Abstract
Background
HIV-1 can develop resistance to antiretroviral drugs, mainly through mutations within the target regions of the drugs. In HIV-1 protease, a majority of resistance-associated mutations that develop in response to therapy with protease inhibitors are found in the protease’s active site that serves also as a binding pocket for the protease inhibitors, thus directly impacting the protease-inhibitor interactions. Some resistance-associated mutations, however, are found in more distant regions, and the exact mechanisms how these mutations affect protease-inhibitor interactions are unclear. Furthermore, some of these mutations, e.g. N88S and L76V, do not only induce resistance to the currently administered drugs, but contrarily induce sensitivity towards other drugs. In this study, mutations N88S and L76V, along with three other resistance-associated mutations, M46I, I50L, and I84V, are analysed by means of molecular dynamics simulations to investigate their role in complexes of the protease with different inhibitors and in different background sequence contexts.
Results
Using these simulations for alchemical calculations to estimate the effects of mutations M46I, I50L, I84V, N88S, and L76V on binding free energies shows they are in general in line with the mutations’ effect on values. For the primary mutation L76V, however, the presence of a background mutation M46I in our analysis influences whether the unfavourable effect of L76V on inhibitor binding is sufficient to outweigh the accompanying reduction in catalytic activity of the protease. Finally, we show that L76V and N88S changes the hydrogen bond stability of these residues with residues D30/K45 and D30/T31/T74, respectively.
Conclusions
We demonstrate that estimating the effect of both binding pocket and distant mutations on inhibitor binding free energy using alchemical calculations can reproduce their effect on the experimentally measured values. We show that distant site mutations L76V and N88S affect the hydrogen bond network in the protease’s active site, which offers an explanation for the indirect effect of these mutations on inhibitor binding. This work thus provides valuable insights on interplay between primary and background mutations and mechanisms how they affect inhibitor binding.
Keywords: Alchemical binding free energy change calculation, Distant site mutations, HIV-1 protease inhibitors, Hydrogen bond network perturbation, Resistance-associated mutations
Background
With around 36.7 million people already infected and 1.8 million being newly infected per year, the human immunodeficiency virus type 1 (HIV-1) (further HIV) remains a global epidemic [1]. Since more than half of the infected individuals receive antiretroviral therapy (ART) [2], acquired immune deficiency syndrome (AIDS)-related deaths have dropped to 1 million per year [1]. For those under treatment, resistance towards drugs is a major cause for the need for switching of the therapy.
HIV protease (Fig. 1), a protein responsible for cleaving HIV polyproteins, is one of the major targets for ART, and protease inhibitors (PIs) are currently recommended as second- or third-line ART treatments [3]. PIs are competitive binders of the protease, occupying the active site of the protein once bound. In line with this, most of the major resistance-associated mutations (RAMs) towards PIs appear in the different structural elements composing the active site pocket, such as in the active site loop (residues D30, V32, and L33), the so-called 80s loop (residues V82 and I84) which together form the sides of the pocket, and the flap region of the protease (residues M46, I47, G48, I50, and I54, Fig. 1). Yet several RAMs are also found in distant to the binding pocket sites, e.g. in the amino acids N88 and L90 in the protease’s -helix or L76 in the protein’s hydrophobic core. The effect of these mutations on inhibitor binding is likely to be not through direct interactions with PIs.
Fig. 1.

HIV protease structure. Flap region in cyan, 80s loop in brown, active-site proximate loop in olive colours. Mutations analysed in this study (red), catalytic site residue (blue) and bound inhibitor (magenta) are shown in sticks model
Some mutations exhibit opposite effects on binding of some PIs, e.g. L76V is associated with resistance towards Amprenavir (APV), Indinavir (IDV), Darunavir (DRV), and Lopinavir (LPV), but increases sensitivity towards Atazanavir (ATV) and Saquinavir (SQV) [4, 5]. Similarly, N88S is a RAM towards IDV and Nelfinavir (NFV), but increases susceptibility towards APV [6–8] or its prodrug Fosamprenavir (FPV) [9].
Numerous studies have addressed the molecular effects of RAMs. Some studies analyse the effects of selected major RAMs on binding of different inhibitors [10–17] and others the effect of different RAMs on binding of the same inhibitor [18–27]. Most of the studies are however focused on a single mutation-inhibitor combination, particularly for major RAMs outside of the binding pocket, and thus offer only a limited perspective on molecular mechanisms of the protease resistance. To the best of our knowledge, the mechanism of action of a mutation on binding of different inhibitors has not been investigated in the aforementioned cases, where the same mutation is known to cause resistance to certain inhibitors, while making the protein sensitive towards other inhibitors [4–9]. In clinical practice of HIV treatment, cases like this potentially provide an opportunity for combined treatment: a combination of PIs that are associated with opposite effects of a particular mutation put the virus in a situation where either mutation variant in this position will render the protease susceptible to one of the drugs [5, 6, 28]. Understanding the underlying molecular phenomena could potentially provide important insights into HIV inhibitor resistance, as well as a possibility to transfer this knowledge to treatment of other viruses, and for inhibitor design.
In an experimental setting, resistance of mutant proteins towards PIs, such as in the studies above, is typically measured in terms of (concentration required to inhibit viral activity by 50%). Thus, the ratio between in mutant and the same measurement for the wildtype protease (typically with the consensus sequence from the strain HXB2), also called resistance factor (RF), is a useful descriptor for resistance of different mutated proteins. RF is directly related to the free energy of inhibitor binding, , and the protease enzymatic activity, [29].
We have previously shown that the effect of mutations in the HIV protease on inhibitor binding, , can be accurately predicted in silico using alchemical methods based on molecular dynamics (MD) simulations [17]. Additionally, analysis of the underlying trajectories from the MD simulations can also reveal the mechanisms underlying the effects of mutations on protein-inhibitor interactions, protein structural changes, as well as on their altered dynamics.
In this study, we apply alchemical calculations based on MD simulations to estimate the effect of RAMs M46I, I50L, I84V, N88S, and L76V, both in the binding pocket and outside of it, on the binding of PIs APV, IDV, LPV, and SQV: in total 19 different PI-mutation combinations. We demonstrate that we can faithfully reproduce the previously reported effects of the first four mutations on the resistance, including previously investigated sensitising and desensitising effects of the mutation N88S on binding of APV and IDV [6, 8]. We show that changes of the hydrogen bonding network of the protease that involve D30, located in the active site pocket, can explain these effects.
The data on M46I, I50L, I84V, and N88S were acquired from the HIVdb database [30]. Measurements can be paired such that the protease sequences are identical with the exception of the mutated site. Moreover, for all of these mutations wildtype and mutant protein background sequences were identical across the different inhibitors, with the exception of N88S, where complexes with FPV had a background mutation L77I and complexes with IDV had a R57G background mutation present. Both of R57G and L77I mutations are found next to each other on two parallel -strands at a distant site of the protease and close to major RAM sites I54 and L76, respectively. However, unlike in case of the latter residues, the side chains of residues 77 and 57 are pointing away from the protease binding pocket. Nevertheless, the mutation R57G has been suggested to be a protease-inactivating mutation [31]. L77I on the other hand, has been reported to be a compensatory mutation for I84V, restoring protein stability [32]. M46I, I84V, and N88S are all considered to be major RAMs against the corresponding inhibitors as reported in the HIVdb. In addition to that N88S has been previously reported to increase susceptibility against APV/FPV [6–9]. I50L on the other hand has been reported to induce resistance against ATV while increasing sensitivity against remaining PIs [33–36].
For L76V, located in the protease’s hydrophobic core, we investigate its effect on binding of ATV, IDV, LPV, and SQV in different clinically relevant sequence contexts. In the phenotypic assays we consistently observe a resensitising effect of this mutation towards ATV and SQV as well as its resistance-associated effect for IDV and LPV, which was reported previously [4, 5], and we generally reproduce these effects computationally. We suggest a mechanistic explanation of this effect, demonstrating that these mutations affect the arrangement of residues around the binding pocket of the protease, which, in turn, similarly to N88S, affects the hydrogen bonding network of D30, directly influencing inhibitor binding.
Results and discussion
Estimation of resistance factors from the change in the inhibitor binding free energy
We aimed to assess whether we can reproduce the ratios of experimentally measured resistance factors () between the wildtype and mutant proteins by estimating the change in free energy of inhibitor binding upon mutations in the protease using MD simulation with alchemical methods [37]. For this purpose we selected a dataset of 20 complexes from HIVdb database [30] for which resistance factors towards inhibitors FPV, IDV, LPV, and SQV had been measured experimentally (Table 1 and Additional file 1: Table S1). These complexes could be paired amongst each other such that the RF has been measured for the same inhibitor and the same protease strain with and without the mutation, namely: IDV and FPV with mutation M46I; IDV and FPV with mutation I50L; FPV, IDV, LPV, and SQV with mutation I84V; FPV and IDV with mutation N88S. For all of these pairs, protease with a RAM had a higher RF than the wildtype, except I50L reducing resistance towards FPV and IDV and N88S reducing resistance to FPV (Table 1).
Table 1.
RF values for different mutant and corresponding wildtype sequences from HIVdb [30]
| Mutation | Background polymorphisms | RF | ||||
|---|---|---|---|---|---|---|
| IDV | SQV | LPV | FPV | |||
| M46I | ||||||
| Wildtype [33] | V3I, S37N, A71V | 0.6, 1.0, 1.2 | – | – | 0.3, 0.6, 0.7 | |
| Mutant [83] | 4.4 | – | – | 2.2 | ||
| I50L | ||||||
| Wildtype [33] | V3I, S37N, A71V | 0.6, 1.0, 1.2 | – | – | 0.3, 0.6, 0.7 | |
| Mutant [33] | 0.1, 0.3, 0.3 | – | – | 0.2, 0.2, 0.3 | ||
| I84V | ||||||
| Wildtype [84] | V3I, L10F, S37N | 1.5 | 1.3 | 1.6 | 1.8 | |
| Mutant [84] | 2.1, 3.2 | 2.7, 3.7 | 6.2, 7.7 | 4.6, 8.4 | ||
| N88S | ||||||
| Wildtype [6] | V3I, S37N, L63P, R57G/L77Ia | 1.1 | – | – | 1.0 | |
| Mutant [6] | 2.6 | – | – | 0.1 | ||
First column indicates the mutation analysed, while the second column indicates background polymorphisms that are present in both wildtype and mutant sequences compared to the reference HIV sequence HXB2. Multiple RF measurements for the same protein are separated by comma. M46I wildtype and mutant measurements reported from different studies, but performed using the same susceptibility test method (Phenosense, Monogram, San Francisco, USA)
a Indicates that the R57G background mutation was found in sequences where RF for IDV was measured and L77I in sequences where RF for FPV was measured
We first estimated the effect of the mutation on the free energy of inhibitor binding, . For this purpose, we performed MD simulations for the protease mutants and wildtype in inhibitor-bound and unbound state, where in the bound state the simulations were performed in both alternative protonation states of the catalytic residues of the active site, D25 and D25, to account for asymmetry of this complex. This allows us to identify which protonation state is more likely for both wildtype and mutant complexes, as well as to increase the accuracy of the estimation, as we reported previously [17]. The resulting calculations (Table 2 and Additional file 1: Table S2) overall indicated a good agreement in discriminating resistant and sensitising effects of mutations on the protein–ligand binding, including the opposite effects of N88S towards IDV and APV. An exception to that is M46I, where the mutation had a modest effect on which was within the estimated error range. The mutation of this flap residue, whose side-chain points away from the protease binding pocket, has been associated with resistance towards different PIs, but it typically appears in combination with other RAMs and has been suggested to compensate the decreased catalytic activity of mutant proteases [38–43].
Table 2.
Change of the binding free energy () of inhibitors upon mutation
| Inhibitor | Mutation | |||
|---|---|---|---|---|
| APV | M46I | 3.14–7.33a | 13.13 | |
| IDV | M46I | 3.67–7.33 | 12.69 | |
| APV | I50L | 0.29–1a | 0.74 | |
| IDV | I50L | 0.08–0.5 | 0.62 | |
| APV | I84V | 2.56–4.67a | 33.75 | |
| IDV | I84V | 1.4–2.13 | 7.23 | |
| LPV | I84V | 3.88–4.81 | 6.09 | |
| SQV | I84V | 2.08–2.85 | 1.33 | |
| APV | N88S | 0.1a | 0.3 | |
| IDV | N88S | 2.27 | 41.12 |
All values in kcal/mol, and ± shows bootstrap error estimate. indicates value ranges calculated using from previously reported experimental RF measurements, where "a" stands for measurements for FPV, the prodrug of APV. indicates average calculated value from the distribution described in Eq. 4 with subscript and superscript corresponding to lower and upper bound of 95% credible interval, respectively
The possibility of mutations having an effect on the catalytic activity of the enzyme, , precludes direct comparison of the estimates of mutation effects on inhibitor binding and the corresponding to that mutation. In previous studies of resistance mutations of another enzyme of HIV, reverse transcriptase, was considered to approximate changes in [44–46]. This is at odds with the fact that the majority of the mutations for which this approximation was used, such as L100I, V106A, and Y188L, although not located directly in the active site, have been previously reported to affect the catalytic potential of the enzyme [47–51]. Although some studies show correlation between predicted relative drug binding free energy upon HIV protease mutation and the one approximated by measurements [52], RAMs affecting the catalytic activity of protease have also been reported [39, 40, 53, 54]. In the present study, similarly to the aforementioned computational studies, only the experimentally measured values for mutations are available for the enzyme and different inhibitors. To account for the changes in the binding free energy and , we developed a Bayesian method which combines multiple experimental measurements and estimates to calculate (see “Estimation of resistance factor change from free energy of inhibitor binding change”).
We then compared the estimated values to their experimental measurements (Table 2, Fig. 2). The increase of resistance towards inhibitors () was correctly predicted for M46I, I84V, and N88S (with IDV) mutations, as was the sensitising effect () of I50L towards FPV and IDV as well as N88S towards APV. The experimental values were within the corresponding calculated distributions based on the estimates (Additional file 1: Figure S1).
Fig. 2.

Predicted and experimental RF measurements. Each symbol corresponds to a unique sequence background and colour corresponds to inhibitor. In case of APV, measurements are for its prodrug FPV
While the structural effect of N88S mutation has been previously analysed for NFV, to the best of our knowledge, the opposite effects of this mutation on susceptibility towards APV and IDV have not been previously addressed. It has been suggested that substitution of asparagine with serine creates a hydrogen bond with the residue D30, which in turn affects the interaction between D30 and the inhibitor NFV [55]. A similar observation with regard to the N88S effect on the interaction with D30 has been previously made for L10F/N88S mutant with NFV as well as the unliganded N88S protease [56, 57]. Another mutation at this site that occurs in patients treated with NFV, N88D, has been reported to co-occur with mutation D30N, which coincides with losing water molecules that mediate this site’s interactions with residues T31 and T74 [58]. Seeking to verify whether these effects extend to complexes of N88S with APV and IDV, we performed hydrogen bond network analysis of mutant and wildtype complexes, where we measured the average number of hydrogen bonds over the course of the simulations. Indeed, similar effects were confirmed: S88 formed a hydrogen bond with D30 more frequently than N88, whereas N88 formed a hydrogen bond more frequenly to T31 and T74 compared to S88 (Additional file 1: Table S3).
Estimating resistance factor for clinical samples with the L76V mutation
Among patients in Germany undergoing HIV treatment, individuals who underwent multiple therapy failures against different PI-regimens were identified. Among the group of patients we described in the manuscript of Wiesman et al. [5], there were viral variants which displayed different resistance-determining mutations to ATV, SQV, IDV, and LPV. The extraordinary observation was that a specific amino acid change L76V increased resistance to LPV and IDV, while at the same time giving a clinically relevant re-sensitisation to SQV and ATV. Those variants were observed in the diagnostic procedure, sequenced, and subsequently tested in a phenotypic assay. We analysed variants which showed some of the highest changes in RF upon mutation to evaluate whether we can computationally estimate RF values as measured in the phenotypic assay (Table 3).
Table 3.
Protease RF values for L76V mutation
| Genotype | RF | ||||
|---|---|---|---|---|---|
| ATV | SQV | IDV | LPV | ||
| FB15 | |||||
| L76 | 63 | 74 | – | – | |
| V76 | 0.9, 2.3, 4.5 | 3.6, 4.6, 5.8 | – | – | |
| GH9 | |||||
| L76 | 90 | 18.6 | – | – | |
| V76 | 1.2, 1.9, 2.4, 3.2, 3.6 | 1.2, 1.3, 1.5 | – | – | |
| RU1 | |||||
| V76 | 2.7 | – | – | 157 | |
| L76 | 8.4, 9, 10 | – | – | 27, 46, 47 | |
| iZ2 | |||||
| V76 | 4.1 | – | 59 | 71 | |
| L76 | 7.8, 12, 30 | – | 2.2, 7.9, 10 | 5.5, 11, 12 | |
Multiple RF measurements for the same protein separated by comma. For each genotype the first row represents the wildtype position as in the original sample and the second row represents the mutation introduced at position 76
Just as for mutations M46I, I50L, I84V, and N88S, for the mutation L76V multiple RF measurements for different inhibitors in the same sequence context were available, thus enabling us to computationally predict the values. The sequences of the protease complexes analysed had a large number of background mutations accumulated compared to the reference sequence HXB2, making it difficult to find complexes in the Protein Data Bank (PDB) with sequences matching the studied genotypes. Thus in the protein modelling stage between 11 and 19 mutations had to be introduced to create protein models with sequences corresponding to those for which was measured (see “System preparation”). Including the target mutation L76V as well meant that up to 20% of protease residues had to be modelled in silico.
First, we estimated the effect of the mutation L76V on inhibitor binding in terms of the change of the binding free energy (Table 4 and Additional file 1: Table S4). The increase of the binding free energy, corresponding to the decrease in inhibitor affinity, was predicted for all complexes where mutations were observed to increase the protease RF (RU1 with LPV, iZ2 with IDV and LPV). The decrease of RF, on the other hand, did not always correspond to a negative value of : L76V was predicted to increase the affinity of inhibitor binding for inhibitors ATV and SQV only for the genotype GH9, but not for the genotype FB15, nor for inhibitor ATV in the context of the genotypes RU1 or iZ2. The genotypes FB15 and iZ2 lack the background mutation M46I (the former being wildtype at that position and the latter having mutation M46L), which has been suggested to co-occur with L76V to compensate for its compromising effect on the replication capacity of HIV [41, 42]. This suggests that in this case the dominant effect of the mutation L76V might be exerted through decreasing the protease’s catalytic activity . However for RU1, which has the M46I mutation, we cannot explain the positive value for complexes with ATV using this argument.
Table 4.
Change of the binding free energy () of inhibitors upon mutation L76V
| Inhibitor | Genotype | |||
|---|---|---|---|---|
| ATV | FB15 | 0.01–0.07 | 0.23 | |
| SQV | FB15 | 0.04–0.08 | 0.11 | |
| ATV | GH9 | 0.01–0.04 | 0.23 | |
| SQV | GH9 | 0.06–0.08 | 0.03 | |
| ATV | RU1 | 0.27–0.32 | 3.52 | |
| LPV | RU1 | 3–5 | 3.94 | |
| ATV | iZ2 | 0.14–0.53 | 35.61 | |
| IDV | iZ2 | 5.9–26.82 | 88.25 | |
| LPV | iZ2 | 5.91–12.9 | 10.95 |
All values in kcal/mol, and ± shows bootstrap error estimate. indicates value ranges calculated using from experimental RF measurements, indicates average calculated value from the distribution described in Eq. 4 with subscript and superscript corresponding to lower and upper bound of 95% credible interval, respectively
The estimates were used to calculate in the same fashion as for mutations M46I, I50L, I84V, and N88S (Table 4, Fig. 3). For most of the complexes we correctly predicted whether the mutation made the protein more resistant or more sensitive towards the inhibitor. This included the prediction of sensitising effect of mutation in the genotype FB15 for both ATV and SQV for which the inhibitor affinity increased based on the estimates. Sensitisation towards ATV was, on the other hand, not observed for genotypes RU1 and iZ2. The experimental values were however within the corresponding calculated distributions based on the estimates (Additional file 1: Figure S2). Overall, just like in the case for M46I, I50L, M84I, and N88S mutations, estimates converged roughly after half of simulation time (Additional file 1: Figure S3).
Fig. 3.

Predicted and experimental RF measurements. Each symbol corresponds to a unique sequence background and colours correspond to inhibitor
Effect of L76V on molecular interactions
Next, we focused on the structural changes in the protease upon the mutation L76V, for which purpose we first analysed the hydrogen bond network of the protein. It was consistently seen across all of the different genotypes that the mutation L76V increases the probability of observing a hydrogen bond between residues D30 and K45 (Table 5). Previous studies found a significant correlation between mutations in these sites [59, 60], potentially indicating the importance of the interaction between these two oppositely charged residues. Seeking whether this was a result of side chain rearrangement, we performed functional mode analysis (FMA) based on partial least-squares (PLS) regression, a supervised machine learning technique which builds models that distinguish behaviour in MD trajectories between the wildtype and mutant protease complex based on their Cartesian atoms coordinates. These models are interpretable in terms of protein conformational changes associated with the mutation. In analysing FMA models, we could see that the mutation L76V caused a tendency of the side chains of residues D30, K45, and Q/E58 to shift towards the binding pocket (Fig. 4 and Additional file 1: Figure S4). This shift is likely the result of fine rearrangement of residues in the region as a consequence of a larger side chain of leucine being replaced by a smaller valine. The effect of L76V on D30–K45 hydrogen bond is thus similar to the effect of the other distant site mutation we analysed in this study, N88S, on hydrogen bonding of D30. Mutations at site N88 have been reported to be correlated to mutations of D30 and K45 [59, 60]. Displacement of Q/E58, on the other hand, is in line with the previously reported co-occurance of mutations L76V and Q58E [61], and both of these mutations were found in the patient sample RU1. L76V has recently been reported to increase the distance between atoms of residues 16 and 62 on the surface of the protease when in complex with DRV [62]. The same effect on the structure was observed in both the resistance-inducing and the sensitising cases in that study, which is in line with the consistent observation of side-chain rearrangement we report here.
Table 5.
Average number of hydrogen bonds between residues D30 and K45 for protease wildtype and mutant complexes
| Drug | Genotype | D30–K45 | D30–K45 | ||
|---|---|---|---|---|---|
| L76 | V76 | L76 | V76 | ||
| ATV | FB15 | ||||
| SQV | FB15 | ||||
| ATV | GH9 | ||||
| SQV | GH9 | ||||
| ATV | RU1 | ||||
| LPV | RU1 | ||||
| ATV | iZ2 | ||||
| IDV | iZ2 | ||||
| LPV | iZ2 | ||||
± Indicates standard error of bond frequency across independent simulations
Fig. 4.
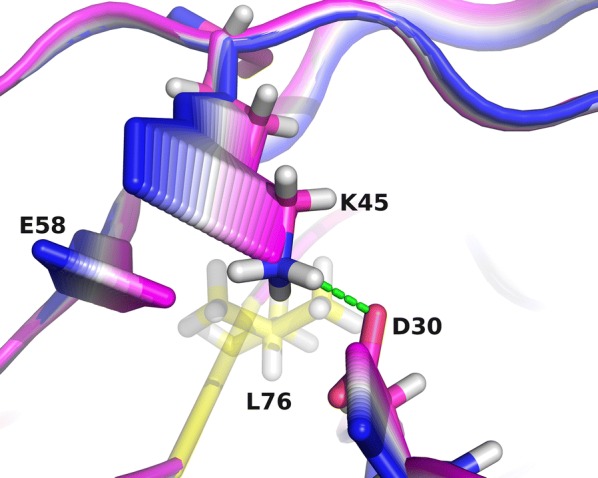
Interpolation between the extremes of the FMA models for the protease (genotype RU1) in complex with LPV. Blue-to-magenta bands correspond to the interpolation along the mode as represented as cartoon for backbone and as sticks for residues 30, 45, and 58, with blue corresponding to L76 state and magenta to V76 state. Green dashed line represents a hydrogen bond between residues D30 and K45. Mutated residue 76, here semi-transparent in yellow, as well as hydrogen atoms, here in gray, were not part of the FMA models and are here for representational purposes only
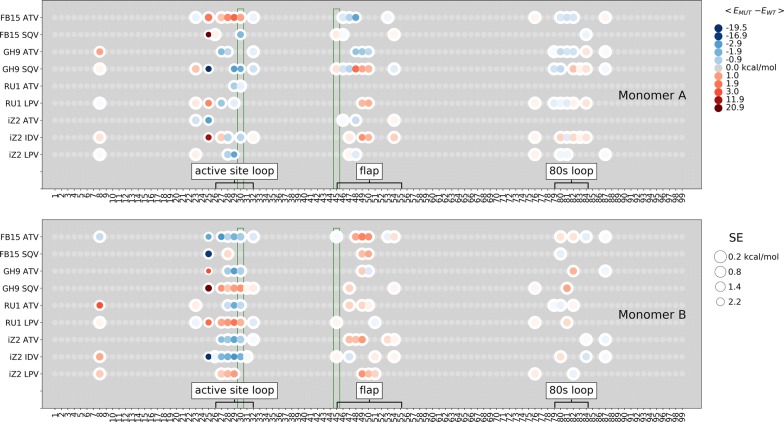
We calculated direct protein-inhibitor interaction energies to see whether the L76V mutation impacted direct interactions of D30, K45, or other residues with the inhibitors (Fig. 5 and Additional file 1: Figure S5). Indeed, changes in the interaction of D30/D30 with the inhibitors are in general in line with changes of : negative, or favourable, interaction energy values correspond to , and positive, or unfavourable, interaction energies correspond to . Exceptions to that are the proteases of the iZ2 genotype in complex with IDV, where a favourable effect on the direct interaction energy of D30/D30 with the inhibitor is observed, and in complex with LPV, where no notable effect on this interaction is seen. The effect of L76V on interaction energies between the inhibitors and K45/K45 was, on the other hand, modest. A number of other residues’ direct interactions with inhibitor were affected. These residues are widely distributed across most of the active site pocket, including the active site loop (including the D30), flap (including the K45), and 80s loop regions. This is in line with our observations from a previous study of the effect of mutations I50V, G48V, and L90M on protein-inhibitor interactions, where interactions of residues in these regions were also affected by the mutations [17]. Interestingly, measurable differences were observed for interactions of the residue at position 76 with the inhibitor for complexes of genotype FB15 with LPV and ATV and genotype iZ2 with IDV. But given that side chain of residue 76 has minimal exposure to the binding pocket, those differences are negligible.
Fig. 5.
Energy differences of non-bonded interaction between protein and inhibitor in wildtype and mutant complexes. Residues, for which the difference () between the wildtype and the mutant complexes is higher than the propagated error (SE) and its absolute value higher than 0.1 kcal/mol, are represented as a colored circle, where the color represents relative interaction energy and the size of the circle relates inversely to the standard error of the estimate. Residues’ 30 and 45 interactions are highlighted in green box
Overall, these results indicate a pathway for how the mutation L76V impacts the inhibitor binding through altering the interactions of other residues with the inhibitor without actual mutations at these sites. A similar observation has been previously made for another pair of an active site loop and distant site mutations, namely for mutation L90M that alters the interactions of the residue at this position with D25, which in turn affects the interactions of D25 with the inhibitor as well as with other residues in the binding pocket [17, 63, 64]. Our observations on energetic and structural consequences of the mutation L76V are also in line with its previously reported effects on the ligand binding affinity for the inhibitor DRV through both changes in protein-inhibitor interactions and changes in the inter-residue distances in the binding pocket [26].
Recently, a study reported experimentally resolved structures of the wildtype and the L76V mutant of the HIV protease in complex with inhibitors DRV, LPV, Tipranavir (TPV), as well as with two experimental compounds, GRL-0519 and GRL-5010 [65]. It has been observed that mutation does not change the backbone structure of the protease, however residue 76 loses contacts with D30 and T74, and, for structures with LPV, there is a slight shift of K45 towards the binding pocket in the mutant structure. Overall, similar interactions were reported between wildtype and mutant proteins with different inhibitors, with the exception of GRL-5010, which interacted with D30 in an altered way. These results thus partially support the observations made in our study on the effects of the L76V mutation on the structure and interactions within the HIV protease.
Conclusions
In this work, we analysed a set of mutations in the HIV protease associated with resistance towards PIs in complex with different clinically used inhibitors. First, we analysed four mutations with resistance factors extracted from the literature, where resistance factor measurements for the same sequence and the same inhibitors were available from multiple experiments. We showed that the effect of the mutation on the resistance factor, both increasing resistance and sensitising, was successfully reproduced using alchemical free energy calculations of affinity of inhibitor binding. Second, we modelled complexes for sequences based on our own clinical samples containing the mutation L76V with four PIs. Even though the sequences in question had a large number of background mutations, we could in most cases reproduce the resistant and sensitising effects of L76V. These calculations gave us insight into whether change in resistance is predominantly the result of change in inhibitor binding affinity or a change in the catalytic activity of the protease, for example for sequences which lacked the compensatory mutation M46I. Further analysis of L76V in different sequence contexts revealed that the effect of this mutation on direct protein residue-inhibitor interactions, including that of D30, is generally line with the changes in the resistance. Potentially causal to the observed changes is the favourable effect of the mutation on the hydrogen bond stability between residues D30 and K45 of the binding pocket. Analysis of another distant site mutation, N88S, also revealed changes in hydrogen bonding of the mutated residue with D30 as well as with T31 and T74, suggesting changes in hydrogen bonding network of the protease as a major pathway for how mutations outside of the binding pocket affect inhibitor binding.
Methods
Computational studies
System preparation
Crystal structures of protease-inhibitor complexes were obtained from the PDB [66] (IDs 1HPV (APV), 1HXB (SQV), 1K6C (IDV), 1MUI (LPV), 2BPX (IDV), 3EKV (APV), 3EL1 (ATV), 3PWR (SQV)). Modeller [67] version 9.12 was used to introduce mutations targeted in this study as well as the background mutations. The following background mutations were introduced in the studied protein from HIVdb dataset: K7Q, R14K, R57G, T82V, and V84I in 1K6C; K7Q, R14K, K41R, and V77I in 3EKV; L10F and S37N in 1MUI; L10F and S37N in 1HPV (84V); S37N and A71V in 1HPV (46V and 50L); L10F and S37N in 2BPX (84V); S37N and A71V in 2BPX (46M and 50L). For the phenotypic assay dataset the following background mutations were introduced: K7Q, I13V, G16E, K20I, I33F, M36L, S37N, I62V, I63H, A67C, A71V, G73S, I84V, L90M, and 95C in 3PWR for the genotype FB15; K7Q, I13V, G16E, K20I, I33F, M36L, K41R, I62V, P63H, V64I, A71V, G73S, I84V, and L90M in 3EL1 for the genotype FB15; K7Q, L10V, I13V, G16E, K20R, I33L, E35D, M36I, S37N, M46I, I54V, Q58E, I62V, I63H, I64V, A67C, V82F, I84V, and A95C in 3PWR for the genotype GH9; L10I, I13V, K20M, E35D, M36I, S37N, R41K, M46I, I54V, Q58E, H69K, V82M, and L89I in 1MUI for the genotype RU1; K7Q, L10V, I13V, R14K, K20M, E35D, M36I, M46I, I54V, Q58E, P63L, V64I, H69K, V82M, and L89I in 3EL1 for the genotype RU1; L10I, I13V, K20M, E35D, M36I, S37N, R41K, M46I, I54V, Q58E, H69K, V82M, and L89I in 1MUI for the genotype RU1; K7Q, L10I, I13V, R14K, L24I, L33F, K46L, I62V, A71V, T82A, V84I, and Q92K in 1K6C for the genotype iZ2; L10I, I13V, L24I, L33F, S37N, M46L, I62V, L63P, A71V, and V82A in 1MUI for the genotype iZ2; K7Q, L10I, I13V, R14K, L24I, L33F, K41R, M46L, I62V, V64I, A71V, V82A, and Q92K in 3EL1 for the genotype iZ2.
In the following, preparation for simulations of all the structures mentioned above is described in both holo and apo states, with the exception of structure 1HXB, 1MUI, and 2BPX for the HIVdb dataset, for which simulations in apo state were not performed (see "Equilibrium MD simulations and free energy calculations" section).
Remaining set up of the system in this study has been performed in the manner as described previously [17]. In short, the Gromacs simulation software package was used to set up (version 4.6.5), carry out, and analyse the MD simulations (versions 5.0.2 and 5.1.2) [68, 69]. The of residues was predicted using Propka [70] and protease was assigned monoprotonated state on either D25/D25, here the prime refers to the second subunit of the protein. The Amber99SB*-ILDN force field was used for parametrisation of the protease. The Chemaxon Calculator [71] was used to determine inhibitor protonation, while Gaussian09 [72] was used to optimise inhibitor geometry and calculate electrostatic potential. Partial charges were assigned by performing restrained electrostatic potential fit [73]. The complex was solvated in TIP3P water molecules with 1.4 nm buffer in each dimension with 0.15 mol/l concentration of and ions [74] to neutralise the system.
Equilibrium MD simulations and free energy calculations
The equilibrium MD simulations and the free energy calculations in this study have been performed in the manner as described previously [17]. In short, each system was subjected to steepest descent energy minimisation. Before equilibrium simulation, in order to avoid too close contacts between atoms, simulated annealing in length of 1 ns was performed for the following complexes: 1MUI (76V for the genotype iZ2) in D25 protonation state, 1MUI (76L for the genotype iZ2) in D25 protonation state, 2BPX (46V and 84I variants) in D25 protonation state, 3EKV (88S variant) in D25 protonation state, 3EL1 (76L and 76V for the genotype iZ2) in D25 protonation state. Ten replicas of 200 ns simulation for each complex were performed at 300 K. For all of the analyses that followed, the first 20 ns of the simulations were considered to be a part of the system equilibration process and thus discarded, with the exception of free energy calculations, where first 10 ns were discarded. The protocol for free energy calculations was adjusted from the non-equilibrium simulation approach used in assessing changes in protein thermal stabilities and protein–protein interactions upon amino acid mutation [75]. For calculating the free energy change upon mutation of apo structures, , for inclusion in estimates for different inhibitors, in case of mutations M46I and I50L, wildtype apo structure 1HPV was used for the simulations as the background sequences for these mutations were the same. Similarly, for wildtype I84V as well as mutant M46I, I50L, and I84V simulations corresponding apo structure 1HPV variants were used for estimates for each mutation.
From each of the equilibrium simulations described above, trajectory frames were extracted equidistantly in time every 10 ns. After generating hybrid structures for every snapshot using the pmx [76] framework, short 20 ps simulations were performed to equilibrate velocities, after which alchemical transitions were carried out in 50 ps. Identical parameters were used for equilibrium simulations, equilibration, and alchemical transitions with soft-core potential for non-bonded interactions [77]. The Crooks Fluctuation Theorem [78] was used to relate the obtained work distributions to the free energy values by employing maximum likelihood estimator [79], with the error estimates obtained by the bootstrap approach. Simulations in both active site protonation states contributed to the free energy estimates, while for the rest of analysis reported in this study only the lowest free energy protonation state was used.
Partial least-squares regression
Partial least-squares regression was performed with the functional mode analysis tool [80] using the heavy atoms of protein as predictors. Wildtype and mutant protein simulation trajectories were labelled using constants 0 and 1 as target values, respectively.
For each mutation and inhibitor combination cross-validation (CV) was performed to verify the models. During CV, all trajectories for wildtype and mutant complexes were concatenated, superimposed to minimise the variance over the ensemble [81], and divided into five equal parts. For every iteration, a model was trained on four parts of labelled input in equal proportions from wildtype and mutant simulations, after which it was used to make prediction for the last part. The Pearson correlation between the actual and predicted labels was used to evaluate the quality of the model. Number of components was tested in each iteration. For the final model, the number of components was chosen from the correlation curve in CV such that choosing a higher number of components only marginally improves the performance of the model.
Estimation of resistance factor change from free energy of inhibitor binding change
Cheng-Prusoff equation [29] relates inhibitor’s and binding affinity , which in turn can be estimated from inhibitor binding free energy :
| 1 |
where [S] is a fixed substrate concentration, is the concentration of the substrate at which the enzyme is at its half-maximal activity, is the Boltzmann constant, and T is the absolute temperature. Thus, given two RF values for two proteases with sequences A and B, and , their ratio can be related to :
| 2 |
We are interested in obtaining a distribution of the values after calculating the double free energy differences :
| 3 |
When there are multiple measurements and calculations available for the wildtype and mutant protein complexed with different ligands, C can be expressed as a function of the available values . This gives the final posterior distribution:
| 4 |
where . The values are sampled from a Gaussian distribution with the mean and standard deviation corresponding to the calculated double free energy difference and estimated error, respectively.
Phenotypic assay for resistance factor value estimation
The experimental data on L76V resistance is based on samples of patients who underwent multiple therapy failures with different PIs. Those variants were observed in the diagnostic procedure, sequenced, and subsequently tested in a phenotypic assay as described by Walter et al. [82]. The tests were carried out after the patient’s variant was cloned into a recombinant derivate of the HIV NL4-3, called pNL4-3-Delta-PRT5. The L76V mutation was reverted to wildtype by site directed mutagenesis. This allowed to determine the effect of the genetic background upon the L76V. These variants were analysed in cell culture experiments where they were exposed to different PIs in different concentrations to estimate their RF values (Table 3). Based on these variants, the clones were specifically modified by site-directed mutagenesis so that different variants of L76V could be tested in different genetic backgrounds. For simplicity, regardless of the residue at position 76 of protease as present in the original clinical samples, in the context of this paper L76 is referred to as wildtype residue and V76 as the mutant residue as per reference sequence HXB2.
Supplementary information
Additional file 1: Table S1. The names of the isolates, whose RF data was used in this study, as reported in HIVdb, and the reference of the study where RF measurements were performed. Table S2. Inhibitor binding free energy change upon switching the proton from the reference protonated active site residue to the active site residue on the opposite subunit for wildtype and mutant proteins. ± shows bootstrap error estimate, all values in kcal/mol. Table S3. Average hydrogen bonds number between residues D30, T31, and T74 with N88 and S88 for wildtype and mutant complexes, respectively. Columns 3 and 4 of the table corresponds to hydrogen bonds within monomer A of protease and columns 5 and 6 of the table corresponds to hydrogen bonds within monomer B of protease (residues marked with prime symbol). ± indicates standard error of bond frequency across independent simulations. Table S3. Inhibitor binding free energy change upon switching the proton from the reference protonated active site residue to the active site residue on the opposite subunit for wildtype and mutant proteins. ± shows bootstrap error estimate, all values in kcal/mol. Figure S3. Convergence of the RFR estimates. The shaded areas show the 95% credible interval. Figure S4. Interpolation between the extremes of the FMA models for the corresponding complexes. Blue-to-magenta bands correspond to the interpolation along the mode as represented as cartoon for backbone and as sticks for residues 30, 45, and 58, with blue corresponding to L76 state and magenta to V76 state. Mutated residue 76 is not part of the model and is represented here as gray dash. Table S4. Inhibitor binding free energy change upon switching the proton from the reference protonated active site residue to the active site residue on the opposite subunit for wildtype and mutant proteins. ± shows bootstrap error estimate, all values in kcal/mol. Figure S5. Energy differences of non-bonded interactions between protein and inhibitor in wildtype and mutant complexes. Only residues, for which the difference between the wildtype and the mutant complexes is higher than the propagated error and its absolute value higher than 0.1 kcal/mol are shown.
Acknowledgements
We are grateful to Alejandro Pironti for providing resistance factor data from HIVdb and to Mazen Ahmad for discussions and technical advice on the simulations. The simulations presented in this work were in part performed on HPC systems of the Max Planck Computing and Data Facility.
Abbreviations
- AIDS
Acquired immune deficiency syndrome
- APV
Amprenavir
- ART
Antiretroviral therapy
- ATV
Atazanavir
- CV
Cross-validation
- DRV
Darunavir
- FMA
Functional mode analysis
- FPV
Fosamprenavir
- HIV-1
Human immunodeficiency virus type 1
- IDV
Indinavir
- LPV
Lopinavir
- MD
Molecular dynamics
- NFV
Nelfinavir
- PDB
Protein Data Bank
- PI
Protease inhibitor
- PLS
Partial least-squares
- RAM
Resistance-associated mutation
- RF
Resistance factor
- SQV
Saquinavir
- TPV
Tipranavir
Authors' contributions
All authors wrote and approved the manuscript. All authors read and approved the final manuscript.
Funding
OVK is grateful to the Klaus Faber Foundation for their financial support. VG and BLdG were supported by the BioExcel CoE (http://www.bioexcel.eu), a project funded by the European Union contracts H2020-INFRAEDI-02-2018-823830, H2020-EINFRA-2015-1-675728.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by the University Clinics Cologne (16-460).
Consent for publication
All study participants consent to publication.
Competing interests
TB is an employee of AstraZeneca and has stock ownership and/or stock options or interests in the company.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12977-020-00520-6.
References
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS): UNAIDS DATA 2017. Geneva; 2017. http://www.unaids.org/sites/default/files/media_asset/20170720_Data_book_2017_en.pdf. Accessed 12 June 2018.
- 2.Joint United Nations Programme on HIV/AIDS (UNAIDS): right to health. 2017. http://www.unaids.org/sites/default/files/media_asset/RighttoHealthReport_Full_webNov.pdf. Accessed 12 June 2018.
- 3.World Health Organization . Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. 2. Geneva: World Health Organization; 2016. [PubMed] [Google Scholar]
- 4.Young TP, Parkin NT, Stawiski E, Pilot-Matias T, Trinh R, Kempf DJ, Norton M. Prevalence, mutation patterns, and effects on protease inhibitor susceptibility of the L76V mutation in HIV-1 protease. Antimicrob Agents Chemother. 2010;54(11):4903–4906. doi: 10.1128/aac.00906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiesmann F, Vachta J, Ehret R, Walter H, Kaiser R, Stürmer M, Tappe A, Däumer M, Berg T, Naeth G, Braun P, Knechten H. The L76V mutation in HIV-1 protease is potentially associated with hypersusceptibility to protease inhibitors atazanavir and saquinavir: is there a clinical advantage? AIDS Res Ther. 2011;8(1):7. doi: 10.1186/1742-6405-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziermann R, Limoli K, Das K, Arnold E, Petropoulos CJ, Parkin NT. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to amprenavir. J Virol. 2000;74(9):4414–4419. doi: 10.1128/JVI.74.9.4414-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resch W, Ziermann R, Parkin N, Gamarnik A, Swanstrom R. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the gag polyprotein precursor aberrantly. J Virol. 2002;76(17):8659–8666. doi: 10.1128/JVI.76.17.8659-8666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeiren H, Craenenbroeck EV, Alen P, Bacheler L, Picchio G, Lecocq P. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J Virol Methods. 2007;145(1):47–55. doi: 10.1016/j.jviromet.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Rhee S-Y, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob Agents Chemother. 2010;54(10):4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou T, Yu R. Molecular dynamics and free energy studies on the wild-type and double mutant HIV-1 protease complexed with amprenavir and two amprenavir-related inhibitors: mechanism for binding and drug resistance. J Med Chem. 2007;50(6):1177–1188. doi: 10.1021/jm0609162. [DOI] [PubMed] [Google Scholar]
- 11.Muzammil S, Armstrong AA, Kang LW, Jakalian A, Bonneau PR, Schmelmer V, Amzel LM, Freire E. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J Virol. 2007;81(10):5144–5154. doi: 10.1128/jvi.02706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcaro S, Artese A, Ceccherini-Silberstein F, Ortuso F, Perno CF, Sing T, Svicher V. Molecular dynamics and free energy studies on the wild-type and mutated HIV-1 protease complexed with four approved drugs: mechanism of binding and drug resistance. J Chem Inf Model. 2009;49(7):1751–1761. doi: 10.1021/ci900012k. [DOI] [PubMed] [Google Scholar]
- 13.Shen CH, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters. FEBS J. 2010;277(18):3699–3714. doi: 10.1111/j.1742-4658.2010.07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittal S, Bandaranayake RM, King NM, Prabu-Jeyabalan M, Nalam MN, Nalivaika EA, Yilmaz NK, Schiffer CA. Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50. J Virol. 2013;87(8):4176–4184. doi: 10.1128/jvi.03486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan R, Lazim R, Zhang D. Understanding the basis of I50V-induced affinity decrease in HIV-1 protease via molecular dynamics simulations using polarized force field. J Comput Chem. 2015;36(25):1885–1892. doi: 10.1002/jcc.24020. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y, Wang J, Shao Q, Shi J, Zhu W. Effects of drug-resistant mutations on the dynamic properties of HIV-1 protease and inhibition by amprenavir and darunavir. Sci Rep. 2015;5(1):10517. doi: 10.1038/srep10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastys T, Gapsys V, Doncheva NT, Kaiser R, de Groot BL, Kalinina OV. Consistent prediction of mutation effect on drug binding in HIV-1 protease using alchemical calculations. J Chem Theory Comput. 2018;14(7):3397–3408. doi: 10.1021/acs.jctc.7b01109. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Boross PI, Wang YF, Tozser J, Louis JM, Harrison RW, Weber IT. Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S. J Mol Biol. 2005;354(4):789–800. doi: 10.1016/j.jmb.2005.09.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovalevsky AY, Tie Y, Liu F, Boross PI, Wang YF, Leshchenko S, Ghosh AK, Harrison RW, Weber IT. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J Med Chem. 2006;49(4):1379–1387. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kožíšek M, Bray J, Řezáčová P, Šašková K, Brynda J, Pokorná J, Mammano F, Rulíšek L, Konvalinka J. Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants. J Mol Biol. 2007;374(4):1005–1016. doi: 10.1016/j.jmb.2007.09.083. [DOI] [PubMed] [Google Scholar]
- 21.Hu G-D, Zhu T, Zhang S-L, Wang D, Zhang Q-G. Some insights into mechanism for binding and drug resistance of wild type and I50V V82A and I84V mutations in HIV-1 protease with grl-98065 inhibitor from molecular dynamic simulations. Eur J Med Chem. 2010;45(1):227–235. doi: 10.1016/j.ejmech.2009.09.048. [DOI] [PubMed] [Google Scholar]
- 22.Cai Y, Schiffer CA. Decomposing the energetic impact of drug resistant mutations in HIV-1 protease on binding DRV. J Chem Theory Comput. 2010;6(4):1358–1368. doi: 10.1021/ct9004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Zhang S, Liu X, Zhang Q. Insights into drug resistance of mutations D30N and I50V to HIV-1 protease inhibitor TMC-114: free energy calculation and molecular dynamic simulation. J Mol Model. 2010;16(3):459–468. doi: 10.1007/s00894-009-0553-7. [DOI] [PubMed] [Google Scholar]
- 24.Kar P, Knecht V. Energetic basis for drug resistance of HIV-1 protease mutants against amprenavir. J Comput Aided Mol Des. 2012;26(2):215–232. doi: 10.1007/s10822-012-9550-5. [DOI] [PubMed] [Google Scholar]
- 25.Meher BR, Wang Y. Interaction of I50V mutant and I50L/A71V double mutant HIV-protease with inhibitor TMC114 (darunavir): molecular dynamics simulation and binding free energy studies. J Phys Chem B. 2012;116(6):1884–1900. doi: 10.1021/jp2074804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragland DA, Nalivaika EA, Nalam MNL, Prachanronarong KL, Cao H, Bandaranayake RM, Cai Y, Kurt-Yilmaz N, Schiffer CA. Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease. J Am Chem Soc. 2014;136(34):11956–11963. doi: 10.1021/ja504096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragland DA, Whitfield TW, Lee S-K, Swanstrom R, Zeldovich KB, Kurt-Yilmaz N, Schiffer CA. Elucidating the interdependence of drug resistance from combinations of mutations. J Chem Theory Comput. 2017;13(11):5671–5682. doi: 10.1021/acs.jctc.7b00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larder B, Kemp S, Harrigan P. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269(5224):696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 29.Yung-Chi C, Prusoff WH. Relationship between the inhibition constant () and the concentration of inhibitor which causes 50 per cent inhibition () of an enzymatic reaction. Biochem Pharmacol. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 30.The Stanford HIV Drug Resistance Database. https://hivdb.stanford.edu/dr-summary/resistance-notes/PI/. Accessed 12 June 2018.
- 31.Ott DE, Coren LV, Chertova EN, Gagliardi TD, Nagashima K, Sowder RC, Poon DTK, Gorelick RJ. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J Virol. 2003;77(10):5547–5556. doi: 10.1128/JVI.77.10.5547-5556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang MW, Torbett BE. Accessory mutations maintain stability in drug-resistant HIV-1 protease. J Mol Biol. 2011;410(4):756–760. doi: 10.1016/j.jmb.2011.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colonno R, Rose R, McLaren C, Thiry A, Parkin N, Friborg J. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J Infect Dis. 2004;189(10):1802–1810. doi: 10.1086/386291. [DOI] [PubMed] [Google Scholar]
- 34.Yanchunas J, Langley DR, Tao L, Rose RE, Friborg J, Colonno RJ, Doyle ML. Molecular basis for increased susceptibility of isolates with atazanavir resistance-conferring substitution I50L to other protease inhibitors. Antimicrob Agents Chemother. 2005;49(9):3825–3832. doi: 10.1128/AAC.49.9.3825-3832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sista P, Wasikowski B, Lecocq P, Pattery T, Bacheler L. The HIV-1 protease resistance mutation I50L is associated with resistance to atazanavir and susceptibility to other protease inhibitors in multiple mutational contexts. J Clin Virol. 2008;42(4):405–408. doi: 10.1016/j.jcv.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Weinheimer S, Discotto L, Friborg J, Yang H, Colonno R. Atazanavir signature I50L resistance substitution accounts for unique phenotype of increased susceptibility to other protease inhibitors in a variety of human immunodeficiency virus type 1 genetic backbones. Antimicrob Agents Chemother. 2005;49(9):3816–3824. doi: 10.1128/AAC.49.9.3816-3824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gapsys V, Michielssens S, Peters JH, de Groot BL, Leonov H. Calculation of binding free energies. In: Kukol A, editor. Molecular modeling of proteins. Methods in molecular biology. New York: Springer; 2015. pp. 173–209. [DOI] [PubMed] [Google Scholar]
- 38.Ho DD, Toyoshima T, Mo H, Kempf DJ, Norbeck D, Chen C-M, Wideburg NE, Burt SK, Erickson JW, Singh MK. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a c2-symmetric protease inhibitor. J Virol. 1994;68(3):2016–2020. doi: 10.1128/JVI.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pazhanisamy S, Stuver CM, Cullinan AB, Margolin N, Rao B, Livingston DJ. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J Biol Chem. 1996;271(30):17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 40.Schock HB, Garsky VM, Kuo LC. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials compensatory modulations of binding and activity. J Biol Chem. 1996;271(50):31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 41.Nijhuis M, Wensing AMJ, Bierman WFW, de Jong D, Kagan R, Fun A, Jaspers CAJJ, Schurink KAM, van Agtmael MA, Boucher CAB. Failure of treatment with first-line lopinavir boosted with ritonavir can be explained by novel resistance pathways with protease mutation 76V. J Infect Dis. 2009;200(5):698–709. doi: 10.1086/605329. [DOI] [PubMed] [Google Scholar]
- 42.Louis JM, Zhang Y, Sayer JM, Wang Y-F, Harrison RW, Weber IT. The L76V drug resistance mutation decreases the dimer stability and rate of autoprocessing of HIV-1 protease by reducing internal hydrophobic contacts. Biochemistry. 2011;50(21):4786–4795. doi: 10.1021/bi200033z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson GJ, Lee S-K, Irlbeck DM, Harris J, Kline M, Pollom E, Parkin N, Swanstrom R. Interplay between single resistance-associated mutations in the HIV-1 protease and viral infectivity, protease activity, and inhibitor sensitivity. Antimicrob Agents Chemother. 2012;56(2):623–633. doi: 10.1128/aac.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizzo RC, Wang D-P, Tirado-Rives J, Jorgensen WL. Validation of a model for the complex of HIV-1 reverse transcriptase with sustiva through computation of resistance profiles. J Am Chem Soc. 2000;122(51):12898–900. doi: 10.1021/ja003113r. [DOI] [Google Scholar]
- 45.Wang D-P, Rizzo RC, Tirado-Rives J, Jorgensen WL. Antiviral drug design: computational analyses of the effects of the L100I mutation for HIV-RT on the binding of NNRTIs. Bioorgan Med Chem Lett. 2001;11(21):2799–802. doi: 10.1016/S0960-894X(01)00510-8. [DOI] [PubMed] [Google Scholar]
- 46.Udier-Blagović M, Tirado-Rives J, Jorgensen WL. Validation of a model for the complex of HIV-1 reverse transcriptase with nonnucleoside inhibitor TMC125. J Am Chem Soc. 2003;125(20):6016–7. doi: 10.1021/ja034308c. [DOI] [PubMed] [Google Scholar]
- 47.Loya S, Bakhanashvili M, Tal R, Hughes SH, Boyer PL, Hiz A. Enzymatic properties of two mutants of reverse transcriptase of human immunodeficiency virus type 1 (tyrosine 181isoleucine and tyrosine 188leucine), resistant to nonnucleoside inhibitors. AIDS Res Hum Retrovir. 1994;10(8):939–946. doi: 10.1089/aid.1994.10.939. [DOI] [PubMed] [Google Scholar]
- 48.Maga G, Amacker M, Ruel N, Hübscher U, Spadari S. Resistance to nevirapine of HIV-1 reverse transcriptase mutants: loss of stabilizing interactions and thermodynamic or steric barriers are induced by different single amino acid substitutions. J Mol Biol. 1997;274(5):738–747. doi: 10.1006/jmbi.1997.1427. [DOI] [PubMed] [Google Scholar]
- 49.Archer RH, Dykes C, Gerondelis P, Lloyd A, Fay P, Reichman RC, Bambara RA, Demeter LM. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of rnase h cleavage that correlate with HIV-1 replication fitness in cell culture. J Virol. 2000;74(18):8390–8401. doi: 10.1128/JVI.74.18.8390-8401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Locatelli GA, Campiani G, Cancio R, Morelli E, Ramunno A, Gemma S, Hübscher U, Spadari S, Maga G. Effects of drug resistance mutations L100I and V106A on the binding of pyrrolobenzoxazepinone nonnucleoside inhibitors to the human immunodeficiency virus type 1 reverse transcriptase catalytic complex. Antimicrob Agents Chemother. 2004;48(5):1570–1580. doi: 10.1128/AAC.48.5.1570-1580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das K, Lewi PJ, Hughes SH, Arnold E. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors (Structure-guided design of AIDs antivirals) Prog Biophys Mol Biol. 2005;88(2):209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Hosseini A, Alibés A, Noguera-Julian M, Gil V, Paredes R, Soliva R, Orozco M, Guallar V. Computational prediction of HIV-1 resistance to protease inhibitors. J Chem Inf Model. 2016;56(5):915–923. doi: 10.1021/acs.jcim.5b00667. [DOI] [PubMed] [Google Scholar]
- 53.Chen Z, Li Y, Schock HB, Hall D, Chen E, Kuo LC. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J Biol Chem. 1995;270(37):21433–21436. doi: 10.1074/jbc.270.37.21433. [DOI] [PubMed] [Google Scholar]
- 54.Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher CA. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS. 1999;13(17):2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 55.Ode H, Matsuyama S, Hata M, Hoshino T, Kakizawa J, Sugiura W. Mechanism of drug resistance due to N88Ss in CRF01\_AE HIV-1 protease, analyzed by molecular dynamics simulations. J Med Chem. 2007;50(8):1768–1777. doi: 10.1021/jm061158i. [DOI] [PubMed] [Google Scholar]
- 56.Vasavi CS, Tamizhselvi R, Munusami P. Drug resistance mechanism of L10F, L10F/N88S and L90M mutations in CRF01\_ae HIV-1 protease: Molecular dynamics simulations and binding free energy calculations. J Mol Graph Model. 2017;75:390–402. doi: 10.1016/j.jmgm.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Bihani SC, Das A, Prashar V, Ferrer J-L, Hosur MV. Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S. Biochem Biophys Res Commun. 2009;389(2):295–300. doi: 10.1016/j.bbrc.2009.08.138. [DOI] [PubMed] [Google Scholar]
- 58.Mahalingam B, Boross P, Wang Y-F, Louis JM, Fischer CC, Tozser J, Harrison RW, Weber IT. Combining mutations in HIV-1 protease to understand mechanisms of resistance. Proteins. 2002;48(1):107–116. doi: 10.1002/prot.10140. [DOI] [PubMed] [Google Scholar]
- 59.Svicher V, Ceccherini-Silberstein F, Erba F, Santoro M, Gori C, Bellocchi MC, Giannella S, Trotta MP, Monforte Ad, Antinori A, Perno CF. Novel human immunodeficiency virus type 1 protease mutations potentially involved in resistance to protease inhibitors. Antimicrob Agents Chemother. 2005;49(5):2015–2025. doi: 10.1128/AAC.49.5.2015-2025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margerison ES, Maguire M, Pillay D, Cane P, Elston RC. The HIV-1 protease substitution K55R: a protease-inhibitor-associated substitution involved in restoring viral replication. Antimicrob Agents Chemother. 2008;61(4):786–791. doi: 10.1093/jac/dkm545. [DOI] [PubMed] [Google Scholar]
- 61.Champenois K, Baras A, Choisy P, Ajana F, Melliez H, Bocket L, Yazdanpanah Y. Lopinavir/ritonavir resistance in patients infected with HIV-1: two divergent resistance pathways? J Med Virol. 2011;83(10):1677–1681. doi: 10.1002/jmv.22161. [DOI] [PubMed] [Google Scholar]
- 62.Whitfield TW, Ragland DA, Zeldovich KB, Schiffer CA. Characterizing protein–ligand binding using atomistic simulation and machine learning: application to drug resistance in HIV-1 protease. J Chem Theory Comput. 2020;16(2):1284–1299. doi: 10.1021/acs.jctc.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong L, Zhang XC, Hartsuck JA, Tang J. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance. Protein Sci. 2000;9(10):1898–1904. doi: 10.1110/ps.9.10.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ode H, Neya S, Hata M, Sugiura W, Hoshino T. Computational simulations of HIV-1 proteases multi-drug resistance due to nonactive site mutation L90M. J Am Chem Soc. 2006;128(24):7887–7895. doi: 10.1021/ja060682b. [DOI] [PubMed] [Google Scholar]
- 65.Wong-Sam A, Wang Y-F, Zhang Y, Ghosh AK, Harrison RW, Weber IT. Drug resistance mutation L76V alters nonpolar interactions at the flap-core interface of HIV-1 protease. ACS Omega. 2018;3(9):12132–12140. doi: 10.1021/acsomega.8b01683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 68.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 69.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 70.Søndergaard CR, Olsson MH, Rostkowski M, Jensen JH. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput. 2011;7(7):2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 71.Chemaxon Calculator 5.3.8. ChemAxon. 2010. http://www.chemaxon.com.
- 72.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JJA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Keith T, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 9 Revision C01. Wallingford: Gaussian Inc.; 2010. [Google Scholar]
- 73.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97(40):10269–10280. doi: 10.1021/j100142a004. [DOI] [Google Scholar]
- 74.Joung IS, Cheatham TE. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gapsys V, Michielssens S, Seeliger D, de Groot BL. Accurate and rigorous prediction of the changes in protein free energies in a large-scale mutation scan. Angew Chem Int Ed. 2016;55(26):7364–7368. doi: 10.1002/anie.201510054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gapsys V, Michielssens S, Seeliger D, de Groot BL. pmx: automated protein structure and topology generation for alchemical perturbations. J Comput Chem. 2014;36(5):348–354. doi: 10.1002/jcc.23804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gapsys V, Seeliger D, de Groot BL. New soft-core potential function for molecular dynamics based alchemical free energy calculations. J Chem Theory Comput. 2012;8(7):2373–2382. doi: 10.1021/ct300220p. [DOI] [PubMed] [Google Scholar]
- 78.Crooks GE. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys Rev E. 1999;60:2721–2726. doi: 10.1103/PhysRevE.60.2721. [DOI] [PubMed] [Google Scholar]
- 79.Shirts MR, Bair E, Hooker G, Pande VS. Equilibrium free energies from nonequilibrium measurements using maximum-likelihood methods. Phys Rev Lett. 2003;91(14):140601. doi: 10.1103/physrevlett.91.140601. [DOI] [PubMed] [Google Scholar]
- 80.Krivobokova T, Briones R, Hub JS, Munk A, de Groot BL. Partial least-squares functional mode analysis: application to the membrane proteins AQP1, Aqy1, and CLC-ec1. Biophys J. 2012;103(4):786–796. doi: 10.1016/j.bpj.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gapsys V, de Groot BL. Optimal superpositioning of flexible molecule ensembles. Biophys J. 2013;104(1):196–207. doi: 10.1016/j.bpj.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walter H, Schmidt B, Korn K, Vandamme A-M, Harrer T, Überla K. Rapid, phenotypic HIV-1 drug sensitivity assay for protease and reverse transcriptase inhibitors. J Clin Virol. 1999;13(1):71–80. doi: 10.1016/S1386-6532(99)00010-4. [DOI] [PubMed] [Google Scholar]
- 83.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44(4):920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prado JG, Wrin T, Beauchaine J, Ruiz L, Petropoulos CJ, Frost SD, Clotet B, Richard TD, Martinez-Picado J. Amprenavir-resistant HIV-1 exhibits lopinavir cross-resistance and reduced replication capacity. AIDS. 2002;16(7):1009–1017. doi: 10.1097/00002030-200205030-00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The names of the isolates, whose RF data was used in this study, as reported in HIVdb, and the reference of the study where RF measurements were performed. Table S2. Inhibitor binding free energy change upon switching the proton from the reference protonated active site residue to the active site residue on the opposite subunit for wildtype and mutant proteins. ± shows bootstrap error estimate, all values in kcal/mol. Table S3. Average hydrogen bonds number between residues D30, T31, and T74 with N88 and S88 for wildtype and mutant complexes, respectively. Columns 3 and 4 of the table corresponds to hydrogen bonds within monomer A of protease and columns 5 and 6 of the table corresponds to hydrogen bonds within monomer B of protease (residues marked with prime symbol). ± indicates standard error of bond frequency across independent simulations. Table S3. Inhibitor binding free energy change upon switching the proton from the reference protonated active site residue to the active site residue on the opposite subunit for wildtype and mutant proteins. ± shows bootstrap error estimate, all values in kcal/mol. Figure S3. Convergence of the RFR estimates. The shaded areas show the 95% credible interval. Figure S4. Interpolation between the extremes of the FMA models for the corresponding complexes. Blue-to-magenta bands correspond to the interpolation along the mode as represented as cartoon for backbone and as sticks for residues 30, 45, and 58, with blue corresponding to L76 state and magenta to V76 state. Mutated residue 76 is not part of the model and is represented here as gray dash. Table S4. Inhibitor binding free energy change upon switching the proton from the reference protonated active site residue to the active site residue on the opposite subunit for wildtype and mutant proteins. ± shows bootstrap error estimate, all values in kcal/mol. Figure S5. Energy differences of non-bonded interactions between protein and inhibitor in wildtype and mutant complexes. Only residues, for which the difference between the wildtype and the mutant complexes is higher than the propagated error and its absolute value higher than 0.1 kcal/mol are shown.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.