Abstract
Purpose of Review
Pathogenesis-related class 10 (PR-10) proteins are highly conserved plant proteins, which are induced in response to abiotic and biotic stress factors. To date, no unique biological function could be assigned to them. Rather a more general role of PR-10 in plant development and defense mechanisms has been proposed. In addition, some PR-10 proteins act as allergens by triggering allergic symptoms in sensitized individuals. Regardless of the diversity of reported activities, all PR-10 proteins share a common fold characterized by a solvent-accessible hydrophobic cavity, which serves as a binding site for a myriad of small-molecule ligands, mostly phytohormones and flavonoids.
Recent Findings
Most of available data relate to the ligand binding activity of allergenic PR-10, particularly for those belonging to Bet v 1 family of allergens. Bet v 1 and its homologues were shown to bind flavonoids with high affinity, but the specificity appears to differ between homologues from different species. The flavonoid Q3O-(Glc)-Gal was shown to specifically bind to hazelnut Cor a 1 but not to Bet v 1. Similarly, Q3OS bound only to the major isoform Bet v 1.0101 and not to other closely related isoforms. In contrast, Bet v 1 and hazelnut Cor a 1 showed very similar binding behavior towards other flavonoids such as quercetin, genistein, apigenin, daidzein, and resveratrol.
Summary
Recent research findings highlighted the importance of more precise knowledge of ligand binding for understanding the functional diversification of PR-10 proteins.
Keywords: PR-10, Allergens, Bet v 1, Ligand, Flavonoid, Cytokinin
Introduction
Pathogenesis-related class 10 (PR-10) proteins comprise a unique class of highly conserved phytoproteins found in both monocotyledonous and dicotyledonous plants. PR-10 are primarily cytosolic proteins, constitutively expressed in several plant tissues including roots, stems, flowering compartments, fruits, and pollen grains from certain plant species. Their expression is upregulated upon abiotic and biotic stress conditions, such as invading pathogenic viruses, bacteria and fungi, cold, salinity, drought, oxidative stress, ultraviolet radiation, and physical wounding [1]. Hence, it has been proposed that PR-10 proteins have no unique function but play a more general role in plant development and defense mechanisms [2, 3].
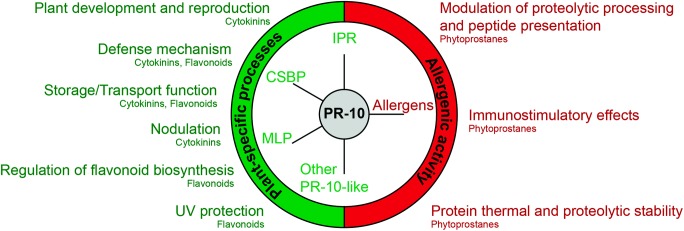
PR-10 proteins are 154 to 163 amino acids long and have a molecular weight of approximately 17 kDa. Their 3-dimensional structure consists of an anti-parallel, seven-stranded β-sheets wrapping around an amphipathic C-terminal α helix (α3) embraced by two short α-helices (α1, α2) forming a V-shape. The main structural feature of the PR-10 fold is a large solvent-accessible, hydrophobic internal cavity spanning the entire protein [4, 5]. This hydrophobic core of PR-10 molecules serves as a binding site for a wide variety of ligands, thus explaining its promiscuous binding behavior. Here we review recent findings on ligand binding of PR-10 proteins with a particular focus on the Bet v 1 family and the impact of bound ligands on the immunological properties of these allergenic proteins. In addition, published data on PR-10 ligand binding in the broad context of plant biology are briefly discussed (Fig. 1).
Fig. 1.
Overview on described functions associated with PR-10 ligand binding; PR-10, pathogenesis-related proteins class 10; IPR, intracellular pathogenesis-related proteins/classic PR-10 proteins; CSBP, cytokinin-specific binding proteins; MLP, major latex proteins; other PR-10-like, not yet classified proteins possessing a PR-10-like fold
Ligand Binding of PR-10 Proteins
Ligand binding has been described for many PR-10 proteins, mostly for allergens derived from plants of the Fagales order, such as birch (Betula verrucosa), hazel (Corylus avellana), and beech (Fagus sylvatica). PR-10 proteins from more distantly related allergenic food sources like peanut (Arachis hypogaea), strawberry (Fragaria ananassa), cherry (Prunus avium), and peach (Prunus persica) were also shown to bind ligands [6••, 7••, 8–10]. Interestingly, not all reported ligand binding PR-10 proteins have been described as allergens. In fact, the taxon of PR-10 proteins can be subdivided into intracellular pathogenesis-related (IPR) proteins/classic PR-10, cytokinin-specific binding proteins (CSBP), and major latex proteins (MLP), although CSBP and MLP only share a sequence identity of about 17 to 25% with the IPR group [1]. In this review, we have additionally included “allergens” as another subgroup of PR-10 because not all proteins displaying the typical features and the canonical fold of classic PR-10 are allergenic. Nevertheless, their common ability to bind low molecular weight compounds within their hydrophobic cavity can be considered a general feature of PR-10 proteins. The promiscuous ligand binding behavior of PR-10 proteins has been extensively documented using a large panel of phytohormones and plant metabolites. Based on these studies, three major chemical classes of PR-10-binding ligands have been defined: (i) cytokinins, (ii) flavonoids, and (iii) sterols [10–13]. A detailed overview of PR-10 ligands described in the literature is given in Table 1.
Table 1.
Protein-ligand interaction of PR-10 proteins
| Protein | Plant (scientific name) | PR-10 class | Protein MW (kDa) | Ligand name | Ligand chemical class | Ligand MW (Da) | Kd (μM) | Method used to confirm ligand binding | Comment ¤ | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Ara h 8.0101 | Peanut (Arachis hypogaea) | Allergens | 16.95 | Quercetin | Flavonoid | 302.23 | -* | ADA | - | [11] |
| Genistein | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Apigenin | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Daidzein | Flavonoid | 254.24 | -* | Direct measurement | - | [11] | ||||
| Resveratrol | Flavonoid | 228.24 | -* | ADA | - | [11] | ||||
| Progesterone | Animal steroid hormone | 314.50 | -* | ADA | - | [11] | ||||
| Caffeic acid | Organic compounds | 180.16 | -* | ADA | - | [11] | ||||
| Palmitic acid | Flavonoid | 256.42 | -* | ADA | - | [11] | ||||
| Arachidic acid | Flavonoid | 312.50 | -* | ADA | - | [11] | ||||
| Ara h 8.0201 | Peanut (Arachis hypogaea) | Allergens | 16.41 | Quercetin | Flavonoid | 302.23 | -* | ADA | - | [11] |
| Genistein | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Apigenin | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Daidzein | Flavonoid | 254.24 | -* | ADA | - | [11] | ||||
| Resveratrol | Flavonoid | 228.24 | -* | Direct measurement | - | [11] | ||||
| Progesterone | Animal steroid hormone | 314.50 | -* | ADA | - | [11] | ||||
| Caffeic acid | Organic compounds | 180.16 | -* | ADA | - | [11] | ||||
| Myristic acid | Flavonoid | 228.37 | -* | ADA | - | [11] | ||||
| Ferulic acid | Organic compounds | 194.18 | -* | ADA | - | [11] | ||||
| Cor a 1.0201 | Hazel pollen (Corylus avellana) | Allergens | 17.41 | Apigenin | Flavonoid | 270.24 | -* | ADA | - | [11] |
| Daidzein | Flavonoid | 254.24 | -* | ADA | - | [11] | ||||
| Resveratrol | Flavonoid | 228.24 | -* | Direct measurement | - | [11] | ||||
| Progesterone | Animal steroid hormone | 314.50 | -* | ADA | - | [11] | ||||
| DOC | Brassinosteroid analog | 414.60 | -* | ADA | - | [11] | ||||
| Caffeic acid | Organic compounds | 180.16 | -* | ADA | - | [11] | ||||
| Myristic acid | Fatty acid | 228.37 | -* | ADA | - | [11] | ||||
| Palmitic acid | Fatty acid | 256.42 | -* | ADA | - | [11] | ||||
| Zeatin | Cytokinin | 219.24 | -* | ADA | - | [11] | ||||
| Quercetin | Flavonoid | 302.23 | -* | ADA | - | [11] | ||||
| Genistein | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Cor a 1.0401 | Hazelnut (Corylus avellana) | Allergens | 17.58 | Q3O-(Glc)-Gal | Flavonoid | 626.53 | Below 5 | NMR | - | [7] |
| Quercetin | Flavonoid | 302.23 | -* | ADA | [11] | |||||
| Genistein | Flavonoid | 270.24 | -* | ADA | [11] | |||||
| Apigenin | Flavonoid | 270.24 | -* | ADA | [11] | |||||
| Daidzein | Flavonoid | 254.24 | -□ | ADA | [11] | |||||
| Epicatechin | Flavonoid | 290.27 | -□ | ADA | [11] | |||||
| Resveratrol | Flavonoid | 228.24 | -* | Direct measurement | [11] | |||||
| Progesterone | Animal steroid hormone | 314.50 | -□ | ADA | [11] | |||||
| DOC | Brassinosteroid analog | 414.60 | -□ | ADA | [11] | |||||
| Caffeic acid | Organic compounds | 180.16 | -□ | ADA | [11] | |||||
| Ferulic acid | Organic compounds | 194.18 | -□ | ADA | [11] | |||||
| 3-Indolybutyric acid | Organic compounds | 203.24 | -□ | ADA | [11] | |||||
| Fag s 1.0101 | European beech (Fagus sylvatica) | Allergens | 17.36 | ANS | Organic compounds | 299.30 | - | ADA | [8] | |
| Dehydroergosterol | Brassinosteroid | 394.60 | - | ADA, NMR | [8] | |||||
| Naringenin | Flavonoid | 272.25 | - | ADA | [8] | |||||
| Kinetin | Cytokinin | 215.21 | - | ADA | [8] | |||||
| SDS | Synthetic detergent | 288.38 | - | ADA | [8] | |||||
| Fra a 1.0101 | Strawberry (Fragaria ananassa) | Allergens | 17.77 | Q3OS | Flavonoid | 626.50 | 5.3 | ITC | - | [10] |
| Fra a 1.0201 | Myricetin (+)-catechin | Flavonoid | 318.23 | 19.5 | ITC | - | [10] | |||
| Fra a 1.0301 | Flavonoid | 290.27 | 8.9 | X-ray, ITC | 4C94, 1:2 | [10] | ||||
| Pru av 1.0101 | Cherry (Prunus avium) | Allergens | 17.66 | Homocastasterone | Brassinosteroid | 478.7 | - | NMR, in silico docking | - | [14] |
| Pru p 1.0101 | Peach (Prunus persica) | Allergens | 17.65 | Zeatin | Cytokinin | 219.24 | 9.4 | ITC | 1:1 | [9] |
| Que a 1.0201 | White oak (Quercus alba) | Allergens | 17.30 | Daidzein | Flavonoid | 254.24 | -* | ADA | - | [11] |
| Quercetin | Flavonoid | 302.23 | -* | ADA | - | [11] | ||||
| Genistein | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Apigenin | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Resveratrol | Flavonoid | 228.24 | -□ | Direct measurement | - | [11] | ||||
| Progesterone | Animal steroid hormone | 314.50 | -□ | ADA | - | [11] | ||||
| Palmitic acid | Fatty acid | 256.42 | -□ | ADA | - | [11] | ||||
| Myristic acid | Fatty acid | 228.37 | -□ | ADA | - | [11] | ||||
| DOC | Brassinosteroid analog | 414.60 | -□ | ADA | - | [11] | ||||
| Que a 1.0301 | White oak (Quercus alba) | Allergens | 17.48 | Daidzein | Flavonoid | 254.24 | -* | ADA | - | [11] |
| Quercetin | Flavonoid | 302.23 | -* | ADA | - | [11] | ||||
| Genistein | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Apigenin | Flavonoid | 270.24 | -* | ADA | - | [11] | ||||
| Resveratrol | Flavonoid | 228.24 | -□ | Direct measurement | - | [11] | ||||
| Progesterone | Animal steroid hormone | 314.50 | -□ | ADA | - | [11] | ||||
| Palmitic acid | Fatty acid | 256.42 | -□ | ADA | - | [11] | ||||
| BpPR-10c | European white birch (Betula verrucosa) | IPR | 17.00 | Kinetin | Cytokinin | 215.21 | - | NMR, in silico docking | 1:1 | [12] |
| Hyperoside | Flavonoid | 464.4 | - | NMR, in silico docking | 1:2 | [12] | ||||
| Rutin | Flavonoid | 610.5 | - | NMR, in silico docking | 1:1 | [12] | ||||
| Emodin | Plant metabolite | 270.24 | - | NMR | - | [12] | ||||
| DOC | Brassinosteroid analog | 414.60 | - | NMR, in silico docking | 1:2 | [12] | ||||
| LlPR-10.1A | Yellow Lupine (Lupinus luteus) | IPR | 16.86 | Zeatin | Cytokinin | 219.24 | - | In silico docking | 2QIM, 1:3 | [15] |
| LlPR-10.1B | 16.66 | Diphenylurea | Cytokinin | 212.25 | - | X-ray | 3E85, 1:4 | [16] | ||
| MtN13 | Barrelclover (Medicago truncatula) | IPR | 18.18 | Zeatin | Cytokinin | 219.24 | - | X-ray | 4JHG, 2:2 | [13] |
| Kinetin | Cytokinin | 215.21 | - | X-ray | 4JHH, 2:3 | [13] | ||||
| IPA | Cytokinin | 203.24 | - | X-ray | 4GY9, 2:2 | [13] | ||||
| N6-benzyladenine | Cytokinin | 225.25 | - | X-ray | 4JHI, 2:2 | [13] | ||||
| SPE16 | Jicama (Pachyrhizus erosus) | IPR | 15.81 | ANS | Organic compounds | 299.30 | - | X-ray | 1TXC, 2:5 | [1] |
| UBP34 | Spreading earthmoss (Physcomitrell a patens) | IPR | 34.00 | azido-CPPU | Cytokinin agonist | 288.69 | 25 | Direct measurement | - | [17] |
| VrCSBP | Mung bean (Vigna radiata) | CSBP | 17.60 | Zeatin | Cytokinin | 219.24 | 106–161 | ADA, ITC, X-ray | 2FLH, 1:1 | [18, 19] |
| ANS | Organic compounds | 299.30 | 32.5 | Direct measurement | - | [18, 19] | ||||
| 4PU30 | Cytokinin | 247.68 | 1.3 | ADA | 1:1 | [18, 19] | ||||
| IPA | Cytokinin metabolite/zeatin precursor | 203.24 | 11.5 | ADA | 1:1 | [18, 19] | ||||
| Kinetin | Cytokinin | 215.21 | 64.1 | ADA | 1:1 | [18, 19] | ||||
| NCS | Yellow meadow-rue (Thalictrum flavum) | Others | 23.34 | 4-Hydroxybenzaldehyde Dopamine | Plant metabolite | 122.12 | - | X-ray | 2VQ5, 2:1 | [20] |
| Catecholamine | 153.18 | - | X-ray | 2VQ5, 2:2 | [20] | |||||
| Hyp-1 | Saint John’s wort (Hypericum perforatum) | Others | 17.81 | Melatonin ANS | Biogenic amine, hormone | 232.28 | - | X-ray | 3IE5, 1:3 | [21] |
| Organic compounds | 299.30 | - | X-ray | 4N3E, 1:4 | [21] |
¤3D structure (PDB code) and binding stoichiometry (Protein:Ligand); *probably strong binding; □probably weak binding
ADA, ANS displacement assay; Allergens, characterized allergens according to the WHO/IUIS allergen nomenclature database; CSBP, cytokinin-specific binding proteins; direct measurement, direct measurement by florescence of ligand molecule; IPR, intracellular pathogenesis-related proteins/classic PR-10; ITC, isothermal titration calorimetry; Kd, equilibrium dissociation constant; MW, molecular weight; NMR, nuclear magnetic resonance spectroscopy; others, not yet classified proteins possessing a PR-10-like fold; X-ray, X-ray crystallography
Cytokinins are phytohormones involved in the regulation of plant development, growth and defense mechanisms, cell division, and the deceleration of senescence [9]. Binding to the naturally occurring cytokinins zeatin and kinetin has been mostly described for CSBPs [8, 9, 12–18, 20–23, 24••]. According to the current hypothesis, CSBPs might have evolved within PR-10 proteins in order to maintain cytokinin homeostasis by specifically binding to these molecules. Thus, the specific PR-10 fold is highly conserved within the CSBP group supporting the notion that their fold enables efficient shuttling and storage of bioactive molecules [18, 19]. Interestingly, the PR-10 from Medicago truncatula (MtN13) was observed to play a role as regulator of free cytokinins during the early phases of nodulation, i.e., the recruitment process of symbiotic nitrogen-fixing bacteria [13]. Based on the fact that zeatin is a nucleoside analog, PR-10 proteins have been proposed to possess nuclease activity [15]. In this respect, several studies focusing on the putative RNase or DNase activity of PR-10 proteins derived from several plant species including birch, cotton, ginseng, peach, and pepper have been reported. However, the biological relevance of these studies has not been further explored and the reported PR-10/DNA/RNA interactions are still a matter of debate [9, 12, 15, 25–30]. Apart from the cytokinins zeatin and kinetin, other phytohormones, such as phytomelatonin, brassinosteroids, and gibberellic acid, were frequently observed to bind PR-10 proteins [14, 31].
Another important class of PR-10 ligands are flavonoids, which are polyphenolic compounds and secondary plant metabolites involved in color and flavor production, UV protection, antioxidation, and pathogen defense [32]. PR-10 allergens, mostly of the Fagales order, were shown to bind flavonoids with high binding affinity within their hydrophobic pocket, but the specificity appears to differ between PR-10 from different species. For instance, the natural ligand of hazelnut Cor a 1, Q3O-(Glc)-Gal is highly similar to the birch Bet v 1 co-purified natural ligand, quercetin-3-O-sophoroside (Q3OS), differing just in the orientation of the hydroxyl group. Nevertheless, the Cor a 1 ligand does not interact with Bet v 1 and vice versa, despite the high structural and sequence similarity between both allergens [7••]. In addition, Q3OS bound only to the major isoform, Bet v 1.0101 and not to other isoforms [33]. These observations are in line with the notion that the presence of many PR-10 isoforms in a plant could be the basis for their functional diversification [34]. In contrast, a study by Mc Bride et al. reported a more general, less discriminative binding of Bet v 1 and hazelnut Cor a 1 to other flavonoids (e.g., quercetin, genistein, apigenin, daidzein, resveratrol), highlighting the relative binding promiscuity of PR-10 allergens for this class of ligand [11]. Accordingly, other PR-10 allergens, such as strawberry Fra a 1, white oak Que. a 1, hazel pollen Cor a 1, and peanut Ara h 8, also bound numerous flavonoids [10, 11, 32]. The exact role of flavonoid binding by PR-10s remains to be elucidated; however, it is speculated that this protein class possesses major regulatory functions in flavonoid biosynthesis via binding and storage of functionally inert glycosylated flavonoids, which are in turn prevented from early activation by enzymatic deglycosylation by glycosyltransferases [7••, 10, 32]. This hypothesis is supported by the observation that the major allergen of beech pollen, Fag s 1, binds with high affinity to naringenin, a metabolic intermediate of flavonoid synthesis, whereas it does not interact with unglycosylated quercetin [8]. Similarly, the binding affinity of Bet v 1 to Q3OS was 60-fold stronger compared with quercetin [4].
Ligand Binding of Bet v 1
IgE sensitization to Bet v 1, the major allergen of birch (Betula verrucosa) pollen, ranges from 53 to 95% among birch pollen allergic patients. Similar sensitization rates were observed for Bet v 1 homologues like Mal d 1 (apple) and Cor a 1 (hazelnut), mostly due to cross-reactivity occurring between structurally similar PR-10 proteins [35, 36]. This results in clinical manifestations described as the oral allergy syndrome (OAS), a very common allergic disorder affecting more than 50% of birch pollen allergic patients [37]. Bet v 1.0101 (formerly designated Bet v 1a) represents the most abundant isoform of Bet v 1 comprising 50 to 70% of the allergen in pollen [4, 5]. The natural function of Bet v 1 is still not fully understood; however, based on the structural similarity with the START domain of the human MLN64 protein, it has been suggested that Bet v 1 play a role in steroid binding [38]. The ability of Bet v 1 to bind a broad spectrum of plant intrinsic ligands, such as fatty acids, cytokinins, or flavonoids, has led to the suggestion of an involvement in different stages of plant reproduction (e.g., protection of pollen DNA from UV-damage, transportation of lipids or flavonoids to the stigmatic surface to support pollen hydration and germination). Bet v 1 may also act as an storage scaffold for such ligands, enabling their rapid release upon seed germination [39]. In addition, the combination of different ligands and Bet v 1 isoforms with differences in their ligand preferences could possibly serve as molecular fingerprints to prevent self-pollination [23, 33, 40].
Bet v 1 has been shown to bind a broad range of hydrophobic to amphipathic ligands, differing in size and shape, to distinct binding sites within its hydrophobic cavity [23]. In general, pollen-derived molecules that interact with Bet v 1 can be grouped into fatty acids, flavonoids, and phytohormones. A detailed description of Bet v 1 ligands reported in the literature is given in Table 2.
Table 2.
Ligand binding of Bet v 1
| Chemical class | Ligand name | MW (Da) | Kd (μM) | 3D structure | (PDB method used to confirm ligand binding code) | Stoichiometry (Protein:Ligand) | References |
|---|---|---|---|---|---|---|---|
| Brassinosteroids | 24-Epicastasteron | 464.70 | - | - | MS | 1:1 | [41] |
| Brassinolide | 480.70 | - | - | MS, in silico docking studies | 1:1 and 1:2 | [41] | |
| Dehydroergosterol | 394.60 | - | - | ADA | - | [23] | |
| Stigmasterol | 412.70 | - | - | ADA | - | [11] | |
| Cytokinins | IPA | 203.24 | 64.3 | - | ADA | - | [23] |
| Kinetin | 215.21 | 84.1 | 4A85, 4A86 | ADA, X-ray | 1:1 | [23, 42] | |
| Zeatin | 219.24 | - | - | ADA | - | [11] | |
| Phytoprostanes | Phytoprostane B1 | 308.40 | 1 | SAW | [6••] | ||
| Phytoprostane E1 | 356.50 | 0.5 | NMR, SAW | [6••] | |||
| Phytoprostane F1 | 328.40 | 2.4 | SAW | [6••] | |||
| Fatty acids | Arachidic acid | 312.50 | 26.9 | ADA | [23] | ||
| Myristic acid | 228.37 | 8.7 | ADA | [23] | |||
| Palmitic acid | 256.42 | 7.7 | ADA | [23] | |||
| Stearic acid | 284.50 | 4.2 | ADA | [23] | |||
| Flavonoids | Apigenin | 270.24 | - | - | ADA | - | [11] |
| Daidzein | 254.24 | - | - | ADA | - | [11] | |
| Flavone | 222.24 | 33.2 | - | ADA | - | [23] | |
| Genistein | 270.24 | - | - | ADA | - | [11] | |
| Naringenin | 272.25 | 28.6 | 4A87 | ADA, X-ray | 1:1 | [23, 42] | |
| Q3OS | 626.50 | 0.56 to 1.5 | - | NMR, MS, SAW | - | [4, 6••] | |
| Quercetin | 302.23 | 9.2 to 33 | - | NMR, ADA | - | [4, 23] | |
| Resveratrol | 228.24 | - | - | ADA | - | [11] | |
| Extrinsic and non-physiological ligands | ANS | 299.30 | 18.5 | 4A80, 4A8V, 4A86 | Direct measurement, NMR, X-ray | 1:1 | [23, 42] |
| DOC | 414.60 | 58.8 | 1FM4, 4A81, 4A83 | X-ray, MS | 1:2 | [6••, 41, 42] | |
| NDSB-256 | 257.35 | - | 4A8G | ADA, X-ray | 1:1 | [42] | |
| P303 | 304.06 | - | 4MNS | X-ray | 1:1 | [43] | |
| Progesterone | 314.50 | - | - | ADA | - | [11] | |
| SDS | 288.38 | 7* and 100□ | 4QIP | ADA, X-ray | 1:2 | [23, 44] | |
| STS | 316.43 | 1* and 20□ | - | NMR | 1:2 | [44] |
*Inner binding site; □outer binding site
ADA, ANS displacement assay; direct measurement, direct measurement by florescence of ligand molecule; Kd, equilibrium dissociation constant; MS, mass spectrometry; MW, molecular weight; NMR, nuclear magnetic resonance spectroscopy; SAW, surface acoustic wave measurement; X-ray, X-ray crystallography
The affinity of Bet v 1 towards fatty acids depends on their chain length and reaches a maximum at 14 to 18 carbon atoms including stearate, palmitate, and myristate. Among flavonoids, flavone, naringenin, apigenin, genistein, quercetin, and daidzein were shown to interact with Bet v 1. The cytokinins N6-(2-isopentenyl)adenine (IPA), kinetin, and zeatin bind to Bet v 1 with lower affinities, whereas the phytohormones indole-3-acetic acid, gibberellic acid, and abscisic acid showed no interaction [11, 23].
Other phytohormones reported to bind Bet v 1 in vitro are the brassinosteroids, brassinolide, and 24-epicastasteron [41]. However, the interaction was analyzed using Bet v 1.0107 (formerly designated Bet v 1l), a hypoallergenic isoform constituting only up to 7% of the total Bet v 1 content in birch pollen [45, 46]. Dehydroergosterol, a frequently used cholesterol model compound, as well as progesterone and the plant sterol stigmasterol were demonstrated to bind Bet v 1 [11, 23]. Recently, phytoprostane E1 (PPE1) and its derivatives B1 (PPB1) and F1 (PPF1) were shown to bind to Bet v 1.0101 with dissociation constants (Kds) of 0.5, 1.0, and 2.4 μM, indicating high binding affinity [6••]. Phytprostanes were detected in relevant quantities in birch pollen and shown to be associated with the induction of immune responses relevant for allergic sensitization [47, 48].
In 2014, Seutter von Loetzen et al. isolated the glycosylated flavonoid Q3OS in complex with Bet v 1 directly from birch pollen extracts. Its binding to this PR-10 protein was confirmed using nuclear magnetic resonance (NMR) spectroscopy and surface acoustic wave (SAW) technology, and is now considered a physiological ligand of Bet v 1 [4, 6••]. Q3OS is a catechol derivative able of binding iron ions [49]. In vitro, the presence of iron strongly affected the affinity of Q3OS to Bet v 1, potentially impacting its allergenicity [50].
Bacterial toll-like receptor (TLR) agonists were often suggested as ligands of Bet v 1 able to modulate its allergenicity [51, 52]. However, this notion has been challenged by a recent study from our laboratory in which very high dissociation constants were measured for the TLR4 agonist lipopolysaccharide (LPS) and the TLR2 agonist lipoteichoic acid (LTA) (199.8 and 185.0 μM, respectively), indicating no physiologically relevant binding [6••].
Besides plant-derived ligands, several artificial model ligands and extrinsic molecules binding Bet v 1 were investigated including the bile acid sodium deoxycholate (DOC), a structural analog of brassinosteroids able to bind Bet v 1 at two distinct sites [6••, 41–43, 53]. Other non-physiological ligands include the detergents sodium dodecyl sulfate (SDS), sodium tetradecyl sulfate (STS), and 3-[Benzyl(dimethyl)ammonio]propane-1-sulfonate (NDSB-256) [6••, 23, 42, 44]. Studies using these surrogate compounds mainly focused on the effects (e.g., fold stability, dynamics, conformation) induced by ligand binding.
Influences on Physicochemical Properties and Proteolytic Processing of Bet v 1
Several ligand binding-induced effects on the physicochemical properties of Bet v 1 were reported with a focus on the influences on the protein’s thermal stability and proteolytic susceptibility [6••, 53]. In general, ligand binding was observed to be associated with rigidification and compactness of the 3D structure of Bet v 1, while the conformational dynamics and flexibility were reduced. No significant alterations regarding the secondary structural element composition due to ligand binding were observed [6••]. Changes in structural dynamics of Bet v 1 affect the accessibility of its proteolytic cleavage sites to lysosomal proteases leading to a reduction in cleavage efficiency and consequently affecting the availability of Bet v 1 peptides for optimal peptide presentation to T cells via the major histocompatibility complex class II (MHC-II) receptor [54, 55]. Antigen presentation facilitated by antigen presenting cells is a necessary aspect for Th2 polarization, a key mechanism mandatory for the development of IgE-mediated allergic immune response. The presentation of allergen-derived peptides via the MHC-II pathway requires the processing of the allergen by lysosomal proteases. The influence of Bet v 1 fold stability on antigen presentation was elucidated by studying several fold stabilized mutants of Bet v 1. In comparison with wildtype Bet v 1, a mutant with optimal proteolytic stability to lysosomal proteases was able to stimulate a pronounced allergy-associated Th2 response in mice, suggesting that fold stability is a contributing factor for allergenicity [56].
In our recent study, describing a higher proteolytic resistance of Bet v 1 due to ligand binding, we have provided a mechanistic explanation for the increased allergenicity of the stabilized Bet v 1 mutant. The study also revealed a newly identified high affinity Bet v 1 ligand known as PPE1, which not only stabilized Bet v 1 from proteolytic degradation by lysosomal proteases but also modulated the proteolytic lysosomal activity of cysteine cathepsins [6••]. This dual-role of the ligand consequently influenced the presentation of the immunodominant Bet v 1 T cell epitope via MHC-II loading. Hence, ligand binding can regulate the processing of Bet v 1 by lysosomal proteases subsequently affecting availability of peptides for MHC-II presentation due to alterations in the frequency of proteolytic processing cleavage sites.
Methods Used to Determine Ligand Binding
The ANS displacement assay (ADA) for assessing the binding of various hydrophobic ligands to Bet v 1 was first described by Mogensen et al. [23]. The assay is based on the ability of the fluorophore 8-anilino-1-naphthalenesulfonic acid (ANS) to interact with PR-10 proteins. ANS behaves as a weak fluorophore in solution, but upon binding to the hydrophobic patches of a protein, its fluorescence intensity dramatically increases [18]. ANS binds to Bet v 1 via its extended hydrophobic surface patches within its prominent cavity [42]. If the ANS binding sites are already occupied with other ligands, this results in a decrease of ANS fluorescence; thus, ligand-induced ANS displacement can be fluorescently monitored. Due to its convenience, ADA is usually the first method of choice to investigate ligand binding of PR-10 proteins, as indicated in Tables 1 and 2. However, the assay has the disadvantage of providing only an indirect measurement that relies on the capacity of ligands to share the same binding sites and to displace ANS. The analysis of the observed fluorescence changes can be complicated by several factors, such as inner filter effects or fluorescence of by the competing ligand. In the first case, a decrease would artificially indicate binding of the tested ligand where the decrease was mostly due to a reabsorption of the ANS fluorescence signal; in the second case, ligand binding could increase the fluorescence signal. Additionally, ligand interaction with ANS and/or Bet v 1 may tune the emission maximum of ANS, e.g., by inducing structural rearrangements which may also result in additional ANS interaction sites in the respective PR-10 molecule. Similarly to ANS, the naturally occurring polyphenol resveratrol was also observed to increase its intrinsic fluorescence upon binding to the hydrophobic patches of Bet v 1 [11].
Alternatives to ADA for the investigation of PR-10 ligand binding include direct measuring techniques, such as isothermal titration calorimetry (ITC), surface acoustic wave (SAW), and microscale thermophoresis (MST), which enable accurate determination of binding affinity and Kd [6••, 9, 18].
NMR and X-ray crystallography also proved to be powerful tools for the identification of structural rearrangements induced by ligand binding compared with the apo-protein, and for information on the binding stoichiometry [4, 6••, 7••, 42]. Both techniques can be used to precisely map the amino acid residues involved in ligand interaction at the respective binding sites. Interestingly, attempts to crystallize some PR-10 proteins without ligand were not successful, consistent with a superordinate role of ligands in protein rigidification and reduction of structural dynamics necessary for successful crystallization [13]. Besides the aforementioned techniques, in silico docking experiments were performed in several studies to evaluate the binding of ligands [15].
Conclusions
Recent studies emphasize that PR-10 proteins’ structural and functional properties cannot be understood by their proteinogenic properties only. A structurally diverse spectrum of hydrophobic ligands can bind selectively to the PR-10 protein family, thereby tuning their physiologic and immunologic functions. Importantly, the organic cargo of PR-10 protein can exhibit secondary functions by addressing distant protein targets. The dual functions of protein ligands are exemplified by the phytoprostane PPE1, which affects protein stability of Bet v 1 and at the same time covalently blocks papain-related cysteine proteases via a Michael reaction warhead.
While the dual proteinogenic and organic composition shapes the physiological functions of PR-10 proteins, it as much accounts for their allergenic properties. The selective binding of ligands provides a rational for hypo- versus hyper-allergenic properties of allergen isoforms, which were previously explained by their differences in amino acid composition and 3d structure. In the light of the new findings, it may become possible to convert an allergen into a hypoallergen by differential ligand loading.
Abbreviations
- ANS
8-Anilinonapthalene-1-sulfonic acid
- CSBP
Cytokinin-specific binding proteins
- DOC
Sodium deoxycholate
- IPR
Intracellular pathogenesis-related proteins
- Kd
Equilibrium dissociation constant
- LPS
Lipopolysaccharide
- LTA
Lipoteichoic acid
- MHC II
Major histocompatibility complex class II
- MLP
Major latex proteins
- IPA
N6-(2-isopentenyl)adenine
- MW
Molecular weight
- NDSB-256
Dimethylbenzylammonium propane sulfonate
- NMR
Nuclear magnetic resonance
- PPA1, PPB1, PPE1, PPF1
Phytoprostane A1, B1, E1, and F1
- PR-10
Pathogenesis-related proteins class 10
- Q3OS
Quercetin 3-O-sophoroside
- SAW
Surface acoustic wave
- SDS
Sodium dodecyl sulfate
- STS
Sodium tetradecyl sulfate
- TLR
Toll-like receptor
Funding Information
Open access funding provided by Paris Lodron University of Salzburg. The work of the authors has been financially supported by the Austrian Science Funds (Projects P27589, P32189, and W1213), by the University of Salzburg priority program “Allergy-Cancer-BioNano Research Centre”, and by the Doctoral School Program “Biomolecules” of the University of Salzburg. Wai Tuck SOH is a recipient of the Kishimoto Foundation Fellowship.
Compliance with Ethical Standards
Conflict of Interest
Dr. Ferreira reports personal fees from HAL Allergy, Swiss Institute of Allergy and Asthma Research, and AllergenOnline, outside the submitted work. The other authors declare no conflicts of interest relevant to this manuscript.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Allergens
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Fernandes H, Michalska K, Sikorski M, Jaskolski M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013;280(5):1169–1199. doi: 10.1111/febs.12114. [DOI] [PubMed] [Google Scholar]
- 2.Flores T, Alape-Giron A, Flores-Diaz M, Flores HE. Ocatin. A novel tuber storage protein from the andean tuber crop oca with antibacterial and antifungal activities. Plant Physiol. 2002;128(4):1291–1302. doi: 10.1104/pp.010541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ukaji N, Kuwabara C, Takezawa D, Arakawa K, Fujikawa S. Accumulation of pathogenesis-related (PR) 10/Bet v 1 protein homologues in mulberry (Morus bombycis Koidz.) tree during winter. Plant Cell Environ. 2004;27(9):1112–1121. doi: 10.1111/j.1365-3040.2004.01216.x. [DOI] [Google Scholar]
- 4.Seutter von Loetzen C, Hoffmann T, Hartl MJ, Schweimer K, Schwab W, Rosch P, et al. Secret of the major birch pollen allergen Bet v 1: identification of the physiological ligand. Biochem J. 2014;457(3):379–390. doi: 10.1042/bj20130413. [DOI] [PubMed] [Google Scholar]
- 5.Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.•• Soh WT, Aglas L, Mueller GA, Gilles S, Weiss R, Scheiblhofer S, et al. Multiple roles of Bet v 1 ligands in allergen stabilization and modulation of endosomal protease activity. 2019;74(12):2382–93 The study showed that pollen-derived phytoprostanes bind to Bet v 1 inhibit lysosomal cysteine cathepsin and affects the allergen processing and presentation to T cells. [DOI] [PMC free article] [PubMed]
- 7.Jacob T, von Loetzen CS, Reuter A, Lacher U, Schiller D, Schobert R, et al. Identification of a natural ligand of the hazel allergen Cor a 1. Sci Rep. 2019;9(1):8714. doi: 10.1038/s41598-019-44999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moraes AH, Asam C, Almeida FCL, Wallner M, Ferreira F, Valente AP. Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1. Sci Rep. 2018;8(1):10512. doi: 10.1038/s41598-018-28358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zubini P, Zambelli B, Musiani F, Ciurli S, Bertolini P, Baraldi E. The RNA hydrolysis and the cytokinin binding activities of PR-10 proteins are differently performed by two isoforms of the Pru p 1 peach major allergen and are possibly functionally related. Plant Physiol. 2009;150(3):1235–1247. doi: 10.1104/pp.109.139543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanal A, Zander U, Munoz C, Dupeux F, Luque I, Botella MA, et al. The strawberry pathogenesis-related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J Biol Chem. 2013;288(49):35322–35332. doi: 10.1074/jbc.M113.501528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride JK, Cheng H, Maleki SJ, Hurlburt BKJF. Purification and characterization of pathogenesis related class 10 panallergens. 2019;8(12):609. [DOI] [PMC free article] [PubMed]
- 12.Koistinen KM, Soininen P, Venalainen TA, Hayrinen J, Laatikainen R, Perakyla M, et al. Birch PR-10c interacts with several biologically important ligands. Phytochemistry. 2005;66(21):2524–2533. doi: 10.1016/j.phytochem.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Ruszkowski M, Szpotkowski K, Sikorski M, Jaskolski M. The landscape of cytokinin binding by a plant nodulin. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 12):2365–2380. doi: 10.1107/s0907444913021975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neudecker P, Schweimer K, Nerkamp J, Scheurer S, Vieths S, Sticht H, Rösch P. Allergic cross-reactivity made visible: solution structure of the major cherry allergen Pru av 1. J Biol Chem. 2001;276(25):22756–22763. doi: 10.1074/jbc.M101657200. [DOI] [PubMed] [Google Scholar]
- 15.Biesiadka J, Bujacz G, Sikorski MM, Jaskolski M. Crystal structures of two homologous pathogenesis-related proteins from yellow lupine. J Mol Biol. 2002;319(5):1223–1234. doi: 10.1016/S0022-2836(02)00385-6. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes H, Bujacz A, Bujacz G, Jelen F, Jasinski M, Kachlicki P, Otlewski J, Sikorski MM, Jaskolski M. Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein. FEBS J. 2009;276(6):1596–1609. doi: 10.1111/j.1742-4658.2009.06892.x. [DOI] [PubMed] [Google Scholar]
- 17.Gonneau M, Pagant S, Brun F, Laloue M. Photoaffinity labelling with the cytokinin agonist azido-CPPU of a 34 kDa peptide of the intracellular pathogenesis-related protein family in the moss Physcomitrella patens. Plant Mol Biol. 2001;46(5):539–548. doi: 10.1023/a:1010693213437. [DOI] [PubMed] [Google Scholar]
- 18.Pasternak O, Bujacz GD, Fujimoto Y, Hashimoto Y, Jelen F, Otlewski J, Sikorski MM, Jaskolski M. Crystal structure of Vigna radiata cytokinin-specific binding protein in complex with zeatin. Plant Cell. 2006;18(10):2622–2634. doi: 10.1105/tpc.105.037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto Y, Nagata R, Fukasawa H, Yano K, Azuma M, Iida A, Sugimoto S, Shudo K, Hashimoto Y. Purification and cDNA cloning of cytokinin-specific binding protein from mung bean (Vigna radiata) Eur J Biochem. 1998;258(2):794–802. doi: 10.1046/j.1432-1327.1998.2580794.x. [DOI] [PubMed] [Google Scholar]
- 20.Ilari A, Franceschini S, Bonamore A, Arenghi F, Botta B, Macone A, Pasquo A, Bellucci L, Boffi A. Structural basis of enzymatic (S)-norcoclaurine biosynthesis. J Biol Chem. 2009;284(2):897–904. doi: 10.1074/jbc.M803738200. [DOI] [PubMed] [Google Scholar]
- 21.Sliwiak J, Dauter Z, Jaskolski M. Crystal structure of Hyp-1, a Hypericum perforatum PR-10 protein, in complex with melatonin. Front Plant Sci. 2016;7:668. doi: 10.3389/fpls.2016.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes H, Pasternak O, Bujacz G, Bujacz A, Sikorski MM, Jaskolski M. Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin. J Mol Biol. 2008;378(5):1040–1051. doi: 10.1016/j.jmb.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Mogensen JE, Wimmer R, Larsen JN, Spangfort MD, Otzen DE. The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J Biol Chem. 2002;277(26):23684–23692. doi: 10.1074/jbc.M202065200. [DOI] [PubMed] [Google Scholar]
- 24.Sliwiak J, Sikorski M, Jaskolski M. PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine. FEBS J. 2018;285(10):1907–1922. doi: 10.1111/febs.14455. [DOI] [PubMed] [Google Scholar]
- 25.Wu F, Yan M, Li Y, Chang S, Song X, Zhou Z, Gong W. cDNA cloning, expression, and mutagenesis of a PR-10 protein SPE-16 from the seeds of Pachyrrhizus erosus. Biochem Biophys Res Commun. 2003;312(3):761–766. doi: 10.1016/j.bbrc.2003.10.181. [DOI] [PubMed] [Google Scholar]
- 26.Bantignies B, Seguin J, Muzac I, Dedaldechamp F, Gulick P, Ibrahim R. Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol Biol. 2000;42(6):871–881. doi: 10.1023/a:1006475303115. [DOI] [PubMed] [Google Scholar]
- 27.Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 2004;37(2):186–198. doi: 10.1046/j.1365-313x.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- 28.Moiseyev GP, Beintema JJ, Fedoreyeva LI, Yakovlev GI. High sequence similarity between a ribonuclease from ginseng calluses and fungus-elicited proteins from parsley indicates that intracellular pathogenesis-related proteins are ribonucleases. Planta. 1994;193(3):470–472. doi: 10.1007/bf00201828. [DOI] [PubMed] [Google Scholar]
- 29.Chadha P, Das RH. A pathogenesis related protein, AhPR10 from peanut: an insight of its mode of antifungal activity. Planta. 2006;225(1):213–222. doi: 10.1007/s00425-006-0344-7. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X-J, Lu S, Xu Y-H, Wang J-W, Chen X-Y. A cotton cDNA (GaPR-10) encoding a pathogenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci. 2002;162(4):629–636. doi: 10.1016/S0168-9452(02)00002-X. [DOI] [Google Scholar]
- 31.Ruszkowski M, Sliwiak J, Ciesielska A, Barciszewski J, Sikorski M, Jaskolski M. Specific binding of gibberellic acid by cytokinin-specific binding proteins: a new aspect of plant hormone-binding proteins with the PR-10 fold. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 7):2032–2041. doi: 10.1107/s1399004714010578. [DOI] [PubMed] [Google Scholar]
- 32.Munoz C, Hoffmann T, Escobar NM, Ludemann F, Botella MA, Valpuesta V, et al. The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol Plant. 2010;3(1):113–124. doi: 10.1093/mp/ssp087. [DOI] [PubMed] [Google Scholar]
- 33.von Loetzen CS, Jacob T, Hartl-Spiegelhauer O, Vogel L, Schiller D, Spörlein-Güttler C, et al. Ligand recognition of the major birch pollen allergen bet v 1 is isoform dependent. 2015;10(6). [DOI] [PMC free article] [PubMed]
- 34.Lebel S, Schellenbaum P, Walter B, Maillot P. Characterisation of the Vitis viniferaPR10 multigene family. BMC Plant Biol. 2010;10(1):184. doi: 10.1186/1471-2229-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blankestijn MA, Knulst AC, Knol EF, Le TM, Rockmann H, Otten HG, et al. Sensitization to PR-10 proteins is indicative of distinctive sensitization patterns in adults with a suspected food allergy. Clin Transl Allergy. 2017;7:42. doi: 10.1186/s13601-017-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biedermann T, Winther L, Till SJ, Panzner P, Knulst A, Valovirta E. Birch pollen allergy in Europe. Allergy. 2019;74(7):1237–1248. doi: 10.1111/all.13758. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Kim SH, Park HW, Cho SH, Chang YS. Oral allergy syndrome in birch pollen-sensitized patients from a Korean University Hospital. J Korean Med Sci. 2018;33(33):e218. doi: 10.3346/jkms.2018.33.e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller WL. Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim Biophys Acta. 2007;1771(6):663–676. doi: 10.1016/j.bbalip.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Agarwal P, Agarwal PK. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol Biol Rep. 2014;41(2):599–611. doi: 10.1007/s11033-013-2897-4. [DOI] [PubMed] [Google Scholar]
- 40.Kundu S, Roy D. Structural study of biologically significant ligands with major birch pollen allergen Betv1 by docking and molecular dynamics simulation. Bioinformation. 2010;4(7):326–330. doi: 10.6026/97320630004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marković-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, et al. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J Mol Biol. 2003;325(1):123–133. doi: 10.1016/S0022-2836(02)01197-X. [DOI] [PubMed] [Google Scholar]
- 42.Kofler S, Asam C, Eckhard U, Wallner M, Ferreira F, Brandstetter H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen bet v 1. J Mol Biol. 2012;422(1):109–123. doi: 10.1016/j.jmb.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nony E, Bouley J, Le Mignon M, Lemoine P, Jain K, Horiot S, et al. Development and evaluation of a sublingual tablet based on recombinant Bet v 1 in birch pollen-allergic patients. Allergy. 2015;70(7):795–804. doi: 10.1111/all.12622. [DOI] [PubMed] [Google Scholar]
- 44.Grutsch S, Fuchs JE, Freier R, Kofler S, Bibi M, Asam C, et al. Ligand binding modulates the structural dynamics and compactness of the major birch pollen allergen. 2014;107(12):2972–81. [DOI] [PMC free article] [PubMed]
- 45.Swoboda I, Jilek A, Ferreira F, Engel E, Hoffmann-Sommergruber K, Scheiner O, et al. Isoforms of Bet v 1, the major birch pollen allergen, analyzed by liquid chromatography, mass spectrometry, and cDNA cloning. J Biol Chem. 1995;270(6):2607–2613. doi: 10.1074/jbc.270.6.2607. [DOI] [PubMed] [Google Scholar]
- 46.Ferreira F, Hirtenlehner K, Jilek A, Godnik-Cvar J, Breiteneder H, Grimm R, et al. Dissection of immunoglobulin E and T lymphocyte reactivity of isoforms of the major birch pollen allergen Bet v 1: potential use of hypoallergenic isoforms for immunotherapy. J Exp Med. 1996;183(2):599–609. doi: 10.1084/jem.183.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilles S, Mariani V, Bryce M, Mueller MJ, Ring J, Jakob T, et al. Pollen-derived E1-phytoprostanes signal via PPAR-γ and NF-κB-dependent mechanisms. J Immunol. 2009;182(11):6653–6658. doi: 10.4049/jimmunol.0802613. [DOI] [PubMed] [Google Scholar]
- 48.Traidl-Hoffmann C, Mariani V, Hochrein H, Karg K, Wagner H, Ring J, et al. Pollen-associated phytoprostanes inhibit dendritic cell interleukin-12 production and augment T helper type 2 cell polarization. J Exp Med. 2005;201(4):627–636. doi: 10.1084/jem.20041065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho EA, Song HK, Lee SH, Chung BH, Lim HM, Lee MK. Differential in vitro and cellular effects of iron chelators for hypoxia inducible factor hydroxylases. J Cell Biochem. 2013;114(4):864–873. doi: 10.1002/jcb.24423. [DOI] [PubMed] [Google Scholar]
- 50.Roth-Walter F, Gomez-Casado C, Pacios LF, Mothes-Luksch N, Roth GA, Singer J, et al. Bet v 1 from birch pollen is a lipocalin-like protein acting as allergen only when devoid of iron by promoting Th2 lymphocytes. J Biol Chem. 2014;289(25):17416–21. [DOI] [PMC free article] [PubMed]
- 51.Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, et al. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. 2018;175(6):1651–64. e14. [DOI] [PMC free article] [PubMed]
- 52.Bublin M, Eiwegger T, Breiteneder H. Do lipids influence the allergic sensitization process? J Allergy Clin Immunol. 2014;134(3):521–529. doi: 10.1016/j.jaci.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asam C, Batista A, Moraes A, de Paula V, Almeida F, Aglas L, et al. Bet v 1–a Trojan horse for small ligands boosting allergic sensitization? 2014;44(8):1083–93. [DOI] [PubMed]
- 54.Freier R, Dall E, Brandstetter H. Protease recognition sites in Bet v 1a are cryptic, explaining its slow processing relevant to its allergenicity. 2015;5(1):1–9. [DOI] [PMC free article] [PubMed]
- 55.Grutsch S, Fuchs JE, Ahammer L, Kamenik AS, Liedl KR, Tollinger M. Conformational flexibility differentiates naturally occurring Bet v 1 isoforms. Int J Mol Sci. 2017;18(6). 10.3390/ijms18061192. [DOI] [PMC free article] [PubMed]
- 56.Machado Y, Freier R, Scheiblhofer S, Thalhamer T, Mayr M, Briza P, et al. Fold stability during endolysosomal acidification is a key factor for allergenicity and immunogenicity of the major birch pollen allergen. 2016;137(5):1525–34. [DOI] [PMC free article] [PubMed]