Abstract
Olfactory sensory neurons (OSNs) located in the dorsomedial and ventromedial regions of the olfactory epithelium (OE) are distinguished from one another based on their molecular expression patterns. This difference is reflected in the separation of the glomerular layer of the olfactory bulb (OB) into dorsomedial and ventrolateral regions. However, it is unclear whether a complementary separation is also evident in the projection neurons that innervate the OB glomeruli. In this study, we compared the development of the OB between different regions by focusing on the transcription factor, Tbx21, which is expressed by mitral and tufted cells in the mature OB. Examining the OB at different developmental ages, we found that Tbx21 expression commenced in the anteromedial region called the tongue-shaped area, followed by the dorsomedial and then ventrolateral areas. We also showed that the tongue-shaped area was innervated by the OSNs located in the most dorsomedial part of the ventrolateral OE, the V-zone:DM. Interestingly, the generation of OSNs occurred first in the dorsomedial zone including the V-zone:DM, suggesting a correlation between the time course of OSN generation in the OE and Tbx21 expression in their target region of the OB. In contrast, expression of vGluT1, which is also found in all mitral cells in the mature OB, was first detected in the ventrolateral region during development. Our findings demonstrate that the development of projection neurons occurs in a compartmentalized manner in the olfactory bulb; tongue-shaped, dorsomedial, and ventrolateral areas, and that not all projection neurons follow the same developmental pathway.
Keywords: olfactory bulb, mitral cell, development, compartment, tongue-shaped area, RRID:AB_2200219, RRID:AB_778267, RRID:AB_609568, RRID:AB_400326, RRID:AB_2209751, RRID:AB_2149710, RRID:AB_887877, RRID:AB_2315007, RRID:AB_1603148, RRID:AB_141708, RRID:AB_162543, RRID:AB_141514, RRID:AB_141596, RRID:AB_141709, RRID:AB_142672, RRID:AB_141788, RRID:AB_2629482, RRID:IMSR_CRL:22, RRID:IMSR_JAX:024507, RRID:IMSR_JAX:007909, RRID:SCR_014199
Graphical Abstract

Examining the mouse olfactory bulb at different developmental ages, we found that Tbx21 expression commenced in the anteromedial region called the tongue-shaped area, followed by the dorsomedial and then ventrolateral areas. In contrast, expression of vGluT1 was first detected in the ventrolateral region during development. Our findings demonstrate that the development of projection neurons occurs in a compartmentalized manner in the olfactory bulb and that not all projection neurons follow the same developmental pathway.
Introduction
In the olfactory epithelium (OE), odorants are received by the olfactory sensory neurons (OSNs), which convert the olfactory information into an electric signal and transmit it to the olfactory bulb (OB). OSNs expressing a particular odorant receptor are distributed within a specific area in the OE. It was first believed that the OE could be divided into four distinct zones, zone I to IV, along with the dorsomedial-ventrolateral axis. Now, however, it is known that the expression areas specific to each odorant receptor in zones II to IV are arranged in an overlapping and continuous manner in the OE (Miyamichi et al., 2005). Nevertheless, there is a clear difference in molecular profile of OSNs in the zone I, which is also called D-zone of OE, and the other zones, including zones II to IV or V-zone of OE. For example, NQO1 (NAD(P)H quinone dehydrogenase 1) and O-MACS (olfactory-specific medium-chain acyl CoA synthase) are only expressed by the D-zone OSNs while OCAM (olfactory cell adhesion molecule) is exclusively expressed by the V-zone OSNs (Yoshihara et al., 1997; Oka et al., 2003; Gussing and Bohm, 2004). Furthermore, axons of NQO1+ OSNs target the glomeruli in the dorsomedial region of the OB and OCAM+ OSN axons project to the ventrolateral glomeruli. A number of studies have provided evidence that the representation of odors in the OB can be functionally defined. For example, ventral glomerular domains of the OB are associated with attractive behavior, such as food seeking, while signals from the dorsomedial glomeruli to the cortical amygdala are responsible for the innate fear responses caused by predator odors in mice (Kobayakawa et al., 2007; Mori and Sakano, 2011; Root et al., 2014; Kondoh et al., 2016).
In the glomeruli, OSN axons synapse onto the dendrites of mitral cells, a primary OB projection neuron. Of note, each of the ~33,000 mitral cells in the OB has a single primary dendrite that projects to a single glomerulus (Richard et al., 2010). In contrast to the apparent boundary between dorsomedial and ventrolateral glomeruli, no characteristic difference has been identified between mitral cells in dorsomedial and ventrolateral regions. However, we previously reported that early- and late-generated mitral cells were differentially distributed in dorsomedial and ventrolateral regions of the mouse OB, respectively (Imamura et al., 2011). Thus, we hypothesize that mitral cells in different regions develop with different time courses.
It was previously suggested that D-zone OSNs developed earlier than V-zone OSNs; therefore, the dorsal OB formed earlier than the ventral part (Takeuchi et al., 2010) and that OSN axons first reach the anteromedial region of the OB called the tongue-shaped area (Yoshihara et al., 2005). In contrast, synaptogenesis and glomerular formation are reported to begin in the ventromedial region of the developing OB (Blanchart et al., 2008). These apparently contradictory results emphasize the importance of a detailed study of mitral cell development in different OB regions. We previously reported that Tbr1 expression preceded Tbr2 in mitral cells (Imamura and Greer, 2013). However, regional or topographic analyses were not performed. In this study, we examined the spatiotemporal expression pattern of three T-box transcription factors, Tbr1, Tbr2, and Tbx21, in developing mouse OB. Then, the sequence of Tbx21 expression was examined in detail. Our results show that mitral cells develop along different time courses in at least three different regions in the mouse OB: tongue-shaped area, dorsomedial region, and ventrolateral region. The results have implications for understanding the functional organization of odor representation in the OB and the mechanisms shaping the organization of OB circuits.
Materials and Methods
Animals
CD1 (Charles River; Wilmington, MA; strain code 022; RRID:IMSR_CRL:22) or the offspring of CD1 female mated with the Tbx21Cre x tdTomato male mouse line were used in this study. The Tbx21Cre x tdTomato line was created by crossing B6;CBA-Tg (Tbx21-cre)1Dlc/J mice (The Jackson Laboratory; Bar Arbor, ME; stock #024507; RRID:IMSR_JAX:024507) with B6.Cg-Gt(ROSA)26Sortm9 (CAG-tdTomato) Hze/J reporter mice (The Jackson Laboratory; stock #007909; RRID:IMSR_JAX:007909). The day on which we found a copulation plug was called E0, and the succeeding days of gestation were numbered in order. Prenatal embryos were harvested and fixed in 4% paraformaldehyde (PFA) overnight after pregnant mothers were euthanized with CO2 inhalation. Postnatal pups younger than P10 were sacrificed with decapitation and fixed in 4% PFA overnight. Mice older than P10 were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) and were transcardially perfused with phosphate-buffered saline (PBS) followed by 4% (wt/vol) PFA in PBS. The heads were removed and placed in the same fixative at 4°C for 2 hours. All protocols were approved by and all methods were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Penn State College of Medicine.
Thymidine analogue injection
Briefly, 5-chloro-2′-deoxyuridine (CldU; Sigma; St. Louis, MO) or 5-iodo-2′-deoxyuridine (IdU; Sigma) (XdU) was intraperitoneally injected into pregnant mothers at E10, E11, E12, E13, or E16 (50mg/kg). Injections were performed between 10am and noon.
NeuroVue retrograde labeling
To label OSNs in the OE, P0 or P1 mice were decapitated and fixed in 4% PFA for 2 hours. The surface of the OB was exposed, and a piece of NeuroVue Orange filter (Polysciences; Warrington, PA) was inserted into the anteromedial region. Then, the brain attached with the OE was postfixed overnight with 4% PFA followed by incubation in PBS at 20–25°C for 3–4 weeks. The tissue was cryopreserved in 30% sucrose (wt/vol) in PBS and embedded in optimal cutting temperature compound (Sakura Finetek; Torrance, CA). The olfactory tissues were cut on a cryostat into 20-μm slices, and cells labeled with NeuroVue were visualized by fluorescence microscopy.
Immunohistochemistry
The fixed brains were cryopreserved in 30% sucrose (wt/vol) in PBS and embedded in optimal cutting temperature compound. The olfactory tissues were cut on a cryostat into 20μm slices and stored at −80°C until use. The slices were pretreated for 30min in 0.025 M HCl at 65°C and rinsed with 0. 1M borate buffer (pH 8.5), PBS and TBS-T (10mM Tris-HCl (pH 7.4), 100 mM NaCl with 0.3% Triton X-100 (vol/vol)). The slices were then blocked with blocking buffer (5% normal donkey serum (vol/vol) in TBS-T) at 20 – 25°C for 1 hour and incubated with primary antibodies diluted in blocking buffer overnight at 4°C. Sections were washed with TBS-T and then incubated with secondary antibodies with 4’6-diamino-2-phenylindole dihydrochloride (DAPI; Thermo Fisher Scientific; Waltham, MA; RRID:AB_2629482) for nucleus staining for 1 hour. The immunoreacted sections were washed and coverslipped with Fluoro-Gel mounting medium (Electron Microscopy Science; Hatfield, PA).
Primary antibodies used in this study are summarized in Table 1. Goat or donkey anti-species IgG conjugated with Alexa 488 or Alexa 555 (Thermo Fisher Scientific; 1:300; RRID:AB_141708, RRID:AB_162543, RRID:AB_141514, RRID:AB_141596, RRID:AB_141709, RRID:AB_142672, RRID:AB_141788) were used as secondary antibodies.
Table 1.
Antibodies: Sources, Specificities, Dilutions
| Antigen, Host (Dilution) |
Source, catalog # Immunogen, Specificity |
|---|---|
| Tbx21 rabbit (1:5000) |
Source: Dr. Y. Yoshihara (RIKEN Brain Science Institute, Saitama, Japan) (RRID: not found) Immunogen: Synthetic peptide, 20 aa from C-terminal of mouse Tbx21 (Yoshihara et al., 2005) Specificity: Recognizes Tbx21 (58 kDa) by immunoblot assay (Mizuguchi et al., 2012). Immunohistochemical staining pattern of cellular morphology and distribution in the olfactory bulb was identical to previous report (Mitsui et al., 2011). |
| Tbr1 rabbit (1:5000) |
Source: abcam (Cambridge, MA), ab31940 (RRID:AB_2200219) Immunogen: Synthetic peptide within mouse Tbr1 aa 50–150 conjugated to keyhole limpet haemocyanin. Specificity: Immunohistochemical staining pattern of cellular morphology and distribution in the olfactory bulb was identical to previous report (Imamura and Greer, 2013). |
| Tbr2 rabbit (1:5000) |
Source: abcam (Cambridge, MA), ab23345 (RRID:AB_778267) Immunogen: Synthetic peptide conjugated to KLH derived from within residues 650 to the Cterminus of mouse Tbr2/Eomes. Specificity: Immunohistochemical staining pattern of cellular morphology and distribution in the olfactory bulb was identical to previous report (Imamura and Greer, 2013). |
| CldU/BrdU rat (1:200) |
Source: Accurate Chemical & Scientific Corp (Westbury, NY), OBT0030 (RRID:AB_609568) Immunogen: Chemical BrdU; Clone: BU1/75 (ICR1) Specificity: Recognizes BrdU incorporated into single stranded DNA, attached to a protein carrier and free BrdU. Cross reacts with CldU but does not cross react with thymidine or IdU (Imamura et al., 2011). |
| IdU/BrdU mouse (1:200) |
Source: BD Biosciences (San Jose, CA), 347580 (RRID:AB_400326); Clone: B44 Preparation: Derived from hybridization of mouse Sp2/0-Ag14 myeloma cells with spleen cells from BALB/c mice immunized with IdU-conjugated ovalbumin. Specificity: Recognizes BrdU and IdU incorporated into single stranded DNA (Imamura et al., 2011). |
| tdTomato rabbit (1:200) |
Source: Rockland Immunochemicals Inc. (Limerick, PA), 600-401-379 (RRID:AB_2209751) Immunogen: Recombinant RFP fusion protein corresponding to the full length amino acid sequence (234aa) derived from the mushroom polyp coral Discosoma. Specificity: Recognizes a single band on western blot of around 27 – 30 kDa (manufacturer’s information). |
| OCAM goat (1:200) |
Source: R&D Systems (Minneapolis, MN), AF778 (RRID:AB_2149710) Immunogen: Mouse myeloma cell line NS0-derived recombinant mouse OCAM aa 20 – 700 Specificity: Detects mouse OCAM in direct ELISAs and Western blots. In direct ELISAs, less than 1% cross-reactivity with recombinant mouse NCAM-1 is observed (manufacturer’s information). |
| vGluT1 rabbit (1:1000) |
Source: Synaptic Systems (Goettingen, Germany), 135–302 (RRID:AB_887877) Immunogen: Recombinant protein corresponding to aa 456 – 560 from rat vGluT1 Specificity: Recognizes a single band on western blot of around 55 kDa on synaptic vesicle fraction of rat brain (manufacturer’s information). The vGluT1 staining in the current study is consistent with the staining pattern in the olfactory bulb previously described (Mizuguchi et al., 2012). |
| OMP goat (1:1000) |
Source: Wako Chemicals USA (Richmond, VA), 544–10001 (RRID:AB_2315007) Immunogen: Purified natural rat OMP Specificity: Immunohistochemical staining pattern of cellular morphology and distribution in the olfactory bulb was identical to previous report (Treloar et al., 1999). |
| Reelin mouse (1:200) |
Source: EMD Millipore (Burlington, MA), ab78540 (RRID:AB_1603148); Clone: G10 Immunogen: Recombinant fusion protein, corresponding to aa 164 – 496 of mouse Reelin Specificity: Recognizes a single band on western blot of around 388 kDa on rat/mouse brain lysate (manufacturer’s information). The reelin staining in the current study is consistent with the staining pattern in the olfactory bulb previously described (Imamura et al., 2006). |
Image acquisition, quantification, and statistics
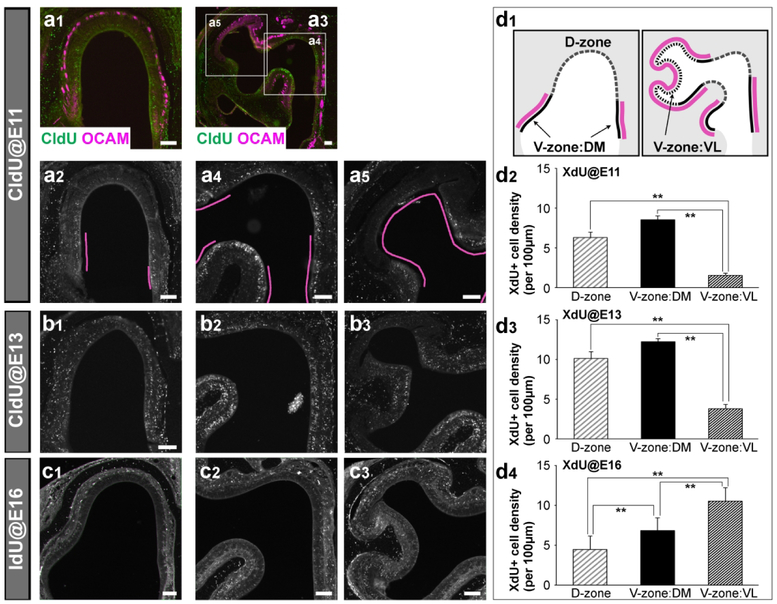
Quantification was performed using images acquired with a digital camera attached to the microscope (Carl Zeiss AG; Oberkochen, Germany). Levels were adjusted in Photoshop software (Adobe; RRID:SCR_014199), but the images were otherwise unaltered. To calculate the percentages of TBX21+ or tdTomato+ cells labeled with XdU, we collected coronal sections of the OB every 120 μm (E15 for Tbx21) or 160 μm (E17 for tdTomato) and all XdU-labeled cells within the OB region were analyzed. The numbers of XdU-labeled, Tbx21+, or tdTomato+ cells in the embryonic OB were manually counted (n=3 animals per experimental condition). To quantify the distributions of XdU-labeled cells in the P0 or P1 OE, three coronal sections, including anterior, middle, and posterior (160 μm apart), were selected and subdivided the OE into D-zone, V-zone:DM, and V-zone:VL based on the OCAM signal and structure (see Fig. 5d1). The numbers of XdU-labeled cells were manually counted, and the density was measured by calculating the number of XdU-labeled cells in region 100 μm length (n=3 animals per experimental condition). All results are presented as mean ± standard error of the mean (SEM). The results were statistically analyzed using one-way ANOVA followed by Tukey HSD test.
Figure 5. Distributions of olfactory sensory neurons generated at different embryonic days.
(a-c) Coronal sections of anterior (a1, a2, b1, c1) and posterior (a3, a4, a5, b2, b3, c2, c3) OE of neonatal mice in which CldU or IdU were injected into pregnant mothers at E11 (a), E13 (b), or E16 (c). XdU were immunohistochemically detected with anti-CldU or IdU antibodies (green in a1, a3; and white in a2, a4, a5, b, c). OSNs in the V-zone were labeled with an anti-OCAM antibody (magenta in a1, a3), and the V-zone OE is indicated by magenta bars in a2, a4, and a5. (d) The OE was separated into D-zone, V-zone:DM, and V-zone:VL according to the definition shown in d1. Magenta bars indicate the V-zone where OCAM+ OSNs localize. Densities of XdU+ cells were measured separately in D-zone, V-zone:DM, and V-zone:VL in the P0 or P1 OE after XdU injection at E11 (d2), E13 (d3), or E16 (d4). OSNs in the V-zone:DM are generated earlier than V-zone:VL. **p<0.01 (one-way ANOVA followed by Tukey HSD test). Scale bars, 200 μm.
Map construction
Coronal sections OB of offspring of CD1 female mated with Tbx21Cre-tdTomato male mouse were stained with anti-OCAM antibody. Images were taken from each OB every 80 μm using a 20X objective lens. All tdTomato+ cells in the OB were detected by intrinsic tdTomato signal and plotted on the mitral cell layers (MCL) of OB sections, which were defined by DAPI signal. Boundaries between dorsomedial and ventrolateral MCL were marked based on the boundaries of OCAM+ and OCAM− glomeruli in the glomerular layer. To quantify the density of tdTomato+ cells in the OB, the MCLs of each coronal section analyzed was unrolled from the ventral side then divided into 200-μm compartments, and the number of tdTomato+ cells in each compartment was manually counted. The unrolled MCLs were aligned from anterior to posterior using the dorsal edge to build an MCL map. Percentages of tdTomato+ cells in the compartments of each OB were represented by a gradient density bar from 0% to 5% on the constructed MCL map. OCAM+ regions of the OB were superimposed on the MCL map with dashed lines. Three OBs were analyzed at each time point.
Results
Spatiotemporal expression pattern of Tbr1, Tbr2, and Tbx21 in the developing mouse olfactory bulb
The Tbr1 subfamily of T-box transcription factors regulating brain development includes Tbr1, Tbr2/Eomes, and Tbx21/T-bet. In the mouse olfactory system, mitral/tufted cells express all three of these transcription factors during development (Bulfone et al., 1999; Faedo et al., 2002). However, the onset of their expression is different (Imamura and Greer, 2013). Many Tbr1-positive (Tbr1+) cells were found in the OB at embryonic day (E) 13 (Fig. 1a1). The number of Tbr2+ cells was reduced compared with Tbr1+ cells (Fig. 1b1), and even fewer cells expressed Tbx21 (Fig. 1c1). At E15, similar numbers of Tbr1+ and Tbr2+ cells were found both in the outer layer, intermediate zone, and the inner layer, the ventricular zone, of the developing OB (Fig. 1a2, 1a3, 1b2, and 1b3). However, the number of Tbx21+ cells was still significantly lower compared with Tbr1+ and Tbr2+ cells, and these cells were found only in the intermediate zone, largely confined to the anterodorsomedial region (Fig. 1c2 and 1c3).
Figure 1. Spatiotemporal expression patterns of Tbr1, Tbr2, and Tbx21 in embryonic olfactory bulbs.
(a-c) Embryonic OBs were stained with anti-Tbr1 (a), Tbr2 (b), or Tbx21 (c) antibody. Horizontal sections of E13 OBs (a1, b1, c1) and both horizontal (a2, b2, c2) and coronal sections of E15 OBs (a3, b3, c3) were stained. Clusters of Tbx21+ cells are found in the anterodorsomedial and posterolateral regions. (d) Anterodorsomedial region of E15 OB in which CldU and IdU were injected at E10 and E12, respectively. Many Tbx21+ cells (magenta) were colabeled with CldU (d1, green) while few Tbx21+ cells were colabeled with IdU (d2, green). (e) Graph showing percentages of XdU-labeled cells expressing Tbx21 in the E15 OB. XdU was injected into pregnant mothers at indicated time points. A significantly higher percentage of Tbx21+ cells are colabeled with XdU injected at E10 than E11 or E12; **p<0.01, *p<0.05 (one-way ANOVA followed by Tukey HSD test). Scale bars: 200 μm.
We previously reported that greater than 90% of mitral cells are generated in embryonic mouse brain between E10 and E12 (Imamura et al., 2011). To determine the birthdate of cells expressing Tbx21 in E15 OB, the thymidine analog, XdU (CldU or IdU), was intraperitoneally injected into pregnant mothers at different time points, and the percentage of XdU labeled cells that were also Tbx21+ was examined. When XdU was injected at E10 or E11, 60.6 ± 8.6% or 24.5 ± 1.3% of Tbx21+ cells were colabeled with XdU+, respectively (Figs. 1d1 and 1e). In contrast, only 1.2 ± 1.0% of Tbx21+ cells were colabeled following the E12 injection of XdU (Figs. 1d2 and 1e). Therefore, Tbx21+ cells in E15 OB are mostly generated early from E10 to E11. Importantly the Tbx21+ cells at E15 are found almost exclusively in the dorsal domains. There was almost no evidence of colabeled cells in the ventrolateral domain following the E10 and E11 XdU injections. These results suggest that Tbx21 expression in the OB is dependent upon both neuronal age and OB domain.
Expression of tdTomato in Tbx21Cre-tdTomato mice
Tbx21+ cells appear first in the anterodorsomedial region in developing OB. As neuronal age is a regulator of Tbx21 expression onset, it is reasonable to assume that mitral cells in this region mature earlier than other regions. Furthermore, we can examine the developmental timing of mitral cells in different OB regions by tracking the onset of Tbx21 expression across the entire OB. However, the number of Tbx21+ cells in the OB rapidly increased after E15, and no obvious regional difference was found by E17. This feature makes it difficult to track the onset of Tbx21 expression in different regions. In this study, we created a transgenic mouse line Tbx21Cre-tdTomato in which expression of the red fluorescent protein, tdTomato, is controlled by the Tbx21 promoter (Fig. 2a). As expected, all Tbx21+ mitral/tufted cells expressed tdTomato in adult OB (Fig. 2b). Similar to Tbx21 expression, only a small number of tdTomato+ cells were found in anterodorsomedial region of the E15 OB. Then, tdTomato+ cells increased mainly in the dorsomedial region. However, not all Tbx21+ cells expressed tdTomato while Tbx21 was already widely expressed in mitral cell layer (MCL) at E17 and postnatal day (P) 0 (Fig. 2c). Finally, most of the Tbx21+ mitral cells became tdTomato+ by P4. The discrepancy between Tbx21 and tdTomato expression may conceivably reflect a delay due to the extra steps, such as production of active Cre recombinase and recombination between LoxP sites. A similar dissociation was observed in the AP2εCre transgenic mice. When the AP2εCre mice were crossed with Rosa26 mice that only express β-galactosidase (β-Gal) following Cre-mediated recombination, the first evidence of β-Gal activity in mitral cells occurred exclusively around E16.5, while AP2ε expression was detected in the OB from E11.5 onwards (Feng and Williams, 2003; Feng et al., 2009).
Figure 2. Expression of tdTomato in Tbx21Cre-tdTomato mice.
(a) Schematic diagram for Tbx21Cre-tdTomato mice generation. The mice were created by crossing the Ai9 reporter mice with the Tbx21Cre mice in which cre recombinase is expressed under the regulation of the Tbx21 promoter. (b) Expression of tdTomato in P21 OB of Tbx21Cre-tdTomato mouse. tdTomato (magenta) is expressed by all mitral and tufted cells expressing Tbx21 (green). (c) Expression of tdTomato in developing OBs of the Tbx21Cre-tdTomato mice. The tdTomato expression starts from the anterodorsomedial region. In the immature OB, while all tdTomato+ cells express Tbx21, not all Tbx21+ cells are tdTomato+. (d) Anterodorsomedial region of E17 OB of Tbx21Cre-tdTomato mouse in which IdU and CldU were injected at E10 and E12, respectively. Many tdTomato+ cells (magenta) were colabeled with IdU (d1, green) while few tdTomato+ cells were colabeled with CldU (d2, green). (e) Graph showing percentages of XdU-labeled cells expressing tdTomato in the E17 OB. XdU were injected into pregnant mother at indicated time points. Significantly higher percentage of tdTomato+ cells are colabeled with XdU injected at E10 and E11 than E12; **p<0.01 (one-way ANOVA followed by Tukey HSD test). Scale bars: 200 μm.
To confirm that the early-generated mitral cells start to express tdTomato earlier than late-generated mitral cells, we injected XdU into pregnant Tbx21Cre-tdTomato mice at E10, E11, or E12 and determined the birthdate of tdTomato+ cells observed in E17 OB. When XdU was injected at E10, 59.8 ± 3.3% of Tbx21+ cells were labeled with XdU (Figs. 2d1 and 2e). The percentage was lower (40.9 ± 7.1%) with the E11 injection, and only 2.2 ± 0.7% of Tbx21+ cells were colabeled with the E12 injection (Figs. 2d2 and 2e). These results indicate that expression of tdTomato in the Tbx21Cre-tdTomato follows the onset of Tbx21 expression, the overall development following neurogenesis at either E10, E11, or E12 is comparable and topographic or domain organization is identical for both markers. Thus, we can conveniently use the onset of tdTomato expression to track the onset of Tbx21 in different regions of the OB.
Sequence of the appearance of tdTomato+ cells in the developing olfactory bulb
In the E17 OB of Tbx21Cre-tdTomato mice, a prominent cluster of tdTomato+ cells was observed in the dorsomedial regions of the anterior sections (Fig. 3a). Of note, this cluster was located immedeately below the OCAM+ glomeruli in the anteromedial OB, which is a rostromedial extension of OCAM+ glomeruli called “a tongue-shaped area” (Nagao et al., 2000). To schematically show the spatiotemporal change of tdTomato+ mitral cell distributions in the developing OB, we created tdTomato maps from E15, E17, P0, and P2 OBs of Tbx21CretdTomato mice. First, the location of tdTomato+ cells was plotted on an unrolled map of the MCL, and then the percentages of cells among total tdTomato+ cells were calculated in each 200-μm linear compartment. The spatial arrangement of OCAM+ and OCAM− glomeruli were also superimposed on the map (see Material and Methods and Supplementary Figure 1).
Figure 3. Distributions of tdTomato+ cells in the developing olfactory bulbs of Tbx21Cre-tdTomato mice.
(a) Coronal sections of the E17 OB of the Tbx21Cre-tdTomato mouse. The sections were stained with anti-tdTomato (magenta) and OCAM (green) antibodies. All nuclei were stained with DAPI (blue). (b) Distribution of tdTomato+ in E17 OB. Cells expressing tdTomato were marked with magenta dots in the coronal sections taken every 160 μm. MCLs were defined based on DAPI signals. Short bars indicate the boundaries of OCAM+ and OCAM− glomeruli in the glomerular layer. (c-e) MCLs unrolled from coronal sections of E17 (c), P0 (d), and P2 (e) OBs were aligned from anterior to posterior using the dorsal edge (black triangles). Percentages of tdTomato+ cells located in each compartment were superimposed onto the unrolled MCL maps. Dashed lines indicated the boundary of OCAM+ and OCAM− glomeruli in the glomerular layer. (f) Schematic diagram of the spatiotemporal expression pattern of tdTomato+ cells in the OB during development. The tdTomato+ cells first appear in the tongue-shaped area followed by the dorsomedial then ventrolateral areas. Scale bars: 200 μm.
In the E17 OB, in addition to the tongue-shaped area, a significant number of tdTomato+ cells were found below the cluster of OCAM− glomeruli in the dorsomedial region (Fig. 3c). Among the dorsomedial region, a higher percentage of tdTomato+ cells tended to be localized in the medial region. In contrast, tdTomato+ cells were sparsely distributed below the ventral OCAM+ glomeruli at these ages. At P0, as the number of tdTomato+ cells in the ventral area increased, a higher percentage was still evident in the dorsal area (Fig. 3d). Finally, tdTomato+ mitral cells were distributed throughout the entire MCL by P2, and no specific regional differences in tdTomato expression were found (Fig. 3e).
These results show the following temporal/regional sequence of tdTomato+ expression onsets in developing OB of Tbx21Cre-tdTomato mouse: 1) tdTomato+ mitral cells appear in the tongue-shaped area; 2) the mitral cells in the dorsomedial area start to express tdTomato; and 3) tdTomato expression occurs in the mitral cells in the ventrolateral area (Fig. 3f).
Innervation of the tongue-shaped area by early-generated olfactory sensory neurons
The tongue-shaped area is innervated by the OCAM+ OSNs. In the OE, OCAM+ OSNs are found in the ventrolateral region callled the V-zone. To determine the location of OCAM+ OSNs innervating the tongue-shaped area, a lipophilic tracer, NeuroVue dye, was placed on the anteromedial region of the fixed neonatal OB to retrogradely label the OSN cell bodies (Fig. 4a). After incubating 3–4 weeks in PBS, the dye-labeled OSN cell bodies were observed in the OE, mostly at the boundary of OCAM− D-zone and OCAM+ V-zone (Fig. 4b, c). In contrast, the dye-labeled OSNs were rarely observed in the lateral OE. Of particular note, the sections from anterior OE tended to contain more dye-labeled OSNs than posterior OE. These results indicate that the tongue-shaped area in the OB is innervated by the OCAM+ OSNs located at the most dorsomedial part of V-zone (V-zone:DM) of the anterior OE (Fig. 4d).
Figure 4. Olfactory sensory neuron innervation to the tongue-shaped area of the olfactory bulb.
(a) Image of the left OB and OE from P1 mouse. A piece of NeuroVue® Orange Filter was placed at the anteromedial portion of the OB after PFA fixation. The image was taken after incubating the brain in PBS for three weeks. (b, c) Distributions of NeuroVue-labeled (b) and OCAM+ (c) OSNs in the coronal sections of OE. Positions of NeuroVue-labeled OSNs and the area where OCAM+ OSNs are located are indicated with magenta dots (b) and green bars (c), respectively. (d) Illustration showing that the tongue-shaped area in the OB receives axons from OSNs in the dorsomedial part of V-zone OE. Scale bars, 200 μm.
Recent studies suggested that OCAM+ OSNs in the lateral OE were generated later than OCAM− OSNs located in the dorsomedial OE (Takeuchi et al., 2010). Moreover, within the V zone, the majority of OSNs generated at the early stage of development were located in the V-zone:DM, while OSNs generated at the later stage of development were preferentially located in the ventrolateral part called the V-zone:VL (Eerdunfu et al., 2017). In this study, we injected XdU into pregnant females at E10 (data not shown), E11 (Fig. 5a), E13 (Fig. 5b), or E16 (Fig. 5c) and calculated the density of XdU-labeled cells separately in the D-zone, V-zone:DM, and V-zone:VL OE at P0 (Fig. 5d). Significant XdU-labeling was not observed in any OE region with the injection at E10. When the XdU was injected at E11, many XdU-labeled cells were found at the V-zone:DM (8.5 ± 0.5 cells/100 μm), where we found the dye-labeled OSNs after NeuroVue injection into the tongue-shaped area (Fig. 5a, d). In contrast, fewer XdU-labeled cells were observed in the D zone (6.3 ± 0.7 cells/100 μm), and almost no XdU-labeled cells were found in V-zone:VL (1.5 ± 0.2 cells/100 μm) following XdU at E11. When XdU injection was performed at E13, we observed many XdU-labeled OSNs in the V-zone:DM (12.2 ± 0.4 cells/100 μm) and D-zone (10.1 ± 0.8 cells/100 μm), while few labeled cells were present in the V-zone:VL (3.8 ± 0.6 cells/100μm) (Fig. 5b, d). We finally observed a significant number of XdU-labeled cells in the V-zone:VL (10.5 ± 1.7 cells/100 μm) with the E16 XdU injection (Fig. 5c, d). In contrast, XdU-labeled cells in the D-zone (4.5 ± 1.6 cells/100 μm) and V-zone:DM (6.8 ± 1.6 cells/100 μm) became less dense. These results indicate that OCAM+ OSNs in the V-zone:DM that innervate the tongue-shaped area of the OB are generated in a timeframe comparable to that of D-zone OSNs and earlier than OCAM+ OSNs in the V-zone:VL. Taken together, the sequence of OSN generation in the developing OE corresponds with the sequence of Tbx21 expression in their target OB areas.
Different spatiotemporal expression patterns of Tbx21, vGluT1, and reelin in the developing olfactory bulb
In contrast to our results showing that Tbx21 expression in mitral cells starts from the tongue-shaped area in the developing OB, a previous study suggested that synaptogenesis commenced in the ventral region followed by the medial, lateral, and dorsal regions (Blanchart et al., 2008). To assess this further, we immunohistochemically examined the distribution of OMP+ axons in developing OB. At E13 and E15, a thick OMP+ layer was observed in the medial and anterolateral parts of the OB, whereas OMP+ axons formed a thinner layer and did not penetrate deep into the anteromedial region where Tbx21+ cells were present (Fig. 6a, b). The spatiotemporal expression of vesicular glutamate transporter 1 (vGluT1), a synaptic protein expressed in mitral/tufted cells, was also examined. First, no vGluT1 signal was detected in the E13 OB. At E15, vGluT1 signal was observed in the intermediate zone of the OB especially in the lateral region while there was no significant vGluT1 expression in the medial region, including the Tbx21+ tongue-shaped area (Fig. 6b, c). Of note, the vGluT1+ region corresponded with the region where OMP+ OSN axons penetrated into the intermediate zone to form protoglomeruli (Fig. 6c2) (Treloar et al., 1999). While the vGluT1 signal appeared in the dorsomedial region of E17 and P0 OBs, stronger signals were still seen in the ventrolateral region (Fig. 6d, e). At these ages, penetration of OMP+ OSN axons into the deeper layer of the OB was also observed in the dorsomedial region, whereas distinct glomeruli were exclusively formed in the ventrolateral region. At P2, regional differences in vGluT1 expression were no longer obvious, and glomeruli were formed all around the OB (Fig. 6f). Therefore, the vGluT1 expression pattern in mitral cells differed from that of Tbx21 and corresponded with the spatiotemporal pattern of glomerular formation in developing OB. In addition, no regional difference was observed in the expression of reelin, another established mitral cell marker (Fig. 6g).
Figure 6. Different spatiotemporal expression patterns of Tbx21, vGluT1, and reelin in the developing olfactory bulb.
(a, b) Horizontal section of E13 (a) and coronal section of E15 (b) OBs stained with anti-Tbx21 (green) and anti-OMP (magenta) antibodies. A cluster of Tbx21+ cells is observed in the anteromedial portion where less OMP+ OSN axons innervate. (c-f) Coronal sections of E15 (c), E17 (d), P0 (e) and P2 f) OBs stained with anti-vGluT1 (green) and anti-OMP (magenta) antibodies. Expression of vGluT1 in the OB starts from the lateral portion where OMP+ OSN axons penetrate into deeper layers (c2). (g) Coronal section of E15 OB stained with anti-reelin (green) and anti-OMP (magenta) antibodies. No obvious regional difference is observed in reelin expression. All nuclei were stained with DAPI (blue). Scale bars, 200 μm in a, b, c1, d-g; 100 μm in c2.
Taken together, Tbx21 expression may be regulated largely by intrinsic factors and occur independently of primary afferent input via the olfactory nerve. However, afferent input that would occur following the contact between mitral cell dendrites and OSN axons may largely contribute to vGluT1 expression. These results also show that while both Tbx21 and vGluT1 are well-known mitral cell markers in the OB, the onset of their expressions occurs independently. These results indicate that multiple temporal and regional pathways may contribute to mitral cell development/maturation.
Discussion
Testing our hypothesis that mitral cells in different regions develop via different mechanisms, we examined the expression time course of Tbx21, a transcription factor expressed by all mitral cells in mature OB. Our study showed that Tbx21 expression in mitral cell precursors starts from the tongue-shaped area followed by dorsomedial and then ventrolateral areas during development. The segregation of mitral cells based on the Tbx21 expression onset corresponds well with the topographical segregation of glomeruli, which is believed to be a key feature for the olfactory information processing.
What regulates the sequence of Tbx21 expression in developing OB? Since early-generated mitral cells preferentially localize in the dorsomedial area (Imamura et al., 2011), we first thought that Tbx21 might be expressed in mitral cell precursors in the order of timing of neurogenesis. This thought was partly true because many Tbx21+ cells were found in the tongue-shaped area at E15 and these cells are mostly E10- or E11-generated cells, but not E12-generated cells. However, E10-generated cells outside the tongue-shaped area were mostly Tbx21-negative. Therefore, while time after generation may be a factor regulating Tbx21 expression in mitral cell precursors, there should be an extrinsic factor that regulates the timing of Tbx21 expression in tongue-shaped, dorsomedial, and ventrolateral areas in the developing OB.
Is the arrival of OSN axons necessary for inducing the expression of Tbx21 in mitral cell precursors? Our results show that Tbx21 expression started from the OB region where OMP+ axons did not deeply penetrate. There are several transgenic mouse lines in which OSN axons do not reach the OB and form a tangled sphere of axons between the OE and OB (Yoshihara et al., 2005; Hirata et al., 2006; Besse et al., 2011). Agenesis of OB was observed in these transgenic mice, and the OB-like structure was formed in the prefrontal cortex instead. Of particular note, Tbx21 expression was observed in this olfactory-bulb like structure (Yoshihara et al., 2005; Hirata et al., 2006; Besse et al., 2011). These evidences suggest that Tbx21 expression in developing mitral cells occurs independently of the arrival of OSN axons. That said, we cannot eliminate the possibility that some pioneer axons might reach the OB-like structure and induce the Tbx21 expression in these transgenic lines.
The role of Tbx21 in mitral cell development is not fully understood at this time. Given that Tbx21 appears in developing mitral cells later than Tbr1 and Tbr2, which are involved in generation/differentiation of glutamatergic neurons (Bulfone et al., 1998; Arnold et al., 2008; Sessa et al., 2008), it might regulate the later steps of neuronal development. Since Tbx21 expression seems to occur independent of OSN innervation, Tbx21 may be involved in synaptogenesis in the olfactory cortex that occurs perinatal stages in rodents (Kunkel et al., 1987; Newman-Gage et al., 1987). A possible role of Tbx21 is that Tbx21 regulates the expression of molecules involved in axon guidance and/or synaptogenesis, and differences in Tbx21 expression onset may be a regulator of different innervation patterns in the olfactory cortex from dorsomedial and ventrolateral OB areas.
Despite its unique structure, the functional significance of tongue-shaped area is largely unknown. A previous study raised the possibility that the tongue-shaped area is a region innervated by pioneer olfactory axons that induce the evagination of the OB by regulating the cell cycle kinetics of neuronal progenitors (Gong and Shipley, 1995; Hebert et al., 2003; Yoshihara et al., 2005). The sequence of Tbx21 expression may reflect that mitral cells in this area mature earlier than other regions. Another study suggested that the musk odorants muscone activated several glomeruli in the tongue-shaped area (Shirasu et al., 2014). They also showed that the anterodorsal bulbar lesions caused muscone anosmia. Revealing the property of mitral cells in the tongue-shaped area and their axonal projection patterns are essential to understand the muscone perception.
We also showed that the tongue-shaped area of the OB received an axonal projection from the OSNs in the most dorsomedial part of the V zone, V-zone:DM, especially the anterior, OE. The V-zone:DM corresponds to zone II or to the zone where OSNs expressing an odorant receptor P2 are distributed (Ressler et al., 1993; Eerdunfu et al., 2017). Our result is consistent with the study showing that an odorant receptor expressed by zone II OSNs represented a glomerulus in the tongue-shaped area (Nagao et al., 2000). Surprisingly, our result showed that the OSNs in this region are generated earlier than OSNs in D-zone OE, while the OSN generation in lateral OE starts later than D-zone. Therefore, interestingly, the sequence of OSN generation in the developing OE corresponded with that of Tbx21 expression in the areas they innervate. It was suggested that sequential arrival of axons from D-zone OSNs to V-zone OSNs was critical for the formation of glomerular topography in the OB (Takeuchi et al., 2010). Our results suggest that the sequential arrival of OSN axons may also regulate the topographical segregation of MCL.
In contrast to Tbx21, vGluT1 expression in mitral cells starts from the ventrolateral OB where OMP+ OSN axons first deeply penetrated into the intermediate zone. The spatiotemporal vGluT1 expression pattern in the developing OB is correlated with synaptogenesis and glomerular formation (Blanchart et al., 2008). Correlation between synaptogenesis and upregulation of vGluT1 expression was also observed in the retinal and cerebral cortex (Sherry et al., 2003; Berry et al., 2012). Given that vGlut1 is a synaptic protein, contact between OSN axons and mitral cell dendrites is likely necessary for the expression of vGluT1 in developing mitral cells. Therefore, the spatiotemporal expression patterns of Tbx21 and vGluT1 in the developing OB likely differ depending on the regulatory factors.
Despite the contact between OSN axons and dendrites of OB neurons in the glomeruli, glomerular segregation is mostly defined by the molecules expressed by OSN subsets. To date, no molecule expressed by mitral cells can define the glomerular subsets. Also in this study, there was not a clear boundary for the onset of Tbx21 expression in mitral cell precursors. Nevertheless, beyond the data reported here, there are other lines of evidence that support the notion that mitral cells are not a homogeneous population: 1) several ion channels are expressed by subsets of mitral cells in a mosaic pattern (Panzanelli et al., 2005; Padmanabhan and Urban, 2010); 2) mitral cells have diverse intrinsic biophysical properties (Padmanabhan and Urban, 2010; Angelo and Margrie, 2011; Angelo et al., 2012); and 3) mitral cells have different somatic shapes, such as triangle, round, or fusiform (Kikuta et al., 2013). However, it remains a challenge to identify a mechanism for generating heterogeneous mitral cells. A previous study suggested several subdomains in the nascent external plexiform layer based on the molecular heterogeneity (Williams et al., 2007). In this study, we showed that Tbx21 expression precedes that of vGluT1 in some mitral cells and vice versa, while both Tbx21 and vGluT1 are expressed by all mature mitral cells. This finding clearly demonstrates that not all mitral cells follow the same developmental pathway, which may be an underlying mechanism for the production of subpopulations in mitral cells.
Supplementary Material
Supplementary Figure 1. Distributions of tdTomato+ cells in the developing olfactory bulbs of Tbx21Cre-tdTomato mice. MCLs unrolled from coronal sections of E17, P0, and P2 OBs were aligned from anterior to posterior using the dorsal edge (black triangles). Percentages of tdTomato+ cells located in each compartment were superimposed onto the unrolled MCL maps. Dashed lines indicated the boundary of OCAM+ and OCAM− glomeruli in the glomerular layer.
Acknowledgments
We thank Dr. Yoshihiro Yoshihara for the antibody to Tbx21. This work was supported by NIH grant R01DC016307 (F.I.).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Angelo K, Margrie TW. 2011. Population diversity and function of hyperpolarization-activated current in olfactory bulb mitral cells. Sci Rep 1:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo K, Rancz EA, Pimentel D, Hundahl C, Hannibal J, Fleischmann A, Pichler B, Margrie TW. 2012. A biophysical signature of network affiliation and sensory processing in mitral cells. Nature 488(7411):375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnar Z, Robertson EJ, Groszer M. 2008. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev 22(18):2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry CT, Sceniak MP, Zhou L, Sabo SL. 2012. Developmental up-regulation of vesicular glutamate transporter-1 promotes neocortical presynaptic terminal development. PLoS One 7(11):e50911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse L, Neti M, Anselme I, Gerhardt C, Ruther U, Laclef C, Schneider-Maunoury S. 2011. Primary cilia control telencephalic patterning and morphogenesis via Gli3 proteolytic processing. Development 138(10):2079–2088. [DOI] [PubMed] [Google Scholar]
- Blanchart A, Romaguera M, Garcia-Verdugo JM, de Carlos JA, López-Mascaraque L. 2008. Synaptogenesis in the mouse olfactory bulb during glomerulus development. Eur J Neurosci 27(11):2838–2846. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Martinez S, Marigo V, Campanella M, Basile A, Quaderi N, Gattuso C, Rubenstein JL, Ballabio A. 1999. Expression pattern of the Tbr2 (Eomesodermin) gene during mouse and chick brain development. Mech Dev 84(1–2):133–138. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Wang F, Hevner R, Anderson S, Cutforth T, Chen S, Meneses J, Pedersen R, Axel R, Rubenstein JL. 1998. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron 21(6):1273–1282. [DOI] [PubMed] [Google Scholar]
- Eerdunfu, Ihara N, Ligao B, Ikegaya Y, Takeuchi H. 2017. Differential timing of neurogenesis underlies dorsal-ventral topographic projection of olfactory sensory neurons. Neural Dev 12(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faedo A, Ficara F, Ghiani M, Aiuti A, Rubenstein JL, Bulfone A. 2002. Developmental expression of the T-box transcription factor T-bet/Tbx21 during mouse embryogenesis. Mech Dev 116(1–2):157–160. [DOI] [PubMed] [Google Scholar]
- Feng W, Simoes-de-Souza F, Finger TE, Restrepo D, Williams T. 2009. Disorganized olfactory bulb lamination in mice deficient for transcription factor AP-2epsilon. Mol Cell Neurosci 42(3):161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Williams T. 2003. Cloning and characterization of the mouse AP-2 epsilon gene: a novel family member expressed in the developing olfactory bulb. Mol Cell Neurosci 24(2):460–475. [DOI] [PubMed] [Google Scholar]
- Gong Q, Shipley MT. 1995. Evidence that pioneer olfactory axons regulate telencephalon cell cycle kinetics to induce the formation of the olfactory bulb. Neuron 14(1):91–101. [DOI] [PubMed] [Google Scholar]
- Gussing F, Bohm S. 2004. NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci 19(9):2511–2518. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Lin M, Partanen J, Rossant J, McConnell SK. 2003. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development 130(6):1101–1111. [DOI] [PubMed] [Google Scholar]
- Hirata T, Nakazawa M, Yoshihara S, Miyachi H, Kitamura K, Yoshihara Y, Hibi M. 2006. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development 133(8):1433–1443. [DOI] [PubMed] [Google Scholar]
- Imamura F, Ayoub AE, Rakic P, Greer CA. 2011. Timing of neurogenesis is a determinant of olfactory circuitry. Nat Neurosci 14(3):331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura F, Greer CA. 2013. Pax6 regulates Tbr1 and Tbr2 expressions in olfactory bulb mitral cells. Mol Cell Neurosci 54:58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura F, Nagao H, Naritsuka H, Murata Y, Taniguchi H, Mori K. 2006. A leucine-rich repeat membrane protein, 5T4, is expressed by a subtype of granule cells with dendritic arbors in specific strata of the mouse olfactory bulb. J Comp Neurol 495(6):754–768. [DOI] [PubMed] [Google Scholar]
- Kikuta S, Fletcher ML, Homma R, Yamasoba T, Nagayama S. 2013. Odorant response properties of individual neurons in an olfactory glomerular module. Neuron 77(6):1122–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, Mori K, Sakano H. 2007. Innate versus learned odour processing in the mouse olfactory bulb. Nature 450(7169):503–508. [DOI] [PubMed] [Google Scholar]
- Kondoh K, Lu Z, Ye X, Olson DP, Lowell BB, Buck LB. 2016. A specific area of olfactory cortex involved in stress hormone responses to predator odours. Nature 532(7597):103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel DD, Westrum LE, Bakay RA. 1987. Primordial synaptic structures and synaptogenesis in rat olfactory cortex. Synapse 1(2):191–201. [DOI] [PubMed] [Google Scholar]
- Mitsui S, Igarashi KM, Mori K, Yoshihara Y. 2011. Genetic visualization of the secondary olfactory pathway in Tbx21 transgenic mice. Neural Syst Circuits 1(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. 2005. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci 25(14):3586–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Naritsuka H, Mori K, Yoshihara Y. 2012. Tbr2 deficiency in mitral and tufted cells disrupts excitatory-inhibitory balance of neural circuitry in the mouse olfactory bulb. J Neurosci 32(26):8831–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sakano H. 2011. How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci 34:467–499. [DOI] [PubMed] [Google Scholar]
- Nagao H, Yoshihara Y, Mitsui S, Fujisawa H, Mori K. 2000. Two mirror-image sensory maps with domain organization in the mouse main olfactory bulb. Neuroreport 11(13):3023–3027. [DOI] [PubMed] [Google Scholar]
- Newman-Gage H, Westrum LE, Bertram JF. 1987. Stereological analysis of synaptogenesis in the molecular layer of piriform cortex in the prenatal rat. J Comp Neurol 261(2):295–305. [DOI] [PubMed] [Google Scholar]
- Oka Y, Kobayakawa K, Nishizumi H, Miyamichi K, Hirose S, Tsuboi A, Sakano H. 2003. O-MACS, a novel member of the medium-chain acyl-CoA synthetase family, specifically expressed in the olfactory epithelium in a zone-specific manner. Eur J Biochem 270(9):1995–2004. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K, Urban NN. 2010. Intrinsic biophysical diversity decorrelates neuronal firing while increasing information content. Nat Neurosci 13(10):1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzanelli P, Perazzini AZ, Fritschy JM, Sassoe-Pognetto M. 2005. Heterogeneity of gamma-aminobutyric acid type A receptors in mitral and tufted cells of the rat main olfactory bulb. J Comp Neurol 484(1):121–131. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. 1993. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73(3):597–609. [DOI] [PubMed] [Google Scholar]
- Richard MB, Taylor SR, Greer CA. 2010. Age-induced disruption of selective olfactory bulb synaptic circuits. Proc Natl Acad Sci U S A 107(35):15613–15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Denny CA, Hen R, Axel R. 2014. The participation of cortical amygdala in innate, odour-driven behaviour. Nature 515(7526):269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. 2008. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60(1):56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry DM, Wang MM, Bates J, Frishman LJ. 2003. Expression of vesicular glutamate transporter 1 in the mouse retina reveals temporal ordering in development of rod vs. cone and ON vs. OFF circuits. J Comp Neurol 465(4):480–498. [DOI] [PubMed] [Google Scholar]
- Shirasu M, Yoshikawa K, Takai Y, Nakashima A, Takeuchi H, Sakano H, Touhara K. 2014. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron 81(1):165–178. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Inokuchi K, Aoki M, Suto F, Tsuboi A, Matsuda I, Suzuki M, Aiba A, Serizawa S, Yoshihara Y, Fujisawa H, Sakano H. 2010. Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell 141(6):1056–1067. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Purcell AL, Greer CA. 1999. Glomerular formation in the developing rat olfactory bulb. J Comp Neurol 413(2):289–304. [PubMed] [Google Scholar]
- Williams EO, Xiao Y, Sickles HM, Shafer P, Yona G, Yang JY, Lin DM. 2007. Novel subdomains of the mouse olfactory bulb defined by molecular heterogeneity in the nascent external plexiform and glomerular layers. BMC Dev Biol 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara S, Omichi K, Yanazawa M, Kitamura K, Yoshihara Y. 2005. Arx homeobox gene is essential for development of mouse olfactory system. Development 132(4):751–762. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K. 1997. OCAM: A new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci 17(15):5830–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Distributions of tdTomato+ cells in the developing olfactory bulbs of Tbx21Cre-tdTomato mice. MCLs unrolled from coronal sections of E17, P0, and P2 OBs were aligned from anterior to posterior using the dorsal edge (black triangles). Percentages of tdTomato+ cells located in each compartment were superimposed onto the unrolled MCL maps. Dashed lines indicated the boundary of OCAM+ and OCAM− glomeruli in the glomerular layer.