Abstract
Receptor activity-modifying proteins (RAMPs) interact with G-protein-coupled receptors (GPCRs) to modify their functions, imparting significant implications upon their physiological and therapeutic potentials. A resurging interest in identifying RAMP-GPCR interactions has recently been fueled by coevolution studies and orthogonal technological screening platforms. These new studies reveal previously unrecognized RAMP-interacting GPCRs, many of which expand beyond Class B GPCRs. The consequences of these interactions on GPCR function and physiology lays the foundation for new molecular therapeutic targets, as evidenced by the recent success of Erenumab. Here, we highlight recent papers that uncovered novel RAMP-GPCR interactions, human RAMP-GPCR disease-causing mutations, and RAMP-related human pathologies, paving the way for a new era of RAMP-targeted drug development.
Keywords: Receptor activity-modifying proteins (RAMPs), G-protein-coupled receptor, Erenumab, CGRP, Adrenomedullin, Coevolution
Historical Paradigm of RAMP-GPCR Signaling
G-protein-coupled receptors (GPCRs) are the most tractable and druggable class of proteins, comprising approximately 30% of all approved therapeutics [1-3]. These seven-transmembrane pass receptors mediate intercellular communication through the binding of endogenous or exogenous ligands eliciting subsequent receptor conformational changes to release bound G-proteins. Over the past decade, the field of GPCR biology has experienced a dramatic revitalization with the discovery of biased signaling, spawning a new generation of allosteric drugs. Similarly, in the last three years, exciting evidence has emerged for novel GPCR interactions with a class of single-transmembrane proteins called receptor activity-modifying proteins (RAMPs), ushering in a new RAMPage that will revolutionize the GPCR field (BOX 1) [4].
BOX 1: Structural determinants of RAMP function.
RAMPs are single-pass transmembrane proteins that consist of an ~100 amino acid N-terminal extracellular domain (ECD) and a short ~9 amino acid C-terminal intracellular domain. Amino acid multiple alignment indicates that RAMP1, RAMP2, and RAMP3 are approximately 31% homologous, yet 56% similar, which supports both redundant and independent functions between RAMP family members [4, 16]. Functional differences between RAMPs can be attributed to differences within their N-terminal and C-terminal domains to alter ligand binding and G-protein coupling, respectively [91, 92]. For example, the RAMP2 N-terminal domain contains an additional 28 amino acids not found in either RAMP1 or RAMP3 [16]. Conversely, the RAMP3 C-terminal domain contains a type-1 PDZ motif (DTLL) that mediates interactions with the NA+/H+ exchange regulatory factor (NHERF) and N-ethylmaleimide-sensitive factor (NSF) to alter GPCR pharmacology [63, 64]. Important novel insights into GPCR ligand affinity and selectivity, as well as the role of RAMPs in modulation of GPCR pharmacology, are being reported through structural analysis of crystal and cryo-EM structures.
Dynamic regulation of GPCR function by RAMPs involves receptor-dependent alterations in GPCR trafficking, recycling, signal transduction, post-translational modifications, and G-protein coupling. Consequently, RAMPs impart pharmacological and physiological diversity to GPCR function [4-9]. Recent reports, reviewed herein, provide compelling evidence for RAMP-GPCR pairings within most GPCR subfamilies, but the pharmacologic, physiological, and pathological consequences of these interactions remain largely unknown. With the discovery of two disease-associated human mutations in the AM-CLR-RAMP2 signaling axis and the accumulated wealth of physiologic knowledge gleaned from genetic mouse models, the recent groundbreaking RAMP pharmacologic advancements are primed to drive new translational explorations of RAMP-GPCR functions in human biology and disease.
Previous reviews have comprehensively described the pharmacology, physiology, and protein structure of RAMP-GPCR interactions [10-12]. However, considering the rapid progression of new discoveries related to RAMPs, the purpose of this review is to i) highlight recent evidence supporting a global RAMP-GPCR interactome, ii) illuminate the substantial physiological functions imparted by RAMP-GPCR interactions in animal models and humans, iii) speculate on the therapeutic implications of such interactions, and iv) provide future perspectives as to the trajectory of the multi-faceted study of RAMP-GPCR interactions.
RAMP-GPCR Coevolution: Emerging Evidence of a Global RAMP-GPCR Interactome
As of 2016, there were only 11 reported RAMP-GPCR interactions [12]. Further, 9 of these 11 known RAMP-GPCR pairings involved Class B GPCRs, with the remaining 2 RAMP-GPCR pairs involving Class A and Class C receptors [13-15]. Considering Class B GPCRs only account for approximately 5% of the non-odorant GPCRs, one can speculate that the breadth of RAMP-GPCR pairings is more extensive than previously thought.
In support of this hypothesis, Barbash et al. 2017 provided compelling evidence for widespread RAMP-GPCR pairings [16, 17]. They utilized genomic and transcriptomic data to perform coevolution (see Glossary) and co-expression analyses to interrogate global RAMP-GPCR interactions. A protein phylogenetic analysis, based upon the supposition that protein-protein interactions which convey a fitness advantage will coevolve, was employed by this study [18]. This approach is advantageous in predicting RAMP-GPCR interactions because it is unburdened by technical limitations such as heterologous overexpression systems or antibody specificity, problems that have plagued previous biochemical studies of RAMP-GPCR pairs [12].
From their analysis, Barbash et al. 2017 concluded that RAMPs and GPCRs showed substantial evidence of coevolution. Specifically, they found that RAMPs and GPCRs had orthologs present within the same species and had correlated phylogenetic trees [16]. Further, Barbash et al. hypothesized that RAMP1 and RAMP3 have redundant GPCR interactions due to similar N-terminal sequence homology and hydrophobicity [4, 16]. To this end, the authors report a positive correlation, in the same species, between the expression of specific GPCRs and RAMP1 and the same GPCRs and RAMP3. Additionally, they report an inverse relationship for RAMP2 compared to RAMP1 or RAMP3, suggesting that RAMP2 coevolved with a distinct group of GPCRs. To provide further support of global RAMP-GPCR interactions, the authors analyzed gene expression data from human tissues to test the hypothesis that interacting proteins will exhibit similar expression patterns. The authors found a positive correlation between RAMP and GPCR expression patterns which was significantly higher than expected by chance. The authors conclude that RAMPs and GPCRs showed substantial evidence of coevolution, and that RAMP1 and RAMP3 may have redundant functions, in contrast to independent roles of RAMP2. However, this protein coevolution analysis is limited by the inability to distinguish direct versus indirect protein interactions; coevolved proteins may be members of the same complex or broadly associated within the same signaling pathway [18].
To expand upon their bioinformatic findings, Barbash et al. 2019 developed an experimental approach to validate several predicted RAMP-GPCR pairings. Here, the authors overexpressed RAMP2 in HEK293T cells and employed a modified multiplexed error-correcting fluorescence in situ hybridization (MERFISH) assay and whole exome expression profiling to measure GPCR transcript levels [17, 19]. This work was based on the hypothesis that overexpression of RAMP2 would result in expression changes of putative-interacting-GPCRs [20-22]. The authors focused on RAMP2 as it is unplagued by the possible functional redundancy between RAMP1 and RAMP3. Use of these RAMPs may be complicated by cell compensatory mechanisms which would confound data analysis. Using the modified MERFISH assay, the authors found that 5 of the 14 GPCRs analyzed resulted in significant expression changes upon RAMP2 over-expression. Interestingly, all five responsive GPCRs were Class A GPCRs. A correlation was found between RAMP2 overexpression induced changes in GPCR gene expression and the extent of coevolution [16, 17]. Next, the authors used whole exome expression profiling to probe global GPCR expression changes in response to RAMP2 overexpression and concluded that there was a global downregulation of GPCR expression upon RAMP2 expression. These results support the 2017 phylogenetic analysis and overarching hypothesis of global RAMP-GPCR interactions.
Expanding the RAMP Repertoire
The early groundwork for RAMP-GPCR interactions focused on Class B GPCR receptors, which revealed that RAMPs interact with calcitonin receptor (CT) [23], calcitonin receptor-like receptor (CLR) [4], corticotropin-releasing factor receptors (CRF) [8], glucagon receptor [24, 25], parathyroid hormone receptors [25], secretin receptor [26], and pituitary adenylate cyclase activating peptide (PACAP) receptors [8, 25]. The characterization of these interactions uncovered diverse functions of RAMPs including alteration of ligand binding, specificity, and potency, best exemplified by CT [23] and CLR [4]. RAMPs were also shown to alter receptor trafficking and modulate G-protein coupling and secondary messengers, exemplified by VPAC1 [25]. Additional studies evaluated the interactions of RAMPs with the Class A receptor GPER/GPR30 (described in the ‘Physiological and Pathophysiological Roles of RAMPs’ section below) [27], and the Class C receptor, calcium-sensing receptor (CaSR) [13]. These seminal papers identifying RAMP-GPCR interactions relied heavily on over-expression of both a RAMP and a GPCR followed by detection of an increase in either RAMP or GPCR cell surface expression or their colocalization at the plasma membrane. Detection was primarily achieved using fluorescent-activated cell sorting (FACs), immunofluorescence microscopy, and ELISA assays [4, 7, 8, 13, 25, 26]. Later studies began to implement proximity-based approaches to detect direct RAMP-GPCR interactions, such as bioluminescence resonances energy transfer (BRET) assays [26, 27]. Commonly, these results were validated using a combination of radioligand binding, coimmunoprecipitation and western blot analysis, and interrogation of downstream G-protein signaling [4, 7, 8, 13, 23, 25-27].
While early studies were successful in interrogating specific RAMP-GPCR interactions, the coevolution studies described above prompt the need to systematically screen all GPCRs for putative interactions with RAMPS using biochemical, pharmacological, and physiological assays. Recently, two independent studies developed screening platforms to identify previously unrecognized RAMP-GPCR interactions. In the first study, Lorenzen et al. 2019 adapted a multiplexed suspension bead array (SBA) immunoassay to screen for putative interactions between RAMPs and Class B GPCRs, as well as a small cohort of non-Class B GPCRs [28]. Interactions were identified between at least one RAMP and members of the adhesion family of receptors, several orphan GPCRs (i.e. GPR182 and GPR4), members of the chemokine receptor family (part of Class A GPCRs), and Class B GPCRs. However, the functional and physiological consequences of these interactions were not explored.
A second study by Mackie et al. 2019 screened 24 chemokine receptors within the broader Class A family for possible interactions with RAMPs [29]. Leveraging a BRET screening methodology and FACs-based cell surface expression screening, the authors identified multiple putative chemokine receptor-RAMP interactions, including interactions with the sub-family of atypical chemokine receptors (ACKRs) (TABLE 1). Further, the authors chose to investigate the cellular consequences of one of the newly discovered RAMP-GPCR interactions between the atypical chemokine receptor ACKR3 and RAMP3. ACKR3 functions as a decoy receptor for the ligands CXCL12/SDF-1, CXCL11, and adrenomedullin [30-33]. The authors found that RAMP3 alters the decoy activity of ACKR3 through a Rab4-dependent recycling mechanism to promote plasma membrane re-sensitization of ACKR3. The exciting physiological consequences of this interaction is discussed in the following ‘Physiological and Pathophysiological Roles of RAMPs’ section.
Table 1:
Currently identified RAMP-interacting GPCRs, methods of interaction, identification, and FDA approved drugsa
| Official IUPHAR Receptor Name |
Class | RAMP | Cell Line | Identification Method |
FDA Approved Drugsb |
Clinical Indication |
Ref. |
|---|---|---|---|---|---|---|---|
| Chemerin receptor 1 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/Ac | --- | [29] |
| CCR1 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CCR2 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CCR3 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CCR4 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | mogamuli zumab | Cutaneous T-cell Lymphoma (Mycosis Fungoides, Sézary Syndrome) | [29] |
| plerixafor | non-Hodgkin lymphoma, multiple myeloma | ||||||
| CCR5 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | maraviroc | HIV/AIDS | [29] |
| ibalizuma b | HIV/AIDS | ||||||
| CCR6 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CCR7 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| CCR8 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CCR9 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CCR10 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| CXCR1 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CXCR2 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| CXCR3 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | [28] | ||||
| CXCR4 | A | RAMP1, RAMP3 | HEK293T, COS-7 | BRET assay | plerixafor | non-Hodgkin lymphoma, multiple myeloma | [29] |
| CXCR5 | A | RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| CXCR6 | A | RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| CX3CR1 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| XCR1 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| ACKR1 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| ACKR2 | A | RAMP1, RAMP3 | HEK293T, COS-7 | BRET assay, FACS | N/A | --- | [29] |
| ACKR3 | A | RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS, PLA, Confocal microscopy | plerixafor | non-Hodgkin lymphoma, multiple myeloma | [29] |
| RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | [28] | ||||
| ACKR4 | A | RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay | N/A | --- | [29] |
| ACKR5 | A | RAMP3 | COS-7 | FACS | N/A | --- | [29] |
| GPR4 | A | RAMP1, RAMP2, RAMP3 | HEK293T, HEK293 FreeStyle | SBA immunoassay, PLA | N/A | --- | [28] |
| GPR182 | A | RAMP1, RAMP2, RAMP3 | HEK293T, HEK293 FreeStyle | SBA immunoassay, PLA | N/A | --- | [28] |
| GPER/GPR30 | A | RAMP3 | HEK293T | BRET assay, Co-IP, Confocal microscopy | estradiol (estrogen receptor agonist; naturally occurring) | Oral contraceptives, treatment of menopausal and perimenopausal symptoms, and hypoestrogenism | [27] |
| CT receptor | B | RAMP1, RAMP2, RAMP3 | COS-7, CHO-P | Radioligand binding, Crosslinking analysis, Confocal microscopy | pramlintide | Type I & Type II Diabetes | [23] |
| RAMP1, RAMP2, RAMP3 | HEK293T, HEK293 FreeStyle | SBA immunoassay | [28] | ||||
| Calcitonin receptor like receptor | B | RAMP1, RAMP2, RAMP3 | Xenopus oocytes, HEK293T | Radioligand binding, FACS, Crosslinking analysis | eptinezumab, fremanez umab, glacanezumab, erenumab | Chronic Migraine | [4] |
| RAMP1, RAMP2, RAMP3 | HEK293T, COS-7 | BRET assay, FACS, PLA, Confocal microscopy | [25, 29] | ||||
| RAMP1, RAMP2, RAMP3 | HEK293T, HEK293 FreeStyle | SBA immunoassay, PLA | [28] | ||||
| CRF1 receptor | B | RAMP2 | HEK293S, CHO-K1 | ELISA | N/A | --- | [8] |
| RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | N/A | --- | [28] | ||
| CRF2 receptor | B | RAMP3 | HEK293 FreeStyle | SBA immunoassay | N/A | --- | [28] |
| GHRH receptor | B | RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | sermorelin | Growth hormone deficiency or growth failure, prevention of HIV-induced weight loss | [28] |
| GIP receptor | B | RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | N/A | --- | [28] |
| GLP-1 receptor | B | RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | exenatide, sorafenib, lixisenatide, mecaserm in rinfabate, dulaglutide, albiglutide, conivaptan, lenalidomide | Type II Diabetes | [28, 89] |
| GLP-2 receptor | B | RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | teduglutide | Short Bowel Syndrome | [28] |
| Glucagon receptor | B | RAMP2 | HEK293, COS-7 | Confocal microscopy, radioligand binding | glucagon recombinant, glucagon hydrochloride, oxyphenb utazone, chlordiaze poxide | Type II Diabetes | [7, 8, 25] |
| RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | N/A | --- | [28] | ||
| Secretin receptor | B | RAMP3 | COS-7, CHO-K1 | Bimolecular fluorescence complementation, BRET assay | secretin synthetic porcine, ezetimibe, pegfilgrastim | Treat High Blood Cholesterol, Lipid Abnormalities | [26] |
| RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | [28] | ||||
| PTH1 receptor | B | RAMP2 | HEK293, COS-7 | Confocal microscopy | teriparatide, abaloparatide | Osteoporosis | [25, 89] |
| RAMP1, RAMP2, RAMP3 | HEK293T, HEK293 FreeStyle | SBA Immunoassay, PLA | [28, 89] | ||||
| PTH2 receptor | B | RAMP3 | HEK293, COS-7 | Confocal microscopy | teriparatide, recombin ant parathyroid hormone | Osteoporosis | [25, 89] |
| RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA Immunoassay | [28, 89] | ||||
| PAC1 receptor | B | RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA Immunoassay | N/A | --- | [28] |
| VPAC1 receptor | B | RAMP1, RAMP2, RAMP3 | HEK293, COS-7 | Confocal microscopy | N/A | --- | [25] |
| RAMP2, RAMP3 | HEK293 FreeStyle | SBA Immunoassay | [28] | ||||
| VPAC2 receptor | B | RAMP1, RAMP2, RAMP3 | HEK293S, CHO-K1 | ELISA | N/A | --- | [8] |
| RAMP2, RAMP3 | HEK293 FreeStyle | SBA Immunoassay | [28] | ||||
| CaS receptor | C | RAMP1, RAMP3 | COS-7, HEK293 | Confocal microscopy, Co-IP | etelcalcetide | Secondary Hyperparathyroidism | [13] |
| ADGRF 5 | Adhesion Family | RAMP1, RAMP2, RAMP3 | HEK293 FreeStyle | SBA immunoassay | N/A | --- | [28] |
IUPHAR: The International Union of Basic and Clinical Pharmacology, RAMP: Receptor activity-modifying protein, BRET: Bioluminescence Resonance Energy Transfer, FACS: Fluorescenceactivated cell sorting, SBA: Suspension bead array, PLA: Proximity ligation assay, Co-IP: Coimmunoprecipitation.
To determine whether each RAMP-interacting GPCR listed in the table had an associated FDA approved drug, each GPCR was cross referenced against the public resource DrugBank [90], a database which combines drug data with drug target information, and a recent review profiling trends in GPCR drug discovery through 2017.
N/A acronym in the Drug column stands for None Approved and is meant to denote that for the given GPCR there are no currently approved therapies directed at this GPCR in DrugBank.
These studies highlight the breadth of putative RAMP-GPCR interactions and accentuate the current lack of studies aimed at validating or characterizing the functional significance of these interactions in vitro and in vivo. Screening platforms, like those described above, provide an invaluable tool to further interrogate RAMP-GPCR interactions on a large scale. Of note, both groups (Lorenzen and Mackie) utilized proximity ligation assay (PLA) to validate RAMP-GPCR interactions [28, 29]. This technique has traditionally been used over the last decade in the GPCR field to validate ex vivo GPCR heterodimers [34-36]. Future identification of novel RAMP-GPCR interactions can leverage existing PLA protocols and other non-traditional in vitro approaches for new exploratory and validation studies. As new RAMP-GPCR interactions are identified and biochemically validated, the implications of RAMP regulation of GPCR pharmacology and cellular functions will continue to be unraveled.
Physiological and Pathophysiological Roles of RAMPs
The aforementioned bioinformatic and biochemical screening approaches revealed exciting new RAMP-GPCR interactions. However, despite the diverse and widespread tissue expression of RAMPs, the field has been largely unsuccessful in linking specific RAMP-GPCR pairings to physiological and pathological phenotypes [37]. This is due in large part to a lack of rigorous in situ detection approaches. To date, much of our current understanding of the physiological role of RAMPs has been gleaned through global and conditional RAMP knockout mice. These studies consistently show that RAMPs play essential roles in the cardiovascular, lymphatic, immune, endocrine, and central and peripheral nervous systems [38-44]. Global genetic knockout of RAMP2 results in embryonic lethality marked by excessive fluid accumulation in the embryo [45-49], while global knockout of RAMP1 or RAMP3 leads to viable offspring with mild phenotypes[46]. Further, studies using haploinsufficient RAMP2 mice linked RAMP2 to the endocrine and skeletal systems [46]. In the following sections, we have chosen to highlight new discoveries made since 2018 that spotlight the influence of RAMP-GPCR interactions on physiology and pathology.
RAMP1
RAMP1 has been most intensely studied due to its role in the CGRP signaling axis. CGRP is a neuropeptide that signals through the CGRP receptor (RAMP1-CLR) [4]. The CGRP receptor is expressed on a variety of cell types [50, 51] (Figure 1A-B) and activation via CGRP results in potent vasodilation that has been clinically linked to migraine pathology [52] (Figure 1C). The successful therapeutic targeting of the CGRP receptor for the treatment of migraines is discussed in a later section titled ‘Therapeutic Targeting of RAMP-GPCRs’. The recent generation of sophisticated genetically engineered mouse models, such as the inducible neuronal overexpression of human RAMP1 (hRAMP1/Nestin-Cre) model, have proven invaluable to explore the physiological roles of RAMPs. Sabharwal et al. 2019 utilized the hRAMP1/Nestin mice to examine whether the known antihypertensive effects of CGRP were mediated primarily through neuronal or vascular CGRP receptors [53]. Cardiovascular phenotyping revealed that hRAMP1/Nestin mice display no baseline phenotypes, but in two hypertension models showed a reduction in the development of hypertension [53]. This paper highlighted an underappreciated neuronal role for the RAMP1-CLR signaling axis in mediating protection against hypertension.
Figure 1: Role of RAMP1-CLR in disease.
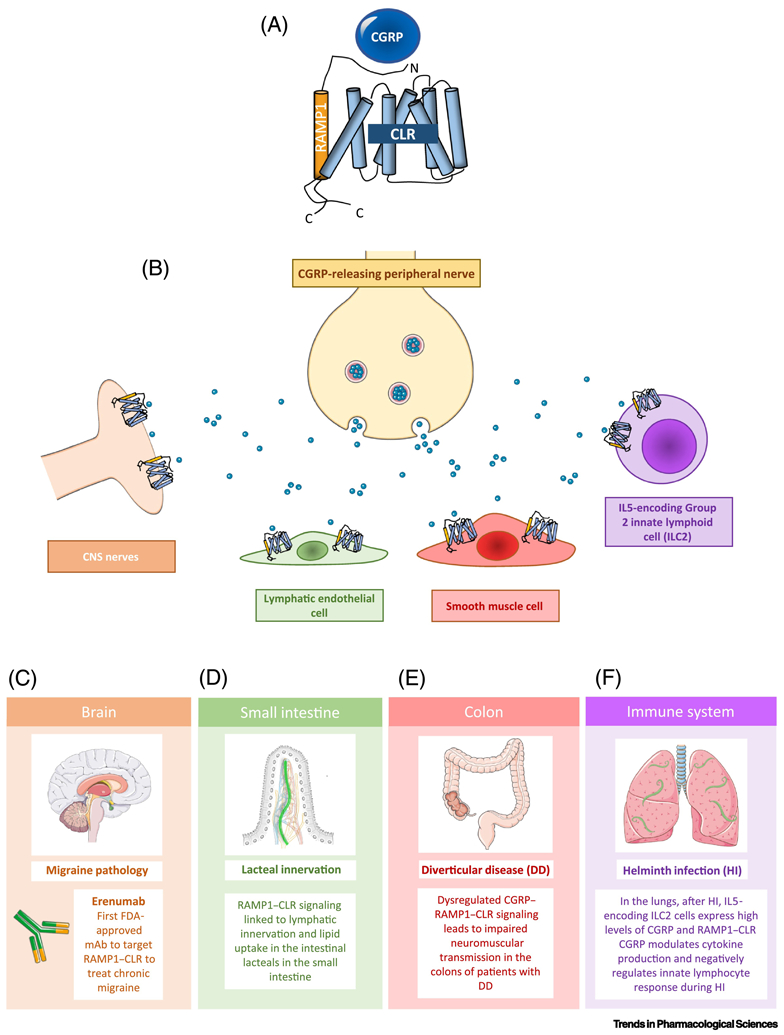
(A) Graphical representation of the CGRP receptor components CLR and RAMP1. (B) Schematic depicting release of CGRP from peripheral nerves and its potential targets including lymphatic endothelial cells, CNS nerves, and smooth muscle cells. (C) CGRP can also stimulate CNS nerves, and other cell types in the CNS, to incite migraines. Erenumab, the first FDA-approved monoclonal antibody against a GPCR, targets the RAMP1-CLR receptor to help treat migraine. (D) CGRP can bind to its receptor on lymphatic endothelial cells, which is linked to lacteal innervation and lipid uptake in the small intestine. (E) CGRP can also bind to receptors on smooth muscle cells, which can have physiological effects in the colon. Human studies have shown dysregulation of CGRP signaling in patients with diverticulitis disease (DD), highlighting the importance of this signaling axis in human disease. (F) CGRP is specifically upregulated in a distinct population of Type 2 Innate Lymphoid Cells found in the lungs in response to specific cytokine cues released by lung-infiltrated N. brasiliensis worms during helminth infection and constrains the magnitude of the innate immune response.
Recently, the role of the RAMP1-CLR signaling axis in peripheral neurons has been expanded. An established link between CGRP and the digestive system indicates that CGRP-responsive neurons innervate multiple intestinal cells, including the epithelia, vasculature, smooth muscle, and enteric nerves [54]. Expanding upon these findings, Davis et al. 2019 linked RAMP1-CLR signaling to lymphatic innervation and lipid uptake in the intestinal lacteals [55]. This was accomplished using two different mouse models, 1) an inducible lymphatic-specific deletion of Calcrl (Calcrlfl/fl/Prox1-CreERT2), and 2) global deletion of RAMP1 (RAMP1−/−). The authors found that mice with lymphatic deletion of CLR, subjected to a high fat diet, displayed defective lacteal lipid transport, suppressed fat accumulation, and reduction and mis-patterning of the enteric nerve cage surrounding the lacteals. Similar results were found in the RAMP1 −/− mice, which presented with dilated lymphatics and similar enteric nerve mis-patterning and density reduction (Figure 1D).
The dysregulation of the CGRP-RAMP1-CLR signaling axis in peripheral neurons and the gastrointestinal system has recently been linked to human disease. Pauza et al. 2019 associated dysregulation of CGRP-RAMP1-CLR signaling with diverticular disease (DD) pathology. DD, a common large bowel disease, is characterized by impaired colonic motility accompanied with expansive remodeling of the enteric nervous system. An imbalance in neuromuscular transmission has been recognized as a major contributing factor in the development of DD [56-58]. Considering the link between CGRP innervation and colonic motility, the authors investigated changes in CGRP and CLR-RAMP1 expression on enteric ganglia from colon biopsies from healthy individuals, asymptomatic and symptomatic DD patients. The authors hypothesized that changes in CGRP signaling within discrete colon structures would result in motility impairments in DD patients. Interestingly, DD samples showed a decrease in CGRP peptide expression with an associated increase in CLR expression [58]. This study demonstrated that pathologic alterations to CGRP signaling leads to neuro-muscular signaling imbalances in DD colons, and that targeting this signaling axis may prove to be an effective treatment strategy for DD (Figure 1E).
Neuropeptides, such as Neuromedin U (NMU) and CGRP, have garnered much attention in the immunology field through the discovery that neuro-immune crosstalk can influence allergic inflammation and shape innate lymphocyte responses during infection [59, 60]. Both NMU and CGRP are specifically upregulated in distinct populations of Type 2 innate lymphoid cells (ILC2) during an innate immune response and can modulate the inflammatory response of these cells during allergy or helminth infection [60]. Similar to NMU, Nagashima et al. 2019 found that CGRP and the CGRP receptor (RAMP1-CLR) were upregulated in discrete ILC2 populations in response to helminth infection (Figure 1F) [59]. Specifically, CGRP was found to modulate the cytokine production of these cells and constrain the magnitude of the innate immune response. The authors showed that inhibition of CGRP signaling, through the use of RAMP1 deficient mice and cells, resulted in improved ILC2 responses to helminth infection [59]. This suggests that the CGRP-RAMP1-CLR signaling axis is a viable pharmacological target during helminth infection.
RAMP2
A groundbreaking paper by Mackie et al. 2018 identified the first instance of a disease-causing mutation that disrupts a RAMP-GPCR interaction. Specifically, they identified a recessive mutation in human Calcr, which impairs the interaction between CLR and RAMP2 and is required for adrenomedullin signaling. [61]. The mutation was identified in a consanguineous family who presented with a high incidence of fetal demise due to excessive fluid accumulation associated with lymphatic insufficiency, a condition known as nonimmune hydrops fetalis (NIHF). This mutation is an in-frame deletion of the highly conserved valine 205 (V205del) within the first extracellular loop of CLR (Figure 2A). Homozygous V205del resulted in NIHF, while the heterozygous carrier exhibits female subfertility (Figure 2B). In vitro, CLR V205del showed defective RAMP2-CLR receptor oligomerization, plasma membrane localization of RAMP2-CLR, and cAMP signaling. A prior role for the AM-CLR-RAMP2 signaling axis in the development of NIHF was established utilizing murine global knockout models of AM, RAMP2 and CLR [45-49]. To further define the role of this signaling axis in lymphatics, Mackie et al. generated two independent murine models of lymphatic Calcrl loss: a constitutive Calcrlfl/fl;Lyve1-Cre and inducible Calcrlfl/fl;Prox1-CreERT2. These models recapitulated clinical phenotypes: lymphatic growth arrest, hypoplastic jugular lymph sacs, and dilated dermal lymphatics. Further, loss of RAMP2, modeled by rescue of the global RAMP2 knockout mouse with an endothelial expressed RAMP2 transgene (RAMP2−/−Tg), was shown to recapitulate the lymphatic and NIHF phenotypes of the Calcrl knockout mouse models. This study successfully translated clinical identification of a Calcrl mutation to a structurally and biochemically validated AM-CLR-RAMP2 signaling axis in lymphatic development.
Figure 2. Recently identified human mutations in the AM-RAMP2-CLR signaling axis.
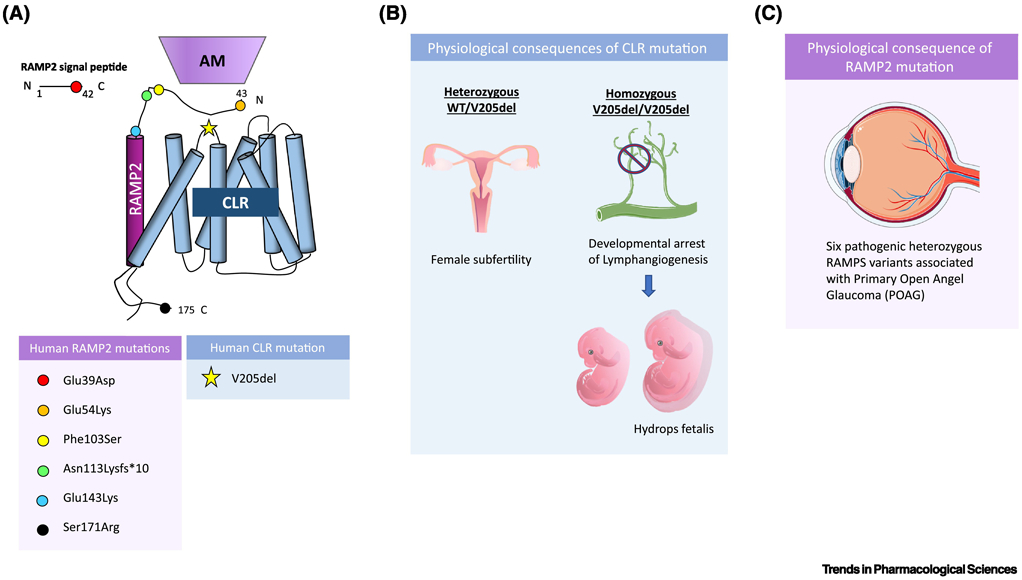
(A) As of 2019, in humans, a single pathologic mutation was identified in CLR, specifically on extracellular loop 1 of the receptor, and 6 heterozygous pathologic RAMP2 variants were identified, with mutations on the cleaved signaling peptide of RAMP2, and both the extracellular and cytosolic portion of the membrane-localized protein, with no mutations identified in the transmembrane domain. These mutations are spatially displayed on a schematic of CLR bound to RAMP2 and adrenomedullin, with each colored circle corresponding to one of the six identified RAMP2 variants and the star corresponding to the identified CLR mutation. (B) In humans, homozygous deletion of Valine 205 in CLR results in developmental arrest of lymphangiogenesis associated with lethal fluid accumulation, known as non-immune hydrops fetalis, while heterozygous carriers display female subfertility. Lymphatic deletion of CLR in mice results in similar phenotypes as seen in humans. (C) 6 pathologic RAMP2 variants were identified in an exome sequencing study of patients with primary open angle glaucoma (POAG) and each of the 6 variants were linked to functional impairments in AM-RAMP2-CLR signaling and deleterious effects on retinal ganglion nerve health.
In 2019, Gong et al. identified novel mutations within RAMP2 linked to primary open-angle glaucoma (POAG) [62]. POAG is one of the most common types of glaucoma and irreversible blindness. Taking advantage of the rapidly advancing repertoire of genome profiling tools, Gong et al. sought to identify novel POAG-causing genes and variants through exome sequencing analysis of 398 patients with POAG and 2010 control individuals. From their analysis, they identified 6 heterozygous pathologic variants in Ramp2 (Figure 2A). The authors probed the functional effects of these variants in vitro and found that mutant RAMP2 formed intracellular aggregates, indicative of protein trafficking defects. Further, they found decreased cAMP expression, a known output of functional adrenomedullin-RAMP2-CLR-cAMP signaling. Gong et al. utilized a heterozygous RAMP2 knockout mouse to analyze the effects of RAMP2 haploinsufficiency in the eye. These mice displayed retinal ganglion death and reduced sensitivity of the retina to produce cAMP in response to adrenomedullin stimulation [62]. This study identified and linked Ramp2 mutations to impaired AM-RAMP2-CLR cAMP signaling in retinal ganglion cells, thereby laying the groundwork for development of RAMP2 targeting strategies for the treatment of POAG (Figure 2C).
RAMP3
Recent years have witnessed significant advances in defining the role of RAMP3 in physiology and pathology. RAMP3 is unique amongst the RAMPs and differs from RAMP1 and RAMP2 in several ways. First, RAMP3 is capable of trafficking to the plasma membrane even in the absence of over-expression of a cognate GPCR [25, 29, 63]. Secondly, unlike RAMP1 and RAMP2, the N-terminus of RAMP3 contains four N-glycosylation sites and its intracellular C-terminus harbors a PDZ binding motif which has been shown to regulate receptor recycling [63-65]. Finally, as described below, the repertoire of RAMP3-assocaited GPCRs appears to be quite broad and impart evident physiological functions.
For example, one of the first studies to describe a RAMP interaction beyond the Class B receptors was performed by Barrick et al. 2012. Here, the authors describe an interaction between RAMP3 and the Class A GPCR GPER/GPR30, whose ligand is estradiol (Figure 3A) [27]. The effect of RAMP3 on GPER/GPR30 signaling was investigated in vivo in the context of heart disease by crossing RAMP3 knockout mice onto a cardiac disease-prone genetic background (RenTgMK;RAMP3−/−). These mice were treated with a GPER/GPR30 agonist, resulting in a significant reduction in cardiac hypertrophy and perivascular fibrosis that was both RAMP3- and sex-dependent (Figure 3B).
Figure 3: Pathophysiological roles of RAMP3.
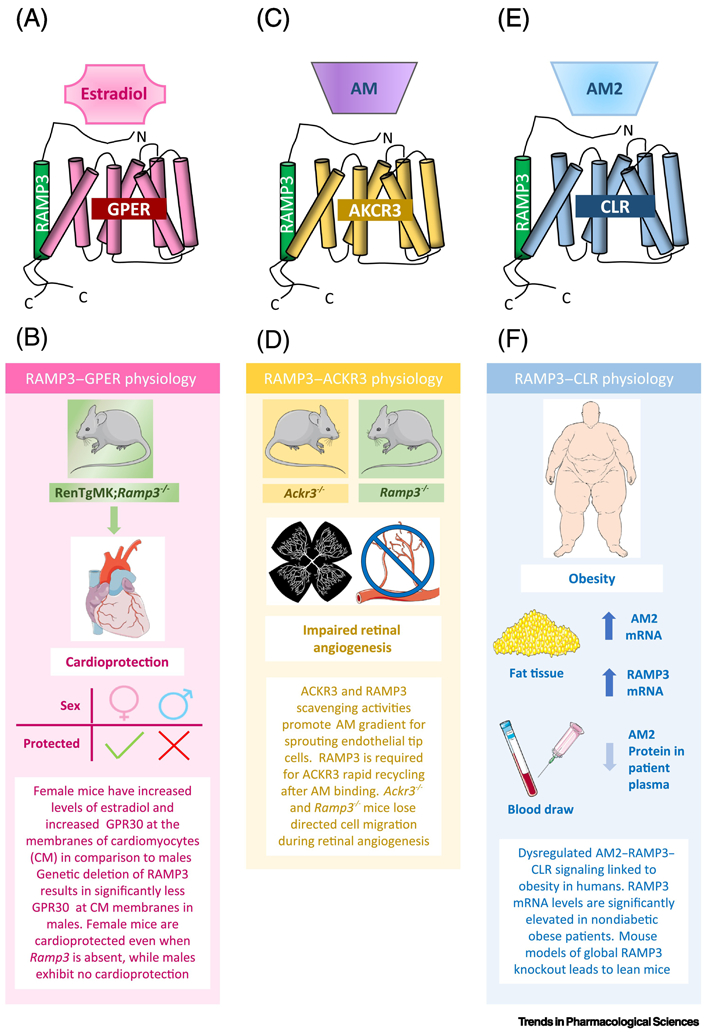
(A) Depiction of the receptor complex RAMP3-GPER/GPR30 and its ligand estradiol. (B) Murine studies looked at the link between RAMP3-GPER/GPR30 and heart disease by crossing RAMP3 knockout mice onto a heart disease-prone genetic background. This in vivo activation of GPER/GPR30 resulted in significant reduction in heart disease parameters that was both RAMP3 and sex dependent. (C) The RAMP3-ACKR3 receptor is a decoy-receptor for the ligand adrenomedullin (AM), in that it binds AM, but does not result in G-protein signaling. (D) Recently, RAMP3 was shown to alter the decoy activity of ACKR3 through a recycling mechanism, which promoted plasma membrane re-sensitization of ACKR3. This decoy activity was shown to be important during guided cell migration in murine retinal angiogenesis. (D) RAMP3 can also interact with CLR to form a receptor for adrenomedullin 2 (AM2). (E) This signaling axis was investigated in obese and non-obese human patients, where it was found that RAMP3 mRNA levels were increased in obese patients, which correlates with RAMP3 knockout mice phenotypes. These studies highlight the importance of continuing to study RAMP3 in human disease, particularly metabolic disorders.
Recently, Mackie et al. 2019 further demonstrated the consequences of deleting RAMP3 during development [29]. Using a BRET screening assay, RAMP3 was found to interact with the atypical chemokine receptor, ACKR3, which is a decoy receptor for the chemokine ligands CXCL12/SDF1 and CXCL11 as well as adrenomedullin (Figure 3C). The authors showed in vitro that RAMP3 was required for ACKR3 rapid endosome recycling and its subsequent re-sensitization at the plasma membrane. In addition, the developmental consequences of this interaction were shown using knockout mouse models of the AM-ACKR3-RAMP3 decoy signaling axis (ACKR3−/− and RAMP3−/−). In the absence of RAMP3, the chemotactic gradients established by ACKR3 decoy functions fail to be established, thereby disrupting normal guided cell migration during angiogenesis in murine retinal development (Figure 3D).
The initial generation of RAMP3 knockout mice was reported by Dackor et al. 2007, where Ramp3−/− mice were surprisingly viable and overtly phenotypically normal [46]. However, aged mice displayed decreased weight, suggesting an age-dependent induction of RAMP3 in normal metabolism. A dozen years later, two studies evaluated the levels of adrenomedullin 2/intermedin (AM2), a ligand for RAMP3-CLR, in the context of obesity (Figure 3E). The authors found an inverse relationship between AM2/intermedin plasma levels and the degree of adiposity in both obese mice and humans [66, 67]. Expanding on these findings, an additional paper probed the AM2-RAMP3-CLR signaling axis by examining the expression of AM2/intermedin, RAMP3, and CLR in obese patients with or without type 2 diabetes [68]. In this work, the authors profiled the mRNA expression of Adm2, Calcrl, Ramp1, Ramp2, and Ramp3 in adipose tissue. They found a significant increase in Adm2 mRNA expression in adipose tissue from non-diabetic and diabetic obese patients compared to controls, in contrast to the decreased plasma concentration of AM2/intermedin previously found in obese individuals. Interestingly, significantly higher levels of Ramp1 and Ramp3 mRNA were noted in the non-diabetic obese samples in comparison to both the controls and diabetic obese samples. In vitro experiments using human preadipocytes exposed to a fat microenvironment showed an increase in AM2 mRNA expression but a decrease in AM2/intermedin secretion, recapitulating clinical findings. RAMP3 was not profiled in these in vitro experiment. Taken together, these studies provide a link between AM2-RAMP3-CLR perturbations and obesity and specifically identify significant downregulation of Ramp3 mRNA in non-diabetic obesity (Figure 3F).
Another recent clinical study examined RAMP3 gene polymorphisms in women. The authors looked at bone density and body mass composition and found that amongst women with RAMP3 polymorphisms, fat mass tended to be higher only in the elderly women in this group[69]. This suggests that variations in RAMP3 expression may contribute to age related changes in body composition.
Therapeutic Targeting of RAMP-GPCRs
Considering the rapidly-expanding cohort of GPCRs that functionally interact with RAMPs and the breadth of physiological systems affected by RAMP signaling axes, it stands to reason that the protein-protein interface of a RAMP-GPCR interaction could be exploited for therapeutic benefit. Indeed, the therapeutic tractability of targeting RAMP-GPCR pairs has been recently established through the generation of monoclonal antibodies targeting CGRP or its receptor (RAMP1-CLR) for the treatment of migraines. CGRP has been implicated in migraine pathology due to its location in peripheral and central neurons, and its role as a potent vasodilator and nociception transmitter [70, 71]. Although the pathophysiology of migraines is multifactorial, it is generally thought that the activation and sensitization of the trigeminal system, which regulates blood flow and pain transmission in the head, contributes heavily to migraine symptomology [72, 73]. CGRP is the most abundant neuropeptide released in the trigeminal nerve and due to its short plasma half-life, likely exerts its effects near its release site at the vessel wall [73]. The CGRP receptor (RAMP1-CLR) is expressed throughout the trigeminal system, including in neurons and endothelial cells [73-75]. Migraines are thought to be the result of neurogenic inflammation which triggers the release of CGRP, leading to vasodilation of the peripheral and central nervous system and subsequent pathologic activation of the trigeminal system [75]. In support of this model of migraine pathophysiology, blood levels of CGRP are elevated during active migraine episodes and are further elevated in patients who experience chronic migraine in comparison to patients with episodic migraine [52, 76-78]. Consequently, recent therapies aimed at blocking CGRP activity, either using the small molecule inhibitors classified as “-gepants” [79] or using monoclonal antibodies against CGRP, have experienced great success and received FDA approval by effectively reducing the number of migraine days experienced by chronic migraine sufferers [80].
The essential role of RAMP1 in migraine pathology has been demonstrated in vivo using pre-clinical RAMP1 overexpression transgenic animals. While there are many methods used to simulate migraines in rodents, including adding an inflammatory stimulus directly onto the dura mater of the meninges, there are few genetic models which mimic the clinical symptoms associated with migraines [81]. One such genetic mouse model was designed to overexpresses human RAMP1 in glia and dura under the inducible Nestin-Cre promoter (hRAMP1/Nestin mice), thereby increasing the availability of CGRP receptor to bind CGRP and induce migraine [82]. Intracerebroventricular injection of CGRP into hRAMP1/Nestin mice resulted in various migraine phenotypes including photophobia and decreased motor activity in the dark [83]. Collectively, these pre-clinical animal studies solidified the importance of RAMP1 in migraine pain.
The groundbreaking therapy Erenumab, used as a treatment for migraines, is the first FDA-approved monoclonal antibody targeting a GPCR (Figure 1C). Specifically, the epitope was strategically designed to target the interface of the CGRP receptor RAMP1-CLR rather than RAMP2- or RAMP3-CLR, which form adrenomedullin receptors [84]. Thus, Erenumab is highly selective for the CGRP receptor, with no measurable activity for other CGRP family receptors [85]. Targeting of the RAMP1-CLR interface is supported by the crystal structure of RAMP1-CLR ectodomains bound with a CGRP antagonist (olcegepant or telcagepant) at a peptide-binding cleft within the RAMP1-CLR interface, which disrupts the RAMP1-CLR interaction to impair CGRP signaling [86]. In fact, the first structure of a full-length RAMP-GPCR in complex with its Gs-protein and ligand was CGRP in complex with RAMP1-CLR (BOX 2). These structures support that RAMP-mediated ligand specificity can be attributed to stabilization of receptor complexes [87]. Future studies analyzing the structure of RAMP1-CLR bound to Erenumab would both clarify and underscore the importance of RAMPs in the stabilization of activated GPCR conformations and provide a structural framework for the development of new biologics exploiting tractable RAMP-GPCR interfaces.
BOX 2: New Insights into RAMP-GPCR structure.
Over the past decade, insights into how RAMPs impart pharmacological diversity to GPCR function and ability to bind cognate ligands has been elucidated through structural analyses. However, until recently, these conclusions have been limited due to partial structures consisting of GPCRs and RAMP ECD domains. While valuable in that they allowed insight into how RAMPs and GPCR interact, these structural advancements were not equipped to depict how RAMPs impart peptide selectively. For a thorough review of class B GPCR structure along with commentary on structures of RAMP1-CLR and RAMP2 ECD domains, we recommend Hay and Pioszak 2016 [12].
Recently, Liang et al. 2018 reported a full cryo-EM structure depicting active, G-protein-coupled complex; a RAMP-GPCR pair bound to endogenous ligand and in complex with heterotrimeric G-proteins [87]. Specifically, using Volta phase plate cryo-electron microscopy, they obtained a 3.3 A structure of human CGRP receptor (RAMP1-CLR) bound to its ligand CGRP and the Gs-protein heterotrimer. Regarding the RAMP1-CLR interface, the authors found that 23% of the surface of RAMP1 is buried within the interface of CLR. Structural, mutagenesis, and molecular dynamic stimulations highlight the key function of RAMP1 is in the stabilization of the CLR extracellular loop 2. Regarding the RAMP1-CGRP interface, the authors found that while the CGRP peptide interacts extensively with the RAMP1-CLR complex with 61.5% of CGRP’s surface buried, there are limited direct interactions between RAMP1 and CGRP peptide. These interactions are between the C-terminus of the CGRP peptide and RAMP1 residues F83R-P85R [93]. Further, mutagenesis and modeling experiments accentuated the importance of the ECL2 conformation for CGRP activation of its receptor [9]. Taken together, the authors conclude that RAMP1 likely acts as an allosteric regulator of CLR and functions to stabilize the ECD and ECL2 of CLR to make CLR conformationally amendable for GCRP binding and presentation to the CLR core.
The clinical success of Erenumab to target novel RAMP-GPCR interfaces to treat disease illustrates a massive therapeutic and commercial opportunity to target other RAMP-GPCRs dysregulated in disease. GPCRs are the most tractable and druggable class of proteins and represent the largest family of targets for approved drugs, which have classically been targeted using small molecules [1-3]. Yet in 2018, 5 of the top 10 best-selling prescription drugs were monoclonal antibodies, not small molecules [88]. This demonstrates the current disconnect between historical GPCRs targeting strategies, and the growing shift in the pharmaceutical market towards biologic therapies. Recent reports exploring the global diversity of RAMP-GPCR interactions coupled with the established role that RAMPs dynamically regulate GPCRs necessitates that drug discovery efforts must take into consideration RAMP-GPCR interactions. TABLE 1 includes FDA approved drugs that target RAMP-interacting GPCRs. It is striking to note that 66% of RAMP-interacting GPCRs listed in TABLE 1 have no approved FDA drug. It will be interesting to see how new RAMP-GPCR pairings will guide future drug development efforts.
Concluding Remarks and Future Perspectives
The last several years has marked an exciting time for the RAMP-GPCR field, where RAMP-GPCR coevolution studies hypothesized a global RAMP-GPCR interactome. Importantly, in 2019, we witnessed the experimental validation of these bioinformatic studies with the identification of previously unrecognized RAMP-GPCR interactions using two screening platforms (BRET and SBA). These studies effectively expanded the list of RAMP-interacting GPCRs from 11 to 44 receptors that span Class A, B, C and Adhesion family of GPCRs (TABLE 1). The newly expanded repertoire of RAMP-GPCR interactions alludes to a much more pronounced regulation of the entire GPCR family by RAMPs. In the coming years, it will be exciting to see how others expand upon these findings to further validate the global RAMP-GPCR interactome hypothesis. Such findings are likely to have broad physiological and pathological consequences considering the already established role of RAMPs in human health. This is most apparent with the recent success of Erenumab, a new anti-CGRP therapy, that was designed to specifically target the RAMP1-CLR interface, for the treatment of migraine. Further explorations into RAMP-GPCR signaling in human pathological conditions may help reveal new disease mechanisms and pharmacologically tractable targets for a variety of diseases (see Outstanding Questions). Moving forward, it has become both apparent and essential to consider RAMP-GPCR pairings for the development of more efficacious, selective, and context dependent therapies.
Outstanding Questions:
Will the global RAMP-GPCR interactome hypothesis be validated by biochemical assays?
Do RAMPs interact with discrete GPCRs or is there broad redundancy in their respective interactomes?
In cells that express multiple RAMPs and RAMP-interacting GPCRs, how are cell surface presentation and signaling selectively determined, coordinated, and regulated?
Are there are other proteins associated with RAMP-GPCR pairs?
Are RAMP1 and RAMP3 redundant or will future research reveal context dependent signaling biases?
Are validated RAMP-GPCR interactions cellular, tissue, and context-specific?
Disease causing mutations in RAMP2 have set a precedence. Are there yet unidentified mutations or polymorphisms in RAMPs with physiological consequences?
How will insights gained from RAMP-GPCR high resolution atomic structures influence future drug discovery efforts?
Will RAMP-GPCR interactions be more tractable via biologies or small molecules? Is there a role for allosterism?
Is the failure to account for RAMP-GPCR interactions during target identification and drug development influencing the rampant attrition rate of GPCR-targeted pharmaceutical therapies?
Will context-dependent identification of RAMP-GPCR interactions allow for development more of targeted therapies?
Highlights:
RAMPs interact with GPCRs to regulate receptor function.
RAMP-GPCR coevolution studies suggest that RAMPs globally interact with GPCRs.
In the past year, two studies have expanded the RAMP-GPCR interactome using both SBA and BRET methodologies.
Human mutations in RAMP-GPCR pairings have been identified and linked to human disease, including a mutation in CLR, associated with hydrops fetalis, and mutations in RAMP2, associated with glaucoma.
For the first time, the FDA has approved a GPCR-directed antibody against RAMP1-CLR, which reduces the number of migraine days patients experience.
The expanded RAMP-GPCR interactome and pivotal success of the first targeted therapy of RAMP-GPCR pairs has ushered in a new RAMPage.
GLOSSARY
- ACKR3:
The atypical chemokine receptor 3, also known as C-X-C chemokine receptor type 7 (CXCR7). It is a decoy receptor that binds the chemokines CXCL11 and CXCL12 to induce β-arrestin recruitment and ligand internalization. ACKR3 is also a decoy receptor for adrenomedullin (AM) to cause β-arrestin recruitment.
- Adrenomedullin (AM):
It is a 52-amino acid peptide and member of the calcitonin/calcitonin gene-related peptide (CGRP) family. It is a potent vasodilator that plays a regulatory role in the cardiovascular and lymphatic systems through activation of CLR paired with RAMP2 or RAMP3.
- AM2:
A member of the calcitonin/calcitonin gene-related peptide (CGRP) family, also known as intermedin, that is expressed in both the peripheral and central nervous systems.
- BRET:
Bioluminescence resonances energy transfer is a method to measure protein-protein interactions. It is based upon the resonance energy transfer from a bioluminescent-taggeddonor protein to a fluorescently-tagged-acceptor protein.
- CaSR:
The calcium-sensing receptor is a class C GPCR that regulates calcium homeostasis through the binding of extracellular calcium. It is expressed primarily in the parathyroid and kidney.
- CGRP:
It is a 37 amino acid neuropeptide and member of the calcitonin/calcitonin gene-related peptide (CGRP) family. It is a potent vasodilator that functions though binding to its cognate CGRP receptor (RAMP1-CLR). It is expressed in both the peripheral and central nervous system.
- CLR/Calcrl-
Calcitonin receptor-like receptor is a member of the class B GPCRs.
- Coevolution analysis:
A method to identify putative protein-protein interactions that is based upon the supposition that two proteins that interact will coevolve due to evolutionary pressure that preserves the interaction. The analysis looks for co-acquisition of mutations in interacting proteins and correlations in protein phylogenetic trees.
- Erenumab:
A CGRP-receptor antagonist approved by the FDA in 2018 for the treatment of migraines. It is a monoclonal antibody selective for the CGRP receptor.
- GPER/GPR30:
Originally known as the orphan G-protein-coupled receptor 30 (GPR30) and later re-named the G-protein-coupled estrogen receptor 1 (GPER) when it was found to bind Estradiol to cause cyclic AMP, inositol trisphosphate (IP3) and calcium signaling.
- HEK293T:
Transformed Human embryonic kidney cell line that have very low endogenous RAMP expression. They are commonly used to investigate RAMP-GPCR interactions due to their high transfection efficiency.
- NIHF:
Nonimmune hydrops fetalis is a condition characterized by fluid accumulation in the extravascular and body cavity of a fetus.
- PDZ motif:
Regarding RAMP3, it is a four amino acid sequence (DTLL) on the RAMP3 C-terminus that promotes interactions with RAMP3 and other PDZ proteins, such as NHERF1. These interactions can affect receptor internalization and trafficking of RAMP3-GPCR pairs.
- SBA:
A suspension bead array (SBA) immunoassay, which consists of magnetic, bar-coded beads conjugated to specific antibodies, to capture and detect target protein epitopes from complex cell lysates.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Garland SL (2013) Are GPCRs still a source of new targets? Journal of biomolecular screening 18 (9), 947–66. [DOI] [PubMed] [Google Scholar]
- 2.Sriram K and Insel PA (2018) G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Molecular pharmacology 93 (4), 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatakrishnan AJ et al. (2013) Molecular signatures of G-protein-coupled receptors. Nature 494 (7436), 185–194. [DOI] [PubMed] [Google Scholar]
- 4.McLatchie LM et al. (1998) RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393 (6683), 333–339. [DOI] [PubMed] [Google Scholar]
- 5.Christopoulos A et al. (2003) Novel receptor partners and function of receptor activity-modifying proteins. Journal of Biological Chemistry 278 (5), 3293–3297. [DOI] [PubMed] [Google Scholar]
- 6.Parameswaran N and Spielman WS (2006) RAMPs: the past, present and future. Trends in Biochemical Sciences 31 (11), 631–638. [DOI] [PubMed] [Google Scholar]
- 7.Weston C et al. (2015) Modulation of Glucagon Receptor Pharmacology by Receptor Activity-modifying Protein-2 (RAMP2). Journal of Biological Chemistry 290 (38), 23009–23022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wootten D et al. (2013) Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. British journal of pharmacology 168 (4), 822–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woolley MJ et al. (2013) The role of ECL2 in CGRP receptor activation: a combined modelling and experimental approach. Journal of the Royal Society, Interface 10 (88), 20130589–20130589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay DL (2019) CGRP Receptor Biology: Is There More Than One Receptor? Handb Exp Pharmacol 255, 13–22. [DOI] [PubMed] [Google Scholar]
- 11.Hay DL et al. (2018) Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Br J Pharmacol 175 (1), 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hay DL and Pioszak AA (2016) Receptor Activity-Modifying Proteins (RAMPs): New Insights and Roles. Annu Rev Pharmacol Toxicol 56, 469–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouschet T et al. (2005) Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci 118 (Pt 20), 4709–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai AJ et al. (2014) Role of Receptor Activity Modifying Protein 1 in Function of the Calcium Sensing Receptor in the Human TT Thyroid Carcinoma Cell Line. In PLoS One. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenhart PM et al. (2013) G-protein Coupled Receptor 30 Interacts with Receptor Activity Modifying Protein 3 and Confers Sex-Dependent Cardioprotection. J Mol Endocrinol 51 (1), 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbash S et al. (2017) GPCRs globally coevolved with receptor activity-modifying proteins, RAMPs. Proceedings of the National Academy of Sciences of the United States of America 114 (45), 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbash S et al. (2019) Detection of Concordance between Transcriptional Levels of GPCRs and Receptor-Activity-Modifying Proteins. iScience 11, 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pazos F et al. (2005) Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J Mol Biol 352 (4), 1002–15. [DOI] [PubMed] [Google Scholar]
- 19.Chen KH et al. (2015) Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348 (6233), aaa6090–aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black JB et al. , Feedback regulation of G protein-coupled receptor signaling by GRKs and arrestins, Academic Press, 2016, pp. 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefkowitz RJ and Shenoy SK, Transduction of receptor signals by β-arrestins, 2005, pp. 512–517. [DOI] [PubMed] [Google Scholar]
- 22.Tsao P et al. (2001) Role of endocytosis in mediating downregulation of G-protein-coupled receptors. Trends in pharmacological sciences 22 (2), 91–6. [DOI] [PubMed] [Google Scholar]
- 23.Christopoulos G et al. (1999) Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol 56 (1), 235–42. [DOI] [PubMed] [Google Scholar]
- 24.Weston C et al. (2015) Modulation of Glucagon Receptor Pharmacology by Receptor Activity-modifying Protein-2 (RAMP2). J Biol Chem 290 (38), 23009–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christopoulos A et al. (2003) Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem 278 (5), 3293–7. [DOI] [PubMed] [Google Scholar]
- 26.Harikumar KG et al. (2009) Molecular Basis of Association of Receptor Activity-Modifying Protein 3 with the Family B G Protein-Coupled Secretin Receptor. Biochemistry 48 (49), 11773–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenhart PM et al. (2013) G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. Journal of Molecular Endocrinology 51 (1), 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenzen E et al. (2019) Multiplexed analysis of the secretin-like GPCR-RAMP interactome. Sci Adv 5 (9), eaaw2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackie D.l. et al. (2019) RAMP3 determines rapid recycling of atypical chemokine receptor-3 for guided angiogenesis. Proceedings of the National Academy of Sciences, 201905561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boldajipour B et al. (2008) Control of chemokine-guided cell migration by ligand sequestration. Cell 132 (3), 463–73. [DOI] [PubMed] [Google Scholar]
- 31.Burns JM et al. (2006) A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med 203 (9), 2201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein KR et al. (2014) Decoy Receptor CXCR7 Modulates Adrenomedullin-Mediated Cardiac and Lymphatic Vascular Development. Dev Cell 30 (5), 528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naumann U et al. (2010) CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One 5 (2), e9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonaventura J et al. (2014) L-DOPA-treatment in primates disrupts the expression of A(2A) adenosine-CB(1) cannabinoid-D(2) dopamine receptor heteromers in the caudate nucleus. Neuropharmacology 79, 90–100. [DOI] [PubMed] [Google Scholar]
- 35.Borroto-Escuela DO et al. (2013) G protein-coupled receptor heterodimerization in the brain. Methods Enzymol 521, 281–94. [DOI] [PubMed] [Google Scholar]
- 36.Trifilieff P et al. (2011) Detection of antigen interactions ex vivo by proximity ligation assay: endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. Biotechniques 51 (2), 111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlen M et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347 (6220), 1260419. [DOI] [PubMed] [Google Scholar]
- 38.Barrick CJ et al. (2012) Loss of receptor activity-modifying protein 3 exacerbates cardiac hypertrophy and transition to heart failure in a sex-dependent manner. Journal of Molecular and Cellular Cardiology 52 (1), 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadmiel M et al. (2011) Research resource: Haploinsufficiency of receptor activity-modifying protein-2 (Ramp2) causes reduced fertility, hyperprolactinemia, skeletal abnormalities, and endocrine dysfunction in mice. Molecular Endocrinology 25 (7), 1244–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadmiel M et al. (2017) Loss of receptor activity-modifying protein 2 in mice causes placental dysfunction and alters PTH1R regulation. PLoS ONE 12 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurashige C et al. (2014) Roles of receptor activity-modifying protein 1 in angiogenesis and lymphangiogenesis during skin wound healing in mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 28 (3), 1237–47. [DOI] [PubMed] [Google Scholar]
- 42.Li M et al. (2014) Deficiency of RAMP1 attenuates antigen-induced airway hyperresponsiveness in mice. PloS one 9 (7), e102356–e102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikami N et al. (2014) Calcitonin gene-related peptide regulates type IV hypersensitivity through dendritic cell functions. PloS one 9 (1), e86367–e86367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsujikawa K et al. (2007) Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proceedings of the National Academy of Sciences of the United States of America 104 (42), 16702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caron KM and Smithies O (2001) Extreme hydrops fetalis and cardiovascular abnormalities in mice lacking a functional Adrenomedullin gene. Proc Natl Acad Sci U S A 98 (2), 615–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dackor R et al. (2007) Receptor activity-modifying proteins 2 and 3 have distinct physiological functions from embryogenesis to old age. J Biol Chem 282 (25), 18094–9. [DOI] [PubMed] [Google Scholar]
- 47.Dackor RT et al. (2006) Hydrops fetalis, cardiovascular defects, and embryonic lethality in mice lacking the calcitonin receptor-like receptor gene. Mol Cell Biol 26 (7), 2511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fritz-Six KL et al. (2008) Adrenomedullin signaling is necessary for murine lymphatic vascular development. J Clin Invest 118 (1), 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichikawa-Shindo Y et al. (2008) The GPCR modulator protein RAMP2 is essential for angiogenesis and vascular integrity. J Clin Invest 118 (1), 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amara SG et al. (1982) Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298 (5871), 240–244. [DOI] [PubMed] [Google Scholar]
- 51.Rosenfeld MG et al. (1983) Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature 304 (5922), 129–135. [DOI] [PubMed] [Google Scholar]
- 52.Goadsby PJ et al. (1990) Vasoactive peptide release in the extracerebral circulation of humans during migraine headache. Ann Neurol 28 (2), 183–7. [DOI] [PubMed] [Google Scholar]
- 53.Sabharwal R et al. (2019) Increased receptor activity-modifying protein 1 in the nervous system is sufficient to protect against autonomic dysregulation and hypertension. J Cereb Blood Flow Metab 39 (4), 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sternini C et al. (1992) Calcitonin gene-related peptide neurons innervating the canine digestive system. Regul Pept 42 (1-2), 15–26. [DOI] [PubMed] [Google Scholar]
- 55.Davis RB et al. (2019) Calcitonin-Receptor-Like Receptor Signaling Governs Intestinal Lymphatic Innervation and Lipid Uptake. ACS Pharmacology & Translational Science 2 (2), 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Giorgio R and Camilleri M (2004) Human enteric neuropathies: morphology and molecular pathology. Neurogastroenterol Motil 16 (5), 515–31. [DOI] [PubMed] [Google Scholar]
- 57.Di Nardo G et al. (2008) Review article: molecular, pathological and therapeutic features of human enteric neuropathies. Aliment Pharmacol Ther 28 (1), 25–42. [DOI] [PubMed] [Google Scholar]
- 58.Pauza AG et al. (2019) Alterations in enteric calcitonin gene-related peptide in patients with colonic diverticular disease: CGRP in diverticular disease. Auton Neurosci 216, 63–71. [DOI] [PubMed] [Google Scholar]
- 59.Nagashima H et al. (2019) Neuropeptide CGRP Limits Group 2 Innate Lymphoid Cell Responses and Constrains Type 2 Inflammation. Immunity 51 (4), 682–695 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallrapp A et al. (2017) The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549 (7672), 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackie DI et al. (2018) hCALCRL mutation causes autosomal recessive nonimmune hydrops fetalis with lymphatic dysplasia. J Exp Med 215 (9), 2339–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong B et al. (2019) Mutant RAMP2 causes primary open-angle glaucoma via the CRLR-cAMP axis. Genet Med 21 (10), 2345–2354. [DOI] [PubMed] [Google Scholar]
- 63.Bomberger JM et al. (2005) Novel function for receptor activity-modifying proteins (RAMPs) in post-endocytic receptor trafficking. J Biol Chem 280 (10), 9297–307. [DOI] [PubMed] [Google Scholar]
- 64.Bomberger JM et al. (2005) Receptor activity-modifying protein (RAMP) isoform-specific regulation of adrenomedullin receptor trafficking by NHERF-1. J Biol Chem 280 (25), 23926–35. [DOI] [PubMed] [Google Scholar]
- 65.Flahaut M et al. (2003) N-Glycosylation and conserved cysteine residues in RAMP3 play a critical role for the functional expression of CRLR/RAMP3 adrenomedullin receptor. Biochemistry 42 (34), 10333–41. [DOI] [PubMed] [Google Scholar]
- 66.Lv Y et al. (2016) Adrenomedullin 2 Enhances Beiging in White Adipose Tissue Directly in an Adipocyte-autonomous Manner and Indirectly through Activation of M2 Macrophages. J Biol Chem 291 (45), 23390–23402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H et al. (2016) Intermedin/adrenomedullin 2 polypeptide promotes adipose tissue browning and reduces high-fat diet-induced obesity and insulin resistance in mice. Int J Obes (Lond) 40 (5), 852–60. [DOI] [PubMed] [Google Scholar]
- 68.Kim J et al. (2019) Altered Expression of Adrenomedullin 2 and its Receptor in the Adipose Tissue of Obese Patients. J Clin Endocrinol Metab. [DOI] [PubMed] [Google Scholar]
- 69.Prakash J et al. (2019) Analysis of RAMP3 gene polymorphism with body composition and bone density in young and elderly women. Gene: X 2, 100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brain SD et al. (1985) Calcitonin gene-related peptide is a potent vasodilator. Nature 313 (5997), 54–6. [DOI] [PubMed] [Google Scholar]
- 71.McCulloch J et al. (1986) Calcitonin gene-related peptide: functional role in cerebrovascular regulation. Proc Natl Acad Sci U S A 83 (15), 5731–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noseda R and Burstein R (2013) Migraine pathophysiology: anatomy of the trigeminovascular pathway and associated neurological symptoms, CSD, sensitization and modulation of pain. Pain 154 Suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edvinsson L and Warfvinge K (2019) Recognizing the role of CGRP and CGRP receptors in migraine and its treatment. Cephalalgia 39 (3), 366–373. [DOI] [PubMed] [Google Scholar]
- 74.Lennerz JK et al. (2008) Calcitonin receptor-like receptor (CLR), receptor activity-modifying protein 1 (RAMP1), and calcitonin gene-related peptide (CGRP) immunoreactivity in the rat trigeminovascular system: differences between peripheral and central CGRP receptor distribution. J Comp Neurol 507 (3), 1277–99. [DOI] [PubMed] [Google Scholar]
- 75.Malhotra R (2016) Understanding migraine: Potential role of neurogenic inflammation. Ann Indian Acad Neurol 19 (2), 175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cernuda-Morollon E et al. (2013) Interictal increase of CGRP levels in peripheral blood as a biomarker for chronic migraine. Neurology 81 (14), 1191–6. [DOI] [PubMed] [Google Scholar]
- 77.Goadsby PJ and Edvinsson L (1993) The trigeminovascular system and migraine: studies characterizing cerebrovascular and neuropeptide changes seen in humans and cats. Ann Neurol 33 (1), 48–56. [DOI] [PubMed] [Google Scholar]
- 78.Goadsby PJ et al. (1988) Release of vasoactive peptides in the extracerebral circulation of humans and the cat during activation of the trigeminovascular system. Ann Neurol 23 (2), 193–6. [DOI] [PubMed] [Google Scholar]
- 79.Negro A and Martelletti P (2019) Gepants for the treatment of migraine. Expert Opin Investig Drugs 28 (6), 555–567. [DOI] [PubMed] [Google Scholar]
- 80.Giamberardino MA et al. (2017) Calcitonin gene-related peptide receptor as a novel target for the management of people with episodic migraine: current evidence and safety profile of erenumab. J Pain Res 10, 2751–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wattiez AS et al. (2019) CGRP in Animal Models of Migraine. Handb Exp Pharmacol 255, 85–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Z et al. (2007) Sensitization of calcitonin gene-related peptide receptors by receptor activity-modifying protein-1 in the trigeminal ganglion. J Neurosci 27 (10), 2693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Recober A et al. (2010) Induction of multiple photophobic behaviors in a transgenic mouse sensitized to CGRP. Neuropharmacology 58 (1), 156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.King CT et al. (2019) Discovery of the Migraine Prevention Therapeutic Aimovig (Erenumab), the First FDA-Approved Antibody against a G-Protein-Coupled Receptor. ACS Pharmacology & Translational Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi L et al. (2016) Pharmacologic Characterization of AMG 334, a Potent and Selective Human Monoclonal Antibody against the Calcitonin Gene-Related Peptide Receptor. J Pharmacol Exp Ther 356 (1), 223–31. [DOI] [PubMed] [Google Scholar]
- 86.ter Haar E et al. (2010) Crystal structure of the ectodomain complex of the CGRP receptor, a class-B GPCR, reveals the site of drug antagonism. Structure 18 (9), 1083–93. [DOI] [PubMed] [Google Scholar]
- 87.Liang YL et al. (2018) Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature 561 (7724), 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Technology P (2019) The top selling prescription drugs by revenue. (accessed). [Google Scholar]
- 89.Hauser AS et al. (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16 (12), 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Law V et al. (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42 (Database issue), D1091–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Udawela M et al. (2006) A critical role for the short intracellular C terminus in receptor activity-modifying protein function. Mol Pharmacol 70 (5), 1750–60. [DOI] [PubMed] [Google Scholar]
- 92.Udawela M et al. (2008) The effects of C-terminal truncation of receptor activity modifying proteins on the induction of amylin receptor phenotype from human CTb receptors. Regul Pept 145 (1-3), 65–71. [DOI] [PubMed] [Google Scholar]
- 93.Booe JM et al. (2015) Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor. Molecular Cell 58 (6), 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]


