Abstract
Background
TERMINAL FLOWER 1 (TFL1) is a member of phosphatidylethanolamine-binding protein (PEBP) family, which plays an important role in the determination of floral meristem identity and regulates flowering time in higher plants.
Results
Five BnaTFL1 gene copies were identified in the genome of Brassica napus. The phylogenetic analysis indicated that all five BnaTFL1 gene copies were clustered with their corresponding homologous copies in the ancestral species, B. rapa and B. oleracea. The expression of the BnaTFL1s were confined to flower buds, flowers, seeds, siliques and stem tissues and displayed distinct expression profiles. Knockout mutants of BnaC03.TFL1 generated by CRISPR/Cas9 exhibited early flowering phenotype, while the knockout mutants of the other gene copies had similar flowering time as the wild type. Furthermore, knock-out mutants of individual BnaTFL1 gene copy displayed altered plant architecture. The plant height, branch initiation height, branch number, silique number, number of seeds per silique and number of siliques on the main inflorescence were significantly reduced in the BnaTFL1 mutants.
Conclusions
Our results indicated that BnaC03.TFL1 negatively regulates flowering time in B. napus. BnaC03.TFL1 together with the other BnaTFL1 paralogues are essential for controlling the plant architecture.
Keywords: TERMINAL FLOWER 1, PEBP, CRISPR/Cas9, Early flowering, Plant architecture, Brassica napus
Background
The transition from vegetative to reproductive stage is strictly controlled by both environmental and developmental signals. Plant needs to achieve a certain stage of developmental competence to respond to environmental factors such as day length (photoperiod), winter temperature (vernalization) and water stress [1]. Endogenous signals such as phytohormones, notably gibberellins, also influence the vegetative to floral transition [2, 3]. Rapeseed (Brassica napus L., AACC, 2n = 38) is a major source of edible oil and biofuel, which emerged from natural crossing between its progenitors, B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18) ~ 7500 years ago [4]. Flowering time in rapeseed not only has a crucial impact on yield, but also influences the sowing time of other rotation crops [5]. The timing of flowering and plant requirement for and responsiveness to vernalization are major factors in regional climatic adaptation of elite germplasm. In rapeseed, quantitative trait locus (QTL) analysis has been used to identify candidate flowering time genes. For example, one major QTL related to the BnaA10.FLC gene has been demonstrated to be an important regulator that represses winter type rapeseed flowering in winter [6].
In Arabidopsis, a model plant for eudicots, flowering time is regulated by the photoperiod, autonomous, vernalization, endogenous gibberellin (GA), age and ambient temperature-dependent pathways [7, 8]. The integrated induction signals from these flowering pathways are transmitted via floral integrator genes such as FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and TERMINAL FLOWER1 (TFL1) [9–12] to induce the expression of floral meristem identity genes LEAFY (LFY) and APETALA1 (AP1) at the shoot apical meristem to activate floral transition [13, 14]. The photoperiod pathway regulates flowering time depending on day length [1]. The key regulators in the photoperiod pathway encode proteins with homology to phosphatidylethanolamine binding proteins (PEBPs) [15], which is highly conserved and presented in both animal and plant kingdoms [16]. The PEBP gene family encompasses two particularly important genes, FT and TFL1, which have been found in Arabidopsis and soybean [17–19]. Interestingly, FT and TFL1 share 98% amino acid sequence similarity, but their functions are in antagonistic manners. FT is a flowering activator which promotes flowering [19–22], while TFL1 is a flowering inhibitor and controls the identity of shoot meristem during the plant life span [17, 19, 23, 24].
In Arabidopsis, tfl1 mutant exhibits a shorter vegetative phase, produces fewer leaves, reduces the number of flower buds, branches and flowers and controls the conversion of inflorescence meristem to terminal flower [17, 25, 26]. In contrast, overexpression of the TFL1 gene promotes secondary inflorescence production and delayed flowering [27, 28]. Similarly, mutation of CENTRORADIALIS (CEN), an Antirrhinum TFL1 ortholog, results in the conversion of the normally indeterminate inflorescence to a determinate inflorescence [17, 29]. Both TFL1 and CEN are expressed in the subapical region of the shoot meristem. TFL1 is expressed in both vegetative and inflorescence shoot meristems, whereas CEN is only expressed in the inflorescence meristem [17, 29]. The Determinate stem (Dt1) mutant exhibited determinate growth and terminal flower formation in soybean [30]. Overexpression of RCN1 and RCN2, the TFL1 homologs in rice, rendered more branched, denser panicles and delayed transition to reproductive phase [31]. Mutations in BnaA10.TFL1 have no large effects on flowering time but affect some plant architecture related traits in rapeseed [32, 33] .
Three genome-editing tools have been well developed, including Zinc-Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs) and Clustered Regularly Interspaced Palindromic Repeat (CRISPR)-associated protein 9 system (CRISPR/Cas9), which have been widely used in improving traits for productivity and nutrition in crop plants [34]. Among these genome editing tools, CRISPR/Cas9 system is considered the most efficient and simple, which has been rapidly and widely applied for genome editing [35–37]. The CRISPR/Cas9 system has been applied to target mutations in many plant species including B. napus [38–47].
In rapeseed, the genetic mechanism underlying the control of flowering time has not been fully understood yet. Thus, one of the objectives of rapeseed breeding is to look for new strategies to alter the flowering behavior. Introducing determinate type of inflorescence in the crop will result in shorter flowering time, earlier and consistent maturation which will greatly facilitate harvest in a short time. There are five paralogs of TFL1 in the allotetraploid B. napus due to the whole genome triplication events occurred in the genomes of B. rapa and B. oleracea, the two diploid progenitors of B. napus. In the present study we aimed to evaluate the role of TFL1 gene copies in controlling flowering time and plant architecture in B. napus. We generated mutants of five paralogues of TFL1s in B. napus using the CRISPR/Cas9 technology. Knockout mutants of BnaC03.TFL1 exhibited earlier flowering. In addition, knockout mutant of individual BnaTFL1 gene copy displayed reduced plant height, branch initiation height, branch number, silique number, seed number per silique and number of siliques on the main inflorescence at different degrees. Our study showed that BnaC03.TFL1 negatively regulates the conversion of inflorescence meristem to floral meristem in B. napus. The BnaTFL1 gene copies are involved in the determination of plant architecture, and are promising targets for crop improvement in rapeseed.
Results
Isolation and identification of TFL1 genes in B. napus
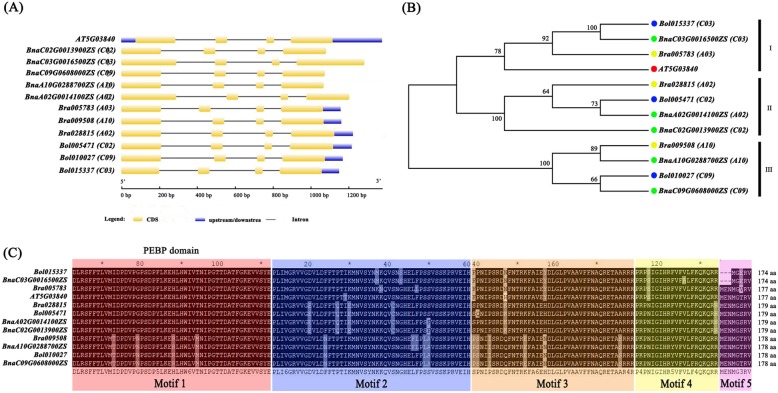
To retrieve TFL1 genes from the genomes of B. napus, B. rapa and B. oleracea, the amino acid sequence of Arabidopsis TFL1 was used as a query to search the B. napus (http://rice.hzau.edu.cn/cgi-bin//bnapus/gb2/gbrowse/ZS11v0/) as well as the B. rapa and B. oleracea databases (http://brassicadb.org/brad/blastPage.php). Five BnaTFL1 gene copies were obtained, including two (BnaA02G0014100ZS and BnaA10G0288700ZS) from the A sub-genome, and three (BnaC02G0013900ZS, BnaC03G0016500ZS and BnaC09G0608000ZS) from the C sub-genome. These five gene copies were named as BnaA02.TFL1, BnaA10.TFL1, BnaC02.TFL1, BnaC03.TFL1 and BnaC09.TFL1, respectively. We also obtained three TFL1 gene copies (Bra005783, Bra028815 and Bra009508) from B. rapa and three TFL1 gene copies (Bol015337, Bol005471 and Bol010027) from B. oleracea. All these gene copies contain four exons, with the lengths of open reading frames (ORFs) ranging from 522 bp to 540 bp (Fig. 1a and Table S2).
Fig. 1.
Phylogenetic, gene structure, and domain analyses of TFL1 genes in B. napus. a Exon-intron structures of the 12 TFL1 genes from B. napus, B. rapa, B. oleracea, and A. thaliana. b Phylogenetic relationship of the 12 TFL1 genes. Genes from B. napus, B. rapa, B. oleracea, and A. thaliana were indicated by green, yellow, blue and red circle, respectively. c Multiple alignment of amino acid sequences of 12 TFL1 genes. Motif compositions of PEBP were identified using the MEME tool. Each motif is represented by a colored block. The sizes of amino acid (aa) are shown on the right side of the figure
The phylogenetic tree was constructed based on their amino acid sequences. The result indicated that Brassica TFL1 gene copies are divided into three clusters which are consistent with their evolutionary relationship (Fig. 1b). The paralogous gene copies in B. napus and their corresponding homologous copies in B. rapa and B. oleracea are grouped in the same clusters (Fig. 1b). For example, BnaA02G0014100ZS and BnaC02G0013900ZS are clustered with Bra028815 from A02 of B. rapa and Bol005471 from C02 of B. oleracea. BnaC03.TFL1 (BnaC03G0016500ZS) and Arabidopsis TFL1 are cluster I, suggesting that BnaC03.TFL1 is more closer to Arabidopsis TFL1 than the other BnaTFL1s (Fig. 1b). Multiple alignment of amino acid sequences indicated all the Brassica TFL1 proteins display very high homology to the Arabidopsis TFL1, with identity ranging from 83.4 to 100%. All TFL1 proteins contain five highly conserved motifs including the PEBP domain, suggesting that these TFL1 genes may have conserved functions (Fig. 1c).
Expression pattern of BnaTFL1
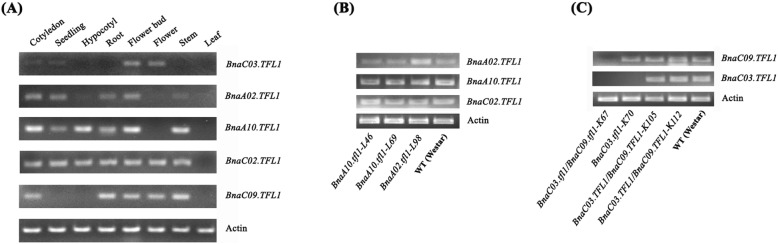
To gain insights into the putative functions of the five TFL1 gene copies in B. napus, we investigated their expression in cotyledons, seedlings, hypocotyls, roots, flower buds, flowers, stems and leaves by RT-PCR. The five BnaTFL1 gene copies displayed distinct expression patterns (Fig. 2a). All genes are expressed in flower buds but not in leaves. BnaC02.TFL1 expressed in all tissues except for leaves. BnaA10.TFL1 and BnaA02.TFL1 showed similar expression pattern. Both gene copies are expressed in cotyledons, seedlings, hypocotyls, roots, flower buds and stem, but the expression levels of BnaA02.TFL1 are lower than that of BnaA10.TFL1 in all these tissues. BnaC03.TFL1 is preferentially expressed in flower buds and flowers and weakly expressed in cotyledons and seedlings. BnaC09.TFL1 is expressed in cotyledons, roots, stems, flower buds and flowers but not in seedlings, hypocotyls and leaves (Fig. 2a). Comparison of promoter sequences of these five BnaTFL1 gene copies indicated that they are divergent, which is consistent with their distinct expression patterns (Fig. S1).
Fig. 2.
Expression pattern of BnaTFL1.a RT-PCR analyses of BnaTFL1 genes in wild type Westar. Expression patterns of the five BnaTFL1s in cotyledons, seedlings, hypocotyls, roots, flower buds, flowers stem and leaves. b The expression of BnaTFL1s in flower buds of sgRNA1/2 mutagenized mutants. c The expression of BnaTFL1s in flower buds of sgRNA3/4 mutagenized mutants. WT stands for wild type Westar
Targeted mutagenesis of TFL1 induced by CRISPR/Cas9
In order to uncover the functions of these BnaTFL1 gene copies, the CRISPR/Cas9 gene editing technology was used to create knockout mutants of the five paralogous BnaTFL1s. Because we could not identify conserved sgRNAs that can target all five BnaTFL1 gene copies simultaneously, we designed four sgRNAs to target the five BnaTFL1s, with sgRNA1 and sgRNA2 targeting to the conserved regions of BnaA02.TFL1, BnaA10.TFL1 and BnaC02.TFL1, and sgRNA3 and sgRNA4 targeting to the conserved regions of BnaC03.TFL1 and BnaC09.TFL1 (Fig. 3a). The constructs containing sgRNA1/sgRNA2 and sgRNA3/sgRNA4 were independently transformed into oilseed callus following standard procedures [43]. A total of 101 T0 transgenic plants were obtained for sgRNA1/2 and 130 T0 transgenic plants for sgRNA3/4. Among them, 40 plants were found to be Cas9-positive for sgRNA1/2 transgenic plants, while 60 plants to be Cas9-positive for sgRNA3/4 transgenic plants (Table S3 and S4). Mutations occurred in these five BnaTFL1 gene copies were screened from these Cas9-positive transgenic plants by ACT-PCR (annealing at critical temperature PCR) [48]. A pair of specific primers were designed for each target sgRNA of the five gene copies and used to screen the Cas9-positive plants. Totally, six T0 plants were identified to have mutations at sgRNA1 and/or sgRNA2 target sites and 12 T0 plants were identified to have mutations at sgRNA3 and/or sgRNA4 target sites by ACT-PCR.
Fig. 3.
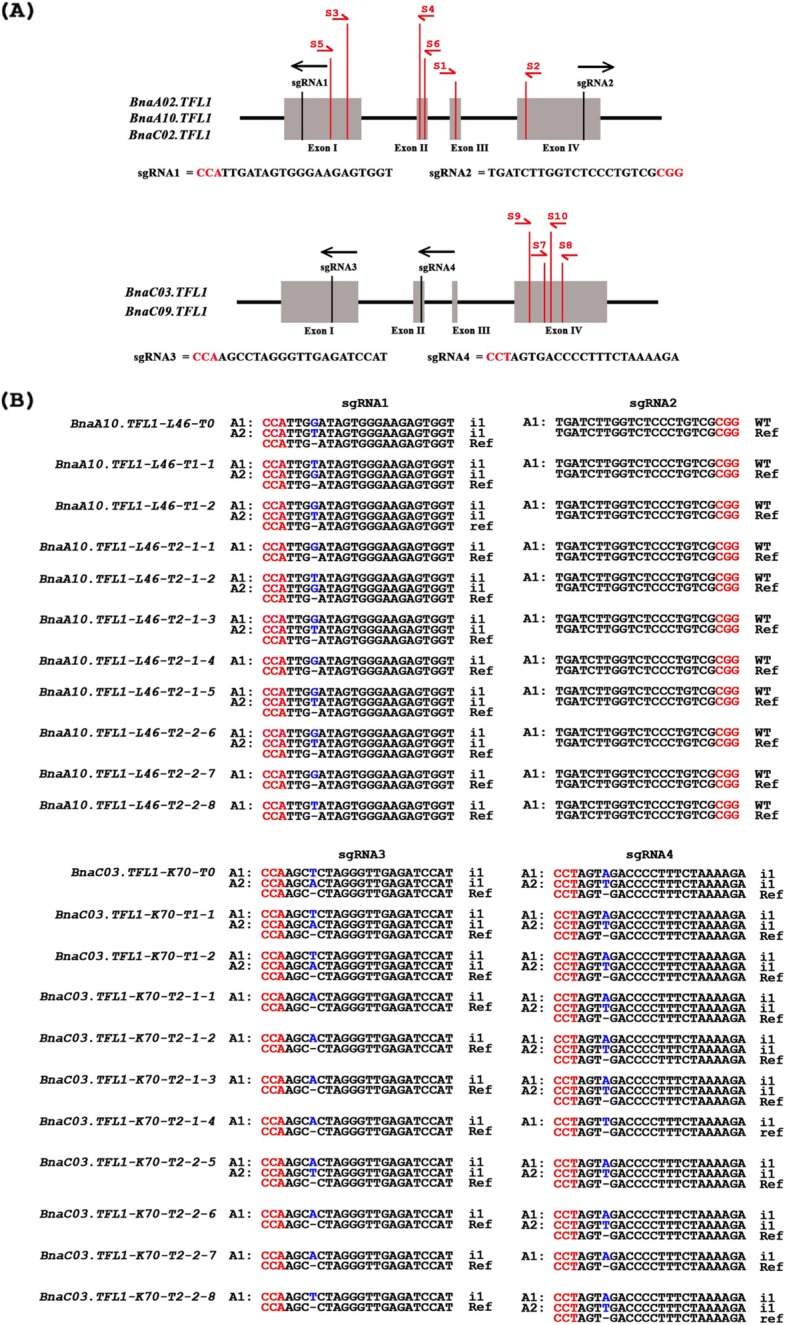
Sequence variations in CRISPR/Cas9-induced BnaTFL1s mutants. a The structure of BnaTFL1 genes including four exons (Gray box) separated by three introns (represented by the solid line). The vertical lines in the genes indicate the target sites of sgRNAs, and the arrows indicate directions of the sgRNAs. The red characters indicate the PAM sites in the target sequences. The single-side arrows indicate primers for RT-PCR. The primer pairs of S1/S2, S3/S4, S5/S6, S7/S8 and S9/S10 were used to amplify BnaC02.TFL1, BnaA10.TFL1, BnaA02.TFL1, BnaC03.TFL1 and BnaC09.TFL1, respectively. b Sequences variations at the gRNA target sites of BnaTFL1.A10 (L46) and BnaTFL1.C03 (K70) in the T0, T1 and T2 generations. Blue characters indicate nucleotide insertions, and i1 means one nucleotide insertion. Ref stands for reference sequence. WT stands for wild-type allele. “A1” and “A2” represent the mutated alleles of the target Bnatfl1 genes
The mutations of individual sgRNA target sites were further confirmed by Sanger sequencing. Among the six T0 mutant plants generated by sgRNA1/2, three plants (L43, L46 and L69) had mutations only in BnaA10.TFL1, two plants (L84 and L98) contained mutations only in BnaA02.TFL1, and one plant (L93) had mutations in both BnaA02.TFL1 and BnaC02.TFL1 (Table S5). No plant had mutations in all three target genes. In these six T0 plants, L46 and L69 were loss-of-function mutants of BnaA10.TFL1, and L84 and L98 were loss-of-function mutants of BnaA02.TFL1. No loss-of-function mutant was obtained for BnaC02.TFL1.
As for the 12 mutant plants generated by sgRNA3/4, five plants (K35, K45, K54, K62 and K85) had mutations in BnaC09.TFL1, one plant (K70) had mutation in both BnaC03.TFL1, and the others (K67, K74, K87, K105, K112 and K130) had mutations in BnaC03.TFL1 and BnaC09.TFL1 (Table S6). In these mutant plants, K70 was a loss-of-function mutant of BnaC03.TFL1, and K67 and K87 were loss-of-function mutants of both BnaC03.TFL1 and BnaC09.TFL1. K85 was a loss-of-function mutant of BnaC09.TFL1. K105 and K112 were chimeric containing wild type alleles.
Detection of CRISPR/Cas9 mediated mutations of BnaTFL1 in the T1 and T2 generations
To assess the inheritance of the CRISPR/Cas9-induce mutations in next generations, we self-pollinated two BnaA10.TFL1 mutants (L46 and L69), one BnaA02.TFL1 mutant (L98), one BnaC03.TFL1 mutant (K70) and one BnaC09.TFL1 mutant (K85), three BnaC03.TFL1/BnaC09.TFL1 double mutants (K67, K105 and K112) to obtain their T1 generation. Two T1 plants were randomly selected from each line to check their genotypes by TA cloning followed by Sanger sequencing. The two T1 plants derived from these eight T0 mutants had the same mutations as their parents (Table S7), except for K105 and K112 which generated new mutations in T1 due to the existence of wild-type allele in the T0 plants.
The transmission of mutations from T1 to T2 generation was further investigated by next-generation sequencing of target amplicons. Eight T2 plants were randomly selected from one of the T1 plants. Each target site was amplified using target-specific primers and amplicons were sequenced using Illumina HiSeq 4000 sequencing platform. The T2 plants of L46, L69, L98, K67 and K70 had the same mutations as their parents, and all were loss-of-function mutants with homozygous or heterozygous genotypes of mutant alleles (Table S8 and S9). For examples, four T2 plants of L46 were heterozygous of two mutant alleles (bi-allelic, i1–1/i1–2) and four T2 plants were homozygous (i1–1/i1–1). Of the eight T2 plants of K70, seven were homozygous (i1–1/i1–1) and one was heterozygous (bi-allelic, i1–1/i1–2). Same results were found in other mutant lines (Fig. 3b). All the T2 plants of K85 were chimeric of multiple mutant alleles, while all the T2 plants of K105 and K112 were chimeric of multiple mutant alleles and wild-type allele. These results indicated that most mutations were inheritable from T1 to T2 generations (Tables 1 and 2).
Table 1.
Genotypic analysis of BnaTFL1s mutants (BnaA02.TFL1, BnaA10.TFL1, and BnaC02.TFL1.) mutagenized by sgRNA1/2 in the T1 and T2 generations
| Lines | Generations | BnaA02.TFL1 | BnaA10.TFL1 | BnaC02.TFL1 | ||||
|---|---|---|---|---|---|---|---|---|
| sgRNA1 | sgRNA2 | sgRNA1 | sgRNA2 | sgRNA1 | sgRNA2 | Early flowering | ||
| L46 | T0 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L46–1 | T1 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L46–1-1 | T2 | WT | WT | Homozygous (i1–1/i1–1) | WT | WT | WT | No |
| L46–1-2 | T2 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L46–1-3 | T2 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L46–1-4 | T2 | WT | WT | Homozygous (i1–1/i1–1) | WT | WT | WT | No |
| L69 | T0 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L69–1 | T1 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L69–1-1 | T2 | WT | WT | Homozygous (i1–1/i1–1) | WT | WT | WT | No |
| L69–1-2 | T2 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L69–1-3 | T2 | WT | WT | Bi-allelic (i1–1/i1–2) | WT | WT | WT | No |
| L69–1-4 | T2 | WT | WT | Homozygous (i1–1/i1–1) | WT | WT | WT | No |
| L98 | T0 | WT | Homozygous (d1–1/d1–1) | WT | WT | WT | WT | No |
| L98–1 | T1 | WT | Homozygous (d1–1/d1–1) | WT | WT | WT | WT | No |
| L98–1-1 | T2 | WT | Homozygous (d1–1/d1–1) | WT | WT | WT | WT | No |
| L98–1-2 | T2 | WT | Homozygous (d1–1/d1–1) | WT | WT | WT | WT | No |
| L98–1-3 | T2 | WT | Homozygous (d1–1/d1–1) | WT | WT | WT | WT | No |
| L98–1-4 | T2 | WT | Homozygous (d1–1/d1–1) | WT | WT | WT | WT | No |
d1 one nucleotide deletion; i1 one nucleotide insertion; WT wild type; i1–1 and i1–2 stand for two different alleles
Table 2.
Genotypic analysis of BnaTFL1s mutants (BnaC03.TFL1 and BnaC09.TFL1) mutagenized by sgRNA3/4 in the T1 and T2 generations
| Lines | Generations | BnaC03.TFL1 | BnaC09.TFL1 | |||
|---|---|---|---|---|---|---|
| sgRNA3 | sgRNA4 | sgRNA3 | sgRNA4 | Early flowering | ||
| K67 | T0 | Bi-allelic (d1/d5) | Chimeric (i1–1/i1–2/WT) | Bi-allelic (i1–1/i1–2) | WT | Yes |
| K67–1 | T1 | Bi-allelic (d1/d5) | Chimeric (i1–1/i1–2/WT) | Bi-allelic (i1–1/i1–2) | WT | Yes |
| K67–1-1 | T2 | Bi-allelic (d1/d5) | Chimeric (i1–1/i1–2/WT) | Homozygous (i1–1/i1–1) | WT | Yes |
| K67–1-2 | T2 | Bi-allelic (d1/d5) | Chimeric (i1–1/i1–2/WT) | Homozygous (i1–1/i1–1) | WT | Yes |
| K67–1-3 | T2 | Bi-allelic (d1/d5) | Chimeric (i1–1/i1–2/WT) | Bi-allelic (i1–1/i1–2) | WT | Yes |
| K67–1-4 | T2 | Bi-allelic (d1/d5) | Chimeric (i1–1/i1–2/WT) | Homozygous (i1–1/i1–1) | WT | Yes |
| K70 | T0 | Bi-allelic (i1–1/i1–2) | Bi-allelic (i1–1/i1–2) | WT | WT | Yes |
| K70–1 | T1 | Bi-allelic (i1–1/i1–2) | Bi-allelic (i1–1/i1–2) | WT | WT | Yes |
| K70–1-1 | T2 | Homozygous (i1–1/i1–1) | Bi-allelic (i1–1/i1–2) | WT | WT | Yes |
| K70–1-2 | T2 | Homozygous (i1–1/i1–1) | Bi-allelic (i1–1/i1–2) | WT | WT | Yes |
| K70–1-3 | T2 | Homozygous (i1–1/i1–1) | Bi-allelic (i1–1/i1–2) | WT | WT | Yes |
| K70–1-4 | T2 | Homozygous (i1–1/i1–1) | Homozygous (i1–1/i1–1) | WT | WT | Yes |
| K85 | T0 | WT | WT | Chimeric (i1–1/i1–2/d1/d5) | WT | No |
| K85–1 | T1 | WT | WT | Chimeric (i1–1/i1–2/i1–3) | WT | No |
| K85–1-1 | T2 | WT | WT | Chimeric (i1–1/i1–2/i1–3/d1) | WT | No |
| K85–1-2 | T2 | WT | WT | Chimeric (i1–1/i1–2/i1–3/d1) | WT | No |
| K85–1-3 | T2 | WT | WT | Chimeric (i1–1/i1–2/i1–3/d1) | WT | No |
| K85–1-4 | T2 | WT | WT | Chimeric (i1–1/i1–2/d1) | WT | No |
| K105 | T0 | Chimeric (i1–1/i1–2/i1–3/WT) | WT | Chimeric (i1–1/i1–2/i1–3/i1–4/WT) | WT | No |
| K105–1 | T1 | Chimeric (i1/4d/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K105–1-1 | T2 | Chimeric (i1–1/i1–2/i1–3/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K105–1-2 | T2 | Chimeric (i1–1/i1–2/i1–3/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K105–1-3 | T2 | Chimeric (i1–1/i1–2/i1–3/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K105–1-4 | T2 | Chimeric (i1–1/i1–2/i1–3/WT) | WT | Chimeric (i1–1/i1–2/WT) | WT | No |
| K112 | T0 | Chimeric (i1–1/i1–2/5d) | WT | Chimeric (i1–1/i1–2/i1–3) | WT | No |
| K112–1 | T1 | Chimeric (i1/d5/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K112–1-1 | T2 | Chimeric (i1–1/i1–2/i1–3/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K112–1-2 | T2 | Chimeric (i1–1/i1–2/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K112–1-3 | T2 | Chimeric (i1–1/i1–2/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
| K112–1-4 | T2 | Chimeric (i1–1/i1–2/WT) | WT | Chimeric (i1–1/i1–2/i1–3/WT) | WT | No |
d1, d4 and d5 means one, four and five nucleotides deletion, respectively; i1 one nucleotide insertion; WT wild type; i1–1, i1–2, etc. stands for different alleles
To check the expression of the target genes in their corresponding mutants, we performed RT-PCR to detect their mRNA transcripts in flower buds where all the five BnaTFL1 gene copies are expressed. The expression levels of BnaA02.TFL1, BnaA10.TFL1 and BnaC02.TFL1 did not show significant changes in the knockout mutant lines L98 (BnaA02.tfl1), and L46 and L69 (BnaA10.tfl1) created by sgRNA1/2, suggesting that the insertions or deletions did not affect the transcription of these genes (Fig. 2b). We also checked the expression of BnaC03.TFL1 and BnaC09.TFL1 in flower buds of different mutants induced by sgRNA3/4 and found BnaC03.TFL1 was not detectable in single gene knockout mutant BnaC03.tfl1 (K70) and in double mutant BnaC03.tfl1/BnaC09.tfl1 (K67). BnaC09.TFL1 was not detectable in the double mutant BnaC03.tfl1/BnaC09.tfl1 (K67) (Fig. 2c). These results suggested that insertions or deletions abolish the expression of these two genes. The expression levels of BnaC03.TFL1 and BnaC09.TFL1 were not affected in the chimeric mutant lines K105 and K112 having wild-type alleles (Fig. 2c).
Knockout of BnaC03.TFL1 promotes flowering in B. napus
It is reported that Arabidopsis TFL1 negatively regulates flowering time [17, 23]. To investigate if the knockout of BnaTFL1 gene copies affect the flowering time, we grew the Bnatfl1 mutants in the field under nature growth conditions in two growth seasons (2018 for T1, and 2019 for T2) to observe the agronomic traits. In the T1 generation, single mutants including BnaA10.tfl1 (L46 and L69), BnaC09.tfl1 (K85) and BnaA02.tfl1 (L98) had the same days from sowing to flowering (DTF) as WT (Westar) (Fig. 4a, b and Table S10). Among the four T1 mutant lines having mutations in BnaC03.TFL1, K70 (130 ± 3d) for BnaC03.tfl1 and K67 (133 ± 3d) for BnaC03.tfl1/BnaC09.tfl1 flowered much earlier than Westar (Fig. 4a and b). K85 was a loss-of-function mutant at BnaC09.TFL1 and displayed normal flowering time as wild type Westar (Fig. 4a, b and Table S10). In the T2 generation, the flowering time of these mutants was similar to that in the T1 generation (Fig. 4c and Table S11). The results indicated that the loss-of-function mutants of BnaC03.TFL1 exhibited early flowering and the loss-of-function mutants of the other gene copies did not affect the flowering time, suggesting that BnaC03.TFL1 play an important role in determining the floral transition, while the other paralogues may have obtained new functions or function redundantly in the determination of floral transition.
Fig. 4.
Flowering time of CRISRP/Cas9-induced BnaTFL1s mutants. a Morphology and flowering time of Westar and CRISRP/Cas9-induced BnaTFL1s mutants. b Comparison of flowering time between the WT and BnaTFL1s mutants in the T1 generation (2018). c Comparison of flowering time between the WT and BnaTFL1s mutants in the T2 generation (2019). DTF stands for days from sowing to flowering. Statistically significant differences were revealed using Student’s t-test: *, P < 0.05; **, P < 0.01, and ***, P < 0.001. Values are means ± SD (n = 20)
BnaTFL1 genes regulate plant architecture in B. napus
Arabidopsis TFL1 also determines the formation of shoot apical meristem and thus the plant architecture. tfl1 mutants displayed altered plant architecture such as less leaves, branches and flowers than the wild type. To see if the BnaTFL1 gene copies also regulate plant architecture in rapeseed, we investigated plant height (PH) and branch initiation height (BIN) of these single mutants and double mutants in the T1 and T2 generations (2018 and 2019, respectively). The knockout mutants of BnaA10.TFL1, BnaC03.TFL1 and BnaC09.TFL1 were significantly shorter than WT (150.2 ± 5 cm in 2018 and 149.1 ± 12 cm in 2019) (Fig. 5b and c). Consequently, the branch initiation height of these knockout mutants were also significantly lower than that of the wild type (Fig. 5d and e). Of these knockout mutants, BnaC03.tfl1 (K70) displayed the largest reduction in both plant height and branch initiation height. These results suggested that these four BnaTFL1s are involved in the determination of plant height, with BnaC03.TFL1 having the strongest and BnaA02.TFL1 having the weakest effects on determining the plant height and the position of the first branch.
Fig. 5.
Differences of plant architecture related traits in CRISRP/Cas9-induced BnaTFL1s mutants. a Whole plant architecture of T2 plants of CRISRP/Cas9-induced BnaTFL1s mutants. Differences of plant height (b and c), Branch initiation height (d and e), Branches number (f and g), Number of siliques on the main inflorescence (h and i), Silique length (j and k), and Number of seeds number per siliques (l and m) in 2018 (left panel) and 2019 (right panel). Statistically significant differences were revealed using Student’s t-test: *, P < 0.05; **, P < 0.01, and ***, P < 0.001. Values are means ± SD (n = 20)
We also investigated four yield-related traits including branch number (BN), number of siliques on the main inflorescence (NSMI), siliques length (SL), and number of seeds per silique (NSS) of these mutants in the T1 and T2 generations (2018 and 2019, respectively). We also noticed that BnaA10.tfl1 (L69), BnaA02.tfl1 (L98), BnaC03.tfl1 (K70) and BnaC03.tfl1/BnaC09.tfl1 (K67) had much less branches than the wild type, while BnaC09.tfl1 had similar number of branches to the wild type (Fig. 5f and g). In addition, all the knockout mutants had less siliques on the main inflorescence (NSMI), shorter siliques and less seeds per silique than the wild type (Fig. 5h - m). These results indicated that the yield-related traits in all the Bnatfl1s mutants were affected at different degrees, suggesting that these BnaTFL1 gene copies play roles in the determination of plant architecture.
Discussions
Arabidopsis has only one TFL1 gene which functions in the control of flowering time and floral architecture. While polyploids or paleopolyploid species usually have multiple TFL1 gene copies with distinct expression patterns and divergent functions in the control of flowering time and floral architecture [49]. Of three TFL1 homologs isolated in pea, PsTFL1a corresponds to DETERMINATE (DET) gene and PsTFL1c corresponds to the LATE FLOWERING (LF) gene. DET specifically expresses in the shoot apex after floral initiation and acts to maintain the indeterminacy of the apical meristem during flowering, while LF controls the length of the vegetative phase by delaying floral initiation [50]. In this study, we identified five BnaTFL1 gene copies in the genome of allotetraploid B. napus. These BnaTFL1 gene copies together with TFL1 gene copies from the ancestor species B. rapa and B. oleracea were classified into three clusters (Fig. 1b). The five BnaTFL1s originated from their corresponding ancestral gene copies in B. rapa and B. oleracea and had distinct expression patterns, suggesting that they may have different roles in regulating the flowering time and/or maintaining the inflorescence meristem in B. napus. In the knockout mutants, only BnaC03.tfl1 single mutant and BnaC03.tfl1/BnaC09.tfl1 double mutant exhibited earlier flowering, indicating that BnaC03.TFL1 is involved in regulating flowering time (Fig. 4) [17, 23]. And consistent with this role, BnaC03.TFL1 is preferentially expressed in flower bud and flower tissues (Fig. 2a). In the double mutant BnaC03.tfl1/BnaC09.tfl1, BnaC09.tfl1 did not enhance the early flowering phenotype of BnaC03.tfl1 (Fig. 4a, b and c), suggesting that BnaC09.TFL1 is not redundant to BnaC03.TFL1 because double mutants usually have stronger phenotypes than single mutants. Except for BnaC03.tfl1 and BnaC03.tfl1/BnaC09.tfl1 mutants, the knockout mutants including BnaA02.tfl1, BnaA10.tfl1 and BnaC09.tfl1 did not exhibit early flowering, suggesting that these BnaTFL1 gene copies may be not involved in the control of flowering time. However, we could not exclude the possibility that BnaA02.TFL1, BnaA10.TFL1 and BnaC09.TFL1 redundantly regulate flowering time. Unfortunately, we failed to obtain single mutant of BnaC02.TFL1, and the double and triple mutants of BnaA10.TFL1, BnaA02.TFL1, and BnaC02.TFL1, and their redundancy in controlling flowering time are needed to further investigate.
In addition to early flowering, the knockout mutants of BnaC03.TFL1 (K67 and K70) also displayed altered plant architecture. The plant height, branch initiation height and yield related traits including branch number, silique length, number of seeds per silique and number of siliques on the main inflorescence in the BnaC03.TFL1 mutants were significantly reduced when compared to the wild type. These results demonstrated that, similar to Arabidopsis TFL1, BnaC03.TFL1 plays important roles in determining flower time and plant architecture, which is consistent with the phylogenetic tree in which BnaC03.TFL1 is more closely related to Arabidopsis TFL1 (Fig. 1b). However, we could not exclude that the alteration of plant architecture in BnaC03.tfl1 and BnaC03.tfl1/BnaC09.tfl1 is caused by altered flowering time. Besides BnaC03.TFL1, the mutation of BnaA02.TFL1, BnaA10.TFL1 and BnaC09.TFL1 also altered plant architecture. The plant height, branch initiation height and yield related traits including number of branches, number of siliques on the main inflorescence, silique length and number of seeds per silique in these BnaTFL1s mutants were decreased at different degrees when compared to the wild type. In all of the BnaTFL1s mutants, BnaC03.tfl1 exhibited the strongest alteration of plant architecture. The altered plant architecture was also observed in BnaA10.tfl1 mutants in previous studies [32, 33]. An EMS mutant of BnaA10.TFL1 has altered yield component traits [32]. Bnsdt1, a natural mutant of BnaA10.TFL1, has reduced plant height, but displayed normal flowering time, yield and yield-related traits including silique number, silique density and seeds per silique as the wild type [33, 51]. It is speculated that the lowered expression of BnaA10.TFL1 in Bnsdt1 results in the determinate inflorescence but does not affect yield-related traits and seed yield [33, 51]. These results indicated that BnaA02.TFL1, BnaA10.TFL1 and BnaC09.TFL1, together with BnaC03.TFL1, are important key regulatory genes involved in the control of floral architecture. Although the knockout mutants of BnaTFL1s in this study had deteriorated yield-related traits, it provides us promising targets for plant architecture improvement in rapeseed. We could manipulate the expression level of these BnaTFL1s to obtain mutant lines with determinate inflorescence and similar or better yield and yield-related trait performance as the natural variation Bnsdt1 [51].
Conclusions
In this study, we identified five TFL1 gene copies in the B. napus genome. These gene copies display different expression patterns, suggesting that the functions of the BnaTFL1 genes have sub-functionalized. Knock-out mutants generated by CRISPR/Cas9 technology indicated that BnaC03.TFL1 negatively regulates flowering time in B. napus. In addition, all the BnaTFL1 gene copies are involved in the control of plant architecture related traits including plant height, branch initiation height and branch number. Our findings provide a base for future understanding the functions of BnaTFL1 genes and for modification of plant architecture of rapeseed.
Methods
Identification of TFL1 genes in Brassica species
The sequences of Arabidopsis TFL1 gene was obtained from the TAIR database (http://www.arabidopsis.org) and used as queries for a BLASTP algorithm-based against the B. napus database (http://rice.hzau.edu.cn/cgi-bin//bnapus/gb2/gbrowse/ZS11v0/), and Brassica database (http://brassicadb.org/brad/blastPage.php) with a p-value cutoff of 0.001 to retrieve homologous genes from the B. napus, B. rapa, and B. oleracea, respectively. For all candidate genes, we also examined whether they contain the PEBP domain in the SMART (http://smart.embl-heidelberg.de) and Pfam (http://pfam.sanger.ac.uk) databases. Sequences without a PEBP domain were deleted. Protein information including the length of amino acids (a.a.) was examined using the online web tool Expasy (ProtParam) (https://web.expasy.org/protparam/).
Conserved motifs and gene structure analysis of TFL1 proteins in Brassica species
Conserved motifs in TFL1 proteins of Brassica species were identified using the program SMART (Pfam) (http://smart.embl-heidelberg.de/), HMMAR and MEME (http://meme-suite.org/) by using default parameters [52]. The TFL1 gene structure was predicted using the program of GSDS2.0 (Gene Structure Display Server, http://gsds.cbi.pku.edu.cn/) for both genome and coding domain sequences.
Phylogenetic tree construction and protein conserved domain sequence alignment
The TFL1 amino acid sequences of B. napus together with A. thaliana, B. rapa and B. oleracea were aligned using Clustalx 2.0 with default settings. The phylogenetic tree was constructed with MEGA7.0 software using the neighbor-joining method [53]. The bootstrap test was executed by 1000 replications. The resulting phylogenetic tree was prepared in MEGA7.0 software.
Vector construction and plant transformation
The genomic sequence of BnaTFL1 genes were subjected to the online CRISPR-P software (http://crispr.hzau.edu.cn/CRISPR2/) to search for guide RNA (gRNA) targets. Two gRNAs were designed in the conserved phosphatidylethanolamine-binding protein (PEBPs) domain for each TFL1 gene. The gRNA sequences (20 bp) were followed by NGG (PAM, protospacer adjacent motif) at the 3′ end of the forward or reverse strands. Finally, the two AtU6 promoter-sgRNA-AtU6 terminator cassettes in template plasmid pCBC-DT1T2 were amplified using the primers shown in Table S1. The PCR fragments were inserted into the pKSE401 vector by Golden Gate Assembly [54].
For plant transformation, a commonly used B. napus cultivar, Westar (An original Canada rapeseed cultivar introduced by our lab) was used as the transformation host in this study [55]. The agrobacterium strain, GV3101 harboring pKSE401-sgRNA vector containing sgRNAs was used to infect the hypocotyls of B. napus as previously described [43]. The transgenic calli resistant to kanamycin (50 mg/ml) were allowed to grow until the development of roots and shoots under controlled temperature of 23 °C and a photoperiod of 16 h/8 h (day/night). The kanamycin resistant plants (Kan+) were then transferred to soil in the greenhouse for seed harvesting.
ACT-PCR assay
The ACT-PCR assay was performed as previous described [48]. One primer target on the sgRNA sites, and the other primer targets on the gene specific region. Genomic DNA was extracted from young leaves of transgenic T0 plants using the cetyltrimethyl ammonium bromide (CTAB) method. The positive transgenic lines were first screened by Cas9 specific primers. Then, the ACT-PCR program was followed as 94 °C for 5 min, 35 cycles at 94 °C for 30 s, gradient 60 °C to 66 °C for 30 s, and 72 °C for 40 s, and 10 min at 72 °C for final extension. All primers were listed in Table S1.
The genotyping of transgenic lines
To analyze the mutations caused by CRISPR/Cas9, genomic DNA was extracted from each transgenic plant using the CTAB method. The flanking sequence around the CRISPR target sites was amplified by PCR using gene-specific primers. In the T0 and T1 generations, most of the amplicons were directly sequenced to analyze the mutations by DSDECODE (http://skl.scau.edu.cn/dsdecode). For the complex mutations, the amplicons were first sub-cloned into the pGEM-T easy vector, and about 10 clones of each amplicon were individually sequenced by Sanger sequencing. All primers were listed in Table S1.
In the T2 generation, the mutation sites were genotyped by next-generation sequencing of target amplicons [56], which includes two rounds of PCR amplification. The first round PCR profiles were 94 °C for 5 min, 30 cycles at 94 °C for 30 s, gradient 57 °C to 59 °C for 30 s, and 72 °C for 40 s, and 10 min at 72 °C for final extension. The second round profiles were 95 °C for 3 min, 20 cycles at 95 °C for 15 s, 65 °C for 30 s, and 72 °C for 15 s, and 5 min at 72 °C for final extension. All primers were listed in Table S1. PCR products were sequenced by the GenoSeq Company, Wuhan, China.
Survey of flowering time and architecture traits
The flowering time of the T1 and T2 mutant plants were recorded as the days from the sowing to the appearance of first flower on the main inflorescence. Flowering time and architecture traits including plant height (PH), branches number (BN), branch initiation height (BIN), number of siliques on the main inflorescence (NSMI), siliques length (SL), and number of seeds per siliques (NSS) were measured with at least 20 plants per mutant line. Plant height (PH) was measured as the length of the plant from the base of the stem to the tip of the main inflorescence. Branches number (BN) was measured as all the number of branches arising from the main inflorescence. Branch initiation height (BIN) was measured as the length from the base of the stem to the first primary branch base. Numbers of siliques on the main inflorescence (NSMI) was measured all siliques on the main inflorescence. Siliques length (SL) and number of seeds per siliques (NSS) were measured based on twenty well-developed siliques from of the main inflorescence. Statistical analyses were performed using the student’s t-test with R-software to compare the differences of phenotypes between the mutant and wild-type plants at the P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001probability levels.
RNA extraction and RT-PCR
Total RNA from different plant tissues were extracted using TRIzol reagent (Aidlab, Wuhan, China) following the manufacturer’s instructions. Approximately 2 μg of total RNA was used for cDNA synthesis using a PrimeScript RT reagent kit (ThermoFisher Scientific). Reverse transcription PCR (RT-PCR) programme was 94 °C for 3 min, 35 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and 5 min at 72 °C for final extension. The BnActin gene (GenBank: AF111812.1) served as the internal control. All primers were listed in Table S1.
Supplementary information
Additional file 1 Table S1 List of PCR primers and their applications.
Additional file 2 Table S2 list of 12 TFL1 genes identified in B. napus, B. oleracea, B. rapa, and A. thaliana.
Additional file 3 Table S3 Percentage of mutated plants in the T0 generation in Brassica napus.
Additional file 4 Table S4 Percentage of mutated plants in the T0 generation in Brassica napus.
Additional file 5 Table S5 The mutation of sgRNAs (sgRNA1 and sgRNA2) of T0 plants found with mutations in the target sequence.
Additional file 6 Table S6 The mutation of sgRNAs (sgRNA3 and sgRNA4) of T0 plants found with mutations in the target sequence.
Additional file 7 Table S7 The detection of CRISPR/Cas9 mediated mutations in the T1 generation.
Additional file 8 Table S8 The detection of CRISPR/Cas9 mediated mutations in the T2 generation.
Additional file 9 Table S9. Genotypic analysis of BnaTFL1 mutants and their transmission to T1 and T2 generations.
Additional file 10 Table S10 The survey and student t-test resulted for days to flowering and architecture of bnatfl1 in T1 (2018) generation.
Additional file 11 Table S11 The survey and student t-test resulted for days to flowering and architecture of bnatfl1 in T2 (2019) generation.
Additional file 12 Fig. S1 The promoter sequences alignment between five BnaTFL1s; BnaTFL1.A02, BnaTFL1.A10, BnaTFL1.C02, BnaTFL1.C03 and BnaTFL1.C09.
Acknowledgements
We thank Prof. Chen Qijun at China Agricultural University for providing the CRISPR/Cas9 vector.
Abbreviations
- TFL1
Terminal Flower 1
- FT
Flowering Locus T
- SOC1
Suppressor of Overexpression of Constans1
- FLC
Flowering Locus C
- LEF
Leafy
- AP1
Apetala1
- Cen
Centroradialis
- RCN1
Rice Centroradialis 1
- RCN2
Rice Centroradialis 2
- Dt1
Determinate stem
- DET
Determinate
- LF
Late flowering
- Bnsdt1
Brassica napus Determinate
- Bna
Brassica napus
- Bra
Brassica rapa
- Bol
Brassica oleracea
- GA
Gibberellin acid
- QTL
Quantitative trait locus
- PEBPs
Phosphatidylethanolamine binding proteins
- MEGA
Molecular Evolutionary Genetics Analysis
- GSDS
Gene Structure Display Server
- Pfam
Protein families database
- CRISPR/Cas9
Clustered Regulatory Interspersed Short Palindromic Repeat associated protein
- SMART
Simple Modular Architecture Research Tool
- TAIR
The Arabidopsis Information Resource
- aa
Amino acid
- MEME
Multiple Expectation Maximization for Motif Elicitations
- ACT-PCR
Annealing at critical temperature Polymerase Chain Reaction
- sgRNA
Single-guide RNA
- CTAB
Cetyl trimethylammonium bromide
- DNA
Deoxyribonucleic acid
- mRNA
Messenger RNA
- DTF
Days to flowering
- PH
Plant height
- BN
Branches number
- BIN
Branch initiation height
- NSMI
Number of siliques on the main inflorescence
- SL
Silique length
- NSS
Number of seed per siliques
- WT
Wild type
- DH
Double haploid
- RT-PCR
Reverse transcription polymerase chain reaction
- ORFs
Open reading frames
- EMS
Ethyl methanesulfonate
Authors’ contributions
S.S. conceived the research and performed the experiments. H.T.L. helped to perform the experiments. C.C.G. conducted next-generation sequencing analysis. T.S. helped to conducted phenotype analysis. C.D. and K.D.L. designed the experiments. S.S., C.D., and K.D.L. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0101007). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The data sets supporting the results of this article are included in this manuscript and its additional information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12863-020-00857-z.
References
- 1.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 2.Zeevaart JAD. Physiology of flower formation. Annu Rev Plant Physiol. 1976;27:321–348. [Google Scholar]
- 3.Pharis RP, King RW. Gibberellins and reproductive development in seed plants. Annu Rev Plant Physiol. 1985;36:517–568. [Google Scholar]
- 4.Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, et al. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science. 2014;345:950. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Long Y, Zhang L, Dalton-Morgan J, Batley J, Yu L, et al. Genome wide analysis of flowering time trait in multiple environments via high-throughput genotyping technique in Brassica napus L. PLoS One. 2015;10:e0119425. doi: 10.1371/journal.pone.0119425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long Y, Shi J, Qiu D, Li R, Zhang C, Wang J, et al. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics. 2007;177:2433–2444. doi: 10.1534/genetics.107.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell. 2004;16:S18. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickland Daniel P, Hanzawa Y. The flowering locus t/terminal flower 1 gene family: functional evolution and molecular mechanisms. Mol Plant. 2015;8:983–997. doi: 10.1016/j.molp.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Wolabu TW, Zhang F, Niu L, Kalve S, Bhatnagar-Mathur P, Muszynski MG, et al. Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 2016;210:946–959. doi: 10.1111/nph.13834. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Luo L, Fu Q, Niu L, Xu Z-F. Identification and characterization of the FT/TFL1 gene family in the biofuel plant Jatropha curcas. Plant Mol Biol Report. 2015;33:326–333. [Google Scholar]
- 11.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 12.Ding L, Wang Y, Yu H. Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the Orchid Dendrobium Chao Parya smile. Plant Cell Physiol. 2013;54:595–608. doi: 10.1093/pcp/pct026. [DOI] [PubMed] [Google Scholar]
- 13.Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- 14.Goslin K, Zheng B, Serrano-Mislata A, Rae L, Ryan PT, Kwaśniewska K, et al. Transcription factor interplay between LEAFY and APETALA/CAULIFLOWER during floral initiation. Plant Physiol. 2017;174:1097. doi: 10.1104/pp.17.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci U S A. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Zhou Z, Liu Y, Liu T, Li Q, Ji Y, et al. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell. 2015;27:323–336. doi: 10.1105/tpc.114.135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Fan C, Zhang X, Wang X, Wu F, Hu R, et al. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS One. 2014;9:e99642. doi: 10.1371/journal.pone.0099642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, et al. Activation tagging of the floral inducer FT. Science. 1999;286:1962. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 20.Yasushi Kobayashi HK, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 21.Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci. 2009;106:8392. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann K, Venail J, Berbel A, Domenech MJ, Money T, Conti L, et al. Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture. J Exp Bot. 2015;66:4769–4780. doi: 10.1093/jxb/erv247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conti L, Bradley D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell. 2007;19:767–778. doi: 10.1105/tpc.106.049767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon R, Igeño MI, Coupland G. Activation of floral meristem identity genes in Arabidopsis. Nature. 1996;384:59. doi: 10.1038/384059a0. [DOI] [PubMed] [Google Scholar]
- 25.Shannon S, Meeks-Wagner DR. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell. 1991;3:877. doi: 10.1105/tpc.3.9.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz EA, Haughn GW. Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development. 1993;119:745. [Google Scholar]
- 27.Jensen CS, Salchert K, Nielsen KK. A TERMINAL FLOWER1-Like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol. 2001;125:1517. doi: 10.1104/pp.125.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 2011;23:3172. doi: 10.1105/tpc.111.088641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. doi: 10.1038/379791a0. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, et al. The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010;153:198–210. doi: 10.1104/pp.109.150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y, Hans H, Christian J, Molina C. Mutations in single FT- and TFL1-paralogs of rapeseed (Brassica napus L.) and their impact on flowering time and yield components. Front. Plant Sci. 2014;5:282. doi: 10.3389/fpls.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li K, Yao Y, Xiao L, Zhao Z, Shaomin G, Fu Z, et al. Fine mapping of the Brassica napus Bnsdt1 gene associated with determinate growth habit. Theor Appl Genet. 2017;131:193–208. doi: 10.1007/s00122-017-2996-z. [DOI] [PubMed] [Google Scholar]
- 34.Chen K, Gao C. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 2014;33:575–583. doi: 10.1007/s00299-013-1539-6. [DOI] [PubMed] [Google Scholar]
- 35.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khatodia S, Bhatotia K, Passricha N, Khurana SMP, Tuteja N. The CRISPR/Cas genome-editing tool: application in improvement of crops. Front Plant Sci. 2016;7:506. doi: 10.3389/fpls.2016.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L, Fan X-D. CRISPR–Cas system: a powerful tool for genome engineering. Plant Mol Biol. 2014;85:209–218. doi: 10.1007/s11103-014-0188-7. [DOI] [PubMed] [Google Scholar]
- 38.Macovei A, Sevilla NR, Cantos C, Jonson GB, Slamet-Loedin I, Čermák T, et al. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to rice tungro spherical virus. Plant Biotechnol J. 2018;16:1918–1927. doi: 10.1111/pbi.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, et al. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao A, Chen H, Chen L, Chen S, Hao Q, Guo W, et al. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019;19:131. doi: 10.1186/s12870-019-1746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barman HN, Sheng Z, Fiaz S, Zhong M, Wu Y, Cai Y, et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019;19:109. doi: 10.1186/s12870-019-1715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, Liu C, Zhao R, Wang L, Chen L, Yu W, et al. CRISPR/Cas9-mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019;19:38. doi: 10.1186/s12870-018-1627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Wu J-J, Tang T, Liu K-D, Dai C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep. 2017;7:7489. doi: 10.1038/s41598-017-07871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braatz J, Harloff H-J, Mascher M, Stein N, Himmelbach A, Jung C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus) Plant Physiol. 2017;174:935. doi: 10.1104/pp.17.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Q, Lin L, Liu D, Wu D, Fang Y, Wu J, et al. CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in brassica napus L. Int J Mol Sci. 2018;19:2716. doi: 10.3390/ijms19092716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng M, Zhang L, Tang M, Liu J, Liu H, Yang H, et al. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.) Plant Biotechnol J. 2019;18:644. doi: 10.1111/pbi.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuzaki A, Ogawa T, Koizuka C, Kaneko K, Inaba M, Imamura J, et al. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol Biochem. 2018. [DOI] [PubMed]
- 48.Hua Y, Wang C, Huang J, Wang K. A simple and efficient method for CRISPR/Cas9-induced mutant screening. Genet Genom. 2017;44:207–213. doi: 10.1016/j.jgg.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Liu Z, Adams KL. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr Biol. 2007;17:1669–1674. doi: 10.1016/j.cub.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 50.Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, et al. DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell. 2003;15:2742–2754. doi: 10.1105/tpc.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia Y, Li K, Liu H, Zan L, Du D. Characterization of the BnA10.tfl1 gene controls determinate inflorescence trait in Brassica napus L. Agronomy. 2019;9:722. [Google Scholar]
- 52.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucl Acids Res. 2006;34:247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Gao X, Yan P, Shen W, Li X, Zhou P, Li Y. Modular construction of plasmids by parallel assembly of linear vector components. Anal Biochem. 2013;437:172–177. doi: 10.1016/j.ab.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 55.Klassen AJ, Downey RK, Capcara JJ. WESTAR SUMMER RAPE. Can J Plant Sci. 1987;67:491–493. [Google Scholar]
- 56.Campbell NR, Harmon SA, Narum SR. Genotyping-in-thousands by sequencing (GT-seq): a cost effective SNP genotyping method based on custom amplicon sequencing. Mol Ecol Resour. 2015;15:855–867. doi: 10.1111/1755-0998.12357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1 List of PCR primers and their applications.
Additional file 2 Table S2 list of 12 TFL1 genes identified in B. napus, B. oleracea, B. rapa, and A. thaliana.
Additional file 3 Table S3 Percentage of mutated plants in the T0 generation in Brassica napus.
Additional file 4 Table S4 Percentage of mutated plants in the T0 generation in Brassica napus.
Additional file 5 Table S5 The mutation of sgRNAs (sgRNA1 and sgRNA2) of T0 plants found with mutations in the target sequence.
Additional file 6 Table S6 The mutation of sgRNAs (sgRNA3 and sgRNA4) of T0 plants found with mutations in the target sequence.
Additional file 7 Table S7 The detection of CRISPR/Cas9 mediated mutations in the T1 generation.
Additional file 8 Table S8 The detection of CRISPR/Cas9 mediated mutations in the T2 generation.
Additional file 9 Table S9. Genotypic analysis of BnaTFL1 mutants and their transmission to T1 and T2 generations.
Additional file 10 Table S10 The survey and student t-test resulted for days to flowering and architecture of bnatfl1 in T1 (2018) generation.
Additional file 11 Table S11 The survey and student t-test resulted for days to flowering and architecture of bnatfl1 in T2 (2019) generation.
Additional file 12 Fig. S1 The promoter sequences alignment between five BnaTFL1s; BnaTFL1.A02, BnaTFL1.A10, BnaTFL1.C02, BnaTFL1.C03 and BnaTFL1.C09.
Data Availability Statement
The data sets supporting the results of this article are included in this manuscript and its additional information files.