Abstract
As one of the most abundant DNA methylation form in prokaryotes, N6-methyladenine nucleotide (6 mA) was however only recently identified in eukaryotic genomes. To explore the implications of N6-adenine methylation in adipogenesis, genomic N6-adenine methylation was examined across adipocyte differentiation stages of 3T3-L1 cells. When the N6-adenine methylation profiles were analyzed and compared with the levels of gene expression, a positive correlation between the density of promoter 6 mA and gene expression level was uncovered. By means of in vitro methylation and gene knockdown assay, METTL4, a homologue of Drosophila methylase CG14906 and C. elegans methylase DAMT-1, was demonstrated to be a mammalian N6-adenine methylase that functions in adipogenesis. Knockdown of Mettl4 led to altered adipocyte differentiation, shown by defective gene regulation and impaired lipid production. We also found that the effects of N6-adenine methylation on lipid production involved the regulation of INSR signaling pathway, which promotes glucose up-taking and lipid production in the terminal differentiation stage.
Subject terms: DNA, DNA, Differentiation, Differentiation, DNA methylation
Introduction
As a major epigenetic modification, genomic DNA methylation is of critical importance to many cellular processes such as expressional regulation, imprinting, X chromosome inactivation, and tumorigenesis1,2. Until very recently, 5-methylcytosine (5mC) has been known to be the only DNA methylation form in eukaryotic genomes3. This is however different from that of prokaryotes, in which 6 mA is another major DNA methylation form and involved in expressional regulation, genome replication as well as restriction-modification systems4,5. The presence of 6 mA was recently revealed in diverse eukaryotic genomes, including Chlamydomonas reinhardtii, Caenorhabiditis elegans, Drosophila melanogaster, zebrafish, pig, Arabidopsis thaliana, mouse and humans6–13. ALKBH1 was identified in 2016 as a 6 mA demethylase involved in the regulation of long interspersed nuclear element1 (LINE-1)13. More recently, 6 mA was found to be involved in tumorigenesis, and its effects were mediated by ALKBH1 and N6AMT1. N6AMT1 was reported in 2018 to be the first mammalian 6 mA methylase14. Despite those progresses, many essential aspects of the metabolism and biological consequences of 6 mA remain to be known.
Adipocyte differentiation of 3T3-L1 preadipocytes has been used as a classic in vitro model system of adipogenesis. Induced by an MDI cocktail consisting of insulin, dexamethasone and 3-isobutyl-1-methyxanthine, the preadipocytes exit from cell proliferation and enter into terminal differentiation stage, to generate matured adipocytes. During this process, the cells undergo stages of growth-arrest’, hormone induction, mitotic clonal expansion, and terminal differentiation. Widespread epigenetic modifications have been recently revealed during the differentiation, shown by dynamic chromosome modifications and complex gene regulation15. Genomic 5mC methylations have been found to be involved in this process, regulating the expression of many adipogenic factors such as Fabp4, Glut4 and Lep16–18. However, whether N6-adenine methylation is also involved in the MDI-induced maturation of adipocytes is still unknown. In the present study, we examined the genomic distribution of 6 mA by means of immunoprecipitation/sequencing assay (6mA-IP-seq), and explored its roles in adipocyte differentiation of 3T3-L1 cells and the underlying mechanisms.
Results
Dynamic changes of genomic N6-adenine methylation during adipocyte differentiation
To explore the involvement of N6-adenine methylation in adipogenesis, the extents of genome-wide modification were determined during the differentiation of 3T3-L1 cells. Upon induction with MDI, proliferating 3T3-L1 cells undergo the process of differentiation, which recapitulates the major events occurring in vivo. To facilitate the description of the process, the day to perform MDI induction is called day(0). Accordingly, the cells grew to confluence on day(−2) and entered into the “growth-arrest” stage from day(−2). After MDI induction on day(0), the cells proceeded to a mitotic clonal expansion stage and entered into a terminal differentiation stage from day(+2) on.
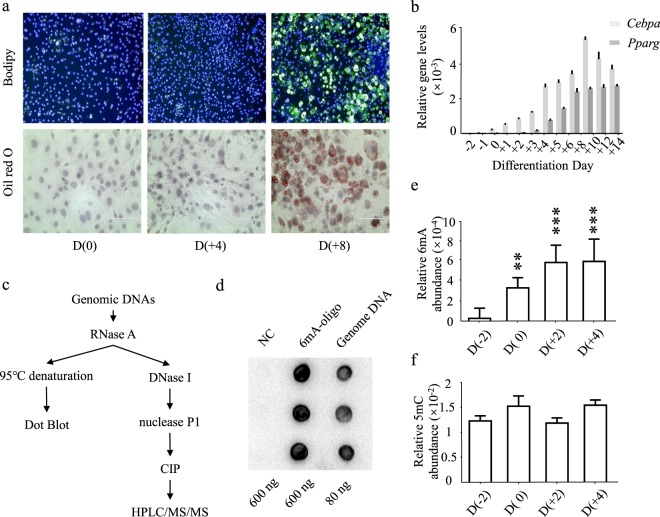
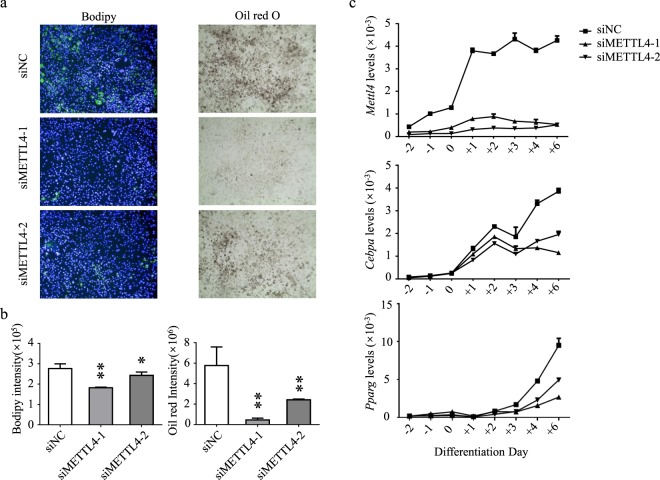
To monitor the progress of differentiation, Bodipy and Oil red O staining were performed (Fig. 1a). In addition to checking the production of cellular lipids, regulation of the major adipogenic factors were also examined by gene expression assay (Fig. 1b). By means of dot blot assay, the presence of genomic N6-adenine methylation was confirmed in proliferating 3T3-L1 cells (Fig. 1c,d). To rule out potential contamination of bacteria or mycoplasma, assays were performed with universal 16 S primer sets (Supplementary Fig. S1a)10, and a commercial mycoplasma detection kit (Supplementary Fig. S1b).
Figure 1.
Profiling of N6-adenine methylation in adipocyte differentiation of 3T3-L1 cells. (a) Staining of cellular lipid of preadipocytes on Day(0), differentiating adipocytes on Day(+4), and mature adipocytes on Day(+8). Lipid staining was performed with Bodipy 493/503 (Green) and Oil red O (Red), nuclei were co-stained with Hoechst 33342 (Blue). (b) Expression profiles of Cebpa and Pparg were determined by qRT-PCR, in relative to the expression of β-actin. (c) Workflow to quantify 6 mA abundance of genome DNA sample. (d) Dot blot assay of N6-adenine methylation. NC, a DNA oligo without N6-adenine methylation. 6mA-oligo, a DNA oligo containing one N6-adenine methylation. Genome DNA, genome DNAs extracted from 3T3-L1 preadipocytes. (e,f) Abundances of 6 mA and 5mC methylation were quantified using HPLC-MS/MS analysis. Molar ratios of 6 mA to dA were calculated, and molar ratios of 5mC to dC were calculated. Data are presented as mean ± s.d, n > 3 independent assay. *P < 0.05; **P < 0.01; ***P < 0.001.
To quantitatively determine the presence of 6 mA, genomic DNAs were extracted from the cells collected on day(−2) before MDI treatment and day(0), day(+2) and day(+4) after MDI treatment, representing proliferating preadipocytes, growth-arresting cells, differentiating adipocytes, and matured adipocytes. DNA samples were then sequentially degraded with RNase, DNase I and nuclease P1. The resulting nucleoside products were separated and quantified using a LC-MS/MS system19, with dA, dC, 5mC and 6 mA as the standards. The abundance of genomic 6 mA was presented as the molar ratio to dA. In proliferating preadipocytes, a relatively low level (0.01%) of 6 mA was found. With the progress of differentiation, 6 mA levels gradually increased and reached a peak of 0.06% at the terminal stage (Fig. 1e). Comparing with 6 mA, 5mC showed a relatively stable profile during the process (Fig. 1f).
Genomic N6-adenine methylation detected by immunoprecipitation and sequencing
Genomic N6-adenine methylation was further investigated by immunoprecipitation and sequencing (6mA-IP-seq). 6mA-IP-seq was performed with DNA samples of preadipocytes and matured adipocytes. The genomic DNAs were sonicated into fragments of 100–500 bp, ligated to adaptors with unique sequence index, denatured to single-stranded DNA, and immuno-precipitated to obtain fragments carrying N6-adenine methylation. 6mA-containing fragments were PCR amplified, and sequenced on an Illumina HiSeq. 2000 platform. From a total of 118 million sequencing reads, 114 million clean reads were obtained. This led to a clean read ratio of 0.9689, much higher than the threshold ratio of 0.75 that indicated a high-quality sequencing data. Sequencing reads were then mapped to mouse reference genome (NCBI37/mm10), using TopHat algorithm. By means of a peak-finding algorithm20, 6 mA sites were individually identified. This led to the identification of ~10,000 high-confidence 6 mA sites in preadipocytes and mature adipocytes, with FDR < 0.01. Furthermore, 6 mA immunoprecipitation/qPCR (6mA-IP-qPCR) was performed to confirm the methylation status of 7 randomly selected positive Peaks and 2 negative Peaks (Supplementary Fig. S2).
Genome-wide distribution of N6-adenine methylation
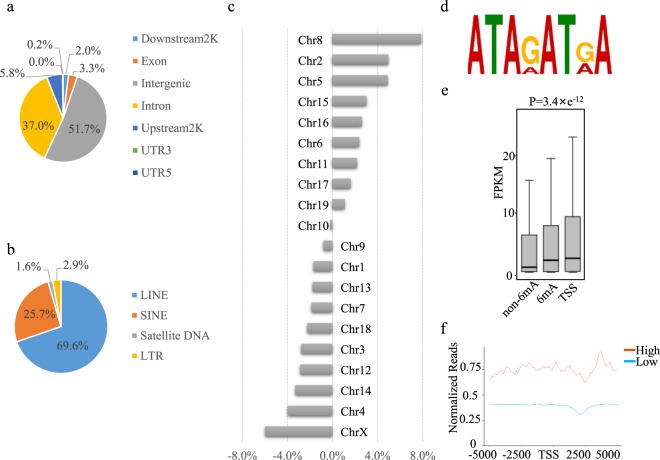
To understand the genome-wide distribution of 6 mA, locus-specific enrichment of 6 mA sites was examined in terms of gene exons, introns, 5’-UTR, 3’-UTR, upstream2K regions (within 2 kb upstream of TSS), downstream 2 K regions (within 2 kb downstream of TES), and intergenic regions. In mature adipocyte cells, 51.7% of the methylations were found within intergenic region, much higher than the background ratio calculated from the input (Fig. 2a). Our results were consistent with the reported profiles for mouse brain and embryonic stem cells13,21. For the 6 mA sites in intergenic regions, 69.6% was found within long interspersed nuclear elements (LINEs). However, no locus-specific preference was observed to distribute within the major TEs, including short interspersed nuclear elements (LINEs), long terminal repeated (LTR) and satellite DNA (Fig. 2b).
Figure 2.
6mA-IP-seq and RNA-seq of mature adipocytes. (a) Genome-wide distribution of N6-adenine methylation. (b) Distribution profiles of 6 mA in transposon elements. (c) Chromosome-wide distribution of 6 mA. For each chromosome, a chromosome length ratio was calculated by dividing its length in nucleotide against the total length of the genome, a 6 mA ratio was calculated by dividing the number of its 6 mA sites against the number of the total 6 mA sites. Then, an 6 mA abundance index was calculated by subtracting the 6 mA ratio by its chromosome length ratio. (d) Sequence motifs of 6 mA sites. (e) Comparison of the median and interquartile range of gene expression between genes without 6 mA sites (Non-6mA), genes with 6 mA sites (6 mA) and genes with 6 mA sites around TSS region (TSS), in 3T3-L1 cell of Day(+4). (f) 6 mA profile around TSS region, for genes with high (FPKM > 50) and low (FPKM < 50) expression level. Reads densities were normalized by RPKM algorithm.
Further examination found that 6 mA sites were enriched on chromosome 8, 2 and 5 (Fig. 2c), which was different from that found in embryonic stem cells where enrichment was found on chromosome X13. On the other hand, similar 6 mA distribution profiles were found for preadipocytes and mature adipocytes (Supplementary Fig. S3a–c).
Among the 6 mA sites identified, ATARATRA(R = A or G) was shown to be the most enriched motif using DREME software, with an E-value of 1.0e−038 (Fig. 2d). This motif shares the sequence of AAATA with another 6 mA motif GRAATA identified in mouse ESCs13.
A positive correlation between 6 mA abundance and gene expression
To examine the implications of N6-adenine methylation in differentiation, further analyses were performed with gene expression data obtained by RNA-seq. Based on the presence of 6 mA sites in a genomic region, protein-coding genes were divided into 6mA-containing group and 6mA-absent group. As shown in Fig. 2e, the median and interquartile FPKM (fragments per kilobase of transcript per million mapped reads) values of the 6mA-containing group were much higher than those of the 6mA-absent group. This result demonstrated that the expression levels of 6mA-containing genes were much higher than that of 6mA-absent genes. When the analyses were performed with genes carrying at least one 6 mA site at their TSS (400-bp window centered on the TSS), this correlation was much more remarkable.
To confirm the observation in another way, genes were divided into high expression (FPKM > 50) and low expression groups (FPKM < 50). The abundances of 6 mA sites in these two groups were analyzed and compared. As expected, genes of the high expression group displayed higher 6 mA abundance at TSS regions, comparing with the genes of low expression (Fig. 2f, Supplementary Fig. S3d). Taken together, a positive correlation was concluded between N6-adenine methylation of a gene and its expression.
Characterization of a mammalian N6-adenine methylase
Potential 6 mA metabolic enzymes were investigated subsequently. To this aspect, DNA 6 mA methylases and demethylases characterized in the other species are summarized in Supplementary Fig. S4. Mammalian homologues of these enzymes were searched by querying Uniprot database. In addition to N6AMT1 that was characterized as the first mammalian N6-adenine methylase12, METTL4 was identified as a candidate of N6-adenine methylase, due to its high similarity to Drosophila methylase CG14906 and C. elegans methylase DAMT-1. Additionally, TETs and ALKBH4 were found homologous to Drosophila demethylase DMAD and C. elegans demethylase NMAD.
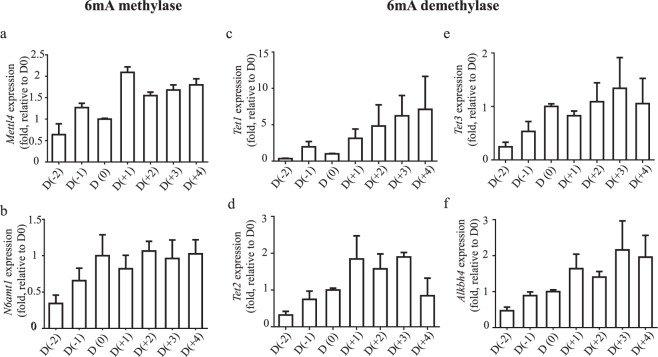
To examine their involvement in adipocyte differentiation, expression profiling was performed for Mettl4, N6amt1, Tets and Alkbh4. Similar expression profiles were found for Mettl4 and N6amt1 (Fig. 3), which were consistent with the increased formation of 6 mA in the genomic DNA. Phylogenetic tree analysis indicated that METTL4 protein is highly conserved in mammals (Fig. 4a).
Figure 3.
Expression regulation of 6 mA methylase and demethylase. During adipocyte differentiation of 3T3-L1 cells, gene expression of Mettl4 (a), N6amt1 (b), Tet1 (c), Tet2 (d), Tet3 (e) and Alkbh4 (f) were determined by quantitative RT-PCR and presented.
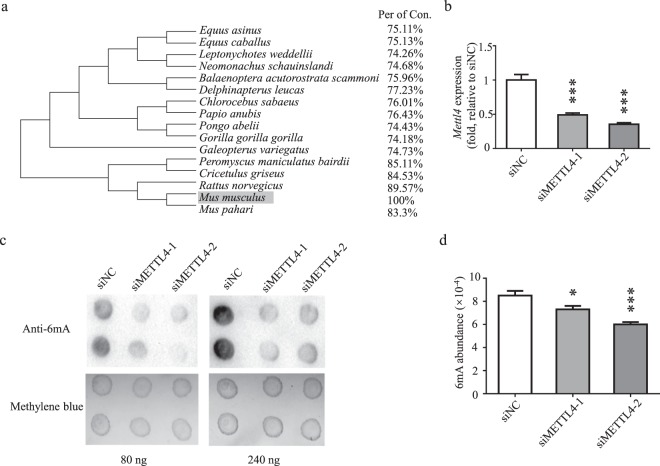
Figure 4.
Knock-down of Mettl4 leads to decreased 6 mA levels. (a) Phylogenetic tree analysis of METTL4 in mammals. (b) 3T3-L1 cells were transfected with Mettl4-targeting siRNAs. With the cells, gene expression of Mettl4 was determined by quantitative RT-PCR, in relative to control cells treated by a sequence-irrelevant siRNA. (c,d) Genomic 6 mA levels were determined by dot blot (c) and HPLC-MS/MS. Full-length blots are presented in Supplementary Fig. S5 (d). Data are analyzed by Student’s t-test and presented as mean ± s.d., n = 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
To examine the role of Mettl4 in N6-adenine methylation, gene knockdown assay was carried out with Mettl4-targeting siRNAs (Fig. 4b). Two days after siRNA transfection, genomic DNAs were extracted from siRNA-treated and untreated cells. The abundances of 6 mA were examined by dot blot assay. It was shown that knock-down of Mettl4 led to significantly decreased 6 mA levels (Fig. 4c, Supplementary Fig. S5a,b). Consistently, HPLC-MS/MS assay found that the abundance of 6 mA decreased by ~30% in Mettl4-knockdown cells (Fig. 4d).
Catalytic activity of METTL4 in vitro
To assess the catalytic activity of METTL4, in vitro methylation assays were performed with recombinant METTL4 protein, according to the reported methylation procedures22. Recombinant METTL4 protein was produced using a prokaryotic expression system23. In vitro methylation assays were performed with two single-stranded oligo substrates, and the products were analyzed by dot blot assay and HPLC-MS/MS analysis.
Dot blot assay indicated the formation of 6 mA modified products (Fig. 5a,b, Supplementary Fig. S6a,b). HPLC-MS/MS analysis found that about 0.1% of the adenine nucleotides were converted to 6 mA after incubated with 0.2 μg METTL4 for 12 h. The conversion efficiency was comparable to that of N6AMT-112.
Figure 5.
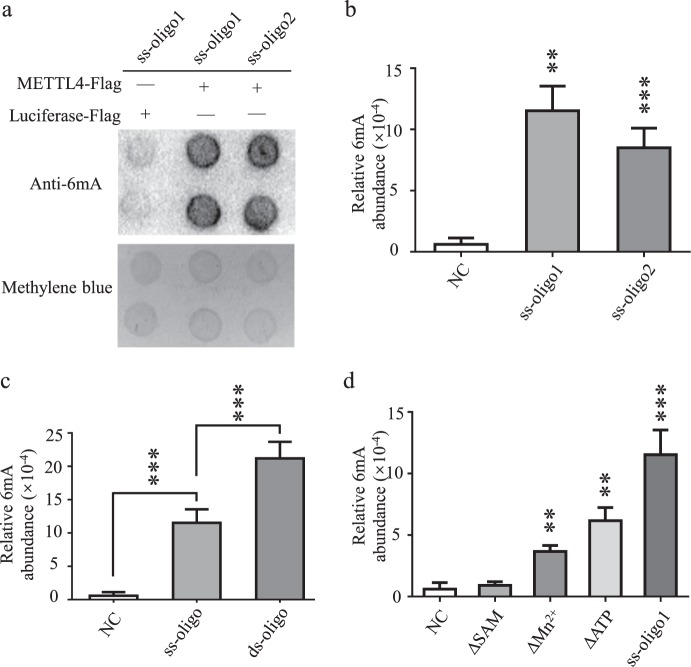
Catalytic activity of METTL4 in vitro. (a,b) Catalytic activity of N6-adenine methylation of recombinant METTL4. Two single-stranded DNA oligos were used as reaction substrates for recombinant METTL4-Flag protein. An irrelevant Firefly-luciferase was included as a negative control. DNA substrates were subjected to dot blotting assay using an anti-6mA antibody (a) and LC-MS/MS assay (b). For dot blotting assay, 600 ng of DNA was loaded for each group and full-length blots are presented in Supplementary Fig. S6. (c) Methylation efficacies on single-stranded (ss-oligo) and double-stranded (ds-oligo) substrates. (d) Optimization of methylation reaction. Data are analyzed by Student’s t-test and presented as mean ± s.d., n = 3 independent experiments. **P < 0.01; ***P < 0.001.
To explore the substrate preference of METTL4, methylation was performed with single- or double-stranded oligo substrates. Results showed that the methylation efficiency of double-stranded substrates was about twice as high as that of single-stranded substrates (Fig. 5c). Moreover, the effects of methyl donor SAM, ATP and Mn2+ on the methylation reaction were examined (Fig. 5d). Among these factors, SAM was found to be the most critical factor, exhibiting the greatest influence on N6-adenine methylation. The second most critical factor was Mn2+, and the absence of Mn2+ led to decreased catalytic activity by about 70%. The absence of ATP decreased the catalytic activity by about 50%.
These in vivo and in vitro investigations demonstrated that METTL4 was a second mammalian N6-adenine methylase, in addition to the firstly reported N6AMT-112.
The effects on adipocyte differentiation
To examine the roles of METTL4 in adipocyte differentiation, gene knockdown assays were performed. 3T3-L1 cells were firstly transfected with gene-specific siRNA on day(−2), treated with MDI cocktail on day(0), transfected again with gene-specific siRNA on day(+1), and harvested on day(+4) or day(+8). Bodipy and Oil Red O staining were performed to check the progress of differentiation. Comparing with control cells, Mettl4 knock-down cells showed greatly altered differentiation and decreased lipid production as much as 70% (Figs. 6a,b, Supplementary Fig. S7).
Figure 6.
Knockdown of Mettl4 leads to altered adipocyte differentiation. (a) The effects of Mettl4 knockdown were monitored by Bodipy staining on day(+4) (Left panel, 10×), and Oil Red O staining on day(+8) (Right panel, 10×). (b) Quantitative analyses of Bodipy (Left panel) and Oil Red O staining (Right panel). (c) The effects on gene expression of the major adipogenic factors. Data are analyzed by Student’s t-test and presented as mean ± s.d., n = 3 independent experiments. **P < 0.01; ***P < 0.001.
The effects on adipocyte differentiation were also confirmed by altered major adipogenic factors. Consistent with defective differentiation phenotype, expressions of Pparg and Cebpa in Mettl4 knock-down cells decreased to 41% and 30% of the normal levels (Fig. 6c).
N6-adenine methylation in the gene promoters by METTL4
We hypothesized that METTL4 exerts its action by increasing N6-adenine methylation in the promoters of downstream adipogenic genes during adipocyte differentiation, which increases their expression and facilitates the progress of differentiation. On the contrary, knockdown of Mettl4 decreases promoter methylation of the genes, resulting in decreased gene expression and altered cell differentiation.
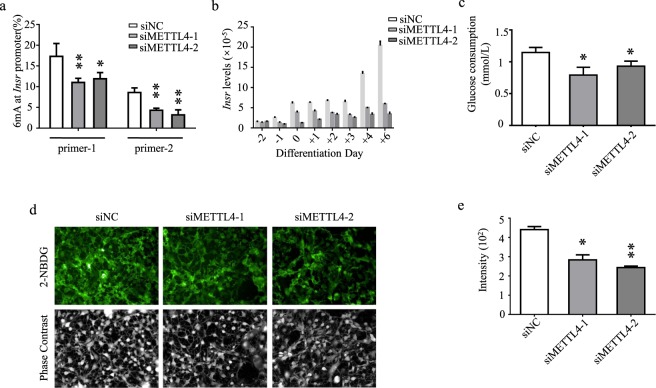
To confirm this hypothesis, we examined the N6-adenine methylation status of a group of adipogenic factors, including CEBPα, CEBPβ, PPARγ, WNT4, INSR, FAS, and PDE-3B in wild-type and Mettl4 knocked-down cells. Genomic DNAs were extracted on day(+2) and day(+4). For each of the genes, promoter 6 mA levels were determined using 6mA-IP-qPCR. Anti-IgG antibody was included in the assay as a negative control, and at least two independent primer sets were checked for each 6 mA site (Supplementary Fig. S8a). Results showed that promoter methylation was decreased by 30–50% for Insr gene in Mettl4 knockdown cells (Fig. 7a, Supplementary Fig. S9a). This was in contrast with the relatively stable levels of 6 mA in the promoters of Cebpa, Cebpb, Pparg, Wnt4, Fas and Pde-3b (Supplementary Figs. 8b–g, 9b–g). When gene expression was examined, a decrease of 65% was found with Insr expression (Fig. 7b).
Figure 7.
The effects of Mettl4 knockdown on glucose metabolism. (a) Promoter 6 mA abundance of Insr gene on day(+2) of the differentiation was determined by 6mA-IP-qPCR. (b) Expressional profiles of Insr in wild-type and Mettl4 knockdown cells. Gene expression levels were determined by quantitative RT-qPCR, in relative to that of β-actin. (c) Knockdown of Mettl4 led to decreased consumption of glucose. (d) Fluorescence images of 2-NBDG uptake. (e) Quantitative analyses of 2-NBDG uptake. Data are analyzed by Student’s t-test and presented as mean ± s.d., n = 3 independent experiments. *P < 0.05; **P < 0.01.
The effects of METTL4 on glucose metabolism
We proposed that INSR signaling pathway is involved in the process. In adipocyte differentiation, INSR pathway is involved in GLUT4 translocation from transport vesicles to cell membrane, therefore promoting transmembrane absorption of glucose. When the translocation of GLUT4 is repressed, both glucose absorption and consumption will be affected, resulting in decreased lipid production24,25.
To investigate the roles of METTL4 in glucose metabolism, glucose consumption (GC) assays were performed with wild-type and Mettl4 knock-down cells. On day(+4) of differentiation, culture medium was replaced with DMEM containing 1000 mg/L glucose and 0.2% BSA. The glucose levels were examined daily by means of glucose oxidase assay26. Comparing with control cells treated with a sequence-irrelevant siRNA, knockdown of Mettl4 led to decreased glucose consumption by 20–30% (Fig. 7c).
To investigate the roles of METTL4 in glucose uptake, glucose absorption assay was also performed27. On day(+4) of differentiation, an equal amount of 2-NBDG, a glucose transport probe, was added to each well of wild-type and Mettl4 knockdown cells. After incubation for 30 minutes, cellular distribution of 2-NBDG was examined. It was shown that knockdown of Mettl4 resulted in significantly reduced up-take of glucose probe by the cells (Fig. 7d). Comparing with control cells, fluorescence intensity decreased by about 50% in Mettl4 knockdown cells (Fig. 7e). Therefore, Mettl4 knockdown led to decreased glucose up-taking and consumption, decreased lipid production and altered adipocyte differentiation.
Discussion
DNA methylation plays critical roles in many biological processes including differentiation of adipocytes. In this study, we investigated the roles of N6-adenine methylation in the differentiation of 3T3-L1 preadipocytes. Comparing with the relatively stable levels of 5mC, dynamic changes of N6-adenine methylation were found, suggesting its involvement in the differentiation process of adipocytes. Genomic-wide distribution of 6 mA was examined by 6mA-IP-seq and 51.7% of the N6-adenine methylation sites was found within the intergenic regions. Chromosomes 2 and 8 were found to be the most enriched ones, accounting for 12.6% and 11.6% of the total N6-adenine methylation. In contrast, chrX was accounted for only 0.3% of the methylation sites. Compared with the 6 mA profile reported for mouse embryonic stem cells13, cell-type-specific distribution patterns were revealed. For ESCs, 74.7% of the 6 mA sites were distributed within the intergenic regions, chromosome X was the most 6mA-enriched chromosome and chromosome 8 was the least enriched chromosome.
A positive correlation between gene expression and the levels of N6-adenine methylation, in particularly at the promoter regions, was found. Highly expressed genes were found to be associated with increased promoter N6-adenine methylation, while genes of low expression were found to be associated with decreased 6 mA levels. Although the levels of 6 mA were found positively correlated with gene expression in general, locus- or sequence-specific roles needs to be further elucidated, considering the added effects of genomic 5mC methylation. 5mC methylations are usually found within CpG islands of promoter regions, functioning in gene inactivation28. However, when the methylations were located in coding region of a gene, an opposite effect on gene transcription was reported29.
During the differentiation of 3T3-L1 cells, the abundance of genome 6 mA was found to increase gradually. METTL4, a homologue of Drosophila methylase CG14906 and C. elegans methylase DAMT-1, was demonstrated to be a mammalian N6-adenine methylase. Knockdown of Mettl4 not only led to decreased 6 mA levels, but also altered the differentiation process of 3T3-L1 cells. We found that knockdown of Mettl4 led to decreased N6-adenine methylation at the promoter region of Insr gene, down-regulated gene expression and inactivated the INSR pathway. With the progress of adipocyte differentiation, expression of 6 mA methyltransferase Mettl4 starts to increase. This leads to up-regulation of promoter methylation and expression of downstream genes, including Insr. In the case of Insr gene, up-regulation of its expression activates INSR signaling pathway, increases glucose up-taking of the cells, and promotes lipid production of adipocytes. In summary, we identified the second mammalian N6-adenine methylase METTL4 and showed N6-adenine methylation played critical roles in the differentiation of adipocytes.
Taken together, these data indicated that INSR pathway is involved in the process. However, learning from the study on 5mC methylation, activities of 6 mA are speculated to be complex and diverse. In the differentiation of 3T3-L1 cells, very likely INSR pathway is one of the mechanisms affected by N6-adenine methylation. The effects on the major adipogenic regulators, such as PPARγ and CEBPβ, might contribute to the process to a larger extent.
Besides Mettl4, gene expression profiling found that a similar profile was also shared by N6amt1, another 6 mA methylase characterized in mammalian. Active regulation was also revealed for the other potential 6mAenzymes examined, likely suggests their involvement in the process. However, to the effects on methylation of specific gene, a complicated DNA methylation process will be implicated. Furthermore, different modification enzymes likely have their distinguishable downstream genes, which process may be guided by a variety of lncRNAs30.
Methods
Cell culture and adipocyte differentiation
3T3-L1 preadipocytes were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Scientific) supplemented with 10% calf serum (Invitrogen) at 37 °C and under 5% CO2 air. Two days after reaching confluence, cells were induced to differentiation by replacing the medium with DMEM containing 10% fetal bovine serum (Invitrogen), 500 μM isobutyl-methylxanthine (Sigma), 1 μM dexamethasone (MP) and 1 μg/mL insulin (Invitrogen). Cells were then incubated for 6–8 days with medium replacement every 2 days31.
BODIPY and Oil Red O staining
For BODIPY staining, cells were fixed in 4% paraformaldehyde for 15 min, permeabilized in PBS containing 1% Triton X-100 for 15 min, and stained with PBS containing BODIPY 493/503 (Invitrogen) and Hoechst 33342 (M&C) for 45 min at 37 °C. Analyses were performed with a High Content Analysis (Operetta).
For Oil Red O staining, cells were fixed with 70% ethanol for 10 min, stained with Oil Red O (Solarbio) in isopropanol for 30 min, and washed three times with 70% ethanol. For quantitative measurements of lipid accumulation, cells were washed with PBS to remove excess of stain solution, dried, and dissolved in 100% isopropanol. Absorbance was measured spectrophotometrically with FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices) at 520 nm31.
Gene silencing assay
Cells were plated in complete growth medium with 10% serum. siRNAs (Ribobio) and Lipofectamine RNAiMAX reagent (Invitrogen) were added to serum-free medium, and incubated at room temperature for 15 min. The transfection complex mixture was then added to the cells and incubated for 24–48 hr before analysis31.
RNA isolation and RT-qPCR
RNAs were isolated using TRIzol reagent (Invitrogen). cDNAs were synthesized using HiscriptII Q RT SuperMix for qPCR kit (Vazyme, Nanjing). qPCR was performed using GoTaq qPCR Master Mix (Promega). Data were normalized to the expression of β-actin gene31.
RNA-seq
RNAs were collected from preadipocyte cells on Day(−2) and matured adipocytes Day(+4) and the amount and quality of RNAs were determined by NanoPhotometer®(IMPLEN, CA, USA), Qubit®3.0 Flurometer (Life Technologies, CA, USA) and 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, CA, USA)10. RNA-seq assays were performed on an Illumina Hiseq 2500 platform and clean reads were mapped to mouse genome (GRCm38/mm10) using HISAT2 v2.1.032. Value of FPKM was calculated to represent the expression level of genes by HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html)33.
Dot Blot assay
Genomic DNAs were isolated using TIANamp Genomic DNA Kit (TIANGEN) and diluted to a concentration of 80 ng/μL. DNA samples were heated at 95 °C for 10 min to denature DNA, placed immediately on ice for 5 min, loaded at an amount of 120 ng or 240 ng per dot on a Hybond+ membrane. Membranes were air dried, irradiated by UV for 2 min to auto-crosslink DNA, blocked for 1 hr in 5% milk TBST, and probed with primary antibody (1:2000, Synaptic Systems) in 5% milk TBST at 4 °C overnight. After washing for five times with TBST, the membranes were probed with secondary antibody in 5% milk for 3 hr at room temperature, and washed for five times with TBST. Hybridization signals were detected with an Immobilon Western Chemilum HRP Substrate (Merck Millipore)7.
Enzymatic hydrolysis of genomic DNA
Genomic DNAs were isolated and treated first by RNase A for 12 hr at 50 °C, purified using TIANquick Maxi Purification Kit (TIANGEN). Purified DNA of 1 μg was sequentially treated by DNase I (8 U) for 12 hr at 37 °C, Nuclease P1 (8 U) for 12 hr at 50 °C, and calf intestinal alkaline phosphatase (1 U) for 12 hr at 37 °C. The hydrolysis products were dried in a vacuum centrifugal concentrator and washed by acetonitrile for 2 times. The resulting nucleoside-containing fractions were reconstituted in ultrapure water to a final concentration of 2 mg/mL, before HPLC-MS/MS assay19.
HPLC-MS/MS assay
Nucleosides sample of 10 μL was added to a 10 μL 6-Cl-Purine solution (3 mg/mL) and filtered through a 0.22 mm filter. 5 μL of the solution was subjected to LC-Ion Trap assay. DNA methylation was analyzed with a LC-ESI-MS/MS system consisting of a Shimadzu LC-20A HPLC system (Shimadzu, Kyoto, Japan) and an ABSciex QTRAP 5500 (AB Sciex, Canada). Data acquisition and processing were performed using AB SCIEX Analyst 1.5.2 Software (Applied Biosystems, CA). LC separation was performed using an Atlantis T3 column (4.6 mm * 150 mm, 5 mm, Waters) (WatersCorp., MA, USA) with a flow rate of 0.3 mL/min at 40 °C. 3 mM ammonium formate (solvent A) and methanol (solvent B) were used as mobile phases. A gradient of 2 min 2% B, 4 min 2–5% B, 2 min 5–10% B, 4 min 10% B, 0.1 min 10–95% B, 2 min 95% B, 0.1 min95–2% B, 4.9 min 2% B was used. Positive electrospray ionization mode was used to perform the mass spectrometry detection. Target analytes were monitored by multiple reaction monitoring (MRM) mode using the mass transitions (precursor ions/productions) of dC (228.1/112.1), dT (243.2/127.1), dA (252.2/136.0), dG (268.1/152.1), 5mC (242.0/126.0), 6 mA (266.1/150.1), 6-Chloropurine (6-CIP, 287.0/155.0). 6-CIP was spiked in as the internal standard in all the assays, due to its optimal detection sensitivity. Using MRM mode, the assays revealed a lineal relationship for tested nucleosides, with a lineal range of 10–10000 ng/mL for dA, 20–1000 ng/mL for dC, 2–1000 ng/mL for 5mC, and 0.1–100 ng/mL for 6 mA. The coefficients of correlation were determined to be higher than 0.98. The MRM parameters of all nucleosides were optimized to achieve maximal detection sensitivity. Quantification was performed by comparing with standard curves. A relative ratio was calculated for each nucleoside, based on the calculated molar concentrations19.
6mA-IP-sequencing
Using a Bioruptor, genomic DNAs were sonicated to fragments of 200–500 bp. DNA adaptors were then ligated to DNA fragments following the Illumina protocol. After denaturing at 95 °C for 5 mins, the resulted single-stranded DNA fragments were immunoprecipitated with 6 mA antibodies (Synaptic Systems) at 4 °C overnight. Together with input DNAs, 6mA-enriched DNA fragments were purified according to Active Motif hMeDIP protocol, PCR amplified using Illumina indexing primers, subjected to library construction and sequencing using an Illumina HiSeq 2000. The experiments, including 6 mA IP, library construction and sequencing, were performed at Annoroad Gene Technology(Beijing, China). Low-quality reads were removed from raw data by using the Trimmomatic package (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-Src-0.35.zip) and clean data were aligned to the mouse genome (UCSC, mm10), using Bowtie2. Peaks calling of 6 mA were performed by means of Macs2 software20 and the p-value was set as<e−5 32.
6mA-IP-qPCR
Using a Vibra-Cell Ultrasonic Liquid Processor (Sonics & Materials Inc, Newtown, USA), genomic DNA was sheared to an average of 200–1000 bp and denatured. 1 mg of denatured DNA was incubated with 1 mg anti-6mA antibody (Synaptic Systems) in IP buffer (1 mM sodium phosphate buffer pH 7.0, 0.14 M NaCl, 0.05% Triton X-100) for 3 hr at 4 °C. Antibody-bound DNA was collected with 10 μL of anti-mouse IgG dynabeads (Invitrogen) overnight at 4 °C with a rotating wheel, washed three times with IP buffer, recovered in 200 μL digestion buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 0.5% SDS, 40 μg proteinase K), and incubated at 56 °C for 2 hr with occasional mixing by vortexing. Recovered DNA was purified and quantified by qPCR assay. The ratio of genes in 6mA-IP group to input was calculated to reach the 6 mA abundance in specific locus. Sequences of the primers for gene detection are included as Supplementary Information31.
Cloning and Expression of METTL4
Full-length coding sequence of mouse METTL4 (GenBank: NP_001344064.1) was synthesized and subcloned into a pMCSG19 vector, generating a plasmid named pMCSG19-His-METTL4. METTL4 proteins were expressed in BL21 (DE3) E. coli strain, and purified for activity characterization.
Glucose consumption assay
3T3-L1 cells were grown in DMEM (4500 mg/L glucose) containing 10% fetal bovine serum and were plated into 96-well plates. Two days after confluence, the cells were introduced to differentiation, and transfected with siRNA on day(−2) and day(+1) for two times. Glucose consumption assays were performed by replacing culture medium with DMEM containing 1000 mg/L glucose and 0.2% BSA. After the assay, culture medium was collected and the glucose concentration was determined by the glucose oxidase method. Comparing with the glucose concentrations of blank wells, glucose consumption (GC) of plated wells was calculated.
Glucose uptake assay
Cells were plated into 6-well plates and induced to differentiation two days after reaching confluence. Four days after induction, culture medium was removed from the culture wells, the cells were washed with PBS for 3 times, and fresh PBS containing 2-NBDG of1mmol/L was added. The cell plate was incubated for 30 min at 37 °C. The cells were washed for 3 times before High Content Analysis using Operetta.
In vitro methylation assay
A methylation reaction of 25 μL contained 10 mM HEPES (pH7.9), 10 mM MgCl2, 160 mM KCl, 5 mM SAM, 20 μM ATP, 0.02 μg protein and 2 ng DNA oligo. Assays were performed by incubating at 37 °C for 12 hours, and stopped by heating to 95 °C for 5 min. Sequences of DNA oligos are included in the Supplementary Information.
Statistical analysis
Analyses were performed with GraphPad Prism5 software. Data was presented as mean ± SD. ANOVA and Student’s t-test were performed to evaluate the statistical significance. The significance level was set at p < 0.05.
Supplementary information
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (31571403),Beijing Natural Science Foundation (2171001) and the National High-tech R&D Program of China (2014AA021103).
Author contributions
Zhenxi Zhang contributed conception of the work, acquisition, analysis of data and had drafted the manuscript. Yingzi Hou contributed design of the work and acquisition of data. Yao Wang and Tao Gao contributed acquisition of data. Ziyue Ma and YingYang helped design the work. Pei Zhang, FanYi, Jun Zhan and Hongquan Zhang interpreted of data. Quan Du contributed conception of the work.
Data availability
The data of 6mA-seq and RNA-seq had been uploaded to SRA database and the accession was PRJNA588576.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64873-w.
References
- 1.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. NAT REV GENET. 2013;14:204. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 2.Schubeler D. Function and information content of DNA methylation. NATURE. 2015;517:321. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. NAT REV GENET. 2012;13:484. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. NAT REV MICROBIOL. 2006;4:183. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. BIOESSAYS. 2006;28:309. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo GZ, Blanco MA, Greer EL, He C, Shi Y. DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol. 2015;16:705. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer EL, et al. DNA Methylation on N6-Adenine in C. elegans. CELL. 2015;161:868. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, et al. N6-methyladenine DNA modification in Drosophila. CELL. 2015;161:893. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. CELL. 2015;161:879. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang Z, et al. DNA N(6)-Adenine Methylation in Arabidopsis thaliana. DEV CELL. 2018;45:406. doi: 10.1016/j.devcel.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. NAT COMMUN. 2016;7:13052. doi: 10.1038/ncomms13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao CL, et al. N(6)-Methyladenine DNA Modification in the Human Genome. MOL CELL. 2018;71:306. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Wu TP, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. NATURE. 2016;532:329. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao CL, et al. N(6)-Methyladenine DNA Modification in the Human Genome. MOL CELL. 2018;71:306. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Ge K. Epigenetic regulation of adipogenesis by histone methylation. Biochim Biophys Acta. 2012;1819:727. doi: 10.1016/j.bbagrm.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noer A, Boquest AC, Collas P. Dynamics of adipogenic promoter DNA methylation during clonal culture of human adipose stem cells to senescence. BMC CELL BIOL. 2007;8:18. doi: 10.1186/1471-2121-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokomori N, Tawata M, Onaya T. DNA demethylation during the differentiation of 3T3-L1 cells affects the expression of the mouse GLUT4 gene. DIABETES. 1999;48:685. doi: 10.2337/diabetes.48.4.685. [DOI] [PubMed] [Google Scholar]
- 18.Melzner I, et al. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J BIOL CHEM. 2002;277:45420. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- 19.Liang D, et al. The decreased N(6)-methyladenine DNA modification in cancer cells. Biochem Biophys Res Commun. 2016;480:120. doi: 10.1016/j.bbrc.2016.09.136. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) GENOME BIOL. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao B, et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. NAT COMMUN. 2017;8:1122. doi: 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimba S, Bokar JA, Rottman F, Reddy R. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. NUCLEIC ACIDS RES. 1995;23:2421. doi: 10.1093/nar/23.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. MOL CELL. 2013;49:18. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favaretto F, et al. GLUT4 defects in adipose tissue are early signs of metabolic alterations in Alms1GT/GT, a mouse model for obesity and insulin resistance. PLOS ONE. 2014;9:e109540. doi: 10.1371/journal.pone.0109540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepherd PR, et al. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J BIOL CHEM. 1993;268:22243. [PubMed] [Google Scholar]
- 26.Yin J, Gao Z, Liu D, Liu Z, Ye J. Berberine improves glucose metabolism through induction of glycolysis. Am J Physiol Endocrinol Metab. 2008;294:E148. doi: 10.1152/ajpendo.00211.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Zhang J, Zhang XY. 2-NBDG as a marker for detecting glucose uptake in reactive astrocytes exposed to oxygen-glucose deprivation in vitro. J MOL NEUROSCI. 2015;55:126. doi: 10.1007/s12031-014-0385-5. [DOI] [PubMed] [Google Scholar]
- 28.Taberlay PC, et al. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. CELL. 2011;147:1283. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. SCIENCE. 2007;315:1141. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 30.Yi F, et al. Long non-coding RNA slincRAD functions in methylation regulation during the early stage of mouse adipogenesis. RNA BIOL. 2019;16:1401. doi: 10.1080/15476286.2019.1631643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou Y, et al. 5mC profiling characterized TET2 as an anti-adipogenic demethylase. GENE. 2020;733:144265. doi: 10.1016/j.gene.2019.144265. [DOI] [PubMed] [Google Scholar]
- 32.Zhou C, et al. Identification and analysis of adenine N(6)-methylation sites in the rice genome. NAT PLANTS. 2018;4:554. doi: 10.1038/s41477-018-0214-x. [DOI] [PubMed] [Google Scholar]
- 33.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. BIOINFORMATICS. 2015;31:166. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of 6mA-seq and RNA-seq had been uploaded to SRA database and the accession was PRJNA588576.