Understanding microbe-mediated plant resistance to drought is important for sustainable agriculture. We performed 16S rRNA gene amplicon sequencing and culture-dependent functional analyses of Alhagi sparsifolia rhizosphere and root endosphere microbiomes and identified key endophytic bacterial taxa and their genes facilitating drought resistance in wheat. This study improves our understanding of plant drought resistance and provides new avenues for drought resistance improvement in crop plants under field conditions.
KEYWORDS: Alhagi sparsifolia, drought resistance, Pseudomonas, root endosphere
ABSTRACT
Drought is among the most destructive abiotic stresses limiting crop growth and yield worldwide. Although most research has focused on the contribution of plant-associated microbial communities to plant growth and disease suppression, far less is known about the microbes involved in drought resistance among desert plants. In the present study, we applied 16S rRNA gene amplicon sequencing to determine the structure of rhizosphere and root endosphere microbiomes of Alhagi sparsifolia. Compared to those of the rhizosphere, endosphere microbiomes had lower diversity but contained several taxa with higher relative abundance; many of these taxa were also present in the roots of other desert plants. We isolated a Pseudomonas strain (LTGT-11-2Z) that was prevalent in root endosphere microbiomes of A. sparsifolia and promoted drought resistance during incubation with wheat. Complete genome sequencing of LTGT-11-2Z revealed 1-aminocyclopropane-1-carboxylate deaminases, siderophore, spermidine, and colanic acid biosynthetic genes, as well as type VI secretion system (T6SS) genes, which are likely involved in biofilm formation and plant-microbe interactions. Together, these results indicate that drought-enduring plants harbor bacterial endophytes favorable to plant drought resistance, and they suggest that novel endophytic bacterial taxa and gene resources may be discovered among these desert plants.
IMPORTANCE Understanding microbe-mediated plant resistance to drought is important for sustainable agriculture. We performed 16S rRNA gene amplicon sequencing and culture-dependent functional analyses of Alhagi sparsifolia rhizosphere and root endosphere microbiomes and identified key endophytic bacterial taxa and their genes facilitating drought resistance in wheat. This study improves our understanding of plant drought resistance and provides new avenues for drought resistance improvement in crop plants under field conditions.
INTRODUCTION
Drought stress is among the most destructive abiotic stresses; as its intensity and frequency increase with climate change, drought is expected to threaten more than 50% of the Earth’s arable land by 2050 (1, 2). Because water scarcity is a rapidly growing sustainability problem worldwide, it is impossible to combat drought by simply increasing irrigation infrastructure (3). Climate change and an increasing global population will further worsen this condition; therefore, there is an urgent need to improve plant resistance to drought under limited water resource availability. To date, the creation of drought-tolerant cultivars has been the predominant approach for mitigating the negative effects of drought stress on crop growth and yield (4, 5). Although conventional breeding techniques and genetic engineering have promoted the development of drought-tolerant crop varieties, each method has disadvantages and neglects the complex ecological context of the plant growth environment (5–7).
In nature, plants harbor a diverse bacterial community in the rhizosphere that affects plant growth and health (2, 8). Some rhizobacteria can transcend the endodermis barrier and colonize internal tissues to thrive as endophytes in roots, stems, leaves, and other organs (9, 10). Endophytic bacteria can also originate from the phyllosphere or be transmitted through seeds (11). Because endophytic bacteria are relatively protected from the competitive and high-stress soil environment and achieve intimate contact with plant tissues, they are considered to have major interactions with host plants (9, 11, 12). Although the functional capacities of rhizospheric and endophytic bacteria in plant growth promotion and disease control have been widely reported (11, 13, 14), their roles in protecting plant resistance to abiotic stresses such as drought are only beginning to gain attention (3, 5, 7). Recent studies have shown that some plant growth-promoting (PGP) bacteria can increase drought resistance in crop plants such as wheat, maize, tomato, lettuce, chickpea, and beans (12, 15–17).
Northwestern China, which has typical arid and semiarid regions, accounts for approximately 38% of the Chinese territory. Water deficiency is a major factor limiting the yield of Triticum aestivum L. (winter wheat), which is among the most important crops grown in semiarid areas of northwestern China (18). Alhagi sparsifolia Shap. (Leguminosae), a major drought-tolerant plant in the desert ecosystem of northwestern China (19), is an ideal host species for the discovery of novel microbial symbionts that confer drought resistance in crop plants for local agriculture. Hence, the aims of this study were (i) to decipher the root endosphere microbiome of the desert plant A. sparsifolia to identify drought resistance-promoting microbes and (ii) to obtain a better understanding of the mechanisms by which these bacteria colonize plants and contribute to drought stress mitigation. To meet these objectives, we applied a combination of culture-dependent and -independent approaches to identify key bacterial taxa in the root endosphere microbiome of A. sparsifolia that showed the ability to increase drought resistance in wheat. We then performed genome sequencing and comparative genomics analysis of a drought resistance-promoting strain to investigate the potential mechanisms of bacterial colonization and enhancement of drought resistance in wheat.
RESULTS
Alpha and beta diversity.
We applied Illumina HiSeq 2500 high-throughput sequencing of the V3 and V4 regions of 16S rRNA genes to analyze the diversity of rhizospheric and endophytic bacterial communities associated with A. sparsifolia. After deletion of chloroplast- and mitochondrion-derived 16S rRNA gene amplicons, high-quality reads were assembled (see Table S1 in the supplemental material). The rarefaction curves (see Fig. S1 in the supplemental material) were close to saturation, indicating that the sequencing depth was sufficient to cover the diversity of microbial populations in the rhizosphere soil and plant tissue samples. We observed clear variation between the rhizosphere soil samples and root endosphere samples in terms of richness and diversity (see Table S2 in the supplemental material). The observed amplicon sequence variants (ASVs) and diversity, indicated by the Chao1, abundance-based coverage estimator, and Shannon and Simpson indices, were much lower in the root endosphere than in the rhizosphere (P < 0.05) (Table S2).
We evaluated beta diversity in terms of ASV composition and relative abundance. High dissimilarity between rhizospheric and endophytic microbial communities was revealed by principal-coordinate analysis (PCoA), which showed clear separation of rhizosphere soil samples from endosphere samples (see Fig. S2 in the supplemental material). A PCoA plot based on phylum abundance showed that principal coordinate 1 (PCo1) explained 73.02% of the Bray-Curtis dissimilarity. Compared to rhizosphere samples, endosphere samples were closer to each other, resulting in much greater dissimilarity among microbial communities in the rhizosphere than among endosphere microbial communities (Fig. S2).
Microbial community composition.
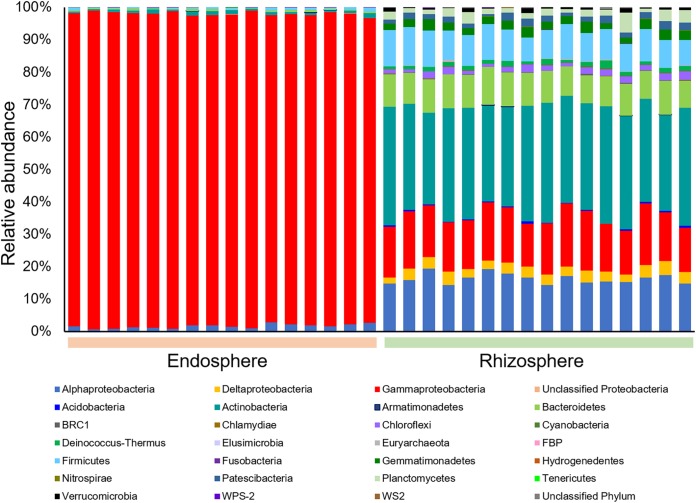
Only a small number of sequences in each sample (<0.1%) were assigned to the phylum Euryarchaeota (Archaea). Within the domain Bacteria, Proteobacteria were further classified into class level. Most of the sequences in the endosphere samples was dominated by Gammaproteobacteria (94.8 to 98.2%) (Fig. 1), while in rhizosphere samples, Actinobacteria (28.2 to 37.1%) was the most abundant phylum, followed by Alphaproteobacteria (14.3 to 19.5%), Gammaproteobacteria (13.2 to 19.0%), Bacteroidetes (8.2 to 11.7%), and Firmicutes (7.4 to 11.6%) (Fig. 1). We evaluated the phylum abundance differences between the rhizosphere and the endosphere by two-tailed Student’s t test followed by Benjamini-Hochberg correction (see Table S3 in the supplemental material). All phyla with relative abundances greater than 0.02% in the rhizosphere were more abundant in the rhizosphere than that in the endosphere, except for Gammaproteobacteria, which comprised 96.39% of the endosphere community (Table S3).
FIG 1.
Community structures at the phylum level. The detailed relative abundances of the phyla and differences between root rhizosphere and endosphere are listed in Table S3 in the supplemental material. Data analysis was performed using DADA2 v1.4.0 implemented in QIIME2.
We further examined microbial community composition at the genus level (see Fig. S3 in the supplemental material). A large proportion of rhizosphere sequences could not be unambiguously classified at the genus level (18.56 to 21.42%); however, only a small proportion of sequences from the root endosphere remained unclassified (<0.78%; Fig. S3). We evaluated the top 30 abundant genera by two-tailed Student’s t test followed by Benjamini-Hochberg correction to test the effects of the plant compartment (rhizosphere versus endosphere) on their relative abundances (see Table S4 in the supplemental material). We detected significantly higher abundances (P < 0.05) of the genera Kocuria (18.802%), Halomonas (2.545%), Pseudomonas (2.504%), Truepera (1.079%), and Planococcus (1.050%) in the rhizosphere than in the endosphere (see Table S4 in the supplemental material). In contrast, the genera Pseudomonas (80.00%), Stenotrophomonas (5.816%), Achromobacter (3.589%), Undibacterium (2.099%), and Providencia (1.912%) were significantly more abundant in the root endosphere than in the rhizosphere (P < 0.05; Table S4).
A Pseudomonas strain improved drought resistance in wheat.
In parallel to the culture-independent study, we isolated bacteria from A. sparsifolia roots for PGP activities and drought resistance promotion. Based on the endophytic bacterial community composition determined by the culture-independent approach, seven strains (Serratia marcescens LTGR-2, Stenotrophomonas maltophilia LTGR-2-1Z, Pseudoxanthomonas wuyuanensis LTGR-13Z, “Candidatus Rhizobium massiliense” LTGR-20, Pantoea dispersa LTGPAF-12F, Acinetobacter oleivorans LTGT-10, and Pseudomonas sp. strain LTGT-11-2Z) were selected and characterized for PGP activities in vitro. To this end, Pseudomonas sp. LTGT-11-2Z showed growth promotion when incubated with wheat. In addition, activities related to PGP were examined, including siderophore production, exopolysaccharide production, 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity,growth in 5% NaCl, growth in 20% polyethylene glycol (PEG), and growth at 42°C, which further supported the PGP of LTGT-11-2Z (see Table S5 in the supplemental material).
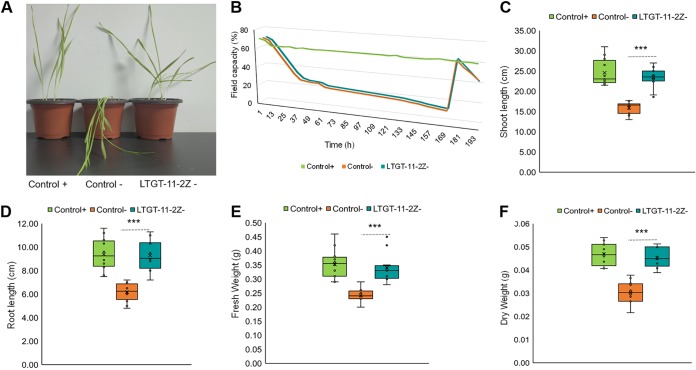
We performed additional experiments to confirm the ability of LTGT-11-2Z to improve drought resistance in wheat. After 7 days of water deprivation, noninoculated control plants were severely affected, whereas plants inoculated with LTGT-11-2Z were healthier and better hydrated (Fig. 2A). Under similar field capacity (Fig. 2B), inoculated plants showed significantly higher shoot length, root length, total plant fresh weight, and dry weight compared with those of the noninoculated stressed control (Fig. 2C to F). In addition, the colonization of LTGT-11-2Z on wheat roots was observed by confocal microscopy, and bacterial cells were clearly detected in wheat root cells. LTGT-11-2Z was observed to have adhered to or colonized on wheat root surfaces (see Fig. S4 in the supplemental material).
FIG 2.
Pseudomonas sp. LTGT-11-2Z improved wheat resistance to drought. +, irrigated at soil water-holding capacity throughout the experiment; −, grown without water for 7 days and watered for 1 day. (A) Representative images of plants inoculated with LTGT-11-2Z compared with those of noninoculated plants under water stress conditions. (B) Field capacity (%). (C) Shoot length (cm). (D) Root length (cm). (E) Plant fresh weight (g). (F) Plant dry weight (g). Statistical analysis between control (−) and LTGT-11-2Z (−) was performed using Student’s t test. ***, P ≤ 0.001.
Genomics analysis of Pseudomonas sp. LTGT-11-2Z.
More than 0.8% of the 16S amplicon sequencing reads of A. sparsifolia root endosphere microbiota were assigned to the 16S sequence of LTGT-11-2Z, which showed 100% similarity with an abundant ASV, suggesting that this bacterium was abundant in endosphere microbial communities. After BLASTn searching against gene sequences in NCBI-Nr database, the 16S rRNA gene of LTGT-11-2Z showed 100% similarity with that of Pseudomonas fluorescens 2P24, which was isolated from wheat roots and has been demonstrated to show PGP ability (20); 2P24 also produces several antifungal compounds, including 2,4-diacetylphloroglucinol (2,4-DAPG), hydrogen cyanide, and siderophores (20).
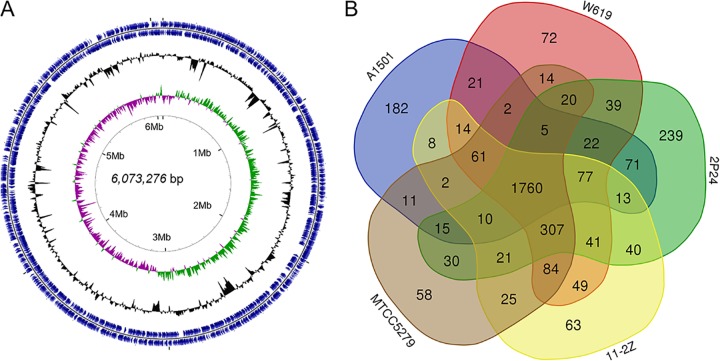
To understand the mechanisms by which LTGT-11-2Z colonizes plants and promotes mitigation of drought stress, we performed complete genome sequencing of the Pseudomonas strain. The genome of Pseudomonas sp. LTGT-11-2Z consisted of a circular chromosome of 6,073,276 bp, with an overall GC content of 61.68%; its circular chromosome contained 5,702 genes, including 77 tRNA-coding genes, 17 rRNA genes, and 5,608 protein-coding genes (see Table S6 in the supplemental material). Despite the high similarity between 16S rRNA gene sequences of LTGT-11-2Z and 2P24, the average nucleotide identity (ANI) between their genomes was only 78.38%. The genome map of LTGT-11-2Z is shown in Fig. 3A.
FIG 3.
Complete genome sequencing and comparative analysis of Pseudomonas sp. LTGT-11-2Z. (A) Circular representation of the genome; illustration is based on visualization of sequence feature information by the CGView Server (http://stothard.afns.ualberta.ca/cgview_server/). (B) Whole-genome comparison between Pseudomonas sp. LTGT-11-2Z and four other endophytes: Pseudomonas fluorescens 2P24, Pseudomonas putida MTCC5279, Pseudomonas putida W619, and Pseudomonas stutzeri A1501. Overlapping regions indicate the number of Kyoto Encyclopedia of Genes and Genomes (KEGG) genes conserved within the specified genomes. Numbers in nonoverlapping portions of each ring indicate the number of KEGG genes unique to each strain.
To gain a better understanding of the functional profiles of LTGT-11-2Z, we compared its genome to those of previously reported plant endophytes, including P. fluorescens 2P24 (20), Pseudomonas putida MTCC5279 (17), Klebsiella sp. strain LTGPAF-6F (21), Serratia proteamaculans 568 (22), Burkholderia phytofirmans PsJN (23, 24), Azospirillum sp. strain B510 (25), Klebsiella pneumoniae 342 (26), Methylobacterium populi BJ001 (24), P. putida W619 (22), Enterobacter sp. strain 638 (27), Pseudomonas stutzeri A1501 (28), Azoarcus sp. BH72 (29), and Gluconacetobacter diazotrophicus Pal5 (30) (see Table S7 in the supplemental material). These reference strains have been reported to play roles in the promotion of plant growth and drought resistance. For example, P. putida MTCC5279 promotes growth and drought stress alleviation of Cicer arietinum L. (chickpea) (17), Klebsiella sp. LTGPAF-6F improves the growth and drought tolerance of wheat (21), and a series of genes involved in bacterial adaptation to plant tissue conditions, such as the limitation of amino acid and carbon source concentrations, were discovered in K. pneumoniae 342 (26). Functions of the reference strains relevant to rhizosphere competence, plant colonization, plant growth promotion, and stress resistance were analyzed and compared with those of LTGT-11-2Z. The results revealed that a number of these functions were present in LTGT-11-2Z; it possessed complete pathways encoding flagellar assembly and chemotaxis-related proteins, as well as genes for curli fiber biosynthesis, which may contribute to plant adhesion (24). A total of 222 glycoside hydrolase genes were detected, several of which may allow bacterial penetration of the plant cell wall and colonization of plant tissues (24). We also annotated biosynthetic genes relevant to plant growth promotion or abiotic stress resistance, which were represented by ACC deaminases and genes for the synthesis of siderophores and spermidine (see Table S8 in the supplemental material), consistent with its activities revealed by experiments (Table S5).
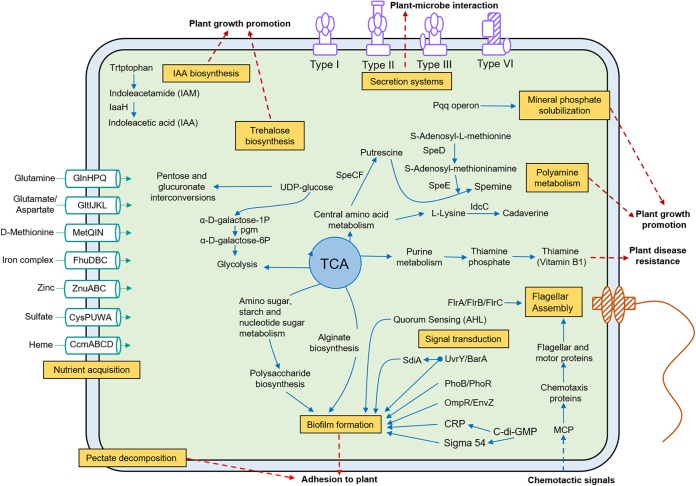
We then sought to understand the specific functions of LTGT-11-2Z that mediate interactions with A. sparsifolia. The genome of this strain was compared with those of the following four previously reported plant endophytes also belonging to the genus Pseudomonas: P. putida W619, an endophytic bacterium of poplar trees (22); P. stutzeri A1501, a rice root-associated bacterium (28); P. putida MTCC5279, a positive PGP rhizobacteria in chickpea (17); and the phylogenetically close strain P. fluorescens 2P24 (20). The resulting Venn diagram revealed that 1,760 Kyoto Encyclopedia of Genes and Genomes (KEGG) genes were shared by LTGT-11-2Z, 2P24, W619, A1501, and MTCC5279 (Fig. 3B). LTGT-11-2Z possessed 63 unique KEGG genes (Fig. 3B; see also Table S9 in the supplemental material). In particular, the colanic acid biosynthesis glycosyl transferase wcaI was only present in LTGT-11-2Z; this gene is involved in colanic acid synthesis, which contributes to biofilm architecture and allows voluminous biofilm formation (31). In addition, two type VI secretion system (T6SS) encoding genes (vasI and impI) were only present in LTGT-11-2Z. An overview of predicted metabolic properties and important transport pathways for interactions between the strain and the host plant is summarized in Fig. 4.
FIG 4.
Overview of predicted metabolism and transport pathways in Pseudomonas sp. LTGT-11-2Z. The metabolic pathways were constructed based on the genes of LTGT-11-2Z annotated using the KEGG database (http://www.genome.ad.jp). The red dashed arrows indicated putative functions or metabolism processes mediating microbe-host interactions.
DISCUSSION
Studies of the contribution of plant-associated microbiomes to plant drought resistance are rare. In the present study, we performed high-throughput 16S rRNA gene amplicon sequencing to describe the rhizosphere and endosphere prokaryotic microbiomes of A. sparsifolia, a typical desert plant that inhabits poor and extremely dry soil environments in northwestern China. We integrated our high-throughput-based assessment of bacterial diversity in the endosphere and culture-dependent functional analyses to identify drought resistance-promoting endophytic bacteria and further mined its genes involved in endophytic colonization and promotion of plant drought resistance.
Bacterial ASV diversity and richness were higher in the rhizosphere than in the root endosphere of A. sparsifolia, indicating that only a limited number of bacteria can adapt to an endophytic lifestyle. A similar result was observed in a previous study (10), which demonstrated a great loss of bacterial diversity and richness from rhizosphere soil to the endosphere compartments of poplar trees. Fitzpatrick et al. (32) found that the rhizosphere exhibited higher diversity and greater evenness of abundance than the endosphere across 30 plant species of 14 families (Amaranthaceae, Apocynaceae, Asparagaceae, Asteraceae, Brassicaceae, Convolvulaceae, Fabaceae, Onagraceae, Plantaginaceae, Poaceae, Polygonaceae, Rosaceae, Solanaceae, and Asteraceae). The rhizosphere of Glaux maritima (Primulaceae), a typical halophytic plant, was discovered to have greater richness and diversity than the endosphere microbial community (33). These results can be explained by the general perspectives that the soil-root interface acts as a selective barrier to determine endosphere community composition and that plant endophytic colonization is limited to specific bacterial species (34).
Members of the phylum Proteobacteria have been found to be enriched in the rhizosphere and root endosphere of a wide range of desert plants, including G. maritima (33) and Phoenix dactylifera (35). Microbes isolated from the root tissues of P. dactylifera significantly increased plant growth under controlled drought stress (35). Consistently, our results revealed that the root endosphere of A. sparsifolia was dominated by Proteobacteria species, which also dominated rhizosphere soil. However, unlike G. maritima (33), the relative abundance of Alphaproteobacteria was lower in the endosphere than in the rhizosphere of A. sparsifolia, and the relative abundance of Gammaproteobacteria was lower in the rhizosphere than in the endosphere, suggesting that members of Gammaproteobacteria, but not members of Alphaproteobacteria, are more effective endophytic colonizers of A. sparsifolia. Moreover, the root endosphere of G. maritima and P. dactylifera were reported to have an abundance of Actinobacteria in the endosphere (33, 35), which is also different from the case of A. sparsifolia. Together, the differences and similarities among the microbes of different desert plant species suggest that both drought environment and plant species influence the recruitment of bacterial endophyte communities in the rhizosphere.
At the genus level, rhizosphere communities were dominated primarily by Kocuria, which has been isolated from rhizosphere soil of various plants inhabiting different environments (2, 5). In contrast, endophytic assemblages were dominated by Pseudomonas, followed by Stenotrophomonas and Achromobacter, all of which are members of the phylum Proteobacteria. The endophytic bacteria Pseudomonas, Stenotrophomonas, and Achromobacter have been isolated from a variety of plant species and may provide beneficial effects for plant growth and health (35, 36). For example, Pseudomonas isolates from Arabidopsis roots showed the ability to adhere and colonize on Arabidopsis and grapevine rhizoplanes (36), and Pseudomonas isolates from Suaeda salsa increased salt stress tolerance and plant growth in cucumber and rice plants (37). These results may indicate common functions among certain microbial taxa in plant roots.
The drought resistance-promoting strain LTGT-11-2Z possesses broad PGP potential, as revealed by comparative genome analysis. Siderophore production is a well-known PGP property (38); polyamine production may contribute to the improvement of plant growth under water stress conditions (39); and lowering plant ethylene levels through ACC deaminase activity is among the major mechanisms employed by PGP bacteria to protect plants against a wide range of environmental stresses (40, 41). LTGT-11-2Z also possesses genes responsible for flagellum biosynthesis, chemotaxis, curli fiber production, and plant cell wall-degrading enzymes, which may be involved in plant adhesion and colonization (24, 27). Certain genes, including T6SS and biofilm formation-related genes, are specific for LTGT-11-2Z to a greater extent than for closely related Pseudomonas strains, implying unique mechanisms that mediate interactions between LTGT-11-2Z and plants. Therefore, our results suggest that LTGT-11-2Z has the potential for use as a biotechnological agent to improve drought resistance in crop plants for arid land agriculture.
Conclusion.
In the present study, we revealed the structure of rhizosphere and root endosphere microbiomes of the desert plant A. sparsifolia. Endosphere microbiomes showed lower diversity than those of the rhizosphere but contained several microbial taxa that were also present in the roots of previously reported desert plants. The Pseudomonas strain LTGT-11-2Z, isolated from A. sparsifolia roots, improved drought resistance in wheat, likely due to synergistic effects of multiple activities related to plant growth promotion and stress resistance. Comparative genomics analysis revealed a subset of genes involved in rhizosphere competence, plant colonization, plant growth promotion, and plant protection. Taken together, these results provide a basis for more detailed studies of the molecular mechanisms responsible for bacterially mediated drought resistance in plants. Further research is needed to evaluate whether drought resistance improvement in crop plants can be achieved by these bacteria under field conditions, which is a prerequisite for their application in agricultural practice to combat drought.
MATERIALS AND METHODS
Sampling.
A. sparsifolia Shap. was collected in August 2014 from Taklamakan Desert, Xinjiang Uyghur Autonomous Region, northwest China. Sixteen independent plant samples with soil attached to the roots were collected at the sampling site and placed in sterile bags. After being transported to the laboratory within 48 h, the rhizosphere soil was collected and stored at −80°C, while the plant material was carefully washed in running water to remove external soil and debris. After drying at room temperature, the roots were separated and subjected to a five-step surface sterilization procedure as described previously (42). Sterility checks were performed by plating the final wash water and placing pieces of the surface-sterilized tissues on Trypticase soy agar (TSA) plates. Sterilized tissues were used immediately for the isolation of endophytic bacteria and stored at −80°C for molecular analysis.
DNA isolation and high-throughput 16S rRNA gene amplicon sequencing.
Rhizosphere soil from 16 A. sparsifolia individuals was separately subjected to DNA extraction by using the E.Z.N.A. soil DNA kit (Omega Bio-Tek Inc., USA) according to the manufacturer’s instructions. Extraction of endobacterial DNA from the root tissues of the 16 independent plants was carried out using a modified bacterial cell enrichment method according to Nissinen et al. (43). For each rhizosphere soil and plant root sample, DNA was extracted in triplicate, and the resulting DNA extracts were mixed together and stored at −20°C for downstream manipulation. Subsequently, 32 DNA samples, including 16 biological replicates, were subjected to PCR amplification. The V3 to V4 hypervariable region of bacterial 16S rRNA gene was amplified using primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GGACTACHVGGGTATCTAAT-3′) (44). The PCR was performed in 50 μl reaction mixture containing 100 ng of template DNA, 1.5 μl of primers (5 μM), 1 μl of KOD DNA polymerase (2.5 U · μl−1) (Toyobo, Osaka, Japan), 5 μl of 10 × KOD buffer, and 5 μl of deoxynucleoside triphosphates (dNTPs) (2.5 mM). The PCR amplification was performed under the following cycling conditions: initial denaturation at 95°C for 2 min, followed by 27 cycles at 98°C for 10 s, 62°C for 30 s, and 68°C for 30 s, and a final extension at 68°C of 10 min. Each DNA sample was amplified in triplicate, mixed into one PCR product, examined by 2% agarose gel, and purified using the AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA). The purified PCR amplicons were quantified with a QuantiFluor-ST fluorometer (Promega Corporation, Madison, WI), pooled at equimolar concentrations, and finally sequenced on an Illumina HiSeq PE250 platform at Gene Denovo Ltd., Co. (Guangzhou, China).
Sequence processing and statistical analysis.
Quality control of the paired-end 16S rRNA amplicon reads was conducted using the NGS QC Toolkit (45). The cutoff value for high-quality filtering was 20. Reads containing more than 30% low-quality bases or unpaired reads were removed. Quality-filtered reads were assembled into error-corrected ASVs using the DADA2 v1.4.0 software (46) to represent unique bacterial taxa. Merged reads were aligned to the SILVA database (47) implemented in the QIIME2 package (46). Taxonomic annotation at different taxonomic levels ranging from phylum to species was performed based on ASV composition and relative abundance. Chimeric sequences and mitochondrial and chloroplast ASVs were removed from all samples. Community richness and diversity indices and rarefaction curves were determined using the “qiime diversity core-metrics-phylogenetic” command for alpha and beta diversity analysis in the QIIME2 package. Relationships between communities were tested using PCoA implemented in the PAST software package based on Bray-Curtis distances (48). Differences in pairwise comparisons between the endosphere and rhizosphere were evaluated using the two-tailed Student’s t test, and all the phyla and the top 30 abundant genera showing significant differences were identified; P values of Student’s t tests were corrected using the Benjamini-Hochberg method.
A. sparsifolia root strain isolation and identification.
After surface sterilization, A. sparsifolia root tissues were cut into small fragments and macerated using a sterile mortar and pestle in sterile distilled water. Macerated samples were serially diluted, spread-plated onto 10% tryptic soy agar (TSA) and Reasoner’s 2A agar (Difco) plates supplemented with 50 μg · ml−1 cycloheximide, and incubated at 28°C for 1 week. After incubation, colonies were picked from the plates and subcultured to obtain pure isolates. For 16S rRNA gene sequencing, genomic DNA was extracted, and the 16S rRNA gene sequence was amplified using the bacterial universal primers 27F and 1492R, as previously described (49). The obtained 16S rRNA gene sequences were compared with available 16S rRNA gene sequences from the EzBioCloud server using the BLASTn online tool (https://www.ezbiocloud.net/) (49). All isolates were assigned to the genus level based on the closest match in the NCBI online database.
Screening of strains improving drought resistance in wheat and related experiments.
Seeds of the winter wheat T. aestivum were surface sterilized and germinated as previously described (50). Three-day-old seedlings of uniform size were selected and planted in a sterilized soil mixture (3:1 soil to sand) in a 14-cm plastic pot. Seedlings were maintained in a growth chamber under a 14-h/10-h light/dark photoperiod. One wheat individual was grown in each pot. After 7 days of growth under normal watering conditions, seedlings were fertilized with single bacterial cultures in sterilized tap water at 108 CFU · g−1 soil, and subjected to normal watering for 10 days prior to drought stress by withholding water irrigation. After observation for up to 7 days, the PGP effects of seven isolated bacterial strains were noted. ACC deaminase activity was determined according to the methods by Penrose and Glick (51), which measures the amount of α-ketobutyrate produced when ACC is cleaved by ACC deaminase; the abundance of α-ketobutyrate was determined by comparing the absorbance at 540 nm of a sample to a standard curve. Exopolysaccharide production and siderophore production, and resistance to abiotic stresses (temperatures and osmotic stress), were determined as described in our previous study (50).
After the selection of Pseudomonas sp. LTGT-11-2Z for further study, additional experiments with more replicates were performed to confirm its PGP and drought resistance promotion. Four wheat individuals were grown in each pot and each treatment included three pots as three replicates. After growth at under normal watering conditions for 7 days, inoculated plants and noninoculated control plants were regularly watered for 10 days and subjected to drought stress by withholding water irrigation for up to 7 days. Plants that were properly irrigated throughout the experiment were also used as a positive control. When the noninoculated plants had become severely wilted, water irrigation was resumed for 1 day, and then plant health was assessed and photographed. Plants exposed to each treatment were then harvested for biomass and length measurement. Statistical analysis was performed using a two-tailed Student’s t test to compare data from plants inoculated with LTGT-11-2Z and from noninoculated plants under drought treatment.
To visualize whether the bacteria could adhere and colonize on the wheat root, confocal microscopy was conducted. After 7 days of water deprivation, only inoculated wheat root (17 days old) was harvested. The strain LTGT-11-2Z was labeled with green fluorescent protein (GFP) transformed with the pKEN-GFP-mut3 plasmid and inoculated on wheat roots. Wheat roots were gently washed to remove weakly bound bacteria and then stained with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific, USA) by incubation in the dye (300 nM in phosphate-buffered saline) for 1 min. Bacterial cells adhering to the wheat root were observed using a laser scanning confocal microscope (LSM710; Carl Zeiss, Germany). The excitation/emission wavelengths of the DAPI and GFP channels were 358/461 nm and 488/520 nm, respectively.
Whole-genome sequencing and genome analysis of the selected Pseudomonas strain.
Genomic DNA of Pseudomonas sp. LTGT-11-2Z was extracted as described previously (52). Whole-genome sequencing was performed using a PacBio RS II and Illumina HiSeq X Ten system. In the PacBio sequencing, a 10-kb insert size library was constructed, and in the Illumina sequencing, a 350-bp short insert library was constructed. The genome coverage in the PacBio sequencing was >100×, and that in the Illumina sequencing was 500×. De novo assembly of the genome was conducted using SPAdes (version 3.12.0) (53), which assembles PacBio and Illumina sequences together, using the options “–pacbio” and “–pe.” After assembly, each genome contained a single contig, and no plasmid sequences were identified.
Prediction of protein-coding genes was performed using the software Prodigal (version 2.6.3) (53). All of the reference genomes used for comparison were downloaded from NCBI GenBank. The endophyte genomes used as references were selected by referring to a previous work (24). The numbers of genes in the functional categories were calculated based on the annotations by searching against the Clusters of Orthologous Genes (COG) (54), KEGG (55), Carbohydrate-Active enZYmes (CAZy) (56), and NCBI nr databases on a local server. Protein-coding sequences were BLASTp searched against the KEGG database using an E value cutoff 1E−7. The unique and common KEGG genes for the four Pseudomonas genomes were identified using Venn analysis (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Accession number(s).
The raw 16S rRNA gene amplicon sequences have been deposited in the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA515584. The genome sequence of LTGT-11-2Z has been deposited in GenBank under accession number CP033104.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 31770121 and 31725003) and by the Fundamental Research Funds for the Central Universities (grants Z111021801 and 2452018154).
We thank the Teaching and Research Core Facility at College of Life Sciences, NWAFU for their support in this work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Vinocur B, Altman A. 2005. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Vurukonda SS, Vardharajula S, Shrivastava M, SkZ A. 2016. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol Res 184:13–24. doi: 10.1016/j.micres.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Rubin RL, van Groenige KJ, Hungate BA. 2017. Plant growth promoting rhizobacteria are more effective under drought: a meta-analysis. Plant Soil 416:309–323. doi: 10.1007/s11104-017-3199-8. [DOI] [Google Scholar]
- 4.Eisenstein M. 2013. Discovery in a dry spell. Nature 501:S7–S9. doi: 10.1038/501S7a. [DOI] [PubMed] [Google Scholar]
- 5.Coleman-Derr D, Tringe SG. 2014. Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Front Microbiol 5:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.East R. 2013. Soil science comes to life. Nature 501:S18–S19. doi: 10.1038/501S18a. [DOI] [PubMed] [Google Scholar]
- 7.Ngumbi E, Kloepper J. 2016. Bacterial-mediated drought tolerance: current and future prospects. Appl Soil Ecol 105:109–125. doi: 10.1016/j.apsoil.2016.04.009. [DOI] [Google Scholar]
- 8.Kaushal M, Wani SP. 2016. Rhizobacterial-plant interactions: strategies ensuring plant growth promotion under drought and salinity stress. Agric Ecosyst Environ 231:68–78. doi: 10.1016/j.agee.2016.06.031. [DOI] [Google Scholar]
- 9.Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Ait Barka E. 2005. Endophytic colonization of Vitis vinifera L. by a plant growth-promoting bacterium, Burkholderia sp. strain PsJN. Appl Environ Microbiol 71:1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckers B, Op De Beeck M, Weyens N, Boerjan W, Vangronsveld J. 2017. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5:25. doi: 10.1186/s40168-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. 2008. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett 278:1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 12.Gagné-Bourque F, Bertrand A, Claessens A, Aliferis KA, Jabaji S. 2016. Alleviation of drought stress and metabolic changes in Timothy (Phleum pratense L.) colonized with Bacillus subtilis B26. Front Plant Sci 7:584. doi: 10.3389/fpls.2016.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker P, Raaijmakers JM. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 14.Dutkiewicz J, Mackiewicz B, Lemieszek MK, Golec M, Milanowski J. 2016. Pantoea agglomerans: a mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann Agric Environ Med 23:206–222. doi: 10.5604/12321966.1203879. [DOI] [PubMed] [Google Scholar]
- 15.Marasco R, Rolli E, Vigani G, Borin S, Sorlini C, Ouzari H, Zocchi G, Daffonchio D. 2013. Are drought-resistance promoting bacteria cross-compatible with different plant models? Plant Signal Behav 8:e26741. doi: 10.4161/psb.26741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naveed M, Mitter B, Reichenauer TG, Wieczorek K, Sessitsch A. 2014. Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD17. Environ Exp Bot 97:30–39. doi: 10.1016/j.envexpbot.2013.09.014. [DOI] [Google Scholar]
- 17.Tiwari S, Lata C, Chauhan PS, Nautiyal CS. 2016. Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol Biochem 99:108–117. doi: 10.1016/j.plaphy.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Hou X, Jia Z, Han Q, Li R, Wang W, Li Y. 2011. Effects of rotational tillage practices on soil water characteristics and crop yields in semi-arid areas of north-west China. Soil Res 49:625–632. doi: 10.1071/SR11143. [DOI] [Google Scholar]
- 19.Wu H, Zhang Y, Zhang W, Pei X, Zhang C, Jia S, Li W. 2015. Transcriptomic analysis of the primary roots of Alhagi sparsifolia in response to water stress. PLoS One 10:e0120791. doi: 10.1371/journal.pone.0120791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei HL, Wang Y, Zhang LQ, Tang W. 2004. Identification and characterization of biocontrol bacterial strain 2P24 and CPF-10. Acta Phytopathol Sin 34:80–85. [Google Scholar]
- 21.Zhang L, Zhong J, Liu H, Xin K, Chen C, Li Q, Wei Y, Wang Y, Chen F, Shen X. 2017. Complete genome sequence of the drought resistance-promoting endophyte Klebsiella sp. LTGPAF-6F. J Biotechnol 246:36–39. doi: 10.1016/j.jbiotec.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, van der Lelie D. 2009. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microbiol 75:748–757. doi: 10.1128/AEM.02239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weilharter A, Mitter B, Shin MW, Chain PS, Nowak J, Sessitsch A. 2011. Complete genome sequence of the plant-growth promoting endophyte Burkholderia phytofirmans strain PsJN. J Bacteriol 193:3383–3384. doi: 10.1128/JB.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitter B, Petric A, Shin MW, Chain PSG, Hauberg-Lotte L, Reinhold-Hurek B, Nowak J, Sessitsch A. 2013. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci 4:120. doi: 10.3389/fpls.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko T, Minamisawa K, Isawa T, Nakatsukasa H, Mitsui H, Kawaharada Y, Nakamura Y, Watanabe A, Kawashima K, Ono A, Shimizu Y, Takahashi C, Minami C, Fujishiro T, Kohara M, Katoh M, Nakazaki N, Nakayama S, Yamada M, Tabata S, Sato S. 2010. Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res 17:37–50. doi: 10.1093/dnares/dsp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouts DE, Tyler HL, De Boy RT, Daugherty S, Ren Q, Badger JH, Durkin AS, Huot H, Shrivastava S, Kothari S, Dodson RJ, Mohamoud Y, Khouri H, Roesch LF, Krogfelt KA, Struve C, Triplett EW, Methé BA. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsielle pneumoniae 342 and virulence predictions verified in mice. PLoS Genet 4:e1000141. doi: 10.1371/journal.pgen.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taghavi S, van der Lelie D, Hoffman A, Zhang YB, Walla MD, Vangronsveld J, Newman L, Monchy S. 2010. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet 6:e1000943. doi: 10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Y, Yang J, Dou Y, Chen M, Ping S, Peng J, Lu W, Zhang W, Yao Z, Li H, Liu W, He S, Geng L, Zhang X, Yang F, Yu H, Zhan Y, Li D, Lin Z, Wang Y, Elmerich C, Lin M, Jin Q. 2008. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci U S A 105:7564–7569. doi: 10.1073/pnas.0801093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause A, Ramakumar A, Bartels D, Battistoni F, Bekel T, Boch J, Böhm M, Friedrich F, Hurek T, Krause L, Linke B, McHardy AC, Sarkar A, Schneiker S, Syed AA, Thauer R, Vorhölter F-J, Weidner S, Pühler A, Reinhold-Hurek B, Kaiser O, Goesmann A. 2006. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat Biotechnol 24:1384–1391. doi: 10.1038/nbt1243. [DOI] [PubMed] [Google Scholar]
- 30.Bertalan M, Albano R, de Padua V, Rouws L, Rojas C, Hemerly A, Teixeira K, Schwab S, Araujo J, Oliveira A, França L, Magalhães V, Alquéres S, Cardoso A, Almeida W, Loureiro MM, Nogueira E, Cidade D, Oliveira D, Simão T, Macedo J, Valadão A, Dreschsel M, Freitas F, Vidal M, Guedes H, Rodrigues E, Meneses C, Brioso P, Pozzer L, Figueiredo D, Montano H, Junior J, Filho GS, Flores VMQ, Ferreira B, Branco A, Gonzalez P, Guillobel H, Lemos M, Seibel L, Macedo J, Alves-Ferreira M, Sachetto-Martins G, Coelho A, Santos E, Amaral G, Neves A, Pacheco AB, Carvalho D, Lery L, Bisch P, Rössle RC, Ürményi T, Pereira AR, Silva R, Rondinelli E, von Krüger W, Martins O, Baldani JI, Ferreira P. 2009. Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450. doi: 10.1186/1471-2164-10-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 32.Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson M. 2018. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci U S A 115:E1157–E1165. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K, Shiwa Y, Ishige T, Sakamoto H, Tanaka K, Uchino M, Tanaka N, Oguri S, Saitoh H, Tsushima S. 2018. Bacterial diversity associated with the rhizosphere and endosphere of two Halophytes: Glaux maritima and Salicornia europaea. Front Microbiol 9:2878. doi: 10.3389/fmicb.2018.02878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T. 2015. Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53:403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- 35.Cherif H, Marasco R, Rolli E, Ferjani R, Fusi M, Soussi A, Mapelli F, Blilou I, Borin S, Boudabous A, Cherif A, Daffonchio D, Ouzari H. 2015. Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ Microbiol Rep 7:668–678. doi: 10.1111/1758-2229.12304. [DOI] [PubMed] [Google Scholar]
- 36.Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R, Pierotti Cei F, Borin S, Sorlini C, Zocchi G, Daffonchio D. 2015. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol 17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 37.Yuan Z, Druzhinina IS, Labbé J, Redman R, Qin Y, Rodriguez R, Zhang C, Tuskan GA, Lin F. 2016. Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci Rep 6:32467. doi: 10.1038/srep32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Heejung C, Ulas K, Dominique L, Benjamin PB, Mary KF, Trent RN, Eoin LB. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhou C, Ma Z, Zhu L, Xiao X, Xie Y, Zhu J, Wang J. 2016. Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. Int J Mol Sci 17:976. doi: 10.3390/ijms17060976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glick BR. 2014. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Bouffaud ML, Renoud S, Dubost A, Moënne-Loccoz Y, Muller D. 2018. 1-Aminocyclopropane-1-carboxylate deaminase producers associated to maize and other Poaceae species. Microbiome 6:114. doi: 10.1186/s40168-018-0503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Wang Y, Wei L, Wang Y, Shen X, Li S. 2013. Taibaiella smilacinae gen. nov., sp. nov., an endophytic member of the family Chitinophagaceae isolated from the stem of Smilacina japonica, and emended description of Flavihumibacter petaseus. Int J Syst Evol Microbiol 63:3769–3776. doi: 10.1099/ijs.0.051607-0. [DOI] [PubMed] [Google Scholar]
- 43.Nissinen RM, Männistö MK, van Elsas JD. 2012. Endophytic bacterial communities in three arctic plants from low arctic fell tundra are cold-adapted and host-plant specific. FEMS Microbiol Ecol 82:510–522. doi: 10.1111/j.1574-6941.2012.01464.x. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Lee C, Kim J, Hwang S. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 45.Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammer Ø, Harper DAT, Ryan PD. 2008. PAST—palaeontological statistics, ver. 1.89. Paleontological Museum, University of Oslo, Oslo, Norway: http://folk.uio.no/ohammer/past/index.html. [Google Scholar]
- 49.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Xin K, Liu H, Cheng J, Shen X, Wang Y, Zhang L. 2017. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci Rep 7:41564. doi: 10.1038/srep41564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penrose DM, Glick BR. 2003. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 52.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D, Chen GL, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.