Since there is an increasing number of coronavirus disease 2019 (COVID-19) cases confirmed in multiple countries around the world, resulting in alarmingly high mortality and morbidity [1], the World Health Organization has declared a global pandemic [2]. To our knowledge, most current research on COVID-19 focuses on the epidemiology, clinical features and treatment, but not on viral RNA shedding. Here, we discuss COVID-19 viral RNA conversion time and the risk factors associated with viral RNA shedding in a large patient cohort.
Short abstract
The median time of SARS-CoV-2 viral RNA shedding was 19 days. Patients with coronary heart disease comorbidity or decreased albumin levels experienced delays in clearance of SARS-CoV-2 RNA. https://bit.ly/2zjQu64
To the Editor:
Since there is an increasing number of coronavirus disease 2019 (COVID-19) cases confirmed in multiple countries around the world, resulting in alarmingly high mortality and morbidity [1], the World Health Organization has declared a global pandemic [2]. To our knowledge, most current research on COVID-19 focuses on the epidemiology, clinical features and treatment, but not on viral RNA shedding. Here, we discuss COVID-19 viral RNA conversion time and the risk factors associated with viral RNA shedding in a large patient cohort.
A total of 410 laboratory-confirmed COVID-19 patients were recruited to this study, who were discharged from Wuhan Tongji hospital, Wuhan Jin Yin-tan hospital and Wuhan Union hospital main district (all in Wuhan, China) between 1 February 2020 and 20 February 2020. Throat-swab samples were collected and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA was detected using real-time reverse transcription PCR (rRT-PCR). Data were extracted for demographic characteristics, comorbidities, symptoms, laboratory values and treatments using electronic medical records. All patients who were hospitalised received a standard diagnosis and treatment based on the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) [3]. Follow-up throat-swab specimens were obtained at intervals of 4±2 days from when symptoms began to improve, including fever reduction or improvement in chest computed tomography scans. The date of symptom onset was defined as the day when initial symptoms were noticed. T1 referred to the duration from the symptom onset to when SARS-CoV-2 RNA was negative for two consecutive times, which indicated the duration of viral RNA shedding. The date of fever resolution was defined as the first day after admission when the axillary temperature of a patient was no longer >37.3°C. T2 was defined as the duration from the date of fever resolution to rRT-PCR conversion. A hazard ratio (HR) of <1 indicated prolonged viral RNA shedding. This study was approved by the ethics commission of all hospitals (S2020-055, S2020-056, 2020-YJ-046.01) where patients were observed, and the requirement for informed consent was waived.
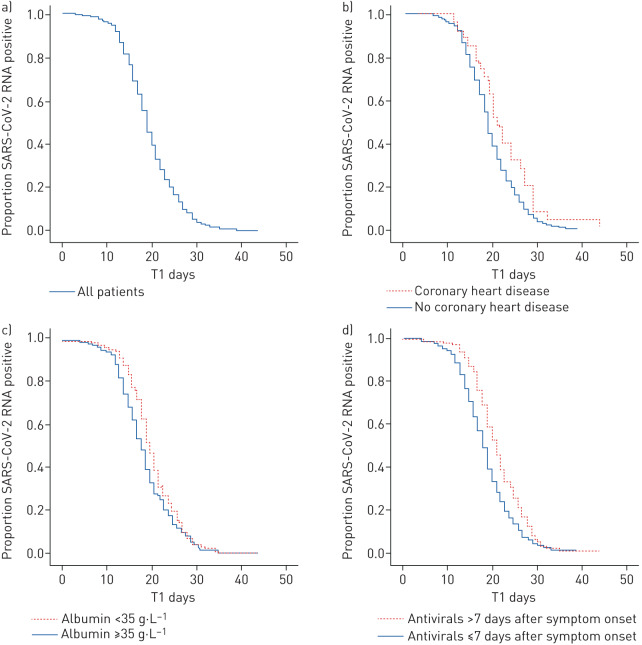
Out of the 410 patients, there were 198 (48.3%) men and the median age for all patients was 52 years (interquartile range (IQR) 38–66 years). The patients received an average of three rRT-PCR tests after admission. A total of 58 out of 410 patients had one negative test followed by one positive test. The median time of viral RNA shedding was 19 days (IQR 16–23 days), ranging from 3 to 44 days (figure 1a). Most patients (96%) tested negative for SARS-CoV-2 RNA within 30 days after symptom onset. The median time of T2 was 7 days (IQR 4–10 days). However, 36 patients tested negative for viral RNA and then achieved fever resolution and four patients still had a fever when discharged.
FIGURE 1.
Kaplan–Meier curves showing the proportions of SARS-CoV-2 RNA-positive patients. T1 refers to the duration from the symptom onset to the day when SARS-CoV-2 RNA was negative for two consecutive times, which was considered to indicate an undetectable viral RNA level. a) All patients (n=410). b) Patients with (n=27) and without (n=383) coronary heart disease; p=0.029. c) Patients with albumin levels <35 g·L−1 (n=193) and ≥35 g·L−1 (n=217); p=0.032. d) Patients who started antiviral therapy >7 days (n=128) and ≤7 days (n=278) after symptom onset; p<0.001.
Additionally, we explored the risk factors involved in prolonged viral RNA shedding using a multivariable Cox proportional hazards model. Coronary heart disease (CHD) (HR 0.619, 95% CI 0.411–0.933; p=0.022), albumin levels (HR 1.027, 95% CI 1.005–1.049; p=0.014) and the initial time of antiviral treatment (HR 1.467, 95% CI 1.187–1.815; p<0.001) were independent factors associated with SARS-CoV-2 RNA shedding. The median times for viral RNA shedding in patients with or without CHD were 21 days and 19 days, respectively (figure 1b). Patients with CHD showed prolonged viral RNA shedding (p=0.029). Patients with albumin ≥35 g·L−1 had a shorter duration of viral RNA shedding compared to those with albumin <35 g·L−1 and the median times were 18 days and 20 days, respectively (figure 1c). The median times for patients who started antiviral therapy within or later than 7 days after symptom onset were 18 days and 21 days, respectively (figure 1d).
Our results first revealed that the median time from symptom onset to rRT-PCR conversion was 19 days, which was similar to what was uncovered in previous data reported by Zhou et al. [4], where the median time was 20 days for 137 cases. In this study, most patients showed a negative PCR for viral RNA within 30 days, and the longest time observed was 44 days. Moreover, many patients had a fever during the course of the disease, and the median time from fever resolution to rRT-PCR conversion was 7 days, with a range of 0–28 days. However, owing to the fact that PCR tests sometimes show false-negative results, it is best to repeat this at least twice. Based on the results, it is recommended that patients continue to follow prophylactic measures, including wearing masks, even after being discharged from the hospital.
Based on these data, patients with decreased albumin levels experienced prolonged viral RNA shedding. As previously reported, hypoalbuminaemia has a negative impact on the immune response system and increases short-term mortality in hospitalised patients with acute infectious diseases [5, 6]. However, further studies are needed to address the mechanism underlying any link between albumin and viral RNA shedding. In addition, patients with comorbidities showed prolonged viral RNA shedding. Nevertheless, only CHD was proven to be an independent risk factor for prolonged viral RNA shedding. Similarly, major comorbidities including cardiovascular diseases, diabetes and malignancies have previously been found to be related to slower viral clearance in patients with SARS or influenza A [7, 8]. As previously reported, patients with pre-existing cardiovascular diseases were at a higher risk for COVID-19 and experienced worse outcomes, showing a 5–10-fold increase in mortality [9]. Hence, patients with chronic comorbidities, especially CHD, had more difficultly in achieving viral clearance.
It is worth noting that only throat-swab specimens were used for viral RNA detection, since this is a retrospective study. Evidence has shown that SARS-CoV-2 could be detected in stool samples as well. However, a recent study declared that infectious virus was predominantly derived from the respiratory tract and not from stools [10]. One limitation of our work is that we detected virus by rRT-PCR but not by virus isolation. As rRT-PCR detects both viable and nonviable virus, more convincing studies are needed to investigate whether people just with persistent positive rRT-PCR results are capable of transmission or not.
In conclusion, the median time from symptoms onset to viral RNA shedding was 19 days. Patients with CHD comorbidity, decreased albumin levels and delayed antiviral therapy experienced delays in clearance of SARS-CoV-2 RNA. A better understanding of viral RNA shedding provides guidance for public health measures and contributes to identifying an appropriate plan for viral RNA detection.
Shareable PDF
Supplementary Material
Acknowledgements
We acknowledge all of the medical workers who participated in the diagnosis and treatment of patients in Wuhan. We thank all the patients involved in this study.
Footnotes
Conflict of interest: Y. Fu has nothing to disclose.
Conflict of interest: P. Han has nothing to disclose.
Conflict of interest: R. Zhu has nothing to disclose.
Conflict of interest: T. Bai has nothing to disclose.
Conflict of interest: J. Yi has nothing to disclose.
Conflict of interest: X. Zhao has nothing to disclose.
Conflict of interest: M. Tao has nothing to disclose.
Conflict of interest: R. Quan has nothing to disclose.
Conflict of interest: C. Chen has nothing to disclose.
Conflict of interest: Y. Zhang has nothing to disclose.
Conflict of interest: Q. He has nothing to disclose.
Conflict of interest: M. Jing has nothing to disclose.
Conflict of interest: X. Xiong has nothing to disclose.
Conflict of interest: D. Tian has nothing to disclose.
Conflict of interest: W. Yan has nothing to disclose.
Support statement: This study is funded by the National Natural Science Foundation of China (grant numbers 81570501, 81770554, 81772607, 81974383) and Key Projects in the Natural Scientific and Technology Program (2018ZX10302204-002-003). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.World Health Organization Coronavirus disease (COVID-19) outbreak situation. www.who.int/emergencies/diseases/novel-coronavirus-2019 Date last accessed: 25 April 2020.
- 2.World Health Organization WHO Director-General's opening remarks at the media briefing on COVID-19 – 11 March 2020. www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 Date last updated: 11 March 2020. Date last accessed: 25 April 2020.
- 3.National Health Commission of the People's Republic of China Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) www.gov.cn/zhengce/zhengceku/2020-03/04/content_5486705.htm Date last updated: 3 March 2020. Date last accessed: 25 April 2020.
- 4.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395: 1054–1062. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziedzic T, Pera J, Klimkowicz A, et al. Serum albumin level and nosocomial pneumonia in stroke patients. Eur J Neurol 2006; 13: 299–301. doi: 10.1111/j.1468-1331.2006.01210.x [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Sun Y, Xing Y, et al. Functional impairment and serum albumin predict in-hospital mortality in nonagenarians with acute infection: a retrospective cohort study. BMC Geriatr 2019; 19: 269. doi: 10.1186/s12877-019-1301-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Tang F, Fontanet A, et al. Long-term SARS coronavirus excretion from patient cohort, China. Emerg Infect Dis 2004; 10: 1841–1843. doi: 10.3201/eid1010.040297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee N, Chan PK, Hui DS, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis 2009; 200: 492–500. doi: 10.1086/600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu PP, Blet A, Smyth D, et al. The science underlying COVID-19: implications for the cardiovascular system. Circulation 2020; in press [ 10.1161/CIRCULATIONAHA.120.047549]. [DOI] [PubMed] [Google Scholar]
- 10.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581: 465–469. doi: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01190-2020.Shareable (271.6KB, pdf)