Abstract
Heterozygous mutations in the X-linked gene CASK are associated with intellectual disability, microcephaly, pontocerebellar hypoplasia, optic nerve hypoplasia and partially penetrant seizures in girls. The Cask+/− heterozygous knockout female mouse phenocopies the human disorder and exhibits postnatal microencephaly, cerebellar hypoplasia and optic nerve hypoplasia. It is not known if Cask+/− mice also display seizures, nor is known the molecular mechanism by which CASK haploinsufficiency produces the numerous documented phenotypes. 24-hour video electroencephalography demonstrates that despite sporadic seizure activity, the overall electrographic patterns remain unaltered in Cask+/− mice. Additionally, seizure threshold to the commonly used kindling agent, pentylenetetrazol, remains unaltered in Cask+/− mice, indicating that even in mice the seizure phenotype is only partially penetrant and may have an indirect mechanism. RNA sequencing experiments on Cask+/− mouse brain uncovers a very limited number of changes, with most differences arising in the transcripts of extracellular matrix proteins and the transcripts of a group of nuclear proteins. In contrast to limited changes at the transcript level, quantitative whole-brain proteomics using iTRAQ quantitative mass-spectrometry reveals major changes in synaptic, metabolic/mitochondrial, cytoskeletal, and protein metabolic pathways. Unbiased protein-protein interaction mapping using affinity chromatography demonstrates that CASK may form complexes with proteins belonging to the same functional groups in which altered protein levels are observed. We discuss the mechanism of the observed changes in the context of known molecular function/s of CASK. Overall, our data indicate that the phenotypic spectrum of female Cask+/− mice includes sporadic seizures and thus closely parallels that of CASK haploinsufficient girls; the Cask+/− mouse is thus a face-validated model for CASK-related pathologies. We therefore surmise that CASK haploinsufficiency is likely to affect brain structure and function due to dysregulation of several cellular pathways including synaptic signaling and cellular metabolism.
Keywords: CASK, EEG, Proteomics, iTraq, microcephaly, synapse, mitochondria, ribosome
Introduction
Heterozygous mutations in the X-linked gene CASK are associated with mental retardation and microcephaly with pontine and cerebellar hypoplasia in girls (MICPCH; OMIM #300749) ([1-5]). CASK mutations in boys can produce neurodevastating conditions with epileptic encephalopathies [6,7], but some CASK missense mutations in boys are milder and are commonly found in cases of X-linked intellectual disability in normocephalic boys [8]. The molecular function of CASK and the mechanism of CASK-linked disorders remain unknown.
CASK (calcium/calmodulin activated serine kinase) is a multi-domain protein belonging to the MAGUK (membrane associated guanylate kinase) class of scaffolding proteins. From N-to C-terminus, CASK is comprised of a CaMK (calcium/calmodulin dependent kinase) domain, two L27 (lin-2/lin-7) domains, a PDZ (PSD95, dig-1 and ZO1) domain, SH3 (Src homology 3) domain and a GuK (guanylate kinase) domain [9-11]. Proposed functions for CASK have included, among others, a role as a presynaptic scaffolding molecule [9], involvement in Tbr1-mediated transcription [12], establishing cell polarity [13], and trafficking of ion channels [14, 15]. Deletion of CASK does not alter active or passive membrane properties of neurons or cell polarity [16-18], indicating that there may be molecular redundancy in these CASK functions. Additionally, Cask mutations do not alter neuronal migration in mice, suggesting that the Tbr-1/reelin pathway is not the cause of the observed phenotypes [17,19,18,20]. Although synapse formation itself remains unaltered in Cask mutant mice [17], we have observed a reduction in the number of active zones within the large retinogeniculate synapses of Cask+/− mice [21]. Biochemically, CASK interacts with and phosphorylates the cytosolic tail of neurexins via its PDZ domain. CASK also links neurexins to presynaptic scaffolding molecules such as Mint1, Caskin and the active zone organizer liprins-α [22]. In addition to neurexins, the PDZ domain of CASK may also interact with other proteins such as syndecan2, syncam/IGSF4 and parkin, as well as phospholipids [23-26].
CASK is an evolutionarily conserved protein [27,28], and an unbiased analysis of CASK-interacting proteins in Drosophila melanogaster revealed that CASK is also likely to interact with a set of mitochondrial proteins [29]. Using a murine model, we have previously demonstrated that the heterozygous deletion of CASK in female mice is sufficient to produce postnatal progressive microcephaly, cerebellar hypoplasia and optic nerve hypoplasia [21,20], thereby phenocopying the human disorder. Here we use this face-validated model to examine underlying mechanisms of the observed phenotypes. Constitutive CASK deletion in mice does not affect overall core neuronal functions but performs a selectively essential role [17]. Specifically, CASK seems to play an evolutionarily conserved role in postnatal brain growth in mammals. To uncover the molecular role/s of CASK, in this study we performed unbiased analyses of electrographic changes and molecular changes present in the whole brain of CASK haploinsufficient adult mice. We also performed an unbiased protein interaction mapping of CASK to understand the mechanism/s of these molecular changes. Our data indicate that CASK may participate in multiple cellular pathways, including mitochondrial, synaptic and protein metabolism. Dysfunction of these cellular functions is likely to underlie the complex neurological condition associated with CASK loss-of-function.
Materials and Methods
Study Approval - Procedures Involving Animals
All procedures involving the use of animals were approved by the Institutional Animal Care and Use Committee of the University of Virginia and the Institutional Animal Care and Use Committee of Virginia Tech.
Sexually mature adult animals were used for RNA sequencing (ages 2-4 months) and proteomics (P40), since the postnatal progressive microcephaly associated with CASK heterozygosity has stabilized by this age. EEG experiments were conducted at either P7-P14 for pups or between 2 and 4 months of age for adults. The age range for pups (P7-P14) was chosen since this stage of murine development is considered equivalent to human infancy; between 2 and 4 months was chosen for adult mice so as to observe changes between genotypes once the microcephalic phenotype has plateaued. Cask+/− mutants and Cask+/+ wild-type (WT) littermates are both from a C57BL/6J background and were backcrossed for at least 25 generations. Mice were genotyped using polymerase chain reaction followed by gel electrophoresis with primers targeted to the exon 1-intron 1 boundary of CASK.
Electrode Implantation Surgery and EEG Data Acquisition
Pups
Pups (P7-P14) were anesthetized using 5% isoflurane for induction and 2-2.5% isoflurane for maintenance adapted from a previously published protocol [31]. Unipolar insulated stainless steel depth electrodes were stereotaxically implanted in the right hippocampus and right parietal cortex, with two reference electrodes on the left parietal cortex and the cerebellum. After the surgery, pups were allowed to recover for 24 hours with the mother. The next day, the pups were connected to a video-EEG monitoring system via a flexible cable connected to the amplifier. The output signal was amplified using a Grass amplifier (Model1, Natus Neurology Incorporated-Grass Products, Warwick, RI), digitized and recorded for later review using Labchart software (ADInstruments, Dunedin, New Zealand).
Adults
Female C57BL/6J mice of each genotype (four WT and three Cask+/−) between 68 and 93 days old were initially anesthetized using 5% isoflurane and head-fixed in a stereotaxic apparatus with continual 3% isoflurane administration throughout the surgery. Burr holes were drilled free-handed through the skull at 3mm anteroposterior and 3mm mediolateral from bregma to expose the dura. Two stainless steel electrodes were lowered into the hippocampus and overlying cortex; electrode positions were verified in post-hoc analysis by the presence of a characteristic theta peak recorded from hippocampal electrodes. Mice were allowed to recover for at least 6 days post-surgery before recordings began.
PTZ Experiments
Adult mice received an intraperitoneal (IP) injection of 40mg/kg of PTZ. If the animal did not seize for approximately 10 minutes after the first injection, then 20mg/kg of PTZ was injected every approximately 10 minutes until the animal seized. The highest dose received in the study was 80mg/kg of PTZ. Latency to seizure was calculated from the time of the first injection to electrographic seizure onset, and duration was calculated by the time length of electrographic seizure.
EEG Data Analysis
Raw data were visually inspected within the GRASS Technologies software for epileptiform activity by two trained observers. Upon identification of a seizure, the video recording was inspected to rule out artifacts due to movement or physical alterations in the recording setup. While blinding was not possible during recording due to the noticeably smaller size of the Cask+/− mice, subsequent spectral analyses were conducted by an independent experimenter blinded to the genotypes corresponding to each recording. Power spectra were calculated using the EEGlab toolbox in MATLAB2017a. Data were sampled at 400Hz. 20-hour-long epochs spanning the full day and majority of the night were used for spectral analysis, including periods of sleep and wake as well as all typical locomotor behaviors. No raw data during this 20-hour period was excluded from downstream analysis. The raw data were filtered from 0-50Hz using a finite impulse response (FIR) filter, DC offset was removed, and mean power spectral densities were calculated using the spectopo() function in EEGlab with a window length of 400 samples, FFT length of 400, and 0 overlap for each channel independently over 20 hours of recording. The mean power spectral density for hippocampal and cortical electrode placements were then averaged within each genotype. These genotype averages were then compared between WT and (Cask+/− conditions. Subsequently, the 0-50Hz filtered data was manually binned into frequency ranges of 0-4Hz, 4-8Hz, 8-12Hz, 12-15Hz, 15-20Hz, 20-30Hz, and 30-50Hz. Mean power spectral densities for each genotype in each frequency bin were calculated and compared as described above. Data were analyzed for statistical significance (p<0.05) using an f-test to determine homogeneity of variation and then a two-tailed Student’s t-test with equal variance for all comparisons; data were collected from three Cask+/− mice and four WT mice.
RNA Sequencing Analysis
Whole brain RNA was extracted from four Cask+/− and four WT adult mice (age 2-4 months) using Aurum™ Total RNA Fatty and Fibrous Tissue kit from Biorad following manufacturer’s instructions. PolyA mRNA was extracted and fragmented into 180-200 nucleotide fragments. RNA sequencing experiments were performed at the Biocomplexity Institute of Virginia Tech on an Illumina Nextseq 500 platform. Strand-specific libraries were constructed using Illumina’s TruSeq Stranded mRNA HT Sample Prep Kit (Illumina, RS-122-2103). Generated libraries were validated using an Agilent 2100 Bioanalyzer and clustered. Single-stranded, paired-end sequencing was performed on an Illumina NextSeq 500 (sequencing reagent kit was NextSeq 500/550 High Output kit V2; P/N FC-404-2005) with 75-bp read lengths (1 x 75; 400 million clusters). Illumina NextSeq Control Software v2.1.0.32 with Real Time Analysis RTA v2.4.11.0 was used to provide the management and execution of the NextSeq 500 and to generate BCL files. The BCL files were converted to FASTQ files using bcl2fastq Conversion Software v2.20, and sequences were trimmed of adapters and demultiplexed using ea-utils and Btrim [32,33]. Sequencing reads were then aligned to the genome (Ensembl Mus_musculus.GRCm38.78) using Tophat2/HiSat2 [34] and counted via HTSeq [35]. Quality control summary statistics were examined to identify any problematic samples (e.g. total read counts, quality and base composition profiles (+/− trimming)). Following successful alignment and counting, mRNA differential expression was determined using contrasts of reads from the two groups and tested for significance using the false discovery rate (FDR) (Benjamini-Hochberg) corrected Likelihood Ratio Test (LRT) in the R-package DESeq2 [36]. The gene ontology classification of significant changes (padjusted <0.05) was performed on the NIH DAVID web interface (https://david.ncifcrf.gov/).
iTRAQ Proteomics
Three C57BL/6J mice from each genotype (WT and Cask+/−) were sacrificed by cervical dislocation, and the brains were hemisected. One hemisphere of each brain was used for isobaric Tagging for Relative and Absolute Quantitation (iTRAQ) Liquid Chromatography Tandem Mass Spectroscopy (LC-MS/MS) [37]. iTRAQ experiments were performed commercially by MtoZ Biolabs (Boston, MA, USA). Total brain protein was extracted by physical homogenization in lysis buffer and subjected to trypsin digestion at a concentration of 0.025μg/μL overnight at 37°C. The TMT10plex Isobaric Label Reagent Set from Thermofisher Scientific (USA) dissolved in 41μL anhydrous acetonitrile was used to label peptides. The three WT samples were labelled with TMT10-126, TMT10-129N, and TMT-129C respectively; the three Cask+/− samples were labelled with TMT10-130N, TMT10-130C, and TMT10-131N, respectively. Peptides were subjected to tandem LC-MS/MS using an Easy-nLC1000 chromatograph (Thermofisher Scientific, USA) and an Orbitrap™ Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermofisher Scientific, USA).The raw MS/MS data were searched against the UniProt Mus musculus protein database using Proteome Discover 2.1 (Thermofisher Scientific, USA) using the following parameters: protein modifications were set to carbamidomethylation (C) (fixed), oxidation (M) (variable); enzyme set to trypsin; maximum missed cleavages set to 2; precursor ion mass tolerance set to 10ppm; MS/MS tolerance set to 20ppm. The resulting proteomic changes between WT and CASK+/− were selected based on significance (p<0.05) using a one-tailed Student’s t-test assuming equal variance comparing abundance levels for each peptide between the three Cask+/− and three WT samples. Only changes with p<0.05 between groups were analyzed. Significant changes were categorized into subcellular compartments by using the GO_CC_DIRECT algorithm (NIH DAVID). The changes were also categorized into functional pathways using the KEGG_PATHWAY algorithm to sort them into functional categories (NIH DAVID). We also performed R spider analysis and Profcom GO analysis using the bioprofiling.de web interface [38,39].
Co-Immunoprecipitation Mass Spectroscopy
These experiments were done using methodology described previously [29]. Briefly, lysates of HEK 293 cells expressing either GFP or GFP-CASK were incubated overnight at 4°C with whole rat brain lysate, to allow in vitro complex formation. Both cellular and rat brain lysates were made using a solubilizing buffer consisting of 20mM HEPES-KOH (pH7.2), 150mM NaCl, 2mM EDTA, 1% Triton-X 100, 1% sodium deoxycholate and protease inhibitor cocktail. The lysate mixture was filtered through 0.2 micron filter and pre-cleared with glutathione agarose for 2 hours. GFP-Trap beads were subsequently incubated with the lysate mixtures for an hour to precipitate GFP-tagged proteins. Beads were washed thrice with ice cold solubilizing buffer without protease inhibitors. The GFP-Trap beads were then treated with trypsin, and peptide mixtures were then separated by nano-liquid chromatography (1260 Infinity from Agilent Technologies, Santa Clara, CA) and analyzed by tandem mass spectrometry with an LTQ XL™ linear ion trap mass spectrometer (ThermoFisher, Waltham, MA). Separation columns were prepared in-house using 100 μm i.d. fused silica capillaries (Polymicro Technologies Phoenix, AZ) and 5 μm Zorbax SB-C18 particles (Agilent technologies, Santa Clara, CA). The length of the column ranged from 10 cm to 12 cm. Mobile phases A and B consisted of 0.01% TFA in 4% and 90% acetonitrile, respectively. The peptides were eluted at a flow rate of ~160-180 nL/min by using a 200 min long gradient with increasing concentrations of solvent B [40]. MS data were acquired via a data-dependent acquisition strategy by performing Zoom/MS2 scans on the 5 most intense peaks in each MS scan. Data were acquired over a mass range of 500 – 1600 m/z. Collision-induced dissociation (CID) was performed at a normalized collision energy of 35%, activation Q = 0.250, and activation time of 30 msec. The threshold for triggering MS2 scans was set as 100 counts. The data-dependent parameters were set for exclusion mass width ±1.5 m/z, zoom scan width ± 5 m/z, dynamic exclusion repeat count 1, repeat duration 30 sec, and exclusion duration 60 sec, and exclusion list size of 200. The Thermo Proteome Discoverer 1.4 software package was employed to search the MS/MS spectra against the Rattus norvegicus (Rat) protein database (SwissProt/TrEMBL, combined 36081 entries) extracted from UniProt on November 7, 2017 (https://www.uniprot.org) (rat brain homogenate was used for protein interactions). The database search used the Sequest HT algorithm and included fully tryptic peptides with a maximum of 2 missed cleavages, minimum/maximum peptide length of 6/144, precursor ion mass ranging from 500-5000 Da with a precursor ion tolerance of 2 Da and fragment tolerance of 1 Da, no posttranslational modifications, and a stringent FDR of <1 % and relaxed of <3 %. In the case of 0 peptides coming down with GFP, a threshold of at least 2 peptide spectrum matches from a protein coming down with CASK-GFP was qualified as a putative interaction. In cases with background binding with GFP-beads, 3 times more peptides coming down with CASK over GFP alone was used as a threshold to identify putative interactors.
Results
Cask+/− mice display minimal electrographic changes
Mutations in CASK may co-occur with partially penetrant epilepsy with the majority not showing any seizures [2]. Mutations specifically in boys are associated with epileptic encephalopathies during infancy such as Ohtahara syndrome and West syndrome [6,7]. The mechanism of the seizures remains unknown. Although an imbalance in excitatory and inhibitory synaptic transmission has been noted, these changes are limited to spontaneous action potential-independent release events [17,18]. Evoked excitatory or inhibitory synaptic responses and neuronal excitability remain unchanged in CASK-null neurons [17]. The effect of spontaneous release of single vesicles on larger field potentials is uncertain. The electroencephalographic (EEG) signal emerges as a measure for large-scale extracellular voltage changes which includes both post-synaptic potentials as well as pre-synaptic currents and action potentials [41].
To better characterize CASK-associated epileptic encephalopathies, we therefore performed video EEG (vEEG) recordings in infant mice at postnatal day 7, 8, 13, and 14. The vEEG recordings in infant CASK+/− mice did not reveal any interictal discharges or seizures (data not shown). We next performed two weeks of 24-hour vEEG recording of adult Cask+/− mice, which only revealed a single hippocampal seizure in a single mouse identified visually by two trained observers (Figure 1A). Because we did not observe frequent seizures or interictal discharges, we next performed a power spectral analysis of the EEG signal to determine whether electrographic activity was altered in a non-ictal manner. No significant differences in mean power were found in the entire 0-50Hz range or when binned into biologically relevant frequency bands of delta, theta, alpha, beta, and low gamma (Figure 1B,C). Overall, the Cask+/− mice displayed a qualitatively and quantitatively normal EEG signature (Figure 1D,E). Although we failed to see more than one readily observable epileptic episode or resting power spectral differences at the height of the phenotype in the young and adult mice that we recorded, it may still be that the epileptic threshold in these animals is lowered. We therefore performed a kindling experiment with pentylenetetrazol (PTZ) [42]. Our data suggest that CASK+/− mice do not differ from wildtype mice with respect to epileptic threshold (Figure 1F). Overall our data suggest that although seizures may occur in Cask+/− mice, overall electrographic signals remain unaltered at the ages of the Cask+/− mice that we recorded. Any cellular or molecular changes are thus unlikely to be the result of large-scale changes in activity of neuronal ensembles. Furthermore, our data indicate that an imbalance in spontaneous action potential independent release does not translate into changes in field potentials, and the observed seizures in some cases of MICPCH cannot be accounted for simply by a change in baseline synaptic activity in isolation.
Figure 1: Electroencephalographic analysis of Cask+/− mice.
A) The only spontaneous hippocampal (Hc) seizure recorded with vEEG, with corresponding cortical (Ctx) recording. No behaviorally apparent phenotype was observed in the video during this spectrographic ictal event. B) Mean power in the 0-50Hz frequency range for cortical and hippocampal electrode placements. C) Mean power binned into biologically relevant frequency bands including delta, theta, alpha, beta, and low gamma. D) Qualitative power spectral density over 15 minutes comparing a single Cask+/− and WT mouse. E) Shape of power spectral density versus frequency curve for each individual mouse (n=4 WT, n=3 Cask+/− for B, C, and E). F) Latency to PTZ-induced seizure and length of PTZ-induced seizures for each genotype (n=3 wild-type, 5 Cask+/−). Comparisons for B, C, and F were made using a two-tailed Student’s t-test for each frequency band; data represent mean±SEM.
Transcriptional changes in CASK+/− mice
RNA-Sequencing analysis was performed on three Cask+/− mice and four wildtype littermates to determine transcriptional changes. In all three Cask+/− mice, CASK mRNA was reduced to a similar degree, confirming that the X-chromosome inactivation was not skewed. Significant changes were seen in only a very few transcripts, with 105 transcripts in addition to the CASK transcript reaching statistical significance (p-adjusted value of lower than 0.05). Out of the altered transcripts, 43 transcripts were reduced in amount and 62 transcripts showed an increase. CASK has been proposed to affect transcription via the Tbr1 pathway [12], but we did not see changes in transcription of RNAs downstream of Tbr1, such as reelin or NR2b [12,43,18]. This is consistent with our previous biochemical analysis of MICPCH-associated missense mutations, in which we found that the CASK-Tbr1 interaction does not underlie CASK-associated brain pathology [5]. Only 11 transcripts are ascribed functions related to core neuronal function such as synaptic transmission or membrane excitability (Figure 2; category:synapse). Approximately 50% of the reduced transcripts fall within the Gene Ontology category 0005737 (cytoplasm), which includes “all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures” Other than that, the largest subcellular class of decreased transcripts are identified as nuclear, encoding transcription factors or proteins responsible for mRNA processing or export (Figure 2). Most of the transcription factors that are reduced are known to regulate cell cycle rather than neuronal or synaptic function. The remaining intracellular transcript changes include proteins in metabolic pathways, with five of them belonging to the oxidative phosphorylation pathway in mitochondria and five belonging to the solute carrier family. Intriguingly, the majority of the transcripts in the mitochondrial pathway that are upregulated are not nuclear but rather mitochondrially encoded. Strikingly, the largest class of increased transcripts (24 out of 62) belong to proteins of the extracellular matrix (Figure 2). The relatively small number of changes precluded further meaningful bioinformatics analysis, such as functional pathway analysis. R-spider or KEGG-spider analysis did not reach significance, and KEGG pathway analysis included less than a third (~30%) of the genes identified in our study as having altered transcript levels. Overall, the most notable changes which rise to a level of significance are found in Gene Ontology groups that have the rather generic descriptions of “extracellular region”, “membrane fraction” and “cytoplasmic fraction”. Our transcriptomic data thus do not support an overall nuclear reprogramming in the brains of CASK heterozygous knockout mice, and we do not observe any specific cellular pathway alteration induced by haploinsufficiency of CASK. The changes noted in transcripts for extracellular proteins are more likely to reflect increased ECM turnover, a phenomenon observed in many developmental disorders [30].
Figure 2: Transcriptomic changes in brains of Cask+/− mice.
Results from RNA-seq were analyzed. 105 genes with p<0.05 were observed. A pie chart depicts enrichment in particular pathways and relative contribution to total changes. Redness indicates negative-fold change and blackness reflects positive-fold change. The fold change of genes in individual groups has been plotted as indicated by arrows. Comparisons were made using the false discovery rate (FDR) (Benjamini-Hochberg) corrected Likelihood Ratio Test (LRT) in the R-package DESeq2. Note that the GO:0005737 classification “cytoplasm” includes all genes which do not fall under nuclear, transmembrane, or extracellular; therefore, we manually curated genes into their respective subcellular compartments, leaving those transcripts which could not be otherwise grouped in the “cytoplasm” category.
Proteomic changes in CASK+/− mice
Changes at the transcript level may not always be representative of changes at the protein level [44,45]. An additional factor which complicates interpretation of changes at the transcript level specifically in the setting of CASK reduction is that CASK has been implicated as a scaffolding protein that facilitates the localization and binding of protein complexes in situ; if CASK’S scaffolding function is reduced or eliminated in a cell, it is possible that this will impact turnover of its binding partners. We therefore decided to examine changes in the levels of brain proteins in Cask+/− mice, using sensitive and unbiased iTRAQ mass spectrometry. Comparisons were made between three wild-type (WT) and three Cask+/− mice at postnatal day 40. In contrast to the limited number of changes observed at the transcript level, we observed significant changes in 525 different proteins, excluding CASK. CASK was again found to be reduced in all three mutant mice, confirming previous literature [17,18,20]. Of all the changes observed, only a single one correlated with a change also noted at the level of transcription: the CART (Cocaine- and Amphetamine-Regulated Transcript) peptide. CART peptide acts as a neurotransmitter and a hormone. It plays a role in body energy metabolism and body weight [46], and its promoter is known to be regulated by cyclic adenosine monophosphate response element [47]. All other alterations at the protein level appear to be post-transcriptional in nature. Of the 525 proteins, 246 proteins are decreased and 279 proteins are increased (Figure 3). The largest single organelle group (99 out of 525) identified as having altered protein levels is the mitochondrion; this is a ~2-fold higher number of proteins changed than the number of synaptic protein changes (49 (excluding CASK) out of 525) (GO analysis in Supplementary Table 1). KEGG pathway analysis included approximately half of the proteins observed to be altered but did identify oxidative phosphorylation and cardiac contractility as the top cellular pathways that are disturbed (Supplementary Table 2), and the proteins involved with these pathways were overlapping. Of the proteins that were increased in the setting of lower CASK levels, many were membrane and cytoplasmic proteins (Figure 3A). In addition to the expected adhesion molecules such as CASK’S known interactors, neurexins and syndecan2, other proteins found at increased levels include synaptic proteins, many members of ribosomal, cytoskeletal, proteasome protein families and metabolic proteins (Figure 3A,B and Supplementary Table 3). Intriguing changes were also found in ion channels and proteins known to be involved in axonal guidance, long-term plasticity and glutamatergic synapse (Supplementary Table 3 and 4). Of the proteins found to be increased, only 86 were able to be mapped in the R spider analysis [39], and the only significant pathway alteration identified by this analysis as significant was in the protein degradation pathway (Figure 3B). Less significant, but notable nonetheless, were changes seen in synaptic pathways. GO analysis of proteins found to be decreased in the brains of Cask+/− mice again identified the mitochondrion as the most significant group, with 65 proteins (Figure 3C and Supplementary Table 5 and 6) from the mitochondrial matrix, inner membrane and respiratory chain complex III KEGG pathway analysis was possible on nearly 50% of the proteins with altered levels and revealed enrichment in mostly metabolic pathways including oxidative phosphorylation, glycolysis/gluconeogenesis, biosynthesis of antibiotics, and amino acids (Supplementary Table 2). R spider analysis included only 67 proteins from the decreased expression group, and several pathways were identified that may be significantly impeded in the Cask+/− mouse brain, including the electron transport chain, NADH metabolic processes and oxaloacetate metabolism (Figure 3D). The population of proteins found to be decreased in the Cask+/− mouse brain was enriched with proteins that play a role in intracellular protein transport and mRNA processing (Figure 3D). The proteomic data thus explains at a protein expression level the mitochondrial dysfunction we previously observed in the Cask+/− mouse brain [20].
Figure 3: Protein changes in Cask+/− mouse brain.
iTraq proteomic results were analyzed using bioprofiling.de web interface for ProfCom and R spider analysis [38,39]. A) Top 12 GO changes noted upon analysis of the proteins that were found at increased levels in Cask+/− mouse brain. The full table can be found as Supplemental Table 3. B) Top 12 functional changes noted upon analysis of the proteins that were found at increased levels in Cask+/− mouse brain. The full table can be found as Supplemental Table 4. C) Top 12 GO changes in proteins that were found at decreased levels in Cask+/− mouse brain. The full table can be found as Supplemental Table 5. D) Top 12 top functional changes in proteins that were found to be decreased in Cask+/− mouse brain. The full table can be found as Supplemental Table 6. IA is the ratio of changes within the particular GO group to total changes (enrichment observed). IB is the ratio of the total number of proteins in the specific GO group in the database to the total number of proteins in the database (enrichment by chance). The odds ratio strength of association is calculated as IA/IB.
CASK can phosphorylate the presynaptic adhesion molecule neurexin1 and link it to the active zone organizers liprins-α. CASK has also been speculated to play a role in the trafficking of post-synaptic molecules and to participate in Tbr1-mediated gene regulation, particularly that of reelin and NR2b [48]. Recently it has been reported that CASK null cells in Cask+/− mice may express lower amounts of NR2b [17,18]. A mitochondrial and trafficking function of CASK has also been suggested [14,29,20]. To gain a more comprehensive understanding of the changes induced in the Cask+/− mouse brain, we therefore did a heat map analysis on the three functional groups with the largest change (synaptic, protein biosynthesis, and mitochondrial; Figure 3) to examine which of the above pathways are most affected in Cask+/− mice.
First we examined bona fide presynaptic and postsynaptic proteins. We observed changes in many categories of presynaptic proteins. Five major active zone proteins (liprin-α4, Rims1, piccolo, bassoon and munc13) are all decreased in the Cask+/− mice. We also found changes in proteins involved in synaptic vesicle cycling, including decreases in Munc18c, synaptotagmin-12, rabphilin and SNAP25 and an increase in tomosyn (Figure 4). Overall the changes in these proteins suggest that loss of CASK should affect synaptic vesicle recycling negatively. On the postsynaptic side, postsynaptic density proteins are almost entirely increased. In a striking contrast to expected and previously published data [18], levels of NR2b protein are increased in the setting of lower CASK expression (Figure 4). Within the protein biosynthetic pathway, most of the RNA processing proteins are decreased, whereas ribosomal subunits are increased in abundance. Mitochondrial proteins operating in the oxidative phosphorylation pathway are mostly decreased, with the exception of an increase in a subset of mitochondrial proteins (Figure 4). We also observed changes in several cytoskeletal molecules, including tubulins. Overall these data confirm a synaptic function for CASK and also indicate that CASK additionally participates in mitochondrial and protein biosynthetic pathways.
Figure 4. Protein changes encompassing protein synthetic, synaptic and mitochondrial pathways in Cask+/− mouse brain.
iTraq proteomic results from Figure 3 were analyzed and subdivided into subcellular compartments using NIH DAVID and manual curation. 526 proteins, including CASK, are altered. Pathways altered were identified using NIH DAVID functional annotation database. Heat maps of three major pathways were plotted using heatmapper.ca. Neuron and organelle diagrams are for representational purposes only since subcellular proteomics was not conducted.
Putative protein complexes in which CASK participates
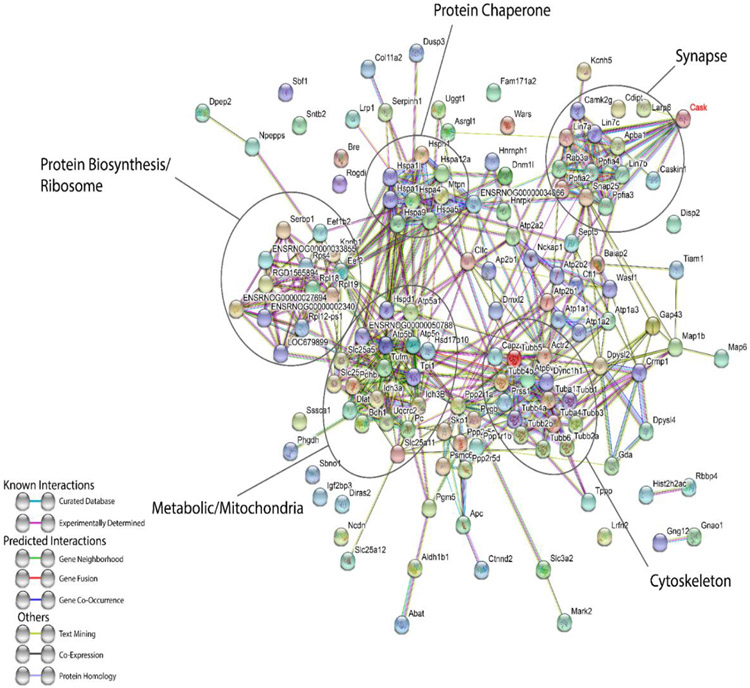
Previous experiments examining protein interactions in Drosophila melanogaster were done using transgenic flies expressing a transgene of CASK fused with YFP (yellow fluorescence protein); a GFP-Trap matrix was employed to capture interacting partners. YFP-CASK can rescue CASK loss-of-function in Drosophila ([49] and unpublished observations), and GFP-CASK maintains all of its known interactions [50,4,5,22,29]. In these Drosophila experiments, both synaptic and mitochondrial proteins were pulled down by CASK [29]. Because both of these categories of protein (synaptic and mitochondrial) were identified in the proteomic study described here (Figure 4), and since the protein changes cannot be explained by changes in either electrical activity (Figure 1) or nuclear reprogramming (Figure 2), we investigated CASK interacting partners in a mammalian system with a focus on non-plasma membrane proteins. Because we have previously shown that using antibodies to internal epitopes of CASK for immunoprecipitation sometimes inhibits binding of crucial interactors due to steric hindrance [22], we once again used a recombinant GFP-CASK pulldown approach with GFP-Trap beads. GFP-CASK expressed in HEK293 cells was used to precipitate all CASK-interacting proteins from rat brain, and mass spectrometry was used for identification. GFP alone was employed as a negative control; 640 GFP peptides were identified in the GFP-beads sample, while 224 GFP peptides were identified in GFP-CASK beads. Additionally, 2018 CASK peptides were identified in GFP-CASK beads while no CASK peptides were identified on GFP-beads. Based on our candidate inclusion criteria (minimum of two peptides identified and at least 3 times more peptides with GFP-CASK over GFP alone), we identified 170 candidate proteins that interact with CASK either directly or indirectly. A complete list of interactors which met our inclusion criteria can be found in Supplemental Table 7. Many of the peptides belonged to previously known interactors such as veli, Mint1 and liprins-α. The dataset of putative interacting proteins were mapped using known interactions in the STRING database (Figure 5, Supplemental Table 7; [51]). Five major clusters emerged: presynaptic proteins, mitochondrial proteins, ribosomal proteins, chaperone proteins, and cytoskeletal proteins (Figure 5, Supplemental Table 7). Based on the STRING (database) mapping derived from previously known interactions on which we have overlaid the list of proteins from our pulldown experiment and the design of the experiment itself, it is likely that proteins pulled down in these experiments may represent both direct and indirect binding partners. In the future it will be critical to validate specific interactions that are described here. However, the similarity of these clusters with the proteomic changes in the brain suggests that CASK most likely forms complexes within these pathways. Furthermore, multiple new interacting partners identified here exhibit changes in their abundance in the iTraq proteomic study such as Nedd4, CamKIIa, Ppp2r1a, Slc25a5, Wars, Uqcrc2 and Tufm (Figure 6A), increasing our confidence in these interactions. Since CASK is a scaffolding protein, lack of CASK may destabilize interacting molecules and reduce their amount. Strikingly, analysis of 27 direct interacting partners revealed that interacting proteins can either increase or decrease in amount in the Cask+/− brain (Figure 6A), suggesting that CASK likely regulates these protein complexes dynamically.
Figure 5. Brain proteins identified to be interacting with GFP-CASK through GFP-Trap mass spectrometry analysis are plotted using the STRING database interface.
Protein clusters belonging to a specific pathway are indicated with ellipses. Color of strings between proteins indicates whether an interaction is experimentally determined or predicted as per the legend. Solved structures are indicated within each sphere for a given protein.
Figure 6. CASK interacting proteins exhibit both increase and decrease in Cask+/− mouse brain.
A) Heat map of proteins found to interact with CASK that are also significantly changed in the iTRAQ proteomics data. B) A model indicating three cellular pathways that are likely to be altered by CASK haploinsufficiency. Our model highlights the likelihood that changes seen in mRNA processing and ribosomal proteins themselves impact other pathways.
Discussion
Mutations in the gene encoding CASK associate with MICPCH, ONH, growth retardation and often seizures. Seizures are, in fact, a common accompaniment of a multitude of neurodevelopmental disorders [52]. As CASK is a putative synaptic molecule [9], one could argue that a shift in steady state (excitatory/inhibitory) E/I balance induced by loss of CASK might lead to epileptiform activity. We were able to record at least one apparent spontaneous seizure, but an extensive analysis of EEG data from adult and young mice failed to reveal significant electrographic changes in Cask+/− mice. Overall these data indicate that epilepsy in rodents is likely to have low penetrance with CASK haploinsufficiency; these findings are consistent with clinical data, since many CASK mutation subjects do not exhibit seizures [53,4,5,54]. Typical epileptic disorders are characterized by epileptiform activity in less than 1% of total brain activity [55], indicating that ictogenic triggers may play a role in such cases. Cask+/− mice do not display a decreased threshold for seizures after exposure to the kindling agent PTZ, a GABA antagonist. These data suggest that in Cask+/− mice, E/I imbalance is unlikely to be the major ictogenic trigger. Our data thus indicate that previously described changes in spontaneous vesicular release are insufficient to precipitate hypersynchronous neuronal activity required for ictogenesis.
In mammals, CASK was discovered due to its ability to interact with the cytosolic tail of the presynaptic adhesion molecule neurexin [10] and was proposed to be a scaffolding molecule involved in presynaptic assembly [9]. This idea was challenged since CASK deletion did not affect synapse formation and assembly [17] and also because the CASK-neurexin interaction is not needed for neurexin’s synaptogenic activity [56]. CASK, however, interacts with other probable presynaptic scaffolds such as Mint1 [9], Caskin1 [57], liprin-α [58], and rabphilin [59], and participates in presynaptic specialization [60]. We have previously demonstrated that CASK is critical for linking presynaptic adhesion molecules to the active zone via interaction with the liprins-α [22]. Furthermore, we have demonstrated that CASK haploinsufficiency reduces the number of release sites (active zones) in the large retinogeniculate synapses [21]. Despite our and others’ numerous studies, CASK’S role in the assembly of the presynaptic active zone remains a subject of debate. The increase in the level of neurexin1 and concomitant decrease in five major presynaptic active zone proteins in the Cask+/− mouse brain provide strong evidence for the role of CASK in active zone assembly, formation, and maintenance. Specifically, lack of CASK decreases the levels of: 1) piccolo and 2) bassoon, two large proteins involved in synaptic vesicle clustering [61]; 3) RIM1, a molecule involved in docking, presynaptic short term plasticity and positioning of calcium channels [62,63]; 4) Munc13, a protein involved in the priming of synaptic vesicles [64,65]; and 5) liprin-α4, which is involved in active zone organization [66,67]. In summary, lack of CASK affects the biochemical composition of the active zone. In addition to the changes seen in active zone protein levels, we also observe a decrease in a number of proteins that are involved in synaptic vesicle cycling, including the SNARE protein SNAP25 and a number of SNARE fusion-regulating proteins such as Munc18c [68], complexin-2 [69], rabphilin [70] and synaptotagmin-12 [71]. The only presynaptic molecule which is increased is tomosyn, a protein shown to inhibit vesicle fusion by inducing formation of non-fusogenic SNARE complexes [72]. Based on decreases in the many vital active zone and fusion-related proteins we observe, neurotransmitter release should be negatively impacted in the Cask+/− brain.
Cask+/− mouse brain also displays an increase in a number of postsynaptic proteins (Figure 4) like the NR2B subunit of the NMDA receptor and the scaffolding molecules PSD95, SHANK3, SYNGAP1, CaMKII and Homer. Since a parallel increase in mRNA production is not observed (Fig. 2), we suspect that these protein-level increases are not a nuclear response at the level of transcription but rather a change in turnover rate. Proteins associated with the postsynaptic density might, for example, be stabilized by changes in presynaptic function, thus decreasing the turnover rate. In fact, RIM1 knockout mice also display a modest increase in postsynaptic density proteins [73]. Reduction or deprivation of presynaptic release also often results in an observed increase in levels of postsynaptic density proteins [74,75]. Multiple studies have shown that neurons adjust synaptic strength in response to chronically increased or decreased activity in a process called homeostatic synaptic scaling, utilizing mechanisms independent of those traditionally associated with long-term potentiation/depression [76,77]. As such, decreases observed here in active zone proteins resulting from loss of CASK may induce compensatory changes in protein turnover at the post-synapse. Further, it has recently been shown that changes to presynaptic neurexin composition can differentially affect postsynaptic glutamatergic responses [78,79], and an increase in presynaptic neurexin may also increase Homer1 in positive excitatory post-synapses [80]. A caveat to this argument is that the reported changes are global and may arise from different synapses and indeed different kinds of neurons. The experimental approach described here does not have the resolution to provide a more detailed description of neuron-specific changes. Furthermore, at a cellular level, both the MICPCH disorder and its animal model presented here are mosaic in regard to the expression of the X-linked gene CASK in the female population. Approximately 50% of neurons express normal levels of CASK while approximately 50% express no CASK. Some of the observed changes may also represent compensatory changes in CASK-expressing cells.
Indeed, CASK has also been proposed to be present in the postsynaptic compartment, with evidence suggesting that it interacts with parkin [25], syndecan2 [24], SAP-97 [16,81] and CaMKII [82,83,29], as well as potentially playing a role in the trafficking of NMDA receptors [84]. Therefore, an alternative and equally intriguing possibility is that CASK plays a cell-autonomous, but negative, role in formation of the postsynaptic density via interaction with postsynaptic molecules, specifically CaMKII. Herein, we, for the first time, demonstrate that this critical interaction with CaMKII first described in the Drosophila literature is evolutionarily conserved [82,83,29]. CaMKII plays a crucial role in the postsynaptic compartment in the process of long-term potentiation (LTP). LTP may involve interaction with and phosphorylation of the NR2b subunit of the NMDA receptor by CaMKII [85,86]. The increase noted in NR2b and other postsynaptic density proteins may thus be explained by the CASK-CaMKII interaction. Our results suggest that mammalian CASK via interaction with CaMKII may participate in postsynaptic plasticity.
We also demonstrate that CASK interacts with mitochondrial proteins and that a lack of CASK alters levels of mitochondrial proteins. We have previously demonstrated that the CASK+/− mouse brain exhibits reduced mitochondrial respiration [20], but the mechanism for this observation remained unclear. Our proteomic data thus provides a biochemical explanation for our prior observation. Overall, we find that the mitochondrial proteome is disproportionately affected in the brain of CASK+/− mice (99 of the 525 total proteins found to be altered are mitochondrial; Figure 4). The majority of these changes were decreases and would negatively impact mitochondrial function. Our data suggest a critical role of CASK in regulating mitochondrial function. Strikingly, in our RNA sequencing analysis, we observed increases in mitochondrial DNA encoded genes, indicative of a compensatory response. Future studies are required to determine where and how CASK interacts with the mitochondrial proteins and regulates their function. Interestingly, it also remains unclear how a reduction in mitochondrial function in CASK mutant mice affects overall neurophysiology. Finally, we found protein-level changes suggestive of a role for CASK in protein biosynthetic and degradation pathways, including proteins involved in mRNA processing, ribosomal subunits and proteins belonging to proteasomal pathways. This function of CASK was hitherto unknown and also needs further investigation. Furthermore, a function within protein biosynthesis or degradation pathways by itself may contribute directly to post-transcriptional alterations described here (Figure 6B).
CASK is not a neuron-specific or indeed even a brain-specific molecule [10], and in Caenorhabdites elegans (C. elegans), deletion of CASK does not produce a neuronal phenotype [87]. Could CASK have a more central cellular function that may explain many of the observed changes reported here? CASK forms a well-characterized and evolutionarily highly conserved tripartite complex with Mint1/X11-α/lin-10 and veli/Mals/lin-7 [9,88,29]. Orthologs of all these genes were discovered in a vulval precursor cell-fate patterning screen in C. elegans as early as 1980, which suggests a functional relevance of this complex [89,90]. The vulval differentiation function of these three genes in C. elegans is dependent on proper membrane targeting of the tyrosine kinase LET-23. One possibility is that CASK/Mint-1/veli complex may also be involved in polarized and regulated trafficking of proteins from the endoplasmic reticulum/golgi complex in other animal clades. In fact Mint1 and CASK have both been shown to participate in such trafficking [91,92], which could explain changes in both pre-and post-synaptic molecules. Furthermore recent evidence suggests mitochondrial protein import also involves the endoplasmic reticulum [93], and veli has been shown to localize to mitochondria [94]. Indeed CASK has been isolated from endoplasmic reticulum-mitochondria contact sites [95,96]. Future experiments need to address these ideas.
There are number of limitations in this study, including lack of spatial and temporal resolution in the molecular changes, and number and age of animals used. Interpretation of findings here must also acknowledge the possibility that species differences may play a role. Furthermore, the study has been done in a highly homogeneous genotypic background; while this does allow us to draw statistical significance from a smaller group of animals, it may also fail to represent the spectrum of CASK-linked disorders in the highly heterogeneous human subject population. This issue is particularly pertinent in incompletely penetrant phenotypes such as seizures, which may derive from a complex interaction of CASK dysfunction and genotypic background. While it may be possible for us to document more seizures in the C57Bl6 background by increasing number and time periods of recordings, it is more likely that we would observe higher seizure numbers in a different strain of mice. These deficiencies will be addressed in future studies. Overall, the data presented here elucidate several biochemical functions of the CASK protein in multiple aspects of cellular physiology (Figure 6B). Pathology of the CASK-linked brain phenotype is highly complex and involves the entire brain, therefore here we have taken an approach which is highly sensitive, but does not address specific molecular or cell-type roles of CASK. This is to delineate identifiable cellular pathways that are affected by loss of CASK. In the future, we will investigate specific molecular interactions and region and cell-type specific changes to obtain mechanistic insights into this multifaceted neurodevelopmental disorder. Our data here confirm the canonical synaptic role of CASK while furthering the discussion of effects of heterozygous deletion at the whole brain level. Furthermore, non-canonical phenotypic observations such as effects on mitochondrial respiration have been explained at the level of mRNA, protein, and protein-protein interactions. In doing so, these data enhance our understanding of the diverse functions of CASK and the microcephaly produced by clinically relevant heterozygous CASK mutations at a whole brain level.
Supplementary Material
Heterozygous deletion of murine Cask phenocopies human cases of CASK-linked MICPCH.
Global electrographic activity remains normal in Cask+/− brains.
Transcriptional changes are largely limited to extracellular matrix related mRNAs.
Proteomic analysis reveals altered synaptic, metabolic and proteostasis pathways.
Protein interaction mapping reveal CASK binds with proteins in these same pathways.
Acknowledgements
We thank Vrushali Chavan for technical assistance.
Funding
This work was supported by the NIH National Eye Institute (R01EY024712 to KM) and a UVA-VTC seed fUnd to KM and HG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Consent for Publication
Not applicable.
Data Availability
The full datasets generated and analyzed by the experiments described here will be submitted to respective repositories and are available upon reasonable request from the corresponding author.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Burglen L, Chantot-Bastaraud S, Garel C, Milh M, Touraine R, Zanni G, Petit F, Afenjar A, Goizet C, Barresi S, Coussement A, Ioos C, Lazaro L, Joriot S, Desguerre I, Lacombe D, des Portes V, Bertini E, Siffroi JP, de Villemeur TB, Rodriguez D (2012) Spectrum of pontocerebellar hypoplasia in 13 girls and boys with CASK mutations: confirmation of a recognizable phenotype and first description of a male mosaic patient. Orphanet J Rare Dis 7:18. doi:1750-1172-7-18 [pii] 10.1186/1750-1172-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moog U, Kutsche K, Kortum F, Chilian B, Bierhals T, Apeshiotis N, Balg S, Chassaing N, Coubes C, Das S, Engels H, Van Esch H, Grasshoff U, Heise M, Isidor B, Jarvis J, Koehler U, Martin T, Oehl-Jaschkowitz B, Ortibus E, Pilz DT, Prabhakar P, Rappold G, Rau I, Rettenberger G, Schluter G, Scott RH, Shoukier M, Wohlleber E, Zirn B, Dobyns WB, Uyanik G (2011) Phenotypic spectrum associated with CASK loss-offunction mutations. J Med Genet 48 (11)741–751. doi:jmedgenet-2011-100218 [pii] 10.1136/jmedgenet-2011-100218 [DOI] [PubMed] [Google Scholar]
- 3.Najm J, Horn D, Wimplinger I, Golden JA, Chizhikov VV, Sudi J, Christian SL, Ullmann R, Kuechler A, Haas CA, Flubacher A, Charnas LR, Uyanik G, Frank U, Klopocki E, Dobyns WB, Kutsche K (2008) Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat Genet 40 (9):1065–1067. doi: 10.1038/ng.194 [DOI] [PubMed] [Google Scholar]
- 4.LaConte LEW, Chavan V, DeLuca S, Rubin K, Malc J, Berry S, Sunnnners CG, Mukheijee K (2019) An N-terminal heterozygous missense CASK mutation is associated with microcephaly and bilateral retinal dystrophy plus optic nerve atrophy. Am J Med Genet A 179 (1):94–103. doi: 10.1002/ajmg.a.60687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaConte LEW, Chavan V, Elias AF, Hudson C, Schwanke C, Styren K, Shoof J, Kok F, Srivastava S, Mukherjee K (2018) Two microcephaly-associated novel missense mutations in CASK specifically disrupt the CASK-neurexin interaction. Hum Genet 137 (3):231–246. doi: 10.1007/s00439-018-1874-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moog U, Bierhals T, Brand K, Bautsch J, Biskup S, Brune T, Denecke J, de Die-Smulders CE, Evers C, Hempel M, Henneke M, Yntema H, Menten B, Pietz J, Pfrndt R, Schmidtke J, Steinemann D, Stumpel CT, Van Maldergem L, Kutsche K (2015) Phenotypic and molecular insights into CASK-related disorders in males. Orphanet J Rare Dis 10:44. doi: 10.1186/s13023-015-0256-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitsu H, Kato M, Osaka H, Moriyama N, Horita H, Nishiyama K, Yoneda Y, Kondo Y, Tsurusaki Y, Doi H, Miyake N, Hayasaka K, Matsumoto N (2012) CASK aberrations in male patients with Ohtahara syndrome and cerebellar hypoplasia. Epilepsia 53 (8):1441–1449. doi: 10.1111/j.1528-1167.2012.03548.x [DOI] [PubMed] [Google Scholar]
- 8.Hackett A, Tarpey PS, Licata A, Cox J, Whibley A, Boyle J, Rogers C, Grigg J, Partington M, Stevenson RE, Tolinie J, Yates JR, Turner G, Wilson M, Futreal AP, Corbett M, Shaw M, Gecz J, Raymond FL, Stratton MR, Schwartz CE, Abidi FE (2010) CASK mutations are frequent in males and cause X-linked nystagmus and variable XLMR phenotypes. Eur J Hum Genet 18 (5):544–552. doi:ejhg2009220 [pii] 10.1038/ejhg.2009.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butz S, Okamoto M, Sudhof TC (1998) A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell 94 (6)773–782 [DOI] [PubMed] [Google Scholar]
- 10.Hata Y, Butz S, Sudhof TC (1996) CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci 16 (8):2488–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Sudhof TC, Wahl MC (2008) CASK Functions as a Mg2+-independent neurexin kinase. Cell 133 (2):328–339. doi: 10.1016/j.cell.2008.02.036S0092-8674(08)00287-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsueh YP, Wang TF, Yang FC, Sheng M (2000) Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature 404 (6775):298–302. doi: 10.1038/35005118 [DOI] [PubMed] [Google Scholar]
- 13.Caruana G (2002) Genetic studies define MAGUK proteins as regulators of epithelial cell polarity. Int J Dev Biol 46 (4):511–518 [PubMed] [Google Scholar]
- 14.Leonoudakis D, Conti LR, Radeke CM, McGuire LM, Vandenberg CA (2004) A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels. J Biol Chem 279 (18):19051–19063. doi: 10.1074/jbc.M400284200M400284200 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Olsen O, Liu H, Wade JB, Merot J, Welling PA (2002) Basolateral membrane expression of the Kir 2.3 channel is coordinated by PDZ interaction with Lin-7/CASK complex. Am J Physiol Cell Physiol 282 (1):C183–195. doi: 10.1152/ajpcell.00249.2001 [DOI] [PubMed] [Google Scholar]
- 16.Lozovatsky L, Abayasekara N, Piawah S, Walther Z (2009) CASK deletion in intestinal epithelia causes mislocalization of LIN7C and the DLG1/Scrib polarity complex without affecting cell polarity. Mol Biol Cell 20 (21):4489–4499. doi: 10.1091/mbc.E09-04-0280E09-04-0280 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, Zhang W, Mukherjee K, Nosyreva ED, Fernandez-Chacon R, Missler M, Kavalali ET, Sudhof TC (2007) Deletion of CASK in mice is lethal and impairs synaptic function. Proc Natl Acad Sci U S A 104 (7):2525–2530. doi:0611003104 [pii] 10.1073/pnas.0611003104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori T, Kasem EA, Suzuki-Kouyama E, Cao XS, Li X, Kurihara T, Uemura T, Yanagawa T, Tabuchi K (2019) Deficiency of calcium/calmodulin-dependent serine protein kinase disrupts the excitatory-inhibitory balance of synapses by downregulating GluN2B (vol 24, pg 1079, 2019). Mol Psychiatr 24 (7):1093–1093. doi: 10.1038/s41380-019-0362-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang TNaH YP (2016) Calcium/calmodulin-dependent serine protein kinase (CASK), a protein implicated in mental retardation and autism-spectrum disorders, interacts with T-Brain-1 (TBR1) to control extinction of associative memory in male mice. Journal of Psychiatry and Neuroscience. doi: 10.1503/jpn.150359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava S, McMillan R, Willis J, Clark H, Chavan V, Liang C, Zhang H, Hulver M, Mukherjee K (2016) X-linked intellectual disability gene CASK regulates postnatal brain growth in a non-cell autonomous manner. Acta Neuropathol Commun 4:30. doi: 10.1186/s40478-016-0295-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang C, Kerr A, Qiu Y, Cristofoli F, Van Esch H, Fox MA, Mukherjee K (2017) Optic Nerve Hypoplasia Is a Pervasive Subcortical Pathology of Visual System in Neonates. Invest Ophthalmol Vis Sci 58 (12):5485–5496. doi: 10.1167/iovs.17-22399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaConte LEW, Chavan V, Liang C, Willis J, Schonhense EM, Schoch S, Mukheijee K (2016) CASK stabilizes neurexin and links it to liprin-alpha in a neuronal activity-dependent manner. Cell Mol Life Sci 73 (18)3599–3621. doi: 10.1007/s00018-016-2183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297 (5586)4525–1531. doi: 10.1126/science.1072356297/5586/1525 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Cohen AR, Woods DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM (1998) Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol 142 (1)129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fallon L, Moreau F, Croft BG, Labib N, Gu WJ, Fon EA (2002) Parkin and CASK/LIN-2 associate via a PDZ-mediated interaction and are co-localized in lipid rafts and postsynaptic densities in brain. J Biol Chem 277 (1):486–491. doi: 10.1074/jbc.M109806200 [DOI] [PubMed] [Google Scholar]
- 26.Ivarsson Y, Wawrzyniak AM, Kashyap R, Polanowska J, Betzi S, Lembo F, Vermeiren E, Chiheb D, Lenfant N, Morelli X, Borg JP, Reboul J, Zimmermann P (2013) Prevalence, Specificity and Determinants of Lipid-Interacting PDZ Domains from an In-Cell Screen and In Vitro Binding Experiments. Plos One 8 (2). doiARTN e54581 10.1371/joumal.pone.0054581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaConte L, Mukheijee K (2013) Structural constraints and functional divergences in CASK evolution. Biochem Soc Trans 41 (4)1017–1022. doi: 10.1042/BST20130061 [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee K, Sharma M, Jahn R, Wahl MC, Sudhof TC (2010) Evolution of CASK into a Mg2+-sensitive kinase. Sci Signal 3(119):ra33 Doi 10.1126/scisignal.20008003/119/ra33 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee K, Slawson JB, Christmann BL, Griffith LC (2014) Neuron-specific protein interactions of Drosophila CASK-beta are revealed by mass spectrometry. Front Mol Neurosci 7:58. doi: 10.3389/fnmol.2014.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu PF, Takai K, Weaver VM, Werb Z (2011) Extracellular Matrix Degradation and Remodeling in Development and Disease. Csh Perspect Biol 3 (12). doiARTN a005058 10.1101/cshperspect.a005058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanelli S, Goodkin HP, Kowalski S, Kapur J (2014) Impact of transient acute hypoxia on the developing mouse EEG. Neurobiol Dis 68:37–46. doi: 10.1016/j.nbd.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aronesty E(2011) ea-utils : "Command-line tools for processing biological sequencing data".
- 33.Kong Y (2011) Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98 (2):152–153. doi: 10.1016/j.ygeno.2011.05.009 [DOI] [PubMed] [Google Scholar]
- 34.Kim D, Landmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. NatMethods 12 (4):357–U121. doi: 10.1038/Nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon Anders PT P, Wolfgang Huber (2014)HTSeq — A Python framework to work with high-throughput sequencing data Bioinformatics. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15 (12). doi:ARTN 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannu NS, Hemby SE(2006) Methods for proteomics in neuroscience. Prog Brain Res 158:41–82. doi: 10.1016/S0079-6123(06)58003-3 [DOI] [PubMed] [Google Scholar]
- 38.Antonov AV, Schmidt T, Wang Y, Mewes HW (2008) ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res 36:W347–W351. doi: 10.1093/nar/gkn239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antonov AV, Schmidt EE, Dietmann S, Krestyaninova M, Hermjakob H (2010) R spider: a network-based analysis of gene lists by combining signaling and metabolic pathways from Reactome and KEGG databases. Nucleic Acids Res 38 W78–W83. doi: 10.1093/nar/gkq482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazar IM, Hoeschele I, de Morais J, Tenga MJ (2017) Cell Cycle Model System for Advancing Cancer Biomarker Research. Sci Rep 7 (1):17989. doi: 10.1038/s41598-017-17845-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buzsaki G, Anastassiou CA, Koch C (2012) The origin of extracellular fields and currents - EEG, ECoG, LFP and spikes. Nat Rev Neurosci 13 (6):407–420. doi: 10.1038/nrn3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajasekaran K, Zanelli SA, Goodkin HP (2010) Lessons From the Laboratory: The Pathophysiology, and Consequences of Status Epilepticus. Semin Pediatr Neurol 17 (3):136–143. doi: 10.1016/j.spen.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang TN, Chang HP, Hsueh YP (2010) CASK phosphorylation by PKA regulates the protein-protein interactions of CASK and expression of the NMDAR2b gene. J Neurochem 112 (6):1562–1573. doi: 10.1111/j.1471-4159.2010.06569.x [DOI] [PubMed] [Google Scholar]
- 44.Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ (2011) Comparative Analysis of Proteome and Transcriptome Variation in Mouse. Plos Genet 7 (6). doi:ARTN e1001393 10.1371/journal.pgen.1001393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haider S, Pal R (2013) Integrated Analysis of Transcriptomic and Proteomic Data. Curr Genomics 14 (2)91–110. doiDoi 10.2174/1389202911314020003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farzi A, Lau J, Ip CK, Qi Y, Shi YC, Zhang L, Tasan R, Sperk G, Herzog H (2018) Arcuate nucleus and lateral hypothalamic CART neurons in the mouse brain exert opposing effects on energy expenditure. Elife 7. doi:ARTN e36494 10.7554/eLife.36494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Wang SH, Yuan L, Yang YX, Zhang BW, Liu QB, Chen L, Yue W, Li YH, Pei XT (2012) Neuron-restrictive Silencer Factor (NRSF) Represses Cocaine- and Amphetamine-regulated Transcript (CART) Transcription and Antagonizes cAMP-response Element-binding Protein Signaling through a Dual NRSE Mechanism. Journal of Biological Chemistry 287 (51):42574–42587. doi: 10.1074/jbc.M112.376590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang GS, Hong CJ, Yen TY, Huang HY, Ou Y, Huang TN, Jung WG, Kuo TY, Sheng M, Wang TF, Hsueh YP (2004) Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron 42 (1):113–128 [DOI] [PubMed] [Google Scholar]
- 49.Slawson JB, Kuklin EA, Ejima A, Mukherjee K, Ostrovsky L, Griffith LC (2011) Central regulation of locomotor behavior of Drosophila melanogaster depends on a CASK isoform containing CaMK-like and L27 domains. Genetics 187 (1):171–184. doi: 10.1534/genetics.110.123406genetics.110.123406 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaConte LE, Chavan V, Mukherjee K (2014) Identification and glycerol-induced correction of misfolding mutations in the X-linked mental retardation gene CASK. PLoS One 9 (2):e88276. doi: 10.1371/journal.pone.0088276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43 (D1)D447–D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crino PB, Miyata H, Vinters HV (2002) Neurodevelopmental disorders as a cause of seizures: Neuropathologic, genetic, and mechanistic considerations. Brain Pathol 12 (2):212–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLuca SC, Wallace DA, Trucks MR, Mukherjee K (2017) A clinical series using intensive neurorehabilitation to promote functional motor and cognitive skills in three girls with CASK mutation. BMC Res Notes 10 (1)743. doi: 10.1186/s13104-017-3065-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kerr A, Patel PA, LaConte LEW, Liang C, Chen CK, Shah V, Fox MA, Mukherjee K (2019) Non-Cell Autonomous Roles for CASK in Optic Nerve Hypoplasia. Invest Ophthalmol Vis Sci 60 (10):3584–3594. doi: 10.1167/iovs.19-27197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staley K (2015) Molecular mechanisms of epilepsy. Nat Neurosci 18 (3):367–372. doi: 10.1038/nn.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gokce O, Sudhof TC (2013) Membrane-Tethered Monomeric Neurexin LNS-Domain Triggers Synapse Formation. Journal of Neuroscience 33 (36):14617–14628. doi: 10.1523/Jneurosci.1232-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabuchi K, Biederer T, Butz S, Sudhof TC (2002) CASK participates in alternative tripartite complexes in which Mint 1 competes for binding with caskin 1, a novel CASK-binding protein. J Neurosci 22 (11)4264–4273. doi:20026421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsen O, Moore KA, Fukata M, Kazuta T, Trinidad JC, Kauer FW, Streuli M, Misawa H, Burlingame AL, Nicoll RA, Bredt DS (2005) Neurotransmitter release regulated by a MALS-liprin-alpha presynaptic complex. J Cell Biol 170 (7):1127–1134. doijcb.200503011 [pii] 10.1083/jcb.200503011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Luan Z, Liu A, Hu G (2001) The scaffolding protein CASK mediates the interaction between rabphilin3a and beta-neurexins. FEBS Lett 497 (2–3):99–102. doi:S0014-5793(01)02450-4 [pii] [DOI] [PubMed] [Google Scholar]
- 60.Spangler SA, Schmitz SK, Kevenaar JT, de Graaff E, de Wit H, Demmers J, Toonen RF, Hoogenraad CC (2013) Liprin-alpha2 promotes the presynaptic recruitment and turnover of RIM1/CASK to facilitate synaptic transmission. J Cell Biol 201 (6)915–928. doi: 10.1083/jcb.201301011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee K, Yang X, Gerber SH, Kwon HB, Ho A, Castillo PE, Liu X, Sudhof TC (2010) Piccolo and bassoon maintain synaptic vesicle clustering without directly participating in vesicle exocytosis. Proc Natl Acad Sci U S A 107 (14):6504–6509. doi: 10.1073/pnas.10023071071002307107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaeser PS, Deng LB, Wang Y, Dulubova I, Liu XR, Rizo J, Sudhof TC (2011) RIM Proteins Tether Ca2+ Channels to Presynaptic Active Zones via a Direct PDZ-Domain Interaction. Cell 144 (2):282–295. doi: 10.1016/j.cell.2010.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castillo PE, Schoch S, Schmitz F, Sudhof TC, Malenka RC (2002) RIM1 alpha is required for presynaptic long-term potentiation. Nature 415 (6869):327–330. doiDOI 10.1038/415327a [DOI] [PubMed] [Google Scholar]
- 64.Ashery U, Varoqueaux F, Voets T, Betz A, Thakur P, Koch H, Neher E, Brose N, Rettig J (2000) Munc13-1 acts as a priming factor for large dense-core vesicles in bovine chromaffin cells. Embo J 19 (14):3586–3596. doiDOI 10.1093/emboj/19.14.3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhee JS, Betz A, Pyott S, Reim K, Varoqueaux F, Augustin I, Hesse D, Sudhof TC, Takahashi M, Rosenmund C, Brose N (2002) beta phorbol ester- and diacylglycerol-induced augmentation of transmitter release is mediated by Munc13s and not by PKCs. Cell 108 (1):121–133. doiDoi 10.1016/S0092-8674(01)00635-3 [DOI] [PubMed] [Google Scholar]
- 66.Kaufmann N, DeProto J, Ranjan R, Wan H, Van Vactor D (2002) Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron 34 (1):27–38. doiDoi 10.1016/S0896-6273(02)00643-8 [DOI] [PubMed] [Google Scholar]
- 67.Dai Y, Taru H, Deken SL, Grill B, Ackley B, Nonet ML, Jin YS (2006) SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat Neurosci 9 (12):1479–1487. doi: 10.1038/nn1808 [DOI] [PubMed] [Google Scholar]
- 68.Hata Y, Slaughter CA, Sudhof TC (1993) Synaptic Vesicle Fusion Complex Contains Unc-18 Homolog Bound to Syntaxin. Nature 366 (6453):347–351. doiDOI 10.1038/366347a0 [DOI] [PubMed] [Google Scholar]
- 69.Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC (2006) A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell 126 (6):1175–1187. doi: 10.1016/j.cell.2006.08.030 [DOI] [PubMed] [Google Scholar]
- 70.Deak F, Shin OH, Tang J, Hanson P, Ubach J, Jahn R, Rizo J, Kavalali ET, Sudhof TC (2006) Rabphilin regulates SNARE-dependent re-priming of synaptic vesicles for fusion. Embo J 25 (12):2856–2866. doi: 10.1038/sj.embjol.7601165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maximov A, Shin OH, Liu XR, Sudhof TC (2007) Synaptotagmin-12, a synaptic vesicle phosphoprotein that modulates spontaneous neurotransmitter release. Journal of Cell Biology 176 (1):113–124. doi: 10.1083/jcb.200607021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatsuzawa K, Lang T, Fasshauer D, Bruns D, Jahn R (2003) The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. Journal of Biological Chemistry 278 (33):31159–31166. doi: 10.1074/jbc.M305500200 [DOI] [PubMed] [Google Scholar]
- 73.Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Sudhof TC (2002) RIM1 alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature 415 (6869):321–326. doiDOI 10.1038/415321a [DOI] [PubMed] [Google Scholar]
- 74.Clarkson C, Antunes FM, Rubio ME (2016) Conductive Hearing Loss Has Long-Lasting Structural and Molecular Effects on Presynaptic and Postsynaptic Structures of Auditory Nerve Synapses in the Cochlear Nucleus. Journal of Neuroscience 36 (39):10214–10227. doi: 10.1523/Jneurosci.0226-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guldner FH, Ingham CA (1980) Increase in Postsynaptic Density Material in Optic Target Neurons of the Rat Suprachiasmatic Nucleus after Bilateral Enucleation. Neurosci Lett 17 (1-2)27–31. doiDoi 10.1016/0304-3940(80)90056-7 [DOI] [PubMed] [Google Scholar]
- 76.Chowdhury D, Hell JW (2018) Homeostatic synaptic scaling: molecular regulators of synaptic AMPA-type glutamate receptors. F1000Res 7:234. doi: 10.12688/f1000research.13561.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turrigiano G (2012) Homeostatic Synaptic Plasticity: Local and Global Mechanisms for Stabilizing Neuronal Function. Csh Perspect Biol 4 (1). doi:ARTN a005736 10.1101/cshperspect.a005736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC (2013) Presynaptic Neurexin-3 Alternative Splicing trans-Synaptically Controls Postsynaptic AMPA Receptor Trafficking. Cell 154 (1):75–88. doi: 10.1016/j.cell.2013.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dai J, Aoto J, Sudhof TC (2019) Alternative Splicing of Presynaptic Neurexins Differentially Controls Postsynaptic NMDA and AMPA Receptor Responses. Neuron 102 (5):993-+. doi: 10.1016/j.neuron.2019.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trotter JH, Hao JJ, Maxeiner S, Tsetsenis T, Liu ZH, Zhuang XW, Sudhof TC (2019) Synaptic neurexin-1 assembles into dynamically regulated active zone nanoclusters. Journal of Cell Biology 218 (8)2677–2698. doi: 10.1083/jcb.201812076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanford JL, Mays TA, Rafael-Fortney JA (2004) CASK and Dlg form a PDZ protein complex at the mammalian neuromuscular junction. Muscle Nerve 30 (2):164–171. doi: 10.1002/mus.20073 [DOI] [PubMed] [Google Scholar]
- 82.Hodge JJ, Mullasseril P, Griffith LC (2006) Activity-dependent gating of CaMKII autonomous activity by Drosophila CASK. Neuron 51 (3):327–337. doi: 10.1016/j.neuron.2006.06.020 [DOI] [PubMed] [Google Scholar]
- 83.Lu CS, Hodge JJL, Mehren J, Sun XX, Griffith LC (2003) Regulation of the Ca2+/CaM- responsive pool of CaMKII by scaffold-dependent autophosphorylation. Neuron 40 (6):1185–1197. doiDoi 10.1016/S0896-6273(03)00786-4 [DOI] [PubMed] [Google Scholar]
- 84.Jeyifous O, Waites CL, Specht CG, Fujisawa S, Schubert M, Lin EI, Marshall J, Aoki C, de Silva T, Montgomery JM, Garner CC, Green WN (2009) SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci 12 (8):1011–1019. doi: 10.1038/nn.2362n nn.2362 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Omkumar RV, Kiely MJ, Rosenstein AJ, Min KT, Kennedy MB (1996) Identification of a phosphorylation site for calcium/calmodulindependent protein kinase II in the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem 271 (49):31670–31678. doi: 10.1074/jbc.271.49.31670 [DOI] [PubMed] [Google Scholar]
- 86.Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H (2001) Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature 411 (6839):801–805. doiDoi 10.1038/35081080 [DOI] [PubMed] [Google Scholar]
- 87.Hoskins R, Hajnal AF, Harp SA, Kim SK (1996) The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development 122 (1)97–111 [DOI] [PubMed] [Google Scholar]
- 88.Kaech SM, Whitfield CW, Kim SK (1998) The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94 (6):761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horvitz HR, Sulston JE (1980) Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96 (2):435–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferguson EL, Horvitz HR (1985) Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110 (1):17–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alewine C, Kim BY, Hegde V, Welling PA (2007) Lin-7 targets the Kir 2.3 channel on the basolateral membrane via a L27 domain interaction with CASK. Am J Physiol Cell Physiol 293 (6):C1733–1741. doi: 10.1152/ajpcell.00323.2007 [DOI] [PubMed] [Google Scholar]
- 92.Gross GG, Lone GM, Leung LK, Hartenstein V, Guo M (2013) X11/Mint genes control polarized localization of axonal membrane proteins in vivo. J Neurosci 33 (19)8575–8586. doi: 10.1523/JNEUROSCI.5749-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hansen KG, Aviram N, Laborenz J, Bibi C, Meyer M, Spang A, Schuldiner M, Herrmann JM (2018) An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 361 (6407):1118-+. doi: 10.1126/science.aar8174 [DOI] [PubMed] [Google Scholar]
- 94.Ferrari I, Crespi A, Fornasari D, Pietrini G (2016) Novel localisation and possible function of LIN7 and IRSp53 in mitochondria of HeLa cells. Eur J Cell Biol 95 (8):285–293. doi: 10.1016/j.ejcb.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 95.Poston CN, Krishnan SC, Bazemore-Walker CR (2013) In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J Proteomics 79:219–230. doi: 10.1016/j.jprot.2012.12.018 [DOI] [PubMed] [Google Scholar]
- 96.Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, Ting AY (2017) Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6. doiARTN e24463 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.