RAD23 is able to repair yeast DNA lesions through nucleotide excision in full darkness, a mechanism distinct from photolyase-dependent photorepair of UV-induced DNA damage but functionally unknown in filamentous fungi. Our study unveils that the RAD23 ortholog in a filamentous fungal insect pathogen varies in subcellular localization according to external cues, interacts with a photolyase required for photorepair of cytotoxic (6-4)-pyrimidine-pyrimidine photoproducts in UV-induced DNA lesions, and plays an essential role in conidial UVB resistance and reactivation of UVB-inactivated conidia under visible light rather than in the dark, as required for nucleotide excision repair. Loss-of-function mutations of RAD23 exert pleiotropic effects on radial growth, aerial conidiation, multiple stress responses, virulence, virulence-related cellular events, and phenotype-related gene expression. These findings highlight a novel mechanism underlying the photoreactivation of UVB-impaired fungal cells by RAD23 interacting with the photolyase, as well as its essentiality for filamentous fungal life.
KEYWORDS: biological control, DNA damage repair, UV damage, entomopathogenic fungi, nucleotide excision repair protein, photolyase, photoreactivation
ABSTRACT
RAD23 can repair yeast DNA lesions through nucleotide excision repair (NER), a mechanism that is dependent on proteasome activity and ubiquitin chains but different from photolyase-depending photorepair of UV-induced DNA damages. However, this accessory NER protein remains functionally unknown in filamentous fungi. In this study, orthologous RAD23 in Beauveria bassiana, an insect-pathogenic fungus that is a main source of fungal insecticides, was found to interact with the photolyase PHR2, enabling repair of DNA lesions by degradation of UVB-induced cytotoxic (6-4)-pyrimidine-pyrimidine photoproducts under visible light, and it hence plays an essential role in the photoreactivation of UVB-inactivated conidia but no role in reactivation of such conidia through NER in dark conditions. Fluorescence-labeled RAD23 was shown to normally localize in the cytoplasm, to migrate to vacuoles in the absence of carbon, nitrogen, or both, and to enter nuclei under various stresses, which include UVB, a harmful wavelength of sunlight. Deletion of the rad23 gene resulted in an 84% decrease in conidial UVB resistance, a 95% reduction in photoreactivation rate of UVB-inactivated conidia, and a drastic repression of phr2. A yeast two-hybrid assay revealed a positive RAD23-PHR2 interaction. Overexpression of phr2 in the Δrad23 mutant largely mitigated the severe defect of the Δrad23 mutant in photoreactivation. Also, the deletion mutant was severely compromised in radial growth, conidiation, conidial quality, virulence, multiple stress tolerance, and transcriptional expression of many phenotype-related genes. These findings unveil not only the pleiotropic effects of RAD23 in B. bassiana but also a novel RAD23-PHR2 interaction that is essential for the photoprotection of filamentous fungal cells from UVB damage.
IMPORTANCE RAD23 is able to repair yeast DNA lesions through nucleotide excision in full darkness, a mechanism distinct from photolyase-dependent photorepair of UV-induced DNA damage but functionally unknown in filamentous fungi. Our study unveils that the RAD23 ortholog in a filamentous fungal insect pathogen varies in subcellular localization according to external cues, interacts with a photolyase required for photorepair of cytotoxic (6-4)-pyrimidine-pyrimidine photoproducts in UV-induced DNA lesions, and plays an essential role in conidial UVB resistance and reactivation of UVB-inactivated conidia under visible light rather than in the dark, as required for nucleotide excision repair. Loss-of-function mutations of RAD23 exert pleiotropic effects on radial growth, aerial conidiation, multiple stress responses, virulence, virulence-related cellular events, and phenotype-related gene expression. These findings highlight a novel mechanism underlying the photoreactivation of UVB-impaired fungal cells by RAD23 interacting with the photolyase, as well as its essentiality for filamentous fungal life.
INTRODUCTION
Formulated fungal cells, such as conidia, serve as active ingredients of fungal insecticides and acaricides and are highly sensitive to solar UV irradiation, which comprises UVB (290 to 320 nm) and UVA (320 to 400 nm) wavelengths (1) and restrains wide application of fungal formulations for arthropod pest control (2–4). Fungal cells exposed to UV irradiation may suffer damage to macromolecules, including RNA, DNA, proteins, ribosomes, and biomembranes (5, 6). Therefore, understanding molecular mechanisms involved in fungal UV resistance is of special importance for development and application of fungal pesticides based on fungal insect pathogens.
Nucleotide excision repair (NER) and photoreactivation are major mechanisms involved in the protection of eukaryotic cells from UV damage that occurs in the DNA duplex-forming covalent linkages between adjacent bases for generation of cytotoxic cyclobutane pyrimidine dimers (CPDs) and (6-4)-pyrimidine-pyrimidine photoproducts (6-4PPs) under UV irradiation (7). Such DNA lesions often result in growth defects, gene mutations, and even cell death (8, 9). Many proteins are involved in NER that repair DNA lesions. In Saccharomyces cerevisiae, RAD23 is an accessory NER protein that functions not only in both transcription-coupled and global genomic NER activities (10) but also in DNA damage repair (11). This protein features an N-terminal ubiquitin-like (UBI) domain (namely, the RAD4 interaction domain) and two ubiquitin-associated (UBA) domains separated by a heat shock chaperonin-binding (HSCB) region (12, 13). The UBI domain is required for an interaction of the yeast RAD23 with the 26S proteasome (14, 15), a large protein complex that consists of a 20S core particle and two copies of a 19S regulatory complex and which is involved in the degradation of the ubiquitin pathway-targeted proteins (16, 17) and triggers intracellular proteasome activity and protein degradation through ubiquitin chains (18). The phosphorylation of the UBI domain can regulate the interaction of RAD23 with the proteasome (19). Despite no role in NER activity, the two UBA domains bind ubiquitin for induced inhibition of multiubiquitin chain assembly or prevent RAD23 from proteasomal degradation (20, 21). In the yeast, RAD23 can escape degradation due to a lack of proteasome initiation region (22), and it plays a main role in the NER by direct participation in both the repair biochemistry and stabilization of RAD4, a protein that mediates impaired DNA binding and recognition (23–25). Aside from RAD4, RAD23 interacts with many proteins involved in the proteasome activity and ubiquitin chains, including other Rad and related partners, the CDC48-RAD23/DSK2 axis involved in K48-linked chain specificity of the proteasome (26), and the deubiquitylating enzyme UBP12 that regulates RAD23-dependent proteasomal degradation (27). These intensive studies demonstrate a core role of the yeast RAD23 in multiple cellular processes and events. However, little is known about involvement of the yeast RAD23 in photorepair of DNA lesions.
Putative RAD23 homologs exist widely in filamentous fungi, but none of them have been characterized, leaving it unknown whether such homologs function in cell protection and other cellular events. Beauveria bassiana is a filamentous fungal insect pathogen that serves as a main source of fungal insecticides (28) and has evolved complicated machineries functioning in response and tolerance to all possible types of stresses associated with host immunity defenses and host habitats (3, 4, 29). Two photolyases (PHR1 and PHR2) of B. bassiana have been found to repair UVB-induced DNA lesions by decomposing CPDs and 6-4PPs under visible light and hence to photoreactivate fungal conidia inactivated by a lethal dose of UVB irradiation (30). Overexpression of either the phr1 or the phr2 gene in B. bassiana to a large degree enhances both conidial tolerance to UVB irradiation, a very harmful component in sunlight, and efficiency in photoreactivation of UVB-inactivated conidia (30). While previous study has shed light upon the high potential of both photolyase genes for improved UVB resistance and application strategy of fungal insecticides, no effort has been made to explore the possible roles of many NER proteins in UV resistance and other cellular events of fungal insect pathogens. We speculate that RAD23 orthologs may have unrecognized roles in association with biological control potential of fungal insect pathogens or important cellular processes/events of other filamentous fungi because they share a UBI domain, which enables stimulation of intracellular proteasome activity and protein degradation through ubiquitin chains (18). This study seeks to test the hypothesis by characterization of orthologous RAD23 in B. bassiana. As presented below, the B. bassiana RAD23, which features the same properties as the yeast RAD23, was confirmed to interact with PHR2 and hence to play a crucial role not only in the photoprotection of conidia from UVB damage but also in growth, conidiation, virulence and virulence-related cellular events, and transcriptional activation of many genes involved in asexual development and in multiple stress responses.
RESULTS
Structural, transcriptional, and subcellular properties of RAD23 in B. bassiana.
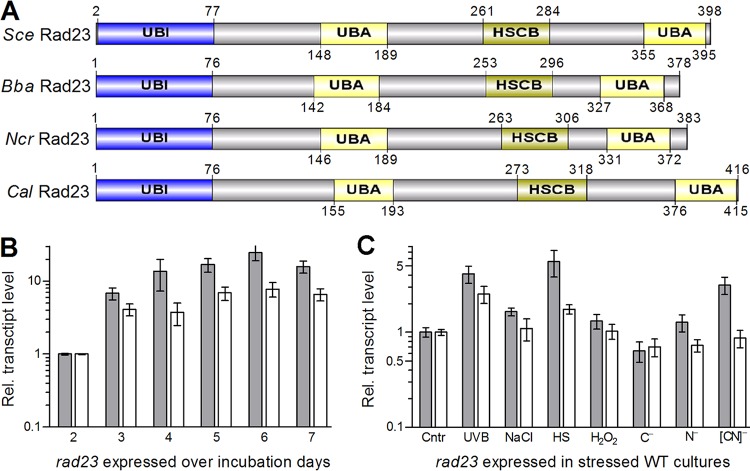
The unique RAD23 (GenPept accession number EJP70161) encoded in the B. bassiana genome (31) consists of 378 amino acids (molecular mass, 39.9 kDa; isoelectric point, 4.48) and features an N-terminal UBI domain (residues 1 to 76) and two UBA domains (residues 142 to 184 and 327 to 368) separated by an HSCB domain at residues 253 to 296 (Fig. 1A). The four typical domains are also present at similar sites of the RAD23 orthologs found in S. cerevisiae, Candida albicans, and Neurospora crassa. The B. bassiana RAD23 shares a higher sequence identity with the counterparts of 13 other filamentous fungi (50 to 84%) than with those of three yeasts or yeast-like fungi (33 to 34%) (Fig. S1).
FIG 1.
Sequence features and transcriptional profile of RAD23 in B. bassiana. (A) Conserved domains of RAD23 homologs found in S. cerevisiae (Sce), B. bassiana (Bba), Neurospora crassa (Ncr), and Candida albicans (Cal). (B, C) Relative transcript (RT) levels of rad23 in the normal WT cultures during a 7-day incubation with respect to the standard on day 2 and in the stressed WT cultures with respect to the nonstressed standard (control [Cntr]). The used cDNA samples were derived from the hyphal cultures incubated for 12 h in CDB free of carbon sources (C−), nitrogen sources (N−), or both ([CN]−), respectively, after collection from 48-h-old SDBY cultures, or triggered for 90 min with 40 mM H2O2, 0.4 M NaCl, and 42°C heat shock (HS) in 1/4 SDBY and exposed to UVB irradiation at a dose of 1 J · cm−2, respectively, after collection from 60-h-old SDBY cultures. The genes tef1 (gray bars) and act1 (white bars) were used as references. Error bars indicate standard deviations (SD) of the mean from three cDNA samples analyzed via quantitative PCR (qPCR).
Two reference genes were used to assess the transcript levels of rad23 in the wild-type strain B. bassiana ARSEF 2860 (here designated WT) during a 7-day incubation on Sabouraud dextrose agar plus yeast extract (SDAY) under the optimal regime of 25°C in a light/dark (L:D) cycle of 12:12 h or in hyphal cultures of the WT exposed to different types of stress cues. The expression of rad23 was upregulated during the normal incubation with respect to a standard at the end of a 2-day incubation (Fig. 1B) and was also upregulated in response to some stress cues, particularly heat shock and UVB irradiation (Fig. 1C). However, the use of the reference gene tef1, which encodes translation elongation factor 1 alpha (30), tended to generate higher transcript levels of rad23 than those generated by the use of the β-actin gene as a reference.
Translocation of RAD23 in response to stress cues.
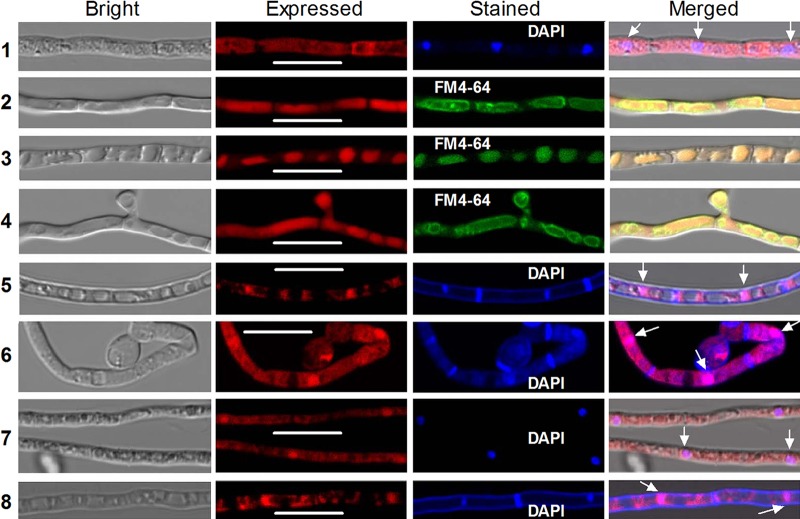
The optimized tef1 promoter Ptef1 was used to drive expression of the red fluorescent protein gene mCherry-tagged rad23 fusion gene in the WT strain. A transgenic strain showing the desired fluorescence signal was incubated for 60 h in SDBY (i.e., agar-free SDAY) at 25°C. In the resultant hyphae stained with the nucleus-specific dye 4′,6′-diamidine-2′-phenylindole dihydrochloride (DAPI) (shown in blue), the expressed fusion protein accumulated mainly in cytoplasm but also very weakly in the nucleus, as shown in laser scanning confocal microscopic (LSCM) images (Fig. 2, row 1). Translocation of the fusion protein occurred in the hyphae exposed to various stress cues for 12 h in a minimal broth. In the hyphae stained with the membrane-specific dye FM4-64 (shown in green), the fusion protein partially migrated to the vacuoles from the cytoplasm in the absence of carbon, completely entered the vacuoles in the absence of nitrogen, and partially moved out of the vacuoles under conditions of both carbon and nitrogen starvation (Fig. 2, rows 2 to 4). Its vacuolar localization under the nitrogen starvation condition was shown by disappearance of both the merged color-defined vacuolar membrane and the expressed color in the cytoplasm. Moreover, the fusion protein accumulated heavily in the nuclei of the hyphae that were exposed for 90 min to oxidative stress at 40 mM H2O2, to osmotic stress at 0.4 M NaCl, to heat shock at 42°C, or to UVB irradiation at 1 J · cm−2 (Fig. 2, rows 5 to 8). Stress-induced nuclear localization is shown by merging the colors of both expressed and DAPI-stained signals. All of these LSCM images indicate that RAD23 localizes mainly in the cytoplasm under normal conditions and can migrate to vacuoles or nuclei under carbon/nitrogen-deficient conditions or external stresses. Thus, conditional translocation of RAD23 in the hyphae suggests its involvement in cytoplasmic, vacuolar, and nuclear events of B. bassiana.
FIG 2.
LSCM images (bars, 10 μm) for translocation of RAD23::mCherry fusion protein (shown in red) in B. bassiana hyphae exposed to stress cues. The observed hyphae were stained with DAPI (shown in blue in rows 1 and 5 to 8) or FM4-64 (shown in green in rows 2 to 4). Row 1: a hypha taken from 60-h-old SDBY culture at 25°C (control). Rows 2 to 4: hyphae incubated for 12 h in CDB free of carbon sources, nitrogen sources, or both, respectively, after collection from 48-h-old SDBY culture. Rows 5 to 7: hyphae triggered for 90 min with 40 mM H2O2, 0.4 M NaCl, and 42°C heat shock in 1/4 SDBY, respectively, after collection from 60-h-old SDBY culture. Row 8: a hypha exposed to UVB irradiation at a dose of 1 J · cm−2 after collection from 60-h-old SDBY culture. Note that the fusion protein localizes mainly in cytoplasm under normal conditions (control), partially or completely migrates to vacuoles under carbon or nitrogen starvation condition, and accumulates heavily in nuclei (see arrows) in response to stress cues.
RAD23 protects B. bassiana from UVB damage by its interaction with PHR2.
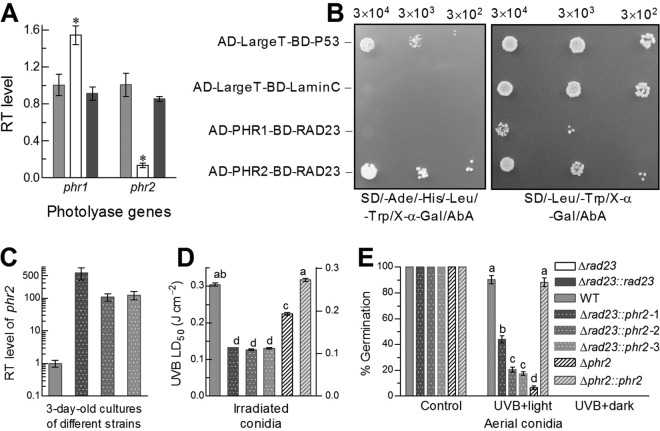
To explore a possible role for RAD23 in B. bassiana, deletion and complementation mutants of rad23 were created based on the WT strain. As a result of the rad23 deletion, the median lethal dose (LD50) indicative of conidial resistance to UVB irradiation decreased by 84% in comparison to the LD50 of 0.31 J · cm−2 estimated from the WT strain (Fig. 3A). Three-hour exposure of UVB-inactivated conidia (irradiated at the lethal dose of 0.5 J · cm−2) to white (visible) light and subsequent 21-h incubation at 25°C in dark resulted in 82% germination for the WT strain but only 4.5% for the Δrad23 mutant (Fig. 3B and C), indicating a 95% loss of photoreactivation capability in the absence of rad23. In contrast, the NER treatment of 24-h dark incubation immediately after the UVB irradiation led to no germination of the inactivated conidia for all tested strains. These changes were restored in the complemented strain, indicating that RAD23 played no role in NER but an essential role in photoprotection of B. bassiana from UVB damage.
FIG 3.
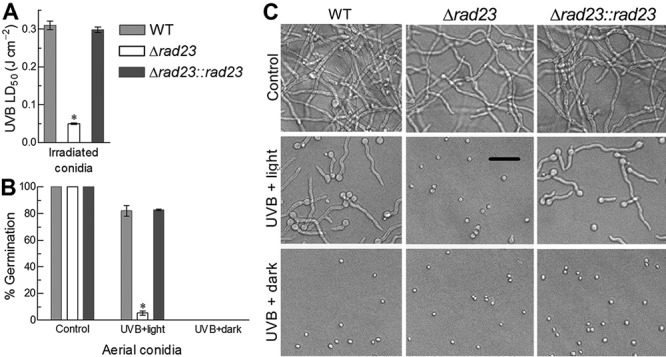
RAD23 protects B. bassiana from UVB damage. (A) LD50 (J · cm−2) estimates indicating conidial UVB resistances of different strains. (B, C) Germination percentages and microscopic images (bar, 20 μm) of the conidia incubated at 25°C for 3 h under white light and for 21 h in full darkness (UVB plus light as photoreactivation treatment) or in the dark for 24 h (UVB plus dark as NER treatment) right after being irradiated at the lethal UVB dose of 0.5 J · cm−2. The conidia not irradiated were incubated for 24 h at 25°C in the dark and used as a control. The asterisked bar in each bar group differs significantly from those that are unmarked (Tukey’s HSD, P < 0.05). Error bars indicate standard deviations from three replicates.
Transcript levels of the photolyase genes phr1 and phr2, which are essential for DNA damage photorepair of UVB-irradiated fungal cells (30), were quantified through real-time quantitative PCR (qPCR) analysis of cDNA samples derived from the 3-day-old cultures of the Δrad23 mutant and control (complemented and WT) strains, which were grown on cellophane-overlaid SDAY plates under the optimal regime. In the Δrad23 mutant, phr2 was downregulated 6.7-fold, contrasting with a 0.5-fold upregulation of phr1, in comparison to similar levels in the control strains (Fig. 4A). Furthermore, a yeast two-hybrid system was used to explore the possible interaction of RAD23 with the CPD photolyase PHR1 or with the 6-4PP photolyase PHR2 based on cell growth on a synthetic defined medium (SD). All types of constructed yeast cells were able to grow on double-dropout SD lacking Leu and Trp (SD/−Leu/−Trp/), as presented in Fig. 4B. In contrast, only the cell type expressing AD-PHR2-BD-RAD23 grew as well as the positive control (AD-LargeT-BD-P53) on quadruple-dropout SD lacking Ade, His, Leu, and Trp (SD/−Ade/−His/−Leu/−Trp), which abolished the growth of those cells expressing AD-PHR1-BD-RAD23 or AD-LargeT-BD-LaminC (negative control). These data indicated a positive interaction of RAD23 with PHR2 rather than with PHR1.
FIG 4.
Photoreactivation of UVB-inactivated conidia by RAD23 interacting with PHR2. (A) Relative transcript (RT) levels of phr1 and phr2 in the 3-day-old SDAY cultures of rad23 mutants versus the WT. (B) Yeast two-hybrid assay for an interaction of RAD23 with PHR1 or PHR2. Note that only the yeast cell type expressing AD-PHR2-BD-RAD23 grew as well as the positive control (AD-LargeT-BD-P53) on quadruple-dropout synthetic defined medium (SD) (left), whereas all four cell types were able to grow on double-dropout SD (right). (C) RT levels of phr2 in the 3-day-old SDAY cultures of three Δrad23::phr2 strains versus the WT. (D) LD50 estimates for conidial UVB resistances of three Δrad23::phr2 strains, WT, and phr2 mutants. (E) Photoreactivated (germinated) percentages of UVB-inactivated conidia. The asterisked bar in each bar group (in panel A) differs significantly from those that are unmarked, and different lowercase letters (in panels D and E) also denote significant differences (Tukey’s HSD, P < 0.05). Error bars indicate standard deviations from three replicates.
Next, we constructed three strains overexpressing phr2 by 110- to 597-fold in the Δrad23 mutant (Fig. 4C) and assayed their UVB resistances and photoreactivation efficiencies in parallel with previous phr2 mutants (30). As a consequence, the extremely severe defect of the Δrad23 mutant in conidial UVB resistance (84% reduction in Fig. 3A) was mitigated by ∼30% in the Δrad23::phr2 strains, which were 30% less tolerant to UVB than the Δphr2 mutant (Fig. 4D). Moreover, the UVB-inactivated conidia of the strains overexpressing phr2 were photoreactivated by 44, 21, and 18% after a 3-h light plus 21-h dark incubation, and the strain expressing phr2 best in the Δrad23 mutant displayed a photoreactivation efficiency secondary to that of the WT strain (Fig. 4E). In contrast, the photoreactivation efficiency was lowered to only 6.7% in the Δphr2 mutant and to 4.5% in the Δrad23 mutant. Again, the UVB-inactivated conidia of all tested strains were unable to be reactivated after a 24-h incubation for NER in the dark.
All of these results indicated an essential role for RAD23 in photoprotection of B. bassiana from UVB damage but no significant role in the NER of UVB-induced DNA lesions. The essential role of RAD23 relied upon its interaction with PHR2, which enabled repair of 6-4PP DNA lesions for partial reactivation of UVB-inactivated conidia under visible light (30).
RAD23 has an important role in hyphal growth, conidiation, and conidial quality.
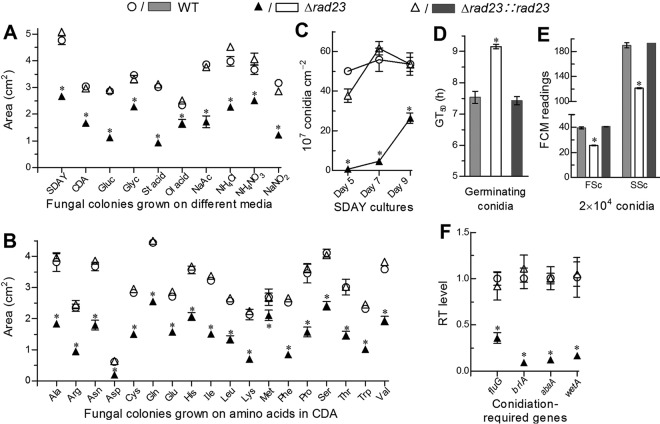
The Δrad23 mutant and its control strains were grown on SDAY, Czapek-Dox agar (CDA), and CDA amended with different carbon or nitrogen (inorganic/organic) sources by spotting 1 μl of a 106 conidia · ml−1 suspension per plate for culture initiation. After an 8-day incubation at the optimal regime, the deletion mutant showed marked growth defects on rich SDAY, minimal CDA, and modified CDAs in comparison to the control strains (Fig. 5A and B). Area reductions of the Δrad23 colonies fell in the range of 40 to 70% on most of the 38 media tested and decreased to 22 to 34% only on the carbon sources of glucose or oleic acid and on the nitrogen sources of NH4NO3 or methionine. Such growth defects demonstrate the involvement of RAD23 in carbon/nitrogen metabolism and nutritional utilization.
FIG 5.
Impacts of rad23 deletion on radial growth, aerial conidiation, and conidial properties. (A, B) Colony sizes of the Δrad23 mutant and control strains grown for 8 days under the optimal regime of 25°C and L:D 12:12 on rich SDAY, minimal CDA, and CDA amended with different carbon sources (3% glucose, glycerol, stearic acid, oleic acid, or sodium acetate) or nitrogen sources (0.3% NH4Cl, NaNO2, NH4NO3, or one of 18 amino acids). Each colony was initiated by spreading 1 μl of a 106 conidia · ml−1 suspension per plate. (C) Conidial yields quantified from the SDAY cultures during a 9-day incubation at the optimal regime. (D) Time (h) for 50% conidial germination (GT50) at 25°C. (E) Conidial size and complexity are indicated by the FSC and SSC readings, respectively, from flow cytometry (FCM) of three samples (2 × 104 conidia per sample). (F) Relative transcript (RT) levels of four genes required for conidiation and conidial maturation. The asterisk marked on a given phenotype of the Δrad23 mutant denotes a significant difference from the same phenotype of two unmarked control strains (Tukey’s HSD, P < 0.05). Error bars indicate standard deviations from three replicates.
The conidiation capacity of each strain was quantified during 9-day incubation period under the optimal regime on SDAY plates, which were spread with 100-μl aliquots of a 107 conidia · ml−1 suspension for culture initiation. Conidial yields quantified from the Δrad23 cultures on days 5, 7, and 9 were reduced by 99, 92, and 51%, respectively, in comparison to those from the WT cultures, which reached a peak yield (∼5.5 × 108 conidia · cm−2 culture) on day 7 (Fig. 5C). The Δrad23 mutant also showed impaired conidial quality. Aside from the drastically reduced UVB resistance as aforementioned, the impaired quality presented as a significant delay in conidial germination at an optimal temperature of 25°C (Fig. 5D) and marked reductions in conidial size and density (complexity) indicated by the respective readings from the forward scatter (FSC) and side scatter (SSC) detectors in the flow cytometry (FCM) of 2 × 104 conidia per sample (Fig. 5E). Revealed by qPCR analysis, three developmental activator genes (brlA, abaA, and wetA) required for conidiation and conidial maturation in B. bassiana (32, 33) were downregulated by 91, 88, and 84% in the 3-day-old SDAY cultures of the Δrad23 mutant grown under the optimal regime, which was followed by transcriptional repression of the upstream transcription factor gene fluG by 65% (Fig. 5F). These data demonstrated that RAD23 was involved in transcriptional activation of the key developmental activator genes and hence played an important role in sustaining conidiation capacity and conidial quality.
RAD23 functions in cellular response to stress cues.
Compared to the control strains, the Δrad23 mutant became significantly (15 to 34%) more sensitive to the stresses of DNA synthesis inhibitor (hydroxyurea), two oxidants (menadione and H2O2), and two hyperosmotic agents (sorbitol and NaCl) despite a null response to topoisomerase I inhibitor (camptothecin) and to two cell wall-perturbing agents (Congo red and calcofluor white) during 8 days of colony growth on CDA at 25°C (Fig. 6A). Additionally, a heat shock of 6 or 9 h at 42°C during the normal incubation at 25°C also suppressed the colony growth recovery of the Δrad23 mutant significantly more than that of the control strains (Fig. 6B).
FIG 6.
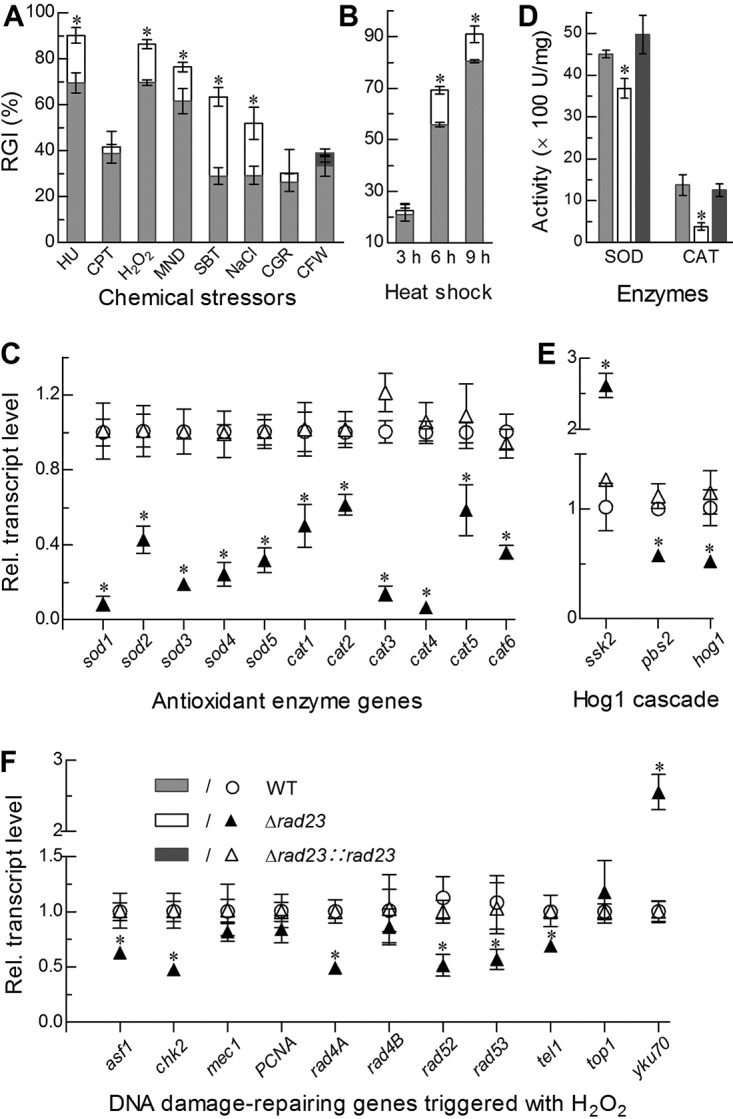
Imapcts of rad23 deletion on cell sensitivities to stress cues and expression levels of stress-responsive enzyme genes. (A, B) Relative growth inhibition (RGI) of fungal colonies by hydroxyurea (HU; 10 mM), camptothecin (CPT; 1 μM camptothecin), H2O2 (2 mM), menadione (MND; 0.02 mM), NaCl (0.4 M), sorbitol (SBT; 1 M), Congo red (CGR; 3 μg · ml−1) or calcofluor white (CFW; 5 μg · ml−1) after an 8-day incubation on CDA at 25°C and by the time of heat shock at 42°C during 8 days of growth on SDAY at 25°C, respectively. (C, D) Relative transcript (RT) levels of all SOD and CAT genes in the 3-day-old SDAY cultures of rad23 mutants versus the WT, and total SOD and CAT activities quantified from the protein extracts of the same cultures. (E) RT levels of hog1 and two upstream genes in the 3-day-old SDAY cultures of rad23 mutants versus the WT. (F) RT levels of DNA damage-repairing genes in the 3-day-old cultures of rad23 mutants versus the WT cocultivated with 2 mM H2O2 on SDAY at 25°C. The asterisk marked on a given phenotype of the Δrad23 mutant denotes a significant difference from the same phenotype of two unmarked control strains (Tukey’s HSD, P < 0.05). Error bars indicate standard deviations from three replicates.
In the Δrad23 mutant, increased sensitivity to oxidative stress correlated well with not only decreased (39 to 91%) transcript levels of all superoxide dismutase (SOD) and catalase (CAT) genes (Fig. 6C) essential or nonessential for antioxidant responses (3) but also reduced SOD (18%) and CAT (72%) activities (Fig. 6D). The increased sensitivity of the Δrad23 mutant to high osmolarity also concurred with differential expression of three genes (Fig. 6E) functioning in the high osmolarity-glycerol (HOG) pathway of B. bassiana (34). For insight into the increased sensitivity of the Δrad23 mutant to the DNA synthesis inhibitor, we assessed the transcript levels of 11 DNA damage repair-related genes in the hyphal cultures of each strain stressed with H2O2. Of those, six genes were suppressed differentially in the Δrad23 mutant versus the WT, including rad4A, rad52, and rad53 (suppressed by 51 to 61%), which are in the same family (Fig. 6F).
All of the above changes in the Δrad23 mutant were restored by targeted rad23 complementation. The results indicated an active role for RAD23 in sustaining cell tolerance to oxidants, DNA synthesis inhibitors, osmotic agents, and heat shock, likely due to its involvement in transcription of stress-responsive genes in B. bassiana.
Importance of RAD23 for fungal virulence and virulence-related dimorphic transition.
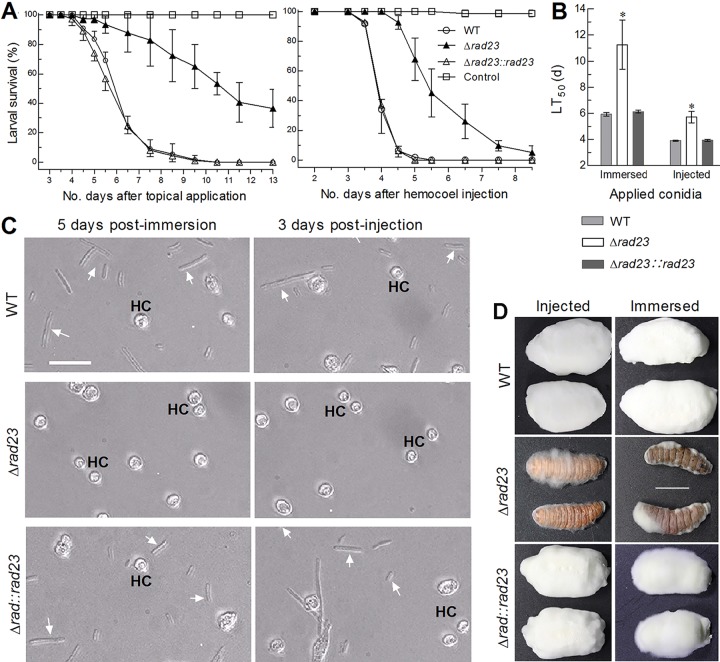
In standardized bioassays, topical application (immersion) of a 107 conidia · ml−1 suspension for normal cuticle infection and intrahemocoel injection of ∼500 conidia per larva for cuticle-bypassing infection resulted in mean (± standard deviation) median lethal time (LT50) estimates of 6.0 (± 0.16) and 3.9 (± 0.06) days, respectively, for the control strains against the larvae of the greater wax moth Galleria mellonella (Fig. 7A and B). In contrast, the mean LT50 for the Δrad23 mutant against the model insect prolonged to 11.3 (± 1.88) days in the normal infection and 5.7 (± 0.43) days in the cuticle-bypassing infection. These LT50 estimates indicated that median lethal actions of the Δrad23 mutant in the two infection modes were delayed by 90 and 46%, respectively, and implied possible blockage of certain infection- and virulence-related cellular events.
FIG 7.
Impact of rad23 deletion on the virulence of B. bassiana. (A, B) Survival trends and LT50 values of G. mellonella larvae inoculated by immersion in a 107 conidia · ml−1 suspension for normal cuticle infection and intrahemocoel injection of ∼500 conidia per larva for cuticle-bypassing infection. The asterisked bar in each bar group differs significantly from those that are unmarked (Tukey’s HSD, P < 0.05). Error bars indicate standard deviations from three replicates. (C) Microscopic images (bar, 10 μm) for hyphal bodies (arrows) and host hemocytes (HC) appearing in the hemolymph samples taken from the larvae surviving 5 days after immersion and 3 days after injection. (D) Images (bar, 10 mm) of fungal outgrowths on surfaces of insect carcasses 6 days postdeath.
Microscopic examination of hemolymph samples taken from the surviving larvae at the end of day 5 postimmersion or of day 3 postinjection revealed abundant hyphal bodies (i.e., blastospores) of the control strains formed in the insect hemolymph (Fig. 7C). However, such hyphal bodies were rare in the samples from the larvae infected by the Δrad23 mutant in either mode, implicating blocked formation of the hyphal bodies, which favor intrahemocoel fungal propagation by yeast-like budding and host mummification to death (35–38). At or near the time of host death, intrahemocoel hyphal bodies must turn back into normal hyphae to penetrate again through the insect cuticle for outgrowth and conidiation on carcasses. The larvae mummified by the control strains were covered with a heavy layer of fungal outgrowth after 7 days of maintenance under optimal conditions, contrasting with very poor growth of the Δrad23 mutant on carcass surfaces, most of which were bald (Fig. 7D). These observations implied that the greatly attenuated virulence of the deletion mutant through the normal infection could be attributable to blockage of both hyphal penetration through the insect cuticle for entry into the host hemocoel and a subsequent dimorphic (hypha-blastospore) transition essential for acceleration of host mummification.
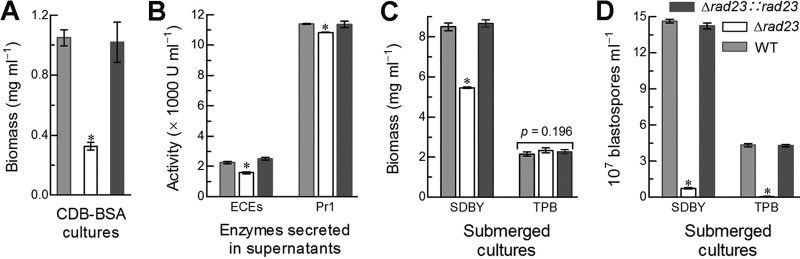
The above speculation was examined by assessing the activities of extracellular (proteolytic, chitinolytic, and lipolytic) enzymes (ECEs) and Pr1 proteases likely involved in cuticle degradation and host infection (39) and the rates of dimorphic transition in vitro in the submerged cultures grown in SDBY and trehalose-peptone broth (TPB) mimicking insect hemolymph. Three-day shaking incubation of a 106 conidia · ml−1 suspension in the CDB (i.e., agar-free CDA) containing the sole nitrogen source of 0.3% bovine serum albumin (BSA) as an inducer of enzyme production resulted in a greater reduction of biomass level (Fig. 8A) than of total ECE or Pr1 activity (Fig. 8B) in the Δrad23 cultures, hinting at a main effect of the rad23 deletion on submerged hyphal growth. In another experiment, the Δrad23 mutant showed a 36% biomass reduction in the 3-day-old SDBY cultures initiated with a 106 conidia · ml−1 suspension and an insignificant change in biomass level of its TPB cultures in comparison to the measurements from the control strains (Fig. 8C). In contrast, blastospore concentrations in the two submerged cultures of the Δrad23 mutant decreased drastically by 95 and 99% (Fig. 8D), and the reduced blastospore yields were equivalent to 92 and 98.6% reductions in dimorphic transition rate per milligram biomass of SDBY and TPB cultures, respectively. Apparently, nearly abolished blastospore formation of the Δrad23 mutant in the TPB cultures correlated with the blocked development of hyphal bodies in the hemolymph samples of the injected larvae.
FIG 8.
Impacts of rad23 deletion on host infection- and virulence-related cellular events. (A, B) Biomass levels and total activities of extracellular enzymes (ECEs) and Pr1 proteases quantified from the 3-day-old cultures of a 106 conidia · ml−1 CDB-BSA. (C, D) Biomass levels and blastospore concentration in the 3-day-old cultures of a 106 conidia · ml−1 SDBY or TPB mimicking insect hemolymph. The asterisked bar in each bar group differs significantly from those that are unmarked (Tukey’s HSD, P < 0.05). Error bars indicate standard deviations from three replicates.
Taken together, the blocked dimorphic transition in vitro and in vivo of the deletion mutant was a main cause of its attenuated virulence through the injection. The much more attenuated virulence of the Δrad23 mutant through normal cuticle infection was likely due to severe defects of both cuticle penetration-required hyphal growth on oligotrophic insect integument close to the tested scant media and dimorphic transition in vivo after entry into hemocoel. Therefore, the mutant virulence was compromised via the normal infection much more than via the cuticle-bypassing infection.
DISCUSSION
In B. bassiana, RAD23 was proven to localize mainly in the cytoplasm under normal culture conditions, to partially or completely migrate to the vacuole under conditions of carbon and/or nitrogen starvation, and to move into the nucleus under oxidative, osmotic, thermal, or UVB-irradiative stress. The conditional nuclear localization of RAD23 is in agreement with a localization of its yeast ortholog in the nucleus, where it functions and interacts with protein partners (40, 41), suggesting a conserved role for RAD23 in the nucleus of both yeast and filamentous fungi. The stress-induced nuclear localization of RAD23 supports its interaction with PHR2, which localizes exclusively in the nucleus like PHR1 in B. bassiana (30), and also implicates its involvement in the fungal transcription-coupled and genomic activities as seen in a model yeast (10). Such a possible involvement helps to understand transcriptional repression of development-required and stress-responsive genes examined in this study. Our finding on the migration of RAD23 to the vacuoles under carbon- or nitrogen-deficient conditions unveils its special role in filamentous fungal adaptation to host and environment because fungal vacuoles are acidic organelles where many cellular events take place, including vacuolar protein sorting, autophagy, cellular homeostasis, signaling, nutrition transport, and stress responses (42, 43). Overall, our experimental data demonstrate not only an important role for RAD23 in the maintenance of multiple phenotypes associated with fungal potential against arthropod pests and expression of phenotype-related genes but also an essential role in protecting conidia from UVB damage via photoreactivation rather than by NER, as discussed below.
Deletion of rad23 in B. bassiana resulted in a hypersensitivity to UVB irradiation and a greatly reduced efficiency in photoreactivation of UVB-inactivated conidia. Such defects concurred with slight upregulation of phr1 but drastic repression of phr2. Previously, PHR1 and PHR2 were shown to reactivate UVB-inactivated conidia by repairing CPD and 6-4PP DNA lesions, respectively, in irradiated cells of B. bassiana under visible light (30). The similar roles of RAD23 and two photolyases in photoreactivation of UVB-inactivated conidia suggest a tight link of RAD23 to PHR1 or PHR2, and the link is clarified by the yeast two-hybrid assay. A positive interaction of RAD23 with PHR2 rather than with PHR1 in the yeast assay correlates well with the sharp repression of phr2 transcription in our Δrad23 mutant. Moreover, overexpression of phr2 in the Δrad23 mutant largely mitigated severe defects of the Δrad23 and Δphr2 mutants in photoreactivation. UVB resistance was more compromised in the strains overexpressing phr2 in the Δrad23 mutant than in the Δphr2 mutant, implying that excessive PHR2 accumulation in the Δrad23 mutant could be cytotoxic, perhaps due to an inability for PHR2 to interact with RAD23. However, our study demonstrated no role of RAD23 in the dark-dependent NER in B. bassiana. These results uncover a novel role for RAD23 in photoreactivation of UVB-inactivated conidia through its interaction with PHR2, which is required for photorepair of 6-4PP DNA lesions in B. bassiana (30); its importance for filamentous fungal adaptation to solar UV irradiation; and a big difference between its role and that of yeast ortholog in NER only (23–25). DNA damage photorepair relies upon rapid breakdown of shorter UV wavelength-induced covalent linkages in DNA lesions through direct transfer of electrons to cytotoxic CPDs or 6-4PPs under longer UV wavelengths or visible light (44, 45). Despite having no role in dark-dependent NER, which works slowly (46, 47), RAD23 could be involved in stabilization of DNA synthesis in B. bassiana. This is revealed by increased sensitivity of the Δrad23 mutant to DNA synthesis-inhibiting hydroxyurea and differential downregulation of several partner genes involved in DNA damage repair, including dsk2 and three of the four other rad genes. In our Δrad23 mutant, expression of rad4A, one of the two rad4 paralogues in B. bassiana, was suppressed significantly, although expression of rad4B was not affected. These suppressed genes suggest an involvement of RAD23 in the proteasome activity of B. bassiana due to the requirement of yeast RAD23 for an interaction with the complicated 26S proteasome (14, 15, 17) and stabilization of RAD4 acting in impaired DNA binding and recognition (24). However, the existence of two RAD4 paralogues implies more complicated RAD23-RAD4 interaction in B. bassiana than in the model yeast. We consider that photoprotection of B. bassiana from UVB damage could be more dependent on an interaction of RAD23 with PHR2 than on its possible involvement in NER-related proteasome activity. Also, more RAD family members and their partner proteins could likely interact with PHR1, PHR2, or both of them and function like RAD23 in photoprotection of filamentous fungal cells. These warrant further studies.
Our results also demonstrate a linkage of RAD23 with transcriptional activation of many genes that function in asexual development and multiple stress responses. First, the delayed conidiation, the reduced conidial yield and the impaired conidial quality in the absence of rad23 correlated with a severe block of the central developmental pathway that is required for conidiation and conidial maturation (32, 33). The blocked pathway is indicated by dramatic repression of brlA, abaA, and wetA in the central pathway of the deletion mutant and the upstream fluG critical for activation of brlA. Second, increased sensitivity of the Δrad23 mutant to high osmolarity correlated with repressed hog1 and pbs2 transcription. Since osmotolerance is one of the hallmark phenotypes regulated by the MAPK Hog1 cascade (4, 34), it was not surprising to see increased sensitivity of the Δrad23 mutant to hyperosmotic stress when hog1 expression was suppressed. Aside from transcriptional suppression of some DNA damage repair-related genes, all SOD and CAT genes essential or nonessential for antioxidant activity of B. bassiana (3) were downregulated in the Δrad23 mutant, resulting in reduced SOD and CAT activities that are obviously responsible for elevated sensitivity of the Δrad23 mutant to oxidative stress of menadione or H2O2.
Furthermore, our Δrad23 mutant was compromised in virulence through the cuticle infection much more than through the cuticle-bypassing infection. The mutant infection via cuticle penetration could be retarded by slower hyphal growth and invasion into the insect due to less efficient use of limited nutrients for hyphal growth on oligotrophic insect integument. This is in evidence with the mutant phenotypes, which include severe growth defects on the scant media with different carbon/nitrogen sources tested and reduced biomass levels in the submerged CDB-BSA and SDBY cultures. After entry into host hemocoel, penetrating hyphae turn into unicellular blastospores to favor intrahemocoel fungal propagation and host mummification (36–38). The postinfection cellular process critical for fungal virulence was blocked in the absence of rad23, as indicated by blocked formation of hyphal bodies in vivo and greatly reduced blastospore production in vitro, particularly in the medium mimicking insect hemolymph. In B. bassiana, therefore, RAD23 is essential for the success of not only host infection but also of dimorphic transition required for acceleration of host mummification and subsequent fungal outgrowth for conidiation on host carcass surfaces.
In conclusion, RAD23 can interact with PHR2, which is critical for photorepair of 6-4PP DNA lesions generated under UVB irradiation (30). This interaction enables RAD23 to take part in photoreactivation of UVB-inactivated conidia for photoprotection of B. bassiana from UVB damage. Nutritional deficiency may trigger RAD23 migration to the vacuoles, where various cellular events take place (42, 43). External stress cues also trigger RAD23 entry into the nuclei, where important nuclear events, including DNA replication and regulation of gene expression and translation, may occur. These help to understand pleiotropic effects of RAD23 in the in vitro and in vivo asexual cycle of B. bassiana. While these findings unveil the significance of RAD23 for the fungal photoprotection and asexual cycle in vitro and in vivo, it remains unclear how RAD23 interacts with other RAD partners to take part in the fungal proteasome activity, warranting further studies in the future.
MATERIALS AND METHODS
Bioinformatic analysis of fungal RAD23 homologs.
The yeast RAD23 sequence (NCBI accession number NP_010877) was used as a query to search through the genomes of B. bassiana (31) under the NCBI accession number NZ_ADAH00000000 and of other fungi by BLAST analysis at http://blast.ncbi.nlm.nih.gov/Blast.cgi/. The sequences of located RAD23 homologs were compared at https://www.ncbi.nlm.nih.gov/Structure/, followed by phylogenetic analysis with the neighbor-joining method in MEGA7 (http://www.megasoftware.net/).
Subcellular localization of RAD23 in B. bassiana.
Our previous plasmid pAN52-Ptef1-C-bar vectoring the optimized tef1 promoter, the C cassette 5′-PmeI-SpeI-EcoRV-EcoRI-BamHI-3′, and the reporter gene bar (30) were used as a backbone for expression of the mCherry-tagged rad23 fusion gene in the WT strain. Briefly, the backbone plasmid was digested with HpaI/XhoI and ligated to mCherry (GenBank accession number KC294599). The coding sequence of rad23 (tag locus BBA_01030) was amplified from the WT cDNA (1,134 bp) with paired primers (see Table S1 in the supplemental material) and ligated to the N terminus of mCherry in pAN52-Ptef1-C-mCherry-bar digested with EcoRV by means of a one-step cloning kit (Vazyme, Nanjing, China). The new plasmid containing the fusion gene rad23::mCherry under the control of Ptef1 was transformed into the WT through blastospore transformation (48). Putative transformants were screened by bar resistance to phosphinothricin (200 μg · ml−1) and examined under a fluorescence microscope. A transformant best expressing red fluorescence signal was chosen for subcellular localization of the expressed fusion protein. The selected transformant was incubated for full conidiation on SDAY (4% glucose, 1% peptone, and 1.5% agar plus 1% yeast extract) at the optimal regime of 25°C and an L:D cycle of 12:12 h. The resultant conidia were suspended in SDBY and incubated for 48 or 60 h on a shaking bed (150 rpm) at 25°C. Hyphae collected from the 60-h-old culture were shaken for 90 min at 25°C in the presence of 0.4 M NaCl (for osmotic stress) or of 40 mM H2O2 (for oxidative stress) in 1/4 SDBY (amended with 1/4 nutrients of SDBY) or shaken for 90 min in chemical-free 1/4 SDBY at 42°C (for heat stress). For UVB stress, the collected hyphae were spread on agar plates, and the plates were then exposed to UVB irradiation at a dose of 1 J · cm−2 in a Bio-Sun++ UV irradiation chamber (Vilber Lourmat, Marne-la-Vallée, France). The hyphae exposed to each stress cue or not exposed (control) were stained with DAPI and rinsed repeatedly in sterile water, followed by LSCM analysis for subcellular localization of the mCherry-tagged RAD23 fusion protein. Alternatively, the hyphae collected from the 48-h-old culture were resuspended in minimal CDB (3% sucrose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4, and 0.001% FeSO4) lacking the carbon source, the nitrogen source or both, followed by a 12-h incubation at 25°C. The resultant hyphae were stained with FM4-64 for LSCM analysis as mentioned above.
Generation of rad23 mutants.
Our previous plasmids p0380-5′x-bar-3′x and p0380-sur-x, in which x denotes a target gene to be deleted (30), were used as backbones to delete rad23 from the WT by homogeneous recombination of its 5′ and 3′ coding/flanking fragments separated by the bar marker and rescue it in an identified Δrad23 mutant by ectopic integration of a cassette consisting of its full-length coding/flanking sequence and the sur marker. Briefly, the 5′ and 3′ fragments (1,355 and 1,552 bp, respectively) of rad23 were amplified from the genomic DNA of the WT with paired primers (Table S1), and the 5′x and 3′x fragments at appropriate enzyme sites were substituted using a one-step cloning kit (Vazyme), forming p0380-5′rad23-bar-3′rad23 for targeted gene deletion. The full-length coding sequence of rad23 with flanking regions (4,767 bp in total) was amplified from the WT DNA and inserted into p0380-sur-x to exchange for the gateway fragment (x) under the action of Gateway BP Clonase II enzyme mix (Invitrogen, Shanghai, China), yielding p0380-sur-rad23 for targeted gene complementation. The two plasmids propagated in Escherichia coli TOP10 and E. coli DH5a (Invitrogen) cells were transformed into the WT and an identified Δrad23 mutant via Agrobacterium-mediated transformation, respectively. Putative mutants grown on a selective medium were screened by bar resistance to phosphinothricin (200 μg · ml−1) or by sur resistance to chlorimuron ethyl (15 μg · ml−1) and identified through PCR and Southern blot analyses with paired primers and a designed probe (Table S1). All genomic DNAs used for Southern blot were digested with SacI. Positive Δrad23 and Δrad23::rad23 mutants (see Fig. S2 in the supplemental material) were evaluated together with the WT strain in phenotypic experiments of three independent cultures or samples taken from the cultures (replicates).
Experiments for phenotypic changes.
The Δrad23 mutant and two control strains were grown on plates of SDAY, CDA (CDB plus 1.5% agar), and CDA amended with different carbon sources (3% glucose, glycerol, stearic acid, oleic acid, or sodium acetate) or nitrogen sources (0.3% NH4Cl, NaNO2, NH4NO3, or one of 18 amino acids) by spotting 1 μl of a 106 conidia · ml−1 suspension per plate for culture initiation. After an 8-day incubation at 25°C and an L:D cycle of 12:12 h, the diameter of each colony was estimated as a growth index with two measurements taken perpendicularly to each other across the center.
The spotting method also was used to initiate colony growth at the same regime on CDA alone (control) or supplemented with a sensitive concentration of each of the following chemical stressors: (i) hydroxyurea (10 mM) or camptothecin (1 μM) for DNA synthesis inhibiting or damaging stress; (ii) H2O2 (2 mM) or menadione (0.02 mM) for oxidative stress; (iii) NaCl (0.4 M) or sorbitol (1 M) for osmotic stress; (iv) Congo red (3 μg · ml−1) or calcofluor white (5 μg · ml−1) for cell wall-disturbing stress. After an 8-day incubation, the diameter of each colony was assessed as mentioned above. Relative growth inhibition of each strain by each chemical stress was calculated as (Sc − St)/Sc × 100 (Sc, control colony area; St, stressed colony area).
SDAY cultures for assessment of conidiation capacity were initiated by spreading 100 μl of a 107 conidia · ml−1 suspension per plate (9-cm diameter). During a 9-day incubation at 25°C and an L:D cycle of 12:12 h, three plugs were taken from each plate of 5-, 7-, and 9-day-old cultures with a cork borer (5-mm diameter). Conidia on each plug were released into 1 ml of 0.02% Tween 80 via ultrasonic vibration. The concentration of the conidial suspension was assessed with a hemocytometer and converted to the yield of number of conidia · cm−2 culture.
Conidia harvested from the 9-day-old SDAY cultures were assayed for quality-related properties. First, conidial viability was quantified as germination time (h) for 50% germination (GT50) on a germination medium (GM; 2% sucrose, 0.5% peptone, and 1.5% agar) at an optimal temperature of 25°C. Second, fluorescence-activated cell sorter analysis was performed to assess conidia size and complexity (density) with the FSC and SSC readings from the flow cytometry of three samples (2 × 104 conidia per sample) per strain, as described previously (49). Third, conidial UVB resistance was quantified following our previous protocols (50, 51). Briefly, 80-μl aliquots of a 107 conidia · ml−1 suspension were spread evenly on GM plates (7-cm diameter). After 10 min of air drying, the uncovered plates were placed in a sample tray of the aforementioned UV irradiation chamber and irradiated with a weighted wavelength of 312 nm at the gradient UVB doses of 0.1 to 0.6 J · cm−2, each of which is automatically adjusted four times per second for an error control of ≤1 μJ · cm−2 (per the manufacturer’s guide). The irradiated plates were covered immediately with lids and incubated for 24 h at 25°C. The germination percentage on each plate was assessed from the counts of germinated and nongerminated conidia in three fields of microscopic view (100× magnification). An LD50 (J · cm−2) indicative of UVB resistance was estimated from the fitted survival trends of conidia over the applied doses.
Assays for photoreactivation of UVB-inactivated conidia were carried out following our previous protocols (30). Briefly, aliquots of standardized conidial suspension were spread on the GM plates and irradiated at the lethal UVB dose of 0.5 J · cm−2 in the UV chamber. The irradiated plates covered with lids were incubated for 3 h at 25°C under white light and then for 21 h in full darkness (photoreactivation treatment) or directly incubated for 24 h at 25°C in the dark (NER treatment). The conidia not irradiated at the lethal UVB dose were incubated for 24 h at 25°C in the dark and used as a control. At the end of dark incubation, conidial germination percentage on each of the plates in each treatment was assessed with microscopic counts as mentioned previously. To explore the role of RAD23-PHR2 interaction in photoreactivation, phr2 amplified from the WT cDNA was transformed into the Δrad23 mutant. Three strains overexpressing phr2 at least 100-fold in the Δrad23 mutant under Ptef1 control were selected for assaying UVB resistances of their conidia and photoreactivation efficiencies of their UVB-inactivated conidia in parallel with the WT and previous phr2 mutants (30), as mentioned previously.
Conidial virulence of each strain was assayed on the fifth-instar larvae of G. mellonella through two infection modes. Briefly, groups of ∼35 larvae were immersed separately for ∼10 s in 40-ml aliquots of a 107 conidia · ml−1 suspension or 0.02% Tween 80 (control) for normal cuticle infection. Alternatively, 5 μl of a 105 conidia · ml−1 suspension or 0.02% Tween 80 (control) was injected into the hemocoel of each larva in each group, resulting in ∼500 conidia injected per larva for cuticle-bypassing infection. All treated groups were maintained in plastic boxes for up to 13 days at 25°C and monitored daily for survival/mortality records. LT50 (in days) was estimated as a virulence index of each strain by probit analysis of the time-mortality trend in each group. During the period of bioassay, hemolymph samples were taken from the larvae surviving for 5 days after immersion or 3 days after injection and examined for the presence or absence of hyphal bodies under a microscope. Images for fungal outgrowths of each strain on carcass surfaces were collected after maintenance under optimal conditions.
To assess the total activities of ECEs and Pr1 proteases involved in cuticle degradation and host infection, 50-ml aliquots of a 106 conidia · ml−1 suspension in CDB containing 0.3% BSA for induction of enzyme production were incubated at 25°C for 3 days on a shaking bed (150 rpm). All cells collected from the cultures were dried in vacuum for assessment of biomass, and the supernatant from each of the cultures was assayed for total activities (U · ml−1 supernatant) of ECEs and Pr1 proteases as described elsewhere (36, 37). In addition, biomass levels and dimorphic transition rates (no. of blastospores · ml−1) were quantified from the 3-day-old submerged cultures of a 106 conidia · ml−1 suspension grown in SDBY and TPB, a CDB amended to mimic insect hemolymph using 3% trehalose as the sole carbon source and 0.5% peptone as the sole nitrogen source (38).
Assays for activities of antioxidant enzymes.
To assay activities of antioxidant enzymes, 100-μl aliquots of a 107 conidia · ml−1 suspension were spread on cellophane-overlaid SDAY plates and incubated for 3 days under the optimal regime, followed by protein extraction from the hyphal cultures. Total SOD and CAT activities were quantified from the protein extracts of each strain using a SOD activity assay kit (Sigma-Aldrich, St. Louis, MO) and a catalase activity assay kit (Jiancheng Biotech, Nanjing, China), respectively, as described previously (52). One unit of enzymatic activity was defined as the SOD amount required for inhibition of 50% pyrogallol autoxidation rate or 1 mM H2O2 consumed per min. The total SOD or CAT activity was expressed as U · mg−1 protein extract.
Yeast two-hybrid assay.
To probe the interaction of RAD23 with PHR1 or PH2, the coding sequences of rad23, phr1, and phr2 were amplified from the WT cDNA and inserted into pGADT7and pGBKT7, followed by verification through sequencing. The verified plasmids were transformed into S. cerevisiae Y2H Gold or Y187 for yeast two-hybrid assay following the Matchmaker GAL4 two-hybrid system 3 and Libraries User Manual (Clontech).
Transcriptional profiling of rad23 and related genes.
To assess the transcript level of rad23 in the WT strain, 100-μl aliquots of a 107 conidia · ml−1 suspension were spread on cellophane-overlaid SDAY plates and incubated at the optimal regime for 7 days. From the end of 2-day incubation onwards, three plate cultures were collected daily for RNA extraction. For transcriptional responses of rad23 to different stress cues, stressed and nonstressed hyphal cultures for RNA extraction were prepared as those used for subcellular localization of the RAD23::GFP fusion protein. For transcriptional profiling of phenotype-related genes, the cultures of Δrad23 and control strains were initiated as mentioned previously on the plates of SDAY alone or supplemented with 2 mM H2O2 (only for profiling of genes involved in DNA damage repair), followed by a 3-day incubation at the optimal regime. Total RNAs were separately extracted from the hyphal cultures of each strain or treatment under the action of an RNAiso Plus reagent (TaKaRa, Dalian, China) and reverse transcribed into cDNAs with a PrimeScript RT reagent kit (TaKaRa) at 37°C. Three cDNA samples (standardized by dilution) derived from the cultures of each strain or treatment were used as templates to assess transcript levels of target genes under the action of SYBR Premix Ex Taq (TaKaRa) via qPCR analysis with paired primers (see Table S2 in the supplemental material). Both tef1 and act1 were used as internal standards for the normalization of rad23 transcripts, whereas only act1 was used for normalization of phenotype-related gene transcripts to avoid overestimation. The threshold cycle (2−ΔΔCT) method was used to compute the relative transcript levels of rad23 in the normal WT cultures during a 7-day incubation with respect to a standard on day 2 or in the stressed WT cultures with respect to a nonstressed standard (control), and that of each phenotype-related gene in the rad23 mutants with respect to the WT standard.
Statistical analysis.
All experimental data from the experiments with three replicates were subjected to one-factor analysis of variance, followed by Tukey’s honestly significant difference (HSD) test for a comparison of the means among the tested fungal strains.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge She-Long Zhang (Core Facilities, College of Life Sciences, ZJU) for technical assistance with LSCM analysis.
Funding of this work was provided by the Ministry of Science and Technology of the People’s Republic of China (grant 2017YFD0201202) and by the National Natural Science Foundation of China (grants 31772218 and 31801795).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Madronich S. 1993. UV radiation in the natural and perturbed atmosphere, p 17−69. In Tevini M. (ed), UV-B radiation and ozone depletion. Lewis Publishers, Boca Raton, FL. [Google Scholar]
- 2.Rangel DEN, Braga GUL, Fernandes ÉKK, Keyser CA, Hallsworth JE, Roberts DW. 2015. Stress tolerance and virulence of insect-pathogenic fungi are determined by environmental conditions during conidial formation. Curr Genet 61:383–404. doi: 10.1007/s00294-015-0477-y. [DOI] [PubMed] [Google Scholar]
- 3.Zhang LB, Feng MG. 2018. Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl Microbiol Biotechnol 102:4995–5004. doi: 10.1007/s00253-018-9033-2. [DOI] [PubMed] [Google Scholar]
- 4.Tong SM, Feng MG. 2019. Insights into regulatory roles of MAPK-cascaded pathways in multiple stress responses and life cycles of insect and nematode mycopathogens. Appl Microbiol Biotechnol 103:577–587. doi: 10.1007/s00253-018-9516-1. [DOI] [PubMed] [Google Scholar]
- 5.Engelberg D, Klein C, Martinetto H, Struhl K, Karin M. 1994. The UV response involving the Ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell 77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths HR, Mistry P, Herbert KE, Lunec J. 1998. Molecular and cellular effects of ultraviolet light-induced genotoxicity. Crit Rev Clin Lab Sci 35:189–237. doi: 10.1080/10408369891234192. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 8.Yasui A, Eker APM, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. 1994. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J 13:6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Laat WL, Jaspers NGJ, Hoeijmakers J. 1999. Molecular mechanism of nucleotide excision repair. Gene Dev 13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 10.Mueller JP, Smerdon MJ. 1996. Rad23 is required for transcription-coupled repair and efficient overall repair in Saccharomyces cerevisiae. Mol Cell Biol 16:2361–2368. doi: 10.1128/MCB.16.5.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas S, Katiyar S, Li G, Zhou X, Lennarz WJ, Schindelin H. 2004. The N-terminus of yeast peptide: N-glycanase interacts with the DNA repair protein Rad23. Biochem Biophys Res Commun 323:149–155. doi: 10.1016/j.bbrc.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 12.Prakash S, Prakash L. 2000. Nucleotide excision repair in yeast. Mutat Res 451:13–24. doi: 10.1016/S0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 13.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol 8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 14.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 15.Dantuma NP, Heinen C, Hoogstraten D. 2009. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair 8:449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Coux O, Tanaka K, Goldberg AL. 1996. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 17.Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. 1999. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell 3:687–695. doi: 10.1016/S1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- 18.Kim HT, Goldberg AL. 2018. UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc Natl Acad Sci U S A 115:E11642–E11650. doi: 10.1073/pnas.1808731115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang RY, Chen L, Ko BT, Shen YH, Li YT, Chen BR, Lin KT, Madura K, Chuang SM. 2014. Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. J Mol Biol 426:4049–4060. doi: 10.1016/j.jmb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Shinde U, Ortolan TG, Madura K. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep 2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heessen S, Masucci MG, Dantuma NP. 2005. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell 18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. 2011. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun 2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortolan TG, Chen L, Tongaonkar P, Madura K. 2004. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res 32:6490–6500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z, Liu S, Zhang Y, Wang Z. 2004. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Res 32:5981–5990. doi: 10.1093/nar/gkh934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong F, Fahy D, Smerdon MJ. 2006. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y. 2017. In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell 66:488–502.e7. doi: 10.1016/j.molcel.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 27.Godderz D, Giovannucci TA, Lalakova J, Menendez-Benito V, Dantuma NP. 2017. The deubiquitylating enzyme Ubp12 regulates Rad23-dependent proteasomal degradation. J Cell Sci 130:3336–3346. doi: 10.1242/jcs.202622. [DOI] [PubMed] [Google Scholar]
- 28.de Faria MR, Wraight SP. 2007. Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol Control 43:237–256. doi: 10.1016/j.biocontrol.2007.08.001. [DOI] [Google Scholar]
- 29.Ying SH, Feng MG. 2019. Insight into vital role of autophagy in sustaining biological control potential of fungal pathogens against pest insects and nematodes. Virulence 10:429–437. doi: 10.1080/21505594.2018.1518089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang DY, Fu B, Tong SM, Ying SH, Feng MG. 2018. Two photolyases repair distinct DNA lesions and reactivate UVB-inactivated conidia of an insect mycopathogen under visible light. Appl Environ Microbiol 85:e02459-18. doi: 10.1128/AEM.02459-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao GH, Ying SH, Zheng P, Wang ZL, Zhang SW, Xie XQ, Shang YF, Zheng HJ, Zhou Y, St Leger RJ, Zhao GP, Wang CS, Feng MG. 2012. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep 2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Shi HQ, Ying SH, Feng MG. 2015. WetA and VosA are distinct regulators of conidiation capacity, conidial quality, and biological control potential of a fungal insect pathogen. Appl Microbiol Biotechnol 99:10069–10081. doi: 10.1007/s00253-015-6823-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang AX, Mouhoumed AZ, Tong SM, Ying SH, Feng MG. 2019. BrlA and AbaA govern virulence-required dimorphic switch, conidiation and pathogenicity in a fungal insect pathogen. mSystems 4:e00140-19. doi: 10.1128/mSystems.00140-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Wang ZK, Sun HH, Ying SH, Feng MG. 2017. Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol 19:1808–1821. doi: 10.1111/1462-2920.13671. [DOI] [PubMed] [Google Scholar]
- 35.Lewis MW, Robalino IV, Keyhani NO. 2009. Uptake of the fluorescent probe FM4-64 by hyphae and haemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology 155:3110–3120. doi: 10.1099/mic.0.029165-0. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Ying SH, Hu Y, Feng MG. 2016. Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ Microbiol 18:1037–1047. doi: 10.1111/1462-2920.13197. [DOI] [PubMed] [Google Scholar]
- 37.Zhang LB, Tang L, Ying SH, Feng MG. 2017. Two eisosome proteins play opposite roles in autophagic control and sustain cell integrity, function and pathogenicity in Beauveria bassiana. Environ Microbiol 19:2037–2052. doi: 10.1111/1462-2920.13727. [DOI] [PubMed] [Google Scholar]
- 38.Tong SM, Zhang AX, Guo CT, Ying SH, Feng MG. 2018. Daylight length-dependent translocation of VIVID photoreceptor in cells and its essential role in conidiation and virulence of Beauveria bassiana. Environ Microbiol 20:169–185. doi: 10.1111/1462-2920.13951. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz-Urquiza A, Keyhani NO. 2013. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects 4:357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elder RT, Song XQ, Chen M, Hopkins KM, Lieberman HB, Zhao Y. 2002. Involvement of rhp23, a Schizosaccharomyces pombe homolog of the human HHR23A and Saccharomyces cerevisiae RAD23 nucleotide excision repair genes, in cell cycle control and protein ubiquitination. Nucleic Acids Res 30:581–591. doi: 10.1093/nar/30.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergink S, Toussaint W, Luijsterburg MS, Dinant C, Alekseev S, Hoeijmakers JH, Dantuma NP, Houtsmuller AB, Vermeulen W. 2012. Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex. J Cell Biol 196:681–688. doi: 10.1083/jcb.201107050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veses V, Richards A, Gow NA. 2008. Vacuoles and fungal biology. Curr Opin Microbiol 11:503–510. doi: 10.1016/j.mib.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 43.Li SC, Kane PM. 2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta Mol Cell Res 1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, Eker AP, Nakajima S, van Steeg H, de Gruijl FR, Yasui A, Hoeijmakers JHJ, van der Horst G. 2005. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol 15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst G, Batschauer A, Ahmad M. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 46.Smerdon MJ, Thoma F. 1990. Site-specific DNA-repair at the nucleosome level in a yeast minichromosome. Cell 61:675–684. doi: 10.1016/0092-8674(90)90479-X. [DOI] [PubMed] [Google Scholar]
- 47.Suter B, Wellinger RE, Thoma F. 2000. DNA repair in a yeast origin of replication: contributions of photolyase and nucleotide excision repair. Nucleic Acids Res 28:2060–2068. doi: 10.1093/nar/28.10.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ying SH, Feng MG. 2006. Novel blastospore-based transformation system for integration of phosphinothricin resistance and green fluorescence protein genes into Beauveria bassiana. Appl Microbiol Biotechnol 72:206–210. doi: 10.1007/s00253-006-0447-x. [DOI] [PubMed] [Google Scholar]
- 49.Wang JJ, Qiu L, Cai Q, Ying SH, Feng MG. 2014. Three α-1,2-mannosyltransferases contribute differentially to conidiation, cell wall integrity, multistress tolerance and virulence of Beauveria bassiana. Fungal Genet Biol 70:1–10. doi: 10.1016/j.fgb.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Huang BF, Feng MG. 2009. Comparative tolerances of various Beauveria bassiana isolates to UV-B irradiation with a description of a modeling method to assess lethal dose. Mycopathologia 168:145–152. doi: 10.1007/s11046-009-9207-7. [DOI] [PubMed] [Google Scholar]
- 51.Yao SL, Ying SH, Feng MG, Hatting JL. 2010. In vitro and in vivo responses of fungal biocontrol agents to gradient doses of UV-B and UV-A irradiation. BioControl 55:413–422. doi: 10.1007/s10526-009-9265-2. [DOI] [Google Scholar]
- 52.Zhang LB, Tang L, Ying SH, Feng MG. 2016. Regulative roles of glutathione reductase and four glutaredoxins in glutathione redox, antioxidant activity, and iron homeostasis of Beauveria bassiana. Appl Microbiol Biotechnol 100:5907–5917. doi: 10.1007/s00253-016-7420-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.