Abstract
Aims/hypothesis
Gestational diabetes mellitus (GDM) affects up to 20% of pregnancies, and almost half of the women affected progress to type 2 diabetes later in life, making GDM the most significant risk factor for the development of future type 2 diabetes. An accurate prediction of future type 2 diabetes risk in the early postpartum period after GDM would allow for timely interventions to prevent or delay type 2 diabetes. In addition, new targets for interventions may be revealed by understanding the underlying pathophysiology of the transition from GDM to type 2 diabetes. The aim of this study is to identify both a predictive signature and early-stage pathophysiology of the transition from GDM to type 2 diabetes.
Methods
We used a well-characterised prospective cohort of women with a history of GDM pregnancy, all of whom were enrolled at 6–9 weeks postpartum(baseline),were confirmed not to have diabetes via 2 h 75 g OGTT and tested anually for type 2 diabetes on an ongoing basis (2 years of follow-up). A large-scale targeted lipidomic study was implemented to analyse ~1100 lipid metabolites in baseline plasma samples using a nested pair-matched case–control design, with 55 incident cases matched to 85 non-case control participants. The relationships between the concentrations of baseline plasma lipids and respective follow-up status (either type 2 diabetes or no type 2 diabetes) were employed to discover both a predictive signature and the underlying pathophysiology of the transition from GDM to type 2 diabetes. In addition, the underlying pathophysiology was examined in vivo and in vitro.
Results
Machine learning optimisation in a decision tree format revealed a seven-lipid metabolite type 2 diabetes predictive signature with a discriminating power (AUC) of 0.92 (87% sensitivity, 93% specificity and 91% accuracy). The signature was highly robust as it includes 45-fold cross-validation under a high confidence threshold (1.0) and binary output, which together minimise the chance of data overfitting and bias selection. Concurrent analysis of differentially expressed lipid metabolite pathways uncovered the upregulation of α-linolenic/linoleic acid metabolism (false discovery rate [FDR] 0.002) and fatty acid biosynthesis (FDR 0.005) and the downregulation of sphingolipid metabolism (FDR 0.009) as being strongly associated with the risk of developing future type 2 diabetes. Focusing specifically on sphingolipids, the downregulation of sphingolipid metabolism using the pharmacological inhibitors fumonisin B1 (FB1) and myriocin in mouse islets and Min6 K8 cells (a pancreatic beta-cell like cell line) significantly impaired glucose-stimulated insulin secretion but had no significant impact on whole-body glucose homeostasis or insulin sensitivity.
Conclusions/interpretation
We reveal a novel predictive signature and associate reduced sphingolipids with the pathophysiology of transition from GDM to type 2 diabetes. Attenuating sphingolipid metabolism in islets impairs glucose-stimulated insulin secretion.
Keywords: Gestational diabetes mellitus, Glucose-stimulated insulin secretion, Lipidomic study, Machine learning, Multiple logistic regression, Pathophysiology, Predictive biomarker, Prospective cohort, Sphingolipid metabolism, Type 2 diabetes
Introduction
Gestational diabetes mellitus (GDM), defined as glucose intolerance first recognised during pregnancy, affects up to14% of pregnancies worldwide [1, 2]. Although the cause remains uncertain, GDM is suspected to arise from the diminished capacity of the pancreas to produce sufficient insulin and impaired insulin action related to pregnancy. GDM pregnancy increases maternal complications [3] and infants of mothers with GDM are at significantly higher risk of obesity, dyslipidaemia and type 2 diabetes [4]. While maternal glucose tolerance generally returns to normal after delivery, GDM is associated with persistent long-term metabolic dysfunction and elevated risk of overt diabetes [5]. Up to 50% of women with GDM may progress to type 2 diabetes within 5–10 years postpartum [6, 7]. These women develop type 2 diabetes at a relatively younger age (e.g. <40 years) than the general population and have a higher risk of cardiovascular disease, non-alcoholic fatty liver, renal disease and early mortality [8–15]. The underlying cause of the transition from GDM to type 2 diabetes and the accurate prediction of this transition are therefore critical.
The ADA recommends that all women with GDM undergo screening for type 2 diabetes via a 2 h 75 g OGTT at 6– 12 weeks postpartum followed by subsequent screening every 1–3 years via fasting plasma glucose (FPG) measurement and 2 h 75 g OGTT [16]. The discriminating power (AUC) of 2 h plasma glucose in the OGTT is at best 65–77% across studies [17–19]. Moreover, the compliance with ADA recommendations among this group for screening via an OGTT is very low (~19%) in many settings [19, 20]. This low compliance could in part be due to the time-consuming and/or unpleasant nature of the tests or healthcare system limitations [19, 21–24]. A simplified and more accurate prognostic test would be desirable to reclassify glucose tolerance after pregnancy and predict future type 2 diabetes progression following GDM pregnancy.
It is well known that the elevation in blood glucose in type 2 diabetes occurs long after the underlying metabolic changes that promote disease development. Thus, discovery-based metabolomics is considered a promising approach for both the early prediction and the identification of underlying pathways of future type 2 diabetes onset. This methodology has led to the identification of several biomarkers for future type 2 diabetes incidence [25–27]. Our group previously identified metabolic biomarkers of subsequent type 2 diabetes onset among women with recent GDM enrolled in the Study of Women, Infant Feeding and Type 2 Diabetes after GDM (SWIFT) prospective cohort [19]. Using clinical variables combined with metabolic biomarkers, including lipid species, we developed a simple four-structure metabolic signature—phosphatidylcholine (PC) aeC40:5, hexoses, branched-chain amino acids (BCAAs) and sphingomyelin (SM) (OH)C14:1—that predicted type 2 diabetes incidence with 83% discrimination power (AUC) in a nested pair-matched (1:1) case–control study of 244 SWIFT participants, where 12% of 1010 women with GDM progressed to type 2 diabetes within about 2 years post-delivery [19]. A smaller nested case–control study of metabolomics (lipidomics), targeting >300 lipid species in blood samples taken from 104 women with GDM at 12 weeks post-delivery, of whom 21 (20%) progressed to type 2 diabetes within 12 years, showed 83.6% accuracy in type 2 diabetes prediction based on three lipids—phosphatidylethanolamine (PE) P-36:2, phosphatidylserine (PS) 38:4 and cholesteryl ester (CE) 20:4—in combination with six other risk factors (age, BMI, pregnancy fasting glucose, postpartum fasting glucose, total triacylglycerols [TAGs] and total cholesterol) [28]. These promising findings provide evidence that novel metabolite markers combined with other factors can facilitate the prediction of type 2 diabetes risk.
Metabolomic studies can also be used to illuminate the pathophysiology of type 2 diabetes and its progression. Both stearoylcarnitine and BCAA levels increased in those who developed type 2 diabetes [29, 30], possibly linked to impaired pancreatic beta cell function [31]. Several specialised lipid metabolites (sphingomyelins [SMs], phosphatidylcholines [PCs] and lysophosphatidylcholines [LPCs]) were inversely associated with type 2 diabetes risk [32]. Our previous metabolomics study in the SWIFT cohort of women with GDM also showed decreased levels of several specialised lipid metabolites (sphingolipids and PCs) in the transition from GDM to type 2 diabetes [19]. These lipid metabolites are known core components of cell membranes and may be linked to type 2 diabetes progression [32, 33].
There is substantial evidence to suggest that lipid imbalances both predict and cause type 2 diabetes. Given the apparent links between lipid biosynthesis, metabolism and beta cell dysfunction leading to type 2 diabetes, the role of lipids has been collectively understudied with respect to diabetes risk. Herein, we used lipidomics to screen a large and broad spectrum of lipid metabolites in relation to subsequent type 2 diabetes development. This lipidomic study sought to identify lipid biomarkers and putative early-stage pathophysiology that may predict and influence future progression to type 2 diabetes in women after GDM pregnancy.
Methods
Study population
The prospective SWIFT cohort enrolled a racially and ethnically diverse group of 1035 women, with GDM (age 20– 45 years), who delivered singleton pregnancies at ≥35 weeks of gestation at Kaiser Permanente Northern California (KPNC) hospitals between 2008 and 2011 [34, 35]. Each participant provided informed consent at the in-person examination at 6–9 weeks postpartum (baseline) before collection of blood specimens from a 2 h 75 g OGTT, completion of surveys, anthropometric and body composition measurements, and annual in-person follow-up examinations for 2 years. The KPNC Institutional Review Board approved the study protocol. The study recruitment, selection criteria, methodologies and other detailed information have been described previously [34–36]. At each 2 h 75 g OGTT, trained research staff collected fasting blood samples and processed and stored plasma samples at −80°C for future studies.
Study design
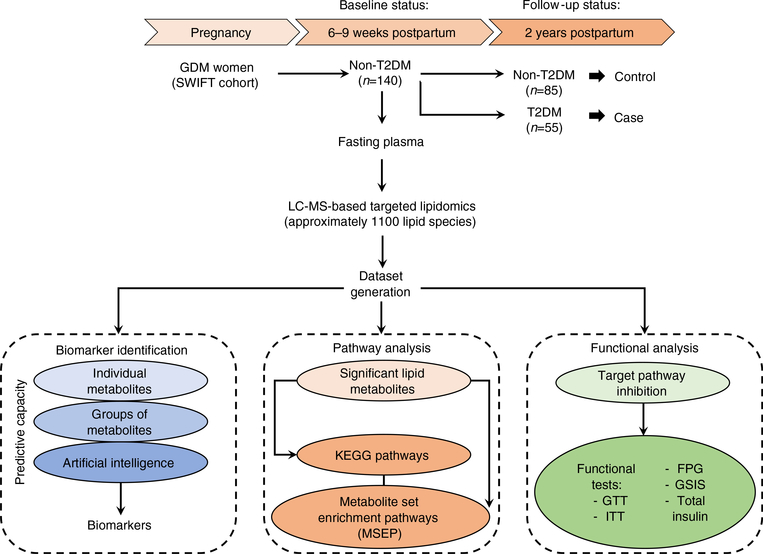
For this study, we selected the incident diabetes cases among Hispanic and Asian groups, and pair-matched (1:1.5) them to control women without progression to diabetes during the 2 year follow-up by age (±2 years), race and ethnicity (completely matched), pre-pregnancy BMI (±0.96 kg/m2) and glucose tolerance at 6–9 weeks postpartum (completely matched). We selected only matched pairs of Hispanic (n = 90) and Asian (n = 50) women to ensure homogeneity of race and ethnic groups. The nested case–control design with pair-matching greater than 1:1 does not allow direct comparisons of incidence rates among the ethnic and racial groups for this subset analysis. The fasting plasma samples were collected from these 140 women at the baseline examination (at 6–9 weeks postpartum), all confirmed not to have-type 2 diabetes at the baseline exam via the 2 h 75 g OGTT. Details of the SWIFT prospective cohort design and follow-up are published elsewhere [30, 37–40]. For women who progressed to type 2 diabetes during the 2 years follow-up period (n = 55), termed here as the ‘follow-up’ time point, the newly diagnosed incident type 2 diabetes was referred to as ‘case’. Women who did not develop type 2 diabetes during the follow-up period (n = 85) are referred to as ‘control’ (Fig. 1). Please see electronic supplementary materials (ESM) Methods for details.
Fig. 1.
The schematic flow diagram of the study design. This was a nested case–control study within the SWIFT study, a prospective cohort of 1035 women diagnosed with GDM and followed up to 2 years postpartum. A total of 140 women were selected out of the 1035 SWIFT participants. These women did not have type 2 diabetesmellitus (T2DM) at 6–9 weeks postpartum(study baseline) based on 2 h 75 g OGTT. Of the 140 selected, 55 women were diagnosed as having T2DM, via 2 h 75 g OGTTs, within 2 years post baseline. This group was termed as ‘case’. The remaining 85 women did not develop T2DMbased on the results of the 2 h 75 g OGTTs within 2 years post baseline. This group was termed ‘control’ (non-T2DM). The fasting plasma from the baseline examination was used for LC-MS-based targeted lipidomics aimed at finding the relation in terms of a predictive signature and the earlier stage pathophysiology of T2DM prospectively within the 2 year follow-up period
Targeted lipid profiling (targeted-lipidomics analysis)
Fasting plasma samples collected at 6–9 weeks postpartum during the SWIFT study were sent to Metabolon (Morrisville, NC, USA) for a single-blind targeted-lipidomics analysis of 1100 lipid species on each plasma sample. For details of lipidomics see ESM Methods.
Data preparation and statistical analysis of the quality of the final dataset
A stringent protocol was followed to prepare the final dataset, which was further scrutinised for quality in terms of the presence of confounding factors and the certainty of the class separation through principal component analysis (PCA) and a partial least squares-discriminant analysis (PLS-DA), respectively, using MetaboAnalyst 3.0 (https://www.metaboanalyst.ca/) in default setting (e.g. tenfold cross-validation). For details of this protocol, see ESM Methods.
Differential expression analysis and pathway analysis
A non-parametric test (Wilcoxon–Mann–Whitney test, α value set at p < 0.05) followed by multiple comparisons with false discovery rate (FDR) analysis (α value set at p < 0.05) was carried out to identify the differentially expressed lipid metabolites between the case and control. These differentially expressed lipid metabolites were used for the pathway analysis by adopting two approaches: (1) a direct approach where differentially expressed lipid metabolites were used in both over-representation pathway analysis using Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa Laboratories, Kyoto, Japan) pathways and metabolite set enrichment pathways (MSEP) analysis and (2) an in silico approach where the interacting proteins with the differentially expressed lipid metabolites were used. All analyses were carried out using one of the following platforms (or a combination of them): MetaboAnalyst 3.0, MBrole 2.0 (Madrid, Spain), and String 10.5 platforms (https://string-db.org). For details of the pathway analyses, see ESM Methods.
Predictive analytics
The biomarker analysis module of MetaboAnalyst 3.0 was used for univariate receiver operating characteristic (ROC) analysis. In the multivariate ROC analysis, the stepwise (both ways) multiple logistic regression (MLR) was carried out in R-studio (Boston, MA, USA) using the ‘glm’ function under the removal of data redundancy protocol and significant contributor calculation (R-script is available in ESM Methods). Machine learning analyses were carried out through WEKA 3.8 (University of Waikato, Hamilton, NZ). The final classifier was further optimised for balancing between the chance of data overfitting, higher ROC possibility and F-score (a measurement of a test’s accuracy based on precision and sensitivity). Optimisation was carried out by applying K-fold cross-validation, confident threshold 1.0 and binary output selection. A series of cross-validation up to K = 100 was conducted to test the stress tolerability of the signature. Forty-five-fold cross-validation (K = 45) was chosen as per ‘one standard error rule’ for final reporting. High confidence threshold (1.0) ensures the proper cleaning of bias from the final signature. Binary output selection further protects the signature from data overfitting and bias selection. The discriminating power of ROC analysis is presented in the form of an AUC. See ESM Methods for details.
In vivo and in vitro functional studies
Animal care
C57BL/6 J male mice were obtained from Charles River (Sherbrook, QB, Canada) at the age of 8 weeks for both in vivo and in vitro islets studies. Mice were housed in the Division of Comparative Medicine facility, University of Toronto. All mouse procedures and maintenance were conducted in compliance with protocols approved by the Animal Care Committee at the University of Toronto and the guidelines of the Canadian Council of Animal Care.
Intraperitoneal injections and monitoring
The mice were injected intraperitoneally either by using 1 mg kg−1 day−1 fumonisin B1 (FB1) (Cayman, Michigan, USA) or vehicle (DMSO–saline [154 mmol/l NaCl]) for 3 weeks. Weight gain and blood glucose were monitored on a weekly basis.
Insulin tolerance test and IPGTT
Both ITTs and GTTs were conducted using standard protocols that are described elsewhere [41].
Sphingolipid profiling and insulin staining of pancreas
After 3 weeks of treatment, mice were euthanised to collect plasma and pancreatic tissue. Plasma samples (n = 3) were subjected to sphingolipid profiling through LC-MS/MS at the Analytical Facility for Bioactive Molecules, SickKids, Toronto. The pancreases (n = 7) were fixed for insulin staining by using the standard protocol [37] of the Centre for Phenogenomics (TCP), Sinai Health System Institute, Toronto. The 40× images of pancreatic slices were produced at TCP and analysed by Aperio ImageScope software package (Wetzlar, Germany).
In vitro glucose-stimulated insulin secretion Glucose-stimulated insulin secretion (GSIS) was assessed, as previously described [41],in both Min6 K8 cells (agift from S. Seino [Kobe University, Kobe, Japan] and J. Miyazaki [Osaka University, Suita, Japan] and isolated male murine C57BL/6 islets in vitro after treatment with either 1 μmol/l FB1 or50 nmol/l myriocin (Cayman, Ann Arbor, MI, USA) for 24 h.
Results
Baseline sociodemographic and clinical characteristics of participants
This nested pair-matched case–control study included a subset of 140 Asian and Hispanic women from the SWIFT cohort (Fig. 1). Sociodemographic and clinical characteristics of case and control groups are summarised in Table 1. There were no statistically significant differences observed in either pre-pregnancy or baseline (6–9 weeks postpartum) BMI, total energy intake or physical activity. Baseline FPG (p < 0.01), 2 h plasma glucose (p < 0.001), fasting insulin (p < 0.01) and fasting TAG (p < 0.05) measurements and median HOMA-IR (p < 0.01) were significantly higher in the type 2 diabetes case group. The case group was more likely than matched control participants to have been treated with insulin or oral medications during pregnancy and were more likely to have a family history of diabetes.
Table 1.
Prenatal and baseline (6– 9 weeks postpartum) characteristics of incident type 2 diabetes cases and matched control (no diabetes) within 2 years postbaseline among women with GDM
| Characteristic | Incident diabetes (n=55) | Controls (n=85) | Unadjusted p value |
|---|---|---|---|
| Prenatal and sociodemographic characteristics | |||
| Age, years | 34.8 (5.1) | 34.3 (4.6) | 0.55 |
| Race/ethnicity, n (%) | 0.05 | ||
| Hispanic | 30 (54.5) | 60 (70.6) | |
| Asian | 25 (45.5) | 25 (29.4) | |
| Parity, n (%) | 0.22 | ||
| Primiparous (1 birth) | 16 (29.1) | 17 (20.0) | |
| Multiparous (>2 births) | 39 (70.9) | 68 (80.0) | |
| GDM prenatal treatment, n (%) | 0.006 | ||
| Diet only | 26 (47.3) | 55 (64.7) | |
| Oral medications | 26 (47.3) | 28 (32.9) | |
| Insulin | 3 (5.5) | 2 (2.4) | |
| Gestational age at GDM diagnosis, weeks | 21.3 (9.1) | 24.1 (7.8) | 0.05 |
| Pre-pregnancy BMI, kg/m2 | 32.0 (5.2) | 32.5 (5.4) | 0.61 |
| Family history of diabetes, n (%) | 40 (72.7) | 48 (56.5) | 0.05 |
| BMI and behaviours at 6–9 weeks postpartum | |||
| BMI, kg/m2 | 32.3 (5.2) | 32.3 (5.2) | 0.99 |
| Smoker (current or past), n (%) | 0 (0.0) | 1 (1.2) | 0.61 |
| Physical activity, met-h/week | 52.8 (35.5) | 45.4 (30.2) | 0.06 |
| Total dietary energy intake, kJ/day | 3609.96 (1590.76) | 3538 (1514.19) | 0.79 |
| Infant feeding status, n (%) | 0.59 | ||
| Exclusive lactation | 8 (14.5) | 19 (22.4) | |
| Mostly lactation | 24 (43.6) | 32 (37.6) | |
| Mostly formula/mixed | 12 (21.8) | 21 (24.7) | |
| Exclusive formula | 11 (20.0) | 13 (15.3) | |
| Plasma variables at 6–9 weeks postpartum | |||
| FPG, mmol/l | 5.68 (0.55) | 5.4 (0.47) | 0.001 |
| 2 h post-load glucose in 75 g OGTT, mmol/l | 7.73 (1.66) | 6.48 (1.7) | <0.001 |
| Fasting insulin, pmol/l | 231.27 (134.73) | 174.32 (100.70) | 0.005 |
| Fasting TAGs, mmol/l | 1.73 (1.11) | 1.32 (0.74) | 0.01 |
| Fasting HDL-cholesterol, mmol/l | 1.25 (0.29) | 1.34 (0.33) | 0.22 |
| HOMA-IR, median (IQR)a | 7.5 (4.5–10.3) | 5.1 (3.6–8.0) | 0.003 |
| HOMA-B, median (IQR)a | 259.0 (191.4–365.8) | 235.9 (170.4–343.6) | 0.15 |
Data are presented as mean (SD) or n (%) unless otherwise noted
Plasma values are from the SWIFT database
p values are for incident diabetes (case) vs no diabetes (control), paired t test
Kruskal–Wallis test applied
IQR, interquartile rang
Statistical analysis of the quality of the final dataset from lipidomics
The final dataset was composed of 626 detectable lipid metabolites. The unsupervised PCA showed two major principal components, with the first comprising 32.8% of the total study population and the second comprising 11.8%. Since the lipidomic analysis was performed at baseline before the earliest diagnosis, it would be overly optimistic to get a higher value for the major principal components. Other components were small contributors in the separation of the study population (ESM Fig. 1a). The supervised PLS-DA, where the groups were pre-identified as control and case, showed distinguishable separation and was presented in a two-dimensional score plot (ESM Fig. 1b). A cross-validation analysis determined that the performance of PLS-DA had a 63% and 64% accuracy for these two clusters, respectively, based on R2 and Q2 (ESM Fig. 1c). Furthermore, the empirical Bayes estimation (here with 1000 random permutations) was applied to confirm that the distinct separation between the two groups found in PLS-DAwas not due to random chance. The empirical p value was significant (0.014; ESM Fig. 1d), indicating that the separation was true for 986 times out of 1000. The distribution of (quantile) normalised and log2-transformed data is showed in ESM Fig. 1e.
Univariate and multivariate ROC analysis and predictive capability of metabolites to predict future type 2 diabetes
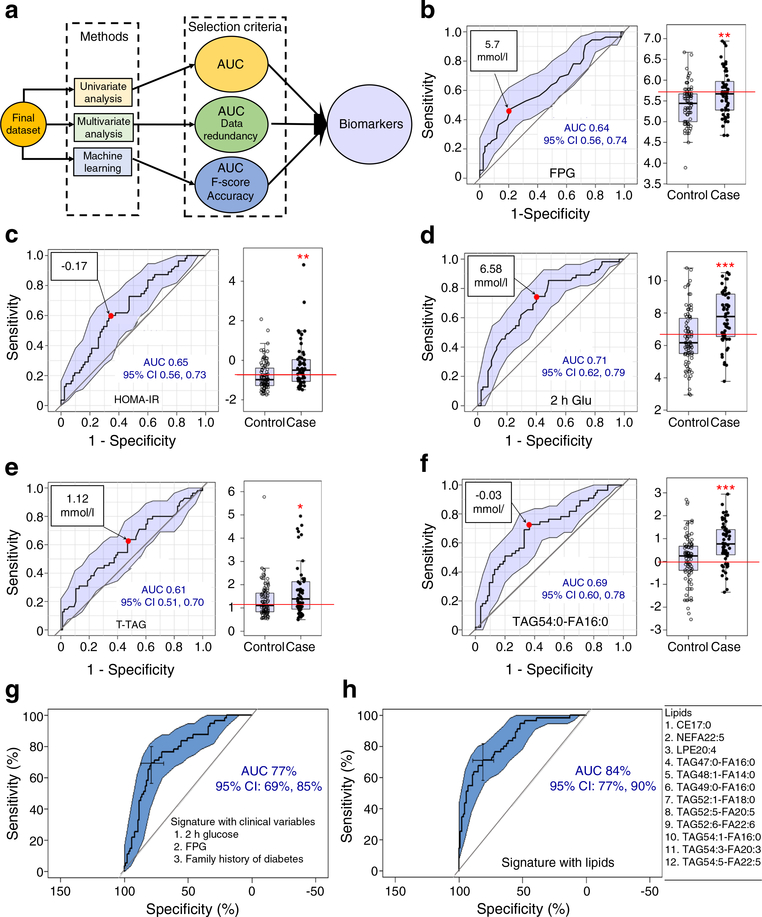
The strategies for predictive biomarker discovery are illustrated in Fig. 2a. FPG, HOMA-IR and 2 h post-load glucose in 75 g OGTT are frequently used for diagnostic purposes and their values for cases vs controls already showed a significant difference at baseline (p < 0.01, p < 0.01 and p < 0.001, respectively). In addition, the total fasting TAG levels were significantly higher in cases vs controls (p < 0.05). However, the ROC-AUCs of FPG, HOMA-IR, 2 h glucose and total fasting TAGs were 0.64, 0.65, 0.71 and 0.61 respectively (Fig. 2b–e, ROC analyses) in classic univariate ROC analyses. These low AUC values indicated a relatively weak ability to predict type 2 diabetes. Although mean differences were statistically significant (p < 0.01, p < 0.01, p < 0.001 and p < 0.05, respectively) (Fig. 2b–e, box plots), low AUC scores led to limitations. Each lipid metabolite was also subjected to classic univariate ROC analysis to find the lipid metabolite with the highest predictive capability for future type 2 diabetes status. Among all lipid metabolites, TAG 54:0-FA 16:0 scored the highest AUC of 0.69 (Fig. 2f). Although its mean difference for cases vs controls was statistically significant (p < 0.001) (Fig. 2f, box plot), its relatively low ROC-AUC score indicated weak predictability. The low ROC-AUC of TAG 54:0-FA 16:0 was in part due to high heterogeneity in the distribution of its concentration within the population. The low AUCs in univariate ROC analyses suggested that one analyte-based diagnostic would not be the best approach to predict type 2 diabetes incidence.
Fig. 2.
Predictive signatures/biomarkers for progression to type 2 diabetes. (a) Schematic flow diagram of the predictive signatures/biomarkers. (b) Univariate ROC analysis and box plot for FPG. The FPG value at 5.7 mmol/l (red circle) is the optimal cut-off for the mean AUC 0.64 within the 95% CI. (c) Univariate ROC analysis and box plot for HOMA-IR. The HOMA-IR value at −0.17 (red circle) is the optimal cut-off and provides the mean AUC 0.65 within the 95% CI. (d) Univariate ROC analysis and the box plot for 2 h post-load glucose in 75 g OGTT (2 h Glu). The 2 h glucose value at 6.58 mmol/l (red circle) is the optimal cut-off and provides the mean AUC 0.71 within the 95% CI. (e) Univariate ROC analysis and box plot for total fasting TAGs (T-TAG). The T-TAG value at 1.12 mmol/l (red circle) is the optimal cut-off and provides the mean AUC 0.61 within the 95% CI. (f) Univariate ROC analysis and box plot for the top AUC exhibiting lipid metabolite TAG54:0-FA16:0. The value at −0.03 mmol/l (red circle) is the optimal cut-off and provides the mean AUC 0.69 within the 95% CI. In the box plots (b–f), the distribution of population (case and control) based on FPG, HOMA-IR, 2 h glucose, T-TAG and TAG54:0-FA16:0 is shown, with the y-axis in mmol/l, except for HOMA-IR (unitless). The bottom and top of the box are the Q1 and Q3 (25th and 75th percentile), respectively, and the central band is the median (Q2 or 50th percentile). The bottom whisker is located within 1.5 IQR of the lower quartile, and the upper whisker is located within 1.5 IQR of the upper quartile. Outliers are presented in the outside of whiskers. The red line in each box plot shows the point that separates the whole population into two groups, case and control, to provide maximum class separation. A two-tailed, paired t test was carried out for each comparison; unadjusted p values: *p<0.05, **p<0.01, ***p<0.001 vs control. (g) In stepwise MLR with clinical variables, the signature with three variables (2 h glucose, FPG and family history of diabetes) provides the mean AUC 77%. (h) In stepwise MLR with lipid metabolites, the signature with 12 variables (lipids, shown on the right) provides the mean AUC 84%
Since type 2 diabetes is a multifactorial disease, multivariate analyses could have better strength in predicting future type 2 diabetes onset. Thus, a popular multivariate ROC analysis, stepwise multiple (both ways) logistic analysis [38, 39], was carried out here to select a signature panel (containing multiple variables) to improve the discrimination power (AUC). In the stepwise MLR analysis with both statistically significant biochemical clinical variables (FPG, 2 h glucose, HOMA-IR and total TAG) and clinical factors (family history of diabetes and type of GDM treatment), a panel of three clinical variables (FPG, 2 h glucose and family history of diabetes) produced an AUC of 77% (95% CI 69%, 85%) (Fig. 2g). In the stepwise MLR analysis with lipids, a panel of 12 lipid metabolites produced an AUC of 84% (95% CI 77%, 90%) (Fig. 2h).
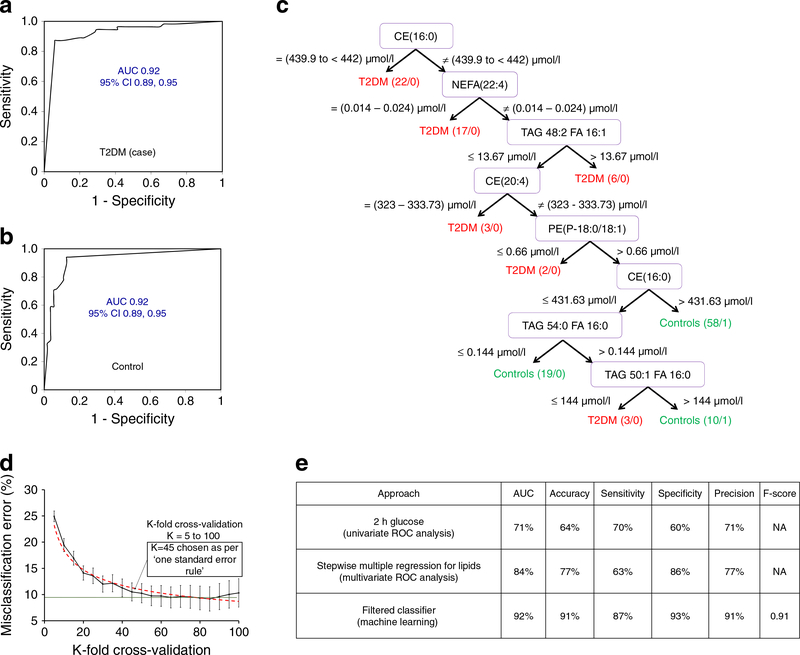
The predictive signatures/biomarkers in machine learning approach and comparison with other methods
The artificial intelligence-assisted machine learning algorithms were further employed using Weka 3.8 to find a predictive signature with a better predictability than the multivariate signature panel. The highest ROC-AUC was found in the filtered classifier algorithm. The ROC-AUC of this panel was 0.92 for both case and control participants (Fig. 3a, b) with 91% accuracy (Fig. 3e). It revealed a predictive signature consisting of seven lipid metabolites with a decision tree having 17 nodes (branching points) and nine leaves (decision points) (Fig. 3c). Although both biochemical and historical clinical variables (total TAGs, FPG, 2 h glucose, HOMA-IR, family history of diabetes and type of GDM treatment) were evaluated with the lipid dataset, they did not appear in the predictive signature, indicating the superior predictive power of lipid metabolites over these clinical variables as well as matching variables (age, race/ethnicity and BMI) in this nested case–control study sample. This signature was validated through a rigorous cross-validation protocol, where a 45-fold cross-validation was selected by adopting one standard error calculation (Fig. 3d). The K = 45 cross-validated model showed no significant difference in misclassification errors in comparison with the K = 20- to 100-fold cross-validated models, having relatively lower standard mean errors and no overfitting due to being outside of the saturation of accuracy (K = 60 to 90). K = 85 cross-validation, which produced the lowest misclassification errors (or highest accuracy), was the most over-fitted model. The K = 45 cross-validated model was further optimised under confidence threshold 1.0 and binary output selection criteria. Altogether, this ensured the signature did not suffer from data overfitting and bias selection. The comparison among the best signatures found using different approaches is summarised in Fig. 3e. Comparisons were made in terms of accuracy, sensitivity, specificity, precision and AUC. The machine learning approach-derived signature had an AUC of 0.92, an accuracy of 91% (correctly predicted 127 out of 140 participants), a sensitivity of 87% (correctly predicted 48 cases out of 55) and a specificity of 93% (predicted 79 controls correctly out of 85).
Fig. 3.
The machine learning approach in predictive signature discovery. (a, b) ROC curve for type 2 diabetes (T2DM) cases (a) and control participants (b) in the filtered classifier algorithm. The mean AUC was 0.92 for both case (a) and control (b) within the 95% CI. (c) The decision tree generated from the filtered classifier algorithm. (d) The selection of cross-validation through the ‘one standard error’ rule where K=45 was selected. (e) Comparison table for the top biomarkers found using the different approaches
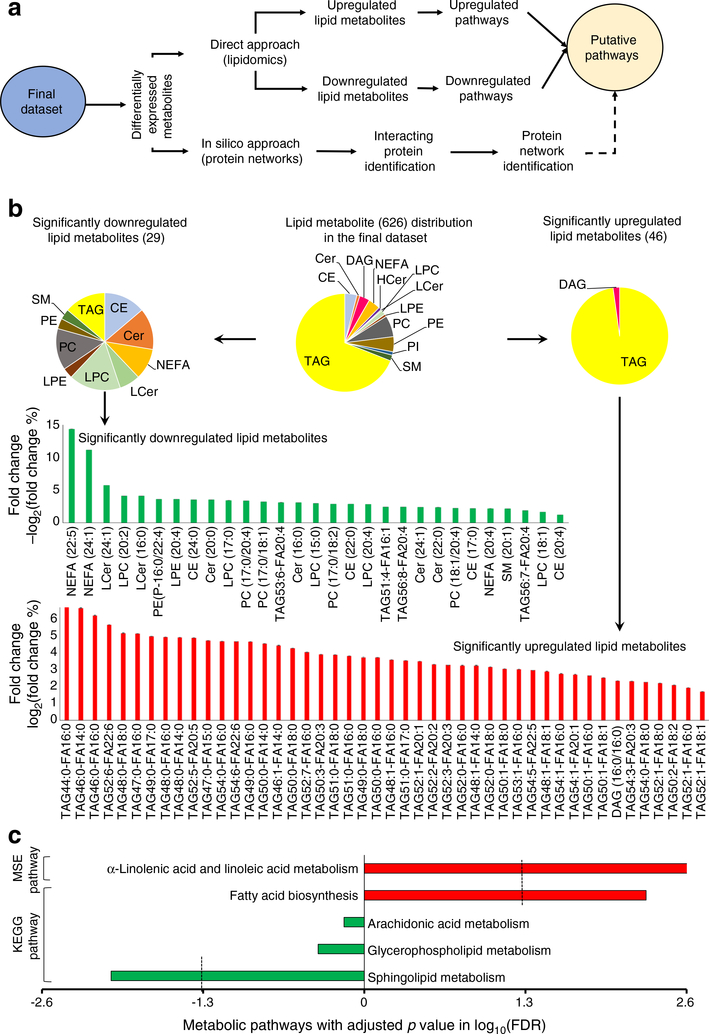
Differential expression and putative pathway analysis based on lipidomics
A total of 75 lipid metabolites were differentially expressed significantly between the case and control groups (Table 2). The putative pathway analysis (Fig. 4a) involved both a direct approach (based on differentially expressed lipids) and an in silico approach (based on the interacting putative proteins of the differentially expressed lipids). In the case group, 46 lipid metabolites were significantly upregulated and 29 were significantly downregulated (Fig. 4b). The significantly upregulated lipid metabolites were predominantly TAG lipid species whereas the significantly downregulated lipid metabolites consisted of CE, ceramide (Cer), NEFA, lactosylceramide (LCer), LPC, lysophosphatidyl-ethanolamine (LPE), PE and SM lipid species (Fig. 4b). The volcano plot for all lipid metabolites and heat map for the differentially expressed lipid metabolites are presented in ESM Fig. 2a, b. The volcano plot showed a subtle fold change between the two groups at this stage before type 2 diabetes development. The heat map of differentially expressed significant lipid metabolites showed the heterogenicity over the studied population.
Table 2.
Significantly altered lipids
| Metabolite | Log2(control) | Log2(case) | p value | FDR | % Fold change |
|---|---|---|---|---|---|
| NEFA(22:5) | 8.78 ± 0.27 | 7.52 ± 0.33 | 0.001515 | 0.033614 | −14.31743265 |
| NEFA(24:1) | 9.15 ± 0.21 | 8.13 ± 0.31 | 0.0017857 | 0.033614 | −11.12784437 |
| LCer(24:1) | 7.78 ± 0.08 | 7.34 ± 0.11 | 0.0018148 | 0.033614 | −5.753291789 |
| LPC(20:2) | 9.06 ± 0.06 | 8.69 ± 0.09 | 0.00052268 | 0.030943 | −4.138335102 |
| LCer(16:0) | 10.57 ± 0.06 | 10.13 ± 0.10 | 0.00088813 | 0.032209 | −4.127533649 |
| PE(P-16:0/22:4) | 10.67 ± 0.07 | 10.28 ± 0.10 | 0.0034702 | 0.036819 | −3.61847002 |
| LPE(20:4) | 8.91 ± 0.06 | 8.59 ± 0.07 | 0.00038527 | 0.030943 | −3.610292601 |
| CE(24:0) | 11.52 ± 0.09 | 11.11 ± 0.11 | 0.0030654 | 0.03489 | −3.547716326 |
| Cer(20:0) | 7.68 ± 0.06 | 7.40 ± 0.07 | 0.0032393 | 0.035451 | −3.546116404 |
| LPC(17:0) | 11.36 ± 0.05 | 10.97 ± 0.08 | 0.00014128 | 0.030943 | −3.457629226 |
| PC(17:0/20:4) | 11.82 ± 0.07 | 11.42 ± 0.09 | 0.002219 | 0.033614 | −3.35741171 |
| PC(17:0/18:1) | 10.47 ± 0.06 | 10.14 ± 0.08 | 0.0023824 | 0.033614 | −3.20304988 |
| TAG53:6-FA20:4 | 7.59 ± 0.05 | 7.35 ± 0.07 | 0.0047343 | 0.045298 | −3.12092987 |
| Cer(16:0) | 8.86 ± 0.05 | 8.58 ± 0.07 | 0.0059226 | 0.049437 | −3.096206958 |
| LPC(15:0) | 10.13 ± 0.05 | 9.83 ± 0.08 | 0.0024855 | 0.033825 | −2.998355538 |
| PC(17:0/18:2) | 12.05 ± 0.05 | 11.70 ± 0.08 | 0.0027807 | 0.034136 | −2.876227347 |
| CE(22:0) | 10.02 ± 0.06 | 9.74 ± 0.08 | 0.0055482 | 0.047578 | −2.875955107 |
| LPC(20:4) | 12.12 ± 0.06 | 11.78 ± 0.08 | 0.00066194 | 0.031875 | −2.833247208 |
| TAG51:4-FA16:1 | 8.74 ± 0.04 | 8.53 ± 0.05 | 0.0029816 | 0.03489 | −2.447692333 |
| TAG56:8-FA20:4 | 11.48 ± 0.07 | 11.20 ± 0.07 | 0.0054754 | 0.047578 | −2.431224594 |
| Cer(24:1) | 11.27 ± 0.05 | 10.99 ± 0.07 | 0.0025927 | 0.034136 | −2.40946479 |
| Cer(22:0) | 10.90 ± 0.05 | 10.64 ± 0.06 | 0.0021259 | 0.033614 | −2.388285567 |
| PC(18:1/20:4) | 14.44 ± 0.06 | 14.12 ± 0.07 | 0.0020069 | 0.033614 | −2.216220196 |
| CE(17:0) | 13.39 ± 0.05 | 13.10 ± 0.07 | 0.0017626 | 0.033614 | −2.180721528 |
| NEFA(20:4) | 13.86 ± 0.06 | 13.56 ± 0.08 | 0.0048608 | 0.045298 | −2.164188978 |
| SM(20:1) | 13.28 ± 0.05 | 12.99 ± 0.07 | 0.0011837 | 0.032216 | −2.158837007 |
| TAG56:7-FA20:4 | 12.64 ± 0.05 | 12.40 ± 0.06 | 0.0037147 | 0.038121 | −1.898035015 |
| LPC(18:1) | 14.57 ± 0.05 | 14.32 ± 0.07 | 0.0036636 | 0.038121 | −1.681348124 |
| CE(20:4) | 18.04 ± 0.05 | 17.82 ± 0.07 | 0.0038012 | 0.03838 | −1.222413179 |
| TAG52:1-FA18:1 | 15.17 ± 0.05 | 15.43 ± 0.07 | 0.0027793 | 0.034136 | 1.713236292 |
| TAG52:1-FA16:0 | 14.67 ± 0.06 | 14.96 ± 0.08 | 0.001687 | 0.033614 | 1.944418393 |
| TAG50:2-FA18:2 | 15.18 ± 0.06 | 15.50 ± 0.09 | 0.0032829 | 0.035451 | 2.114527145 |
| TAG52:1-FA18:0 | 14.30 ± 0.06 | 14.62 ± 0.08 | 0.0010336 | 0.032209 | 2.212793769 |
| TAG54:0-FA18:0 | 9.15 ± 0.05 | 9.36 ± 0.06 | 0.00461 | 0.045091 | 2.279869606 |
| TAG54:3-FA20:3 | 9.32 ± 0.04 | 9.54 ± 0.06 | 0.0010983 | 0.032209 | 2.335706924 |
| DAG(16:0/16:0) | 10.18 ± 0.04 | 10.42 ± 0.07 | 0.0049268 | 0.045298 | 2.353844253 |
| TAG50:1-FA18:1 | 15.73 ± 0.07 | 16.13 ± 0.10 | 0.001065 | 0.032209 | 2.532191884 |
| TAG50:1-FA16:0 | 16.44 ± 0.08 | 16.88 ± 0.11 | 0.0012743 | 0.033238 | 2.672266116 |
| TAG54:1-FA20:1 | 8.42 ± 0.05 | 8.65 ± 0.05 | 0.001354 | 0.033614 | 2.734967661 |
| TAG54:1-FA16:0 | 9.25 ± 0.04 | 9.51 ± 0.05 | 0.00017355 | 0.030943 | 2.783365984 |
| TAG48:1-FA18:1 | 13.51 ± 0.09 | 13.91 ± 0.12 | 0.0042533 | 0.042263 | 2.918331137 |
| TAG54:5-FA22:5 | 9.76 ± 0.05 | 10.05 ± 0.07 | 0.0015687 | 0.033614 | 2.983386082 |
| TAG53:1-FA16:0 | 8.74 ± 0.05 | 9.00 ± 0.06 | 0.0024163 | 0.033614 | 3.049782827 |
| TAG50:1-FA18:0 | 12.11 ± 0.07 | 12.48 ± 0.11 | 0.0030653 | 0.03489 | 3.065694283 |
| TAG52:0-FA18:0 | 11.63 ± 0.06 | 12.00 ± 0.09 | 0.00046749 | 0.030943 | 3.180424891 |
| TAG48:1-FA14:0 | 13.20 ± 0.09 | 13.63 ± 0.12 | 0.0019504 | 0.033614 | 3.263747973 |
| TAG52:0-FA16:0 | 11.01 ± 0.06 | 11.37 ± 0.08 | 0.00059315 | 0.030943 | 3.267105816 |
| TAG52:3-FA20:3 | 10.12 ± 0.06 | 10.45 ± 0.10 | 0.0032846 | 0.035451 | 3.286875028 |
| TAG52:2-FA20:2 | 9.42 ± 0.06 | 9.73 ± 0.08 | 0.0029404 | 0.03489 | 3.325497646 |
| TAG52:1-FA20:1 | 9.30 ± 0.06 | 9.61 ± 0.07 | 0.00046754 | 0.030943 | 3.498632494 |
| TAG51:0-FA17:0 | 8.45 ± 0.06 | 8.76 ± 0.08 | 0.002781 | 0.034136 | 3.554896904 |
| TAG48:1-FA16:0 | 14.10 ± 0.10 | 14.60 ± 0.14 | 0.0023153 | 0.033614 | 3.601714649 |
| TAG50:0-FA16:0 | 13.54 ± 0.08 | 14.04 ± 0.12 | 0.00058382 | 0.030943 | 3.7388893 |
| TAG49:0-FA18:0 | 7.86 ± 0.08 | 8.15 ± 0.14 | 0.0047979 | 0.045298 | 3.753109428 |
| TAG51:0-FA16:0 | 8.98 ± 0.06 | 9.33 ± 0.08 | 0.0024164 | 0.033614 | 3.834911298 |
| TAG51:0-FA18:0 | 8.30 ± 0.07 | 8.62 ± 0.08 | 0.0022509 | 0.033614 | 3.905248088 |
| TAG50:3-FA20:3 | 8.05 ± 0.05 | 8.37 ± 0.17 | 0.0049929 | 0.045298 | 3.908510498 |
| TAG52:7-FA16:0 | 8.48 ± 0.06 | 8.82 ± 0.08 | 0.00091569 | 0.032209 | 4.049985051 |
| TAG50:0-FA18:0 | 12.15 ± 0.09 | 12.67 ± 0.12 | 0.0005839 | 0.030943 | 4.28559829 |
| TAG46:1-FA14:0 | 10.96 ± 0.11 | 11.45 ± 0.15 | 0.0051951 | 0.046459 | 4.462184493 |
| TAG50:0-FA14:0 | 7.94 ± 0.06 | 8.30 ± 0.08 | 0.00042461 | 0.030943 | 4.569745222 |
| TAG49:0-FA16:0 | 10.02 ± 0.09 | 10.49 ± 0.12 | 0.0020077 | 0.033614 | 4.690186781 |
| TAG54:6-FA22:6 | 9.70 ± 0.07 | 10.16 ± 0.11 | 0.00048272 | 0.030943 | 4.693395231 |
| TAG54:0-FA16:0 | 7.06 ± 0.06 | 7.39 ± 0.06 | 0.00012953 | 0.030943 | 4.706367554 |
| TAG47:0-FA15:0 | 8.07 ± 0.08 | 8.45 ± 0.15 | 0.0053337 | 0.047027 | 4.740641233 |
| TAG52:5-FA20:5 | 8.51 ± 0.06 | 8.92 ± 0.10 | 0.0022509 | 0.033614 | 4.912457436 |
| TAG48:0-FA14:0 | 10.32 ± 0.10 | 10.83 ± 0.13 | 0.001115 | 0.032209 | 4.935500085 |
| TAG48:0-FA16:0 | 13.19 ± 0.11 | 13.85 ± 0.16 | 0.0010184 | 0.032209 | 4.962787325 |
| TAG49:0-FA17:0 | 8.85 ± 0.08 | 9.29 ± 0.11 | 0.0021262 | 0.033614 | 5.001577158 |
| TAG47:0-FA16:0 | 9.16 ± 0.09 | 9.63 ± 0.12 | 0.0027423 | 0.034136 | 5.16498652 |
| TAG48:0-FA18:0 | 9.94 ± 0.10 | 10.46 ± 0.13 | 0.0020367 | 0.033614 | 5.194763489 |
| TAG52:6-FA22:6 | 7.90 ± 0.07 | 8.35 ± 0.10 | 0.00074973 | 0.032209 | 5.694221781 |
| TAG46:0-FA16:0 | 11.35 ± 0.14 | 12.06 ± 0.19 | 0.0022508 | 0.033614 | 6.258605708 |
| TAG46:0-FA14:0 | 10.46 ± 0.13 | 11.16 ± 0.17 | 0.001132 | 0.032209 | 6.695044875 |
| TAG44:0-FA16:0 | 9.52 ± 0.14 | 10.17 ± 0.17 | 0.0059229 | 0.049437 | 6.783708462 |
Data are shown as mean ± SE, presented in ascending order of % fold change
DAG, diacylglycerol; FA, fatty acid
Fig. 4.
The putative pathway analysis for the development of type 2 diabetes. (a) Schematic flow diagram of the putative pathway analysis. (b) The distribution of the differentially expressed lipid species (75) within the final dataset (626); the bar graphs show the binary logarithm of fold changes (case/control) of all significant metabolites with ± SEM. (c) Pathway analysis: metabolite set enrichment (MSE) analysis based on FDR <0.05 (−log10 of FDR <1.3) and KEGG pathway analysis based on FDR <0.05 (−log10 of FDR <1.3). Red bars, upregulation; green bars, downregulation. HCer, hexosylceramide
To identify lipid pathways associated with altered lipid metabolites, KEGG pathway analysis was carried out. A significant downregulation of sphingolipid metabolism (FDR 0.009) and upregulation of fatty acid biosynthesis (FDR 0.005) (Fig. 4c) was observed. To understand the predicted consequence of such modulation, metabolite set enrichment analysis was performed. The analysis identified the upregulation of α-linolenic acid and linoleic acid metabolism (FDR 0.002) as the predicted net consequence of upregulated fatty acid biosynthesis (Fig. 4c). The lipid metabolites belonging to the identified different pathways are summarised in ESM Fig. 3a. The upregulated fatty acid synthesis was identified due to the significantly higher concentrations of myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0) and oleic acid (C18:1). The discovery of upregulated α-linolenic acid and linoleic acid metabolism was based on the significantly higher concentrations of linoleic acid (C18:2), dihomo-γlinoleic acid (C20:3), eicosapentaenoic acid (C20:5) and docosahexaenoic acid (C22:5). In the case of downregulated sphingolipid metabolism, a number of significantly decreased ceramides [Cer(16:0), Cer(20:0), Cer(22:0) and Cer(24:1)], lactosylceramides [LCer(16:0), LCer(24:1)] and sphingomyelin [SM(20:1)] species were identified. The specific alterations in these pathways were linked to increased type 2 diabetes risk (ESM Fig. 3a).
Using an in silico approach employing KEGG pathway mapping (ESM Fig. 3b), we identified the upregulation of specific inflammation pathways (loci-1) and the downregulation of sphingolipid metabolism and related pathways (loci-4) as the dominant changes associated with future type 2 diabetes status. Loci-2, the upregulated fatty acid biosynthesis, was found between the connectomes of loci-1 and loci-4. Additionally, the downregulated glycosylphosphatidylinositol (GPI) anchor biosynthesis (loci-3) represents an island locus. GPI proteins are essential for Cer-remodelling and transportation of Cers from the endoplasmic reticulum to the Golgi apparatus where glycosphingolipids and sphingomyelins are formed [40].
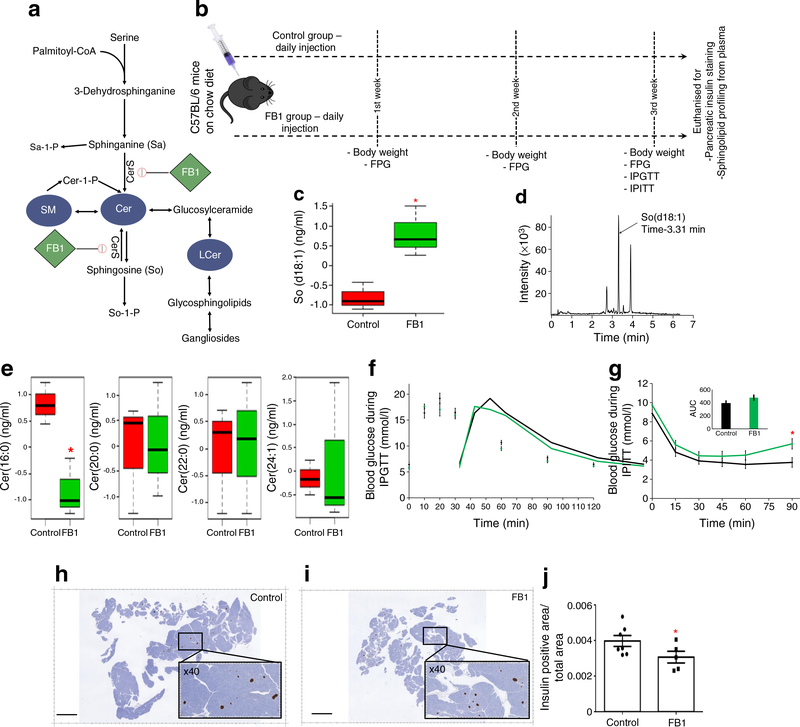
In vivo inhibition of sphingolipid metabolism
Our population-based lipidomics data indicate that a number of Cers, SMs and LCers are significantly downregulated years before type2 diabetes onset(Fig. 4b), suggesting that the down regulation of sphingolipid metabolism could be in part responsible for the future onset of type 2 diabetes among women with previous GDM. To investigate this possibility, an approach was taken to inhibit sphingolipid metabolism. FB1, a pharmacological inhibitor of sphingolipid biosynthesis, was used to induce overall downregulation of sphingolipid metabolism in C57BL/6 mice (n≥ 14). Due to the very short half-life of FB1 (liver 4.07 h, kidney 7.07 h, plasma 3.15 h [42]), our treatment could only transiently block sphingolipid metabolism. This transient downregulation of sphingolipid metabolism was chosen to depict the very early stage of type 2 diabetes pathophysiology. Figure 5a illustrates the sphingolipid metabolism pathway as a target of these inhibitors, with FB1 (1 mg/kg) being delivered intraperitoneally to mice as depicted in Fig. 5b. Serum samples were collected at the end of the treatment and sphingolipid species were profiled by MS (n = 3 per group). The FB1-treated mice showed significant accumulation of sphingosine (So) species So(d18:1) (Fig. 5c, d). In the SWIFT cohort lipidomics study, four Cers—Cer(16:0), Cer(20:0), Cer(22:0) and Cer(24:1)—were found to be significantly downregulated. In the FB1-treated mice, although levels of these four lipid metabolites decreased, the decrease was statistically significant only for Cer (16:0) (Fig. 5e).
Fig. 5.
In vivo functional studies. (a) Schematic flow diagram of the sphingolipid metabolism pathway showing targets of FB1 (pharmacological inhibitor). (b) The in vivo study design (n≥14): the control group of mice was injected with vehicle while the treatment group was injected with FB1 (1 mg/kg) daily. Every week, the weight gain and the FPG were monitored. At the end of the third week, GTT and ITT were performed. Finally, all mice were euthanised to collect whole pancreases and plasma. (c) So concentration in control and FB1-treated mice (n=3). (d) Representative chromatogram of So. (e) Comparison of the four Cer species found to significantly differ in the SWIFT cohort (values were mean-centred [n=3] and divided by the SD of each variable). In the boxplots (c, e), the bottom and top of the box are the Q1 and Q3 (25th and 75th percentile), respectively, and the central band is the median (Q2 or 50th percentile). The bottom whisker is located within 1.5 IQR of the lower quartile, and the upper whisker is located within 1.5 IQR of the upper quartile. (f) GTT single time point comparison between control (black line) and FB1 group (green line) at the end of 3 weeks treatment (n≥7). (g) ITT single time point comparison between control (black line) and FB1 group (green line) at the end of 3 weeks treatment (n≥7); inset shows AUC (mmol/l × min). (h, i) Representative insulin-stained pancreas (5 μm thickness, longitudinally sectioned through the pancreatic headto-tail axis) from control (h) and FB1-treated mice (i); scale bars, 3 mm; insets show ×40 magnification. (j) Insulin-positive area in pancreases of control and FB1-treated mice (n≥5). A two-tailed, unpaired t test was carried out for each comparison. Data are presented as mean ± SEM; unadjusted p values: *p<0.05 vs control
Effects of downregulation of sphingolipid metabolism on glucose homeostasis
At the end of the 3 weeks of treatment, mice (n ≥ 14) were evaluated for weight gain, FPG, fasting insulin and OGTT and ITT were performed. No significant difference were observed between control and treatment groups for weight gain, FPG and fasting insulin (ESM Fig. 4a–c). During the GTT, no difference in blood glucose was observed when comparing control and FB1-treated mice (Fig. 5f). During the ITT, the treatment group (FB1) showed overall reduced responsiveness to insulin in comparison with the control group, most notably (significant) during the later stages of the ITT (Fig. 5g). Interestingly, the islets in the pancreas of FB1-treated mice (n ≥ 5) displayed a small but significant reduction in the insulin-positive area compared with the control mouse islets (Fig. 5h–j).
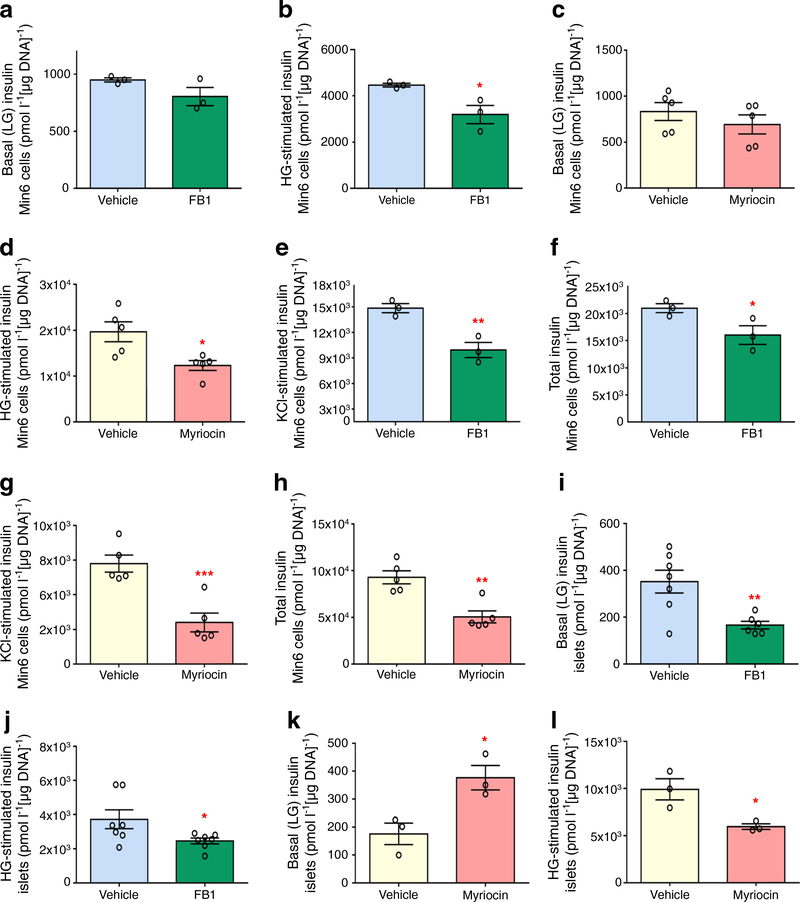
Pancreatic beta cell function in vitro in response to sphingolipid metabolism downregulation
To assess the effects of downregulated sphingolipid metabolism on beta cell function and insulin secretion more directly, murine (C57B/L6) islets and Min6 K8 cells were treated in vitro with either FB1 (1 μmol/l) or a second inhibitor myriocin (50 nmol/l) and GSIS was assessed (Fig. 6). In Min6 K8 cells, both inhibitors significantly decreased GSIS without affecting basal (low glucose) insulin secretion (Fig. 6a–d). The inhibitors also significantly decreased insulin secretion in response to cell depolarisation with KCl (Fig. 6e, g) and decreased total insulin content in Min6 K8 cells (Fig. 6f, h). In murine islets, both inhibitors significantly decreased GSIS (Fig. 6j, l). Moreover, myriocin caused a significant increase in basal insulin secretion (Fig. 6k). In murine islets, neither KCl-stimulated insulin secretion nor total insulin content were significantly altered by either treatment (data not shown).
Fig. 6.
GSIS studies in vitro. (a–h) In Min6 K8 cells, FB1 treatment (green) did not alter basal (LG) insulin secretion (a) but significantly decreased GSIS (high glucose [HG]-stimulated) (b). Myriocin treatment (pink) did not alter basal insulin secretion (c) but significantly decreased GSIS (d). FB1 treatment significantly decreased KCl-stimulated insulin secretion (e) and total insulin (f). Myriocin treatment significantly decreased both KCl-stimulated insulin secretion (g) and total insulin (h). In Min6 K8 cells, 0 mmol/l glucose was used for LG and 10 mmol/l glucose was used in HG stimulation. For KCl stimulation, 25 mmol/l KCl was added to HG solution. (i–l) In murine islets, FB1 treatment significantly decreased both basal insulin secretion (i) and GSIS (j). Myriocin treatment significantly increased basal insulin secretion (k) and significantly decreased GSIS (l). In murine islets, 2.8 mmol/l glucose was used for LG and 16.7 mmol/l glucose was used in HG stimulation. For KCl stimulation, 25 mmol/l KCl was added to HG solution. Vehicle included 0.04% (v/v) DMSO for FB1 treatments (blue) or 0.0001 (v/v) DMSO for myriocin treatments (white). Data are presented as mean ± SEM (n=3 for FB1 in Min6 cells, n=5 for myriocin in Min6 cells, n≥6 for FB1 in C57BL/6 murine islets, n=3 for myriocin in C57BL/6 murine islets). A two-tailed, unpaired t test was carried out for each comparison (unadjusted p values: *p<0.05, **p<0.01, ***p<0.001 vs vehicle)
Discussion
By employing artificial intelligence-based machine learning, we identified a predictive signature with an overall discriminating power (AUC) of0.92with 91% accuracy. The accuracy of this predictive signature is not compromised by either sensitivity (87%) or specificity (93%). This accuracy is better than that provided by well-known clinical diagnostics, including fasting glucose, 2 h post-load glucose in 75 g OGTT, HOMA-IR, family history of diabetes and type of GDM treatment, as well as that reported in some recently published metabolomics-based diagnostic studies [19, 43, 44]. Moreover, unlike other signatures [19, 28], a strength of our predictive signature is that it does not rely on clinical variables since case and control participants were matched on early postpartum glucose tolerance (normal or impaired), age and BMI to reduce confounding of metabolite prediction by these clinical risk factors. The strong suit of the signature was the 45-fold cross-validation under a high confidence threshold (1.0) and binary output, which together minimise the chance ofdata overfitting and bias selection.This protocol ensures the reliability of this signature in making a predictive decision for any unknown blood sample. However, this predictive signature applies specifically to Hispanic and Asian women in predicting early progression to type 2 diabetes within 2 years following GDM pregnancy. Only two racial and ethnic groups were selected for this study, to achieve sample homogeneity. In future, these analyses may be extended to other race groups in the SWIFT cohort, in order to test the signature’s ability to predict progression to overt diabetes after GDM pregnancy within a much longer follow-up period of 10 years.
For the first time in a population-based study, we identified downregulation of sphingolipid metabolism as an antecedent early-stage event in women with previous GDM who developed type 2 diabetes (Fig. 4), together with other known pathways (e.g. upregulated fatty acid biosynthesis and upregulated α-linolenic acid and linoleic acid metabolism). Downregulated sphingolipid metabolism was identified based on a number of significantly downregulated nodes in the pathway (Table 2 and Fig. 4b). However, several cross-sectional clinical studies have shown that Cer levels (a single upstream node of the whole pathway) are higher in obese individuals with type 2 diabetes [45, 46]. These studies evaluated obesity as a covariant in their analyses. However, in this study, obesity was controlled by pair-matching of BMI between groups. Moreover, we employed a prospective postpartum GDM cohort, leaving open the possibility that some nodes of sphingolipid metabolism may arise after disease onset.
To understand the role of sphingolipid metabolism in the early-stage pathophysiology of type 2 diabetes, we used FB1 to inhibit de novo sphingolipid biosynthesis transiently in mice without high-fat diet intervention. The in vivo studies showed that transient inhibition of sphingolipid metabolism has no significant effect on insulin sensitivity, except in the late-phase (indicating disrupted hepatic glucose uptake and/or high gluconeogenesis) in the treatment group. However, this modulation of sphingolipid metabolism appeared to reduce pancreatic beta cell area. Further studies are required to determine whether this impairment of insulin biosynthesis will eventually lead to glucose intolerance in the long term.
The role of downregulated sphingolipid metabolism in overt type 2 diabetes phenotypes has been studied. Park et al [47] showed that Cer synthase 2 null mice with impaired synthesis of sphingolipids C22–24 develop glucose intolerance due to abrogated Akt phosphorylation of the insulin receptor in the liver. Alexaki et al [48] showed that adipocyte-specific Sptlc1-knockout mice exhibit insulin resistance with agedependent loss of adipose tissue, increased macrophage infiltration and tissue fibrosis. Furthermore, Lee et al [49] showed that adipocyte-specific Sptlc2-knockout mice display systemic insulin resistanceandhyperglycaemia. Taken together with our observations, chronic sphingolipid metabolism downregulation could thus potentially interfere with liver, muscle, adipose and beta cell function, contributing to type 2 diabetes onset.
The inhibition of sphingomyelin synthase in INS-1 beta cells significantly reduced insulin exocytosis [50]. Kavishwar and Moore [51] identified sphingolipid patches on the surfaces of pancreatic beta cells as a predictor of their functional capacity; the patches decreased in diabetes, suggesting the importance of sphingolipids in this cell type. In this study, both FB1 and myriocin decreased GSIS. Moreover, myriocin treatment yielded significantly increased basal insulin secretion in murine islets. Furthermore, downregulation of sphingolipid metabolism reduced insulin content. Although both FB1 and myriocin showed similar effects on GSIS in vitro, potential noise from off-target effects of these two inhibitors cannot be ruled out. Stanford et al [52] reported similar results (i.e. decreased GSIS in Min6 cells and murine islets) after inhibiting specific components of sphingolipid metabolism. Recently Ye et al [53] showed that during diet-induced obesity, mice with knockout of pancreatic beta cellspecific LDL receptor-relatedprotein 1 (apleiotropic mediator of cholesterol, insulin, energy metabolism and other cellular processes) were unable to compensate beta cell function partly due todownregulation ofsphingolipidmetabolism. Therefore, downregulated sphingolipid metabolism may play a causal role in pancreatic beta cell dysfunction.
Supplementary Material
Acknowledgments
Funding These studies are supported by Canadian Institutes of Health Research (CIHR), FRN 143219 (MBW). and National Institute of Child Health and Human Development (NICHD) R01 HD050625 (EPG). SRK is supported by a Diabetes Canada post-doctoral fellowship.
Abbreviations
- BCAA
Branched-chain amino acid
- CE
Cholesteryl ester
- Cer
Ceramide
- FB1
Fumonisin B1
- FC
Filtered classifier
- FDR
False discovery rate
- FPG
Fasting plasma glucose
- GDM
Gestational diabetes
- GPI
Glycosylphosphatidylinositol
- GSIS
Glucose-stimulated insulin secretion
- IQR
Interquartile range
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LCer
Lactosylceramide
- LPC
Lysophosphatidylcholine
- LPE
Lysophosphatidylethanolamine
- MLR
Multiple logistic regression
- PC
Phosphatidylcholine
- PCA
Principal component analysis
- PE
Phosphatidylethanolamine
- PLS-DA
Partial least squares-discriminant analysis
- ROC
Receiver operating characteristic
- SM
Sphingomyelin
- So
Sphingosine
- SWIFT
Study of Women, Infant Feeding and Type 2 Diabetes after GDM
- TAG
Triacylglycerol
Footnotes
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Data availability The data are available on request from the authors.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00125-018-4800-2) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations
References
- 1.Koning SH, Hoogenberg K, Lutgers HL, van den Berg PP, Wolffenbuttel BHR (2016) Gestational Diabetes Mellitus:current knowledge and unmet needs. J Diabetes 8(6):770–781. 10.1111/1753-0407.12422 [DOI] [PubMed] [Google Scholar]
- 2.Hunt KJ, Schuller KL (2007) The increasing prevalence of diabetes in pregnancy. Obstet Gynecol Clin N Am 34(2):173–199, vii. 10.1016/j.ogc.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Idris N, Wong SF, Thomae M, Gardener G, McIntyre DH (2010) Influence of polyhydramnios on perinatal outcome in pregestational diabetic pregnancies. Ultrasound Obstet Gynecol 36(3):338–343. 10.1002/uog.7676 [DOI] [PubMed] [Google Scholar]
- 4.Metzger BE, Buchanan TA, Coustan DR et al. (2007) Summary and Recommendations of the Fifth International Workshop-Conference on Gestational Diabetes Mellitus. Diabetes Care 30(Supplement 2): S251–S260. 10.2337/dc07-s225 [DOI] [PubMed] [Google Scholar]
- 5.Gunderson EP, Lewis CE, Tsai AL et al. (2007) A 20-year prospective study of childbearing and incidence of diabetes in young women, controlling for glycemia before conception: the Coronary Artery Risk Development in Young Adults (CARDIA) Study. Diabetes 56(12):2990–2996. 10.2337/db07-1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim C, Newton KM, Knopp RH (2002) Gestational diabetes and the incidence of type 2 diabetes. A systematic review. Diabetes Care 25:1862–1868 [DOI] [PubMed] [Google Scholar]
- 7.Bellamy L, Casas JP, Hingorani AD, Williams D (2009) Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet 373(9677):1773–1779. 10.1016/S0140-6736(09)60731-5 [DOI] [PubMed] [Google Scholar]
- 8.Magee MS, Walden CE, Benedetti TJ, Knopp RH (1993) Influence of diagnostic criteria on the incidence of gestational diabetes and perinatal morbidity. JAMA 269(5):609–615. 10.1001/jama.1993.03500050087031 [DOI] [PubMed] [Google Scholar]
- 9.Beharier O, Shoham-Vardi I, Pariente G et al. (2015) Gestational diabetes mellitus is a significant risk factor for long-term maternal renal disease. J Clin Endocrinol Metab 100(4):1412–1416. 10.1210/jc.2014-4474 [DOI] [PubMed] [Google Scholar]
- 10.Shah BR, Retnakaran R, Booth GL (2008) Increased risk of cardiovascular disease in young women following gestational diabetes mellitus. Diabetes Care 31(8):1668–1669. 10.2337/dc08-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Retnakaran R, Shah BR (2017) Role of type 2 diabetes in determining retinal, renal, and cardiovascular outcomes in women with previous gestational diabetes mellitus. Diabetes Care 40(1):101–108. 10.2337/dc16-1400 [DOI] [PubMed] [Google Scholar]
- 12.Fadl H, Magnuson A, Östlund I, Montgomery S, Hanson U, Schwarcz E (2014) Gestational diabetes mellitus and later cardiovascular disease: a Swedish population based case-control study. BJOG 121(12):1530–1536. 10.1111/1471-0528.12754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunderson EP, Jaffe MG (2017) Pregnancy and subsequent glucose intolerance in women of childbearing age. JAMA Intern Med 177(12):1742–1744. 10.1001/jamainternmed.2017.4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobias DK, Stuart JJ, Li S et al. (2017) Association of history of gestational diabetes with long-term cardiovascular disease risk in a large prospective cohort of US women. JAMA Intern Med 177(12): 1735–1742. 10.1001/jamainternmed.2017.2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ajmera VH, Gunderson EP, VanWagner LB, Lewis CE, Carr JJ, Terrault NA (2016) Gestational diabetes mellitus is strongly associated with non-alcoholic fatty liver disease. Am J Gastroenterol 111(5):658–664. 10.1038/ajg.2016.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.American Diabetes Association (2016) Management of diabetes in pregnancy. Diabetes Care 39(Suppl 1):S94–S98. 10.2337/dc16-S015 [DOI] [PubMed] [Google Scholar]
- 17.Janghorbani M, Zinab Almasi S, Amini M (2015) The product of triglycerides and glucose in comparison with fasting plasma glucose did not improve diabetes prediction. Acta Diabetol 52:781–788 [DOI] [PubMed] [Google Scholar]
- 18.Abdul-Ghani MA, Lyssenko V, Tuomi T, DeFronzo RA, Groop L (2009) Fasting versus postload plasma glucose concentration and the risk for future type 2 diabetes. Results from the Botnia Study. Diabetes Care 32:281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allalou A, Nalla A, Prentice KJ et al. (2016) A predictive metabolic signature for the transition from gestational diabetes to type 2 diabetes. Diabetes 65(9):2529–2539. 10.2337/db151720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blatt AJ, Nakamoto JM, Kaufman HW (2011) Gaps in diabetes screening during pregnancy and postpartum. Obstet Gynecol 117(1):61–68. 10.1097/AOG.0b013e3181fe424b [DOI] [PubMed] [Google Scholar]
- 21.Bennett WL, Ennen CS, Carrese JA et al. (2011) Barriers to and facilitators of postpartum follow-up care in women with recent gestational diabetes mellitus: a qualitative study. J Women’s Health 20(2):239–245. 10.1089/jwh.2010.2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell MA, Phipps MG, Olson CL, Welch HG, Carpenter MW (2006) Rates of postpartum glucose testing after gestational diabetes mellitus. Obstet Gynecol 108(6):1456–1462. 10.1097/01.AOG.0000245446.85868.73 [DOI] [PubMed] [Google Scholar]
- 23.Jones EJ, Roche CC, Appel SJ (2009) A review of the health beliefs and lifestyle behaviors of women with previous gestational diabetes. J Obstet Gynecol Neonatal Nurs 38(5):516–526. 10.1111/j.1552-6909.2009.01051.x [DOI] [PubMed] [Google Scholar]
- 24.Kim C, McEwen LN, Piette JD, Goewey J, Ferrara A, Walker EA (2007) Risk perception for diabetes among women with histories of gestational diabetes mellitus. Diabetes Care 30(9):2281–2286. 10.2337/dc07-0618 [DOI] [PubMed] [Google Scholar]
- 25.Menni C, Fauman E, Erte I et al. (2013) Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62(12):4270–4276. 10.2337/db13-0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang TJ, Larson MG, Vasan RS et al. (2011) Metabolite profiles and the risk of developing diabetes. Nat Med 17(4):448–453. 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen T, Ni Y, Ma X et al. (2016) Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Sci. Rep. 6:20594 10.1038/srep20594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lappas M, Mundra PA, Wong G et al. (2015) The prediction of type 2 diabetes in women with previous gestational diabetes mellitus using lipidomics. Diabetologia 58(7):1436–1442. 10.1007/s00125-015-3587-7 [DOI] [PubMed] [Google Scholar]
- 29.Wang-Sattler R, Yu Z, Herder C et al. (2012) Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol 8:615 10.1038/msb.2012.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aichler M, Borgmann D, Krumsiek J et al. (2017) N-acyl taurines and acylcarnitines cause an imbalance in insulin synthesis and secretion provoking beta-cell dysfunction in type 2 diabetes. Cell Metab 25(6):1334–1347.e1334. 10.1016/j.cmet.2017.04.012 [DOI] [PubMed] [Google Scholar]
- 31.Lynch CJ, Adams SH (2014) Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 10(12): 723–736. 10.1038/nrendo.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floegel A, Stefan N, Yu Z et al. (2013) Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 62(2):639–648. 10.2337/db12-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole LK, Vance JE, Vance DE (2012) Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim Biophys Acta 1821(5): 754–761. 10.1016/j.bbalip.2011.09.009 [DOI] [PubMed] [Google Scholar]
- 34.Gunderson EP, Matias SL, Hurston SR et al. (2011) Study of Women, Infant feeding, and Type 2 diabetes mellitus after GDM pregnancy (SWIFT), a prospective cohort study: methodology and design. BMC Public Health 11:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunderson EP, Hedderson MM, Chiang V et al. (2012) Lactation intensity and postpartum maternal glucose tolerance and insulin resistance in women with recent GDM: The SWIFT cohort. Diabetes Care 35(1):50–56. 10.2337/dc11-1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunderson EP, Hurston SR, Ning X et al. (2015) Lactation and progression to type 2 diabetes mellitus after gestational diabetes mellitus: a prospective cohort study. Ann Intern Med 163(12): 889–898. 10.7326/M15-0807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai FF, Bhattacharjee A, Liu Y et al. (2015) A novel GLP1 receptor interacting protein ATP6ap2 regulates insulin secretion in pancreatic beta cells. J Biol Chem 290(41):25045–25061. 10.1074/jbc.M115.648592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou D, Ye Y, Zou N, Yu J (2017) Analysis of risk factors and their interactions in type 2 diabetes mellitus: A cross-sectional survey in Guilin, China. J Diabetes Investig 8(2):188–194. 10.1111/jdi.12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berk KA, Mulder MT, Verhoeven AJM et al. (2016) Predictors of diet-induced weight loss in overweight adults with type 2 diabetes. PLoS One 11(8):e0160774 10.1371/journal.pone.0160774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gault CR, Obeid LM, Hannun YA (2010) An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol 688:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prentice KJ, Luu L, Allister EM et al. (2014) The furan fatty acid metabolite CMPF is elevated in diabetes and induces beta cell dysfunction. Cell Metab 19(4):653–666. 10.1016/j.cmet.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Larranaga MR, Anadon A, Diaz MJ et al. (1999) Toxicokinetics and oral bioavailability of fumonisin B1. Vet Hum Toxicol 41(6):357–362 [PubMed] [Google Scholar]
- 43.Liu J, Semiz S, van der Lee SJ et al. (2017) Metabolomics based markers predict type 2 diabetes in a 14-year follow-up study. Metabolomics 13(9):104 10.1007/s11306-0171239-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter TC, Rein D, Padberg I et al. (2016) Validation of a metabolite panel for early diagnosis of type 2 diabetes. Metabolism 65(9): 1399–1408. 10.1016/j.metabol.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haus JM, Kashyap SR, Kasumov T et al. (2009) Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes 58(2):337–343. 10.2337/db08-1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez X, Goldfine AB, Holland WL, Gordillo R, Scherer PE (2013) Plasma ceramides are elevated in female children and adolescents with type 2 diabetes. J Pediatr Endocrinol Metab 26(9–10): 995–998. 10.1515/jpem-2012-0407 [DOI] [PubMed] [Google Scholar]
- 47.Park JW, Park WJ, Kuperman Y, Boura-Halfon S, Pewzner-Jung Y, Futerman AH (2013) Ablation of very long acyl chain sphingolipids causes hepatic insulin resistance in mice due to altered detergent-resistant membranes. Hepatology 57(2):525–532. 10.1002/hep.26015 [DOI] [PubMed] [Google Scholar]
- 48.Alexaki A, Clarke BA, Gavrilova O et al. (2017) De novo sphingolipid biosynthesis is required for adipocyte survival and metabolic homeostasis. J Biol Chem 292(9):3929–3939. 10.1074/jbc.M116.756460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S-Y, Lee H-Y, Song J-H et al. (2017) Adipocyte-specific deficiency of de novo sphingolipid biosynthesis leads to lipodystrophy and insulin resistance. Diabetes 66(10):2596–2609. 10.2337/db16-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subathra M, Qureshi A, Luberto C (2011) Sphingomyelin synthases regulate protein trafficking and secretion. PLoS One 6(9):e23644 10.1371/journal.pone.0023644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kavishwar A, Moore A (2013) Sphingomyelin patches on pancreatic beta-cells are indicative of insulin secretory capacity. J Histochem Cytochem 61(12):910–919. 10.1369/0022155413502792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cantrell Stanford J, Morris AJ, Sunkara M, Popa GJ, Larson KL, Özcan S (2012) Sphingosine 1-phosphate (S1P) regulates glucose-stimulated insulin secretion in pancreatic beta cells. J Biol Chem 287(16):13457–13464. 10.1074/jbc.M111.268185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye R, Gordillo R, Shao M et al. (2018) Intracellular lipid metabolism impairs β cell compensation during diet-induced obesity. J Clin Invest 128(3):1178–1189. 10.1172/JCI97702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.