Abstract
Clear cell renal cell carcinoma (ccRCC) is the highest mortality, invasion, and metastasis subtype of renal cell carcinoma. Bone morphogenetic protein (BMP) family has recently emerged as a group of cancer-related proteins in multiple pathogenesis of cancers. Currently, little is known about the prediction role of BMPs in ccRCC. Therefore, we screened The Cancer Genome Atlas Kidney Clear Cell Carcinoma (TCGA-KIRC) database for ccRCC patients with complete clinical information and BMP family expression data. Multivariate analysis showed that high expression of BMP1 was associated with shorter overall survival (OS) (P = 0.001), and shorter disease-free survival (DFS) (P = 0.018). Gene set enrichment analysis (GSEA) showed BMP1 was associated with epithelial–mesenchymal transition (EMT), G2M checkpoint, angiogenesis, hypoxia pathway, and Kirsten rat sarcoma viral oncogene (KRAS) signaling. Knockdown BMP1 suppressed malignancy of ccRCC in vitro and in vivo. Our results indicated that high expressions of BMP1 were poor prognostic factors and gene therapy could be an effective treatment for ccRCC.
Subject terms: Renal cancer, Biomarkers
Introduction
Renal cell carcinoma (RCC) accounts for 3.77% in adult malignancies, is one of the common lethal malignancy in urology with about 65,340 new cases and 14,970 deaths were estimated for 2018 in the United States [1] and with about 403,262 (2.2%) new cases and 175,098 (1.8%) deaths worldwide for all cancers in 2018 [2]. In the past two decades, the incidence of RCC has increased gradually and up to 30% patients have suffered from cancer metastasis when they were primarily diagnosed with RCC [3, 4]. Clear cell RCC (ccRCC) is still an extremely lethal disease and accounts for most cancer-related deaths [5]. Timely detection and early diagnosis is extremely important for the treatment management. Thus, it would be significant to illustrate the potential molecular mechanisms for genesis and development of ccRCC.
Management of different diagnosis and treatment of ccRCC is based on cancer progression. A total of 90% of patient deaths are associated with cancer metastasis and invasion [6, 7]. Tumor recurrence is still a high risk after nephrectomy for local-regional tumor patients [8]. Specific molecular target drugs based on the tyrosine kinase inhibitor (TKI) use benefits metastatic ccRCC patients [9, 10]. Unfortunately, ccRCC patients still develop TKI resistance and lead to poor clinical progression because of intrinsic resistance or acquired resistance of tumor cells [11]. Exploring effective prognostic molecular biomarkers and their molecular mechanisms in tumor progression and metastasis is critical to assessing and managing ccRCC patients.
Bone morphogenetic proteins (BMPs) were originally discovered as secreted cytokines based on their ability to induce bone. BMP was found to be an activator and member of transforming growth factor-beta (TGF-β) family. BMP1 originally identified as a secreted metalloprotease of the astacin metalloproteinase family [12]. TGF-β signaling pathway activation requires BMP1 activity, BMP1 makes the initial cleavage and release TGF-β complex from the matrix. The complex then was cleavage by other metalloproteinases such as MMP2 to free TGF-β. Hence, BMP1 involves in the activation of the TGF-β and BMP signaling pathways [13]. In recent years, BMP1 upregulation may increase cancer invasiveness in gastric cancer [14], lung cancers [15], osteosarcoma [16], and colon cancer [17]. Nevertheless, to the best of our knowledge, the involvement of BMP1 in ccRCC has not understood yet.
In this study, we investigated the expression of BMPs with clinicopathological features and patient survival in TCGA database, gene set enrichment analysis (GSEA) were used to explore the biological role which would be involved in ccRCC. In vitro and in vivo experiments certificated knockdown BMP1 suppressed malignancy of ccRCC. Our study demonstrated that BMP1 acted as an oncogene and high BMP1 expression predicts poor progression. These findings could serve as the foundation for developing new prognostic markers and eventually leading to better treatment efficacy.
Patients and methods
Patient samples
The Cancer Genome Atlas database of Kidney Clear Cell Carcinoma (TCGA-KIRC) contains total 533 patients. Eight patients were excluded as the clinical data were partly missing and 525 patients with complete clinical data were used for univariate and multivariate Cox proportional hazard regression [18]. Detailed clinical and molecular characteristics could be found on the TCGA website (http://cancergenome.nih.gov/). Overall survival (OS) and disease-free survival (DFS) were the primary end points of this study. OS was defined as the time between diagnosis and death or was censored at the last follow-up. DFS was defined as the time between diagnosis or surgery and the recurrence of disease or death (for any reason) at the last follow-up. Surgical specimens of ccRCC patients were obtained from the Department of Urology, the First Affiliated Hospital of Xiamen University between 2016 and 2018. Inform and obtain informed consent to approve the experimental and research procedures from the Xiamen University Institutional Review Board.
RNA extraction and qRT-PCR
Tissue RNA was extracted with TRizol reagent (Thermo, MA, USA) according to the manufacturer’s instructions. The purity and concentration of the RNA was tested by a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). qPCR analysis was performed according to the manufacturer’s instructions (LightCycler 480II; Roche, Basel, Switzerland). Relative expression of BMP1 was calculated by: 2−ΔCt (ΔCt = CtBMP1–CtGAPDH). Primers were purchased from GENEWIZ (GENEWIZ, Suzhou, China):
GAPDH (forward, 5′-GAGTCAACGGATTTGGTCGT-3′; reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′)
BMP1, (forward, 5′-TGGCCGACTACACCTATGAC-3′; reverse, 5′- GGAGGACTTACGAGCTGTGT-3′)
Oligonucleotide, lentivirus, plasmid, and shRNA
Oligonucleotides corresponding to the target sequences were annealed and cloned into the AgeI and EcoRI sites of the plko.l plasmid (Addgene, Cambridge, USA). Primers were listed as follows:
BMP1-1:
5′-CCGGCACCTCCCAGTACAACAACATCTCGAGATGTTGTTGTACTGGGAGGTGTTTTT-3′
5′-AATTAAAAACACCTCCCAGTACAACAACATCTCGAGATGTTGTTGTACTGGGAGGTG-3′
BMP1-2:
5′-CCGGGCGCTACTGTGGCTATGAGAACTCGAGTTCTCATAGCCACAGTAGCGCTTTTT-3′
5′-AATTAAAAAGCGCTACTGTGGCTATGAGAACTCGAGTTCTCATAGCCACAGTAGCGC-3′
Cell culture and transient transfection
HK2, A498, Caki-1, and ACHN and came from The American Type Culture Collection (ATCC, USA) without recently authenticated. Cancer cells were cultured in high glucose DMEM medium (Wuhan Boster Biological Technology, Ltd, Wuhan, China) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and antimycoplasma Plasmocin™ prophylactic (ant-mpp, InvivoGen, Hong Kong) at 37 °C, Under cultivation 5% CO2 incubator.
Cell proliferation, migratory, and invasion assays
The A498 and Caki-1 cells were first transfected with sh-BMP-1, sh-BMP-2, or shRNA negative control plasmid (NC), and then the cells were added to the 96-well plate at a density of 3 × 103/well. Cell proliferation rate (OD value) was determined by Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc., Rockville, MD, USA) according to the manufacturer’s instructions. Homogenized cells without plasma for 24 h, then 105 cells were cultured in a 24-well transwell plate with polycarbonate membrane inserts (Corning, NY, USA) for the migratory assay. A total of 2 × 105 cells were cultured in a 24-well transwell plate with Matrigel (Thermo Fisher Scientific, Waltham, USA) coated polycarbonate membrane inserts for the invasion assay. Incubation for 24 h, cells were fixed with 100% methanol, then stained with 0.05% crystal violet, and five random regions were counted and three independent experiments were conducted in a previous study [19].
Western blotting
Tissues and cells are pyrolyzed in a protein lysis system containing RIPA, PMSF (Wuhan Boster Biological Technology, Ltd, Wuhan, China), and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA). Protein concentration was measured at 562 nm. A total of 30 μg of proteins were presented in SDS-PAGE gel system then separated and transferred to polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Bedford, MA, USA) for 90 min. After transfer to the PVDF membrane, the membrane was blocked in PBS containing 5% skim milk for 1 h and then incubated with antibodies against BMP1 (1:1000; ab205394; Abcam Co., Ltd) and GAPDH (1:2000; BM3876; Wuhan Boster Biological Technology, Ltd, Wuhan, China) at 4 °C overnight. The next day the membranes were washed and incubated with secondary antibodies (1:5000; BA1020; Wuhan Boster Biological Technology, Ltd, Wuhan, China) at room temperature (25 °C) for 2 h. Finally, the membranes were washed and detected by Biosense SC8108 Gel Documentation System with GeneScope V1.73 software (Shanghai BioTech, Shanghai, China).
Immunohistochemistry (IHC)
Five adjacent normal, ccRCC tissues, and six xenograft tumor were fixed in formalin, dehydrated, and embedded, and then the tissue sections were incubated with primary mouse BMP1 (1:100) and Ki67 (1:100; A11005; ABclonal Biotech Co., Ltd), overnight at 4 °C. The sections were washed three times with PBS, and then the sections were incubated with the secondary antibody for 2 h at room temperature.
Xenograft tumor in nude mice
All animal experiments were performed in accordance with animal protocols approved by the Institutional Animal Use and Care Committee of Xiamen University. Six male mice (Beijing Hfk Bioscience, Beijing, China) were randomly divided into two groups. A total of 1 × 106 Caki-1 cells with sh-BMP1-1 or NC were injected subcutaneously into the flanks of mice. The investigator was blinded to the group allocation during the experiment. Tumor size was measured every 3 days. After 30 days, the mice were euthanized and the tumors were harvested. Tumor volumes were calculated as follows: volume = (D × d2)/2.
Bioinformatics analysis
GSEA is used to understand the BMP1 pathway involved in the pathogenesis of TCGA-KIRC [20]. Nominal P < 0.05 and a false discovery rate < 25% had considered to be significantly enriched for enriched gene sets analysis.
Statistical analysis
Data sets were described by median and error bars as s.e.m. The RNA levels of samples were analyzed with unpaired-sample t-test by two-sided and χ2 tests with clinical data. The Kaplan–Meier (KM) curve evaluated survival rate for patients clinical and expression level of BMP1 with log-rank test. Prognostic significance of BMP1 was analyzed by univariate and multivariate Cox proportional hazard regression in ccRCC as previously described [21]. All objects are greater than or equal to three for statistical analysis. The variance is similar between the groups. P < 0.05 was considered to have statistical significance. All statistical analyses were performed by SPSS Statistics 22.0 (IBM SPSS, Chicago, IL).
Results
Relative expression of BMP family in ccRCC
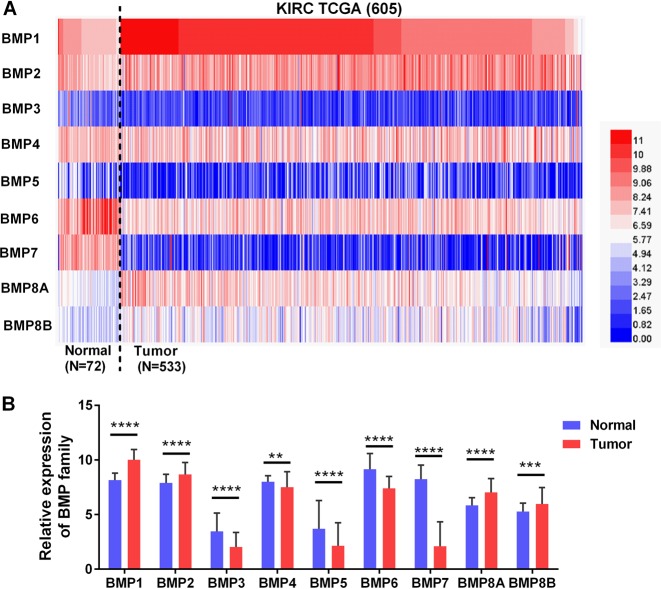
To investigate the expression of BMP family in ccRCC development, we investigated the mRNA expression levels of the nine BMP family members (BMP 1–8 A and B) in TCGA database. The heat map showed the expression levels of BMP family members in Fig. 1a. The expression of each family members in ccRCC tissues and corresponding noncancerous normal tissues is shown in Fig. 1b. Among the nine BMP families, BMP1 exhibited the most obvious high expression in ccRCC (Fig. 1a, b).
Fig. 1.
BMP family expression in TCGA-KIRC microarray datasets. a Heat map depicting BMPs expression in TCGA-KIRC microarray datasets (n = 605). b Relative BMPs expression in TCGA-KIRC. Red indicates high expression; white indicates medium expression; blue indicates low expression. BMP bone morphogenetic protein, TCGA-KIRC The Cancer Genome Atlas kidney renal clear cell carcinoma. **P < 0.01, ***P < 0.001, and ****P < 0.0001
Prognostic significance of BMP family in ccRCC
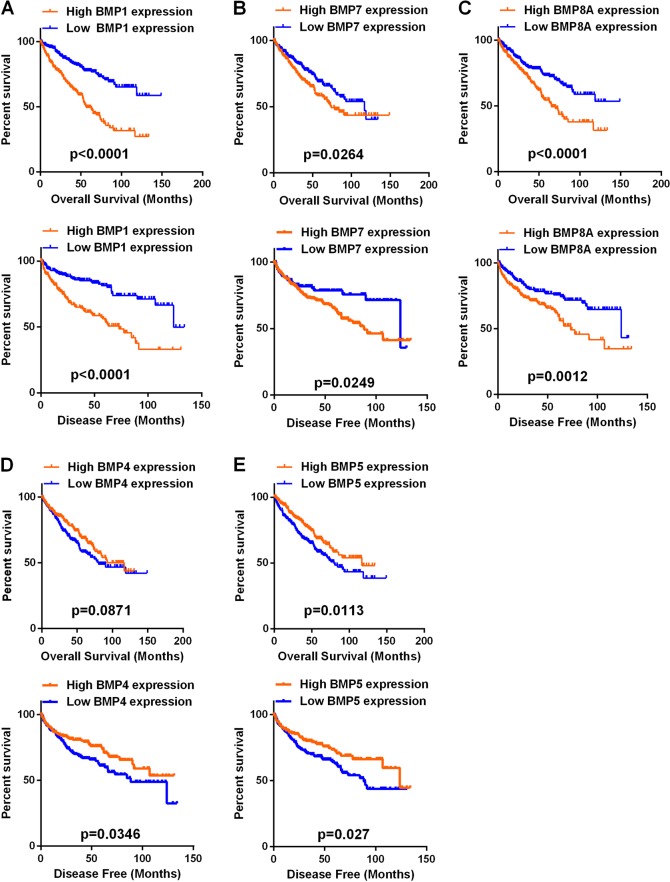
According to the median expression levels of the nine BMP family members, all patients were divided into two groups. Univariate analysis showed that the expression of BMP1, BMP7, or BMP8A was associated with OS and BMP1, BMP4, BMP5, BMP7, or BMP8A was associated with DFS (Table 1). KM analysis demonstrated that the patients with high BMP1, BMP7, and BMP8A expression had shorter OS and DFS than those with low expression (Fig. 2a–c). High BMP4 expression group had longer DFS than the low expression group but no difference in OS (Fig. 2d). The high BMP5 expression group had longer OS and DFS than the low expression group (Fig. 2e). Then we compared the expression of each member in ccRCC tissues and corresponding noncancerous normal tissues, unfortunately we found that BMP4, BMP5, and BMP7 expression was lower in tumor tissues (Fig. 1a, b). Following these initial results, we then focused the statistical analysis on BMP1 and BMP8A to patient’s OS and DFS.
Table 1.
Comparison of OS and DFS between different expression levels of BMP1–8B
| OS | DFS | |||
|---|---|---|---|---|
| Variables | χ2 | P value | χ2 | P value |
| BMP1 (high versus low) | 39.265 | 0.000 | 27.960 | 0.000 |
| BMP2 (high versus low) | 0.822 | 0.365 | 0.035 | 0.851 |
| BMP3 (high versus low) | 0.036 | 0.850 | 0.525 | 0.468 |
| BMP4 (high versus low) | 1.685 | 0.194 | 4.462 | 0.035 |
| BMP5 (high versus low) | 1.356 | 0.244 | 4.891 | 0.027 |
| BMP6 (high versus low) | 3.769 | 0.052 | 0.019 | 0.889 |
| BMP7 (high versus low) | 12.400 | 0.000 | 7.548 | 0.006 |
| BMP8A (high versus low) | 15.423 | 0.000 | 10.554 | 0.001 |
| BMP8B (high versus low) | 1.149 | 0.284 | 0.126 | 0.723 |
OS overall survival, DFS disease-free survival
Fig. 2.
Kaplan–Meier curves of OS and DFS in different expression levels of BMP1, BMP4, BMP5, BMP7, or BMP8A. a–c High BMP1, BMP7, BMP8A expressers had shorter OS and DFS than the low expressers. d Low BMP4 expressers had shorter DFS than the low expressers. e Low BMP5 expressers had shorter OS and DFS than the low expressers. OS overall survival, DFS disease-free survival, BMP bone morphogenetic protein
Clinical and molecular characteristics of the patients
Patients with integrity data (n = 525) were divided by BMP1 and BMP8A median expression levels respectively, detailed clinical pathologic information of these ccRCC patients was presented in Table 2. There was a significant association between high BMP1 expression and tumor stage (T stage), lymphatic metastasis (N stage), distant metastasis (M stage), TNM stages, and grade, but no significant differences in age or sex. Meanwhile, there was a significant association between high BMP8A expression and tumor stage (T stage), distant metastasis (M stage), and TNM stages, but no significant differences in age, sex, N stage, or grade.
Table 2.
Correlation between BMP1 and BMP8A mRNA expression and clinicopathological parameters of ccRCC patients
| BMP1 mRNA expression | BMP8A mRNA expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | Low (n = 263) | High (n = 262) | χ2 | P value | Low (n = 263) | High (n = 262) | χ2 | P value | |
| Age (years) | <=60 | 122 | 138 | 126 | 134 | ||||
| >60 | 141 | 124 | 2.073 | 0.163 | 137 | 128 | 0.550 | 0.485 | |
| Sex | male | 160 | 180 | 96 | 89 | ||||
| female | 103 | 82 | 3.588 | 0.068 | 167 | 173 | 0.369 | 0.584 | |
| T stage | T1 + T2 | 194 | 143 | 192 | 143 | ||||
| T3 + T4 | 69 | 119 | 21.014 | 0.000 | 71 | 117 | 17.807 | 0.000 | |
| N stage | N0 + NX | 260 | 250 | 257 | 253 | ||||
| N1 | 3 | 12 | 5.594 | 0.019 | 6 | 9 | 0.629 | 0.447 | |
| M stage | M0 + MX | 240 | 207 | 235 | 212 | ||||
| M1 | 23 | 55 | 15.563 | 0.000 | 28 | 50 | 7.387 | 0.007 | |
| Grade | G1 + G2 | 147 | 98 | 126 | 119 | ||||
| G3 + G4 | 116 | 164 | 18.027 | 0.000 | 137 | 143 | 0.327 | 0.600 | |
| TNM stage | I + II | 186 | 133 | 184 | 79 | ||||
| III + IV | 77 | 129 | 21.930 | 0.000 | 135 | 127 | 18.709 | 0.000 | |
Multivariate analyses of OS and DFS
To assess the prognostic significance of the aforementioned clinical and molecular characteristics in ccRCC patients, we chose the expression levels of BMP1 and BMP8A (high versus low), age, sex, T stage, N stage, M stage, and grade to construct multivariate analyses of OS and DFS (Table 3 and Table 4). Multivariate analysis demonstrated that age (HR, 1.713; P = 0.001), T stage (HR, 1.6495; P = 0.034), N stage (HR, 2.166; P = 0.020), M stage (HR, 2.522; P = 0.000), Grade (HR, 1.607; P = 0.013), and BMP1 expression (HR, 1.903; P = 0.001) could be considered independent prognostic indicators of OS. Meanwhile, multivariate analysis demonstrated that T stage (HR, 1.856; P = 0.005), N stage (HR, 3.097; P = 0.002), M stage (HR, 5.050; P = 0.000), Grade (HR, 2.172; P = 0.000), and BMP1 expression (HR, 1.692; P = 0.018) could be considered independent prognostic indicators of DFS. Multivariate survival analyses indicated that the BMP1 expression was an independent prognostic factor for OS and DFS in ccRCC patients.
Table 3.
Univariate and multivariate analyses of BMP1 and BMP8A mRNA level and patient overall survival
| Univariate analysis | Multivariate analysisc | |||||
|---|---|---|---|---|---|---|
| Variable | HRa | 95% CIb | P | HR | 95% CI | P |
| Overall survival | ||||||
| Age (years) | ||||||
| ≤60 versus >60 | 1.803 | 1.318–2.468 | 0.000 | 1.713 | 1.252–2.344 | 0.001 |
| Sex | ||||||
| Female versus male | 0.948 | 0.697–1.290 | 0.825 | |||
| T stage | ||||||
| T3 or T4 versus T1 or T2 | 3.120 | 2.306–4.220 | 0.000 | 1.495 | 1.032–2.166 | 0.034 |
| N stage | ||||||
| N1 versus N0 or NX | 3.832 | 2.070–7.061 | 0.000 | 2.166 | 1.132–4.415 | 0.020 |
| M stage | ||||||
| M1 versus M0 or MX | 4.346 | 3.192–5.918 | 0.000 | 2.522 | 1.763–3.610 | 0.000 |
| Grade | ||||||
| G3 or G4 versus G1 or G2 | 2.639 | 1.885–3.697 | 0.000 | 1.602 | 1.106–2.321 | 0.013 |
| BMP1 | ||||||
| High versus low | 2.696 | 1.839–3.951 | 0.000 | 1.903 | 1.324–2.734 | 0.001 |
| BMP8A | ||||||
| High versus low | 1.821 | 1.262–2.626 | 0.001 | 1.194 | 0.842–1.693 | 0.321 |
aHazard ratio, estimated from Cox proportional hazard regression model
bConfidence interval of the estimated HR
cMultivariate models were adjusted for T, N, M classification, age, and gender
Table 4.
Univariate and multivariate analyses of BMP1 and BMP8A mRNA level and patient disease-free survival
| Univariate analysis | Multivariate analysisc | |||||
|---|---|---|---|---|---|---|
| Variable | HRa | 95% CIb | P | HR | 95% CI | P |
| Disease-free survival | ||||||
| Age (years) | ||||||
| ≤60 versus >60 | 1.366 | 0.959–1.945 | 0.084 | |||
| Gender | ||||||
| Female versus male | 1.413 | 0.951–2.100 | 0.087 | |||
| T stage | ||||||
| T3 or T4 versus T1 or T2 | 4.526 | 3.134–6.538 | 0.000 | 1.865 | 1.209–2.877 | 0.005 |
| N stage | ||||||
| N1 versus N0 or NX | 5.942 | 2.983–11.836 | 0.000 | 3.097 | 1.490–6.435 | 0.002 |
| M stage | ||||||
| M1 versus M0 or MX | 8.529 | 5.877–12.379 | 0.000 | 5.050 | 3.316–7.691 | 0.000 |
| Grade | ||||||
| G3 or G4 versus G1 or G2 | 3.376 | 2.236–5.098 | 0.000 | 2.172 | 1.407–3.352 | 0.000 |
| BMP1 | ||||||
| High versus low | 2.696 | 1.839–3.951 | 0.000 | 1.692 | 1.097–2.622 | 0.018 |
| BMP8A | ||||||
| High versus low | 1.820 | 1.262–2.626 | 0.001 | 1.240 | 0.823–1.871 | 0.304 |
aHazard ratio, estimated from Cox proportional hazard regression model
bConfidence interval of the estimated HR
cMultivariate models were adjusted for T, N, M classification, age, and gender
Biological pathogenesis of BMP1 in ccRCC
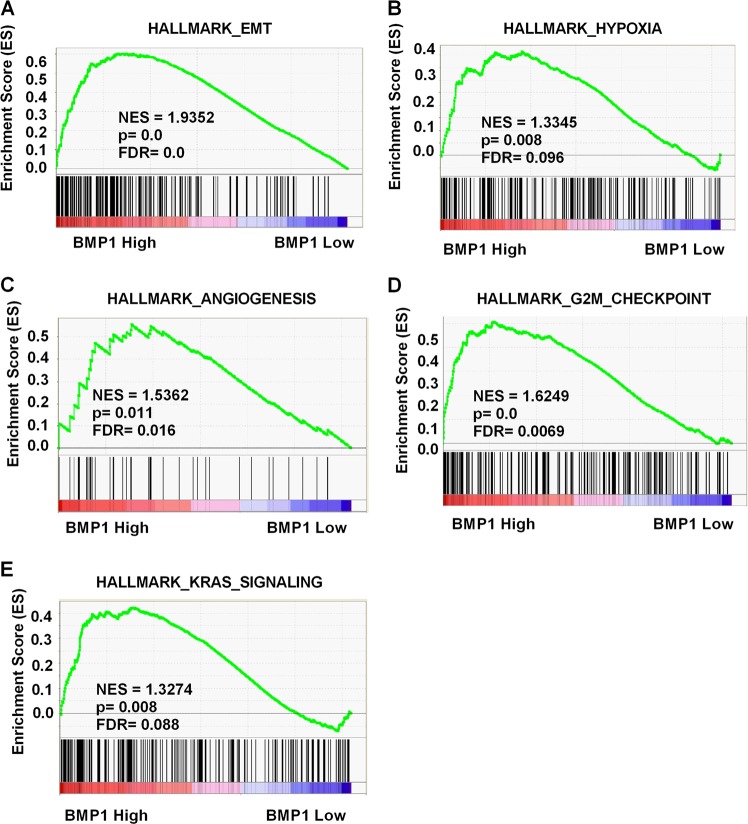
As BMP1 was upregulated and an independent prognostic factor for OS and DFS in TCGA-KIRC database, we were eager to know how BMP1 was involved in ccRCC pathogenesis. Then we used GSEA, a computational tool to obtain statistically significant of the biological pathway in a database to a gene set. The results performed that the expression of BMP1 expression was associated with the gene signatures of epithelial–mesenchymal transition (EMT), hypoxia pathway, angiogenesis, G2M checkpoint, and Kirsten rat sarcoma viral oncogene (KRAS) signaling (Fig. 3).
Fig. 3.
Pathway involved in the pathogenesis of BMP1 in TCGA-KIRC with GSEA. Enrichment curves are shown for activated gene sets related to a EMT, b hypoxia pathway, c angiogenesis, d G2M checkpoint, and e KRAS signaling. BMP bone morphogenetic protein, TCGA-KIRC The Cancer Genome Atlas kidney renal clear cell carcinoma, GSEA gene set enrichment analysis, EMT epithelial–mesenchymal transition, KRAS Kirsten rat sarcoma viral oncogene
Downregulation of BMP1 suppresses malignancy of ccRCC in vitro
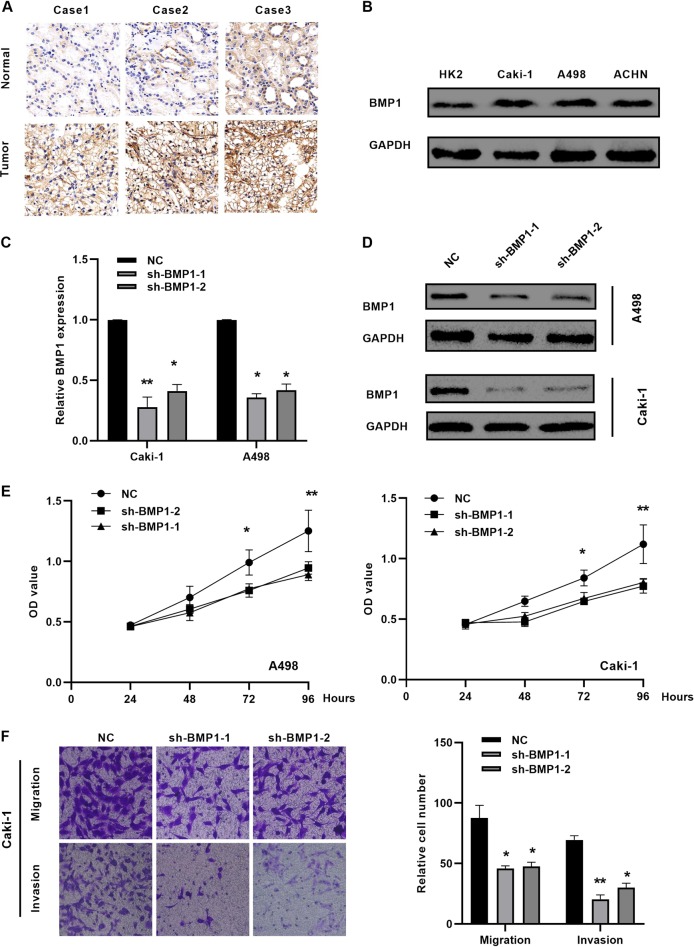
As BMP1 mRNA was upregulated in TCGA-KIRC database, we confirmed the protein level in ccRCC cancer tissues and cells with western blotting (Fig. 4a, b), two BMP1 shRNA plasmids were transfected into the A498 and Caki-1 cells, resulting in a consistent BMP1 knockdown (Fig. 4c, d), CCK-8 and transwell experimental experiments revealed that BMP1 knockdown inhibited the proliferation, cell migration, and invasion in A498 and Caki-1 cells (Fig. 4e, f).
Fig. 4.
Downregulation of BMP1 suppresses malignancy of ccRCC in vitro. a, b BMP1 is upregulated in ccRCC tissues and renal cancer cells by immunohistochemistry. c, d Silence BMP1 with shRNA (sh-BMP1-1 and sh-BMP1-2). e Silence BMP1 significantly repressed the cell proliferation of A498 and Caki-1 cells. f Silence BMP1 significantly repressed the migration and invasion of Caki-1 cells. Data indicate the means ± SEM. *P < 0.05. **P < 0.01
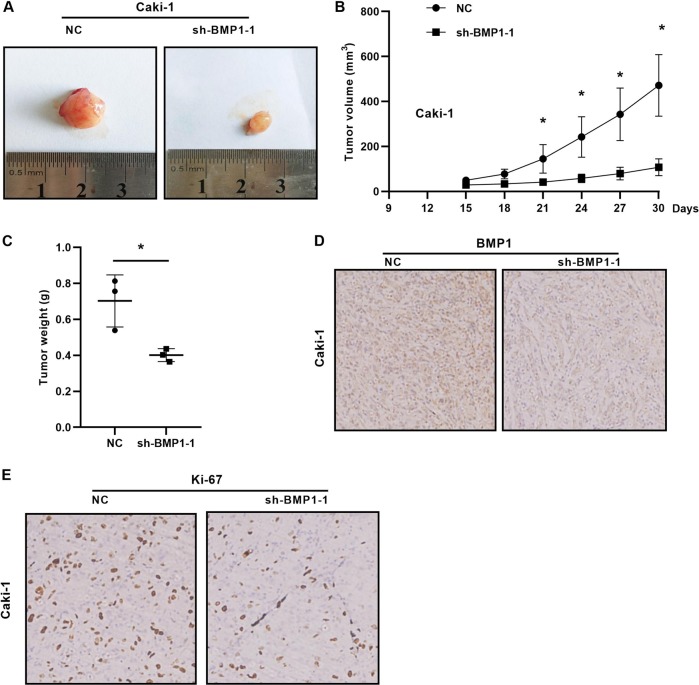
The knockdown of BMP1 levels inhibits tumorigenicity in vivo
We examined the effect of knockdown BMP1 in renal tumor growth in vivo. Caki-1 cells stably infected with lentiviral shRNA-BMP1 and implanted into flank of node mice. The tumor volume was measured every 3 days with the last measurement on day 30. We found that BMP1 knockdown in Caki-1 cells significantly reduced tumor volume and tumor weight (Fig. 5a–c). The expression of BMP1 and Ki67 protein in tumor tissues was shown in Fig. 5d, e, IHC staining indicated that knockdown BMP1 decreased Ki67 protein expression. Taken together, these results demonstrated that BMP1 reducing could inhibit renal cancer growth.
Fig. 5.
The knockdown of BMP1 levels inhibits tumorigenicity in vivo. a–c Tumor size, weight, and volume curves in the xenograft formation assay of BMP1 knockdown by Caki-1 cells. d, e The expression of BMP1 and Ki67 protein in xenograft tumor tissues. Data indicate the means ± SEM. *P < 0.05
Discussion
CcRCC accounted for ~80% of all RCC histological subtypes and had the highest mortality, invasiveness, metastatic rate, resistance to traditional radiotherapy and chemotherapy. This study reported the expression of nine BMP family members in the clinical significance of ccRCC and found that high expressions of BMP1 and BMP8A had prognostic effects on ccRCC, especially the BMP1 and biological functions for the first time.
Wang et al. [22] had shown that BMP2 inhibits tumor-initiating ability in human renal cancer stem cells. But we observed that BMP2 was upregulated in TCGA-KIRC database. This might be explained by the focus on all RCC of the previous study, while our study had specific focus on clear cell subtypes. Markic et al. [23] had shown that all BMPs (BMP2, BMP4, BMP6, BMP7) and their receptors (BMPRIA, BMPRIB, and BMPRII) mRNA have stronger expression levels in RCC, especially BMP2 is elevated strongly in kidney cancer. Lee et al. reported that BMP6 activated interleukin-10-mediated M2 polarization of tumor-associated macrophages to promote RCC growing. BMP6 was a marker signature associated with a poor prognosis in human RCC specimens [24]. BMP1 originally identified as a secreted metalloprotease but not a ligand of the BMP signaling pathway [12]. BMP1 makes the initial cleavage and releases TGF-β complex from the matrix, and then to free TGF-β. Hence, BMP1 involves in the activation of the TGF-β and BMP signaling pathways [13]. In recent years, BMP1 was found to be upregulated in gastric cancer [14], lung cancers [15], osteosarcoma [16], and colon cancer [17]. Nevertheless, the involvement of BMP1 in ccRCC has not been studied yet.
In this study, we investigated the expression of BMPs with clinicopathological features and patient survival in TCGA database, We found that: (I) BMP1 exhibited the most obvious high expression in ccRCC; (II) high BMP1 expression was significantly associated with T stage, N stage, M stage, TNM stages, and grade; (III) the BMP1 expression level was an independent predictor of prognostic of OS and DFS in TCGA-KIRC; (IV) high BMP1 expression was associated with the gene set of EMT, hypoxia pathway, angiogenesis, G2M checkpoint, and KRAS signaling with GSEA; and (V) downregulation of BMP1 suppresses malignancy of ccRCC in vitro and in vivo.
The normal function of von Hippel–Lindau (VHL) tumor suppressor can abolish the hypoxia-inducible transcription factor (HIF) and HIF acts as an important oncogene to promote renal tumor growth [25–28]. BMP1 and HIF1A were positively correlated with the malignant grade of astrocytomas [29], but no research reported their direct or indirect relationship. GSEA demonstrated that high BMP1 expression was associated with the hypoxia signaling pathways in patients with ccRCC, it may be hypothesized that BMP1 could positively regulated by hypoxia signaling, or even upregulated HIF expression in ccRCC, further experiments need to be verified.
Cancer progression by conferring a more motility and aggressive phenotype on cancer cells is a key foundation for EMT [30, 31]. Previous high-throughput screening has identified BMP1 RNA sequences as the most upregulated transcripts in human tumor endothelium associated with angiogenesis [32]. BMP1 was reported to activate TGF-β signaling by cleavage of latent TGF β -binding protein [33]. GSEA demonstrated that high BMP1 expression was associated with the gene signatures of EMT and angiogenesis in TCGA-KIRC database, it may be hypothesized that BMP1 could positively regulate EMT and angiogenesis in ccRCC. GSEA also demonstrated that higher BMP1 expression was associated with the gene signatures of G2M checkpoint and KRAS signaling, it may reveal that BMP1 could positively control cell cycle progression in ccRCC. We proved that knockdown BMP1 suppressed malignancy of ccRCC in vitro and in vivo.
This study is the first study to demonstrate the functional role of BMP1 in ccRCC tumor progression. These results also demonstrate that BMP1 can be used as a potential novel biomarker for predicting the prognosis of ccRCC patients. However, whether BMP1 can promote HIF expression or regulated by HIF does not investigated in ccRCC cells.
In conclusion, we investigated the role of BMP1 in ccRCC progression including TCGA database with univariate and multivariate Cox proportional hazard regression and GSEA. Our studies indicated that BMP1 expression is significantly upregulated and correlated with various clinicopathological parameters. High BMP1 expression is significantly positively correlated with disease progression, which identifies BMP1 as an independent predictor of prognosis in ccRCC patients and is involved in the development of renal cancer. All of these studies provided clues to understanding the potential role of BMP1 in ccRCC and suggested that downregulation of BMP1 may provide a new therapeutic strategy for the management and manipulation of ccRCC patients.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (81602218).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Linehan WM. The genetic basis of kidney cancer: implications for management and use of targeted therapeutic approaches. Eur Urol. 2012;61:896–8. doi: 10.1016/j.eururo.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11:517–25. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Prim. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 7.Campbell S, Uzzo RG, Allaf ME, Bass EB, Cadeddu JA, Chang A, et al. Renal mass and localized renal cancer: AUA guideline. J Urol. 2017;198:520–9. doi: 10.1016/j.juro.2017.04.100. [DOI] [PubMed] [Google Scholar]
- 8.Ravaud A, Motzer RJ, Pandha HS, George DJ, Pantuck AJ, Patel A, et al. Adjuvant sunitinib in high-risk renal-cell carcinoma after nephrectomy. New Engl J Med. 2016;375:2246–54. doi: 10.1056/NEJMoa1611406. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–7. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome. Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarras MP., Jr. BMP-1 and the astacin family of metalloproteinases: a potential link between the extracellular matrix, growth factors and pattern formation. Bioessay. 1996;18:439–42. doi: 10.1002/bies.950180604. [DOI] [PubMed] [Google Scholar]
- 13.Hyytiainen M, Penttinen C, Keski-Oja J. Latent TGF-beta binding proteins: extracellular matrix association and roles in TGF-beta activation. Crit Rev Clin Lab Sci. 2004;41:233–64. doi: 10.1080/10408360490460933. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Deng YF, et al. Upregulation of bone morphogenetic protein 1 is associated with poor prognosis of late-stage gastric Cancer patients. BMC cancer. 2018;18:508. doi: 10.1186/s12885-018-4383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 andp27(kip1) Oncogene. 2014;33:1506–14. doi: 10.1038/onc.2013.108. [DOI] [PubMed] [Google Scholar]
- 16.Garimella R, Tadikonda P, Tawfik O, Gunewardena S, Rowe P, Van Veldhuizen P. Vitamin D Impacts the expression of Runx2 target genes and modulates inflammation, oxidative stress and membrane vesicle biogenesis gene networks in 143B osteosarcoma cells. Int J Mol Sci. 2017;18:642.. doi: 10.3390/ijms18030642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharafeldin N, Slattery ML, Liu Q, Franco-Villalobos C, Caan BJ, Potter JD, et al. A candidate-pathway approach to identify gene-environment interactions: analyses of colon cancer risk and survival. J Natl Cancer Inst. 2015;107:djv160.. doi: 10.1093/jnci/djv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, et al. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res. 2015;43:D812–7. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao W, Wang X, Wang T, Xing J. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging. 2019;11:615–33. doi: 10.18632/aging.101763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao W, Wang X, Wang T, Xing J. TRIM2 downregulation in clear cell renal cell carcinoma affects cell proliferation, migration, and invasion and predicts poor patients’ survival. Cancer Manag Res. 2018;10:5951–64. doi: 10.2147/CMAR.S185270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Park P, La Marca F, Than KD, Lin CY. BMP-2 inhibits tumor-initiating ability in human renal cancer stem cells and induces bone formation. J cancer Res Clin Oncol. 2015;141:1013–24. doi: 10.1007/s00432-014-1883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markic D, Celic T, Grskovic A, Spanjol J, Fuckar Z, Grahovac B, et al. mRNA expression of bone morphogenetic proteins and their receptors in human renal cell carcinoma. Urol Int. 2011;87:353–8. doi: 10.1159/000330797. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Lee GT, Woo SH, Ha YS, Kwon SJ, Kim WJ, et al. BMP-6 in renal cell carcinoma promotes tumor proliferation through IL-10-dependent M2 polarization of tumor-associated macrophages. Cancer Res. 2013;73:3604–14. doi: 10.1158/0008-5472.CAN-12-4563. [DOI] [PubMed] [Google Scholar]
- 25.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metelo AM, Noonan H, Iliopoulos O. HIF2a inhibitors for the treatment of VHL disease. Oncotarget. 2015;6:23036–7. doi: 10.18632/oncotarget.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schodel J, Grampp S, Maher ER, Moch H, Ratcliffe PJ, Russo P, et al. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur Urol. 2016;69:646–57. doi: 10.1016/j.eururo.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Silva R, Uno M, Marie SK, Oba-Shinjo SM. LOX expression and functional analysis in astrocytomas and impact of IDH1 mutation. PLoS ONE. 2015;10:e0119781. doi: 10.1371/journal.pone.0119781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–54. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 31.Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 2016;14:2281–8. doi: 10.1016/j.celrep.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 33.Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. J Cell Biol. 2006;175:111–20. doi: 10.1083/jcb.200606058. [DOI] [PMC free article] [PubMed] [Google Scholar]