Abstract
In the last decades in vitro studies highlighted the potential for crosstalk between Hypoxia-Inducible Factor-(HIF) and glucocorticoid-(GC) signalling pathways. However, how this interplay precisely occurs in vivo is still debated. Here, we use zebrafish larvae (Danio rerio) to elucidate how and to what degree hypoxic signalling affects the endogenous glucocorticoid pathway and vice versa, in vivo. Firstly, our results demonstrate that in the presence of upregulated HIF signalling, both glucocorticoid receptor (Gr) responsiveness and endogenous cortisol levels are repressed in 5 days post fertilisation larvae. In addition, despite HIF activity being low at normoxia, our data show that it already impedes both glucocorticoid activity and levels. Secondly, we further analysed the in vivo contribution of glucocorticoids to HIF activity. Interestingly, our results show that both glucocorticoid receptor (GR) and mineralocorticoid receptor (MR) play a key role in enhancing it. Finally, we found indications that glucocorticoids promote HIF signalling via multiple routes. Cumulatively, our findings allowed us to suggest a model for how this crosstalk occurs in vivo.
Author summary
Hypoxia is a common pathophysiological condition to which cells must rapidly respond in order to prevent metabolic shutdown and subsequent death. This is achieved via the activity of Hypoxia-Inducible Factors (HIFs), which are key oxygen sensors that mediate the ability of the cell to cope with decreased oxygen levels. Although it aims to restore tissue oxygenation and perfusion, it can sometimes be maladaptive and contributes to a variety of pathological conditions including inflammation, tissue ischemia, stroke and growth of solid tumours. In this regard, synthetic glucocorticoids which are analogous to naturally occurring steroid hormones, have been used for decades as anti-inflammatory drugs for treating pathological conditions which are linked to hypoxia (i.e. asthma, rheumatoid arthritis, ischemic injury). Indeed, previous in vitro studies highlighted the presence of a crosstalk between HIF and glucocorticoids. However, how this interplay precisely occurs in an organism and what the molecular mechanism is behind it are questions that still remain unanswered. Here, we provide a thorough in vivo genetic analysis, which allowed us to propose a logical model of interaction between glucocorticoid and HIF signalling. In addition, our results are important because they suggest a new route to downregulate HIF for clinical purposes.
Introduction
Glucocorticoids (GC) constitute a well-characterized class of lipophilic steroid hormones produced by the adrenal glands in humans and by the interrenal tissue in teleosts. The circadian production of glucocorticoids in teleosts is regulated by the hypothalamus-pituitary-interrenal (HPI) axis, which is the equivalent of the mammalian hypothalamus-pituitary-adrenal (HPA) axis. Both are central to stress adaptation [1–4]. Interestingly, both in humans and teleosts cortisol is the primary glucocorticoid and regulates a plethora of physiological processes including glucose homeostasis, inflammation, intermediary metabolism and stress response [5]. In particular, cortisol can exert these functions via direct binding both to the glucocorticoid receptor (Gr) and to the mineralocorticoid receptor (Mr), which bind cortisol with different affinities. [3,6]. Together they act as a transcription factor, which can function either in a genomic or in non-genomic way [5,7–9].
Hypoxia-inducible factor (HIF) transcription factors are key regulators of the cellular response to hypoxia, which coordinate a metabolic shift from aerobic to anaerobic metabolism in the presence of low oxygen availability in order to assure homeostasis [10]. In mammals there are at least three isoforms of HIF-α (HIF-1α, HIF-2α and HIF-3α) and two main isoforms of HIF-1β (ARNT1 and ARNT2). [11]. Interestingly, due to a genome duplication event, there are two paralogs for each of the three Hif-α isoforms (Hif-1αa, Hif-1αb, Hif-2αa, Hif-2αb, Hif-3αa and Hif-3αb) in zebrafish. Among these, Hif-1αb is thought to be the key zebrafish homologue in the hypoxic response [12]. With respect to HIF-1β (ARNT) paralogues, the expression of two genes encoding Arnt1 and Arnt2 proteins has been described in zebrafish [13–16].
Whilst ARNT is constitutively expressed, the cytoplasmic HIF-α subunits are primarily regulated post-translationally via the PHD3-VHL-E3-ubiquitin ligase protein degradation complex. This is believed to occur in order to allow a rapid response to decreasing oxygen levels [12,17–19]. Indeed, hypoxia, is a common pathophysiological condition [20,21] to which cells must promptly respond in order to avert metabolic shutdown and subsequent death [12]. In the presence of normal oxygen levels, a set of prolyl hydroxylases (PHD1, 2 and 3) use the available molecular oxygen directly to hydroxylate HIF-α subunit. Hydroxylated HIF-α is then recognised by the Von Hippel Lindau (VHL) protein, which acts as the substrate recognition part of a E3-ubiquitin ligase complex. This leads to HIF-α proteasomal degradation to avoid HIF pathway activation under normoxic conditions. On the other hand, low O2 levels impair the activity of the PHD enzymes leading to HIF-α stabilisation and subsequent translocation in the nucleus. Here, together with the HIF-β subunit, HIF-α forms a functional transcription complex, which drives the hypoxic response [22]. Although the HIF response is aimed to restore tissue oxygenation and perfusion, it can sometimes be maladaptive and can contribute to a variety of pathological conditions including inflammation, tissue ischemia, stroke and growth of solid tumours [23]. Finally, it is important to note for this study that HIF signalling is able to regulate its own activation via negative feedback, by inducing the expression of PHD genes, in particular prolyl hydroxylase 3 (PHD3) [24,25].
The presence of a crosstalk between glucocorticoids and hypoxia dependent signalling pathways has been reported in several in vitro studies [26–30]. Moreover, synthetic glucocorticoids (ie. betamethasone and dexamethasone), which are analogous to naturally occurring steroid hormones, have been extensively used for decades as anti-inflammatory drugs for treating pathological conditions which are linked to hypoxia (i.e. asthma, rheumatoid arthritis, ischemic injury, etc.) [31–33]. However, due to the presence of adverse effects [34] and glucocorticoid resistance [35,36], their use has been limited. Therefore, extending the research on how precisely this interplay occurs in vivo, may have a wide physiological significance in health and disease.
The first evidence of interaction between HIF and GR was provided by Kodama et al. 2003 [26], who discovered that ligand-dependent activation of glucocorticoid receptor enhances hypoxia-dependent gene expression and hypoxia response element (HRE) activity in HeLa cells. Leonard et al. 2005 [27] subsequently revealed that GR is transcriptionally upregulated by hypoxia in human renal proximal tubular epithelial cells. Furthermore, the hypoxic upregulation of GR was confirmed by Zhang et al 2015 [29]. In contrast, a dexamethasone-mediated inhibition of HIF-1α target genes expression in hypoxic HEPG2 cells was demonstrated by Wagner et al. 2008 [28]. In addition to that, they showed retention of HIF-1α in the cytoplasm, suggesting a blockage in nuclear import. Finally, Gaber et al., 2011 [37] indicated the presence of dexamethasone-induced suppression of HIF-1α protein expression, which resulted in reduced HIF-1 target gene expression.
From these in vitro results it has become clear that the HIF-GC crosstalk is complex and may depend on cell type. In the present study, we have used the zebrafish (Danio rerio) as an in vivo model organism to study how and to what degree hypoxic signalling affects the endogenous glucocorticoids’ response and vice versa. The use of whole animals allows us to show how these signals interact at a more global level than in cell culture, where interactions between different tissues and cell types are not easily modelled. The zebrafish offers an excellent genetic vertebrate model system for endocrine studies, and similar to humans, they are diurnal and use cortisol as the main glucocorticoid hormone [38]. Importantly, unlike other teleosts, zebrafish have only a single glucocorticoid (zGr) and mineralocorticoid receptor (Mr) (zMr) isoform [3]. Moreover, zGr shares high structural and functional similarities to its human equivalent, making zebrafish a reliable model for studying glucocorticoids activity in vivo [39–41]. Additionally, zebrafish share all the components of the human HIF signalling pathway and it has been proved to be a very informative and genetically tractable organism for studying hypoxia and HIF pathway both in physiological and pathophysiological conditions [12,25,42].
In our previous work, we identified new activators of the HIF pathway, e.g. betamethasone, a synthetic glucocorticoid receptor agonist [43]. Counterintuitively, GR loss of function was shown by Facchinello and colleagues to hamper the transcriptional activity linked to immune-response (i.e of cytokines Il1β, Il8 and Il6 and of the metalloproteinase Mmp-13) [5]. Finally, glucocorticoid receptor has been also found to synergistically activate proinflammatory genes by interacting with other signalling pathways [40,44–46].
In the present study, we utilised both a genetic and pharmacological approach to alter these two pathways during the first 120 hours post fertilisation of zebrafish embryos. In particular, we took advantage of two different mutant lines we have generated (hif1βsh544 (arnt1) and grsh543 (nr3c1) respectively), coupled to an already existing vhlhu2117/+;phd3:eGFPi144/i144 hypoxia reporter line [25], to study the effect of HIF activity on GC signalling and vice-versa, via a “gain-of-function/loss-of-function” approach. Phenotypic and molecular analyses of these mutants have been accompanied by optical and fluorescence microscope imaging.
Importantly, we not only confirm that betamethasone is able to increase the expression of phd3:eGFP, a marker of HIF activation in our zebrafish HIF-reporter line, but we also show that BME-driven HIF response requires Hif1β/Arnt1 action to occur.
Furthermore, our results also demonstrate that both Gr and Mr loss of function are able to partially rescue vhl phenotype, allowing us to confirm the importance of glucocorticoids in assuring high HIF signalling levels. This finding may have wider significance in health and disease, as so far it is proven difficult to downregulate HIF signalling.
Our results also demonstrate that in the presence of upregulated HIF pathway (by mutating vhl), both the glucocorticoid receptor activity and the endogenous cortisol levels are repressed in 5 dpf larvae, whereas when the HIF pathway is suppressed (by mutating hif1β) they are significantly increased. Finally, qPCR analysis on GC target genes, in situ hybridisation on the expression of steroidogenic genes and cortisol quantification on the aforementioned mutant lines confirmed our hypothesis.
Taken together, these results allow us to deepen the knowledge of how the crosstalk between HIF and glucocorticoid pathway occurs in vivo and to underscore a new model of interaction between these two major signalling pathways.
Results
Generating arnt1 and arnt1;vhl knockout in zebrafish
To study the interplay between HIF and GC signalling in vivo, using a genetic approach, we required a Hif1β/Arnt1 mutant line (in a phd3:eGFP;vhl+/- background) to enable the downregulation of the HIF pathway. Hif-1β (hypoxia-inducible factor 1 beta, Arnt1) is a basic helix-loop-helix-PAS protein which translocates from the cytosol to the nucleus after ligand binding to Hif-α subunits, after the stabilization of the latter in the cytoplasm. It represents the most downstream protein in the HIF pathway and for this reason it is the most suitable target.
Using CRISPR/Cas9 mutagenesis we obtained a 7 bp insertion in exon 5 (coding bHLH DNA binding domain (DBD) of the Hif-1β protein; allele name sh544) in vhl heterozygote embryos (Fig 1A). The resulting frameshift mutation was predicted to lead to a premature stop codon at the level of the DNA-binding domain, which would result in a severely truncated protein. The resulting line hif1βsh544/+;vhlhu2117/+;phd3:eGFPi144/i144 will be called arnt1+/-;vhl+/-, whereas the vhlhu2117/+;phd3:eGFPi144/i144 line will be called vhl+/- hereafter.
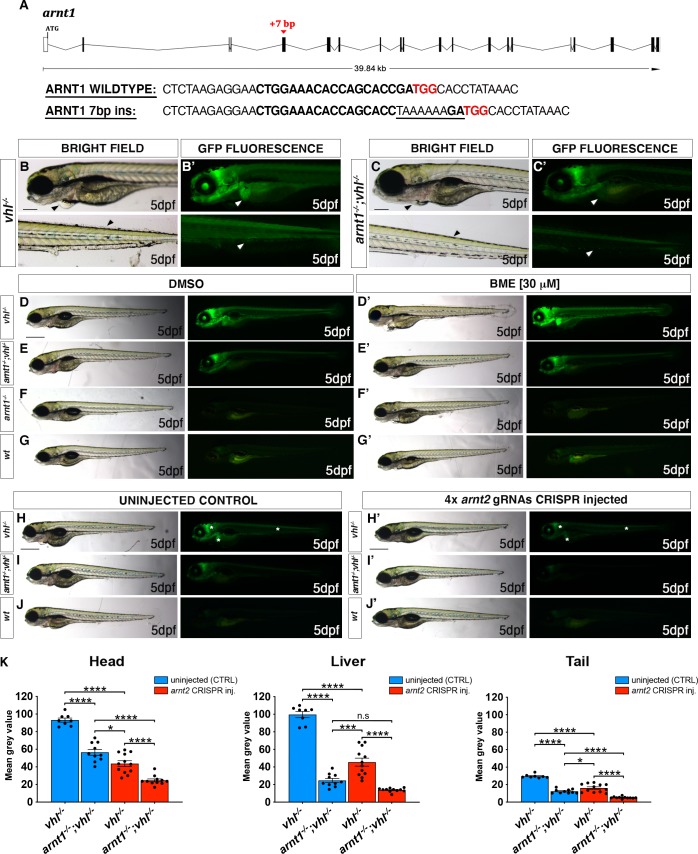
Fig 1. arnt1 and arnt2 have partially overlapping functions and synergistically contribute to HIF signalling.
A. Schematic representation of zebrafish hif1β (arnt1) gene. Exons are shown as black boxes, whereas introns as lines. The red arrowhead shows the position of a +7 bp insertion in exon 5 (encoding the bHLH DNA binding domain). In the arnt1 wt and mutant sequence. CRISPR target site: bold. Protospacer-adjacent-motif (PAM) sequence: red. B-B’. Magnified picture of a representative 5 dpf vhl-/- larva compared to 5dpf arnt1-/-;vhl-/- (C-C’). Among the 120 GFP+ embryos derived from arnt1+/-;vhl+/-(phd3:eGFP) x arnt1-/-;vhl+/-(phd3:eGFP), 15 larvae were characterized by the absence of pericardial oedema, no ectopic extra vasculature at the level of the tail, no bright liver and a reduced brightness in the rest of the body (black and white arrowheads). Genotyping post phenotypic analysis on sorted larvae confirmed genotype-phenotype correlation. Fluorescence, exposure = 2 seconds. Scale bar 200 μm. D-G. Representative picture of phenotypic analysis performed on DMSO and BME [30 μM] treated 5 dpf larvae, derived from arnt1+/-;vhl+/-(phd3:eGFP) x arnt1-/-;vhl+/-(phd3:eGFP) (n = 540). All the genotype combinations observed are represented in the figure. Among the 405 GFP+ larvae, all the 25 arnt1-/-;vhl-/- showed the aforementioned partially rescued vhl phenotype (D). Fluorescence, exposure = 2 seconds. Scale bar 500 μm. H-J. Representative pictures of 5 dpf CRISPANT mutants created by redundantly targeting arnt2 gene via co-injection of 4x gRNAs in arnt1+/-;vhl+/-(phd3:eGFP) x arnt1-/-; vhl+/-(phd3:eGFP) derived embryos (n = 300). Uninjected embryos were used as control (n = 120). White asterisks: head, liver and tail ragions. Fluorescence, exposure = 991,4 ms. Scale bar 500 μm. K. Statistical analysis performed on mean grey values quantification (at the level of the head, liver and tail), after phenotypic analysis on 5 dpf arnt2 4x gRNAs injected and uninjected larvae. vhl-/- uninjected n = 8 larvae: head 93.1 ± 2.33 (mean ± s.e.m); liver 99.65 ± 3.49 (mean ± s.e.m); tail 29.58 ± 0.73 (mean ± s.e.m). arnt1-/-;vhl-/- uninjected n = 10 larvae: head 56.49 ± 3.36 (mean ± s.e.m); liver 24,7 ± 2.36 (mean ± s.e.m); tail 12.39 ± 0,75 (mean ± s.e.m). vhl-/- injected n = 12 larvae: head 43.69 ± 3.25 (mean ± s.e.m); liver 45.54 ± 4.57 (mean ± s.e.m); tail 16.09 ± 1.37 (mean ± s.e.m). arnt1-/-;vhl-/- injected n = 11 larvae: head 24.66 ± 1.63 (mean ± s.e.m); liver 13.88 ± 0.66 (mean ± s.e.m); tail 5.16 ± 0.33 (mean ± s.e.m). Ordinary One-way ANOVA followed by Sidak’s multiple comparison test (*P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001).
Initial analysis performed on arnt1+/-;vhl+/- incross-derived 5 dpf larvae (F1 generation) confirmed the suppressive effect that arnt1 mutation was expected to have on vhl mutants. Overall, arnt1-/-;vhl-/- larvae showed a substantially attenuated vhl phenotype, characterized by a reduced phd3:eGFP related brightness, especially in the liver (Fig 1C’), with the absence of pericardial edema, excessive caudal vasculature and normal yolk usage (Fig 1C) compared to vhl-/- larvae (Fig 1B and 1B’). In particular, this was quantified as a 39% downregulation (P<0.0017) at the level of the head, a 75% downregulation (P<0.0001) in liver and a 58% downregulation (P<0.0001) in the rest of the body (from the anus to the caudal peduncle), in terms of phd3:eGFP-related brightness, compared to vhl-/- larvae (Figs 1C’, 1B’, S1A and S1D).
Furthermore, since homozygous vhl mutants are lethal by 8-10dpf [47], we analysed the efficacy of arnt1 mutation in rescuing vhl phenotype. To this end, we attempted to raise arnt1-/-;vhl-/- after day 5 post fertilization. Notably, double mutants were able to survive beyond 15 dpf, but failed to grow and thrive when compared to their wild-type siblings, which led us to euthanise them due to health concerns at 26 dpf (S1B Fig). Of note, arnt1 homozygotes, in a vhl/+ or wt background, were morphologically indistinct and adults were viable and fertile. In contrast, the previously published arnt2-/- zebrafish larvae were embryonic lethal around 216 hpf [14].
Arnt1 and Arnt2 are mutually required for HIF signalling in zebrafish
As arnt1;vhl double mutants still activate the phd3:eGFP HIF reporter, we examined the importance of Arnt2 isoform in the HIF pathway. Phenotypic analysis was carried out on 5 dpf Arnt2 CRISPANTs, created both in a vhl+/- and arnt1+/-;vhl+/- background, according to the protocol of Wu et al., 2018 [48]. By analysing the expression of the phd3:eGFP transgene, we observed that arnt2 CRISPR injected vhl mutants were characterized by a significant downregulation of phd3:eGFP-related brightness at the level of the head (equals to 53%, P<0.0001), in the liver (equals to 54%, P<0.0001) and in the rest of the body (equals to 46%, P<0.0001), compared to uninjected vhl mutant larvae (Fig 1H’ compared to 1H, white asterisks; Fig 1K).
Furthermore, when both arnt1 and arnt2 isoforms were simultaneously knocked-out (Fig 1I’), the downregulation was even stronger at the level of the head (equals to 74%, P<0.0001), the liver (equals to 86%, P<0.0001) and in the rest of the body (equals to 83%, P<0.0001) (Fig 1I’ compared to 1H; Fig 1K). Of note, phd3:eGFP-related brightness in these mutants was still slightly higher than wildtype, (not shown; these levels are undetectable). Overall, these data show that Arnt1, even if not fundamental for survival, is the main isoform in the zebrafish liver required for HIF signalling, whereas Arnt2 is more expressed in the developing central nervous system (CNS), as reported by Hill et., al 2009. Of note, since both isoforms can form a functional complex with Hif-α isoforms and appear to function in the same organs, this allows us to confirm that they have partially overlapping functions in vivo and to show that they synergistically contribute to the HIF response.
Modulation of HIF signalling affects GR signalling
To investigate the interaction between HIF and glucocorticoid signalling, we quantified the expression of four potential glucocorticoid target genes from mammalian studies (fkbp5, il6st, pck1 and lipca) both in a HIF upregulated (vhl-/-), and downregulated scenario (arnt1-/-) via RTqPCR analysis on 5 dpf larvae. We confirmed that in zebrafish larvae, fkbp5 is the most sensitive and well-established readout of Gr activity [5,49,50], whilst the other aforementioned genes do not directly take part in the GC-GR negative feedback loop. Therefore, we focused this analysis on fkbp5.
Interestingly, our analysis shows that the expression of fkbp5 is downregulated (fold change = 0.1; P = 0.0035) in the presence of an upregulated HIF pathway (vhl-/-) compared to DMSO treated vhl siblings (Fig 2A). Vice versa, when the HIF pathway is suppressed (arnt1-/-), fkbp5 expression is upregulated (fold change = 24.1; P<0.0001), compared to DMSO treated wild-type levels (Fig 2A’).
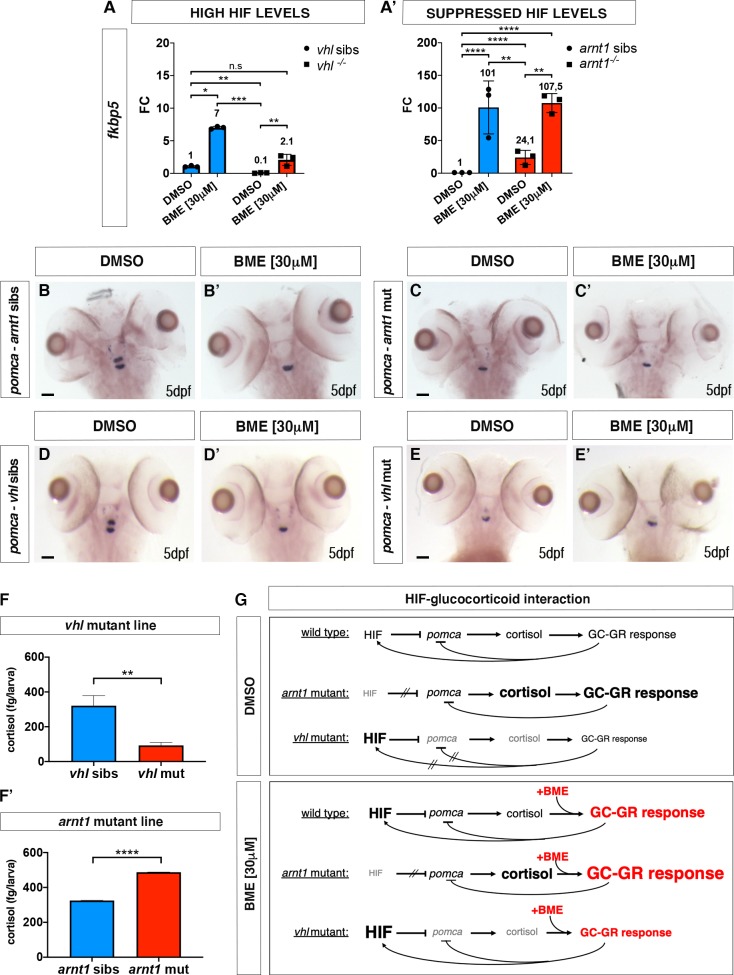
Fig 2. HIF signalling inversely correlate with GC transcriptional activity and cortisol biosynthesis.
A. Schematic view of RTqPCR analysis on fkbp5 expression performed on the vhl+/-(phd3:eGFP) and the arnt1+/-(phd3:eGFP) mutant lines at 5 dpf:. Upregulated (in vhl-/-) HIF signalling repressed Gr activity, whereas arnt1 loss of function derepressed it. Statistical analysis was performed on ΔΔCt values, whereas data are shown as fold change values for RTqPCR analysed samples; ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test (*P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001). B-C’. Representative pictures of WISH performed on DMSO and BME [30 μM] treated arnt1 mutant line, at 5 dpf, using pomca as probe. arnt1 wt DMSO treated (n = 30/30 larvae) showed normal pomca expression; arnt1 wt BME treated (n = 29/30 larvae) showed downregulated pomca expression. In contrast, arnt1-/- DMSO treated (n = 28/30) and arnt1-/- BME treated (n = 30/30) larvae showed downregulated pomca expression. Chi-square test (****P < 0.0001). Scale bar 50 μm. D-E’. Representative pictures of WISH performed on DMSO and BME [30 μM] treated vhl mutant line, at 5 dpf, using pomca as probe. DMSO treated vhl siblings (n = 26/28) showed normal pomca expression; BME treated vhl siblings (n = 28/30) showed downregulated pomca expression. In contrast, vhl-/- DMSO (n = 28/29) and BME (n = 28/28) treated larvae showed downregulated pomca expression. Chi-square test (****P < 0.0001). Scale bar 50 μm. F-F’. Steroid quantification results showed a significantly reduced cortisol concentration (P value <0;0028) in vhl mutants (92.7 fg/larva, in triplicate), compared to vhl siblings (321 fg/larva, in triplicate) at 5 dpf (F). Moreover, a significantly increased cortisol concentration (P value <0;0001) was measured in arnt1 mutants (487.5 fg/larva, in triplicate), compared to arnt1 wild-types (325 fg/larva, in triplicate) at 5 dpf (F’); unpaired t-test (**P < 0.01; ***P <0.001). G. DMSO: Speculative scheme of how the putative HIF-GC crosstalk occurs in wildtypes and how it is affected both in arnt1-/- and in vhl-/- larvae at 5 dpf. In wildtype scenario HIF signalling helps the GC-GR negative feedback to protect the body from an uncontrolled stress response. In particular, we speculate that HIF transcriptional activity is able to inhibit pomca expression when cortisol levels arise over a certain threshold in order to maintain both HIF and GC basal levels. However, in arnt1-/- scenario, the HIF-mediated negative feedback is compromised by the lack of a functional Arnt1. This triggers an initial uncontrolled pomca expression, which increases cortisol levels and subsequently downregulate pomca expression itself. Vice versa, in vhl-/- scenario, the HIF-mediated negative feedback can exert a stronger inhibition of pomca due to the presence of upregulated HIF signalling. This results both in downregulated cortisol levels and in a suppressed GR responsiveness. However, the presence of alternative mechanisms cannot be completely excluded (i.e HIF might interact more directly with GC/GR to impair its function). Finally, the combination high cortisol/low pomca is very rare and this combination may change over the course of development. G. BME: Speculative scheme of how the putative HIF-GC crosstalk occurs in wildtypes and how it is affected both in arnt1-/- and in vhl-/- larvae at 5 dpf, after BME[30μM] treatment. In all the cases, because of betamethasone acts downstream of the HPI axis, by binding directly to Gr, it is able to upregulate glucocorticoid target genes expression. Consequently, since GC are able to stimulate HIF signalling, as expected, we observed an increase phd3:eGFP-related brightness both in wildtypes and in vhl-/-. However, the fact that we did not observed any HIF upregulation both in arnt1-/- and in arnt1-/-;vhl-/-, highlighted the fact that the BME-induced HIF signalling activation is an Arnt1 dependent mechanism.
To further examine the effect of HIF signalling on glucocorticoid responsiveness, we also performed betamethasone (BME) treatment [30 μM] on the aforementioned mutant lines, followed by RTqPCR analysis. Of note, BME was able to increase fkbp5 expression in vhl siblings and was only able to mildly do that in vhl mutants. Indeed, its induction levels appeared not only lower in BME treated vhl mutants (fold change = 2.1) than in BME treated siblings (fold change = 7, P = 0.0286), but also its expression was not significantly different from DMSO treated wild-types (Fig 2A). In contrast, when the HIF pathway was suppressed (arnt1-/-), BME treatment was able to further upregulate the expression of fkbp5 (fold change = 107,5; P = 0.0031), compared to DMSO treated arnt1 mutants (Fig 2A’).
Collectively, we speculate that the upregulated HIF levels are able to repress the glucocorticoid receptor activity and can blunt its responsiveness to an exogenous GR agonist (BME treatment). On the other hand, importantly, although HIF activity is expected to be low in wild-type larvae in a normoxic environment, its function is also detectable with respect to suppression of GR activity. Indeed, if arnt1 gene is knocked-out (arnt1-/-) an increased GR sensitivity is observed (Fig 2A’). To further test whether this had repercussions on steroidogenesis and/or cortisol levels, we analysed them both in a HIF upregulated (vhl-/-) and downregulated scenario (arnt1-/-).
HIF signalling acts as negative regulator of steroidogenesis
To investigate the relationship between HIF signalling and steroidogenesis, we initially performed in situ hybridization on larvae obtained from the arnt1+/- mutant line, using both pro-opiomelanocortin (pomca) and Cytochrome P450 family 17 polypeptide 2 (cyp17a2) as probes. Expression of pomca, at the level of the anterior part of the pituitary gland, is a well-established readout of GR function in zebrafish larvae. Pomca is negatively regulated by increased blood cortisol levels via GC-GR signalling, as part of the HPI axis feedback loop [4,51]. Previous work also suggested that HIF promotes POMC activity in the mouse hypothalamic region [52]. On the other hand, Cyp17a2 is an enzyme involved in steroid hormone biosynthesis at the level of the interrenal gland, which is activated upon ACTH stimulation [53–55].
We found that 5 dpf arnt1-/- larvae, which were characterized by an upregulated GC responsiveness, showed upregulated cyp17a2 expression (S3C–S3C’ Fig) coupled to downregulated pomca (Fig 2C). As expected, arnt1 siblings showed normally expressed cyp17a2 (S3A–S3A’ Fig) and pomca (Fig 2B), which were observed to be downregulated only as a consequence of BME treatment (S3B–S3B’ and S2B’ Figs). Therefore, we speculate that in the absence of arnt1 (HIF suppressed scenario), pomca downregulation is most likely to occur as a consequence of GC-GR induced negative feedback loop, triggered by putative high cortisol levels (Fig 2A and 2G, DMSO, arnt1 mutant).
We subsequently examined both pomca and cyp17a2 expression in the opposite -HIF upregulated- scenario, by performing WISH analysis on the vhl mutant line. Interestingly, 5 dpf vhl-/- larvae, which were characterized by a downregulated GR activity, displayed downregulated cyp17a2 expression (S3G–S3G’ Fig), coupled to downregulated pomca expression (Fig 2E). On the other hand, vhl siblings showed normally expressed pomca (Fig 2D), which was observed to be downregulated after BME treatment, as expected (Fig 2D’). Consequently, we speculate that in the absence of vhl (HIF upregulated scenario), pomca downregulation is most likely to occur as a consequence of HIF-mediated downregulation of pomca expression. (Fig 2G, DMSO, vhl mutant).
Cumulatively, if this is true, we predicted to observe reduced levels of endogenous cortisol in vhl-/- larvae and normal or even increased levels in arnt1-/- larvae at 5 dpf.
Steroidogenesis is repressed in vhl-/- and derepressed in arnt1-/-
To confirm this hypothesis, we performed cortisol quantification on the aforementioned vhl and arnt1 mutant lines. Interestingly, cortisol concentration was significantly reduced (P value <0.0028) in vhl mutant larvae (92,7 fg/larva), compared to vhl siblings (321 fg/larva) (Fig 2F). Conversely, cortisol was significantly increased (P value <0.0001) in arnt1 mutants (487.5 fg/larva), compared to arnt1 siblings (325 fg/larva) (Fig 2F’).
Taken together, these data confirmed our hypothesis and showed for the first time that HIF signalling can act as negative regulator both of GR transcriptional activity and of steroidogenesis. Indeed, if only GR transcriptional activity was blocked by HIF, cortisol levels would be expected to be high in vhl mutants. This is because by blocking GR (i.e as occurs in gr-/-), the GC-GR mediated negative feedback cannot occur, making larvae hypercortisolemic [3,5]. Interestingly, since vhl-/- larvae are characterized both by downregulated cortisol levels and GR transcriptional activity, this strongly suggests that HIF signalling can act both at the hypothalamic level (to inhibit pomca expression) and intracellularly to block GR transcriptional activity itself.
Generating gr and gr;vhl knockout in zebrafish
Conversely, to investigate the role of glucocorticoids on the HIF response, we created a novel glucocorticoid receptor (gr, nr3c1) mutant line and we crossed it with the vhlhu2117/+;phd3:eGFPi144/i144 hypoxia reporter line (this line will be called gr+/-;vhl+/- hereafter). We created this line because the existing grs357 allele may still have some activity via non-genomic pathways or tethering, promoting HIF activation upon GC treatment [4,43,51]. Of note, gr mutants are hypercortisolemic [3,5]. This is due to the inability of glucocorticoids to bind to a functional receptor (GR). As a result, they fail to provide negative feedback and are not able to shut down GC biosynthesis [3,5]. We generated an 11 bp deletion at the level of gr exon 3, which is predicted to truncate the DNA binding domain, lacks the C-terminal ligand binding domain and is predicted to be a true null (Fig 3A). The homozygous gr/nr3c1 mutants, characterized during the first 5dpf, were morphologically similar to control siblings and adult fish were viable and fertile, as predicted [5].
Fig 3. gr mutation partially rescues vhl phenotype.
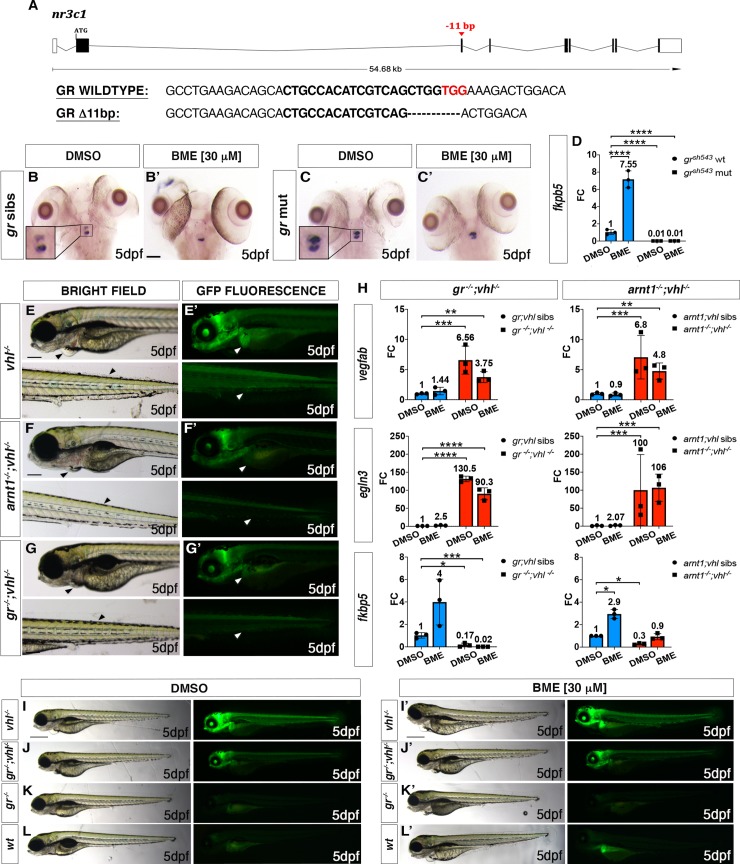
A. Schematic representation of zebrafish gr (nr3c1) gene. Exons are shown as boxes, introns as lines. The red arrowhead shows the position of a -11 bp deletion in exon 3 (encoding the DNA binding domain). gr wt and mutant sequence. CRISPR target site: bold. PAM sequence: red. B-C’. Representative pictures of WISH performed on DMSO and BME [30 μM] treated gr mutant line, at 5 dpf, using pomca as probe. Scale bar 100 μm. gr siblings DMSO treated (n = 30/30 larvae) showed normal expression; gr siblings (n = 29/30 larvae) showed downregulated pomca expression after BME treatment. Both DMSO treated (n = 30/30) and BME treated (n = 30/30) gr-/- larvae showed upregulated pomca expression. D. RTqPCR analysis performed on gr wt (n = 10; 3 repeats) and gr-/- (n = 10; 3 repeats) larvae at 5 dpf, using fkbp5 as probe. Statistical analysis was performed on ΔΔCt values, whereas data are shown as fold change values. Ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test (****P < 0.0001). E-G. Magnified picture of representative gr-/-; vhl-/- larvae compared to arnt1-/-;vhl-/- and vhl-/- larvae. Both double mutants are characterized by the absence of pericardial oedema, no ectopic extra vasculature at the level of the tail, no bright liver and a reduced brightness in the rest of the body (white and black arrowheads), compared to vhl-/- larvae. Fluorescence, exposure = 2 seconds. Scale bar 200 μm. H. RTqPCR analysis performed both on HIF and GC target genes expression carried out on gr-/-; vhl-/- and sibling at 5 dpf, (n = 10 larvae, per group, in triplicate) compared to arnt1-/-;vhl-/- larvae and siblings, at 5dpd (n = 10 larvae, per group, in triplicate). Both vegfab and egln3 are HIF target genes, whereas fkbp5 is a GC target gene. Statistical analysis was performed on ΔΔCt values, whereas data are shown as fold change values, Ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test. I-L. Representative picture of phenotypic analysis performed on DMSO and BME [30 μM] treated gr+/-; vhl+/-(phd3:eGFP) incross-derived 5 dpf larvae (n = 600). All the genotype combinations observed are represented in the figure. Among the 450 GFP+ larvae analysed, 28 showed a partially rescued vhl phenotype which resembled the arnt1’s one. Three experimental repeats. In all panels: *P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001. Fluorescence, exposure = 2 seconds. Scale bar 500 μm.
To confirm loss-of-function, we initially subjected larvae to a visual background adaptation (VBA) test, as it is linked to impaired glucocorticoid biosynthesis and action [4,56]. Larvae derived from gr+/- incross were VBA tested and sorted according to melanophore size at 5 dpf. PCR-based genotyping on negative VBA-response sorted samples revealed that most larvae were homozygous for the gr allele, whereas positive VBA-response samples were always gr siblings.
Furthermore, WISH analysis performed on 5 dpf DMSO and BME treated gr+/- incross derived larvae using pomca as probe, showed the presence of upregulated pomca expression in DMSO treated gr-/- at the level of the anterior part of the pituitary gland (Fig 3C), compared to wild-type siblings (Fig 3B). Of note, BME treatment was not able to downregulate pomca levels in gr-/-(Fig 3C’), as it occurs in BME treated siblings (Fig 3B’) via GC-GR mediated negative feedback loop, due to the absence of a functional gr allele. Finally, the loss of function was also determined in 5 dpf gr mutants by the strong downregulation of fkbp5 mRNA levels quantified via RTqPCR, both in the presence (fold change = 0.01; P<0.0001) and in the absence of BME treatment (DMSO treated, fold change = 0.01; P<0.0001), compared to DMSO treated wildtypes (Fig 3D).
gr mutation partially rescues vhl phenotype
We next analyzed the effect of gr loss of function on vhl phenotype. Phenotypic analysis carried out on 5dpf larvae, derived from gr+/-;vhl+/- incross, revealed that nr3c1 mutation was able to cause an efficient, but not complete rescue of vhl phenotype, in a way which resembled arnt1 mutation (Fig 3F’ and 3G’).
In particular, 5dpf gr-/-;vhl-/- larvae showed a 43% downregulation at the level of the head (P<0.0001), a 66% downregulation in the liver (P<0.0001) and a 51% downregulation in the tail (from the anus to the caudal peduncle) (P = 0.0020), in terms of phd3:eGFP-related brightness, compared to vhl-/- larvae (Figs 3G’ compared to 3E’ and S4A). As expected, 5 dpf double mutant larvae were unable to respond to BME [30 μM] treatment (Figs 3J–3J’ and S4A), as also confirmed via RTqPCR analysis on HIF (vegfab and egln3) and GC target genes (fkbp5) (Fig 3H).
Rescue was also apparent by morphology. Indeed, even if gr-/-;vhl-/- showed reduced yolk usage, they displayed a reduction in ectopic vessel formation at the level of the dorsal tailfin, no pericardial edema, and developed air-filled swim bladders (Fig 3G and 3E). Moreover, whilst vhl mutants are inevitably deceased by 10 dpf [47], we were able to raise all selected double mutants beyond 15 dpf, but then (similarly to arnt1-/-;vhl-/-) they failed to grow and thrive when compared to their siblings. This led us to euthanise them due to health concerns at 21 dpf (S4B Fig). Together, these data indicate for the first time, in our in vivo animal model, that GR function is essential for HIF signalling in zebrafish larvae, particularly at the level of the head and the liver.
gr loss of function can further reduce HIF signaling in arnt1;vhl double mutants
The similarity of gr and arnt1 mutations could mean they work in a single linear “pathway”. If true, mutation of both genes should not lead to a further attenuation of the reporter expression. To test this, we bred the gr mutant line with the arnt1;vhl double mutant line and we crossed gr+/-;arnt1+/-;vhl+/- triple carriers. Phenotypic analysis carried out on 5 dpf phd3:eGFP positive larvae (n = 488) showed a small class of larvae with an even more rescued phenotype and a stronger downregulation of phd3:eGFP related brightness compared both to arnt1-/-;vhl-/- (Fig 4B–4B’) and gr-/-;vhl-/- double mutants (Fig 4C–4C’). Of note, 7 putative very weak GFP+ larvae were selected and genotypic analysis confirmed that 5 out of 7 were indeed gr-/-; arnt1-/-;vhl-/-. In particular, these triple mutants showed a 54% downregulation at the level of the head, a 71% downregulation in the liver and a 72% downregulation in the tail region, in terms of phd3:eGFP-related brightness compared to vhl-/- (Figs 4D–4D’ and S5). Thus, these data suggest that glucocorticoids are likely to act on both Arnt1 and Arnt2 mediated HIF signalling pathway.
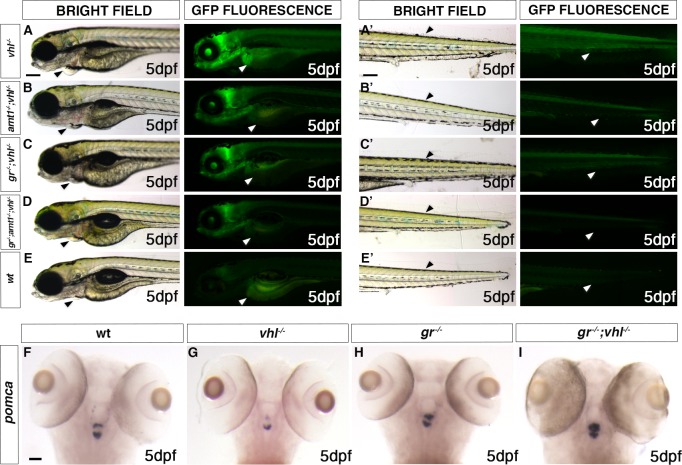
Fig 4. gr loss of function effect is stronger when HIF-signalling is moderately upregulated.
A-E. Representative picture of the main differences between vhl-/-, arnt1-/-;vhl-/-, gr-/-;vhl-/- and triple gr-/-;arnt1-/-;vhl-/- larvae at 5 dpf. Among the 488 phd3:eGFP expressing larvae analysed, 7 larvae were characterized by the absence of pericardial oedema (black arrowheads, left), no ectopic extra vasculature at the level of the tail (black arrowheads, right), no visible phd3:eGFP HIF reporter in the liver (white arrowheads, left) and even more reduced levels of this marker in the head and in the rest of the body (white arrowheads, right). Genotypic analysis allowed to confirm the presence of a genotype-phenotype correlation in 5 out 7 samples and to prove that they were triple mutants. Fluorescence, exposure = 2 seconds. Scale bar 200 μm. F-I. Representative pictures of WISH performed on gr+/-; vhl+/- incross derived larvae, at 5 dpf, using pomca as probe. Of note, gr-/-;vhl-/- showed upregulated pomca expression (20/20 larvae), as observed in gr-/- (20/20 larvae); vhl mutants showed downregulated pomca expression (20/20 larvae), whereas wildtypes showed normal pomca expression (19/20). Chi-square test (****P < 0.0001). Scale bar 50 μm.
The BME-induced HIF response is Arnt1 dependent
To further examine the effect of glucocorticoids on HIF signalling, we performed BME [30 μM] treatment on all the available mutant lines. Of note, unlike cortisol, betamethasone has a very high affinity for Gr, but an insignificant affinity for Mr [57,58]. As expected, 5 dpf wild-types larvae showed a mild upregulation of phd3:eGFP-related brightness at the hepatic level, compared to untreated controls (Figs 1G’ and 3L’). BME treatment was also able to further increase phd3:eGFP-related brightness at the level of the head and the liver of 5 dpf vhl-/-, as also confirmed by WISH, using both lactate dehydrogenase A (ldha) (Fig 5B–5B’, black arrowheads) and prolyl hydroxylase 3 (phd3) as probes (Fig 5D–5D’, black arrowheads). As predicted, both gr-/- and gr-/-;vhl-/- mutants were unaffected due to the absence of functional Gr (Fig 3K–3K’ and 3J–3J’). Interestingly, in both the arnt1-/-(Fig 1F) and also arnt1-/-;vhl-/- (Fig 1E) the phd3:eGFP-related brightness did not change after BME treatment (Fig 1E’ and 1F’). This was also confirmed via, RTqPCR analysis carried out on HIF target gene egln3/phd3 in the arnt1 mutant (S1C Fig; see also arnt1-/-;vhl-/- Fig 3H)
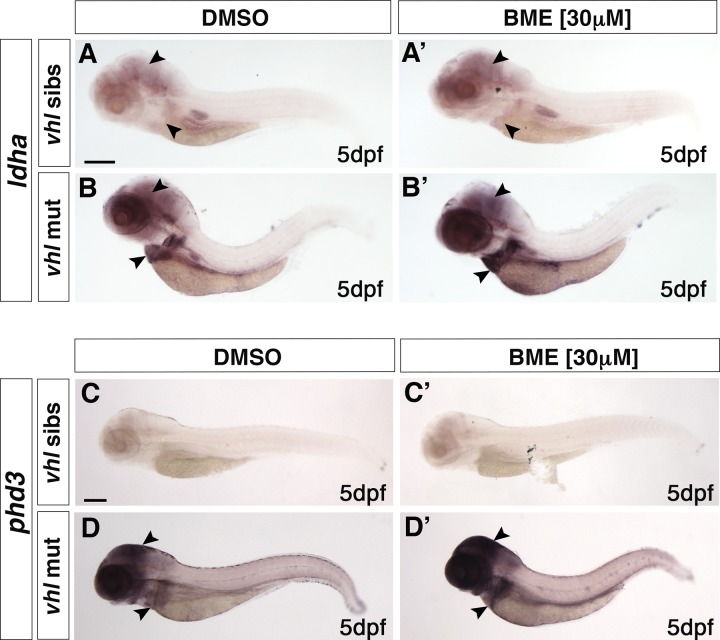
Fig 5. BME treatment is able to upregulate HIF signalling in vhl-/-.
A-B’. Representative pictures of WISH performed on DMSO (A-B) and BME [30 μM] (A’-B’) treated vhl+/- incross derived larvae, at 5 dpf, using ldha as probe. DMSO treated vhl siblings showed basal ldha expression (34/35 larvae), which showed to be upregulated after BME treatment (33/35 larvae). On the other hand, DMSO treated vhl-/- showed upregulated ldha expression (32/35 larvae), which was further upregulated after BME treatment (34/35 larvae). (black arrowhead: head and liver) Chi-square test (****P < 0.0001). Scale bar 200 μm. C-D’. Representative pictures of WISH performed on DMSO (C-D) and BME [30 μM] (C’-D’) treated vhl+/- incross derived larvae, at 5 dpf, using phd3 (egln3) as probe. As expected, vhl siblings DMSO treated (n = 30/30 larvae) showed basal phd3 expression, which was mildly increased after BME treatment (n = 27/30 larvae). Vhl-/- DMSO treated (n = 28/30 larvae) showed upregulated phd3 expression, which was further increased after BME treatment (n = 26/30 larvae). (black arrowhead: head and liver) Chi-square test (****P < 0.0001). Scale bar 200 μm.
Taken together these data suggest that in vhl-/- larvae, BME treatment can upregulate HIF signalling by “bypassing” HIF-mediated pomca negative regulation. By directly binding to Gr, it can compensate for the repressed cortisol levels in vhl mutants. (Fig 2F). On the other hand, both in arnt1-/- and also arnt1-/-;vhl-/- larvae, even if BME can act downstream of pomca, it can only upregulate GR responsiveness, but cannot upregulate HIF signalling due to arnt1 loss of function. Cumulatively, we speculate that even if Arnt2 can interact with the HIF-α isoforms to maintain a moderately upregulated HIF levels (arnt1-/-;vhl-/-), the BME-mediated HIF upregulation is Arnt1 dependent.
gr mutation overrides HIF-mediated pomca suppression in a vhl deficient background
To examine the effect of gr loss of function on steroidogenesis in gr-/-;vhl-/-, we performed WISH analysis on 5 dpf gr+/-;vhl+/- incross derived larvae, using pomca as probe. As expected, vhl-/- showed downregulated pomca expression (Fig 4G), whereas gr -/- displayed upregulated pomca (Fig 4H), compared to wildtypes (Fig 4F). Notably, since a strong upregulation of pro-opiomelanocortin a was observed in the double mutants (Fig 4I), this suggests that gr mutation overrides HIF-mediated pomca inhibition. PCR-analysis performed post-WISH confirmed this genotype-phenotype correlation.
These data, in accordance with our hypothesis, suggest that in gr-/-;vhl-/- mutants the upregulation of pomca, triggered by the absence of functional Gr (and of the GC-Gr mediated negative feedback), cannot be inhibited with the same efficiency by HIF activity at the hypothalamic level. In gr-/-;vhl-/- mutants, we speculate that the upregulated endogenous cortisol interacts with Mr to stimulate the HIF pathway, resulting in a mildly upregulated phd3:EGFP expression, in-between the levels seen in vhl mutants and wild-type larvae (Fig 3J and 3I). To test this assumption, we set up to block the mr gene in a gr-/-;vhl-/- background, in order to check the importance of Mr in the HIF signaling pathway.
Both Gr and Mr are directly required in the HIF signalling pathway
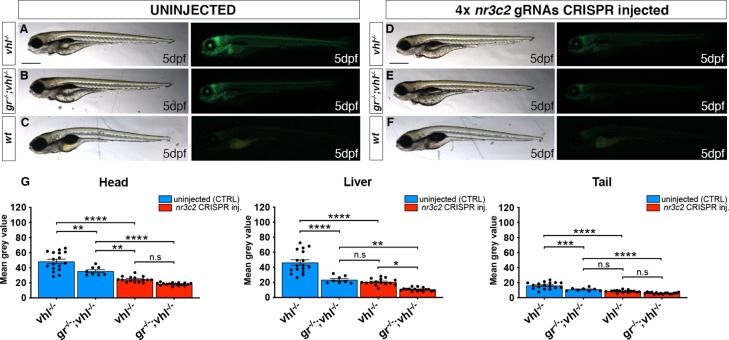
Cortisol has high affinity both for Gr and Mr and they have been recently shown to be differentially involved in the regulation of stress axis activation and function in zebrafish [3]. Therefore, we analysed the role of Mr on the HIF signaling pathway. To achieve this, we knocked-out mr in gr+/-;vhl+/-;phd3:eGFP incross-derived embryos, using CRISPant technology [48,59]. Interestingly, phenotypic analysis performed on 5 dpf injected and uninjected larvae revealed that mr CRISPR injected vhl mutants were characterized by a significant downregulation of phd3:eGFP-related brightness at the level of the head (equals to 49%, P<0.0001), in the liver (equals to 56%, P<0.0001) and in the rest of the body (equals to 47%, P<0.0001), compared to vhl-/- mutant uninjected larvae (Fig 6D compared to 6A). Moreover, when both gr and mr were knocked-out, the downregulation was even stronger at the level of the head (equals to 62%, P<0.0001), in the liver (equals to 77%, P<0.0001) and in the rest of the body (equals to 63%, P<0.0001) compared to vhl-/- mutant uninjected larvae (Fig 6E compared to 6A). Of note, mr injection in vhl-/- larvae was more efficient in downregulation of phd3:eGFP expression compared to uninjected gr-/-;vhl-/- larvae at the level of the head (equals to 31%, P = 0.0087) (Fig 6D compared to 6B).
Fig 6. Both Gr and Mr are directly required in the HIF signalling pathway.
A-F. Representative pictures of 5 dpf CRISPANT mutants created by redundantly targeting nr3c2 (mr) gene via co-injection of 4x gRNAs in gr+/-;vhl+/-(phd3:eGFP) x gr-/-; vhl+/-(phd3:eGFP) derived embryos (n = 344). Uninjected embryos were used as control (n = 170). Fluorescence, exposure = 991,4 ms. Scale bar 500 μm. G. Statistical analysis performed on mean grey value quantification (at the level of the head, liver and tail), after phenotypic analysis, on 5 dpf mr 4x gRNAs injected and uninjected larvae. vhl-/- uninjected n = 17 larvae: head 48.28 ± 2.99 (mean ± s.e.m); liver 46.47 ± 3.55 (mean ± s.e.m); tail 16.15 ± 1.06 (mean ± s.e.m). gr-/-;vhl-/- uninjected n = 8 larvae: head 35.48 ± 2.03 (mean ± s.e.m); liver 23.56 ± 1.72 (mean ± s.e.m); tail 10.98 ± 0.75 (mean ± s.e.m). vhl-/- injected n = 15 larvae: head 24.62 ± 0.97 (mean ± s.e.m); liver 20.67 ± 1.1 (mean ± s.e.m); tail 8.57 ± 0.39 (mean ± s.e.m). gr-/-;vhl-/- injected n = 16 larvae: head 18.33 ± 0.46 (mean ± s.e.m); liver 10.71 ± 0.56 (mean ± s.e.m); tail 6.07 ± 0.26 (mean ± s.e.m); ordinary One-way ANOVA followed by Sidak’s multiple comparison test.
To test the reliability of CRISPant method, we chose to knock-out a gene (which was not involved in the HIF pathway) into vhl+/- incross derived embryos, to test whether it was able to affect HIF signalling. Laminin, beta 1b (lamb1b), which codes for an extracellular matrix glycoprotein, was injected as CRISPR-injection control in vhl+/-incross derived embryos at 1 cell stage. Genotypic analysis carried out on these larvae confirmed that these guides were effective. Finally, quantification of phd3:eGFP-related brightness performed on 5 dpf injected and uninjected vhl-/- larvae, showed no significant differences between the two groups (S6A and S6C Fig). Overall, these data corroborated the efficiency of the CRISPant method and, at the same time, confirmed that both glucocorticoid and mineralocorticoid receptor play a pivotal role in the HIF signalling in vivo.
Discussion
Both HIF and glucocorticoid mediated transcriptional responses play a pivotal role in tissue homeostasis, glucose metabolism and in the regulation of cellular responses to various forms of stress and inflammation [60–62]. Previous in vitro studies highlighted the potential for crosstalk between HIF and glucocorticoid pathways, however there are still conflicting data on how this interaction occurs in vivo and there is no information on Mr contribution to HIF signalling. In this regard, we have presented a novel in vivo study using zebrafish larvae, focusing on the crosstalk between these two pathways. In contrast to in vitro cell culture studies, a whole animal study allows us to consider the interactions that occur between various tissues and provide novel insights. To this end, we generated arnt1 and gr null mutants to downregulate HIF and GR signalling respectively, as a basis for a genetic analysis of this crosstalk.
As a prelude to this, we had to establish the relative importance of arnt1 and arnt2 in the overall HIF response. To achieve this, a discriminative test was devised to place them in a vhl mutant background, where HIF signaling is strongly upregulated [25,42]. Phenotypic analysis performed on 5 dpf arnt1-/-;vhl-/- larvae showed reduced phd3:eGFP related brightness, normal yolk usage, properly developed and air-filled swim bladder as well as by the absence of pericardial oedema and excessive caudal vasculature. However, beyond 5 days, these double mutants exhibited only partial recovery from the vhl phenotype. Indeed, they developed well till 15 dpf, but subsequently failed to grow and thrive when compared to their siblings. In addition, arnt1 homozygous mutants were found to be viable and fertile, in contrast to both homozygous vhl and arnt2 mutants, which are embryonic lethal by 8–10 dpf [14,47].
Even though Arnt1 is not fundamental for survival, we found that it is required in the liver and in organs outside the central nervous system for HIF−α function. Conversely, using CRISPant technology [48,59], we established that Arnt2 is mainly required in the developing central nervous system (CNS), as also reported by Hill et al. in 2009 [14]. However, the similarities observed in terms of phd3:eGFP-induced brightness in both arnt1-/-;vhl-/- and arnt2 CRISPR injected vhl mutants, suggest there is no strong functional separation. Therefore, both Arnt2 and Arnt1 have partially overlapping functions in vivo and both contribute to the HIF response.
The effect of HIF signalling on the glucocorticoid pathway
We next investigated the effect of HIF signalling and glucocorticoid responsiveness, by performing RTqPCR analysis on 5 dpf larvae. Collectively, we show that strong activation of HIF signalling (in vhl-/-) is able to blunt glucocorticoid receptor transcriptional regulation as judged by fkbp5 expression, whereas arnt1 loss of function derepressed it. As our experiments are done at normal atmospheric oxygen levels, we conclude from the latter result that normoxic HIF activity nevertheless suffices to attenuate GR transcriptional regulation.
We checked whether HIF signalling affects steroidogenesis. To this end, we quantified the expression of steroidogenesis-related genes (pomca and cyp17a2) both in vhl-/- and in arnt1-/- larvae, via whole-mount in situ hybridization. Surprisingly, both lines showed downregulation of pomca expression. However, arnt1-/- larvae showed upregulated cyp17a2 expression, whereas vhl-/- larvae, were characterized by downregulated cyp17a2.
Considering our results with GR-target fkbp5 in these mutants, we assume that in an arnt1 knock-out scenario, pomca downregulation occurs as a consequence of the GC/GR-mediated negative feedback loop aimed to control cortisol biosynthesis. This is also consistent with a significant upregulated basal cortisol levels quantified in these mutants. Vice versa, when HIF signalling is upregulated (in vhl mutants) we speculate that pomca and cyp17a2 downregulation may occur via HIF-mediated activity, leading to the observed low cortisol levels coupled to suppressed GR activity.
Indeed, glucocorticoids regulate a plethora of physiological processes, act on nearly every tissue and organ in the body to maintain homeostasis and are characterized by a potent anti-inflammatory and immunosuppressive actions. For these reasons, their secretion must be finely controlled by the HPA/I axis [63].
As previous work in our laboratory showed that glucocorticoids also act as HIF activators [25,43], we infer that HIF can in turn control GC levels by acting on pomca. This would enable HIF signalling not only to control its own levels, but also to assure homeostasis. Finally, since HIF signalling is a master regulator of cellular pro-inflammatory responses to hypoxia [64–66], which would counteract the anti-inflammatory glucocorticoid activity, we speculate that the simultaneous expression of both upregulated HIF and GC pathway would be detrimental to homeostasis.
Our data would also be in accordance with a previous study showing that hypoxia exposure resulted in downregulation of steroidogenic genes (StAR, cyp11c1, hmgcr, hsd17b2, cyp19a, cyp19b) in 72 hpf larvae, whereas zHIF-α loss of function triggered the upregulation specifically of StAR, cyp11b2 and cyp17a1 [67].
Cumulatively, if this is true, we predicted to observe reduced levels of endogenous glucocorticoids in vhl-/- and normal or even increased levels in arnt1-/-. Importantly, the fact that cortisol levels were lowered in vhl mutants and were upregulated in arnt1 mutants is consistent with our hypothesis.
As a consequence of the above considerations, the HIF-mediated pomca negative regulation seems to be a logic homeostatic interaction: Increased HIF reduces GR activity, which in turn should lead to less HIF signalling.
The effect of glucocorticoids on the HIF signaling pathway
To further investigate the role of glucocorticoids on the HIF signalling, we initially analyzed the effect of gr loss of function on vhl phenotype. Surprisingly, we observed that gr mutation was able to cause an efficient, but not complete rescue of the vhl phenotype. Notably, gr-/-;vhl-/- survived much longer than vhl-/- (> = 21 dpf compared to max. 10 dpf), but then similar to arnt1-/-;vhl-/-, they failed to grow and thrive when compared to siblings. Our previous work [43] established that activation of GR signalling negatively regulates VHL protein in human liver cells. Our current genetic analysis shows that in zebrafish larvae, there must be an additional point of interaction between these two pathways, as we observed further activation of our HIF reporter after GC treatment even in the absence of VHL. Cumulatively, we showed for the first time in an in vivo animal model that Gr is fundamental to allow high HIF signalling levels.
We next analysed the effect of betamethasone treatment in arnt1-/-. Although BME activated the GR target fkbp5, as expected, it failed to activate HIF signaling [43]. This was unexpected and would be best explained by assuming that a Gr-BME complex would preferentially interact with a HIFα/ARNT1 complex but not a HIFα/ARNT2 complex. Whether this holds up in mammalian cells would be interesting to address.
Evaluation of mineralocorticoid receptor contribution to HIF signalling
Recent work published by Faught and Vijayan, 2018 showed that both Gr and Mr are involved in the regulation of zebrafish stress axis activation and function [3]. Nothing is known about mineralocorticoid receptor contribution to HIF signalling. Therefore, we tested the effect of mr knock-out in gr+/-;vhl+/-;phd3:eGFP incrossed derived embryos. Interestingly, in mr injected- vhl-/- we observed a significant reduction of phd3:eGFP-related brightness, compared to uninjected vhl-/- larvae. Moreover, a further reduction of phd3:eGFP expression was found at the level of the head in mr injected- vhl-/- compared to gr-/-;vhl-/- larvae. Finally, the additional removal of mr in a gr-/-;vhl-/- background reduced the hypoxia reporter expression even further.
Therefore, we were able to show that both the glucocorticoid receptor and mineralocorticoid receptor play a pivotal role in promoting HIF signaling in zebrafish. In contrast to mammals, teleosts lack aldosterone and cortisol is the primary glucocorticoid hormone that can interact both with Gr and Mr to assure a correct HPI axis activity [68,69]. Of note, Mr was shown to not have a role in rapid non-genomic behaviors that required HPI axis signaling in zebrafish [70]. However, our hypothesis is consistent with Faught and Vijayan, 2018 elegant work, showing that both Gr and Mr signalling are involved in the GC negative feedback regulation. Importantly, this outcome may have a wider significance in health and disease. This is because so far, HIF signalling, which plays a key role in tumour growth, is proven difficult to downregulate. In this regard, our study suggests that modulation of Gr and Mr might be a potential avenue. In conclusion, although Mr contribution to HIF response in other organisms remains unclear, our work suggests that research into its function is warranted.
Conclusion
Our present study stresses the importance of the glucocorticoid pathway in driving HIF signalling. In addition, we uncovered a negative regulatory role played by HIF in regulating both GR responsiveness and steroidogenesis as demonstrated via RTqPCR and steroid hormone quantification. We also identified a mineralocorticoid receptor contribution to HIF-GC crosstalk. Finally, we presented novel gr+/-;vhl+/-, arnt1+/-;vhl+/- and arnt1+/-;gr+/-;vhl+/- zebrafish mutant lines which helped to better understand how the interplay between HIF and glucocorticoids occurs in vivo. For these reasons, we believe that this work could pave the way for further in vivo analysis to precisely identify the extensive crosstalk behind these two major signalling pathways.
Materials and methods
Ethics statement
Zebrafish (Danio rerio) lines were raised and maintained under standard conditions (14 hours of light and 10 hours of dark cycle, at 28°C) in the Aquaria facility of the University of Sheffield. Zebrafish embryos used for experiments were reared in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM MgCl2, 0.33 mM CaCl2, pH 7.2) with or without methylene blue (Sigma-Aldrich) and staged according to standard methods [71] for up to 5,2 days post fertilisation (dpf) in accordance with UK Home Office legislation. Our studies conform with the UK Home Office guidelines (ASPA), Licence No. PC9C3D4CB and PB2866ED0. Ethics approval was obtained from the University of Sheffield Ethics committee AWERB.
Zebrafish strains
The following zebrafish lines were used: wild-type (wt) strain AB (ZDB-GENO-960809-7), vhlhu2117/+;phd3:eGFPi144/i144 (ZDB-GENO-090611-18), hif1βsh544/+; hif1βsh544/+;vhlhu2117/+, grsh543/+, grsh543/+;vhlhu2117/+, grsh543/+;hif1βsh544/+; and grsh543/+;hif1βsh544/+;vhlhu2117/+ lines were generally maintained in a phd3:eGFPi144/+ background. The following 4x gRNAs CRISPR-injected G0 null mutant lines were created according to Wu et al, 2018 [48] protocol and raised up to to 5,2 dpf: mr;grsh543/+;vhlhu2117/+;phd3:eGFPi144/+, hif1β2;1hif1βsh544/+;vhlhu2117/+;phd3:eGFPi144/+ and lamb1b;vhlhu2117/+;phd3:eGFPi144/+.
Generation of gr (nr3c1) and hif1β (arnt1) null zebrafish lines
Both nr3c1 mutant line (grsh543/+) and arnt1 mutant line (hif1βsh544/+) were generated using the CRISPR/Cas9-based mutagenesis method. A gene-specific guide RNA (sgRNA) sequence was identified using the CHOPCHOP website [72,73]. To design both gr and arnt1 sgRNA, an 18 nucleotides sequence upstream to a selected PAM site (grsh543: CCAGCTGACGATGTGGCAG; hif1βsh544: TCGGTGCTGGTGTTTCCAG) was inserted into a scaffold sequence [74], containing a promoter for the T7 Polymerase. The sgRNA was amplified via PCR, purified from agarose gel and in vitro transcribed using MEGAshortscript T7 kit (Ambion). 1 nl of CRISPR mixture containing 2,4 μg/μl of gRNA and 0.5 μl Cas9 protein (NEB) was injected in one-cell stage embryos and raised for 24 hours. Wild-type (wt), strain AB embryos were used to generate the gr mutant line, whereas vhlhu2117/+;phd3:eGFPi144/+ incross-derived embryos were used to create the hif1β mutant line. Efficiency was verified via whole-embryo PCR-based genotyping, by a diagnostic restriction digest. Injected embryos were raised to adulthood. Embryos collected from transmitting G0 founders crossed with WT(AB) fish were raised and genotyped to confirm germline transmission of the mutation (F1 generation). Heterozygous mutants, carrying the same mutation, were selected and crossed to obtain homozygous mutant embryos (F2 generation).
Generation of CRISPR/Cas9-mediated mutants (CRISPANTs)
To generate G0 knockout embryos we used the method developed by Burger et al 2016 [59] and improved by Wu et al., 2018 [48]. In short, a pool of four guide-RNAs (25μM each, Sigma Aldrich) were co-injected with 0.5 μl Cas9 protein (NEB, M0386, 20μM), diluted 1:10) and 1 μl tracrRNA (100μM) in one-cell stage embryos. This method was used to create G0 CRISPANTs for the following genes of interest: mineralocorticoid receptor (mr, nr3c2), aryl hydrocarbon receptor nuclear translocator 2 (arnt2, hif1β2) and laminin, beta 1b (lamb1b). The latter was used as CRISPR-injection control. The gRNA target sequences used in this study are as follows: arnt2: gRNA1-ACGGCGCCTACAAACCCTCC (exon 5), gRNA2-GGCCGATGGCTTCTTGTTCG (exon6), gRNA3-TTCACGCCACAATTCGGATG (exon11), gRNA4-GTCGCAGGTGCGTAAAAACA (exon 14); nr3c2: gRNA1-GCATTGTGGGGTCACCTCCA (exon 2), gRNA2-AAGGGGATTAAACAGGAAAC (exon 2), gRNA3-CAACCAGCTCGCCGGAAAAC (exon 5), gRNA4-ATATCTGACGCCGTCCGTCT (exon 5); lamb1b gRNA1-TTGTTAATAGCATAGTACATTGG (sequence upstream 5’UTR), gRNA2-GGAGAACAAGCAAAACGATGAGG (ATG), gRNA3- GCGTGGTGCAGGGTTTGTAG (5’UTR), gRNA4- TCACAATGACATGTGTGCG (exon 2). The success of the injection was determined via phenotypic analysis, followed by quantification of phd3:eGFP related brightness and whole-embryo PCR-based genotyping performed on a fraction of injected embryos at 5 dpf.
Whole-mount in situ hybridisation
Whole-mount in situ hybridization (WISH) was performed according to standard protocols [75]. The following antisense RNA probes were used: proopiomelanocortin a (pomca) created as previously described [76]; Cytochrome P450 family 17 polypeptide 2 (cyp17a2), created as previously described [55], both prolyl hydroxylase 3 (phd3; BC066699), and lactate dehydrogenase A (ldha1; BC067188) probes, generated as previously described [25,47].
Embryos harvesting, drug treatment and fixation for WISH
Embryos intended for whole-mount in situ hybridisation were treated with 16,8 μl of 1-phenyl 2-thiourea (PTU, stock concentration 75mg/ml) diluted into 35 ml E3 medium to inhibit melanogenesis, according to Karlsson et al., 2001 [77]. GR agonist treatment was performed on batches of 15 embryos each, at 4 dpf, treated in 6-well plates, with 30 μM Betamethasone 17,21-dipropanoate (BME) and with 1% DMSO (Sigma-Aldrich), as control, for 24 hours [4]. Inside the 6-well plates, embryos were incubated in 3 ml total volume of E3 medium, without methylene blue. Afterwards, up to 30 embryos at 5 dpf were collected in 1,5 ml Eppendorf tubes and anaesthetized using Tricaine Solution (MS-222, Sigma Aldrich) prior to fixation in 1 ml 4% PFA solution overnight, at 4°C. Embryos were then washed twice for 10 minutes in PBST and post-fixed in 1 ml 100% MeOH. Finally, samples were stored at -20°C.
grsh543 mutants sorting by visual background adaptation (VBA)
Visual background adaptation (VBA) is a glucocorticoid receptor-dependent neuroendocrine response which causes zebrafish melanocytes to shrink when exposed to bright illumination [78,79]. To identify grsh543 mutants from siblings and to confirm the absence of a functional VBA response, 5dpf larvae were exposed to 30 minutes darkness and then transferred onto a white background under bright, whole-field illumination, using a 30W fluorescent lamp mounted 50 cm above the dish [80,81].
Cortisol extraction and quantification
Three biological replicates of 150 larvae at 5 dpf each of hif1βsh544 mutants, hif1βsh544 siblings, vhlhu2117 mutants and vhlhu2117 siblings, respectively, were used for steroid hormone extraction and quantification. vhl-/- larvae were sorted among siblings at 4 dpf according to both their phenotype and phd3:eGFP-related brightness. Because of the lack of visible phenotype, arnt1-/- larvae where derived from arnt1-/- fish incrossed, whereas siblings were from arnt1+/- fish crossed with arnt1+/+ ones. Cortisol quantification was carried out according to the protocol published by Eachus et al., 2017 [55], based on the use of an Acquity UPLC System (Waters, Milford, CT) coupled to a Xevo TQ-S tandem mass spectrometer (Waters).
RNA isolation, cDNA synthesis and qPCR analysis
Transcript abundance of target genes was measured by quantitative real-time PCR (RTqPCR). Three biological replicates of 10 larvae at 4 dpf each, were treated for 24 hours with 30 μM Betamethasone 17,21-dipropanoate and with 1% DMSO, used as control, prior to RNA isolation. Total RNA was extracted from pools of 10 larvae at 5dpf with TRIzol reagent (Invitrogen by Thermo Fisher Scientific, 15596026). RNA extracted was quantified using a Nanodrop ND-1000 spectrophotometer. cDNA was then synthesized from 1μg RNA template through reverse transcription using Protoscript II First Strand cDNA Synthesis Kit (New England Biolabs), as recommended by manufacturer’s instructions. All RTqPCR reactions were performed in triplicate using TaqMan probes in combination with CFX96 Touch Real-Time PCR Detection System (BioRad), paired with CFX Maestro Analysis Software.
Each reaction mixture (20 μl) reaction mixture containing 1 μl cDNA template (100ng/ml), 1 μl FAM probe and 10 μl TaqMan Universal Master Mix (Applied biosystems by Thermo Fisher Scientific, Epsom, UK) was amplified as follows: denaturation at 95°C for 10 minutes and 39 cycles at 95°C for15 seconds, 60°C for 30 seconds. Four hypoxia-inducible factor dependent genes (egln3: Dr03095294_m1, pfkfb3: Dr03133482_m1, vegfab: Dr03072613_m1 and slc2a1a: Dr03103605_m1) and four glucocorticoid dependent genes (fkbp5: Dr03114487_m1, il6st: Dr03431389_m1, pck1: Dr03152525_m1 and lipca: Dr03113728_m1) were quantified in the present study (Applied biosystems by Thermo Fisher Scientific, Epsom, UK).
Expression levels for each gene were normalized to eef1a1 (Dr03432748_m1) and/or rps29 (Dr03152131_m1) and fold change values were generated relative to wild-type DMSO treated control levels, according to ΔΔCT method [82]. All data were expressed as fold change mean ± s.e.m and P ≤ 0.05 was considered statistically significant.
Quantifying phd3:eGFP-related brightness
Images were acquired using Leica Application Suite version 4.9, which allowed the capture both of bright-field and GFP fluorescent images. To quantify the phd3:eGFP-related brightness of live embryos derived from each incrossed mutant line used in this project, Fiji (Image J) software v.2.0.0 was used. Images were converted into a grey scale 8-bit format and subsequently analysed by the software, by summing the grey values of all the pixels in the selected area, divided by the number of pixels. By default, since values equal to 0 are assigned to black and values equal to 255 to white, the quantified mean grey values are proportional to the intensity of the eGFP-related brightness expressed in the embryos. In particular, head, liver and tail (from the anus to the caudal peduncle) related brightness were selected and measured in all the mutant lines used in this study (S1D Fig). Genotyping post phenotypic analysis on phd3:eGFP sorted larvae confirmed the genotype-phenotype correlation.
Statistical analysis
GraphPad Prism version 8.0 for MacOS (GraphPad Software, La Jolla, California, USA, www.graphpad.com) was used to perform statistical analysis on all the samples analysed. Unpaired t tests were used to test for significant differences between two sample groups (i.e cortisol quantification). One-way ANOVA was used for assessing mean grey values data quantification, whereas two-way ANOVA was used to evaluate qPCR data. As post-hoc correction tests, Sidak’s method for multiple comparisons was used on normally distributed populations following one-way ANOVA, while Dunnett’s correction was used for comparing every mean to a control mean, on normally distributed populations following two-way ANOVA.
Supporting information
A. Statistical analysis performed on mean gray value quantification (at the level of the head, liver and tail), after phenotypic analysis on 5dpf DMSO and BME [30μM] treated arnt1+/-;vhl+/-(phd3:eGFP) x arnt1-/-; vhl+/-(phd3:eGFP) derived larvae (n = 540). vhl-/- DMSO treated n = 17 larvae: head 166.67 ± 9.63 (mean ± s.e.m); liver 138.61 ± 12.05 (mean ± s.e.m); tail 50.31 ± 4.51 (mean ± s.e.m). arnt1-/-;vhl-/- DMSO treated n = 13 larvae: head 121.05 ± 6.99 (mean ± s.e.m); liver 49.61 ± 3.88 (mean ± s.e.m); tail 21.75 ± 1.12 (mean ± s.e.m). vhl-/- BME treated n = 18 larvae: head 199.88 ± 7.71 (mean ± s.e.m); liver 222.57 ± 8.72 (mean ± s.e.m); tail 57.57 ± 4.11 (mean ± s.e.m). arnt1-/-;vhl-/- BME treated n = 12 larvae: head 153.71 ± 8.66 (mean ± s.e.m); liver 62.58 ± 5.16 (mean ± s.e.m); tail 25.82 ± 1.54 (mean ± s.e.m). Ordinary One-way ANOVA followed by Sidak’s multiple comparison test (*P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001). B. Kaplan-Meier survival curves of the zebrafish arnt1+/-; vhl+/-(phd3:eGFP) genotype analysed in this study. Time is shown in days. Siblings n = 30; arnt1-/-; vhl-/-(phd3:eGFP) n = 8. The Log-rank (Mantel-Cox) test was used for statistical analysis. arnt1-/-; vhl-/-(phd3:eGFP) vs. siblings: **P < 0.0027. C. RTqPCR analysis performed on arnt1 siblings (n = 10; 3 repeats) and arnt1-/- (n = 10; 3 repeats) larvae at 5 dpf, using egln3 as probe. Statistical analysis was performed on ΔΔCt values, whereas data are shown as fold change values. Ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test (***P <0.001;****P < 0.0001). D. Representative picture of head, liver and tail areas selected in each larva to quantify the phd3:eGFP-related brightness via mean grey value quantification (Fiji, ImageJ software).
(TIF)
Schematic view of RTqPCR analysis on il6st, pck1 and lipca (GC target genes) expression performed on the following mutant lines: vhl+/-(phd3:eGFP), arnt1+/-;vhl+/-(phd3:eGFP) and arnt1+/-(phd3:eGFP). Statistical analysis performed on ΔΔCt values; data are shown as fold change values for RTqPCR analysed samples; ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test (*P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001).
(TIF)
A-D’. Representative pictures of WISH performed on DMSO and BME [30 μM] treated arnt1 mutant line, at 5 dpf, using cyp17a2 as probe. A-A’) arnt1 wt DMSO treated larvae (n = 26/28) showed normal cyp17a2 expression, whereas 2/28 larvae showed a weaker one; B-B’) arnt1 wt BME treated larvae (n = 28/30) showed downregulated cyp17a2 expression, whereas 2/30 larvae showed a normal one. C-C’) In contrast, arnt1-/- DMSO treated larvae (n = 24/28) showed upregulated cyp17a2 expression, whereas 4/28 larvae showed a weaker one. D-D’) arnt1-/- BME treated larvae (n = 25/29) showed downregulated cyp17a2 expression, whereas 4/29, showed a normal one. Chi-square test (****P < 0.0001). Scale bar 200 μm. E-H’. Representative pictures of WISH performed on DMSO and BME [30 μM] treated vhl mutant line, at 5 dpf, using cyp17a2 as probe. E-E’) DMSO treated vhl siblings (n = 18/21) showed normal cyp17a2 expression, whereas 3/21 larvae showed a weaker one; F-F’) BME treated vhl siblings (n = 28/30) showed downregulated cyp17a2 expression, whereas 2/30 larvae showed a normal one. G-G’) On the other hand, vhl-/- DMSO treated larvae (n = 27/28) showed weak cyp17a2 expression, whereas 1/28 larvae showed a normal one. H-H’) vhl-/- BME treated larvae (n = 30/30) showed downregulated cyp17a2 expression. Chi-square test (****P < 0.0001). Scale bar 200 μm. I-I’. Representative picture of the colour threshold area calculation method (ImageJ software’s tool) used to quantify the area occupied by the cyp17a2 WISH staining both in arnt1 siblings (n = 9) and arnt1-/- (n = 9). I’. unpaired t-test (****P <0.0001).
(TIF)
A. Statistical analysis performed on mean gray value quantification (at the level of the head, liver and tail), after phenotypic analysis on 5dpf DMSO and BME [30μM] treated gr+/-;vhl+/-(phd3:eGFP) x gr-/-; vhl+/-(phd3:eGFP) derived larvae (n = 600). vhl-/- DMSO treated n = 9 larvae: head 186 ± 15.12 (mean ± s.e.m); liver 177.01 ± 20.85 (mean ± s.e.m); tail 62.34 ± 7.27 (mean ± s.e.m). gr-/-;vhl-/- DMSO treated n = 7 larvae: head 106.96 ± 3.21 (mean ± s.e.m); liver 60.75 ± 2.56 (mean ± s.e.m); tail 30.67 ± 1.27 (mean ± s.e.m). vhl-/- BME treated n = 14 larvae: head 224.32 ± 6.83 (mean ± s.e.m); liver 244.07 ± 5.31 (mean ± s.e.m); tail 80.51 ± 5.49 (mean ± s.e.m). gr-/-;vhl-/- BME treated n = 9 larvae: head 125.85 ± 3.6 (mean ± s.e.m); liver 63.56 ± 2.91 (mean ± s.e.m); tail 33.67 ± 1.02 (mean ± s.e.m). Ordinary One-way ANOVA followed by Sidak’s multiple comparison test (*P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001). B. Kaplan-Meier survival curves of the zebrafish gr+/-; vhl+/-(phd3:eGFP) genotype analysed in this study. Time is shown in days. Wild-types n = 20; gr+/-; vhl+/- n = 20; gr-/-; vhl-/-(phd3:eGFP) n = 5. The Log-rank (Mantel-Cox) test was used for statistical analysis. gr-/-; vhl-/-(phd3:eGFP) vs. gr+/-; vhl+/-, ****P < 0.0001; gr-/-; vhl-/-(phd3:eGFP) vs. wt, ****P < 0.0001.
(TIF)
Statistical analysis performed on mean gray values quantification (at the level of the head, liver and tail), after phenotypic analysis on 5dpf gr+/-;arnt1+/-vhl+/-(phd3:eGFP) incross-derived GFP+ larvae (n = 488). vhl-/- n = 5 larvae: head 125.82 ± 13.05 (mean ± s.e.m); liver 98.52 ± 3.8 (mean ± s.e.m); tail 37.43 ± 2.45 (mean ± s.e.m). gr-/-;arnt1-/-;vhl-/- n = 5 larvae: head 40.24 ± 2.46 (mean ± s.e.m); liver 26.07 ± 1.31 (mean ± s.e.m); tail 11.22 ± 0.47 (mean ± s.e.m); unpaired t-test (***P = 0.0002; ****P < 0.0001).
(TIF)
A-D. Representative pictures of 5 dpf CRISPANT mutants created by redundantly targeting lamb1b gene via co-injection of 4x gRNAs in vhl+/-(phd3:eGFP) incross-derived embryos (n = 400). Uninjected embryos were used as control (n = 470). Fluorescence, exposure = 991,4 ms. Scale bar 500 μm. E. Statistical analysis performed on mean grey values quantification (at the level of the head, liver and tail), after phenotypic analysis on 5 dpf lamb1b 4x gRNAs injected and uninjected vhl+/-(phd3:eGFP) incross-derived larvae. vhl-/- uninjected n = 24 larvae: head 54.83 ± 3.68 (mean ± s.e.m); liver 77.86 ± 6.46 (mean ± s.e.m); tail 19.56 ± 1.43 (mean ± s.e.m). vhl-/- injected n = 25 larvae: head 59.74 ± 4.05 (mean ± s.e.m); liver 83.23 ± 5.92 (mean ± s.e.m); tail 19.9 ± 1.38 (mean ± s.e.m); unpaired t-test (all panels: *P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001).
(TIF)
Acknowledgments
We thank the University of Sheffield aquarium staff for the excellent care of fish stocks. We are grateful to Rosemary Kim, Helen Eachus, Emily Noël, Chris Derrick, Jack Paveley and Dheemanth Subramanya for useful discussions contributing to this study and for sharing chemical compounds. We finally thank Elisabeth Kugler for her technical support for image analysis.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
DM was awarded by a University of Sheffield, Biomedical Science department studentship (https://www.sheffield.ac.uk/) and supported by two BBRSC grants to FVE (BB/R015457/1; BB/M02332X/1). FVE was funded by two Biotechnology and Biological Sciences Research Council (BBRSC: https://bbsrc.ukri.org/) grants (code 1: BB/R015457/1; code 2: BB/M02332X/1). NK was funded by the Deutsche Forschungs Gesellschaft (https://www.dfg.de/) grant (KR3363/3-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alsop D, Vijayan M. The zebrafish stress axis: Molecular fallout from the teleost-specific genome duplication event. Gen Comp Endocrinol. 2009;161: 62–66. 10.1016/j.ygcen.2008.09.011 [DOI] [PubMed] [Google Scholar]
- 2.Tokarz J, Möller G, Hrabě De Angelis M, Adamski J. Zebrafish and steroids: What do we know and what do we need to know? Journal of Steroid Biochemistry and Molecular Biology. 2013. 10.1016/j.jsbmb.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Faught E, Vijayan MM. The mineralocorticoid receptor is essential for stress axis regulation in zebrafish larvae. Sci Rep. 2018;8: 1–11. 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths BB, Schoonheim PJ, Ziv L, Voelker L, Baier H, Gahtan E. A zebrafish model of glucocorticoid resistance shows serotonergic modulation of the stress response. Front Behav Neurosci. 2012;6: 68 10.3389/fnbeh.2012.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facchinello N, Skobo T, Meneghetti G, Colletti E, Dinarello A, Tiso N, et al. nr3c1 null mutant zebrafish are viable and reveal DNA-binding-independent activities of the glucocorticoid receptor. Sci Rep. 2017;7: 4371 10.1038/s41598-017-04535-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamberger CM, Schulte HM, Chrousos GP. Molecular Determinants of Glucocorticoid Receptor Function and Tissue Sensitivity to Glucocorticoids. Endocr Rev. 1996;17: 245–261. 10.1210/edrv-17-3-245 [DOI] [PubMed] [Google Scholar]
- 7.Mitre-Aguilar IB, Cabrera-Quintero AJ, Zentella-Dehesa A. Genomic and non-genomic effects of glucocorticoids: implications for breast cancer. Int J Clin Exp Pathol. 2015;8: 1–10. Available: https://www.ncbi.nlm.nih.gov/pubmed/25755688 [PMC free article] [PubMed] [Google Scholar]
- 8.Stahn C, Buttgereit F. Genomic and nongenomic effects of glucocorticoids. Nat Clin Pract Rheumatol. 2008;4: 525 Available: 10.1038/ncprheum0898 [DOI] [PubMed] [Google Scholar]
- 9.Panettieri RA, Schaafsma D, Amrani Y, Koziol-White C, Ostrom R, Tliba O. Non-genomic Effects of Glucocorticoids: An Updated View. Trends in Pharmacological Sciences. 2019. 10.1016/j.tips.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semenza GL. Oxygen Sensing, Homeostasis, and Disease. N Engl J Med. 2011;365: 537–547. 10.1056/NEJMra1011165 [DOI] [PubMed] [Google Scholar]
- 11.Dougherty EJ, Pollenz RS. ARNT: A Key bHLH/PAS Regulatory Protein Across Multiple Pathways. Compr Toxicol. 2010; 231–252. 10.1016/B978-0-08-046884-6.00214-1 [DOI] [Google Scholar]
- 12.Elks PM, Renshaw SA, Meijer AH, Walmsley SR, van Eeden FJ. Exploring the HIFs, buts and maybes of hypoxia signalling in disease: lessons from zebrafish models. Dis Model Mech. 2015;8: 1349–1360. 10.1242/dmm.021865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelster B, Egg M. Hypoxia-inducible transcription factors in fish: expression, function and interconnection with the circadian clock. J Exp Biol. 2018;221: jeb163709 10.1242/jeb.163709 [DOI] [PubMed] [Google Scholar]
- 14.Hill AJ, Heiden TCK, Heideman W, Peterson RE. Potential Roles of Arnt2 in Zebrafish Larval Development. Zebrafish. 2009;6: 79–91. 10.1089/zeb.2008.0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasch AL, Tanguay RL, Mehta V, Heideman W, Peterson RE. Identification of Zebrafish ARNT1 Homologs: 2,3,7,8-Tetrachlorodibenzo-<em>p</em>-dioxin Toxicity in the Developing Zebrafish Requires ARNT1. Mol Pharmacol. 2006;69: 776 LP– 787. 10.1124/mol.105.016873 [DOI] [PubMed] [Google Scholar]
- 16.Wang WD, Wu JC, Hsu HJ, Kong ZL, Hu CH. Overexpression of a Zebrafish ARNT2-like Factor Represses CYP1A Transcription in ZLE Cells. Mar Biotechnol (NY). 2000;2: 376–386. Available: http://europepmc.org/abstract/MED/10960127 10.1007/s101260000001 [DOI] [PubMed] [Google Scholar]
- 17.Köblitz L, Fiechtner B, Baus K, Lussnig R, Pelster B. Developmental Expression and Hypoxic Induction of Hypoxia Inducible Transcription Factors in the Zebrafish. PLoS One. 2015;10: e0128938 10.1371/journal.pone.0128938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berra E, Roux D, Richard DE, Pouysségur J. Hypoxia-inducible factor-1α (HIF-1α) escapes O 2 -driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep. 2001;2: 615–620. 10.1093/embo-reports/kve130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moroz E, Carlin S, Dyomina K, Burke S, Thaler HT, Blasberg R, et al. Real-Time Imaging of HIF-1α Stabilization and Degradation. PLoS One. 2009;4: e5077 Available: 10.1371/journal.pone.0005077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8: 967–75. 10.1038/nrc2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123: 3664–3671. 10.1172/JCI67230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148: 399–408. 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch Eur J Physiol. 2005;450: 363–371. 10.1007/s00424-005-1413-7 [DOI] [PubMed] [Google Scholar]
- 24.Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landázuri MO, et al. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem J. 2005;390: 189–197. 10.1042/BJ20042121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santhakumar K, Judson EC, Elks PM, McKee S, Elworthy S, Van Rooijen E, et al. A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res. 2012;72: 4017–4027. 10.1158/0008-5472.CAN-11-3148 [DOI] [PubMed] [Google Scholar]
- 26.Kodama T, Shimizu N, Yoshikawa N, Makino Y, Ouchida R, Okamoto K, et al. Role of the glucocorticoid receptor for regulation of hypoxia-dependent gene expression. J Biol Chem. 2003;278: 33384–33391. 10.1074/jbc.M302581200 [DOI] [PubMed] [Google Scholar]
- 27.Leonard MO, Godson C, Brady HR, Taylor CT. Potentiation of Glucocorticoid Activity in Hypoxia through Induction of the Glucocorticoid Receptor. J Immunol. 2005;174: 2250–2257. 10.4049/jimmunol.174.4.2250 [DOI] [PubMed] [Google Scholar]
- 28.Wagner AE, Huck G, Stiehl DP, Jelkmann W, Hellwig-Bürgel T. Dexamethasone impairs hypoxia-inducible factor-1 function. Biochem Biophys Res Commun. 2008;372: 336–340. 10.1016/j.bbrc.2008.05.061 [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Qiang Q, Jiang Y, Hu L3, Ding X, Lu Y, et al. Effects of hypoxia inducible factor-1α on apoptotic inhibition and glucocorticoid receptor downregulation by dexamethasone in AtT-20 cells. BMC Endocr Disord. 2015;15: 1–9. 10.1186/1472-6823-15-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Fang L, Wu HM, Ding P, Shen QY, Liu R. Down-regulation of GR?? expression and inhibition of its nuclear translocation by hypoxia. Life Sci. 2016;146: 92–99. 10.1016/j.lfs.2015.12.059 [DOI] [PubMed] [Google Scholar]
- 31.Busillo JM, Cidlowski JA. The five Rs of glucocorticoid action during inflammation: Ready, reinforce, repress, resolve, and restore. Trends in Endocrinology and Metabolism. 2013. 10.1016/j.tem.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolaus S, Fölscn U, Schreiber S. Immunopharmacology of 5-aminosalicylic acid and of glucocorticoids in the therapy of inflammatory bowel disease. Hepato-gastroenterology. 2000. [PubMed] [Google Scholar]
- 33.Neeck G, Renkawitz R, Eggert M. Molecular aspects of glucocorticoid hormone action in rheumatoid arthritis. Cytokines Cell Mol Ther. 2002;7: 61–69. 10.1080/13684730412331302081 [DOI] [PubMed] [Google Scholar]
- 34.Moghadam-Kia S, Werth VP. Prevention and treatment of systemic glucocorticoid side effects. Int J Dermatol. 2010;49: 239–248. 10.1111/j.1365-4632.2009.04322.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnes PJ, Adcock IM. Glucocorticoid resistance in inflammatory diseases. Lancet. 2009;373: 1905–1917. 10.1016/S0140-6736(09)60326-3 [DOI] [PubMed] [Google Scholar]
- 36.Barnes PJ. Glucocorticosteroids: current and future directions. Br J Pharmacol. 2011;163: 29–43. 10.1111/j.1476-5381.2010.01199.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaber T, Schellmann S, Erekul KB, Fangradt M, Tykwinska K, Hahne M, et al. Macrophage Migration Inhibitory Factor Counterregulates Dexamethasone-Mediated Suppression of Hypoxia-Inducible Factor-1α Function and Differentially Influences Human CD4<sup>+</sup> T Cell Proliferation under Hypoxia. J Immunol. 2011;186: 764 LP– 774. 10.4049/jimmunol.0903421 [DOI] [PubMed] [Google Scholar]
- 38.Weger BD, Weger M, Görling B, Schink A, Gobet C, Keime C, et al. Extensive Regulation of Diurnal Transcription and Metabolism by Glucocorticoids. PLOS Genet. 2016;12: e1006512 Available: 10.1371/journal.pgen.1006512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatzopoulou A, Roy U, Meijer AH, Alia A, Spaink HP, Schaaf MJM. Transcriptional and metabolic effects of glucocorticoid receptor α and β signaling in zebrafish. Endocrinology. 2015;156: 1757–1769. 10.1210/en.2014-1941 [DOI] [PubMed] [Google Scholar]
- 40.Xie Y, Tolmeijer S, Oskam J, Tonkens T, Meijer AH, Schaaf MJM. Glucocorticoids inhibit macrophage differentiation towards a pro-inflammatory phenotype upon wounding without affecting their migration. bioRxiv. 2019; 473926 10.1101/473926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alsop D, Vijayan MM. Development of the corticosteroid stress axis and receptor expression in zebrafish. Am J Physiol Regul Integr Comp Physiol. 2008;294: R711–R719. 10.1152/ajpregu.00671.2007 [DOI] [PubMed] [Google Scholar]
- 42.van Rooijen E, Santhakumar K, Logister I, Voest E, Schulte-Merker S, Giles R, et al. A Zebrafish Model for VHL and Hypoxia Signaling. Methods Cell Biol. 2011;105: 163–190. 10.1016/B978-0-12-381320-6.00007-2 [DOI] [PubMed] [Google Scholar]
- 43.Vettori A, Greenald D, Wilson GK, Peron M, Facchinello N, Markham E, et al. Glucocorticoids promote Von Hippel Lindau degradation and Hif-1α stabilization. Proc Natl Acad Sci. 2017; 201705338 10.1073/pnas.1705338114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR Interaction Code: Predictive Value of Direct/Indirect DNA Recruitment for Transcription Outcome. Mol Cell. 2012;47: 38–49. 10.1016/j.molcel.2012.04.021 [DOI] [PubMed] [Google Scholar]
- 45.Dittrich A, Khouri C, Sackett SD, Ehlting C, Böhmer O, Albrecht U, et al. Glucocorticoids increase interleukin-6–dependent gene induction by interfering with the expression of the suppressor of cytokine signaling 3 feedback inhibitor. Hepatology. 2012;55: 256–266. 10.1002/hep.24655 [DOI] [PubMed] [Google Scholar]
- 46.Langlais D, Couture C, Balsalobre A, Drouin J. Regulatory Network Analyses Reveal Genome-Wide Potentiation of LIF Signaling by Glucocorticoids and Define an Innate Cell Defense Response. PLOS Genet. 2008;4: e1000224 Available: 10.1371/journal.pgen.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Rooijen E, Voest EE, Logister I, Korving J, Schwerte T, Schulte-Merker S, et al. Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood. 2009;113: 6449 LP– 6460. 10.1182/blood-2008-07-167890 [DOI] [PubMed] [Google Scholar]
- 48.Wu RS, Lam II, Clay H, Duong DN, Deo RC, Coughlin SR. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev Cell. 2018;46: 112–125.e4. 10.1016/j.devcel.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 49.Schaaf MJM, Chatzopoulou A, Spaink HP. The zebrafish as a model system for glucocorticoid receptor research. Comp Biochem Physiol—A Mol Integr Physiol. 2009;153: 75–82. 10.1016/j.cbpa.2008.12.014 [DOI] [PubMed] [Google Scholar]
- 50.Chatzopoulou A, Schoonheim PJ, Torraca V, Meijer AH, Spaink HP, Schaaf MJM. Functional analysis reveals no transcriptional role for the glucocorticoid receptor β-isoform in zebrafish. Mol Cell Endocrinol. 2017;447 10.1016/j.mce.2017.02.036 [DOI] [PubMed] [Google Scholar]
- 51.Ziv L, Muto A, Schoonheim PJ, Sebastiaan H, Meijsing, Strasser D, et al. An affective disorder in zebrafish with mutation of the glucocorticoid receptor. 2014;18: 681–691. 10.1038/mp.2012.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Zhang G, Gonzalez FJ, Park S, Cai D. Hypoxia-Inducible Factor Directs POMC Gene to Mediate Hypothalamic Glucose Sensing and Energy Balance Regulation. PLOS Biol. 2011;9: e1001112 Available: 10.1371/journal.pbio.1001112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramamoorthy S, Cidlowski JA. Corticosteroids. Mechanisms of Action in Health and Disease. Rheumatic Disease Clinics of North America. 2016. 10.1016/j.rdc.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weger M, Diotel N, Weger BD, Beil T, Zaucker A, Eachus HL, et al. Expression and activity profiling of the steroidogenic enzymes of glucocorticoid biosynthesis and the fdx1 co-factors in zebrafish. J Neuroendocrinol. 2018;30: 1–15. 10.1111/jne.12586 [DOI] [PubMed] [Google Scholar]
- 55.Eachus H, Zaucker A, Oakes JA, Griffin A, Weger M, Güran T, et al. Genetic disruption of 21-hydroxylase in zebrafish causes interrenal hyperplasia. Endocrinology. 2017;158: 4165–4173. 10.1210/en.2017-00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muto A, Taylor MR, Suzawa M, Korenbrot JI, Baier H. Glucocorticoid receptor activity regulates light adaptation in the zebrafish retina. Front Neural Circuits. 2013;7: 145 10.3389/fncir.2013.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montgomery MT, Hogg JP, Roberts DL, Redding SW. The use of glucocorticosteroids to lessen the inflammatory sequelae following third molar surgery. J Oral Maxillofac Surg. 1990;48: 179–187. 10.1016/s0278-2391(10)80207-1 [DOI] [PubMed] [Google Scholar]
- 58.Fromage G. Steroids: what are they and what is their mechanism of action? J Aesthetic Nurs. 2012;1: 198–201. 10.12968/joan.2012.1.4.198 [DOI] [Google Scholar]
- 59.Burger A, Lindsay H, Felker A, Hess C, Anders C, Chiavacci E, et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development. 2016;143: 2025 LP– 2037. 10.1242/dev.134809 [DOI] [PubMed] [Google Scholar]
- 60.Chrousos G, Kino T. Glucocorticoid Signaling in the Cell: Expanding Clinical Implications to Complex Human Behavioral and Somatic Disorders. 2009;100: 130–134. 10.1016/j.pestbp.2011.02.012.Investigations [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Revollo JR, Cidlowski JA. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann N Y Acad Sci. 2009;1179: 167–178. 10.1111/j.1749-6632.2009.04986.x [DOI] [PubMed] [Google Scholar]
- 62.Wilson KS, Tucker CS, Al-Dujaili EAS, Holmes MC, Hadoke PWF, Kenyon CJ, et al. Early-life glucocorticoids programme behaviour and metabolism in adulthood in zebrafish. J Endocrinol. 2016;230: 125–142. 10.1530/JOE-15-0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oakley RH, Cidlowski JA. The biology of the glucocorticoid receptor: New signaling mechanisms in health and disease. J Allergy Clin Immunol. 2013;132 10.1016/j.jaci.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imtiyaz HZ, Simon MC. Hypoxia-inducible factors as essential regulators of inflammation. Curr Top Microbiol Immunol. 2010;345: 105–120. 10.1007/82_2010_74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eltzschig HK, Carmeliet P. Hypoxia and Inflammation. N Engl J Med. 2011;364: 656 10.1056/NEJMra0910283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41: 518–528. 10.1016/j.immuni.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan T, Yu RMK, Wu RSS, Kong RYC. Overexpression and Knockdown of Hypoxia-Inducible Factor 1 Disrupt the Expression of Steroidogenic Enzyme Genes and Early Embryonic Development in Zebrafish. Gene Regul Syst Bio. 2017;11: 1177625017713193–1177625017713193. 10.1177/1177625017713193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker ME, Katsu Y. 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Evolution of the mineralocorticoid receptor: sequence, structure and function. J Endocrinol. 2017;234: T1–T16. 10.1530/JOE-16-0661 [DOI] [PubMed] [Google Scholar]
- 69.Cruz SA, Lin C-H, Chao P-L, Hwang P-P. Glucocorticoid Receptor, but Not Mineralocorticoid Receptor, Mediates Cortisol Regulation of Epidermal Ionocyte Development and Ion Transport in Zebrafish (Danio Rerio). PLoS One. 2013;8: e77997 Available: 10.1371/journal.pone.0077997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee HB, Schwab TL, Sigafoos AN, Gauerke JL, Krug 2nd RG, Serres MR, et al. Novel zebrafish behavioral assay to identify modifiers of the rapid, nongenomic stress response. Genes Brain Behav. 2019/01/15. 2019;18: e12549–e12549. 10.1111/gbb.12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Dev Dyn. 1995;203: 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 72.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44: W272–W276. 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014/05/26. 2014;42: W401–W407. 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140: 4982–4987. 10.1242/dev.099085 [DOI] [PubMed] [Google Scholar]
- 75.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3: 59 Available: 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- 76.Muthu V, Eachus H, Ellis P, Brown S, Placzek M. Rx3 and Shh direct anisotropic growth and specification in the zebrafish tuberal/anterior hypothalamus. Development. 2016;143: 2651 LP– 2663. 10.1242/dev.138305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karlsson J, von Hofsten J, Olsson P-E. Generating Transparent Zebrafish: A Refined Method to Improve Detection of Gene Expression During Embryonic Development. Mar Biotechnol. 2001;3: 522–527. 10.1007/s1012601-0053-4 [DOI] [PubMed] [Google Scholar]
- 78.Kramer BMR, Kolk SM, Berghs CAFM, Tuinhof R, Ubink R, Jenks BG, et al. Dynamics and plasticity of peptidergic control centres in the retino-brain-pituitary system of Xenopus laevis. Microsc Res Tech. 2001;54: 188–199. 10.1002/jemt.1132 [DOI] [PubMed] [Google Scholar]
- 79.Kurrasch DM, Nevin LM, Wong JS, Baier H, Ingraham HA. Neuroendocrine transcriptional programs adapt dynamically to the supply and demand for neuropeptides as revealed in NSF mutant zebrafish. Neural Dev. 2009;4: 22 10.1186/1749-8104-4-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muto A, Orger MB, Wehman AM, Smear MC, Kay JN, Page-McCaw PS, et al. Forward Genetic Analysis of Visual Behavior in Zebrafish. PLOS Genet. 2005;1: e66 Available: 10.1371/journal.pgen.0010066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatamoto K, Shingyoji C. Cyclical training enhances the melanophore responses of zebrafish to background colours. Pigment Cell Melanoma Res. 2008;21: 397–406. 10.1111/j.1755-148X.2008.00445.x [DOI] [PubMed] [Google Scholar]
- 82.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]