Abstract
The severe acute respiratory coronavirus 2 (SARS-CoV-2), which emerged in December 2019 in Wuhan, China, has spread rapidly to over a dozen countries. Especially, the spike of case numbers in South Korea sparks pandemic worries. This virus is reported to spread mainly through person-to-person contact via respiratory droplets generated by coughing and sneezing, or possibly through surface contaminated by people coughing or sneezing on them. More critically, there have been reports about the possibility of this virus to transmit even before a virus-carrying person to show symptoms. Therefore, a low-cost, easy-access protocol for early detection of this virus is desperately needed. Here, we have established a real-time reverse-transcription PCR (rtPCR)-based assay protocol composed of easy specimen self-collection from a subject via pharyngeal swab, Trizol-based RNA purification, and SYBR Green-based rtPCR. This protocol shows an accuracy and sensitivity limit of 1-10 virus particles as we tested with a known lentivirus. The cost for each sample is estimated to be less than 15 US dollars. Overall time it takes for an entire protocol is estimated to be less than 4 hours. We propose a cost-effective, quick-and-easy method for early detection of SARS-CoV-2 at any conventional Biosafety Level II laboratories that are equipped with a rtPCR machine. Our newly developed protocol should be helpful for a first-hand screening of the asymptomatic virus-carriers for further prevention of transmission and early intervention and treatment for the rapidly propagating virus.
Keywords: Coronavirus, SARS virus, Infectious disease, Diagnostic techniques and procedures, Communicable diseases, Emerging, COVID-19
INTRODUCTION
The severe acute respiratory coronavirus 2 (SARS-CoV-2), which emerged since December 2019 in Wuhan, China, has spread rapidly to over a dozen countries. According to a very recent report, common symptoms include fever (98.6%), fatigue (69.6%), and dry cough (59.4%), lymphopenia (70.3%), prolonged prothrombin time (58%), and elevated lactate dehydrogenase (39.9%) [1]. This virus is reported to spread mainly through person-to-person contact via respiratory droplets generated by coughing and sneezing, or possibly through surface contaminated by people coughing or sneezing on them [2].
More critically, alarming cases of asymptomatic infections have been reported. For example, according to Science Alert, a 20-year-old woman diagnosed with SARS-CoV-2 in one case analysis showed no symptoms during the month when her entire family was classified as confirmed [3]. Of the 126 Germans sent to Germany from Wuhan, China, on February 1, 2020, 114 patients were asymptomatic but found to be positive for SARS-CoV-2, suggesting that a symptom-based screening process may not be sufficiently effective [4]. Asymptomatic transmission has also been reported where an asymptomatic family has spread the virus to other families [5]. These very recent reports clearly indicate that there are virus-carriers who do not show any typical symptoms of SARS-CoV-2, such as fever, cough and sore throat. Furthermore, these reports raise an alarming possibility of this virus to transmit even before a virus-carrying person to show any symptoms.
The recent surge of case numbers in the Republic of Korea has also raised a serious pandemic threat. Over 5,621 virus-carriers have been confirmed along with 34 reported deaths by March 4th, 2020, after a total of 28,414 people were tested for SARS-CoV-2. The current eligibility for Korean government-supported diagnostic test include: 1) People with respiratory symptoms such as fever, cough and sore throat within 14 days of visiting China; 2) Patients with fever or respiratory symptoms within 14 days of intimate contact with the SARS-CoV-2 patient; 3) According to the opinion of doctors for those suspected of SARS-CoV-2 infection [6]. Any person who does not meet the above criteria is denied of the nationally-supported test and is left with a personal option to obtain a commercial test at a personal cost. Due to the surge of the number of virus-carrying patients, there is an immediate demand for diagnostic test among asymptomatic population. However, there is a serious shortage of inspection personnel and test facilities due to the rapidly increasing number of suspected patients [7, 8]. To make the matter worse, once a positive virus-carrying patient is identified, the whole building, institute, business, or school in the vicinity of the identified patient is shut-down, incapacitating the health-care personnel and facilities. Therefore, there is an immediate need for a first-hand screening test procedure, which should be readily available for asymptomatic people, to determine whether the tested person is negative or potentially positive for the presence of SARS-CoV-2.
The currently available detection tools and kits for SARS-CoV-2, which is a RNA carrying virus, are based on a simple concept of indirect detection of RNA by rtPCR via RNA to complementary DNA conversion (cDNA) [9]. In brief, a potential carrier person is subjected to a collection of pharyngeal swab (throat swab) and/or sputum. The collected sample is then processed through a series of procedures to purify viral RNA, convert RNA to cDNA by a reverse transcriptase, and measurement of the copy number by rtPCR using specific primer sets for the genomic sequence of SARS-CoV-2 [9]. The important safety related step in this procedure is the purification of viral RNA step, during which any potential infectious virus is inactivated by addition of phenol-containing extraction agents such as Trizol [10]. The guidelines and specific primer sets for SARS-CoV-2 are now available through Center for Disease Control (CDC), USA [11]. The entire sequence of the genomic RNA of SARS-CoV-2 derived from the first Korean patient has been determined and reported [12]. In addition, the total RNA of SARS-CoV-2 is available from the Korean Center for Disease Control (KCDC), which can be safely used as a positive control. Therefore, using these available resources, it is possible for any researcher in the field of biology can test the presence of SARS-CoV-2 safely in a conventional laboratory, certified for Biosafety Level II [13], minimally equipped with a rtPCR machine.
In this study, we have gathered all the available resources to develop a fast, highly accurate, low-cost, laboratory-ready and laboratory-safe protocol to test asymptomatic subjects as a first-hand screening procedure for detection of SARS-CoV-2. Our protocol is based on a rtPCR and composed of easy specimen collection from a subject via pharyngeal swab, and Trizol-based RNA purification. Our primary goal of developing a new protocol is to focus on determination of the negative (absence of SARS-CoV-2), rather than the positive (presence of SARS-CoV-2), so that a negative person can be free of worries and allowed to work or study or sport freely, while a positive person is recommended for a further diagnosis with certified diagnostic kits.
MATERIALS AND METHODS
Call for volunteers
Human volunteers to collect pharyngeal swab samples were called to participate. A total of 12 volunteers participated in this study. Each volunteer was given a thorough explanation of the purpose of the study and procedure of sample collection through pharyngeal swab, which was approved by the Institutional Review Board of Seoul National University Hospital (IRBY-H-1807-197-966). A written informed consent was obtained from all 12 volunteers.
Pharyngeal swab procedure
The detailed pharyngeal swab procedure was established and adopted based on the previous report describing the equal performance of self-collected and health care worker-collected pharyngeal swabs for Group A Streptococcus testing by PCR [14]. To prevent any possible contact within volunteers and between a volunteer and a health care worker, we adopted the self-collection procedure. The self-collection was performed under a well-ventilated fume hood with a constant air inflow at 0.4 m/s. Each volunteer was asked to strictly follow the instructions as shown in Fig. 1.
Fig. 1.
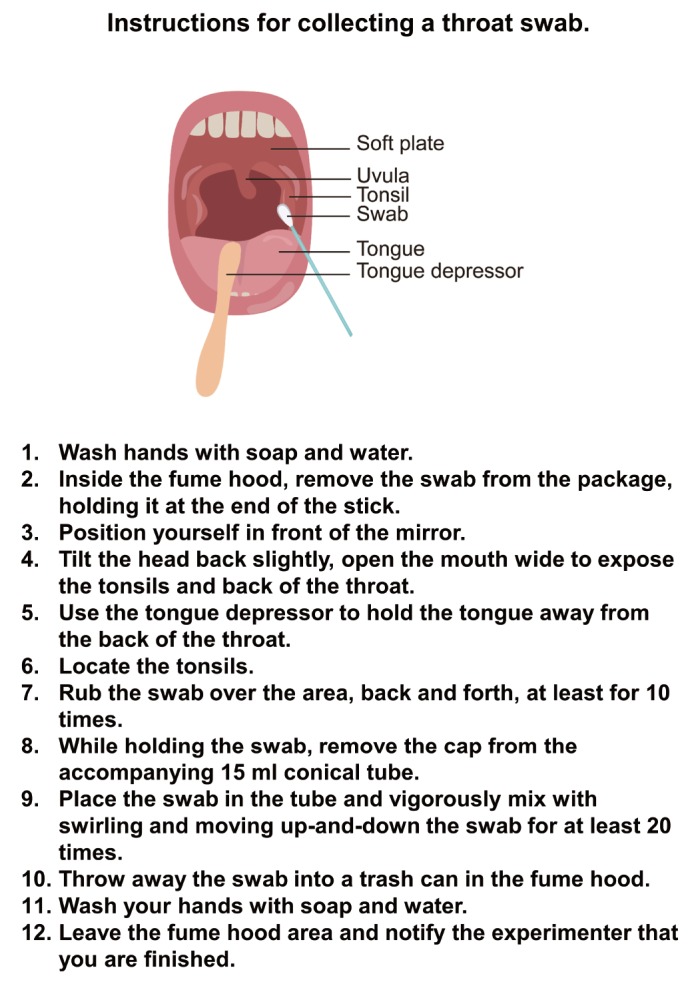
Instructions for collecting a pharyngeal swab.
We selected and utilized a polyester swab with a plastic shaft (6.4×3.4×16.8 mm tip, 163.3 mm length; Catalog #: 6-6587-31; LMS, Japan) over a wood shaft because some wood materials are known to contain chemicals that are toxic to bacteria and viruses. After collection, the swab was immediately placed in a 15 ml conical tube. The collected tissue on swab was then immediately transferred and dissolved into 500 µl of Dulbecco’s Modification of Eagle’s Medium (DMEM, Catalog #: 10-013-CV, Corning, USA) by vigorously mixing with swirling and moving up-and-down the swab for at least 20 times. The swab was then thrown into a biohazard trash can and the cap of the conical tube was closed and placed in a tube rack. After the volunteer was finished with sample collection procedure and had left the collection site, an experimenter opened the cap of the 15 ml conical tube and transferred 300 µl of the dissolved tissue sample in DMEM into a 1.5 ml centrifuge tube containing 700 µl of Trizol (easy-BLUETM Total RNA Extraction Kit; Catalog #17061; iNtRON, Republic of Korea), mixed by inverting tube 5 times. At this point, the 1.5 ml centrifuge tube containing Trizol was considered free of virus and thus non-infectious and safe. This 1.5 ml tube was then moved from the sample collection site to a bench in a certified Biosafety Level II Laboratory.
Total RNA extraction
Total RNA from the collected tissue sample was extracted by Trizol. The tissue sample was incubated in Trizol for 5 minutes in room temperature, followed by an addition of 200 µl chloroform and mixing by inverting the tube 5 times. Then the 1.5 ml tube was incubated for 3 minutes and centrifuged for 15 minutes at 12,000×g at 4°C. The clear upper aqueous layer which contains RNA (approximately 500~600 µl) was transferred to a new 1.5 ml tube and the same amount of isopropanol was added. A gentle mix by inverting 5 times was followed before incubating for 10 minutes on ice. The sample was centrifuged for 10 minutes at 12,000×g at 4°C. The supernatant was discarded and the remaining pellet (when visible) or the possible location of pellet (when not visible) was re-suspended in 500 µl of 75% ethanol. The sample was again centrifuged for 5 minutes at 7,500×g at 4°C. The supernatant was again discarded and the RNA pellet was air dried for 5~10 minutes. To solubilize the RNA pellet, the pellet was re-suspended in 10 µl of RNase-free water and incubated at 37°C for 10~15 minutes. The RNA concentration and purity were determined with NanoDrop (Thermo Fisher Scientific, USA) by calculating the ratio of optical density at wavelengths of 260/280 and 260/230. It took about 1 hour for the entire procedure of total RNA extraction.
Conversion of total RNA to cDNA
For rtPCR to detect SARS-CoV-2 virus infection, total RNA from a volunteer sample was converted to cDNA using SuperScriptTM III First-Strand Synthesis System (Catalog #: 18080051; InvitrogenTM, USA), following the manufacturer’s recommended procedures. For cDNA synthesis, a mixture of 8 µl of extracted RNA, 1 µl of 50 µM of oligo(dT)20, and 1 µl of 10 mM dNTP was prepared before adding the 10 µl of cDNA Synthesis Mix, which contains a reverse transcriptase. It took about 1 hour and 30 minutes for the entire procedure.
Use of the SARS-CoV-2 total RNA as a positive control
The positive control containing SARS-CoV-2 viral RNAs was prepared by extracting total RNA from Vero cells infected with a viral clone, BetaCoV/Korea/KCDC03/2020 at MOI 0.05. Detailed protocol will be published elsewhere. This control total RNA was used to convert to cDNA and the converted cDNA was used in rtPCR run as a positive control for SARS-CoV-2.
Primer sets specific for SARS-CoV-2
Primers were designed based on the recently available full sequence of SARS-CoV-2 virus from the first patient in Korea [12]. We utilized NCBI-Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to design specific primers (Table 1). Some primer sets were taken directly from the primer sets reported by CDC, USA [11] (Table 1). The forward and reverse primer sets were synthesized and delivered by two separate companies, Macrogen (Republic of Korea) and Cosmogenetech (Republic of Korea).
Table 1.
List and information of primer sets for SARS-CoV-2 detection
| Origin | Species target | Target gene | Primer name | Forward sequence (5'-3') | Reverse sequence (5'-3') | Final conc. | Ref. |
|---|---|---|---|---|---|---|---|
| IBS | SARS-CoV-2 | RdRP | SARS-CoV-2_IBS_RdRP1 | CATGTGTGGCGGTTCACTAT | TGCATTAACATTGGCCGTGA | 0.5 μM | Current study |
| 15441-15558 | |||||||
| S | SARS-CoV-2_IBS_S1 | CTACATGCACCAGCAACTGT | CACCTGTGCCTGTTAAACCA | ||||
| 23114-23213 | |||||||
| E | SARS-CoV-2_IBS_E1 | TTCGGAAGAGACAGGTACGTT | CACACAATCGATGCGCAGTA | ||||
| 26259-26365 | |||||||
| N | SARS-CoV-2_IBS_N1 | CAATGCTGCAATCGTGCTAC | GTTGCGACTACGTGATGAGG | ||||
| 28732-28849 | |||||||
| RdRP | SARS-CoV-2_IBS_RdRP2 | AGAATAGAGCTCGCACCGTA | CTCCTCTAGTGGCGGCTATT | ||||
| 15092-15193 | |||||||
| RdRP | SARS-CoV-2_IBS_RdRP3 | TCTGTGATGCCATGCGAAAT | ACTACCTGGCGTGGTTTGTA | ||||
| 14015-14127 | |||||||
| S | SARS-CoV-2_IBS_S2 | GCTGGTGCTGCAGCTTATTA | AGGGTCAAGTGCACAGTCTA | ||||
| 22340-22447 | |||||||
| E | SARS-CoV-2_IBS_E2 | TTCGGAAGAGACAGGTACGTTA | AGCAGTACGCACACAATCG | ||||
| 26259/26374 | |||||||
| N | SARS-CoV-2_IBS_N2 | GCTGCAATCGTGCTACAACT | TGAACTGTTGCGACTACGTG | ||||
| 28736-28855 | |||||||
| Human IPC | RPP30 | IBS_RPP30 | CTATTAATGTGGCGATTGACCGA | TGAGGGCACTGGAAATTGTAT | 0.5 μM | Current study | |
| RPP40 | IBS_RPP40 | CTTGGCATAAAACAGGTTCAGAA | GAGATCTCTCAACGTGCTCAG | ||||
| US CDC | SARS-CoV-2 | N | CDC_N1 | GACCCCAAAATCAGCGAAAT | TCTGGTTACTGCCAGTTGAATCTG | 0.05~20 μM | [11] |
| 28287-28358 | |||||||
| N | CDC_N2 | TTACAAACATTGGCCGCAAA | GCGCGACATTCCGAAGAA | ||||
| 29164-29230 | |||||||
| N | CDC_N3 | GGGAGCCTTGAATACACCAAAA | TGTAGCACGATTGCAGCATTG | ||||
| 28681-28752 | |||||||
| Human IPC | RNase P | CDC_RNAse P | AGATTTGGACCTGCGAGCG | GAGCGGCTGTCTCCACAAGT | 0.05~20 μM | [11] |
IPC, Internal positive control.
Quantitative rtPCR
Quantitative rtPCR was carried out using SYBR Green PCR Master Mix (Catalog #: 4367659; Applied BiosystemsTM, USA). In brief, each reaction buffer consisted of a total volume of 20 µl containing 8 µl of 100 µM forward and reverse primers (4 µl for each primer), 2 µl of cDNA, and 10 µl power SYBR Green PCR Master Mix. rtPCR was performed with Quantstudio 1 Real-Time PCR system (Applied Biosystems, USA). The automatic setting of threshold for calculating the threshold cycle (Ct) value was determined by Quantstudio Design & Analysis software. It took about 1 hour and 10 minutes for the entire rtPCR process.
Determination of detection efficiency and limit using Lentivirus
The efficiency of the virus collection was independently assessed by using a well-known, research-purpose, RNA-containing, recombinant virus, Lentivirus. The Lentivirus used in this study was pSicoR-scrambled shRNA with a mCherry reporter [15]. This Lentivirus was packaged by the IBS Virus Facility (http://www.ibs.re.kr/virusfacility) with a pre-determined titer of 5.91×107 genomic copies/ml. The calibration curve for the genomic copy number versus Ct value was obtained from the rtPCR machine, Quantstudio 1 Real-Time PCR system (Applied Biosystems, USA) using the Lentiviral DNA vector included in qPCR Lentivirus Titration Kit (Catalog #: LV900, ABM, USA).
To compare the efficiency of Trizol-based RNA extraction, we compared it with QIAamp Viral RNA Mini Kit (Lot #: 52904, Qiagen, USA). The efficiency was tested on Lentivirus-infected HEK293T cells, which were infected overnight (12 hours).
RESULTS
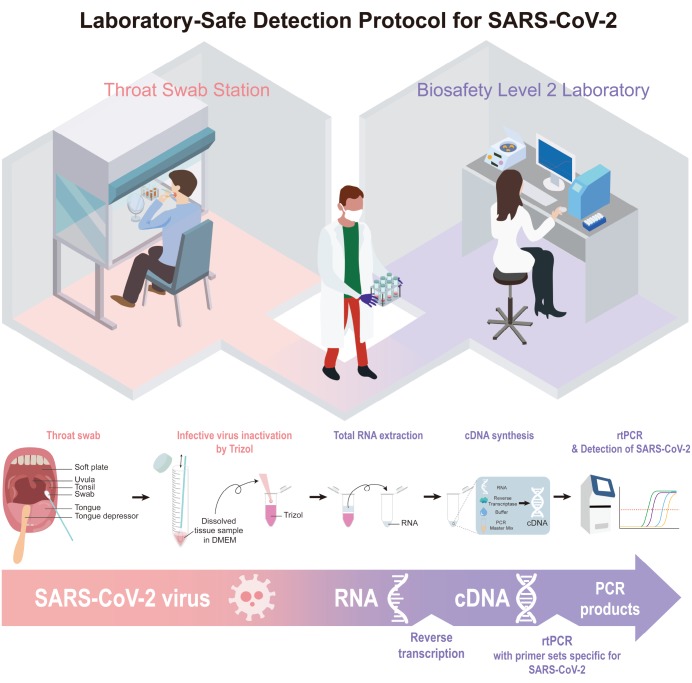
The overall design of the protocol is depicted in the schematic diagram in Fig. 2. In short, each volunteer visits a throat swab station where a tissue sample self-collection is carried out. There is a spatial separation between the throat swab station and the Biosafety Level II laboratory space where the total RNA extraction, RNA to cDNA conversion and rtPCR are carried out.
Fig. 2.
Schematic diagram of the low-cost, laboratory-safe protocol for SARS-CoV2.
Symptomatic history of the volunteers in relation to SARS-CoV-2
Total of twelve asymptomatic volunteers were gathered to self-collect throat swab samples. All volunteers showed normal range of body temperature below 37°C and no sign of fever (Table 6). Ten volunteers showed no obvious symptoms of COVID-19, except for Volunteer 1, who showed symptoms of dry coughing and headache, and fever (which was controlled by asprin), and Volunteer 12, who was recently diagnosed as acute nasopharyngitis but confirmed negative for COVID-19. Even though Volunteer 1 showed somewhat similar symptoms as COVID-19, Volunteer 1 was diagnosed as simple influenza because the fever subsided immediately after taking asprin. Volunteer 1 was denied of government-aided diagnostic test due to her absence of fever. Volunteer 12 served as a perfect negative control because of a recent diagnostic test confirmed negative for SARS-CoV-2. Both Volunteer 1 and 12 were self-isolated for one week before they were released. Volunteer 7 was also self-isolated for two weeks due to her recent visit to Daegu area, where the serious outbreak of COVID-19 occurred. All volunteers had no history of contacting a confirmed SARS-CoV-2 carrier. Two couples, Volunteer 2 and 11, Volunteer 4 and 12 were spouses.
Table 6.
Volunteer information related to COVID-19, RNA quality and SARS-CoV-2 test results
| Volunteer Number | Body Temp. (°C) | Symptoms and history of contact with a COVID19 patient | RNA Conc. (ng/ μl) | RNA Purity A260/A280, A260/A230 | Test Result |
|---|---|---|---|---|---|
| 1 | 36.6 | Previous fever controlled by aspirin, dry coughing and headache, Self-isolated for 1 week, no previous contact | 171.3 | 1.77, 1.95 | Negative |
| 2 | 36.7 | Asymptomatic, no previous contact Spouse of 11 | 204.5 | 1.80, 2.07 | Negative |
| 3 | 36.7 | Asymptomatic, no previous contact | 309.9 | 1.96, 2.21 | Negative |
| 4 | 36.4 | Asymptomatic, no previous contact Spouse of 12 | 580.6 | 1.69, 1.98 | Negative |
| 5 | 36.8 | Asymptomatic, no previous contact | 445.2 | 2.16, 2.30 | Negative (after re-test, Inconclusive at 1sttest) |
| 6 | 36.4 | Asymptomatic, no previous contact | 366.3 | 2.04, 2.24 | Negative |
| 7 | 36.6 | Asymptomatic, Visited Daegu area, self-isolated for 2 weeks, no previous contact | 210.5 | 1.67, 1.87 | Negative |
| 8 | 36.7 | Asymptomatic, no previous contact | 185.7 | 1.72, 1.94 | Negative |
| 9 | 36.6 | Asymptomatic, no previous contact | 349.2 | 1.83, 2.13 | Inconclusive (after re-test) |
| 10 | 36.7 | Asymptomatic, no previous contact | 197.4 | 1.67, 1.98 | Negative |
| 11 | 36.8 | Asymptomatic, no previous contact Spouse of 2 | 219.8 | 1.82, 2.11 | Negative |
| 12 | 36.4 | Diagnosed as acute nasopharyngitis, Negative for COVID19, Self-isolated for 1 week, no previous contact Spouse of 4 | 200.8 | 1.84, 2.10 | Negative (after re-test, Inconclusive at 1sttest) |
Self-collection of tissue sample via pharyngeal swab
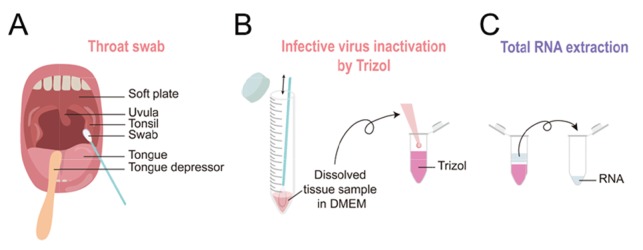
Each volunteer was asked to collect the tissue sample from the tonsil via pharyngeal swab or throat swab (Fig. 3A). To minimize the contact between the volunteer and the experimenter, each volunteer was asked to perform the procedure alone at an isolated and well-ventilated “Throat Swab Station.” Immediately after self-collection, each volunteer was asked to dissolve and tap-off tissue sample in DMEM (Fig. 3B). After placing the tube with tissue sample on a tube rack, each volunteer was asked to call the experimenter to transfer the tissue-sample-containing DMEM into Trizol, which will inactivate any infectious virus and other living organisms (Fig. 3B). At this point the tissue sample was considered to be non-infectious. Because Trizol is known to be highly toxic to human [16], only the experimenter was allowed to handle the Trizol. The experimenter would safely move the Trizol-inactivated tissue sample to the laboratory area (Biosafety Level II), where the tissue sample is further processed for total RNA extraction (Fig. 3C). This procedure took less than 10 minutes.
Fig. 3.
Experimental scheme for human tissue sampling and total RNA extraction. (A) Each volunteer collects his/her tissue sample through pharyngeal swab, following the detailed procedures in Fig. 1. (B) The collected human sample on polyester swab was dissolved in DMEM and transferred to a Trizol containing tube. (C) Following the Trizol-based total RNA extraction, the RNA was further processed.
Total RNA extraction and conversion to cDNA
We expected that if there are SARS-CoV-2 viruses in a tissue sample, the extracted RNA would contain both viral RNA and human RNA. During RNA extraction, we obtained on average 287 ng/µl from each volunteer sample, with relatively low purity, most likely due to a phenol contamination (Table 6). However, the phenol appeared not to influence the rtPCR results. Then the extracted RNA was further converted to cDNA using a kit, SuperScriptTM III First-Strand Synthesis System (Catalog #: 18080051; InvitrogenTM, USA). On average, this procedure took about 60 minutes.
Determination of detection efficiency and limit of rtPCR system using Lentivirus
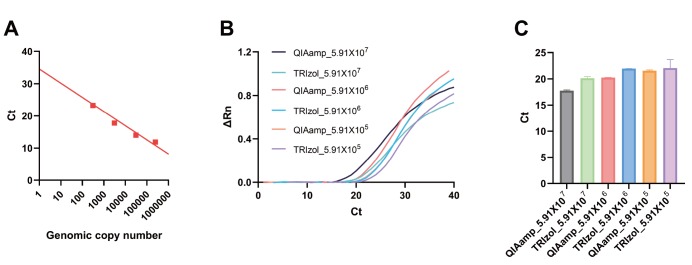
To calibrate our rtPCR system so that we can predict the genomic copy number based on Ct value, we performed SYBR Green-based rtPCR with a known number of genomic copy of Lentiviral Long terminal repeats (LTR) to generate a standard curve (Fig. 4A). The fitted-line showed that Ct value of around 35 was estimated to be around 1 copy of the gene and this Ct value is the detection limit of our rtPCR system. Therefore, we used a stringent limiting Ct value of around 37 to be the detection limit of our rtPCR system. Based on the previous report on molecular diagnosis of SARS-CoV-2 [17], which suggested that 1 copy of the genes (ORF1b or N) is estimated as Ct value of near 37, we estimate that the Ct value of 37 would detect about 1~10 particles of virus, which defines the limit of our detection system. We then tested the effectiveness of Trizol-based RNA extraction using the Lentivirus. We compared it with a well-known kit from Qiagene, QIAamp Viral RNA Mini Kit, which is much more expensive than Trizol-based extraction method. We found that between QIAmpt and Trizol, there was virtually no significant difference in the ability to extract RNA from the cultured HEK293 cells infected with Lentivirus (Fig. 4B and 4C). These results indicate that Trizol was equally sensitive and effective in extracting RNA, compared to the commercially available kit. Therefore, based on these results we decided to continue to use Trizol-based RNA extraction method.
Fig. 4.
Determination of detection efficiency and limit of rtPCR system using Lentivirus. (A) A standard calibration curve of Ct vs. genomic copy number was generated based on rtPCR results from LTR targeting primer set and known genomic copy number of LTR. (B) rtPCR results for RNA extraction by QIAmp kit or Trizol-based method. The known number of Lentivirus was tested with the two extraction methods and compared. (C) The average Ct values are compared between QIAmp and Trizol methods.
Selection of the primer sets and validation of each primer set
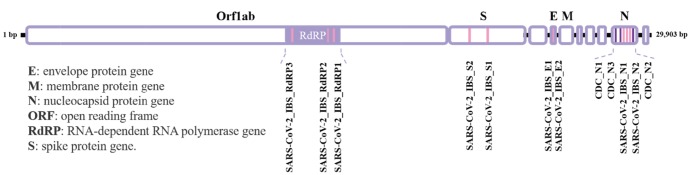
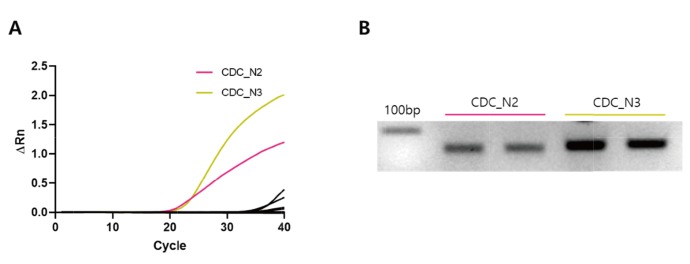
An important key step in this study was to identify the unique primer target sequences that can distinguish the SARS-CoV-2 over other common RNAs from human tissue. We utilized the recently reported full sequence of the genomic RNA for SARS-CoV-2 from the first patient in Republic of Korea (Fig. 5 [12]) to search for the unique sequences to specifically target SARS-CoV-2. We initially found 3 primer sets available from CDC targeting N, nucleocapsid protein gene [11]: CDC_N1, CDC_N2, and CDC_N3 (Fig. 5, Table 1). Unexpectedly, two of these three primer sets, CDC_N2 and CDC_N3, gave false positive signals with significant Ct values of 20. 266 and 21.129 (Ct<37) during SYBR Green-based rtPCR even in the absence of cDNA (no template control condition) (Fig. 6A, Table 2). More surprisingly, we found a clear PCR product band size of less than 100 base pair when we performed electrophoresis with the reaction product after the rtPCR (Fig. 6B). These results were difficult to understand considering the fact that there was no cDNA template to amplify. From these primer validation tests, we decided not to pursuit the primer sets originated from CDC.
Fig. 5.
The domain map of SARS-CoV-2 form the sequence obtained from the first patient in Republic of Korea. The location of each primer set is indicated at corresponding sequence. The primer set from CDC is targeting nucleocapsid protein gene (N) to detect SARS-CoV-2. In-house (IBS) designed primer sets for detecting SARS-CoV-2 target RNA-dependent RNA polymerase gene (RdRP), spike protein gene (S), envelope protein gene (E) and N.
Fig. 6.
Unexpected amplification during rtPCR with CDC primer sets in no-template control condition. (A) Quantitative rtPCR result by using all primer sets in Table 1 in no-template control. All primers in Table 1 were tested, and the data were represented as black traces, except CDC_N2 and CDC_N3 primer sets. (B) Gel electrophoresis result confirming the amplification by CDC_N2 and CDC_N3 primer sets in no-template control.
Table 2.
Ct values of each primer set in in no-template control
| Primer | Ct value |
|---|---|
| IBS_RdRB1 | u.d. |
| IBS_RdRB2 | u.d. |
| IBS_RdRB3 | u.d. |
| IBS_S1 | u.d. |
| IBS_S2 | u.d. |
| IBS_E1 | u.d. |
| IBS_E2 | u.d. |
| IBS_N1 | u.d. |
| IBS_N2 | u.d. |
| IBS_RPP30 | u.d. |
| IBS_RPP40 | u.d. |
| CDC_RNase P | u.d. |
| CDC_N1 | u.d. |
| CDC_N2 | 20.266 |
| CDC_N3 | 21.129 |
u.d., undetermined (Ct>37 cycles).
We next searched for unique primer sequences to target presumably well-conserved regions of SARS-CoV-2 viral gene. We selected total of 9 primer sets, targeting RdRP (RNA-dependent RNA polymerase gene), S (spike protein gene), E (envelope protein gene) and N (nucleocapsid protein gene) (Table 1). Each of these primer set of forward and reverse primer pair would generate a rtPCR product size of 100~120 base pair. We validated each of these primer sets by running rtPCR under no template condition and all of these in-house designed primer sets showed no false positive signals (Undetermined = Ct > 37 cycles) (Fig. 6A, Table 2).
Furthermore, we searched for human RNA specific primer sets to serve as an internal control. We targeted human Ribonuclease P (RNase P) protein, which is a ubiquitous enzyme in all cells and cellular compartments that synthesizes tRNA [18]. We selected the unique primer sets, IBS_RPP30 and IBS_RPP40, targeting RPP subunit 30 and 40, respectively. Additionally, we also utilized the internal control primer set from CDC, CDC_RNAse P, targeting the same RNase P gene). These internal control primer sets also were validated and showed no false positive signals during rtPCR (Fig. 6A, Table 2). The internal control primers were critical for evaluating the validity of negative signals from the SARS-CoV-2 targeting primer sets (see Table 4).
Table 4.
Result interpretation Table
| SARS-CoV-2_IBS_RdRP1 | SARS-CoV-2_IBS_S1 | SARS-CoV-2_IBS_E1 | SARS-CoV-2_IBS_N1 | IBS_RPP30 | IBS_RPP40 | CDC_RNase P | Result interpretation |
|---|---|---|---|---|---|---|---|
| + | + | + | + | ± | ± | ± | SARS-CoV-2 detected |
| At least 1 positive | ± | ± | ± | SARS-CoV-2 detected | |||
| - | - | - | - | + | + | + | SARS-CoV-2 NOT detected |
| - | - | - | - | At least 1 positive | SARS-CoV-2 NOT detected | ||
| - | - | - | - | - | - | - | Inconclusive detection of SARS-CoV-2 (needs re-test) |
Quantitative rtPCR of volunteer samples
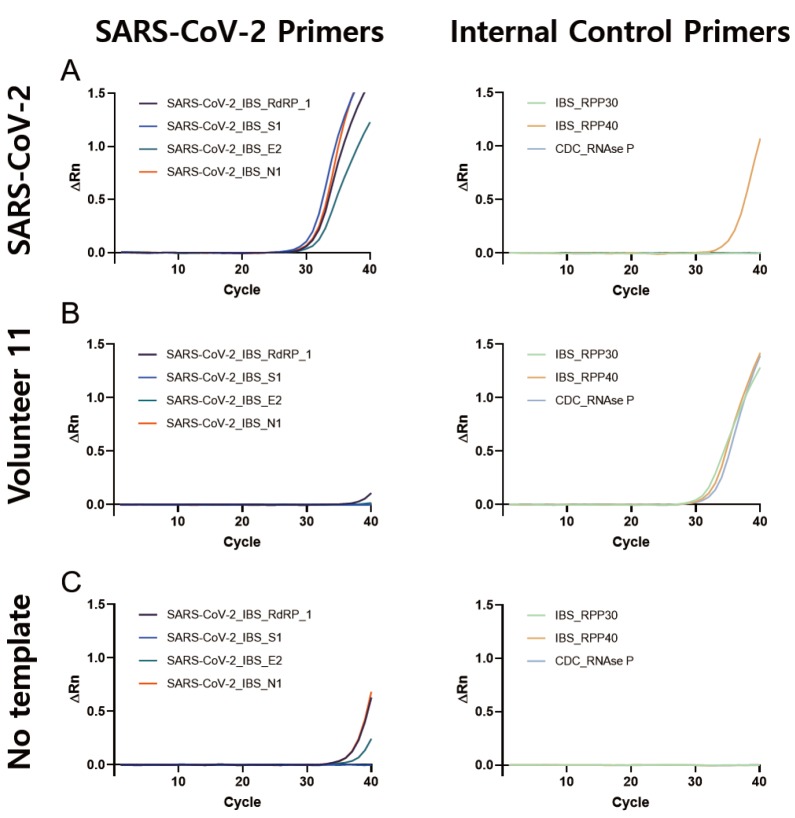
Using the validated primer sets for SARS-CoV-2 and human internal controls, we performed quantitative rtPCR for each volunteer sample. As a positive control, we utilized the purified total RNA of SARS-CoV-2 extracted from BetaCoV/Korea/KCDC03/2020 virus-infected Vero cells. As expected, the SARS-CoV-2 positive control showed significantly positive signals with Ct values under 37 with all four of the SARS-CoV-2 primer sets, SARS-CoV2_IBS_RdRP1, SARS-CoV2_IBS_S1, SARS-CoV2_IBS_E1, and SARS-CoV2_IBS_N1, along with at least one of the internal control primer sets positive (Fig. 7A, Table 3). Most of the volunteer samples showed negative signals for all four of the SARS-CoV-2 primer sets, along with at least one of the three internal control primer sets with positive signal (Fig. 7B, Table 3). Among 12 volunteer samples, three of them (Volunteer 5, 9, and 12) showed all negative signals for SARS-CoV-2 primer sets and internal control primer sets (Table 3). These three volunteer samples were considered inconclusive, which required a re-testing. The volunteers were recalled for additional self-collection of pharyngeal swab and re-test. For a negative control, we used a no-template condition and all primer sets gave negative signals, as expected (Fig. 7C, Table 3).
Fig. 7.
Representative quantitative rtPCR result from IBS-designed SARS-CoV-2 detecting primer sets in (A) SARS-CoV-2 as a positive control; (B) Volunteer 11; and (C) no template. Left panels represent SARS-CoV-2 specific amplifying results and right panels represent human internal control amplifying results.
Table 3.
Results of rtPCR for each volunteer
| Template | SARS-CoV-2_IBS_RdRP1 | SARS-CoV-2_IBS_S1 | SARS-CoV-2_IBS_E1 | SARS-CoV-2_IBS_N1 | IBS_RPP30 | IBS_RPP40 | CDC_RNase P |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | 30.3 | 28.5 | 31.1 | 30.6 | u.d. | u.d. | 35.2 |
| Volunteer 1 | u.d. | u.d. | u.d. | u.d. | 35.4 | u.d. | u.d. |
| Volunteer 2 | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. | 34.5 |
| Volunteer 3 | u.d. | u.d. | u.d. | u.d. | 35.2 | 34.1 | 34.8 |
| Volunteer 4 | u.d. | u.d. | u.d. | u.d. | u.d. | 33.4 | u.d. |
| Volunteer 5 | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. |
| Volunteer 6 | u.d. | u.d. | u.d. | u.d. | 31.4 | u.d. | 32.7 |
| Volunteer 7 | u.d. | u.d. | u.d. | u.d. | 31.9 | u.d. | 32.9 |
| Volunteer 8 | u.d. | u.d. | u.d. | u.d. | 34.8 | u.d. | u.d. |
| Volunteer 9 | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. |
| Volunteer 10 | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. | 34.9 |
| Volunteer 11 | u.d. | u.d. | u.d. | u.d. | 31.2 | 30.8 | 32.9 |
| Volunteer 12 | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. |
| No template | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. | u.d. |
Ct value, u.d.=undetermined (Ct>37 cycles), Results for all u.d. are listed in red font.
To interpret the test results as shown in Table 3, we made an easy-to-follow interpretation table based on the combination of the SARS-CoV-2 primer set results and internal control primer results (Table 4). The primary goal of this interpretation table was to make stringent criteria for a negative (absence of SARS-CoV-2), requiring all four SARS-CoV-2 primer sets to show negative signals. This negative signal must be validated by at least one of the internal control primer sets positive (Table 4). In contrast, criteria for a positive (presence of SARS-CoV-2) was less stringent with at least one of the four SARS-CoV2 primer sets positive. This less stringent criteria for a positive allows first-hand screening for a potential positive for further stringent testing in other certified clinics. Based on the stringent criteria for a negative, we determined 9 out of 12 volunteers to be negative for SARS-CoV-2, whereas other 3 were found to be inconclusive due to negative signals for internal control primer sets (Table 3 in red font). The 3 volunteer samples were then re-tested with a doubled cDNA amount as a template for rtPCR. The re-test rtPCR results showed that Volunteer 5 and Volunteer 12 still showed negative signals for SARS-CoV-2 primer sets, while one of the three internal control primer sets came out positive (Table 5). After the re-test, Volunteer 5 and Volunteer 12 were confirmed to be negative. Volunteer 9 remained for future re-collection of throat swab sample.
Table 5.
Results of re-test
| Template | SARS-CoV-2_IBS_RdRP1 | SARS-CoV-2_IBS_S1 | SARS-CoV-2_IBS_E1 | SARS-CoV-2_IBS_N1 | IBS_RPP30 | IBS_RPP40 | CDC_RNase P |
|---|---|---|---|---|---|---|---|
| Volunteer 5 | u.d. | u.d. | u.d. | u.d. | 35.644 | u.d | u.d |
| Volunteer 9 | u.d. | u.d. | u.d. | u.d. | u.d | u.d | u.d |
| Volunteer 12 | u.d. | u.d. | u.d. | u.d. | u.d | u.d | 36.649 |
Cost and duration of detection protocol
To estimate the cost of the newly developed protocol, we obtained a cost breakdown list (Table 7). The total cost was estimated to be around $15, without taking an account of labor cost. The total duration of the protocol was also estimated and found to be around 4 hours (Table 8). This duration can be shortened if we eliminate the RNA to cDNA conversion step. Currently there are several commercially available kits which contain reverse transcriptase and polymerase together. We can shorten the duration of the protocol by about 1 hour but the cost for each sample will more than quadruple. With a low cost of $15 and short duration of 4 hours, this optimized protocol for detection of SARS-CoV-2 should be the most economical detection protocol available.
Table 7.
Cost breakdown per volunteer sample ($ = US Dollar)
| Material | Unit Price | Quantity | Price per one rxn | Price per one volunteer (8rxn) |
|---|---|---|---|---|
| Swab and tongue presser | $5.00 | 100 each | $0.50 | $0.50 |
| easy-BLUETM Total RNA Extraction Kit (Trizol) | $154.17 | 100 ml | $1.07 | $1.07 |
| SuperScriptTM III First-Strand Synthesis System | $406.67 | 50 reaction | $8.13 | $8.13 |
| Power SYBRTM Green PCR Master Mix | $322.5 | 5 ml | $0.65 | $5.20 |
| Primer set | $0.10 | |||
| Tubes and buffers | $1.00 | |||
| Total Price | $15.00 | |||
Table 8.
Duration of each procedure
| Procedure | Duration |
|---|---|
| Collection of pharyngeal swab | 10 min |
| RNA extraction | 60 min |
| cDNA synthesis | 90 min |
| rtPCR | 70 min |
| Total time | 230 min (3 hour 50 min) |
DISCUSSION
In this study, we have established a general protocol for a detection of the threatening SARS-CoV-2. This protocol should not be limited to this alarming virus, but also for any other future pandemic viruses to come. The highlight of the protocol is the feature to keep the biosafety level high by making self-collection of the tissue sample from a volunteer, eliminating the need for unnecessary contact between the examiner and the volunteer. Even though commercial viral RNA prep kits are used in clinical diagnosis, we adopted the Trizol-based total RNA extraction method for two reasons: bio-safety and economical advantage to reduce the total cost. The Trizol-based RNA extraction makes certain that Trizol instantly inactivates the potentially infectious tissue samples, if viral particles exist, enhancing the safety level further. This safe protocol allows any conventional biology laboratory minimally equipped with a Level II Bio-safety cabinet and a rtPCR machine testing the presence or absence of SARS-CoV-2. The Trizol-based RNA extraction and SYBR Green-based rtPCR both make this protocol at very low-cost. We have confirmed that RNA quality is maintained to the similar level as the commercial viral RNA kit when we processed with Trizol-based method (Fig. 3). Due to the safety feature of our protocol, we expect to turn conventional biological laboratories to an efficient testing stations under emergency situations such as when there is a serious outbreak of SARS-CoV-2.
The primary goal of this study was to develop a novel protocol to be used as a primary screening platform for asymptomatic people to be tested for a real negative. However, the results should not be considered as a clinical diagnosis, which requires medical expertise and staff for proper diagnosis. Instead, this protocol is designed for identifying a virus-negative asymptomatic people to be free of worries and to work, study, and sport normally. To achieve the sensitivity required for detecting a true negative (absence of SARS-CoV-2), we adopted SYBR Green-based rtPCR method over Taqman-based rtPCR method. Taqman-based diagnosis is accepted in the field as more accurate method. To detect a real positive (presence of SARS-CoV-2), the Taqman-based method could be more appropriate in more clinical settings. However, our study is rather focusing on the lowering the threshold for positive signals so that the threshold for negative signals is enhanced. To enhance the threshold for negative signals, we selected multiple target regions in SARS-CoV-2 genome and internal control for human RNA: four target genes in SARS CoV-2 and three targets in human RNAse P. We found a robust positive signal for SARS-CoV-2 primer sets in SARS-CoV-2 positive control using our in-house primer sets. We also tested other primer sets that we have selected and these can be also used in combination to increase further the sensitivity for positive signals.
In the course of our study, we realized that the primer validation step (Fig. 6) is absolutely critical for developing a detection protocol via rtPCR. We found that many previously reported primer sets can give very strong false positive signals (Ct<37). In fact, the CDC originated primer sets gave Ct values of around 20 in the absence of template cDNA. This is a very serious issue when the sensitivity relies on the primer specificity. We strongly suggest that the CDC protocol to be thoroughly re-examined before it is further distributed as a detection kit for SARS-CoV-2. The primer sets that we have designed in-house, IBS_SARS-CoV-2 prime sets (Table 1), were all passed the primer validation test and should be good for sensitive detection of SARS-CoV-2.
In summary, we have developed a laboratory-safe, low-cost, high-sensitivity detection protocol for SARS-CoV-2. We are cautious about the potential for use of this protocol in clinics because there should be much more thorough examinations of our protocol for clinical use. Those who already show obvious symptoms of COVID-19 should not rely on our detection protocol, but resort to certified hospital and health agencies. Our protocol should be useful when the purpose of the testing is to identify the negative people, who need to work, study, and sport normally. Furthermore, the newly developed tools and concepts in this study should be useful for any future outbreak of a novel virus or pathogen.
ACKNOWLEDGEMENTS
This work was supported by financial support by Institute for Basic Science (IBS), Center for Cognition and Sociality (IBS-R001-D2) to C.J.L.
REFERENCES
- 1.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Centers for Disease Control; Atlanta: 2020. How COVID-19 spreads [Internet] Available from: https://www.cdc.gov/coronavirus/2019-ncov/about/transmission.html. [Google Scholar]
- 3.Bendix A. Business Insider; New York: 2020. A person can carry and transmit COVID-19 without showing symptoms, scientists confirm [Internet] Available from: https://www.sciencealert.com/researchers-confirmed-patients-can-transmit-the-coronavirus-without-showing-symptoms. [Google Scholar]
- 4.Hoehl S, Berger A, Kortenbusch M, Cinatl J, Bojkova D, Rabenau H, Behrens P, Böddinghaus B, Götsch U, Naujoks F, Neumann P, Schork J, Tiarks-Jungk P, Walczok A, Eickmann M, Vehreschild MJGT, Kann G, Wolf T, Gottschalk R, Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020 doi: 10.1056/NEJMc2001899. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu P, Zhu J, Zhang Z, Han Y, Huang L. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa077. doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ministry of Health and Welfare. South Korea; Sejong: 2020. [Government support 2019 - novel coronavirus testing condition] Korean. [Google Scholar]
- 7.Ahn S. LawTalk News; Seoul: 2020. [Coronavirus test cost] [Internet] Available from: https://news.lawtalk.co.kr/issues/1848. Korean. [Google Scholar]
- 8.Yu E. dongA.com; Seoul: 2020. [Lack of medical personnel following the corona spread] [Internet] Available from: http://www.donga.com/news/article/all/20200221/99817769/1. Korean. [Google Scholar]
- 9.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blow JA, Dohm DJ, Negley DL, Mores CN. Virus inactivation by nucleic acid extraction reagents. J Virol Methods. 2004;119:195–198. doi: 10.1016/j.jviromet.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Centers for Disease Control and Prevention; Atlanta: 2020. Research use only 2019-novel Coronavirus (2019-nCoV) real-time RT-PCR primer and probe information [Internet] Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. [Google Scholar]
- 12.Park WB, Kwon NJ, Choi SJ, Kang CK, Choe PG, Kim JY, Yun J, Lee GW, Seong MW, Kim NJ, Seo JS, Oh MD. Virus isolation from the first patient with SARS-CoV-2 in Korea. J Korean Med Sci. 2020;35:e84. doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Centers for Disease Control and; Atlanta: 2020. Interim laboratory biosafety guidelines for handling and processing specimens associated with Coronavirus disease 2019 (COVID-19) [Internet] Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html. [Google Scholar]
- 14.Murray MA, Schulz LA, Furst JW, Homme JH, Jenkins SM, Uhl JR, Patel R, Cockerill FC, Myers JF, Pritt BS. Equal performance of self-collected and health care worker-collected pharyngeal swabs for group a streptococcus testing by PCR. J Clin Microbiol. 2015;53:573–578. doi: 10.1128/JCM.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thermo Fisher Scientific. Thermo Fisher Scientific; Waltham: 2020. The manual of TRIzolTM LS Reagent [Internet] Available from: https://www.thermofisher.com/order/catalog/product/10296028. [Google Scholar]
- 17.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular diagnosis of a novel Coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa029. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopalan V, Vioque A, Altman S. RNase P: variations and uses. J Biol Chem. 2002;277:6759–6762. doi: 10.1074/jbc.R100067200. [DOI] [PubMed] [Google Scholar]