SUMMARY
DNA binding proteins, supercoiling, macromolecular crowders, and transient DNA attachments to the cell membrane have all been implicated in the organization of the bacterial chromosome. However, it is unclear what role these factors play in compacting the bacterial DNA into a distinct organelle-like entity, the nucleoid. By analyzing the effects of osmotic shock and mechanical squeezing on Escherichia coli, we show that macromolecular crowders play a dominant role in the compaction of the DNA into the nucleoid. We find that a 30% increase in the crowder concentration from physiological levels leads to a 3-fold decrease in the nucleoid’s volume. The compaction is anisotropic, being higher along the long axes of the cell at low crowding levels. At higher crowding levels, the nucleoid becomes spherical, and its compressibility decreases significantly. Furthermore, we find that the compressibility of the nucleoid is not significantly affected by cell growth rates and by prior treatment with rifampicin. The latter results point out that in addition to poly-ribosomes, soluble cytoplasmic proteins have a significant contribution in determining the size of the nucleoid. The contribution of poly-ribosomes dominates at faster and soluble proteins at slower growth rates.
Keywords: DNA, nucleoid, chromosomal organization, macromolecular crowders, osmotic shock, Escherichia coli
Graphical Abstract
The bacterial chromosome is compacted into a distinct organelle-like entity termed the nucleoid, which occupies about one half of the cytosolic volume. Our studies of Escherichia coli cells using osmotic and mechanical shocks show that the main contribution to the formation of the nucleoid comes from the macromolecular crowders in the cytoplasm. The main crowder species responsible for chromosome compaction are soluble cytoplasmic proteins and poly-ribosomes.
INTRODUCTION
Fully replicated chromosomal DNA in Escherichia coli consists of approximately 4.6 Mbp of DNA and has a contour length of 1.6 mm. Despite being about a thousand times longer than the dimensions of the cell, it is confined to a fraction of the cytosolic volume, which is referred to as the nucleoid (Wang et al., 2013). The cytosolic volume fraction occupied by a nucleoid has been estimated to be in the range of 20–75% in E. coli (Woldringh, 2002, Sherratt, 2003, Fisher et al., 2013) and many other bacterial species (Gray et al., 2019). In contrast, when DNA is released from the cell, it expands to a volume that exceeds the cell volume by about 1000-fold (Cunha et al., 2001, Romantsov et al., 2007, Wegner et al., 2012). What leads to the significant compaction of the DNA within the cell is not completely understood. Answering this question is important because DNA compaction can be expected to affect DNA replication, segregation, repair, and transcription. Via transcription, the size of the nucleoid can exert control over a majority of cellular activities.
Several factors contributing to the size of the nucleoid have been proposed. These include nucleoid associated proteins (NAPs), DNA supercoiling, DNA linkages to the membrane (transertion), transcriptional activity, and molecular crowders (Wang et al., 2013, de Vries, 2010, Jin et al., 2013). How much each of these factors/processes contributes to compacting the millimeter-long DNA into the micron-sized cell has remained unclear. Although several NAPs, which can bridge (H-NS) and bend (HU, Fis, IHF) chromosomal DNA, are abundantly present in E. coli during its lag-phase growth (for review see (Dame et al., 2011, Dillon and Dorman, 2010)) they appear to affect chromosome conformations only at the local and intermediary scales - in regions spanning less than 300 kb (Lioy et al., 2018). Moreover, the removal of most of these binding proteins from the cell one at a time does not change the nucleoid size (Wu et al., 2019, Wang et al., 2013). The exception is a low-abundance of MatP that anchors the replication terminus region to the divisome (Espeli et al., 2012, Männik et al., 2016) and conveys different organization to this ~800 kb long domain (Mercier et al., 2008, Lioy et al., 2018). Also, condensin-like structural maintenance complex MukBEF has been reported to lead to global-scale decondensed chromosomes (Nolivos and Sherratt, 2014) possibly via DNA looping mechanisms (Terakawa et al., 2017).
Along with NAPs, DNA supercoiling can also be expected to compact chromosomal DNA (Woldringh et al., 1995). It has been proposed that individual supercoils can nematically align at densities, 13 g/l, which could lead to a compacted polymer (Odijk, 1998). Although such DNA densities are present in the E. coli nucleoid, correlations between DNA supercoiling levels and nucleoid size were not observed in vivo under inhibition of gyrase activity (Stuger et al., 2002, Cagliero and Jin, 2013). However, supercoiling dependent shrinkage of nucleoid has been reported in vitro (Romantsov et al., 2007).
In addition to the mechanisms that compact the nucleoid, an active process, termed transertion, expands it (Libby et al., 2012, Bakshi et al., 2014, Bakshi et al., 2012). Transertion refers to a process whereby DNA is tethered to the plasma membrane via concurrently occurring transcription, translation, and membrane insertion (Woldringh, 2002, Roggiani and Goulian, 2015). Transertion linkages can form when an integral membrane protein with an N-terminal membrane domain is synthesized. These linkages consist of RNA polymerase, transcribed mRNA, translating ribosomes, and the membrane protein that has inserted its N-terminal domain into the cell membrane before its synthesis has completed. These large and structurally complex linkages are inherently short-lived and limited by the time it takes to transcribe a gene (~10 sec). Evidence for the existence of these linkages has been provided in recent studies (Libby et al., 2012, Bakshi et al., 2014), but the conditions in which the linkages affect nucleoid size have not been mapped out yet.
While all the above processes have been extensively studied, none of them appear to be capable of explaining the extent of the compaction needed to confine DNA into the experimentally observed nucleoid sizes. A number of theoretical (Odijk, 1998) and modeling studies (Mondal et al., 2011, Shendruk et al., 2015, Shin et al., 2014, Kim et al., 2015, Joyeux, 2018) have pointed out that macromolecular crowders can be the main mechanism for the compaction of chromosomal DNA. In the seminal work, Odijk proposed that crowders and DNA separate into two distinct phases in an E. coli cell, a nucleoid and cytosolic phase (Odijk, 1998). The nucleoid phase contains the chromosome and is depleted of cytoplasmic proteins, whereas the cytosolic phase has an excess number of soluble proteins. Several modeling studies have confirmed these predictions (Mondal et al., 2011, Shendruk et al., 2015, Shin et al., 2014, Kim et al., 2015). However, all these results are based on equilibrium thermodynamics and coarse-grained models of the DNA and the cytosol. It is unclear to what degree these approaches are valid for living cells, which are far out of equilibrium and whose DNA has a complex folding structure.
To date, quantitative experimental tests to verify these models have been carried out in vitro, using purified DNA (Zhang et al., 2009) or DNA liberated from cells (Cunha et al., 2001, Pelletier et al., 2012). In these studies, charge-neutral polymers, dextran or polyethylene glycol (PEG), have been used as crowding agents to mimic the cytoplasmic environment even though most cytosolic crowders do not have a neutral charge. All these experiments agree that crowding can lead to significant compaction of the DNA. However, the data are in disagreement if the compaction occurs abruptly via a first order coil-globule phase transition with an observed metastable state (Pelletier et al., 2012) or gradually via a second order transition as the concentration of crowding agents increases (Cunha et al., 2001).
Thus far, there are no quantitative experimental studies on how osmolality and associated changes in macromolecular crowding affect nucleoid size in living bacteria. At a qualitative level, it is known that hyperosmotic shock leads to the compaction of nucleoids (Cagliero and Jin, 2013, Wu et al., 2019). Here we carry out microfluidic experiments to quantitatively study the role of molecular crowders in the compaction of the E. coli nucleoid (i) by rapidly changing the osmolality of the growth media for steady-state growing bacteria in the mother machine device, and (ii) by squeezing individual cells in a device specially designed for such measurements. We show that in vivo E. coli nucleoids respond to the osmolality continuously. Close to physiological crowder concentrations, the nucleoid length and width change linearly. As the crowder concentration exceeds the physiological level by 30% the compressibility significantly decreases. Also, our data show that the compressibility is strongly anisotropic being higher along the long axes of the cell and it is independent of growth conditions (slow and moderately fast growth). The latter finding indicates that the overall crowding level rather than the exact composition of crowders controls the compaction in these growth conditions. Furthermore, our results indicate that poly-ribosomes are the dominant crowder species in fast growth conditions while the soluble cytosolic proteins are dominant in slow growth conditions.
RESULTS
Compaction of the nucleoid under osmotic shock is anisotropic and larger than the compaction of the cytosol
Our first goal was to find a quantitative relationship between crowder concentrations/volume fractions and sizes of the nucleoids. Since altering the number of crowders in the cell is not practically possible because all macromolecules are potential crowders, one has to rely on changing the volume of the cell instead. One possibility to vary the cytosolic volume, and thereby alter the crowder concentration, is to change the osmolality of the growth media. By increasing the external osmolality, cytosolic water leaves the cell and crowder concentration increases, while the decrease in osmolality leads to the opposite result. To vary the external osmolality and observe cellular changes in real time, we image bacteria in microfluidic mother-machine devices (Wang et al., 2010, Yang et al., 2018), which allowed us to quickly change media without perturbing cell imaging (Fig 1A, B, SI Movie M1, M2). To induce osmotic shock, we changed the concentration of NaCl in a chemically defined growth medium. To quantify the changes in both cytoplasmic and nucleoid sizes, the cells carried the tagRFP-T and HupA-mNeonGreen labels (Table S1). The former label diffusively fills the cytosol while the latter binds non-specifically to DNA (Wery et al., 2001). To understand how different macromolecular crowders affect DNA compaction, we studied cells in slow and moderately fast growth conditions. The doubling times in these two conditions at 28°C are Td = 226 ± 103 min and Td = 95 ± 24 min, respectively (Table S2). We excluded fast growth because a much more complicated DNA topology and nucleoid shape in these conditions would have made the interpretation of the results ambiguous. From slow to moderately fast growth, one would expect the ribosome protein ratio in the cells to increase by 1.5–2.0 times (Bremer and Dennis, 2008, Ehrenberg et al., 2013, Dai et al., 2017) allowing differentiation of the effects arising from stable RNA and protein based crowders on the compaction of the nucleoid.
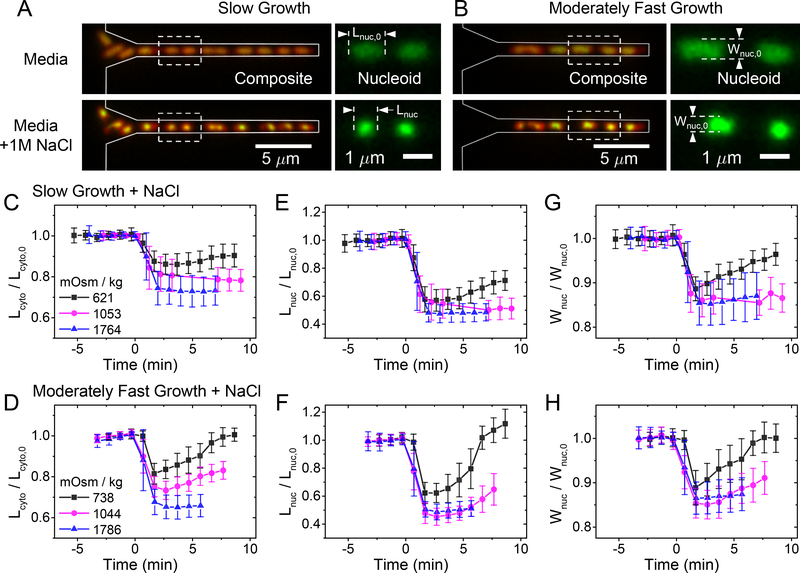
Figure 1. Kinetics of the cytosol and nucleoid compaction during hyperosmotic shocks.
(A) Left: Images of E. coli cells growing in mother machine channels before (top) and 1 min after osmotic shock (bottom). The images are composites of fluorescence images of nucleoid (green) and cytosol (red). Channel contours, which are determined from phase images, are indicated by grey lines. Right: Nucleoid images for the two cells indicated by a dashed box in the left image. The cells were grown in slow growth conditions in M9 glycerol medium. (B) The same strain in moderately fast growth conditions in M9 glucose + CAS medium. (C, D) Relative changes of cytosolic lengths for three hyperosmotic shocks in slow and moderately fast growth conditions, respectively. The shock magnitudes are indicated in lower left. Lcyto,0 is cytosolic length right before the shock. The same for nucleoid length (E, F) and width (G, H). Error bars correspond to std. The number of cells analyzed in each measurement is reported in SI Table S3.
To quantify changes in nucleoid and cell sizes at different crowding levels, we analyzed cells in the early stages of the cell cycle when their nucleoid has an ellipsoidal shape. In the studied growth conditions, such morphology appears in nucleoids that are less than half replicated while in later stages of replication, the nucleoids obtain a characteristic bilobed shape (Bates and Kleckner, 2005, Männik et al., 2016). We limit our study to single-lobed nucleoids because these can be easily characterized by their length and width measured along the long and short axes of the cell, respectively. We followed changes in these two parameters along with changes in cytoplasmic sizes in both hypo- and hyperosmotic conditions. However, changes in cytoplasmic volume were essentially negligible even for the most hypoosmotic shock possible when the regular growth medium was replaced by distilled water (0 Osm). Thus, almost all of our studies cover hyperosmotic conditions.
As expected, the change in external osmolality by NaCl leads to rapid changes in cell volume and nucleoid size for both growth conditions (Fig. 1C–H). These changes occurred during a one-minute period. The rapid changes in cell shape observed here are consistent with earlier reports (Pilizota and Shaevitz, 2013, Rojas et al., 2014). Interestingly, the changes in nucleoid dimensions followed the same time-dependence as changes in cell dimensions. However, dimensions of the nucleoid changed to a larger extent than the overall dimensions of the cell, as will be detailed later. Although the focus of this study is not on recovery processes, in milder osmotic shocks (less than about 0.7 Osm kg−1), the recovery of the cells is visible within 5 minutes after the shock. In these mild shocks, recovery of cell shape and nucleoid shape followed the same time-dependence, which is consistent with the idea that crowding is responsible for the nucleoid compaction.
In some cells, the hyperosmotic treatment resulted in plasmolysis. The majority of these cases showed detachment of the plasma membrane from the cell wall at the cell poles; detachments from the cylindrical parts of the cell body were less frequent, which has also been reported by others (Pilizota and Shaevitz, 2013). In cells that plasmolyzed from their sidewalls, the cell width is poorly defined. Moreover, in such cells, nucleoids become irregularly shaped. For these reasons, we excluded side-wall-plasmolyzed cells from further analyses. We found that in both moderately fast and slow growth conditions, the increase in external osmolality leads to an approximately linear decrease in cell length for osmotic shocks although above ≈1.2 Osm kg−1 the decrease slowed (Fig. 2A, B). Similar to cell length, the nucleoid length also decreased linearly with the osmolality change for smaller osmotic shocks (<0.9 Osm kg−1). At the same time, the relative changes in nucleoid length were about 2.5 times as large as the relative changes in cell length (SI Table S4). At above about 0.9 Osm kg−1 the change of nucleoid length ceased. Nucleoid width behaved qualitatively similar to nucleoid length while the cytoplasmic width did not show significant changes throughout the range of osmolalities studied (Fig. 2 C, D). To rule out that the plateauing of nucleoid dimensions at higher osmolalities is caused by the point-spread-function of the microscope, we measured the diameters of 100 nm fluorescent beads for comparison. The measured diameters of the beads were, by more than a factor of two, smaller than the smallest measured nucleoid dimension at the highest osmotic shock (SI Fig. S1). Thus, these measurements confirmed that the measured dimensions reflect the intrinsic size of the nucleoid in the plateau region. The presence of a plateau region shows that, in vivo, the nucleoid undergoes a transition from a linear high compressibility regime to a non-linear low compressibility regime upon compaction.
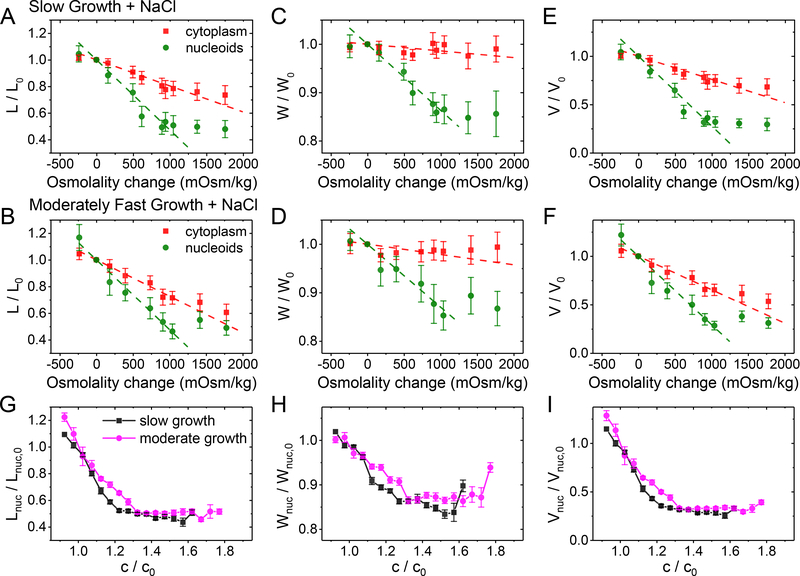
Figure 2. Change in nucleoid dimensions as a function of external osmolality and as a function of crowder concentration.
(A, B) Relative change of nucleoid (green circles) and cytosolic (red squares) lengths at different osmotic shock in slow and moderately fast growth conditions, respectively. Each osmolality value corresponds to a separate measurement. Error bars are std. Dashed lines are linear fits to the experimental data in the range from −0.25 to 1.25 Osm kg−1. The best fit parameters can be found in SI Table S4. The same for nucleoid and cell widths (C, D) and calculated volumes (E, F) in these two growth conditions. (G) Nucleoid length, (H) width, and (I) calculated volume in slow (black squares) and moderately fast (magenta circles) growth conditions as a function crowder concentration. The crowder concentrations (c) are relative to those in normal growth medium without excess NaCl (co). The details of calculating these concentrations can be found from SI Text: Determination of crowder concentration based on the intensity of fluorescent reporters. Error bars represent s.e.m.
A comparison of nucleoid width and length curves showed strongly anisotropic compaction. The changes in nucleoid width at smaller osmolality changes (<0.9 Osm kg−1) was by a factor of 3.7 smaller than the corresponding change in the nucleoid length. The anisotropy was also present in the plateau region where the nucleoid length was compressed to 50%, while its width only compressed to 70% of its original size. The nucleoid volume calculated based on these values decreased to 30% of its original size (Fig. 2 E–F). The anisotropic compaction leads to a spherical nucleoid having an aspect ratio of one at osmolalities of about 0.9 Osm kg−1 and above (SI Fig. S2). The spherical shape indicates the underlying isotropic organization of the nucleoid in the compacted state. Since our measurement time-scale is 1 minute, this conclusion furthermore implies that the native uncompressed nucleoid should also have globally isotropic organization because global rearrangement from anisotropic to isotropic is unlikely to occur during this time-scale.
Nucleoid compaction is a second order phase transition and independent of growth conditions
To relate changes in osmolality to changes in crowding of the cytosolic environment, we calculated the ratio of crowder concentration immediately after the osmotic shock (c) relative to that during regular growth conditions (c0). This concentration ratio (c/c0) applies to any cytosolic molecular species whose diffusion across the plasma membrane during osmotic shock can be neglected. It includes not only macromolecular crowders but also all small molecules and ions. For all these molecules the relative concentration change (c/c0) equals the inverse of their relative cytosolic volume change (Vcyto,0/Vcyto). However, instead of using cytosolic volumes to calculate the latter, we determined the relative concentration of cytosolic tagRFP-T proteins before and after the salt treatment (for details see SI Text), and used this value for c/c0. We chose this approach because even though we only analyzed cells that did not show apparent plasmolysis from their lateral cell walls, we could have overlooked some plasmolyzed regions during a visual inspection of the images. Overall, the two methods yielded comparable results at smaller osmotic shocks but deviated marginally from each other at larger ones (SI Fig. S3).
Our data show that the length, width, and volume of the nucleoid decreased, initially, linearly as the crowder concentrations increased (Fig. 2G–I). The linear relationship corresponds to a constant compressibility of the nucleoid by the crowders. Here the compressibility, κ, is defined as the relative change in volume upon the change in osmotic pressure, ΔP, by . In the lowest order approximation P~c, and in this case, κ is proportional to . The latter corresponds to the slope of the curves in Fig. 2I. Once the crowding level exceeded about 30% of the level in normal growth conditions, the compressibility decreased sharply. The transition to the low compressibility regime was smooth unlike in several previous in vitro studies (Pelletier et al., 2012, Krotova et al., 2010, Yoshikawa et al., 2010). There was no sign of co-existence of collapsed and extended DNA conformations during a visual inspection of the nucleoid images nor in the raw data of nucleoid lengths and widths (SI Fig. S4). Both findings together indicate that the coil-globule transition of the nucleoid is a second rather than first order phase transition in vivo conditions.
Interestingly, we found that the dependence of the nucleoid size on the crowder concentration is almost the same in both studied growth conditions (Fig. 2G–I). This outcome is surprising because concentrations of the main macromolecular crowders, the ribosomes, and the proteins, are expected to be significantly different in these two growth conditions (Ehrenberg et al., 2013, Dai et al., 2017). Based on available published data, we estimate the concentration of soluble cytosolic proteins to be about two-fold higher and ribosomes about two-fold lower in slow compared to moderately fast growth conditions (SI Table S5–S6).
We hypothesized that the insensitivity of nucleoid compaction to the crowder composition could indicate the importance of some other factor than the macromolecular crowders to the compaction process. It is known that E. coli responds to osmotic shock by pumping K+ ions from its surrounding environment (Epstein and Schultz, 1965, Richey et al., 1987). Furthermore, it was reported that an E. coli nucleoid did not compact under osmotic shock in a medium depleted of K+, albeit the presented evidence relied on an image of a single cell (Cagliero and Jin, 2013). To further understand the effect of K+ influx on nucleoid compaction, we substituted K+ in the moderately fast growth medium with Na+ 20 min prior to osmotic shock. Osmotic shock with 0.75 Osm/kg NaCl in K+ depleted medium yielded the same result as the osmotic shock where K+ was present at the regular 24 mM concentrations, although nucleoid recovery was altered for times longer than 5 min (SI Fig. S5). While this finding contradicts the earlier report (Cagliero and Jin, 2013), it is consistent with in vitro findings that monovalent ions such as K+ only modestly influence DNA condensation; unlike tri and higher valent cations (Joyeux, 2016). Also, previous in vivo measurements support the idea that the influx of K+ is relatively minor during shorter time scales of about 5 min to significantly alter (<10%) the intracellular K+ concentration (Epstein and Schultz, 1965). Several past works thus support our findings show that at short time scales (< 5 min) following hyperosmotic shock, the K+ influx into the cytoplasm does not affect nucleoid dimensions.
Mechanical squeezing measurements confirm osmotic shock data
In addition to removing cellular water content via osmosis, we also carried out measurements where we removed water by the mechanical squeezing of the cells. For that purpose, we constructed a microfluidic device, which we refer to as the microanvil (Fig. 3A, for details, see SI Text). This device allowed us to mechanically press out part of the cytosolic content while imaging the cell (Fig. 3B). Since the applied pressure to the cells can be expected to activate mechanosensitive channels (Kung et al., 2010), one could expect in addition to water molecules also ions and small metabolite molecules could leave the cell. Our first proof-of-principle devices had the smallest anvil dimension of about 2 μm. In future designs, this dimension can be reduced. Since 2 μm constitutes a sizeable portion of the cell length (2–4 μm), we studied longer than normal cells. The cells were elongated by inducing SulA expression from an extra plasmid copy, thereby inhibiting cell division (Dajkovic et al., 2008). The elongated cells had lengths in the range of 7–15 μm and had multiple nucleoids. The amount of cytosolic content that can be removed from the cells can be controlled by an external pressure applied by the anvil. However, at higher pressures, the anvil divides the cell into two distinct compartments (Fig. 3B, SI Movie M3). In the region between the two compartments, the cytosolic content of the cell is almost completely removed (SI Fig. S6). Further increases to the externally applied pressure by the anvil do not appreciably change the cytosolic volume of the cell. For this reason, the maximum volume that was removed was limited to about 25% of the initial cytosolic volume in these experiments.
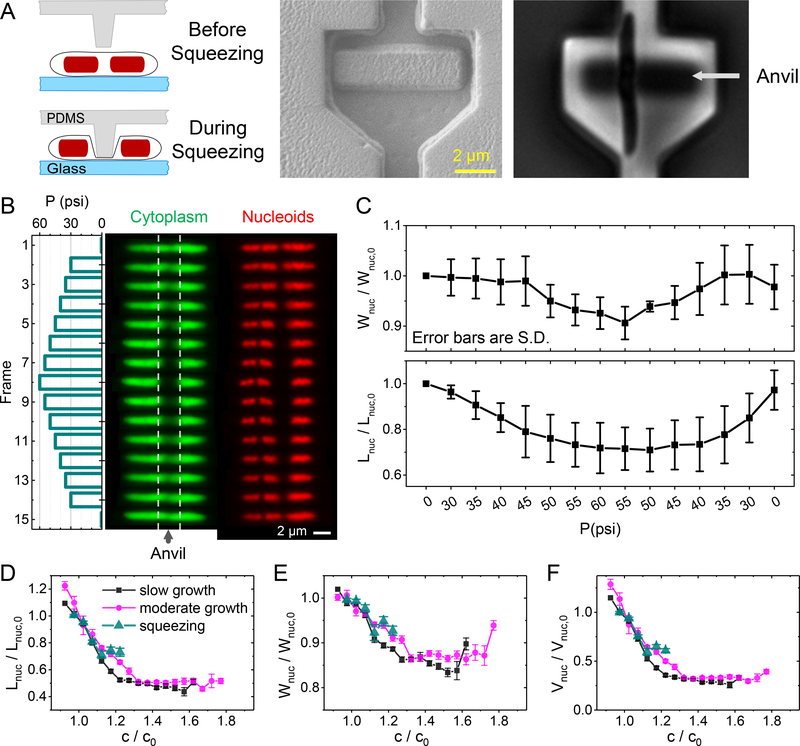
Figure 3. Measurements of nucleoid compaction during cell squeezing.
(A) Left: Schematics of the measurement. A pressure actuated valve is fabricated with a small anvil-like protrusion. Externally applied pressure deflects the anvil downward and squeezes about a 2 μm long portion of the underlying cell. Middle: SEM image of valve and anvil. Right: phase image of cell trapped under the anvil. (B) Response of an elongated E. coli cell to a pressure cycle. Left panel: Change of externally applied pressure during the measurements. In the 1st (bottom) and the last (top) frame the cell is not squeezed. Middle panel: images of cytoplasmic mNG label during this pressure cycle. The region between dashed lines correspond to the portion of the cell where the anvil touches it. Right panel: Nucleoid images for this cell. (C) The average nucleoid widths (top) and length (bottom) for this cell. The average is over three nucleoids in this cell and error bars reflect std. Both the width and length of the nucleoids are relative to those at the beginning of the squeezing cycle (Wnuc,o, Lnuc,o, respectively). (D) Comparison of relative nucleoid length as a function of crowder concentration from squeezing measurements (green triangles) to that from osmotic shock measurements (magenta circles and black squares). The same for nucleoid width (E) and calculated volume (F).
The compaction of the nucleoid increased during stepwise pressure ramps and recovered when the pressure was lowered (Fig. 3C). As with measurements with osmolality variation, efflux and influx of water and other cytosolic components were taking place at a faster time scale than the measurement rate (2 min−1). After one pressure cycle, the majority of cells were capable of resuming growth.
We determined nucleoid dimensions and volumes from these measurements as a function of the changes in crowder concentrations and compared these results with the findings from the osmotic shock measurements (Fig. 3D–F, S7). Within uncertainties of the measurement, the two approaches yielded indistinguishable results, thus further confirming the osmotic shock measurements. The data also implies that increased ion concentration during osmotic shock measurements does not significantly affect nucleoid compaction, because in squeezing measurements, one could expect the ionic strength of the cytosol to not significantly change if the assumption of the opening of mechanosensitive channels is correct.
Transertion linkages and crowding by polysomes have a limited effect on nucleoid size in slow growth conditions
Next, we aimed to understand which of the macromolecular crowders in the bacterial cytosol play the dominant role in compaction of the nucleoid. Several lines of research point out that the main compacting agents among different macromolecular crowders species are poly-ribosomes (polysomes) (Joyeux, 2016, Mondal et al., 2011, Bakshi et al., 2014, Bakshi et al., 2015). This conclusion has been partially drawn from studies of cells treated with rifampicin, a transcription halting drug. The drug appears to affect the nucleoid size via two different mechanisms in fast growth conditions (Bakshi et al., 2014). The measurements by Bakshi et al., which were carried out in EZ Rich Defined Medium at 30°C where the cell doubling times were 48 min, have shown that the nucleoids compact at shorter time scales (0–5 min). The compaction has been explained to be the result of severing transertional linkages between DNA and the inner membrane. These linkages are expected to keep the nucleoid in an expanded state and to resist the compaction effects from the crowders. In longer timescales (>10–15 min), rifampicin leads to the expansion of chromosomal DNA so that it appears to fill the whole cytosolic volume (Bakshi et al., 2015). The expansion has been interpreted as the result of polysomes dissociating into 30S and 50S ribosomal subunits and by the lower ability of these subunits to compact the nucleoid (Mondal et al., 2011, Bakshi et al., 2014, Bakshi et al., 2015). Since the dissociation of the polysomes during rifampicin treatment has such drastic effects on the nucleoid size, one can expect them to be the dominant species in compacting the nucleoid.
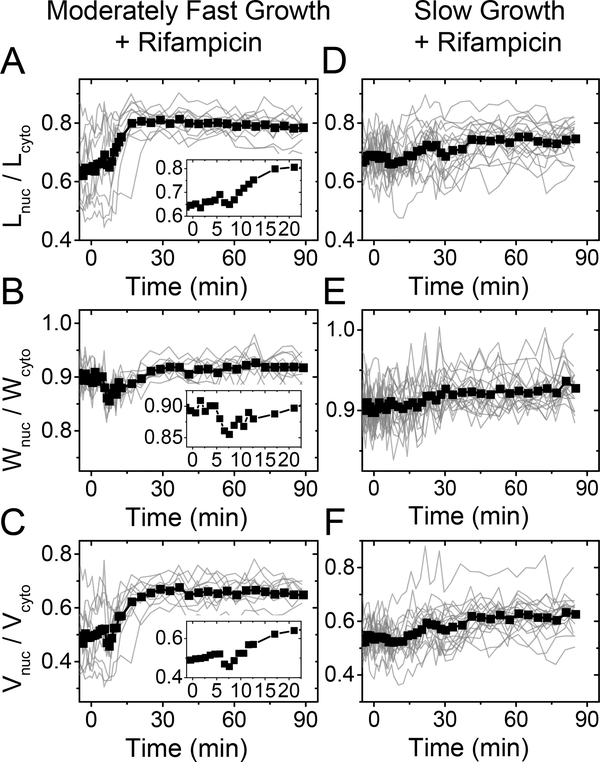
We were interested in understanding if the above described nucleoid behaviors during rifampicin treatment also hold in moderately fast and slow growth conditions where ribosome and polysome concentrations are expected to be lower (Ehrenberg et al., 2013, Dai et al., 2017). Treating moderately fast growing cells with rifampicin, we observed an initial compaction of the nucleoid at about the 5 min timescale (Fig 4 A–C), which is consistent with the earlier reports (Bakshi et al., 2014, Spahn et al., 2018). Although the effect of compaction in our experiments was smaller (3–5%) (Fig. 4 A–C insets) than in the previous reports (about 8%), which were carried out in fast growth conditions, the presence of nucleoid contraction supports the idea of the existence of transertion linkages. However, their effect in determining the size of the nucleoid appears rather modest in this growth condition. At timescales longer than 5 min, we observed the expansion of the nucleoid (Fig. 4 A–C). Note that in these data, nucleoid dimensions have been normalized by cell dimensions instead of nucleoid dimensions at the beginning of the drug treatment. We used this scaling because of nucleoid length and its volume increase approximately proportionally to the corresponding cell dimensions. Since the cell growth does not stop immediately after the drug is introduced to the growth medium (SI Fig S8), normalization using the cell dimensions compensates for the growth related expansion of the nucleoid. Our data show that the expansion of the nucleoid dimensions unaccounted by the cell growth stops about 20 minutes after introduction of the drug (Fig 4A–C). The observed expansion is qualitatively consistent with the earlier reports (Bakshi et al., 2014, Cabrera et al., 2009). At a quantitative level, we find that the expansion leads to a 27% increase in nucleoid length and a 2% increase in nucleoid width in moderately fast growth conditions.
Figure 4. Change in nucleoid dimensions under rifampicin treatment in moderately fast (left) and slow (right) growth conditions.
300 μg ml−1 rifampicin is administered to cells at 0 minutes. There is 2–3 min delay in fluidic lines before the drug reaches the cells. (A, D) Ratio of nucleoid to cell length as a function of time. Traces from individual cells are shown by thin lines, and the population average trace by a thick line with squares. N=20 for slow and N=11 for moderately fast growth conditions. Inset in moderately fast growth conditions is zoomed in from the beginning of the trace. (B, E) The same for nucleoid width. (C, F) The same for the calculated nucleoid volume.
All of the above changes in nucleoid dimensions were significantly diminished in slow growth conditions (Fig. 4 D–F), although rifampicin manifested itself in stopping cell growth (SI Fig. S8). There was no distinctly detectable decrease in nucleoid dimensions at short time scales (< 5 min), indicating that the effect of transertion in determining nucleoid size is negligible in slow growth conditions. This can be expected. Lower growth rate means a proportionally lower total protein synthesis rate in cells while the number of transertion linkages at any given time should scale with this rate. In longer time periods of about 30 minutes, the nucleoid length and width increased by 7% and 2%, respectively. While the change in width was comparable to that found in moderately fast growth conditions, the change in length was four times smaller. The significantly smaller nucleoid expansion could result from about a twice smaller concentration of polysomes in this growth condition (Table S6). Their dissociation to the 30S and 50S subunits would, therefore, lead to the smaller expansion of the nucleoid as observed in the experiments. The latter conclusion leads then to a further inference that polysomes have a minor contribution to the compaction of the nucleoid in slow growth conditions.
Effect of osmotic shock on nucleoids after rifampicin treatment
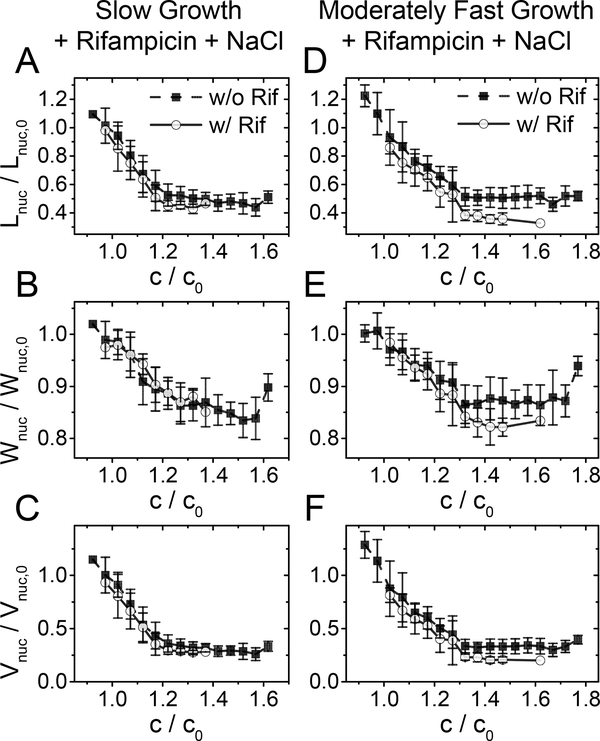
To further understand the role of polysomes in the compaction of the nucleoid, we applied osmotic shocks to cells that were treated for 20–25 min with rifampicin (Fig. 5). The latter period corresponds approximately to the time of nucleoid expansion (cf. Fig. 4 A–C). During this treatment period, the majority of the polysomes should dissociate into 30S and 50S subunits based on single molecule tracking results (Bakshi et al., 2012, Sanamrad et al., 2014) and on estimated the mRNA lifetime in E. coli of about 5 min (Bernstein et al., 2002). Note that the chosen 20–25 min period is short enough for the concentration of the proteins and stable RNA species in the cytosol to not alter significantly. We estimate their change to be less than 10% due to their dilution by residual cell growth while their decrease due to degradation can be expected to be minimal during this period. If ribosomal subunits are less effective in compacting the chromosomes (Bakshi et al., 2014, Bakshi et al., 2015) then we would expect weaker compaction of nucleoids in rifampicin treated cells during osmotic shock measurements. Contrary to this expectation, the nucleoid compaction was somewhat larger in rifampicin treated cells in both growth conditions (Fig. 5). In moderately fast growth conditions, a slight increase in compaction could be assigned to the fact that prior to the osmotic shock, the nucleoid was significantly larger in rifampicin-treated cells than in untreated ones. To compensate for this effect, we extrapolated nucleoid lengths from the beginning of the treatment to the point where salt shock occurred, assuming an increase in nucleoid length would have followed the same increase as during normal growth (SI Fig. S9). The resulting data still showed negligible effects from rifampicin treatment on the nucleoid compaction curves (SI Fig. S10,11). Furthermore, we found the distribution of the absolute, as opposed to normalized, nucleoid widths to be indistinguishable between rifampicin treated and untreated cells after the osmotic shock (SI Fig. S12). Thus, the nucleoid is compacted into the same final width (in micrometers) as a result of hyperosmotic shock irrespective of a prior rifampicin treatment or not. This result is consistent with our earlier inference that polysomes play a minor role in compaction of the nucleoid in slow growth conditions. However, at moderately fast growth, the same explanation would be an apparent contradiction with our prior reasoning for nucleoid expansion upon polysome dissociation. A possible way to reconcile these conflicting explanations is that only part of the nucleoid expansion under rifampicin treatment can be explained by polysome dissociation in moderately fast growth conditions, while the rest may be the result of some other effect rifampicin has on the nucleoid. This idea is also supported by examining the structure of the nucleoids before and after rifampicin treatment. In fast growth conditions (in defined EZ-Rich medium) and to some extent in moderately fast growth conditions, nucleoids display a distinct structure in our images (SI Fig. S13). This structure was completely lost following the 20–25 minute treatment by the drug, indicating that beyond affecting crowding and transertion, rifampicin has some further effects on nucleoid size and structure.
Figure 5. Effect of osmotic shock after rifampicin treatment.
Cells were first treated for 25–30 min with 300 μg ml−1 rifampicin and then the concentration of NaCl in the medium was changed. Rifampicin was also present in the medium during osmotic shock. Nucleoid dimension right after osmotic shock divided by the same dimension right before the shock (solid line open circle). For comparison the same ratio without rifampicin treatment is also shown (black dashed line solid square; from Fig. 2G–F). (A, B) Relative nucleoid length as a function of relative crowder concentration. (C, D) The same for relative nucleoid width, and (E, F) for the calculated volume.
Crowding model qualitatively agrees with experimental findings
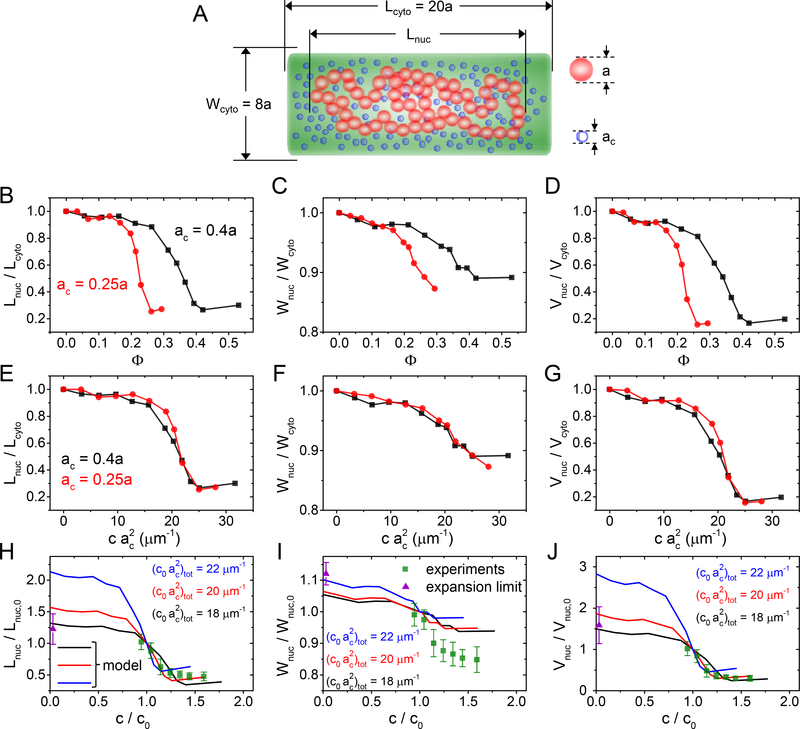
We sought to explain the measured data by using Brownian dynamics simulations of chromosomes in crowded and confined environments. Our computational approach is similar to the one reported by Kim et al. (Kim et al., 2015) (for more details, see SI Text). Extending the previous report, we also calculate the nucleoid width and volume in addition to the nucleoid length to compare these quantities to experimental values. The model represents the chromosome as a chain of linked beads and crowders as smaller-sized unlinked beads. Both species interact via repulsive excluded volume interactions and are confined to a cylindrical volume (Fig. 6A). The model chromosome corresponds to a single fully replicated circular E. coli chromosome.
Figure 6. Comparing a polymer model to the experimental data.
(A) A model of DNA consists of 150 beads with diameter a that are confined to a cylindrical volume. The same volume is also occupied by a variable number of crowders having diameters ac. For details of the model see SI Text: Coarse-grained Brownian Dynamics simulations. (B-D) Calculated nucleoid length, width, and volume for two different size crowders as a function of their volume fraction. (E-G) Calculated nucleoid length, width, and volume as a function of effective “crowding level” defined as . The curves with different crowder diameters collapse into a single one. (H-J) Comparing modelled nucleoid length, width, and volume (solid lines) to experimental data from slow growth conditions (green squares; from Fig. 2G–F). The experimental data is augmented by a data point at the zero crowder concentration limit where the nucleoid is expected to extend over the whole cytosolic volume (violet triangle). A set of curves from the model with “crowding level” 18, 20, 22 μm−1 are plotted. Of those the best agreement between the experiment and the model is at 20 μm−1. The error bars for the data are std.
According to model calculations, the length, width, and volume show approximately sigmoidal dependence on the volume fraction of crowders (Φ) or the concentration of crowders (Fig. 6B–D, SI Fig. S14). The former is defined as the total volume of crowders divided by the volume of the cytoplasm and it is proportional to the concentration of crowders (Rubinstein and Colby, 2003). Similar dependence for nucleoid length on the volume fraction was also observed in earlier models (Shendruk et al., 2015, Kim et al., 2015). The model predicts that in both low and high crowder volume fractions, nucleoid dimensions are insensitive to the number of crowders in the cellular volume. The transition between the two regimes occurs continuously, i.e., the coil-globule transition in this model is second order. All these predictions are consistent with our experimental data on a qualitative level.
We calculate the aforementioned dependencies for two different crowder sizes. The model predicts that the mid-point of nucleoid compaction occurs at larger volume fractions for larger crowders. However, a smaller concentration of larger crowders is needed to compact the nucleoid to the same level (SI Fig. S11). Kim et al. have previously found that multiplying crowder concentration by the square of the crowder diameter, i.e., by , collapses ) for different size crowders to a single curve (Kim et al., 2015). Our model indicated that this scaling applied in addition to the nucleoid length also to its width and volume (Fig. 6 E–G). Furthermore, results by Kim et al. showed that for a polydisperse ensemble of crowders Lnuc vs curves collapsed for different crowder mixtures into a single curve (Kim et al., 2015). The above sum is taken over all crowder species. Thus, the level of crowding in polydisperse samples is, according to the model, characterized by a single parameter which one could calculate knowing the concentrations and diameters of all crowders. Note that even though scaling was found semi-empirically for the above described course grained model, it has a clearer meaning at the microscopic level. The quantity , where LDNA is the contour length and d the effective diameter of chromosomal DNA (≥ 2.2 nm) (Odijk, 1998), is approximately the excluded volume fraction associated with DNA and crowder interactions(Odijk, 1998). So, in leading order is proportional to the relevant volume fraction. This argument applies to small crowders such as proteins. Large crowders, such as polysomes, can be expected to interact predominantly with DNA supercoils. In this case the above quantity is not relevant but the coarse-grained model can be expected to capture the interaction well. Furthermore, the sum rule found by Kim et al. (Kim et al., 2015) indicates that crowder-crowder interactions (crowding between crowders) can be effectively neglected and each crowder species acts independently of each other on DNA via their partial osmotic pressures.
We used the scaling behavior of model curves to compare them to our experimental data. By adjusting the crowding parameter , which corresponds to the crowding level in the regular medium, we found the best match between experiment and model for (Fig. 6 H–J). In this comparison, we also accounted that at zero crowder concentration, the nucleoid must fill the whole cell volume. This extra constraint was determined as the ratio of the measured cytoplasmic size to nucleoid size in regular growth medium (violet triangles in Fig. 6H–J). As can be seen from Fig. 6H, the model predicts the experimentally observed length dependence on the crowding level reasonably well. The agreement is poorer for the nucleoid width, where the experiment shows much larger variations and thus smaller anisotropy in the compressibility (Fig. 6I). Taking the coarse-grained nature of the model, some quantitative discrepancies can be expected; yet the main experimental characteristics are clearly represented in the model.
DISCUSSION
Smooth coil-globule transition of nucleoids in the cellular environment
Our measurements show that under hyperosmotic treatment and during mechanical squeezing, E. coli nucleoids undergo rapid compaction on a timescale of about one minute. The compaction occurs concurrent with the changes in the cytoplasmic volume, but during mild shocks, the nucleoid volume decreases by a factor of about 2.5 times more than the cytoplasm. The shrinkage of the nucleoid can be explained by an increased osmotic pressure from cytosolic macromolecular crowders, concentration of which arises when the cytosolic volume of the cell decreases. Our in vivo findings are in overall agreement with previous in vitro studies where artificial crowding agents or ions have been used. However, several in vitro studies have reported an abrupt transition from an extended state of the liberated nucleoid (coil) to a highly compacted state (globule) (Krotova et al., 2010, Pelletier et al., 2012, Yoshikawa et al., 2010). Here, we find that in vivo, the compaction of the nucleoid is a continuous function of cytoplasmic crowder concentration. Therefore, the associated coil-globule transition of the chromosome is the second rather than the first order phase transition in the cellular environment. The difference between in vivo and in vitro experiments can be related to the different nature of crowders, the different organization of DNA, or the differences in ionic composition of the environment. The in vitro measurements used relatively small crowders (PEG and BSA) compared for example, to polysome particles. Also, there was a reduced level (Pelletier et al., 2012) or no (Krotova et al., 2010, Yoshikawa et al., 2010) supercoiling and DNA cross-linking via DNA binding proteins present in measurements in vitro. Moreover, in measurements by Krotova et al. and Yoshikawa et al., changes in the salt concentration in the medium played an important role in the abrupt change from the coil to the globular configuration of DNA.
The physiological implication of the second order transition is that the nucleoid and cellular processes related to the nucleoid respond to osmolality changes continuously, instead of maintaining a steady nucleoid homeostasis up to a certain shock magnitude and then completely losing this homeostasis in the fashion of an on-off switch.
Anisotropic compressibility of the nucleoid and bottlebrush-like organization of the chromosome
Our data show that the nucleoid compresses anisotropically at lower crowder concentrations. Its longitudinal compressibility (Young’s modulus) is about four times as high as its radial one. At osmotic shocks of about 1 Osm kg−1, nucleoids become spherical and remain spherical at higher shocks (SI Fig. S2 C, D). Based on EM images of lysed cells (Kavenoff and Bowen, 1976), it has been proposed that E. coli nucleoids have a bottlebrush-like organization with supercoiled segments or just DNA loops stretching out radially from a backbone, which is aligned with the long axes of the cell (Wang et al., 2013). This view also has some support from 3C/Hi-C studies of Caulobacter crescentus (Le et al., 2013), Bacillus subtilis (Marbouty et al., 2015), and E. coli (Lioy et al., 2018) chromosomes. These studies all show well-defined chromosomal interaction domains, which could correspond to supercoiled segments that stretch radially out from a common backbone.
It can be expected that bottlebrush like organization leads to anisotropic compressibility. It should be harder to compact plectonemic supercoils along their length, which according to this model, are oriented radially relative to the long axes of the cell. At the same time spacing between supercoils allows them to be easily compacted along the cell length. Although our data appears consistent with the above explanation, the modeling results show that anisotropic compaction can be expected even for the chromosome that lacks supercoiling and is a consequence of cylindrical confinement of the chromosome by the inner membrane. Moreover, it would be unlikely that the anisotropic bottlebrush-like chromosome would be compacted to a spherical entity. The bottlebrush-like structure should retain its anisotropy, and as a result, its aspect ratio should differ from one, which is contrary to what is found in our experiments (SI Fig S2 C, D). One could argue that a bottlebrush-like organization, which is present in the native state of the nucleoid, could be significantly perturbed by osmotic compaction. This seems to be an unlikely scenario for us because plectonemic interactions should increase rather than decrease as the nucleoid becomes more compacted. Therefore one would expect higher rather than lower anisotropy at higher compression levels, i.e., the stiffness along the backbone should be higher. Altogether, our data favor a more disordered organization of plectonemic supercoils than envisioned by the bottlebrush model where supercoils emanate from a single linear backbone.
Interplay of different crowders in compacting the nucleoid
Our data provide new information on how different cytoplasmic macromolecular crowders affect nucleoid compaction. In particular, we find that polysomes cannot be the sole dominant crowder species that leads to nucleoid compaction despite their large volume fraction, high charge state, and prominent exclusion from the nucleoid. These conclusions are based on essentially identical nucleoid compaction curves in two different growth rates, and the insensitivity of these curves to rifampicin treatment. In the following discussions, in order to further rationalize these findings, we estimate the contribution of different macromolecular crowders in compacting the nucleoid based on their literature reported abundances and sizes (SI Table S5). The crowder groups that we consider are cytoplasmic proteins, 30S and 50S ribosomal subunits, tRNA, and poly-ribosomes (polysomes). Not all cytoplasmic proteins qualify as crowders. We exclude DNA binding proteins from this group. Also, we group proteins that are involved in translation together with their rRNA and tRNA counterparts. There are an estimated 3 million proteins in E. coli in fast growth conditions (Milo, 2013). Of those, 20–25% qualify as cytoplasmic crowders that are not part of ribosomes, chromosomes, and envelope layers. In slow growth, where the cell needs to synthesize metabolic components from simpler molecules, the fraction of cytosolic crowders can increase to 40% of the total proteome (Li et al., 2014). 80–85% of the cellular RNA content should be a part of the actively translating ribosomes. These ribosomes form polysomes. The remaining 15% of the RNA mass should be in the form of 30S and 50S subunits in moderately fast growth conditions (Dai et al., 2017). In slow growth conditions, the corresponding numbers are 65% for polysomes and 35% for subunits. tRNA abundance is about nine molecules per one ribosome (Bremer and Dennis, 2008). In slow growth conditions, ribosome to protein mass ratio is estimated to be 1.5–2.0 times smaller than that in moderately fast growth conditions (Ehrenberg et al., 2013). This appears to be the result of a decrease in ribosome concentration in slow growth while its protein concentration remains approximately unchanged.
The results from Kim et al. (Kim et al., 2015) and from our modelling predict that different crowders (indexed by i) contribute to the nucleoid compaction not by their volume fraction but via their crowding level given by . Contributions of different crowder species to the total crowding level can then be characterized by ratio . Using the crowder sizes and abundancies from SI Table S6, we estimate that in moderately fast growth conditions polysome and protein based crowders are the main contributors (54%, and 29%, respectively) while the contributions from rRNA subunits and tRNA are smaller (5% and 12%, respectively) (SI Table S6). In slow growth conditions, we find the roles of cytosolic proteins and ribosomes reversed. We estimate that proteins contribute 65% and polysomes 22% to the total crowding level (SI Table S6). The overall crowding levels, in these two growth conditions are comparable (10% lower in slow growth).
The large contribution of polysomes in the above estimates stems from their large (excluded) volume, which also leads them to be spatially separated from the nucleoid; even at small concentrations. Large contributions of proteins arise from their much larger numbers. Unlike ribosomes, the fraction of proteins that are excluded from the nucleoid needs not to be very large. Even a small fractional difference (1–5%) in protein concentration between the nucleoid and cytosolic phase is sufficient to cause a similar effect on nucleoid compaction than the ribosomes because of their much larger numbers and translational entropy. Altogether, polysomes and protein crowders are distributed very differently between cytosolic and nucleoid phases but they both have significant contributions to nucleoid compaction.
Clearly, the above estimates contain significant uncertainties, as they are based on limited and scattered quantitative data from the concentrations of cellular crowders and their dimensions. Despite these uncertainties, the above analysis points out that cytosolic proteins, beyond polysomes, can be expected to have a significant contribution to nucleoid compaction. Furthermore, the analysis also indicates that the contribution of protein crowders to nucleoid compaction increases as the growth rate slows down. These two broader predictions are consistent with our experimental data.
Role of transcription in determining the global size of the nucleoid
Our experiments indicate that the transcription halting drug rifampicin leads to a significant increase in nucleoid dimensions in moderately fast growth, but much more limited ones in slow growth conditions longer time scales (> 5min). Increased nucleoid expansion at a higher growth rate is consistent with the estimates provided by the crowding level . Upon dissociation of polysomes in the slow growth condition, we estimate the crowding level to decrease by 9% while in moderately fast conditions, the decrease is 25% (Table S6). While associated with significant uncertainties, these estimates are consistent with the overall trend in the data.
We also found that prior treatment of cells with rifampicin did not significantly alter the size of the nucleoid upon osmotic shock in both growth conditions. While this explanation is consistent with the idea that polysomes have a relatively minor contribution to the nucleoid compaction in slow growth conditions, the same appears not to apply to moderately fast growth conditions where the nucleoid expanded significantly under rifampicin treatment. This finding implies that inhibition of transcription likely has some additional effect on nucleoid compaction beyond the one leading to the dissociation of polysomes. It is likely that transcription could increase supercoiling levels, which in turn could result in more compacted nucleoids. However, the link between increased supercoiling levels and nucleoid compaction has so far not found experimental support in vivo (Stuger et al., 2002, Cagliero and Jin, 2013). It has been also reported that at faster growth rates, ribosomal operons (rrnA-G) cluster into a nucleolus-like particle and this clustering might to lead to compaction of the nucleoid (Jin et al., 2015). Nevertheless, the underlying mechanism is not known. Irrespective of the exact mechanism, some transcription related process that is not associated with polysome dissociation appears to play a role in determining the nucleoid size at faster growth rates and alters the nucleoid organization. This effect diminishes at lower growth rates where the total transcription rate (per unit genome) is also lower.
Physiological consequences of nucleoid size
Our measurements bring out the importance of macromolecular crowders in determining the size of the nucleoid. It appears to be the leading factor in determining the nucleoid size at slower growth rates. Furthermore, we find that in slow growth, the contribution from cytosolic proteins dominates over the contribution from polysomes for nucleoid compaction. As the growth rate increases, and with it, the number of polysomes. The latter becomes the dominant crowder species. In all growth conditions, polysomes are essentially excluded from the nucleoid (Bakshi et al., 2014, Bakshi et al., 2012, Mohapatra and Weisshaar, 2018) while the exclusion of proteins is minor (cf. Fig 1 ). Nevertheless, proteins are capable of contributing to the nucleoid compaction because of their much larger number compared to polysomes. Our data also indicates that at higher growth rates, transertion and likely some other processes influenced by transcription also contribute to the size of the nucleoid and to its distinct folding. The latter mechanism remains to be elucidated.
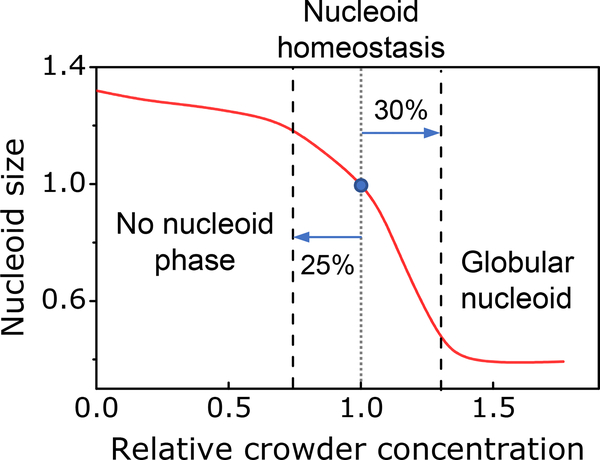
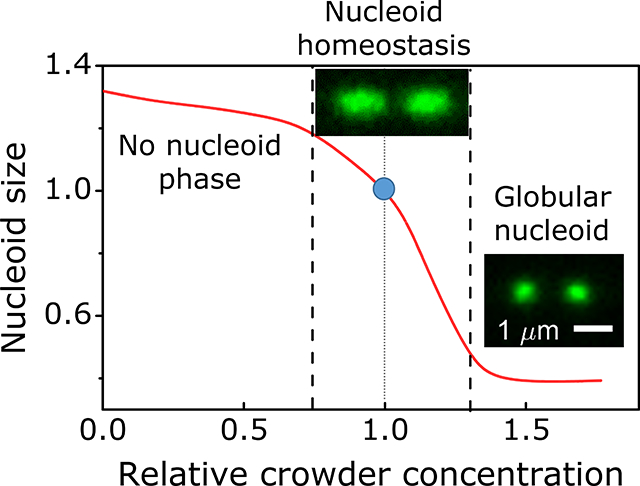
Finally, our data lend strong support to theoretical ideas and prior in vitro measurements that macromolecular crowders confine the nucleoid into a distinct state/phase in E. coli. Our data also show that this phase is limited to a relatively narrow level of crowding (Fig. 7). As predicted by our modeling, a 25% decrease in the crowding level can lead to a completely diffuse chromosome, which fills the whole cytoplasmic volume. This scenario appears in some bacteria. It has been reported, that in C. crescentus the chromosomal DNA extends throughout the cytosolic volume (Montero Llopis et al., 2010), although it may be excluded from the immediate vicinity of the inner membrane (Woldringh, 2010). At the same time, a 30% increase in crowding levels leads to a highly compressed nucleoid according to both experimental and modeling results. Such a nucleoid state would possibly hinder some DNA transactions, such as replication and transcription, as the unwinding of DNA strands requires more energy in a compacted nucleoid. It remains an intriguing question for further research to understand if it is a coincidence that crowding levels are just right to maintain the nucleoid in a sensitive part of the compaction curve or is there some dedicated regulatory mechanisms involved. These mechanisms could possibly control either the cell volume or the number of crowding molecules, although the former seems more plausible.
Figure 7. Nucleoid homeostasis in the presence of crowders.
Distinct nucleoid where DNA is moderately compacted exists in a limited range of crowding levels. 25% reduction in crowding levels leads to abolishment of distinct nucleoid phase from which crowders to some degree are excluded. At the same time, a 35% increase in crowder concentration leads to the formation of globular nucleoid with high compressibility and likely of limited functionality. Red curve is model data at μm−1 matched approximately to experimentally measured nucleoid length. The blue circle corresponds to a nucleoid size in physiological conditions.
EXPERIMENTAL PROCEDURES
Strains and growth conditions
All strains in this study were derivatives of E. coli K-12 MG1655 (SI Table S1). The genomic insertions were constructed using λ-Red recombination and shuffled between strains using P1 transduction as described in (Datsenko and Wanner, 2000). The details of strain construction can be found in SI Text. The bacteria were grown and imaged at 28°C. In slow-growth conditions, cells were cultivated in M9 minimal medium (Teknova Inc., CA) supplemented with 2 mM magnesium sulfate (MgSO4) (Millipore Sigma, MO), 0.3% glycerol (Fisher Scientific), and 100 μg ml−1 leucine (Fisher Scientific). In moderately fast growth conditions M9 minimal medium was supplemented with 2 mM MgSO4, 0.5% glucose (Millipore Sigma, MO), and 0.2% casamino acids (Fisher Scientific). In all experiments, except squeezing measurements, cells were first plated and grown on M9 agar plates supplemented with 2 mM MgSO4 and 0.5% glucose. From the plate, a single colony was inoculated into 2.5 ml of media. The culture was grown overnight and then injected into the microfluidic chips. Further details on handling bacteria during mother machine measurements, and during their hyper/hypo osmotic and rifampicin treatments can be found in SI Text.
In squeezing experiments (strain DY3), cells were first plated and grown on LB agar plates supplemented with 20 μg ml−1 chloramphenicol and 0.2% glucose. A single colony was inoculated into M9 supplemented with 2 mM MgSO4, 0.5% glucose, and 20 μg ml−1 chloramphenicol, then grown overnight. The overnight culture was diluted to an OD600 ~ 0.002 in fresh M9 medium supplemented with 2 mM MgSO4, 0.3% glycerol, and 50 nM IPTG to induce expression of the extra-chromosomal sulA gene under the control of the lac-promoter. After six hours, the cells were filamentous and contained multiple nucleoids. Details on handling cells during squeezing measurements can be found in SI Text.
Microscopy
A Nikon Ti-E inverted epifluorescence microscope (Nikon Instruments, Japan) with a 100X (NA = 1.45) oil immersion phase contrast objective (Nikon Instruments, Japan), was used for imaging the bacteria. Images were captured on an iXon DU897 EMCCD camera (Andor Technology, Ireland) and recorded using NIS-Elements software (Nikon Instruments, Japan). Fluorophores were excited by a 200W Hg lamp through an ND8 neutral density filter. A Chroma 41004 filtercube was used for capturing mCherry and tag-RFP-T images, and a Chroma 41001 (Chroma Technology Corp., VT) for mNeonGreen images. A motorized stage and a perfect focus system were utilized throughout time-lapse imaging.
Image analysis
Image analysis was carried out using Matlab (MathWorks, MA) scripts based on Matlab Image Analysis Toolbox, Optimization Toolbox, and DipImage Toolbox (https://www.diplib.org/). For each cell analyzed, a segment connecting its two poles in the image of the cytoplasmic label was manually identified. Then, this segment defined the longitudinal axis of the cell. The segment was then broadened to nine pixels along the short axes of the cell and it was used to extract intensity line profiles for the cytoplasmic and for the nucleoid labels. Subsequently, these profiles were smoothed by least-square fitting them to the exponential power function. The latter is a type of generalized Gaussian function. From these smoothened curves, inflection points were determined. The distance between the inflection points of respective profiles was used as an estimate for the cell and nucleoid lengths.
To determine the cytoplasmic and nucleoid widths, intensity line profiles were generated perpendicular to the long axes of the cell. These intensity profiles, taken one pixel apart, were each fitted to a (regular) Gaussian function. From the fittings, the mean distance between the inflection points for each profile was determined. Based on earlier modeling results (Männik et al., 2009), the widths were estimated as the distances between the inflection points of the Gaussians to which an additional constant offset of 80 nm was added. Finally, a mean width was calculated from averaging widths over the length of the cell.
In the microanvil measurements, cells had multiple nucleoids. To determine nucleoid lengths, the nucleoids images were first resampled using a nine-pixel-wide polyline. An averaged intensity line profile based on this polyline was then calculated. This intensity profile was then fit to multipeak exponential power functions. Each nucleoid lobe was represented by a single exponential power function. From the exponential power functions, distances between the second inflection points were determined. The latter served as an estimate for the nucleoid length. Cell and nucleoid volumes were calculated, assuming they were cylinders capped with half spheres on each pole. Cell and nucleoid aspect ratios were length to width ratios. The procedure for the determination of the crowder concentration based on the intensity of fluorescent reporters can be found in SI Text.
Details of microchip design, fabrication, and usage, and coarse-grained modeling are given in the SI Text. Raw experimental data is available from (Yang et al., 2020).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Fabai Wu, Cees Dekker, and Rodrigo Reyes-Lamothe for strains and plasmids, and Sriram Tiruvadi Krishnan, Bryant E. Walker, Conrad L. Woldringh, and Arieh Zaritsky for valuable comments. Authors acknowledge technical assistance and material support from the Center for Environmental Biotechnology at the University of Tennessee. This work was supported by National Science Foundation research grant [MCB-1252890], US-Israel Binational Science Foundation research grant [2017004], and National Institutes of Health award under [R01GM127413]. Part of this research was conducted at the Center for Nanophase Materials Sciences, which is sponsored at Oak Ridge National Laboratory by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy. A part of this research is based upon work performed using computational resources supported by the University of Tennessee and Oak Ridge National Laboratory Joint Institute for Computational Sciences. The authors declare no conflict of interest.
REFERENCES
- Bakshi S, Choi H, Mondal J and Weisshaar JC (2014) Time-dependent effects of transcription- and translation-halting drugs on the spatial distributions of the Escherichia coli chromosome and ribosomes. Mol. Microbiol 94: 871–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Choi H and Weisshaar JC (2015) The spatial biology of transcription and translation in rapidly growing Escherichia coli. Frontiers in Microbiology 6: 10.3389/fmicb.2015.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Siryaporn A, Goulian M and Weisshaar JC (2012) Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol 85: 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D and Kleckner N (2005) Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S and Cohen SN (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U. S. A 99: 9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H and Dennis P (2008) Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus: doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- Cabrera JE, Cagliero C, Quan S, Squires CL and Jin DJ (2009) Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: Examining the effect of transcription on nucleoid structure in the absence of transertion. J. Bacteriol 191: 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliero C and Jin DJ (2013) Dissociation and re-association of RNA polymerase with DNA during osmotic stress response in Escherichia coli. Nucleic Acids Res. 41: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha S, Woldringh CL and Odijk T (2001) Polymer-mediated compaction and internal dynamics of isolated Escherichia coli nucleoids. J. Struct. Biol 136: 53–66. [DOI] [PubMed] [Google Scholar]
- Dai XF, Zhu ML, Warren M, Balakrishnan R, Patsalo V, Okano H, Williamson JR, Fredrick K, Wang YP and Hwa T (2017) Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nature Microbiology 2: 16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Mukherjee A and Lutkenhaus J (2008) Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J. Bacteriol. 190: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Kalmykowa OJ and Grainger DC (2011) Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria. PLoS Genet. 7: e1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R (2010) DNA condensation in bacteria: Interplay between macromolecular crowding and nucleoid proteins. Biochimie 92: 1715–1721. [DOI] [PubMed] [Google Scholar]
- Dillon SC and Dorman CJ (2010) Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol 8: 185–195. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M, Bremer H and Dennis PP (2013) Medium-dependent control of the bacterial growth rate. Biochimie 95: 643–658. [DOI] [PubMed] [Google Scholar]
- Epstein W and Schultz SG (1965) Cation transport in Escherichia coli. V. Regulation of cation content. Journal of General Physiology 49: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R and Boccard F (2012) A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31: 3198–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JK, Bourniquel A, Witz G, Weiner B, Prentiss M and Kleckner N (2013) Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153: 882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WT, Govers SK, Xiang YJ, Parry BR, Campos M, Kim S and Jacobs-Wagner C (2019) Nucleoid size scaling and intracellular organization of translation across bacteria. Cell 177: 1632–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Cagliero C, Mata Martin C, Izard J and Zhou YN (2015) The dynamic nature and territory of transcriptional machinery in the bacterial chromosome. Frontiers in Microbiology 6: 10.3389/fmicb.2015.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Cagliero C and Zhou YN (2013) Role of RNA polymerase and transcription in the organization of the bacterial nucleoid. Chemical Reviews 113: 8662–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyeux M (2016) In vivo compaction dynamics of bacterial DNA: A fingerprint of DNA/RNA demixing? Current Opinion in Colloid & Interface Science 26: 17–27. [Google Scholar]
- Joyeux M (2018) A segregative phase separation scenario of the formation of the bacterial nucleoid. Soft Matter 14: 7368–7381. [DOI] [PubMed] [Google Scholar]
- Kavenoff R and Bowen BC (1976) Electron-microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma 59: 89–101. [DOI] [PubMed] [Google Scholar]
- Kim J, Jeon C, Jeong H, Jung Y and Ha BY (2015) A polymer in a crowded and confined space: effects of crowder size and poly-dispersity. Soft Matter 11: 1877–1888. [DOI] [PubMed] [Google Scholar]
- Krotova MK, Vasilevskaya VV, Makita N, Yoshikawa K and Khokhlov AR (2010) DNA compaction in a crowded environment with negatively charged proteins. Phys. Rev. Lett. 105: 128302. [DOI] [PubMed] [Google Scholar]
- Kung C, Martinac B and Sukharev S (2010) Mechanosensitive Channels in Microbes. Annual Review of Microbiology 64: 313–329. [DOI] [PubMed] [Google Scholar]
- Le TBK, Imakaev MV, Mirny LA and Laub MT (2013) High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C and Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Roggiani M and Goulian M (2012) Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc. Natl. Acad. Sci. U. S. A 109: 7445–7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espeli O, Boccard F and Koszul R (2018) Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172: 771–783. [DOI] [PubMed] [Google Scholar]
- Männik J, Castillo D, Yang D, Siopsis G and Männik J (2016) The role of MatP, ZapA, and ZapB in chromosomal organization and dynamics in Escherichia coli. Nucleic Acids Res. 44: 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männik J, Driessen R, Galajda P, Keymer JE and Dekker C (2009) Bacterial growth and motility in sub-micron constrictions. Proc. Natl. Acad. Sci. U. S. A 106: 14861–14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Le Gall A, Cattoni DI, Cournac A, Koh A, Fiche JB, Mozziconacci J, Murray H, Koszul R and Nollmann M (2015) Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59: 588–602. [DOI] [PubMed] [Google Scholar]
- Mercier R, Petit M-A, Schbath S, Robin S, El Karoui M, Boccard F and Espeli O (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135: 475–485. [DOI] [PubMed] [Google Scholar]
- Milo R (2013) What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35: 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra S and Weisshaar JC (2018) Functional mapping of the E-coli translational machinery using single-molecule tracking. Mol. Microbiol 110: 262–282. [DOI] [PubMed] [Google Scholar]
- Mondal J, Bratton BP, Li YJ, Yethiraj A and Weisshaar JC (2011) Entropy-Based Mechanism of Ribosome-Nucleoid Segregation in E. coli Cells. Biophys. J. 100: 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T and Jacobs-Wagner C (2010) Spatial organization of the flow of genetic information in bacteria. Nature 466: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolivos S and Sherratt D (2014) The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. Fems Microbiol. Rev 38: 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odijk T (1998) Osmotic compaction of supercoiled DNA into a bacterial nucleoid. Biophys. Chem 73: 23–29. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Halvorsen K, Ha B-Y, Paparcone R, Sandler SJ, Woldringh CL, Wong WP and Jun S (2012) Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc. Natl. Acad. Sci. U. S. A 109: E2649–E2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilizota T and Shaevitz JW (2013) Plasmolysis and Cell Shape Depend on Solute Outer-Membrane Permeability during Hyperosmotic Shock in E. coli. Biophys. J. 104: 2733–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richey B, Cayley DS, Mossing MC, Kolka C, Anderson CF, Farrar TC and Record MT (1987) Variability of the intracellular ionic environment of Escherichia coli - Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene-expression. Journal of Biological Chemistry 262: 7157–7164. [PubMed] [Google Scholar]
- Roggiani M and Goulian M (2015) Chromosome-Membrane Interactions in Bacteria. Annual Review of Genetics 49: 115–129. [DOI] [PubMed] [Google Scholar]
- Rojas E, Theriot JA and Huang KC (2014) Response of Escherichia coli growth rate to osmotic shock. Proc. Natl. Acad. Sci. U. S. A 111: 7807–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantsov T, Fishov I and Krichevsky O (2007) Internal structure and dynamics of isolated Escherichia coli nucleoids assessed by fluorescence correlation spectroscopy. Biophys. J. 92: 2875–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M and Colby R, (2003) Polymer Physics. Oxford University Press, New York. [Google Scholar]
- Sanamrad A, Persson F, Lundius EG, Fange D, Gynna AH and Elf J (2014) Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc. Natl. Acad. Sci. U. S. A 111: 11413–11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendruk TN, Bertrand M, de Haan HW, Harden JL and Slater GW (2015) Simulating the Entropic Collapse of Coarse-Grained Chromosomes. Biophys. J. 108: 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt DJ (2003) Bacterial chromosome dynamics. Science 301: 780–785. [DOI] [PubMed] [Google Scholar]
- Shin J, Cherstvy AG and Metzler R (2014) Mixing and segregation of ring polymers: spatial confinement and molecular crowding effects. New Journal of Physics 16: 053047. [Google Scholar]
- Spahn CK, Glaesmann M, Grimm JB, Ayala AX, Lavis LD and Heilemann M (2018) A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels. Scientific Reports 8: 14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuger R, Woldringh CL, van der Weijden CC, Vischer NOE, Bakker BM, van Spanning RJM, Snoep JL and Westerhoff HV (2002) DNA supercoiling by gyrase is linked to nucleoid compaction. Molecular Biology Reports 29: 79–82. [DOI] [PubMed] [Google Scholar]
- Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH and Greene EC (2017) The condensin complex is a mechanochemical motor that translocates along DNA. Science 358: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A and Jun S (2010) Robust growth of Escherichia coli. Curr. Biol. 20: 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Llopis PM and Rudner DZ (2013) Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner AS, Alexeeva S, Odijk T and Woldringh CL (2012) Characterization of Escherichia coli nucleoids released by osmotic shock. J. Struct. Biol. 178: 260–269. [DOI] [PubMed] [Google Scholar]
- Wery M, Woldringh CL and Rouviere-Yaniv J (2001) HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie 83: 193–200. [DOI] [PubMed] [Google Scholar]
- Woldringh CL (2002) The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 45: 17–29. [DOI] [PubMed] [Google Scholar]
- Woldringh CL, (2010) Nucleoid Structure and Segregation In: Bacterial Chromatin. Dame RT & Dorman CJ (eds). Dordrecht: Springer, pp. 71–96. [Google Scholar]
- Woldringh CL, Jensen PR and Westerhoff HV (1995) Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces. FEMS Microbiol. Lett. 131: 235–242. [DOI] [PubMed] [Google Scholar]
- Wu F, Swain P, Kuijpers L, Zheng X, Felter K, Guurink M, Solari J, Jun S, Shimizu TS, Chaudhuri D, Mulder B and Dekker C (2019) Cell boundary confinement sets the size and position of the E. coli chromosome. Curr. Biol. 29: 2131–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Jennings AD, Borrego E, Retterer ST and Männik J (2018) Analysis of factors limiting bacterial growth in PDMS mother machine devices. Frontiers in Microbiology 9: 10.3389/fmicb.2018.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Männik J, Retterer ST and Männik J (2020) The data that support the findings of this study are available from the corresponding author upon reasonable request. [Google Scholar]
- Yoshikawa K, Hirota S, Makita N and Yoshikawa Y (2010) Compaction of DNA Induced by Like-Charge Protein: Opposite Salt-Effect against the Polymer-Salt-Induced Condensation with Neutral Polymer. Journal of Physical Chemistry Letters 1: 1763–1766. [Google Scholar]
- Zhang C, Shao PG, van Kan JA and van der Maarel JRC (2009) Macromolecular crowding induced elongation and compaction of single DNA molecules confined in a nanochannel. Proc. Natl. Acad. Sci. U. S. A 106: 16651–16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.