GHH20 (34%, Supplementary Figure 34H). As observed for histatins, this peptide appeared to suffer from coupling inefficiency at sites of sequential histidine incorporation, which is particularly prominent in this peptide consisting of 13 His, 4 Gly, and 4 Pro (essentially four repeats of the sequence GHHPH; a third His in the fourth repeat is erroneously entered in the APD3 as compared with the original paper(16) and is included in the sequence described here). The high ratio of His to other amino acids (62%) further resulted in analytical complications similar to those described above for the smaller histatins with high ratios of positive charge (Supplementary Figure 34D–G, see Footnote 1). Although purified GHH20 migrated as a single peak by LCMS, analytical HPLC on the Luna C18 column suggested both in the crude and in the purified peptides the presence of two, inseparable products (Supplementary Figure 34H–I). It is unclear whether this represents an undesirable byproduct of synthesis or an intrinsic structural property of the peptide given its unique sequence, though the former is presumed for the purposes of the purity calculations presented here.
Synthesis of Salusin p was complicated by probable diketopiperazine (DKP) formation as evidenced by the marked drop in Fmoc deprotection peak area following the initial Pro-Pro dipeptide sequence (Supplementary Figure 38C), resulting in a modest overall yield of 8.0 mg (Table 4). Prior work both in initial optimization of flow synthesizer conditions and in our AMP syntheses have suggested that the ChemMatrix trityl(Trt)-OH resin is not suitable for use in our flow synthesizers, and thus we did not attempt resynthesis of this AMP with a Trt-based resin.
In one instance in this dataset, SgI-29, the desired product was not the major product obtained after flow synthesis and acid cleavage. The major product identified displayed a mass shift of +242 Da over the expected mass (Supplementary Figure 40D), presumably a Trt adduct and thus more likely to represent an issue with the workup than with the synthesis itself.Despite this, ions for the desired product could be detected migrating in a shoulder to the right of this major peak and were successfully purified with RPFC Method 2 with a 98% final purity by HPLC (Supplementary Figure 40E). Similar to histatins, this peptide with 48% positively charged residues appeared to fragment during ESI (Supplementary Figure 40G). The extracted ion chromatogram (EIC) of this purified product further appeared to show product migrating at two points, which could indicate substantial epimerization, though differential migration was not noted by analytical HPLC as above.
Purification either failed or was not completed in two cases in this dataset. While the desired peptide was the major crude product of Salvic synthesis (Supplementary Figure 39D), purification by RPFC Method 2 failed for unclear reasons. This may have been due to inadequate column equilibration and / or aggregation on the column, as the gradient was raised from 5–45% over 3 CV prior to initiation of the shallow gradient, with slow elution of low levels of peptide thereafter that never reached the set limit for collection despite extension of the shallow gradient to approximately 90% B (data not shown). In the second instance, analytical HPLC of Cathepsin G suggested a pure peptide (Supplementary Figure 42H), but in a fashion similar to Neurotensin, evaluation of the mass spectrum revealed prominent, singly-charged ions at 402.25 and 289.17 m/z, likely representing single and double deletions of Ile, respectively (Supplementary Figure 42F). Purification was deferred in this case in favor of future resynthesis.
Despite the complications delineated above, the use of AFPS instrumentation and flash purification methods proved generally successful in this group as well, with an overall average yield among AMPs in the Miscellaneous category of 14 mg with a recovery of 47% for those purified. Like the neuropeptides, final purity was reasonable at 87% overall, though this improves if considering only those peptides purified by RPFC Method 2 (90% overall, 94% if further discounting GHH20).
CONCLUSIONS
We set out to develop methods for the efficient synthesis and purification of human AMPs in order to facilitate the systematic study and engineering of these miniprotein scaffolds for potential therapeutic development. The first major barrier to be overcome was synthesis itself, and as demonstrated here, the application of the automated flow peptide synthesizers previously developed in our lab provided a highly effective solution to this problem. There were no apparent synthetic failures due to the synthesizers themselves across the 43 distinct peptides described, and the average time of synthesis under conditions optimized for longer peptides, which also generally result in high crude purities among shorter peptides, comes to only 2.5 minutes per amino acid coupling.
While this and prior efforts from our lab demonstrate the ability to rapidly synthesize peptides with high crude purity using flow chemistry(11), a similarly rapid approach to purification will help this technology to achieve its full potential. To this end, we describe here methods for the flash purification of AMPs that reduce the purification time to approximately a third of that required for preparative HPLC purification, while retaining an average final purity of 91% under optimized conditions. It is expected that further optimization via correlation of parameters such as predicted retention time or observed analytical HPLC retention time to the prediction of RPFC retention time will facilitate ongoing improvements in the final purity and efficiency achievable across a broad range of peptides. Given the lesser expenses associated with RPFC equipment, wider application of RPFC in peptide purification may also help to reduce the costs of purification compared with standard HPLC.
In summary, we describe here methods for the efficient synthesis and purification of human AMPs using flow chemistry and flash purification. Similar approaches are being applied to the synthesis of cystine-containing peptides, including optimization of oxidative folding conditions to maximize yield and throughput for the extension of our efforts to a broader range of AMPs. These outcomes will be reported separately in a later manuscript.
EXPERIMENTAL
Synthesis
Prior to synthesis, the C-terminal amino acid of each sequence was manually coupled to 100–200 mg of hydroxy (-OH) functionalized 4-(4-hydroxymethyl-3-methoxyphenoxy)butyric acid (HMPB) ChemMatrix resin, loading approximately 0.44 mmol/g, using 10 equivalents of Fmoc- protected amino acid, 5 equivalents of N,N-diisopropylcarbodiimide (DIC), and 0.1 equivalents of 4-(dimethylamino)pyridine (DMAP) in N,N-dimethylformamide (DMF) at room temperature for 8–24 hours.
Fmoc-protected amino acids were purchased from Novabiochem or Creosalus.Activating agents N,N,N’N,-tetramethyluronium hexafluorophosphate (HATU) and (7- azabenzotriazol-1-yloxy) tripyrrolidinophosphonium (PyAOP) were purchased from P3 BioSystems. Peptide synthesis was carried out in AldraAmine-treated DMF with the amino acids and coupling agents above in addition to N,N-diisopropylethylamine (DIEA). Fmoc deprotection was carried out in flow using 40% piperidine with 2% formic acid.
The primary instrument used to synthesize the AMPs described here is a 3rd generation AFPS(12). A picture is shown in the Table of Contents graphic, while a schematic of this instrument is shown in Figure 1.
Most of the AMPs described here were synthesized using settings optimized for speed, conditions similar to those initially described. Later settings are optimized for length to facilitate the chemical synthesis of long peptides, which involves a longer pump head refill time to facilitate accurate delivery of viscous solutions of the above reagents in DMF. Remaining AMPs were synthesized on a 4th Generation flow synthesizer optimized for length. The settings for each peptide are indicated in Tables 1–4 and in the legends to the associated Supplementary Figures 1–43. Note that the differential synthesizer settings represent not optimization for the indicated sequences, but rather optimization for general lab use.
With the exception of the above differences in pump head refill time, conditions for flow synthesis on the 3rd generation synthesizers are as follows:
Solvent: DMF treated as above with AldraAmine trapping agents for at least 24 hours
Amino Acids: 0.4 M stocks prepared from the above commercial sources (diluted 1:2 in flow with DMF for final concentrations of approximately 0.2 M not including the volume of DIEA)
Activators: 0.38 M HATU or 0.38 M PyAOP (diluted 1:2 in flow with DMF for final concentrations of approximately 0.19 M not including the volume of DIEA)
Base: DIEA
Temperature: 90 °C heating loop and reactor
| Coupling conditions: | 400 μL amino acid, 400 μL activator, and 40 μL base per stroke |
| A and S - HATU 21 strokes | |
| N, Q, R, T, V - PyAOP 21 strokes | |
| Remaining amino acids - HATU 8 strokes |
Deprotection conditions: 40% piperidine and 2% formic acid (13 strokes each deprotection solution and DMF, resulting in 1:2 dilution in flow for final concentrations of 20% and 1%, respectively), monitored at 312 nm
Using the above conditions, a typical synthesis starts with a pre-wash step in DMF followed by an initial deprotection (indicated by “_” in the C panels of Supplementary Figures 1–43). Lines are primed (5 strokes), and amino acids are then coupled as above, followed by line washing with DMF (35 strokes each through the amino acid and activator lines), deprotection of the coupled amino acid, and additional line washing as before prior to the next coupling. Following completion of flow synthesis, resin is swollen and washed with DCM, with subsequent drying and storage at room temperature in the dark until the time of further manipulation. Traces shown in the B panels of Supplementary Figures 1–43 were extracted from the raw control software and downsampled as needed to fit into an Excel spreadsheet prior to graphing in Prism. Axes were cut at sites of extended user-initiated pauses to synthesis, which generally reflect time spent restocking the synthesizer or attending to concurrent experiments. The total time along the y-axis of each trace therefore reflects the sum of actual synthesis and optional, user-initiated pauses. Integral calculations as in the C panels of Supplementary Figures 1–43 were carried out in Python on raw data without downsampling(12) prior to processing in Excel and graphing in Prism.
Batch Coupling
Batch coupling was carried out by dissolving 20 equivalents of Fmoc- protected amino acid and 19 equivalents of HATU in approximately 1.25 mL DMF each before mixing with 500 μL DIEA and adding to resin pre-swollen with DMF for coupling at room temperature for approximately 30 minutes. After filtration and washing with DMF, manual deprotection was typically completed by addition of 3 mL 20% piperidine in DMF to resin twice for approximately three minutes each, followed by additional DMF washes as before and drying in DCM as above if no further manipulations were planned.
Resin Cleavage
Acid cleavage of peptides was completed with Reagent K (82.5% TFA, 5% water, 5% thioanisole, 5% phenol, and 2.5% 1,2-ethanedithiol (EDT)) at room temperature, typically for approximately two hours. Cleavage reactions were subsequently triturated with ice cold ether and spun down to isolate precipitated peptide, which was then resuspended in a mixture of 50% water / 50% acetonitrile with 0.1% TFA, flash frozen in liquid nitrogen, and lyophilized. Yields were determined gravimetrically by subtraction of tube mass from the combined mass of tube and lyophilized product, and all reported masses are those of the TFA salts of the individual peptides.
Liquid Chromatography / Mass Spectrometry (LCMS)
One Agilent 6520 and two Agilent 6550 LCMS QTOF instruments were used in the course of these experiments. LCMS methods were carried out as described below in Supplementary Figures 1–43, all with A (water with 0.1% formic acid) and B (acetonitrile with 0.1% formic acid) gradients as follows:
Method 1) Agilent 6550–1 (1290 Infinity HPLC system with iFunnel QTOF MS run in positive ionization mode with a low m/z range 100–1700) with a Phenomenex Jupiter C4 column, 150 × 1.0 mm, 5 μm, 300 Å silica; flow rate 100 μL/minute, 1–61% B gradient over 10 minutes, MS on from 4–12 minutes
Method 2) Agilent 6550–1 (as above) with a Phenomenex Luna C18(2) column, 150 ×5 mm, 3 μm, 100 Å silica; flow rate 50 μL/min, 1–61% B gradient over 12 minutes, MS on from 4–14 minutes
Method 3) Agilent 6550–2 (as in 6550–1 but with m/z range 100–3000) with an Agilent Zorbax 300SB C3 column, 150 × 2.1 mm, 5 pm, 300 Å silica; flow rate 500 pL/minute, 1–61% B gradient from 2–12 minutes, MS on from 4–12 minutes
Method 4) Agilent 6550–2 (as above) with a Phenomenex Kinetex C18 column; not reported in Supplementary Figures 1–43.
Method 5) Agilent 6520 (1290 Infinity HPLC system with QTOF MS run in positive ionization mode with m/z range 100–3000) with an Agilent Zorbax 300SB C3 column, 150 × 2.1 mm, 5 μm, 300 Å silica; flow rate 800 μL/minute, 1–61% B gradient over 9 minutes, MS on from 4–11 minutes.
Analytical High Performance Liquid Chromatography (HPLC)
Analytical HPLC was carried out on an Agilent 1200 series system with UV detection at 214 nm. Methods used are summarized as follows, referenced by number in the text and in figures:
HPLC Method 1: Column - Phenomenex Aeris Widepore C4 column, 150 × 4.6 mm, 3.6 μm, 200 Åsilica; flow rate 0.8 mL/minute; Solvent System - A = water with 0.1% TFA, B = acetonitrile with 0.08% TFA; Gradient − 3 minute hold 1% B, 1–61% B gradient over 60 minutes, 3 minute hold 61% B, 10-minute post run 1% B; Flow Rate − 0.8 mL/minute
HPLC Method 1a: As in Method 1, but with a 1–61% B gradient over 30 minutes.
HPLC Method 2: Column - Phenomenex Luna C18(2) column, 100 × 4.6 mm, 3 μm, 100 Å silica; flow rate 1.0 mL/minute; Solvent System - A = water with 0.1% TFA, B = acetonitrile with 0.08% TFA; Gradient - 3 minute hold 1% B, 1–61% B gradient over 60 minutes, 3 minute hold 61% B, 10-minute post run 1% B
Integrals of HPLC peaks were calculated automatically with Agilent ChemStation software with subsequent manual inspection of the magnified baseline and modification of the automated calls - most commonly removal of erroneous peaks more consistent with background variation or splitting of a major peak to reflect tailing.
Reverse Phase Flash Chromatography (RPFC)
RPFC was completed on a Biotage Selekt flash chromatography system run on reverse phase columns with automated fraction collection as directed by UV trace. Fractions were subsequently pooled according to review of the UV trace as needed and analyzed either by MALDI-TOF MS on a Bruker microflex™ LRF machine run in linear positive ion mode with subsequent confirmation by LCMS or directly by LCMS. Fractions determined to contain the pure product were then pooled and lyophilized for further analysis. With minor variations made while optimizing protocols, the general methods were as follows:
RPFC Method 1: Column - Biotage SNAP Bio C18 10 g, 20 μm, 300 Å; Solvent System - A = water with 0.1% TFA, B = acetonitrile with 0.1% TFA; Gradient - 3 column volumes (CV) hold 1% B, 1–91% B gradient over 20 CV, 3 CV hold 91% B; a less common variation employed the same gradient approach on a Biotage Sfär Bio C18 25 g column, 20 μm, 300 Å or slightly different hold times before or after the gradient; flow rates as automatically determined for the referenced column by Biotage proprietary methods.
RPFC Method 2: Column - Biotage SNAP Bio C18 10 g as above; Solvent System - A = water with 0.1% TFA, B = acetonitrile with 0.1% TFA; Gradient - 3 CV hold 1 or 5% B, 3 CV ramp to start of the target gradient, 20% range B gradient over 30 CV centered on the estimated % B at the time of elution as determined by analytical HPLC (e.g., for an estimated elution at 30% B, one would employ a gradient of 20–40% B over 30 CV), 3 CV ramp to 90% B, 3 CV hold 90% B; flow rates as automatically determined for the referenced column by Biotage proprietary methods, as above. A less common variation employed the same gradient approach on a Biotage Sfär Bio C18 25 g column, slightly different hold times before or after the gradient, or extension of the gradient at the same slope for additional column volumes in the case of later eluting peptides.
Preparative HPLC
β-Amyloid peptides were purified using mass-directed HPLC on an Agilent 1260 Infinity HPLC system coupled to a 6130 quadrupole MS. Column - Agilent Zorbax 300 SB C3 9.4 × 250 mm, 5 μm, 300 Å semi-preparative column heated to 60 °C; flow rate 4 mL/minute; Solvent System - A = water with 0.1% TFA, B = acetonitrile with 0.1% TFA; Gradient − 3 minute hold at 1% B, 1–61% B gradient over 60 minutes, 3 minute hold at 61% B. Fractions were automatically collected at one-minute intervals. In addition to use of the intrinsic mass spectra generated by this approach, fractions were screened as above for RPFC.
Sequences and Alignments
All AMP sequences are derived from the APD3 antimicrobial peptide database(13). Alignments were made in Clustal Omega(17) using APD3 sequences. Additional manual manipulation of the output for Histatins was completed to show a conservative change at the C- terminus of some Histatins (Y or YR) that had otherwise been aligned 5 or 5–6 positions later in in each sequence. Structures shown in the graphical abstract are PyMOL representations of Protein Data Bank 1KJ6 (hBD-3) and 2K6O (LL-37).
Supplementary Material
ACKNOWLEDGEM ENTS
Dedicated to Prof. Paul Alewood and his innumerable contributions to peptide and protein chemistry. We would like to thank Dr. Nick Truex, Dr. Chris Shugrue, Dr. Andrei Loas, Ms. Carly Schissel, and Mr. Alex Callahan for helpful discussions. Funding for this work was provided in part by a seed grant from the Broad Institute and by NIH U19 AI142780. JSA was supported by NIH T32 AI007061 and by a Research Fellowship Award from the Cystic Fibrosis Foundation.
Footnotes
CONFLICTS OF INTEREST
JSA declares no conflicts of interest. BLP is a founder of Amide Technologies and Resolute Bio.
The LCMS in question on which we typically use the Luna C18 column underwent repairs between the runs resulting in panels D and E of Supplementary Figures 14–18, including replacement of the major high voltage component and recalibration. Interval changes in the machine thus likely account for the differences observed.
SUPPLEMENTARY MATERIAL: Supplementary Figures 1–43 include data on the synthesis, characterization, and purification of each AMP. Supplementary Figure 44 shows alignments of related peptides described in this manuscript.
REFERENCES
- 1.Eisenstein BI, Oleson FB Jr, Baltz RH. Daptomycin: From the Mountain to the Clinic, with Essential Help from Francis Tally, MD. Clin Infect Dis [Internet]. 2010. January [cited 2020 Jan 20];50(s1):S10–5. Available from: https://academic.oup.com/cid/article-lookup/doi/10.1086/647938 [DOI] [PubMed] [Google Scholar]
- 2.Luther A, Urfer M, Zahn M, Müller M, Wang SY, Mondal M, et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature. 2019. December 19; [DOI] [PubMed] [Google Scholar]
- 3.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science [Internet]. 2010. February 19 [cited 2018 Aug 30];327(5968):1010–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20167788 [DOI] [PubMed] [Google Scholar]
- 4.Mourtada R, Herce HD, Yin DJ, Moroco JA, Wales TE, Engen JR, et al. Design of stapled antimicrobial peptides that are stable, nontoxic and kill antibiotic-resistant bacteria in mice. Nat Biotechnol [Internet]. 2019. October 19 [cited 2019 Oct 4];37(10):1186–97. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31427820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: An emerging category of therapeutic agents Vol. 6, Frontiers in Cellular and Infection Microbiology. Frontiers Media S.A.; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koo HB, Seo J. Antimicrobial peptides under clinical investigation. Pept Sci [Internet]. 2019. September 3 [cited 2020 Mar 5]; 111(5). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/pep2.24122 [Google Scholar]
- 7.Haney EF, Straus SK, Hancock REW. Reassessing the Host Defense Peptide Landscape. Front Chem [Internet]. 2019. February 4 [cited 2019 Sep 25];7:43 Available from:https://www.frontiersin.org/article/10.3389/fchem.2019.00043/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock REW, Haney EF, Gill EE. The immunology of host defence peptides: beyond antimicrobial activity. Nat Rev Immunol [Internet]. 2016. May 18 [cited 2019 Oct 5];16(5):321–34. Available from: http://www.nature.com/articles/nri.2016.29 [DOI] [PubMed] [Google Scholar]
- 9.Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, et al. An anti-infective peptide that selectively modulates the innate immune response. Nat Biotechnol [Internet]. 2007. April 25 [cited 2019 Oct 4];25(4):465–72. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17384586 [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Hoover DM, Yang D, Boulegue C, Santamaria F, Oppenheim JJ, et al. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human -defensin 3. Proc Natl Acad Sci [Internet]. 2003. July 22 [cited 2019 Apr 1];100(15):8880–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12840147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truex NL, Holden RL, Wang BY, Chen PG, Hanna S, Hu Z, et al. Automated Flow Synthesis of Tumor Neoantigen Peptides for Personalized Immunotherapy. Sci Rep. 2020;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mijalis AJ, Thomas DA, Simon MD, Adamo A, Beaumont R, Jensen KF, et al. A fully automated flow-based approach for accelerated peptide synthesis. Nat Chem Biol [Internet]. 2017. February 28 [cited 2017 Dec 11];13(5):464–6. Available from: http://www.nature.com/doifinder/10.1038/nchembio.2318 [DOI] [PubMed] [Google Scholar]
- 13.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res [Internet]. 2016. January 4 [cited 2018 Oct 9];44(D1):D1087–93. Available from: https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkv1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon MD, Heider PL, Adamo A, Vinogradov AA, Mong SK, Li X, et al. Rapid Flow- Based Peptide Synthesis. ChemBioChem [Internet]. 2014. March 21 [cited 2020 Jan 20];15(5):713–20. Available from: http://doi.wiley.com/10.1002/cbic.201300796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo S, Zhang S, Kreutzer AG, Nowick JS. An Efficient Method for the Expression and Purification of AP(M1– 42). 2018. [cited 2018 Oct 3]; Available from: https://pubs.acs.org/sharingguidelines [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rydengard V, Shannon O, Lundqvist K, Kacprzyk L, Chalupka A, Olsson A-K, et al. Histidine-Rich Glycoprotein Protects from Systemic Candida Infection. Mitchell AP, editor. PLoS Pathog [Internet]. 2008. August 1 [cited 2018 Sep 28];4(8):e1000116 Available from: http://dx.plos.org/10.1371/journal.ppat.1000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.