ABSTRACT
RNA species play host to a plethora of post-transcriptional modifications which together make up the epitranscriptome. 5-methyluridine (m5U) is one of the most common modifications made to cellular RNA, where it is found almost ubiquitously in bacterial and eukaryotic cytosolic tRNAs at position 54. Here, we demonstrate that m5U54 in human mitochondrial tRNAs is catalysed by the nuclear-encoded enzyme TRMT2B, and that its repertoire of substrates is expanded to ribosomal RNAs, catalysing m5U429 in 12S rRNA. We show that TRMT2B is not essential for viability in human cells and that knocking-out the gene shows no obvious phenotype with regards to RNA stability, mitochondrial translation, or cellular growth.
KEYWORDS: Mitochondria, tRNA, rRNA, post-transcriptional modifications, 5-methyluridine
Introduction
The accurate expression of the human mitochondrial genome (mtDNA) is essential for the faithful synthesis of 13 components of the oxidative phosphorylation machinery. In order to achieve this, the 22 tRNAs and 2 rRNAs encoded in the mtDNA undergo significant post-transcriptional modification performed by nuclear-encoded proteins imported into mitochondria [1]. Recent data indicate that defects in the nucleotide modification of mitochondrial (mt-) RNA can frequently lead to human disorders of mitochondrial respiration [2–5]
5-methyluridine (m5U), is found with high occurrence in bacterial and eukaryotic tRNAs at position 54 in the T-loop, including, although significantly less common, human mitochondrial tRNAs. Although first identified in tRNAs, the T-loop motif has now been identified in a wide array of different non-coding RNAs, including the tmRNA of bacteria [6], RNase P RNA [7], self splicing introns [8], and riboswitches [9], implying that this frequently occurring motif is an important structural building block. T-loops have additionally been implicated in stabilizing the tertiary fold of rRNA, this has been suggested to occur either via base pairing/base pairing tertiary interactions or by stabilizing stems by forming a non-interacting caps [10]. The m5U modification is also found in a number of rRNA species, including that of the mitochondrial small rRNA in hamster [11,12] however as before, their exact contribution to the surrounding tertiary structure is uncertain.
The most well studied m5U-methyltransferases are a group of three SAM-dependent enzymes expressed by E. coli: RlmC, RlmD, and TrmA. Both RlmC and RlmD methylate the 23S rRNA of the large subunit, with the former producing m5U747 and the latter producing m5U1939 [13]. The aforementioned m5U54 tRNA modification is catalysed by TrmA, which also methylates the T-loop in E.coli tmRNA [14]. TrmA has additionally been found to methylate bacterial 16S rRNA in vitro, however this modification is not found in vivo [15]. The proposed catalytic mechanism utilized by m5U-methyltransferases has been supported by the crystal structure of the TrmA-tRNA complex in which the sulphhydryl-group of a conserved cysteine acts as a nucleophile to attack C6, leading to the formation of a covalent bond between the two. The resulting negative charge on O4 prompts a second nucleophilic attack by the C5 carbon on the methyl group of SAM, forming m5U. A second highly conserved residue, a glutamate, acts as a base to abstract a proton from C5 leading to the elimination of the covalent adduct and its release from the regenerated active site [16].The identification and characterization of TrmA has not brought the field significantly closer to understanding the exact role of m5U54, as mutations in E.coli TrmA that lead to the complete loss of methyltransferase activity revealed no perceptible difference in growth rate, codon recognition, ribosome interaction, or translation rate in vivo [17]. A growth defect in the TrmA mutant cells was only observed following growth in a mixed culture in which the two strains directly competed. Interestingly, the TrmA protein itself, but not its known enzymatic activity, is found to be essential for viability. An insertion within the trmA gene corresponding to the N-terminus of the protein is lethal, therefore it is proposed that TrmA has a secondary essential function that is separate from its methyltransferase activity [18].
The loss of m5U54 from both cytoplasmic and mitochondrial tRNAs was first observed in eukaryotic cells following the deletion of the trm2 gene in S.cerevisiae [19]. As was the case for bacterial TrmA, the loss of Trm2 showed no physiological defect compared to wild-type cells, however even in cocultures of mixed populations, the wild-type cells demonstrated no selective advantage over the mutant strain after 35 generations. The requirement for trm2 is further contrasted from that of trmA by the nonessential nature of not only m5U54 but of the protein itself, as a complete deletion of trm2 also shows no physiological defect [20]. The large majority of homology between TrmA and Trm2 lies towards the C-terminus containing the known methyltransferase domain, with little homology towards the N-terminus. This, therefore, supports the notion that the unidentified essential function of TrmA is situated towards the N-terminus, and that this function is either not performed by Trm2 or there is a greater degree of functional redundancy in eukaryotic cells. Although the absence of Trm2 in isolation shows no physiological effect, its deletion has been demonstrated to induce lethality in four strains carrying mutations in tRNASer(CGA) [21], indicating that the stabilizing nature of m5U54 may only be observed when the structure of a tRNA is affected in another manner. Intriguingly, although three of the four mutants require catalytically active Trm2, and therefore m5U54, to be present for stability, one mutation described is entirely rescued through the expression of a catalytically inactive Trm2. It is therefore suggested that in addition to introducing the stabilizing role of m5U54, Trm2 itself may possess a chaperone-like function that is separate from its catalytic role. A separate endo-exonuclease activity has been claimed for Trm2, which is preserved following deletion of the C-terminal methyltransferase domain [22]. Through this activity, Trm2 is proposed to contribute to the repair of double-strand breaks by the 5ʹ-3ʹ resection of DNA ends. However, how this additional activity would explain the phenotypes observed with tRNASer(CGA) mutants is unclear.
Efforts to characterize the modification profile of human tRNAs have identified the presence of m5U54 in the majority of cytoplasmic tRNAs as well as a small number of mitochondrial tRNAs. A very recent study demonstrated that human TRMT2A is responsible for introduction of the majority of m5U in human RNA, and that it targets U54 of cytosolic tRNAs [23]. The enzyme responsible for m5U in human mt-tRNA however remains to be characterized, although TRMT2B has been previously suggested as a candidate [24]. Likewise, the methyltransferase responsible for the introduction of m5U429 in mammalian mitochondrial 12S rRNA is yet to be identified. In this work, we aim to identify the ortholog of Trm2 operating in human mitochondria and to study the consequences of its loss.
Results
TRMT2B is a putative methyltransferase localized to human mitochondria
The m5U54 modification has been identified in both cytoplasmic and mitochondrial tRNAs in humans [25], with the former recently identified to be catalysed by TRMT2A[23], one of the two human orthologs of yeast Trm2 along with TRMT2B. As a mitochondrial role for TRMT2A was not ruled out by Carter et al [23]., we set out to identify which of these paralogs may be operating within human mitochondria.An alignment of protein sequences from a range of species shows that the SAM binding site is highly conserved in the TRMT2A and TRMT2B paralogs. Whilst this is also the case for the remainder of the active site in TRMT2A, TRMT2B displays some dramatic divergences in some species (Fig. 1). The proposed common mechanism for SAM-dependent m5U methyltransferases, supported by the crystal structure of TrmA [16], entirely depends on the involvement of a nucleophilic cysteine and a proton extracting glutamate (Figure S1). Whilst these two residues are conserved in all species analysed for TRMT2A, a number of species contain mutations altering the catalytic cysteine in TRMT2B, which would be predicted to very severely impede the enzymes methyltransferase activity. The majority of these species belong to a single subfamily, the Bovinae, in which this otherwise well conserved cysteine is substituted for tyrosine (Fig. 1). The finding that the bovine TRMT2B homolog appears catalytically inactive is in agreement with the initially surprising observation that m5U54, one of the most common tRNA modifications, is absent from all bovine mt-tRNAs [26]. However, m5U54 is highly abundant in bovine cyto-tRNAs [25], it is tempting to speculate therefore, that these cytosolic substrates are methylated by the functional TRMT2A, which does not operate on mt-tRNAs.
Figure 1.
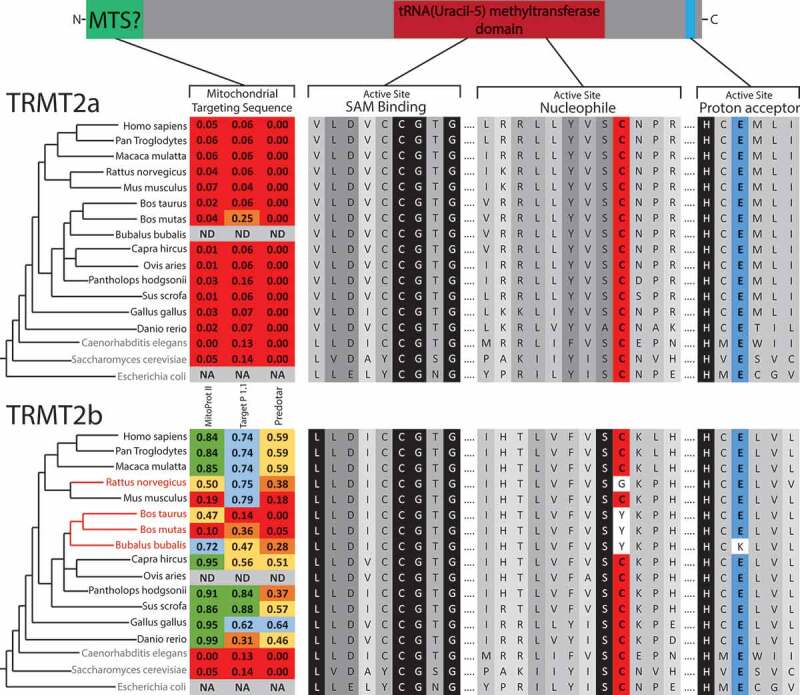
Sequence Analysis of m5U54-methyltransferase homologs.
Schematic of protein domains in both TRMT2A and TRMT2B. Homologous sequences identified through BLAST searches aligned for key catalytic regions, with the degree of shadowing representing the extent of conservation for a given residue. Nucleophilic cysteine shown in red, Proton extracting glutamate shown in blue. Species that contain only a single predicted m5U54-methyltransferase indicated in blue text. Species that contain a TRMT2B sequence that is predicted to be catalytically inactive are indicated in red text. These two paralogs were analysed for the presence of an N-terminal mitochondrial localization signal (MTS) using computational prediction tools. Higher scores (between 0 and 1) indicate a higher probability of mitochondrial localization, which are also colour coded. NA, not applicable; ND, no data
The observation that bovine cyto-tRNAs are methylated at U54, whilst mt-tRNAs are not, could be explained by the solely cytosolic localization of a catalytically active TRMT2A, and the mitochondrial localization of a catalytically inactive TRMT2B. To test this hypothesis, the sequences of both paralogs were analysed for the likelihood of an N-terminal mitochondrial targeting sequence (MTS). In all three of the programs used to predict mitochondrial localization (MitoProt II [27], Target P 1.1 [28], and Predotar [29]), the TRMT2A sequence returned very low scores across all species analysed, consistent with a non-mitochondrial role. These scores were significantly higher for TRMT2B, indicating possible mitochondrial targeting (Fig. 1). To confirm the mitochondrial localization of human TRMT2B, HeLa cells were transiently transfected with a C-terminal GFP-tagged TRMT2B and localized based on fluorescence. Through this analysis, TRMT2b was found to colocalise with the known mitochondrial protein TOM20 (Fig. 2). Taken together these results show TRMT2B is a mitochondria-localized protein.
Figure 2.
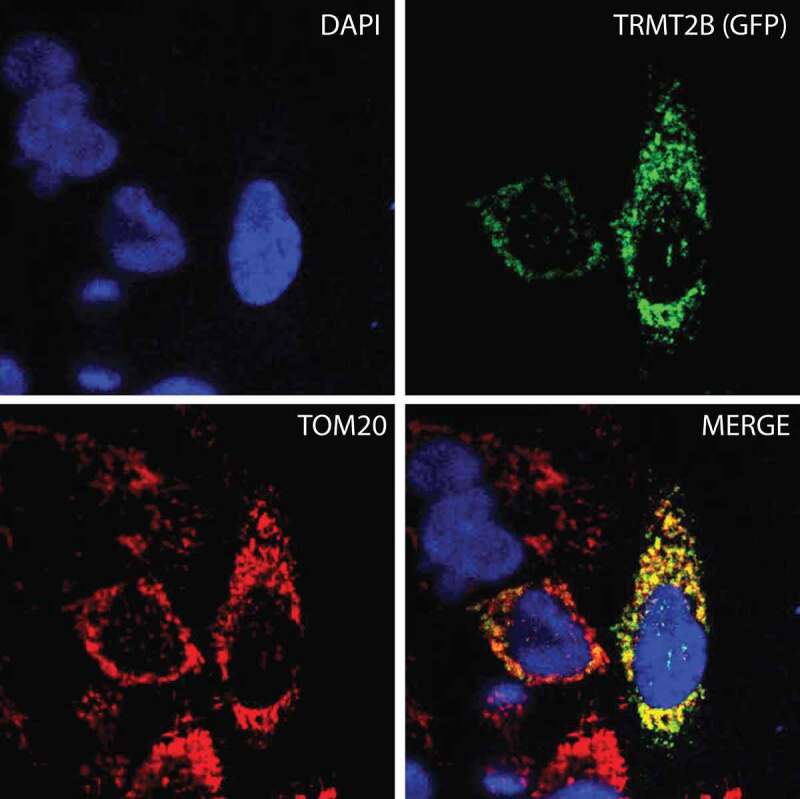
TRMT2B is localized to mitochondria.
TRMT2B-GFP cDNA construct transfected into HeLa cells and detected by fluorescence (top right, green). Nuclei were stained using DAPI (top left, blue). The mitochondrial network was stained using antibodies against the known mitochondrial protein TOM20 (bottom left, red). Colocalisation of TRMT2B and TOM20 appears as yellow in a digitally overlaid image (bottom right).
TRMT2B is a mitochondrial m5U54 tRNA methyltransferase
Next, we set out to determine mtRNA targets for TRMT2B. Due to the location of position 5 on the Hoogsteen, rather than Watson-Crick edge (Figure S2A), its methylation to form m5U does not interfere with the processivity of a reverse transcriptase, and therefore cannot be detected by primer extension without prior treatment. The presence of m5U can be inferred, however, due to its resistance to depyrimidination in the presence of hydrazine, a process which unmodified uracil is susceptible to (Figure S2A). Subsequent treatment with aniline results in phosphodiester bond cleavage at abasic sites, which are then detected due to the accumulation of primers that are unable to extend further (Figure S2B). In order to test the predicted role of TRMT2B, the aforementioned technique was used to test the U54 methylation status following transfection with siRNAs targeted to deplete TRMT2B. For mt-tRNAPro, which has been previously identified as containing m5U54 [30], the degree of stalling at U54, relative to downstream U49, is notably increased following TRMT2B siRNA transfection compared to an untransfected control (Fig. 3A). To confirm the role of TRMT2B in m5U54 methylation the same assay was repeated using RNA extracted from a TRMT2B knock-out HAP1 cell line. Correspondingly, the degree of stalling at U54 was found to be drastically increased in the knock out cell line relative to a control, consistent with the absence of m5U54 (Fig. 3B). This approach was repeated for a selection of other mt-tRNAs containing U54, out of which mt-tRNAAsn and mt-tRNAGln also appeared to contain m5U54 (Figure S3). In agreement with the previously identified role for TRMT2A23, the m5U54 status of cytosolic tRNAs were found to be unaffected following the loss of TRMT2B activity (Figure S4). Collectively, these results show TRMT2B is a mitochondrial protein responsible for the modification of position 54 in mt-tRNAs.
Figure 3.
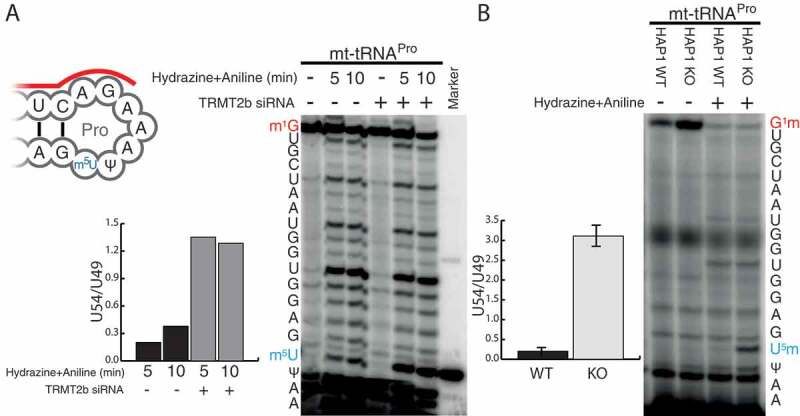
TRMT2B catalyses m5U54 in mt-tRNAPro.
(A) Schematic of mt-tRNAPro T-loop showing annealed primer to be extended (red line) and the position of m5U54 (blue text). HeLa cell derived RNA, either following a 6-day siRNA mediated depletion of TRMT2B or untreated, was subsequently either untreated (-) or treated with hydrazine for either 5 or 10 minutes, followed by aniline, to specifically cleave at unmodified uridine residues. This RNA was subjected to RT-PEx using a [32P]-end labelled primer complementary to the region upstream of m5U54 (red line). The nucleotide sequence of the tRNA, corresponding to stalling events at each position, is shown to the side of the panel. Quantification values represent the ratio between stalling at U54 and stalling at the next uridine residue (U49), after the values in the untreated lanes had been subtracted from both to account for background. (B) RT-PEx reactions as performed above with RNA derived from a HAP1 parental cell line with wild-type (WT) or a HAP1 TRMT2B knockout cell line (KO). Error bars = SEM, n = 3.
TRMT2B is a mitochondrial m5U429 12S rrna methyltransferase
An m5U methylation has also been previously identified at U426 within the small 12S rRNA from hamster mitochondria [11]. A sequence alignment of this region shows a very high degree of conservation across a range of eukaryotic species (Fig. 4A). This sequence conservation is also present in the small 16S rRNA from E.coli where it forms the 790-loop, and whilst the corresponding uracil (U788) is not methylated, mutations within this loop yielded no fully formed 30S subunit [31]. The corresponding human residue, U429, is located within the central domain of 12S rRNA in helix 27 (Fig. 4B), the loop of which is in very close proximity to the codon-anticodon site between bound mRNA and tRNA (Fig. 4C). As this loop displays similarity to the consensus T-loop, and m5U has been detected at this position in hamster mitochondria, 12S U429 may also act as a substrate for TRMT2B, in addition to mt-tRNAs. To test whether TRMT2B is responsible for modification of 12S U429 in human mitochondria, the presence or absence of m5U was tested following an RNA treatment of hydrazine and aniline in wild type and TRMT2B knockout cells. Very little stalling at U429 was observed in wild type cells (Fig. 4D), consistent with a modification conferring resistance to hydrazine. On the contrary, stalling at U429 is significantly increased in RNA extracted from TRMT2B knockout cells (Fig. 4D). These results are consistent with human 12S containing an m5U methylation, as has been shown for hamster, and with this methylation being catalysed by TRMT2B.
Figure 4.
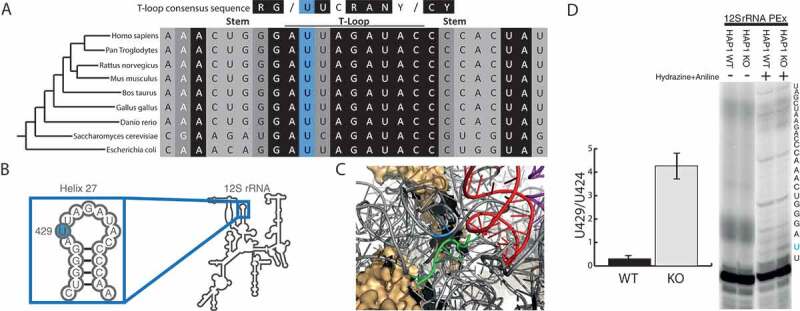
TRMT2B catalyses m5U429 in 12S mitochondrial rRNA.
(A) Alignment of the small ribosomal RNA (rRNA) from a range of species in a region corresponding to helix 27 in human 12S mt-rRNA. The degree of shadowing represents the extent of conservation for a given residue, with U429 (or the corresponding position in other species) shown in blue background. The T-loop consensus sequence is displayed above for comparison. (B) Schematic of human 12S mt-rRNA, helix 27, and the location of m5U429 (blue circle). (C) The structure of the human mitoribosome highlighting U429 (blue), the bound mRNA (green), and the adjacently bound tRNA (red). (D) Separation and detection of RT-PEx products using a [32P]-end labelled primer complementary to the region upstream of m5U429 in 12S rRNA. Extension reactions performed on RNA derived from a HAP1 wild-type (WT) and a HAP1 TRMT2B knockout cell line (KO), with or without hydrazine-aniline treatment. The nucleotide sequence of 12S rRNA is shown to the side of the panel. Quantification values represent the ratio between stalling at U429 and stalling at the next uridine residue (U424). Error bars = SEM, n = 4.
Mitochondrial trna stability and aminoacylation are unaffected by the loss of TRMT2B
Incorrect folding as a consequence of hypomodification has been shown to disrupt the interaction between a tRNA and its cognate aminoacyl-tRNA synthetase [32,33]. In order to assess the impact of m5U54 loss on tRNA charging, RNA extracted from WT and TRMT2B KO cells was subjected to acidic-PAGE, allowing for charged and uncharged tRNAs to be distinguished by their differing electrophoretic mobilities. In this analysis, a selection of tRNAs were assessed including those known to contain m5U54 (mt-tRNAPro (Fig. 3), mt-tRNAGln (Figure S3), mt-tRNASerUCN [34], and mt-tRNALeuUUR [25]), those known to contain unmodified U54 (mt-tRNASerAGY and mt-tRNAMet) [25], and those not containing U54 (mt-tRNAPhe and mt-tRNAArg) [35]. The degree of aminoacylation for three m5U54-containing mt-tRNAs shows no change as a result of the loss of TRMT2B (Fig. 5A). Interestingly, the migration pattern of mt-tRNALeuUUR is altered between the wild-type and knockout cell lines, but this shift is present in both the acylated and deacylated samples, and therefore independent of aminoacylation. The conditions in which acidic-PAGE gels are run may allow for the preservation of certain tRNA secondary structures [36], and therefore this electrophoretic shift is likely an indication of the structural contribution made by m5U54.
Figure 5.
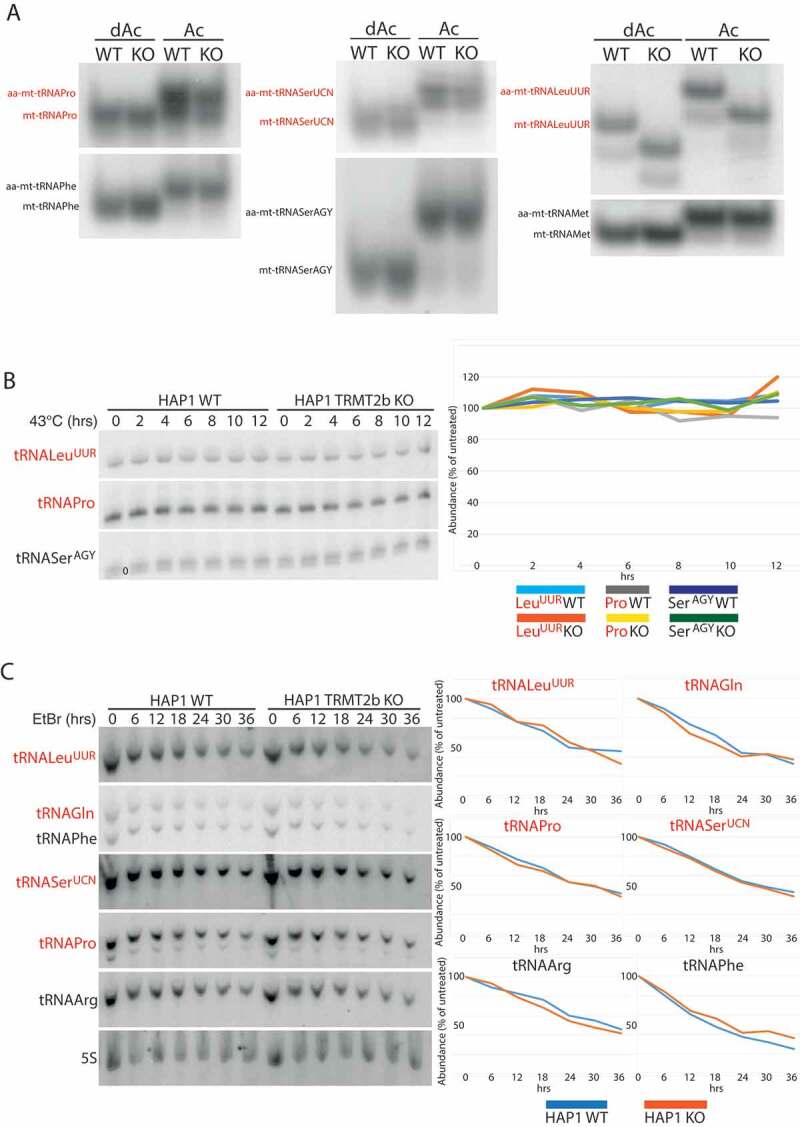
tRNA aminoacylation and stability following the loss of TRMT2B.
(A) High resolution acidic polyacylamide gel electrophoresis (PAGE) northern blot analysis of total RNA extracted from HAP1 wild-type (WT) or TRMT2B knockout cell line (KO). RNA was either maintained in low pH conditions to preserve aminoacylation (Ac), or intentionally deacylated prior to being run on the gel (dAc). The blots were probed with the mt-tRNA-specific riboprobes as indicated, with the tRNAs known to contain m5U54 in red text.(B) High resolution PAGE northern blot analysis of total RNA extracted from HAP1 parental cell line (WT), or a HAP1 TRMT2B knockout cell line (KO), following their growth at 43°C for between 2 and 12 hours. Control cells kept at 37°C shown as ‘0 hrs’. The blots were probed with the mt-tRNA-specific riboprobes as indicated, with the tRNAs known to contain m5U54 in red text. Quantification values were plotted as a percentage of the untreated 0 hr RNA. (C) As above following exposure to 250 μg/mL ethidium bromide (EtBr) for between 6 and 36 hours. Control cells grown in media containing no EtBr shown as ‘0 hrs’. The blots were probed with the mt-tRNA-specific riboprobes as indicated, with the tRNAs known to contain m5U54 in red text. Quantification values were plotted as a percentage of the untreated 0 hr RNA, after they had been normalized to 5S to control for RNA loading.
As in vitro studies have pointed towards a role for m5U54 in tRNA thermal stability [37], the loss of TRMT2B has the potential to induce a temperature-sensitive phenotype. Whilst the stability of mature tRNAs is typically unaffected by heat-stress, a 6 hour heat-shock at 43°C induces a 60% reduction in the levels of initiator cyto-tRNAMet, which coincidentally is one of the few cytoplasmic tRNAs to lack m5U54 [38]. The steady state levels of mt-tRNAs were assessed through northern blotting following RNA extraction from wild-type and knockout cells which were incubated at 43°C for between 2 and 12 hours, or kept at 37°C (0 hrs). No decrease in the steady state levels was observed for m5U54-bearing or m5U54-absent tRNAs, either in the wild-type or the knock-out cells (Fig. 5B). Whilst this suggests no decrease in the stability of m5U54-lacking tRNAs, a constant steady state level could be maintained by a mtDNA transcription rate sufficiently high to replenish any degraded tRNA. Therefore, in order to test if this is the case, cells were treated with ethidium bromide to block mitochondrial transcription [39,40]. RNA was extracted from wild-type and TRMT2B knockout cells at a range of time points between 6 and 36 hours, following the addition of 250 μg/mL of ethidium bromide, along with untreated cells (0 hrs). The rate of degradation for m5U54-bearing or m5U54-absent tRNAs showed no variation between the wild-type and knockout cell line (Fig. 5C). Therefore, these data suggest no difference in the stability of mt-tRNAs following the loss of m5U54.
Mitoribosomestability is not disrupted by the loss of TRMT2B
The disruption of assembly pathways through the loss of an rRNA modification has been previously shown to result in the degradation of subunits that are unable to be incorporated into monosomes [41]. In order to determine if TRMT2B activity contributes towards mitoribosome biogenesis, the steady state levels of protein components of the mitochondrial small 28S subunit were, therefore, assessed by western blotting. However, of the three MRPS proteins determined, the loss of m5U429 appears to have no discernible effect on their steady state levels (Fig. 6A). Likewise, the loss of m5U429 also has no effect on the steady-state levels of 12S mt-rRNA itself relative to 16S mt-rRNA (Fig. 6B). As for mt-tRNAs, the effect of m5U429 on 12S thermal stability was determined by a 43°C heat shock for between 2 and 12 hours. Interestingly, 12S mt-rRNA displayed a significantly greater susceptibility to heat-induced degradation compared to 16S rRNA, however, no difference in the rate of degradation was observed between the wild-type and knockout cell lines (Fig. 6C). An analysis of 12S stability in the absence of replenishment from mitochondrial transcription also identifies no difference in the stability of 12S mt-rRNA resulting from the loss of TRMT2B (Fig. 6D).
Figure 6.
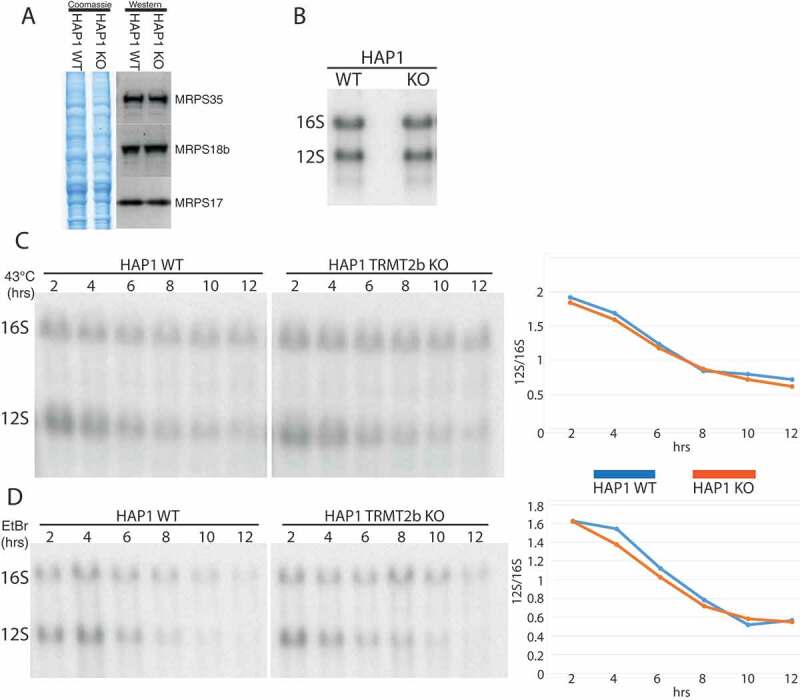
Mitoribosome integrity following the loss of TRMT2B.
(A) Western blot using lysates derived from HAP1 WT and KO probed using antibodies against three protein components on the small subunit. Coomassie gel staining shown as loading control. (B) Northern blot analysis of total RNA extracted from HAP1 parental cell line (WT), or a HAP1 TRMT2B knockout cell line (KO). (C) Northern blot analysis of total RNA extracted from HAP1 WT and TRMT2B KO cell lines following their growth at 43°C for between 2 and 12 hours. Quantification values plotted as the ratio between 12S and 16S. (D) Northern blot analysis of total RNA extracted from HAP1 WT and TRMT2B KO cell lines following their exposure to 250 μg/mL ethidium bromide (EtBr) for between 2 and 12 hours.
The loss of TRMT2B has no significant impact on mitochondrial translation
Although the loss either m5U54 or m5U429 have caused no detectable differences in the stability of mt-tRNAs or 12S mt-rRNA, their absence may instead be detrimental to the efficiency or fidelity of mitochondrial translation. To this end, the steady state levels of representative subunits from respiratory chain complexes were assessed through western blotting, however this displayed no significant difference resulting from the loss of TRMT2B (Fig. 7A). Similarly, the rate of mitochondrial protein synthesis was measured by the incorporation of [35S] labelled methionine into nascent polypeptides. Once again, no difference between the wildtype and knockout cell line was observed (Fig. 7B). Potentially, differences in the mitochondrial translation rate that are too subtle to distinguish through the assays described above, would be expected to provide cells with a growth advantage that would become apparent over a significantly long period of expansion. Wildtype and TRMT2B knockout cells were plated at a low confluency and the cell density measured over a 140 hour time course of growth. These growth curves were performed in either a high glucose medium or low glucose medium, with the latter exacerbating any growth disadvantage stemming from mitochondrial dysfunction, as cells are forced to rely more greatly on oxidative phosphorylation than glycolysis. Of note, growth in medium containing galactose as the sole carbon source, resulted in considerable cell death in both the wildtype and TRMT2B knockout HAP1 cell lines, obscuring accurate measurements. Under both conditions tested, the TRMT2B knockout cell line exhibited no statistically significant decrease in growth rate compared with the wildtype control (Fig. 7C).
Figure 7.
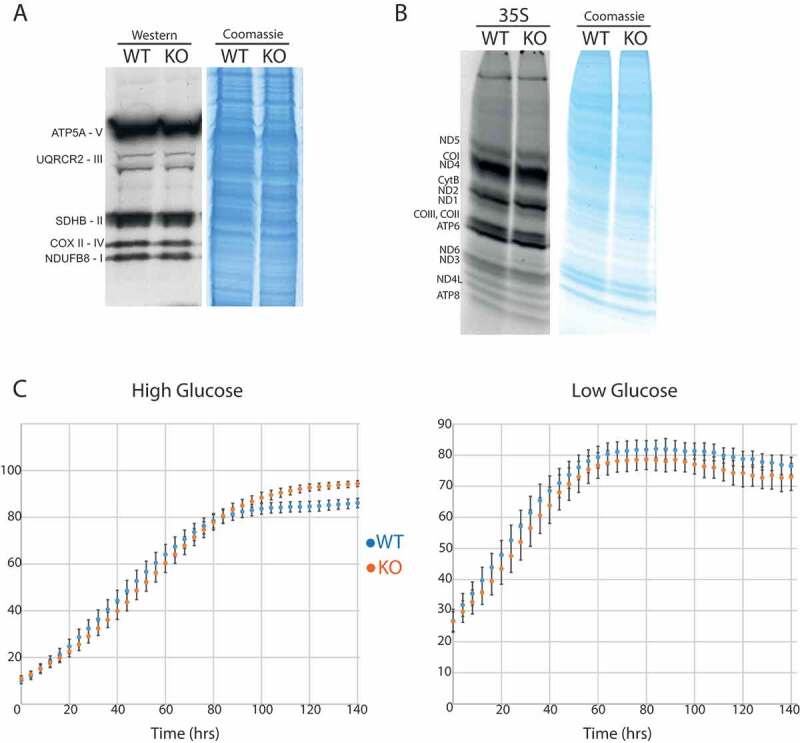
Mitochondrial translation following the loss of TRMT2B.
(A) Western blot analysis of steady-state levels of OXPHOS subunits in HAP1 parental cell line (WT), and the HAP1 TRMT2B knockout cell line (KO). (B) Assessment of de novo mitochondrial protein synthesis through [35S]-methionine incorporation in the HAP1 WT and KO cell lines. Coomassie gel staining was used as a control for protein loading. (C) Growth curves obtained by Incucyte kinetic imaging system of HAP1 WT and KO cell lines. Cells were grown for 140 hours in the presence of ‘high’ (4.5 g/L) or ‘low’ (1 g/L) glucose.
Discussion
TRMT2B catalyses the formation of m5U54 and m5U429 in mt-trnas and 12S rrna, respectively
In S.cerevisiae, the methyltransferase Trm2 is dual localized, catalysing the formation of m5U54 in both cyto-tRNAs and mt-tRNAs [19], however a homology search of the human genome identified two potential orthologs, TRMT2A and TRMT2B. On the basis of mitochondrial localization and the loss of m5U54 in mt-tRNAs in TRMT2B-ablated cells, this work concludes that TRMT2B is responsible for the m5U54-methyltransferase activity in human mitochondria. By extension, this work corroborates an earlier finding that TRMT2A is responsible for this activity in the cytosol [23]. In addition, the presence of m5U429 in 12S mt-rRNA, predicted from its detection in hamster mitochondria, is supported by this study, and also appears to be a product of TRMT2B. This makes TRMT2B the second human mitochondrial methyltransferase found to operate on both mt-tRNAs and mt-rRNA, following the revelation that the mt-tRNA m1A58-methyltransferase TRMT61B also introduces the same modification at position 947 in 16S mt-rRNA [42]. The existence of m5U modifications in both tRNAs and rRNAs is also well known within Archaea and Bacteria, however here they are performed by separate enzymes. For example, E.coli expresses three m5U-methyltransferase paralogs, TrmA catalysing m5U54 in tRNAs, RlmC catalysing m5U747 in 23S rRNA, and RlmD catalysing m5U1939 in 23S rRNA [43]. The data presented herein is corroborated by that of Laptev et al [44]., where the TRMT2B ortholog in mouse is likewise shown to act as a dual specific tRNA and rRNA m5U methyltransferase operating in mitochondria.
How does TRMT2B methylation contribute to mitochondrial translation?
The near-ubiquity in bacterial and eukaryotic tRNA sequences [25],decreased tRNA stability in vitro [37], and a fundamental role in tRNA structure suggested by the convergent evolution of sterically similar m1Ψ54 [45], would lead one to the conclusion that m5U54 holds a pivotal role in translation, with severe consequences resulting from its loss. In reality, the precise biological role for this modified nucleotide has proven difficult to determine since its identification more than 50 years ago [46]. The present study identifies the enzyme responsible for this modification in human mitochondria as TRMT2B, and also shows that the same enzyme additionally catalyses the formation of m5U429 in 12S mt-rRNA. Furthermore, functional studies of TRMT2B appear to show the same apparent lack of phenotype identified following the deletion of yeast Trm2, with no defect observed in mt-RNA stability, mitochondrial translation, or cell growth following the loss of TRMT2B. Correspondingly, neither a reduced mitochondrial translation rate nor decreased 12S rRNA stability were identified in the TRMT2B deficient mouse cell line [44]. However, the aforementioned work did identify a small but significant decrease in the activity of complexes I, III, and IV of the electron transport chain. A reduced translation fidelity rather than translation rate is postulated as to why no concordant observation is made between [35S]-methionine incorporation and oxygen consumption. Further work is required to determine if the decreased activity of electron transport chain complexes is recapitulated in human cells.
As to why this defect may not manifest as a growth phenotype in both yeast and human cells is likely the result of either the effect being too subtle and so below the detection limits of the experiments performed, or the effect is context specific, and the environmental conditions required for the effect to become evident have not yet been applied. An effect such as this is observed for the human cyto-tRNA modifiers NSUN2 and METTL1, where phenotypes are only observed when both enzymes are depleted in conjunction [38]. A degree of functional redundancy may be a common feature of tRNA ‘core modifications’, with the absence of two or more, or their loss together with primary sequence changes, required to induce defects in translation. It is also likely that phenotypes are not being observed simply because the growth conditions are not challenged in the right manner, therefore external environmental changes such as the presence of ribosome inhibitors will also be required to differentiate between the wild type and knockout cell line.
The apparent absence of a catalytically active TRMT2B in both bovine and rat mitochondria also raises doubts surrounding the importance of m5U54 in mitochondrial tRNAs relative to their cytosolic counterparts. Mitochondrial tRNAs are well known for their sometimes dramatic divergences from well conserved features found in ‘canonical tRNAs’ present in bacteria or eukaryotic cytosol [47]. An altered reliance on otherwise well conserved tRNA modifications is likely to also be a product of these significant shifts in the RNA primary sequences. The high degree of sequence conservation evident in the remainder of bovine and rat TRMT2B is not indicative of a product of gene decay. Instead, it appears likely that bovine TRMT2B retains its RNA binding properties, perhaps now acting solely as a tRNA chaperone. If further work is able to uncover a phenotype resulting from the loss of TRMT2B, it will be important to determine the extent to which it can be rescued by a catalytic mutant, in order to unravel the relative contributions made by m5U54 and tRNA binding.
To date, 10 modified bases have been identified in the rRNA of mammalian mitochondria; with this work establishing TRMT2B as responsible for m5U429, the enzymes responsible are now known for all of the modified sites determined thus far. With regard to the other modified sites found alongside m5U429 in the 12S rRNA, the loss of the modification has been clearly demonstrated to have a detrimental effect on mitochondrial translation. In the recent identification of METTL15 as responsible for the catalysis of m4C839, the loss of the enzyme was shown to impair mitoribosome assembly with a concomitant reduction in mitochondrial protein synthesis [48]. Likewise, loss of either NSUN4 catalysing m5C841 [49] and TFB1M catalysing m26A936 and m26A937 [50], have also been shown to impact mitoribosome assembly. Similar effects have also been observed for modifications made to the 16S rRNA. Depletion of MRM2 and MRM3, responsible for the 2ʹ-O-ribose methylations Um1369 and Gm1370, respectively, results in a corresponding disruption to the assembly of the large mitoribosome subunit [51]. Likewise, diminished Ψ1397 resulting from RPUSD4 depletion has been shown to reduce the steady state levels of 16S rRNA, induce aberrant assembly of the mitoribosome large subunit, and severely reduce the rate of mitochondrial translation [52]. Future work, perhaps focusing on specific cell types, developmental stages, or environmental conditions, will be required to uncover the contribution of TRMT2B to mitochondrial translation.
Materials and methods
Cell maintenance
All cell lines were maintained in humidified incubators at 5% CO2 and 37°C, unless otherwise stated. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM), containing 4.5 g/L glucose, 110 mg/L sodium pyruvate, supplemented with 10% foetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin. Wild-type and TRMT2B Knockout (Product ID: HZGHC004364c004) HAP1 cell lines were purchased from Horizon Discovery.
Sirna-mediated protein depletion
Stealth siRNAs (HSS129214, HSS129216, and HSS188304) were obtained from Thermo Fisher Scientific and transfected into HeLa cells using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific).
Subcellular localization by confocal microscopy
The full-length TRMT2B cDNA construct (Source Bioscience, CatNo: IRATp970B1220D) was cloned into a pmaxGFP vector. HeLa cells were grown on coverslips and transiently transfected with the TRMT2B-GFP construct using Lipofectamine 2000 (ThermoFisher Scientific) and after 24 hours cells were visualized by confocal microscopy, with cells treated with DAPI for nuclear staining and mitochondria immuno-stained for TOM20 as described previously [53].
Detection of 5-methyluridine by primer extension
RNA was extracted from cells at 60–80% confluency using TRIzol reagent (ThermoFisher Scientific), following the manufacturer’s instructions. The Aniline-Hydrazine cleavage of total RNA was performed in a two-step procedure: the incubation of RNA with hydrazine which modifies uridines and its derivatives, followed by the cleavage of hydrazine-modified RNA at uridines by aniline. The resulting cleavage is then detected via primer extension (Figure S2). To begin, 5 μg of pelleted RNA was resuspended in 20 μL of chilled 50% hydrazine hydrate and incubated on ice for 5 minutes. The reaction was then stopped with the addition of a 1/10th volume of 3 M NaOAc at pH 5.2 and 3 volumes of 100% ethanol. The sample was then precipitated at −80°C for >12 hours. Following this, the precipitated hydrazine-treated sample was centrifuged and the pellet resuspended in 1 M aniline-acetate pH 4.5. The sample was incubated at 60°C for 20 minutes, and the RNA then precipitated again and resuspended in 30 μL of water.
The reverse transcription primer extension reactions were performed as in (51), with the following primers used:
mt_Pro_m5U54: TGGTCAGAGAAAAAGTC
mt_12S_m5U429: GGGCTAAGCATAGTGGGGTATC
cyto-Asp_m5U54: TGGCTCCCCGTCGGGGAATC
cyto-Glu_m5U54: TGGTTCCCTGACCGGGAAT
Northern blotting
RNA was extracted from cells at 60–80% confluency using TRIzol reagent (ThermoFisher Scientific) and subjected to northern blotting as described previously [54,55]. Briefly, total RNA was resolved on 1% agarose gels containing 0.7 M formaldehyde in 1× MOPS buffer (mt-rRNAs) or on 10% UREA–PAGE in 1× GTB buffer (mt-tRNAs), or on 6.5% UREA–PAGE in 100mM NaOAc pH 5.0 (aminoacyl-mt-tRNAs), transferred to a nylon membrane, ultraviolet-crosslinked to the membrane and hybridized with radioactively labelled T7-transcribed radioactive RNA-probes.
Western blotting
20–30 µg of total extracted protein was diluted to an equal volume and a ⅓ volume of NuPAGE LDS 4 x sample buffer and loaded on SDS-PAGE 4-12% bis-tris gels (Life Technologies) and transferred onto a membrane using iBlot 2 Dry Blotting System (Thermo Fisher Scientific). The following antibodies were used: rabbit anti-MRPS17 (Proteintech 18,881-1-AP, 1:1000), rabbit anti-MRPS18b (Proteintech 16,139-1-AP, 1:1000), rabbit anti-MRPS35 (Proteintech 16,457-1-AP, 1:1000), Total OXPHOS Human WB antibody cocktail (Abcam, ab110411, 1:1000), goat anti-rabbit IgG HRP (Promega W4011, 1:2000), goat, anti-mouse IgG HRP (Promega W4021).
[35S] -methionine metabolic labelling of mitochondrial proteins
The labelling of newly synthesized mtDNA-encoded proteins was performed as in Rorbach et al [51]. Briefly, exponentially growing cells were incubated in methionine/cysteine-free medium for 10 min before the medium was replaced with methionine/cysteine-free medium containing 10% dialysed FCS and emetine dihydrochloride (100 µg/mL) to inhibit cytosolic translation. Following a 20 min incubation, 120 μCi/mL of [35S]-methionine (Perkin Elmer) was added and the cells were incubated for 30 min. Cells were washed with PBS, lysed, and 30µg of protein was loaded on 10-20% Tris-Glycine SDS-PAGE gels. Dried gels were visualized and quantified with a PhosphorImager system with ImageQuant software.
Funding Statement
This work was supported by the Medical Research Council [MC_UU_00015/4].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].Rebelo-Guiomar P, Powell CA, Van Haute L, et al. The mammalian mitochondrial epitranscriptome. Biochim Biophys Acta - Gene Regul Mech. 2019;1862(3):429–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Van Haute L, Dietmann S, Kremer L, et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Garone C, D’Souza AR, Dallabona C, et al. Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum Mol Genet. 2017;26(21):4257–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Powell CA, Nicholls TJ, Minczuk M.. Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease. Front Genet. 2015;6:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boczonadi V, Ricci G, Horvath R. Mitochondrial DNA transcription and translation: clinical syndromes. Essays Biochem. 2018;62(3):321–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bessho Y, Shibata R, Sekine SI, et al. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci. 2007;104(20):8293–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kazantsev AV, Krivenko AA, Harrington DJ, et al. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci. 2005;102(38):13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Toor N, Keating KS, Taylor SD, et al. Crystal Structure of a Self-Spliced Group II Intron. Science. 2008;320(5872):77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458(7235):233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chan CW, Chetnani B, Mondragón A. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip Rev RNA. 2013;4(5):507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dubin DT. Methylated nucleotide content of mitochondrial ribosomal RNA from hamster cells. J Mol Biol. 1974;84(2):257–273. [DOI] [PubMed] [Google Scholar]
- [12].Baer RJ, Dubin DT. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981;9(2):323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Madsen CT, Mengel-Jørgensen J, Kirpekar F, et al. Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003;31(16):4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ranaei-Siadat E, Fabret C, Seijo B, et al. RNA-methyltransferase TrmA is a dual-specific enzyme responsible for C5-methylation of uridine in both tmRNA and tRNA. RNA Biol. 2013;10(4):572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gu X, Ofengand J, Santi DV. In vitro methylation of Escherichia coli 16S rRNA by tRNA (m5U54)-methyltransferase. Biochemistry. 1994;33(8):2255–2261. [DOI] [PubMed] [Google Scholar]
- [16].Alian A, Lee TT, Griner SL, et al. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci U S A. 2008;105(19):6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bjork GR, Neidhardt FC. Physiological and Biochemical Studies on the Function of 5-Methyluridine in the Transfer Ribonucleic Acid of Escherichia coli. J Bacteriol. 1975;124(1):99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Persson BC, Gustafsson C, Bergt DE, et al. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Biochemistry. 1992;89:3995–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hopper AK, Furukawa AH, Pham HD, et al. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell. 1982;28(3):543–550. [DOI] [PubMed] [Google Scholar]
- [20].Nordlund ME, Johansson JO, von Pawel-rammingen U, et al. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA. 2000;6(6):844–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Johansson MJO, Byström AS. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA. 2002;8(3):324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Choudhury SA, Asefa B, Webb A, et al. Functional and genetic analysis of the Saccharomyces cerevisiae RNC1/TRM2: evidences for its involvement in DNA double-strand break repair. Mol Cell Biochem. 2007;300(1–2):215–226. [DOI] [PubMed] [Google Scholar]
- [23].Carter J-M, Emmett W, Mozos IR, et al. FICC-Seq: a method for enzyme-specified profiling of methyl-5-uridine in cellular RNA. Nucleic Acids Res. 2019;47(19):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].De Crécy-Lagard V, Boccaletto P, Mangleburg CG, et al. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res. 2019;47(5):2143–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Machnicka MA, Milanowska K, Osman Oglou O, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41(D1):D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42(11):7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241(3):779–786. [DOI] [PubMed] [Google Scholar]
- [28].Emanuelsson O, Nielsen H, Brunak S, et al. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J Mol Biol. 2000;300(4):1005–1016. [DOI] [PubMed] [Google Scholar]
- [29].Small I, Peeters N, Legeai F, et al. A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4(6):1581–1590. [DOI] [PubMed] [Google Scholar]
- [30].Brulé H, Holmes WM, Keith G, et al. Effect of a mutation in the anticodon of human mitochondrial tRNA Pro on its post-transcriptional modification pattern. Nucleic Acids Res. 1998;26(2):537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Desai PM, Culver GM, Rife JP. Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly. Biochemistry. 2011;50(5):854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Degoul F, Brulé H, Cepanec C, et al. Isoleucylation properties of native human mitochondrial tRNAIle and tRNAIle transcripts. Implications for cardiomyopathy-related point mutations (4269, 4317) in the tRNAIle gene. Hum Mol Genet. 1998;7(3):347–354. [DOI] [PubMed] [Google Scholar]
- [33].Helm M, Giegé R, Florentz C. A Watson−Crick Base-Pair-Disrupting Methyl Group (m1A9) Is Sufficient for Cloverleaf Folding of Human Mitochondrial tRNA Lys. Biochemistry. 1999;38(40):13338–13346. [DOI] [PubMed] [Google Scholar]
- [34].Toompuu M, Yasukawa T, Suzuki T, et al. The 7472insC mitochondrial DNA mutation impairs the synthesis and extent of aminoacylation of tRNASer(UCN) but not its structure or rate of turnover. J Biol Chem. 2002;277(25):22240–22250. [DOI] [PubMed] [Google Scholar]
- [35].Pütz J, Dupuis B, Sissler M, et al. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA. 2007;13(8):1184–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hegg LA, Thurlow DL. Residual tRNA secondary structure in “denaturing” 8M urea/TBE polyacrylamide gels: effects on electrophoretic mobility and dependency on prior chemical modification of the tRNA. Nucleic Acids Res. 1990;18(10):2993–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Davanloo P, Sprinzl M, Watanabet K, et al. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979;6(4April). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Okamoto M, Fujiwara M, Hori M, et al. tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells. 2014;10(9):e1004639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Knight E. Mitochondria-associated ribonucleic acid of the HeLa cell. Effect of ethidium bromide on the synthesis of ribosomal and 4S ribonucleic acid. Biochemistry. 1969;8(12):5089–5093. [DOI] [PubMed] [Google Scholar]
- [40].Yasukawa T, Hino N, Suzuki T, et al. A pathogenic point mutation reduces stability of mitochondrial mutant tRNA(Ile). Nucleic Acids Res. 2000;28(19):3779–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Metodiev MD, Lesko N, Park CB, et al. Methylation of 12S rRNA Is Necessary for In Vivo Stability of the Small Subunit of the Mammalian Mitochondrial Ribosome. Cell Metab. 2009;9(4):386–397. [DOI] [PubMed] [Google Scholar]
- [42].Bar-Yaacov D, Frumkin I, Yashiro Y, et al. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 2016;14(9):e1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Auxilien S, Rasmussen A, Rose S, et al. Specificity shifts in the rRNA and tRNA nucleotide targets of archaeal and bacterial m5U methyltransferases. RNA. 2011;17(1):45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Laptev I, Shvetsova E, Levitskii S, et al. Mouse Trmt2B protein is a dual specific mitochondrial metyltransferase responsible for m 5 U formation in both tRNA and rRNA. RNA Biol. 2019. November;1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pang H, Ihara M, Kuchino Y, et al. Structure of a modified nucleoside in archaebacterial tRNA which replaces ribosylthymine. 1-Methylpseudouridine. J Biol Chem. 1982;257(7):3589–3592. [PubMed] [Google Scholar]
- [46].Zamir A, Holley RW, Marquisee M. Evidence for the occurence of a common pentanucleotide sequence in the structures of transfer ribonucleic acids. J Biol Chem. 1965;240:1267–1273. [PubMed] [Google Scholar]
- [47].Salinas-Giegé T, Giegé R, Giegé P. tRNA Biology in Mitochondria. Int J Mol Sci. 2015;16(3):4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Van HL, Hendrick AG, D’Souza AR, et al. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019;47(19):10267–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Metodiev MD, Spåhr H, Loguercio Polosa P, et al., ed. NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly. Barsh GS. PLoS Genet. 2014;10(2):e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33(1):23–24. [DOI] [PubMed] [Google Scholar]
- [51].Rorbach J, Boesch P, Gammage PA, et al. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol Biol Cell. 2014;25(17):2542–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zaganelli S, Rebelo-Guiomar P, Maundrell K, et al. The pseudouridine synthase RPUSD4 is an essential component of mitochondrial RNA granules. J Biol Chem. 2017;292(11):4519–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Minczuk M, Kolasinska-Zwierz P, Murphy MP, et al. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat Protoc. 2010;5(2):342–356. [DOI] [PubMed] [Google Scholar]
- [54].Rorbach J, Gao F, Powell CA, et al. Human mitochondrial ribosomes can switch their structural RNA composition. Proc Natl Acad Sci. 2016;113(43):12198–12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pearce SF, Rorbach J, Van Haute L, et al. Maturation of selected human mitochondrial tRNAs requires deadenylation. Elife. 2017; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


