Summary
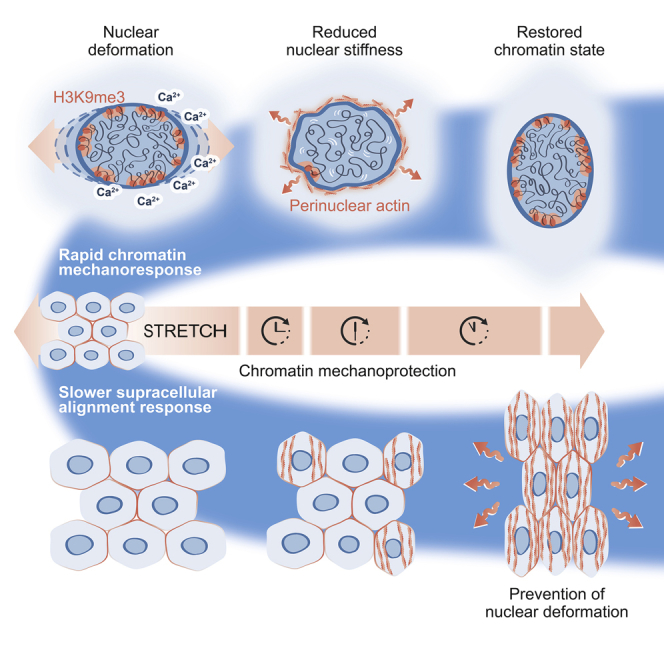
Tissue homeostasis requires maintenance of functional integrity under stress. A central source of stress is mechanical force that acts on cells, their nuclei, and chromatin, but how the genome is protected against mechanical stress is unclear. We show that mechanical stretch deforms the nucleus, which cells initially counteract via a calcium-dependent nuclear softening driven by loss of H3K9me3-marked heterochromatin. The resulting changes in chromatin rheology and architecture are required to insulate genetic material from mechanical force. Failure to mount this nuclear mechanoresponse results in DNA damage. Persistent, high-amplitude stretch induces supracellular alignment of tissue to redistribute mechanical energy before it reaches the nucleus. This tissue-scale mechanoadaptation functions through a separate pathway mediated by cell-cell contacts and allows cells/tissues to switch off nuclear mechanotransduction to restore initial chromatin state. Our work identifies an unconventional role of chromatin in altering its own mechanical state to maintain genome integrity in response to deformation.
Keywords: stem cells, mechanotransduction, chromatin, heterochromatin, mechanoprotection, DNA damage, nuclear lamina, nuclear architecture, nuclear mechanics
Graphical Abstract
Highlights
-
•
Stretch triggers amplitude-dependent supracellular and nuclear mechanoresponses
-
•
H3K9me3 heterochromatin mediates nuclear stiffness and membrane tension
-
•
Nuclear deformation-triggered Ca2+ alters chromatin rheology to prevent DNA damage
-
•
Supracellular alignment redistributes stress to restore chromatin state
When tissues are stretched, cells respond via two distinct mechanosensory mechanisms to protect the genome from damage and maintain tissue homeostasis. First, rapid heterochromatin-mediated mechanosensing, independent of known cellular mechanosensors, drives calcium-dependent nuclear softening. If the mechanical stress persists, a second, tissue-level reorganization occurs, mediated by cell-cell contacts to redistribute mechanical energy to prevent force transmission to the nucleus.
Introduction
Epithelial tissues are load-bearing elements that undergo large-scale, force-driven deformations (Anlaş and Nelson, 2018, Jor et al., 2013). Although accumulation of mechanical stress within tissues can compromise tissue integrity (Casares et al., 2015, Harris et al., 2012), epithelial sheets can sustain extreme deformation and mechanical stress without signs of damage (Latorre et al., 2018, Le et al., 2016). This is in strong contrast to cancer cells, where mechanical deformations have been shown to induce nuclear rupture and DNA damage (Denais et al., 2016, Raab et al., 2016, Xia et al., 2018). Thus, it is likely that noncancerous epithelial cells, such as skin epidermis stem/progenitor cells (EPCs) that are exposed to large-scale, dynamic mechanical forces in their natural microenvironment (Maiti et al., 2016, Obropta and Newman, 2016), have acquired robust mechanisms of genome mechanoprotection.
The nucleus is separated from the cytoplasm by the nuclear envelope (NE), a double membrane continuous with the endoplasmic reticulum (ER). The nuclear lamina, which underlies the NE, is composed of a meshwork of intermediate filament proteins called Lamins, and it functions as a structural and mechanical scaffold for the nucleus (Gruenbaum and Foisner, 2015). The nucleus is mechanically tethered to the extracellular environment, adhesion receptors, and the contractile cytoskeleton through adaptor-protein-mediated interactions at the NE. Additional protein interactions link chromatin to the nuclear lamina (Buchwalter et al., 2019). Inside the nucleus, the genome is packaged and organized nonrandomly to ensure efficient gene expression silencing. The organized chromatin compartments adopt specific positions relative to the nuclear periphery, and a dense layer of heterochromatin can be found underneath the nuclear lamina of most mammalian cells (Buchwalter et al., 2019, van Steensel and Belmont, 2017). These lamina-associated heterochromatin regions are characterized by low gene density, low transcriptional activity, and enrichment of repressive histone modifications H3K9me2, H3K9me3, and H3K27me3 (Buchwalter et al., 2019). Consequently, disruption of methyltransferases that deposit H3K9me2 (Ehmt2/G9a) and H3K9me3 (Setdb1, Suv39H1, and Suv39H2) impair gene silencing and chromatin positioning at the lamina (Bian et al., 2013, Nicetto et al., 2019, Yokochi et al., 2009).
Recent work indicates that extrinsic microenvironment-derived forces are capable of deforming and remodeling chromatin to change global patterns of gene expression (Miroshnikova et al., 2017). However, how mechanical stress is dissipated within the nucleus, and how chromatin responds to and is protected against mechanical stress, are less known. To identify such mechanisms and properties, we exposed EPC monolayers to increasing amplitudes of physiologically relevant cyclic, uniaxial mechanical stretch (Liu et al., 2017, Wessendorf and Newman, 2012). We observed that stretch triggers immediate nuclear deformation that leads to Piezo1-mediated calcium release from the ER to reduce lamina-associated H3K9me3 heterochromatin and subsequent nuclear softening. Inhibiting calcium influx or increasing heterochromatin levels by forced expression of the H3K9me3 methyltransferase Suv39H1 results in failure to modify chromatin mechanics, triggering DNA damage. Long-term exposure to high-amplitude stretch leads to monolayer alignment perpendicular to stretch in a process independent of the nuclear deformation pathway and driven by cell-cell adhesion. This supracellular patterning is essential to redistribute mechanical stress away from the nucleus, allowing the cells to restore their steady-state nuclear and chromatin architecture for long-term mechanoprotection.
Results
Cells Show a Rapid Chromatin and a Slow Supracellular Mechanoresponse with Distinct Amplitude Dependencies
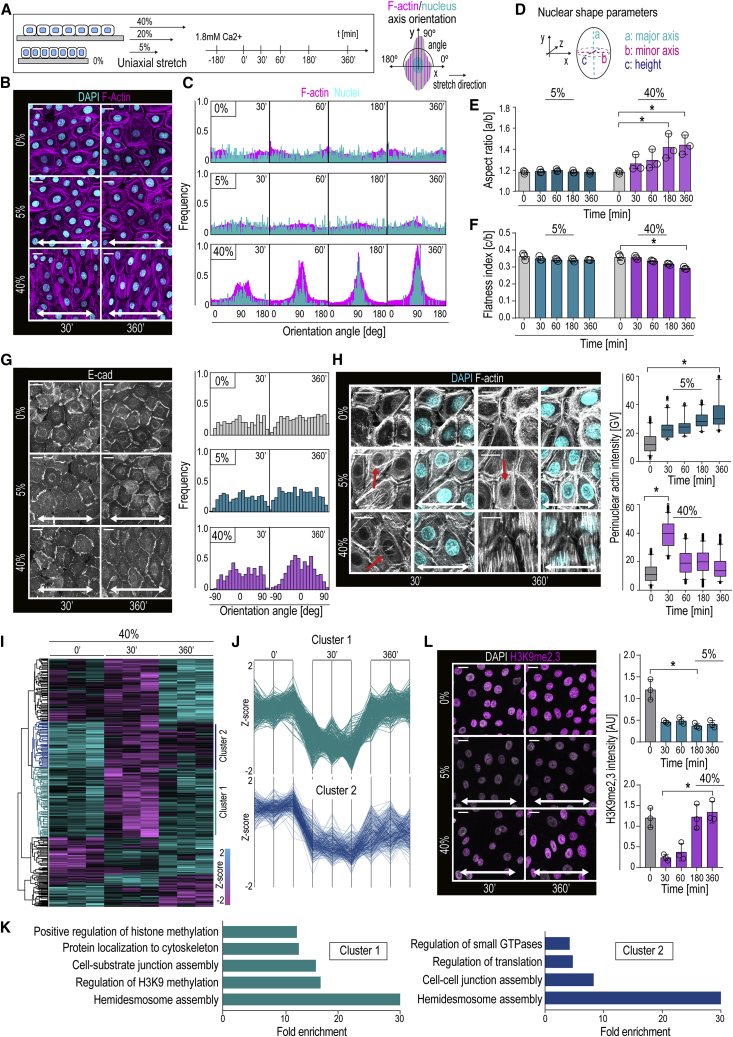
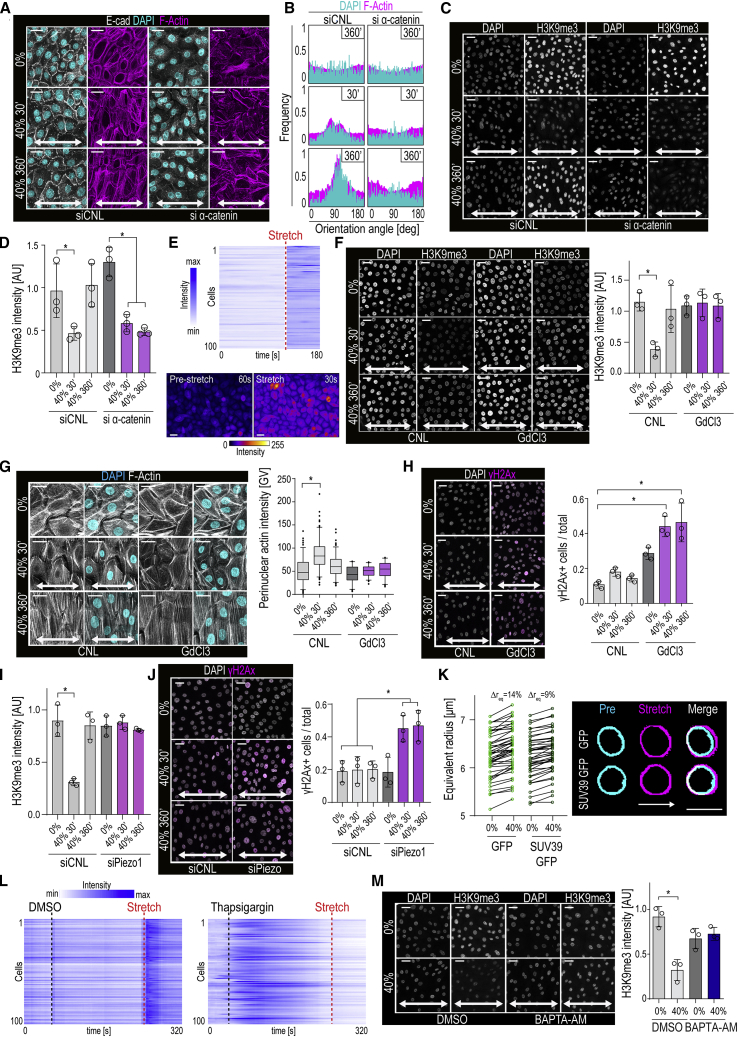
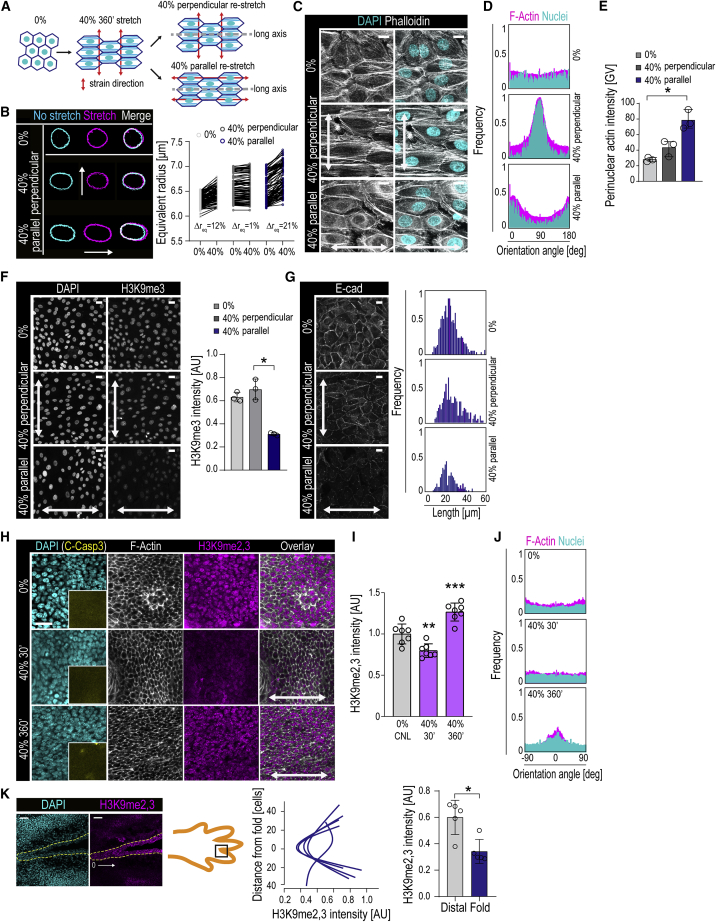
Quantification of EPC monolayer features in response to several uniaxial stretch amplitudes revealed gradual, time-dependent reorientation of filamentous actin (F-actin) and nuclei perpendicular to the direction of stretch (Figures 1A–1C) (De et al., 2008, Faust et al., 2011, Noethel et al., 2018, Wang et al., 2001). Monolayer alignment was initiated at 30 min and completed at 360 min of continuous cyclic (100 mHz) 40% stretch (Figure 1B, 1C). In contrast, 5% stretch was insufficient to trigger alignment, even at longer timescales (Figures 1B, 1C, and S1A), whereas 20% stretch produced an intermediate phenotype (Figure S1B). The stretch-induced reorientation of nuclei was associated with increased nuclear aspect ratio and flattening (Figures 1D–1F), whereas nuclear volume was largely unaltered (Figure S1C). The supracellular monolayer reorganization also included reorientation of adherens junctions into a 45° angle to the stretch axis (Figure 1G).
Figure 1.
Cells Show a Rapid Chromatin and a Slow Supracellular Mechanoresponse with Distinct Amplitude Dependencies
(A) Stretch experiment design and quantification strategy of F-actin and nuclear axis orientation.
(B) Representative F-actin (phalloidin) and DAPI images of cells exposed to uniaxial stretch.
(C) Quantification of images in (B) shows time-dependent reorientation of F-actin and nuclear major axes perpendicular to 40% stretch direction (frequency distribution of >500 cells/condition pooled across three independent experiments).
(D) Parameters measured from 3D nuclear images. Aspect ratio was calculated as a/b and flatness index as c/b.
(E and F) Quantification of nuclear aspect ratio (E) and flatness index (F) (n = 3 independent experiments with > 365 cells/condition/experiment).
(G) Representative E-cadherin (E-cad) images and quantification show time-dependent reorientation of E-cad-positive adherens junctions 45° away from 40% stretch direction (frequency distribution of 380 cells/condition pooled across three independent experiments; ∗p = 0.0390, Friedman/Dunn’s).
(H) Representative F-actin images and quantification illustrating sustained and transient perinuclear actin polymerization (red arrows) at 5% and 40% stretch, respectively (n > 300 cells/condition pooled across three independent experiments; ∗p = 0.0343, Friedman/Dunn’s).
(I) Heatmap and Euclidian distance dendrogram of phosphosites quantified by mass spectrometry at 30 and 360 min of 40% stretch (n = 3 independent experiments; padj cutoff = 0.05).
(J) Distance-based clustering of phosphosites shows a cluster of transiently decreased phosphosites at 30 min (cluster 1) and a cluster with sustained decrease at 30 and 360 min (cluster 2).
(K) GO terms of clusters 1 and 2.
(L) Representative images and quantification show a sustained and transient decrease in H3K9me2,3 in EPC monolayers subjected to 5% and 40% stretch, respectively (n = 3 independent experiments with >200 cells/condition/experiment; ∗p = 0.0451, Friedman/Dunn’s).
Bar graphs show mean ± SD, boxplots show 95% confidence interval, scale bars represent 10 μm, and white arrows indicate stretch direction. AU, arbitrary units; GV, gray values.
See also Figure S1.
Figure S1.
Characterization of EPC Monolayer Changes in Response to Stretch, Related to Figure 1
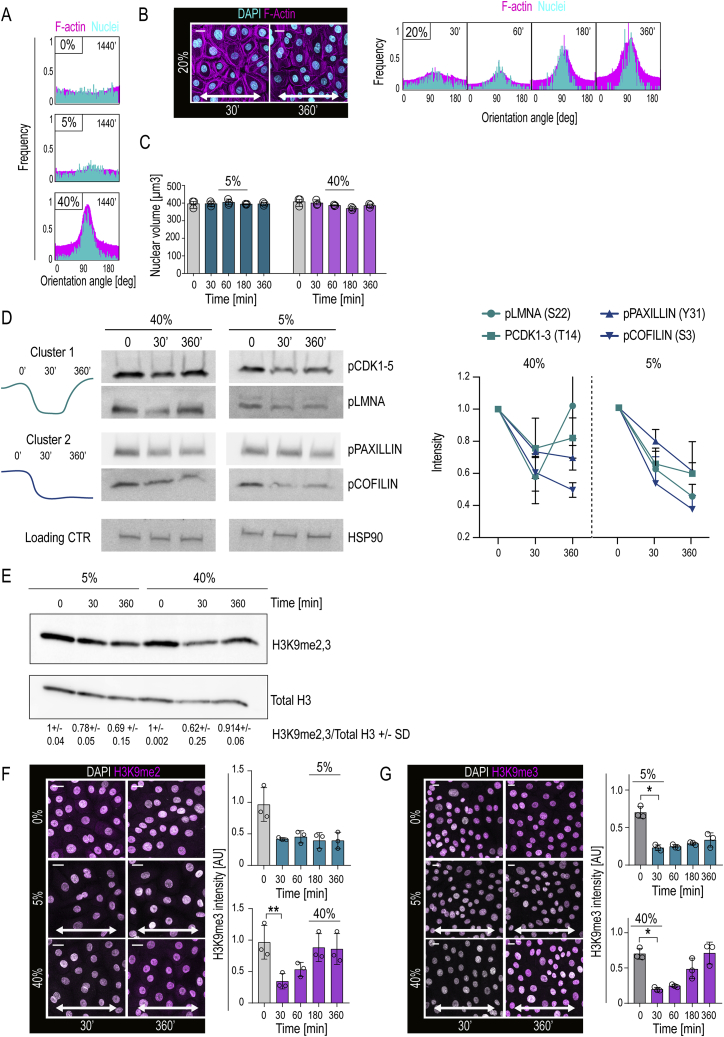
(A) Quantifications of F-actin (phalloidin; magenta) and nuclear (dapi; cyan) orientation after 1440 min of uniaxial cyclic stretch (0.1 Hz) at indicated amplitudes (frequency distribution with n > 300 cells/condition pooled across 3 independent experiments). Note absence of alignment at 5% stretch.
(B) Representative immunofluorescence images and quantification of F-actin (phalloidin; magenta) and nuclear (dapi; cyan) orientation in EPC monolayers exposed to 20% stretch from experiments shown in Figures 1B and 1C (frequency distribution with n > 300 cells/condition pooled across 3 independent experiments).
(C) Quantification of nuclear volume (n = 3 independent experiments with 300 cells/condition/experiment).
(D) Western blot analyses and quantification of selected phosphoproteins found altered in mass spectrometry experiments. Note transient versus sustained downregulation of Cluster 1 and 2 phosphoproteins, respectively, at 40% stretch, whereas all phosphoproteins show sustained response in 5% (n = 3 independent experiments).
(E) Western blot analyses and quantification of H3K9me2,3 levels in EPC monolayers exposed to uniaxial stretch at indicated amplitudes and times (n = 3 independent experiments).
(F) Representative immunofluorescence images and quantification of H3K9me2 in cells exposed to 5% or 40% stretch for indicated times (n = 3 independent experiments with > 200 cells/condition/experiment; ∗∗p = 0.0078, Friedman/Dunn’s).
(G) Representative immunofluorescence images and quantification of H3K9me3 in cells exposed to 5% or 40% stretch for indicated times (n = 3 independent experiments with > 350 cells/condition/experiment; ∗∗p = 0.0393, Friedman/Dunn’s). Bar graphs and dot plots show mean ± SD, scale bars represent 10 μm, white arrows indicate stretch direction, AU = arbitrary units.
Closer inspection of cytoskeletal architecture revealed F-actin specifically around the nucleus (Figure 1H). Intriguingly, although 5% stretch was not sufficient to induce F-actin alignment, this regime did trigger perinuclear actin polymerization. However, in contrast to 40% stretch, the perinuclear actin ring in the 5% condition persisted over time (Figure 1H), suggesting that perinuclear actin polymerization occurs faster and with lower stretch amplitude than supracellular alignment.
To dissect intracellular signaling dynamics that mediate the two distinct patterns of stretch responses, we performed phosphoproteomic analyses on cells exposed to 40% stretch for 30 and 360 min and identified 1,461 differentially regulated phosphosites (Figure 1I; Table S1). K-means clustering revealed dynamic, time-dependent regulation of the phosphoproteome, where the largest cluster of 410 phosphosites showed a transient downregulation at 30 min followed by full recovery at 360 min (Figure 1I, J). Intriguingly, among the most enriched Gene Ontology (GO) terms in this cluster were regulation of histone H3 lysine 9 di- and trimethylation (H3K9me2,3) (Figure 1K). These terms included altered phosphorylation of Lamins A, B1, and B2 and the H3K9 methyltransferase Setdb1 (Table S1). A second large phosphosite cluster that remained downregulated also at 360 min, corresponding to the dynamics of supracellular alignment, contained adhesion and actin regulators such as paxillin, cofilin, and myosin-9 (Figure 1J, K). In addition, both clusters contained hemidesmosome components such as integrin β4, collagen XVII, and plectin (Table S1) (Rottner et al., 2017, Walko et al., 2015). Changes in cluster-specific temporal phosphorylation dynamics were validated by western blotting for representative proteins (Figure S1D).
As the phosphoproteome analysis implicated changes in H3K9me2,3, we next examined changes in constitutive heterochromatin (Becker et al., 2016, Simon and Kingston, 2013) and found that both 40% and 5% stretch induced a strong reduction in H3K9me2,3 levels at 30 min (Figures 1L and S1E). Separate analyses of H3K9 di- and trimethylation revealed alteration of both states (Figures S1F and S1G), with a more substantial effect on H3K9me3, leading us to focus on that modification. Similar to perinuclear actin polymerization, the decrease in H3K9me3 with 40% stretch was transient and reversed after 180–360 min of continuous stretch. In contrast, 5% stretch induced sustained H3K9me3 reduction (Figure 1L, S1F and G). Consistent with the sustained heterochromatin and perinuclear actin changes in 5% stretch, the phosphorylation changes that were reversible in 40% remained downregulated in 5% stretch also at 360 min (Figure S1D).
Collectively, these data indicate that the EPC mechanoresponse under high-amplitude cyclic uniaxial stretch consists of a transient perinuclear actin, NE, and heterochromatin response and a slower, sustained supracellular alignment response. While low-amplitude cyclic stretch is insufficient to induce supracellular alignment, it triggers heterochromatin and NE changes, indicating distinct amplitude dependencies.
Stretch-Triggered Heterochromatin Changes Occur Mainly at Non-coding Regions with No Substantial Correlation with Transcriptional Changes
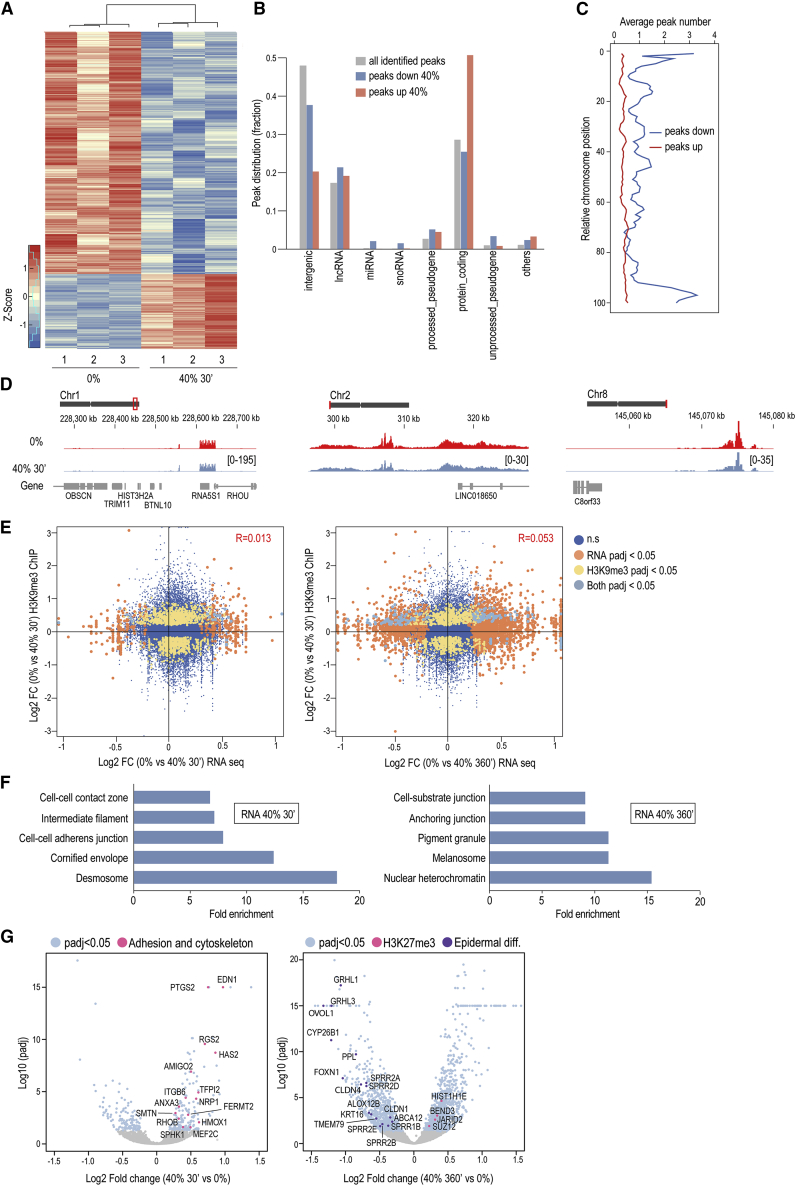
To characterize changes in H3K9me3-heterochromatin, we mapped genome-wide changes in H3K9me3 using chromatin immunoprecipitation sequencing (ChIP-seq) of cells stretched for 30 min at 40%. In addition, we generated corresponding RNA sequencing (RNA-seq) datasets of cells stretched for 30 and 360 min at 40%. Consistent with the phosphoproteome and immunofluorescence data, we observed a substantial overall reduction in H3K9me3 occupancy, as defined by differential peaks (padj < 0.05; Figure 2A; Table S2). Interestingly, reduced H3K9me3 occupancy was found particularly at non-coding regions, whereas the few regions with increased occupancy upon stretch were predominantly at protein-coding genes (Figure 2B). Reduced H3K9me3 occupancy was biased toward chromosome ends (Figures 2C and 2D), consistent with high H3K9me3 levels at subtelomeric regions (Cubiles et al., 2018). In line with their predominant location at non-coding regions, the stretch-regulated H3K9me3 peaks showed no substantial correlation with the stretch-regulated transcriptome (Figure 2E). In contrast, those few regions with increased occupancy overlapped with the transcriptome, with a corresponding decrease in transcript levels of genes with increased H3K9me3 occupancy (Figure S2A), as would be predicted from the repressive function of H3K9me3. Interestingly, these genes with reduced expression were particularly enriched on chromosome 19 (Figure S2B), which is found positioned centrally within the nucleus (Croft et al., 1999).
Figure 2.
Stretch-Triggered Heterochromatin Changes Occur Mainly at Non-coding Regions with No Substantial Correlation with Transcriptional Changes
(A) Distance-based clustering of differential H3K9me3 occupancy from ChIP-seq shows widespread decrease in H3K9me3 upon 40% stretch.
(B) Biotype distribution of all identified H3K9me3 peaks and peaks increased or decreased by stretch.
(C) Differential H3K9me3 peaks plotted according to chromosome position.
(D) Genome browser views of representative H3K9me3 peaks with reduced intensity upon stretch: ribosomal RNA (left panel), non-coding intergenic region (middle panel), and subtelomeric region (right panel). Red boxes show chromosome location, and peak intensity range is in brackets.
(E) Correlation plots and quantification of H3K9me3 and differential gene expression overlap after 30 min (left panel) and 360 min (right panel) of 40% stretch shows no significant correlation between transcriptional changes and altered H3K9me3 occupancy.
(F) GO-term enrichment of significantly altered genes from RNA-seq after 30 min (left) and 360 min (right) of 40% stretch.
(G) Volcano plots of differentially expressed transcripts (padj < 0.05) with examples of cell adhesion, H3K27me3 regulators, and epidermal differentiation genes highlighted.
See also Figure S2.
Figure S2.
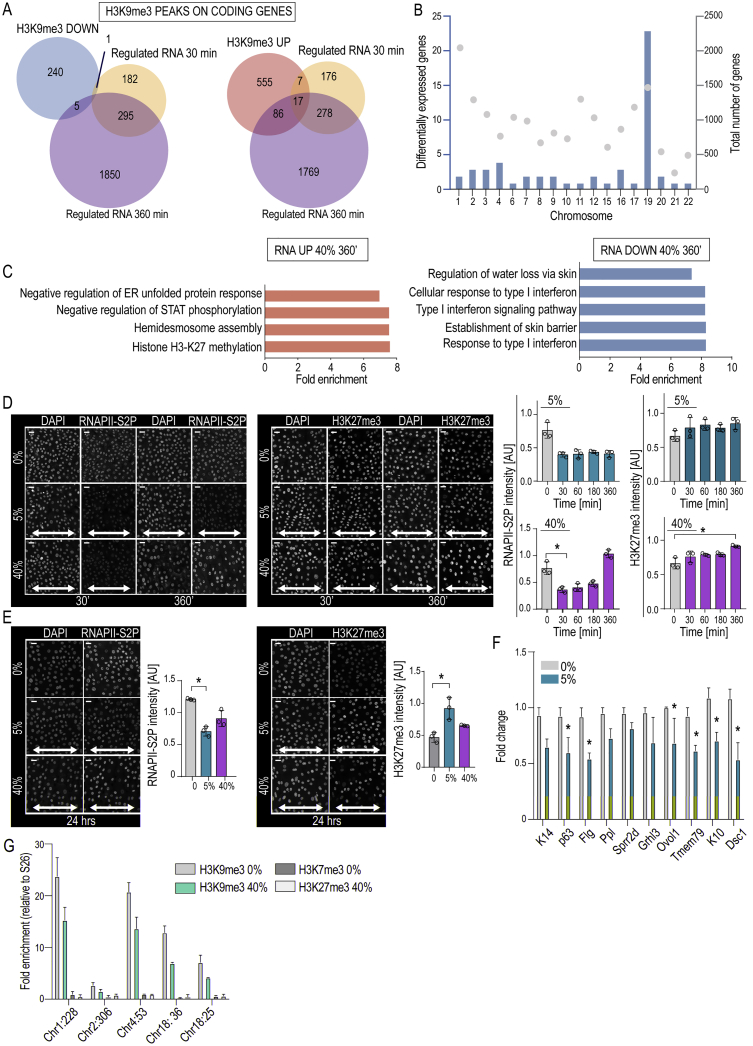
Analyses of Epigenetic and Transcriptional Changes in Stretched Cells, Related to Figure 2
(A) Venn diagrams of overlap between differential H3K9me3 peaks found on protein-coding genes upon 30 min 40% stretch and differentially expressed genes after 30 or 360 min of 40% stretch. Note lack of overlap between H3K9me3 peaks down upon stretch and gene expression changes.
(B) Quantification of chromosome distribution of genes that have decreased levels of H3K9me3 as determined by ChIPseq and corresponding increased levels of mRNA as determined by RNaseq. Grey dots show total amount of genes present in each chromosome.
(C) GO-term analysis of genes upregulated (left panel) or downregulated (right panel) upon 360 min of 40% stretch.
(D) Representative immunofluorescence images and quantification of phosphorylated (Serine2) RNA polymerase 2 (RNAPII-S2P) and H3K9me3 in cells exposed to 5% or 40% stretch for indicated times. Note minor increase in H3K27me3 at 360 min and a stable versus reversible decrease in RNAPII-S2P in 5% and 40% stretch, respectively (mean ± SD; n = 3 independent experiments with > 250 cells/condition/experiment; ∗p = 0.0180, Friedman/Dunn’s).
(E) Representative immunofluorescence images and quantification of RNAPII-S2P and H3K9me3 in cells exposed to 5% or 40% stretch for 24h. Note decrease in RNAPII-S2P and H3K9me3 in 5% but not in 40% stretch conditions (mean ± SD; n = 3 independent experiments with > 250 cells/condition/experiment; ∗p = 0.0286, Friedman/Dunn’s).
(F) Quantitative RT-PCR of selected EPC identity and differentiation genes from cells exposed to 5% stretch for 24 h (mean ± SEM; n = 3 independent experiments; ∗p < 0.05, Student’s t test).
(G) H3K9me3 and H3K7me3 chromatin immunoprecipitation and subsequent quantitative RT-PCR of selected regions with decreased H3K9me3 show lack of compensation with H3K27me3 (mean ± SEM; n = 2 independent experiments with technical replicates). Scale bars represent 20 μm, white arrows indicate stretch direction, AU = arbitrary units.
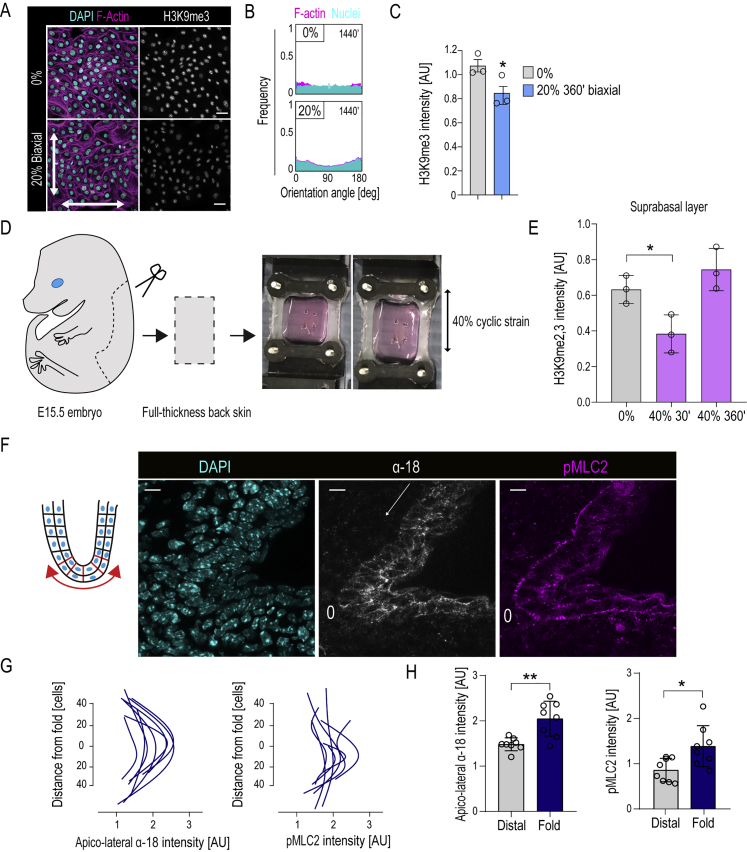
Detailed analysis of the transcriptome revealed that 480 genes were differentially expressed after 30 min of stretch (padj < 0.05), whereas 2,151 genes were altered after 360 min (Figure S2C; Table S3). Among the most enriched GO terms after 30 min were terms related to cell-cell junctions and the cytoskeleton (Figure 2F), consistent with the phosphoproteome data and onset of supracellular alignment. Heterochromatin regulation was the most enriched GO term at 360 min, with strong upregulation of genes related to the facultative heterochromatin mark H3K27me3, and its regulators such as JARID2 and SUZ12 (Margueron and Reinberg, 2011) (Figures 2G and S2C), whereas epidermal differentiation genes were among the most downregulated genes (TGM1, PPL, and SPRR) (Figures 2G and S2C). These results confirm our earlier work, where long-term (12 h) biaxial stretch was observed to decrease global transcription to increase H3K27me3-occupancy and silencing of epidermal differentiation genes (Le et al., 2016). Consistently, we detected reduced levels of the elongating form of RNA polymerase II (RNAPII-S2P) and increased H3K27me3 levels in response to long-term (6–24 h) stretch. This was particularly evident in 5% stretch conditions, where H3K27me3 did not return to steady state upon long-term stretch and differentiation gene expression remained suppressed (Figures S2D–S2F). In line with slower kinetics of H3K27me3 accumulation, ChIP-qPCR of selected chromatin regions with decreased H3K9me3 occupancy did not show corresponding increases in H3K27me3 at these sites after 30 min of 40% stretch (Figure S2G). Collectively, these experiments show that long-term uniaxial stretch, similar to biaxial stretch (Le et al., 2016), triggers transcriptional repression, increased H3K27me3, and silencing of differentiation gene expression. These experiments further reveal that stretch rapidly triggers a widespread reduction in H3K9me3 occupancy, mainly at non-coding regions and chromosome ends, with little immediate transcriptional consequences, raising the question of the biological significance of this change.
Stretch Induces Changes in NE Tension and Nuclear Mechanical Properties
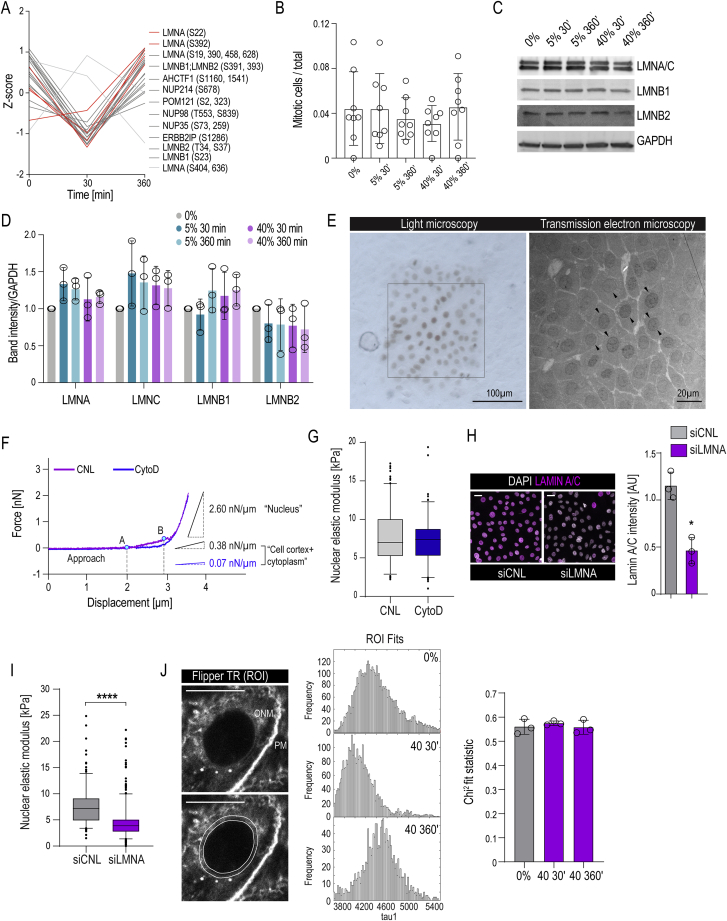
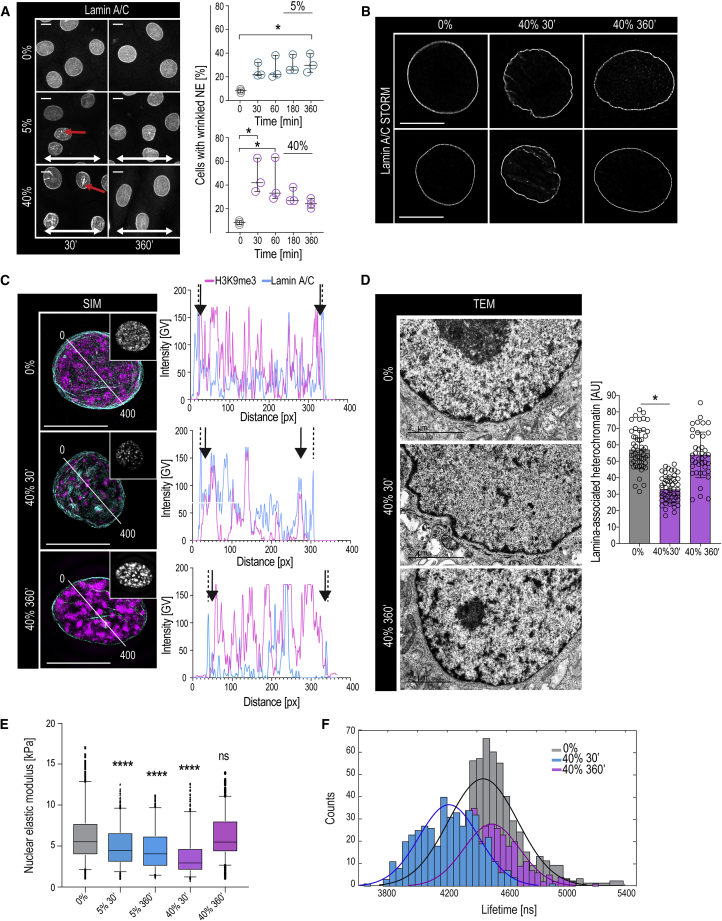
To address the mechanisms and functional relevance of the force-induced reduction in H3K9me3, we further mined the phosphoproteome data and observed that a subset of phosphosites found downregulated upon stretch have been previously reported to be hyperphosphorylated during NE mitotic breakdown, most prominently phosphorylation of Lamin A at S22 and S329 (Güttinger et al., 2009) (Figure S3A), prompting us to examine the NE. Although mitotic frequency was unaltered (Figure S3B), we observed increased NE wrinkling with moderate but sustained effect with 5% stretch and a strong but transient change using 40% stretch (Figure 3A). Superresolution microscopy revealed undulations and wrinkling of the NE/lamina in stretched cells, along with decreased H3K9me3 at the nuclear periphery (Figures 3B and 3C). Interestingly, however, total Lamin levels were unaltered (Figures S3C and S3D). Transmission electron microscopy and chromatin tomography confirmed nuclear wrinkling and revealed a strong reduction in NE-associated condensed heterochromatin (Figures 3D and S3E; Videos S1, S2, and S3).
Figure S3.
Analyses of Nuclear Lamina, Chromatin, and Membrane Tension in Stretched Cells, Related to Figure 3
(A) Profile plots of significantly changed NE-related phosphopeptides with 40% stretch identified by time-resolved phosphoproteomic analysis of stretched EPCs indicate downregulation of phosphosites shown to be hyperphosphorylated during mitotic NE breakdown.
(B) Quantification of mitoses in EPC monolayers exposed to stretch at indicated amplitudes and times (n = 8 independent experiments with > 50 cells/experiment).
(C and D) Representative western blots (C) and quantification (D) of total Lamin A/C, Lamin B1, and Lamin B2 (n = 3 independent experiments).
(E) Representative light microscopy image of DRAQ5 photoconverted nuclei and transmission electron micrograph from the corresponding marked area. Cells analyzed further with electron tomography are indicated with arrowheads in the micrograph.
(F) Representative force - distance curves from AFM force indentation experiments of untreated control cells (CNL) and cells treated with Cytochalasin D (CytoD) to depolymerize the F-actin cytoskeleton shows two components, corresponding to the soft cell cortex and cytoplasm (points A-B) and subsequently the stiffer nucleus. Curve slopes for both components are indicated. Only cortex component is affected by CytoD treatment.
(G) AFM force indentation experiments of cells treated with CytoD (n = 3 independent experiments with > 60 nuclei/condition/experiment; no statistically significant difference found, Mann-Whitney).
(H) Representative immunofluorescence images and quantification Lamin A expression in siLMNA cells show efficient depletion (n = 3 independent experiments with > 200 nuclei/condition/experiment; ∗p = 0.05, Mann-Whitney; scale bars 20 μm).
(I) AFM force indentation experiments of Lamin A depleted cells (siLMNA) (n = 3 independent experiments with > 50 nuclei/condition/experiment; ∗∗∗∗p < 0.0001, Mann-Whitney).
(J) Representative image (lower panel with indicated ROI; PM = plasma membrane, ONM = outer nuclear membrane, scale bars 10 μm), frequency distribution of lifetimes and Chi2 measurements for goodness of fits for the single images of EPC monolayers stained with Flipper-TR membrane tension probe. Bar graphs show mean ± SD, boxplots show 95% confidence interval, AU = arbitrary units.
Figure 3.
Stretch Induces Changes in NE Tension and Nuclear Mechanical Properties
(A) Representative Lamin A/C images and quantification of EPCs exposed to stretch. Increased nuclear wrinkling (red arrows) is observed in 5% and transiently in 40% stretch. White arrows indicate stretch direction (three independent experiments with n > 350 cells/condition/experiment; ∗p = 0.0451, ANOVA/Dunn’s).
(B) Representative STORM images of Lamin A/C immunofluorescence (n > 34 cells/condition visualized from three independent experiments).
(C) Representative structured illumination microscopy (SIM) images and intensity line scans from Lamin A/C and H3K9me3 immunostainings (inset: H3K9me3 alone). Dotted lines mark outer edges of Lamin A/C staining and arrowheads the most peripheral peak of H3K9me (n > 41 cells/condition from three independent experiments).
(D) Representative electron micrographs and quantification of nuclear lamina-associated heterochromatin indicate transient nuclear wrinkling and decreased lamina-associated heterochromatin after 30 min of 40% stretch (n > 40 cells pooled across three independent experiments; ∗p = 0.00284, Kruskal-Wallis/Dunn’s).
(E) Force indentation spectroscopy showing sustained and transient decrease in nuclear elastic modulus in EPC monolayers subjected to 5% and 40% stretch, respectively (n > 140 nuclei/condition pooled across three independent experiments; ∗∗∗∗p = 0.0013, Kruskal-Wallis/Dunn’s).
(F) Distribution of fluorescence lifetimes with Gaussian fits for Flipper-TR membrane tension probe. Note decreased nuclear membrane tension at 30 min of stretch (n > 360 nuclei/condition from three independent experiments; ∗∗p < 0.0001, Kolmogorov-Smirnov).
Scale bars represent 2 μm in (D) and 5 μm in other panels. AU, arbitrary units.
See also Figure S3.
EPC monolayers were subjected to 0% (control) stretch, fixed and processed for electron tomography. Movie shows a model of condensed chromatin (purple) and nuclear envelope (yellow). Tomograms were scaled to a common mean and segmented applying thresholding, smoothing, and size exclusion. High-density compacted chromatin is seen at the nuclear lamina. See also Videos S2 and S3.
EPC monolayers were subjected to 40% stretch for 30 min, fixed, and processed for electron tomography. Movie shows a model of condensed chromatin (purple) and nuclear envelope (yellow). Tomograms were scaled to a common mean and segmented applying thresholding, smoothing, and size exclusion. High-density compacted chromatin at the nuclear lamina is decreased with 30 min of stretch. See also Videos S1 and S3.
EPC monolayers were subjected to 40% stretch for 360 min, fixed, and processed for electron tomography. Movie shows a model of condensed chromatin (purple) and nuclear envelope (yellow). Tomograms were scaled to a common mean and segmented similarly applying thresholding, smoothing, and size exclusion. High-density compacted chromatin is restored to baseline levels at 360 min of stretch. See also Videos S1 and S2.
The increased NE wrinkling without substantial nuclear volume changes pointed to reduced nuclear membrane tension (increased nuclear deformability). Nuclear elasticity and deformability depend on nuclear membrane tension, stiffness of the nuclear lamina and chromatin condensation state (Enyedi and Niethammer, 2017, Stephens et al., 2017, Stephens et al., 2018, Stephens et al., 2019). To quantify nuclear elasticity, we performed atomic force microscopy-based force indentation of the nuclear surface through the cell cortex (Figure S3F). Nuclear force indentation showed decreased stiffness in both 5% and 40% stretch conditions, with full recovery upon long-term 40% stretch (Figure 3E). Importantly, depolymerization of F-actin did not affect nuclear stiffness, indicating that neither the actin cortex nor perinuclear F-actin contributed to the observed nuclear mechanical changes. In contrast, Lamin A depletion, as expected, strongly reduced nuclear stiffness, confirming the validity of the approach (Figures S3F–S3I). To separately assess nuclear membrane tension, we performed fluorescence lifetime imaging (FLIM) using the Flipper-TR membrane tension reporter (Colom et al., 2018). FLIM imaging revealed a decrease in Flipper-TR lifetime at perinuclear membranes after 30 min of 40% stretch, indicative of altered membrane state and reduced nuclear membrane tension (Figures 3F and S3J). Collectively, these experiments indicate that mechanical stretch reduces nuclear membrane tension and increases nuclear deformability.
H3K9me3 Heterochromatin Regulates Nuclear Mechanics and Chromatin Mobility to Prevent DNA Damage
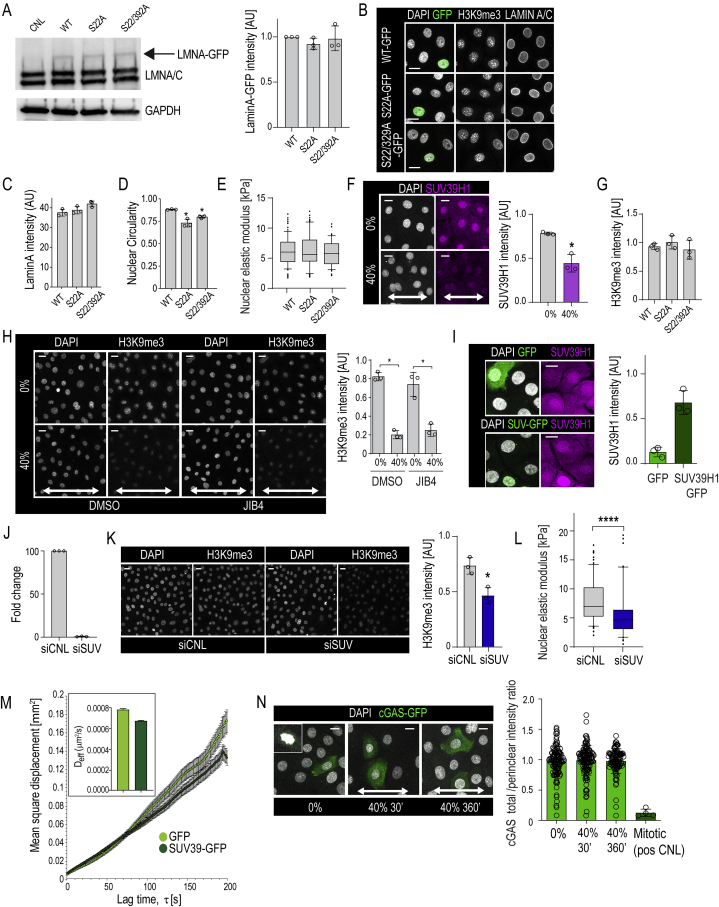
To identify the mechanism by which stretch led to reduced nuclear stiffness and membrane tension, we first investigated the role of changes on Lamin A phosphorylation using phospho-mutant forms of Lamin A (S22A, S22/329A) to mimic the observed decreased phosphorylation of these residues upon stretch. While the phosphorylation-deficient Lamin mutants induced minor nuclear shape abnormalities, they did not impact nuclear elasticity (Figures S4A–S4E) or H3K9me3 (Figures S4B and S4G).
Figure S4.
H3K9me3 Regulates Chromatin Rheology and Nuclear Mechanics, Related to Figure 4
(A) Representative western blots and quantification of GFP-tagged wild-type (WT) Lamin A or Lamin A mutants (Serine 22 to Alanine (S22A) or S22A/S392A double mutant) show equal levels of expression. Lamin A band intensities are normalized to GAPDH (n = 3 independent experiments).
(B) Representative immunofluorescence images of nuclei (DAPI), GFP, Lamin A/C, and H3K9me3 in cells expressing WT Lamin A-GFP or Lamin mutants S22A-GFP; S22A/S392A-GFP.
(C and D) Quantification of Lamin A intensity and nuclear shape in WT Lamin A and mutant expressing cells (n = 3 independent experiments with > 100 nuclei/condition/experiment; ∗p = 0.034, ANOVA/Dunnett’s).
(E) AFM force indentation experiments of cells expressing WT Lamin A-GFP or Lamin mutants S22A-GFP; S22A/S392A-GFP (n = 3 independent experiments with > 40 nuclei/condition/experiment; no statistically significant difference found, Friedman/Dunn’s).
(F) Representative immunofluorescence images and quantification of Suv39H1 levels in cells exposed to uniaxial 40% stretch for 30 min (n = 3 independent experiments with > 100 nuclei/condition/experiment; ∗p = 0.05, Mann-Whitney)
(G) Quantification of H3K9me3 intensity from cells in (B) (n = 3 independent experiments with > 30 nuclei/condition/experiment; no statistically significant difference found, Friedman/Dunn’s).
(H) Representative immunofluorescence images and quantification of H3K9me3 levels in cells treated with JIB4 to inhibit H3K9me3 demethylation and exposed to 30 min uniaxial 40% stretch (n = 3 independent experiments with > 200 nuclei/condition/experiment; ∗p = 0342, Friedman/Dunn’s).
(I) Representative immunofluorescence images and quantification of Suv39H1 levels in cells transfected with Suv39H1-IRES-GFP or GFP only (n = 3 independent experiments with > 24 nuclei/condition/experiment).
(J) Quantitative RT-PCR analyses of Suv39H1 mRNA expression, normalized to B2M in EPC monolayers where Suv39H1 has been silenced by siRNA (siSUV) (n = 3 independent experiments).
(K) Representative immunofluorescence images and quantification of H3K9me3 in siSUV cells cells (n = 3 independent experiments with > 200 nuclei/condition/experiment; ∗p = 0.05, Mann-Whitney).
(L) AFM force indentation experiments of siSUV cells (n = 3 independent experiments with > 40 nuclei/condition/experiment; ∗∗∗∗p < 0.001, Mann-Whitney).
(M) Quantification of chromatin rheology by mean square displacement versus lag time τ of CRISPR-rainbow labeled telomeres. From linear data, material properties of chromatin can be calculated from MSD = Deffτ, where a higher Deff corresponds to less condensed chromatin (n = 40 (GFP)/30 (Suv39-GFP) cells from 3 independent experiments; ∗∗∗p = 0.0001, Student’s t test).
(N) Representative immunofluorescence images and quantification of cGAS-transfected cells to detect nuclear rupture in cells exposed to 30 or 360 min uniaxial 40% stretch. Note lack of perinuclear accumulation of cGAS indicating absence of nuclear rupture upon stretch. Inset shows accumulation of cGAS to chromatin upon mitotic breakdown of NE as positive control (n = 3 independent experiments with > 150 nuclei/condition/experiment). Bar graphs show mean ± SD, boxplots show 95% confidence interval, scale bars represent 10 μm, white arrows indicate stretch direction, AU = arbitrary units.
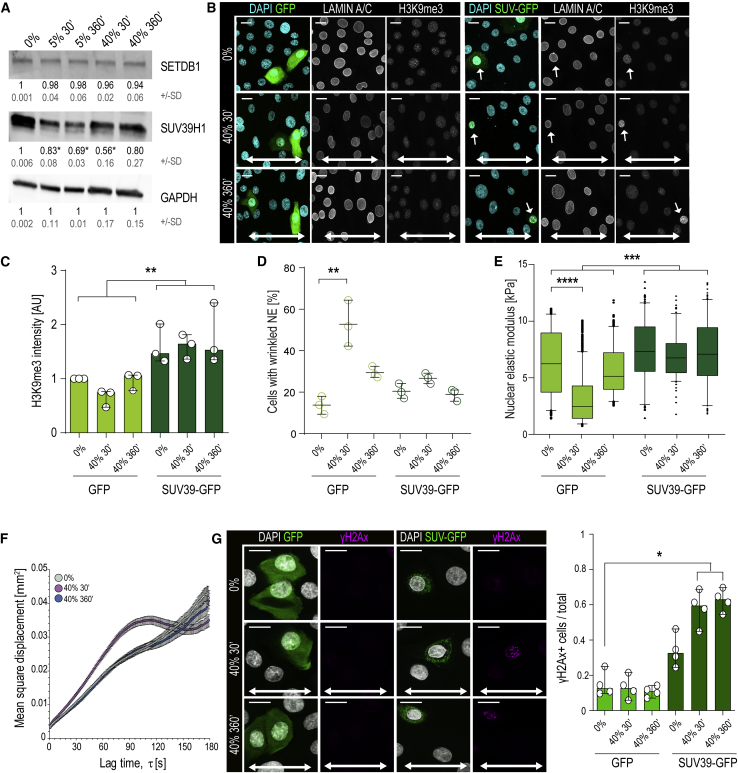
We thus asked if the H3K9me3 heterochromatin itself could determine nuclear elasticity, as shown for bulk heterochromatin (Chalut et al., 2012, Spagnol and Dahl, 2016, Stephens et al., 2018). To this end, we analyzed levels of the H3K9me2 and H3K9me3 methyltransferases Setdb1 and Suv39H1 (Becker et al., 2016) and found that in particular, the expression of Suv39H1 was reduced in stretched cells (Figure 4A), without altering its nuclear localization (Figure S4F), indicating that decreased Suv39H1 and the resulting reduction of net methyltransferase activity could explain the reduced H3K9me3. This was supported by the observation that inhibiting H3K9me3 demethylase activity using the pan-selective Jumonji histone demethylase inhibitor JIB4 (Wang et al., 2013) did not prevent stretch from reducing H3K9me3 (Figure S4H). To directly test the requirement for Suv39H1 activity, we counteracted the observed reduction of Suv39H1 by expressing Suv39H1-IRES-GFP and observed that this indeed prevented H3K9me3 reduction (Figures 4B, 4C, and S4I). Furthermore, this Suv39H1-mediated restoration of H3K9me3 completely prevented nuclear softening (Figure 4E). Vice versa, depletion of Suv39H1 in unstretched cells was sufficient to reduce H3K9me3 and nuclear stiffness (Figures S4J–S4L).
Figure 4.
H3K9me3 Heterochromatin Regulates Nuclear Mechanics and Chromatin Mobility to Prevent DNA Damage
(A) Representative western blots and quantification show reduced levels of Suv39H1 in response to stretch (mean ± SD; n = 3 independent experiments; ∗p = 0.0438, ANOVA/Dunnet’s).
(B–D) Representative images (B) and quantification of H3K9me3 (C) and nuclear wrinkling (D; white arrows) in GFP or Suv39H1-IRES-GFP expressing cells. Note increased H3K9me3 and lack of nuclear wrinkling in Suv39H1 expressing cells (n = 3 independent experiments with > 300 cells/condition/experiment; ∗∗p = 0.0053, Friedman/Dunn’s).
(E) Force indentation experiments show that decreased nuclear stiffness in 40% stretch is prevented by expression of Suv39H1 (n > 100 cells/condition pooled across three independent experiments; ∗∗∗∗p = 0.0015, Kruskal-Wallis/Dunn’s).
(F) Quantification of chromatin rheology by mean square displacement (MSD) versus lag time τ of CRISPR-Rainbow labeled telomeres. Note transient increase in mobility after 30 min of stretch, nonlinear with lag time, consistent with energy-dependent reorganization of chromatin (n > 21 cells/condition with >200 tracks/condition from three independent experiments; control and 360 min are statistically similar; ∗∗p < 0.001 at every lag time, Student’s t test).
(G) Representative images and quantification demonstrating increased γH2AX-positive cells in Suv39-expressing cells upon 40% stretch (n = 4 independent experiments with >250 cells/condition/experiment; ∗p = 0.018, Friedman/Dunn’s).
Bar graphs show mean ± SD, boxplots show 95% confidence interval, white arrows indicate stretch direction, and scale bars represent 10 μm. AU, arbitrary units.
See also Figure S4.
To understand how H3K9me3 could impact nuclear mechanics, we quantified chromatin rheology by live imaging of CRISPRainbow-tagged telomeres (Ma et al., 2016), as H3K9me3 at chromosome ends was particularly reduced by stretch (Figure 2C). We extracted mean square displacement of these regions as a function of lag time (Spagnol and Dahl, 2014) and observed that 30 min of 40% stretch substantially increased telomere mobility, which returned to baseline at 360 min (Figure 4F), consistent with the transient nuclear softening. Conversely, Suv39H1-expressing cells displayed decreased chromatin mobility (Figure S4M), confirming that H3K9me3 levels modulated chromatin rheological properties.
To understand if loss of H3K9me3-marked heterochromatin also contributed to the observed NE wrinkling, we analyzed nuclear morphology in Suv39H1-expressing cells and observed that that Suv39H1 expression prevented stretch-induced NE wrinkling (Figure 4D).
We next asked if the H3K9me3-mediated increased chromatin mobility and decreased nuclear membrane tension function to absorb mechanical energy necessary to prevent DNA damage. Whereas exogenous stretch did not induce any DNA damage in control cells, stretched cells with increased H3K9me3 displayed substantial amounts of DNA damage, as evidenced by increased γH2AX foci detected by immunofluorescence (Figure 4G). Interestingly, no micronuclei or perinuclear accumulation of cGAS-GFP (Raab et al., 2016) was observed (Figures 4B and S4N), indicating that stretch does not induce nuclear rupture. Collectively these results indicate that stretch-induced chromatin softening decreases NE tension, mediated by reduced H3K9me3-marked lamina-associated heterochromatin, and that preventing this heterochromatin remodeling leads to DNA damage.
Chromatin Mechanoresponse Is Mediated by Nuclear-Deformation-Triggered Intracellular Calcium
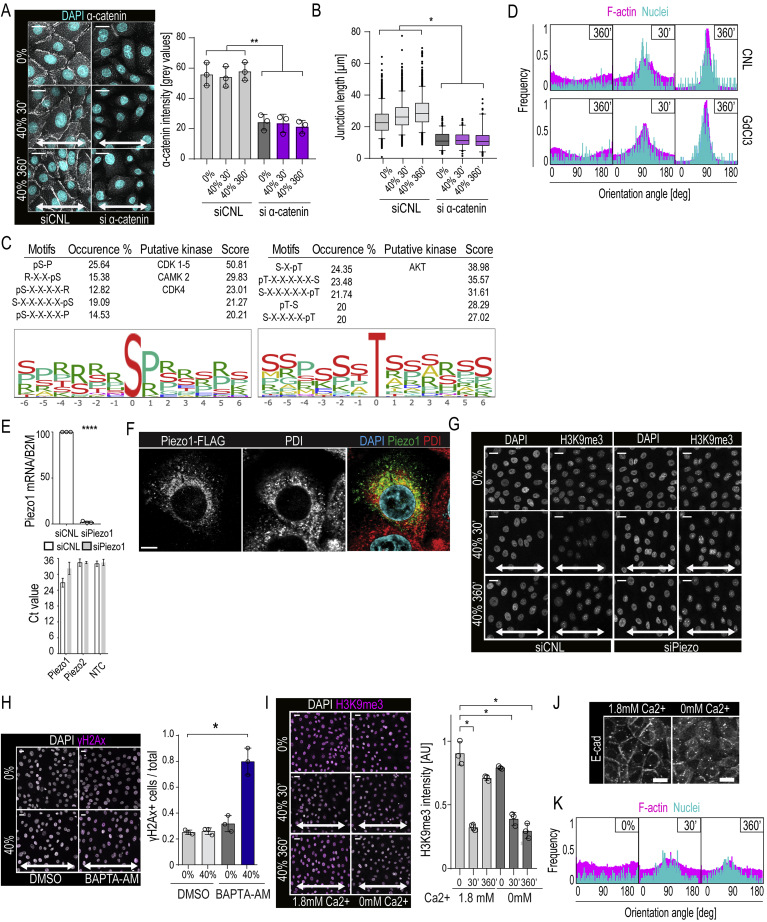
To understand how the supracellular and chromatin mechanoresponses are transduced, we first assessed the role of cell-cell contacts, as cell-cell adhesion was the major phosphoproteome group impacted by stretch (Figures 1I–1L) and the adherens junction component α-catenin is a known mechanotransducer (Yonemura et al., 2010). As expected (Noethel et al., 2018), depletion of α-catenin prevented adherens junction formation and impaired stretch-induced supracellular alignment (Figures 5A, 5B, S5A, and S5B). In contrast, its depletion did not prevent stretch-induced reduction in H3K9me3 (Figures 5C and 5D). However, the H3K9me3 levels failed to recover during long-term stretch of α-catenin-depleted cells (Figures 5C and 5D).
Figure 5.
Chromatin Mechanoresponse Is Induced by Cell/Nuclear-Deformation-Triggered Intracellular Calcium Signaling
(A) Representative F-actin (phalloidin), DAPI, and E-cadherin (E-cad) staining of α-catenin-depleted (siα-catenin) and scrambled control (siCNL) cells exposed to stretch.
(B) Quantification of images in (A) shows absence of F-actin and nuclear alignment in siα-catenin cells (frequency distribution of >500 cells/experiment pooled across three independent experiments).
(C and D) Representative images (C) and quantification (D) of H3K9me3 in α-catenin-depleted cells. Note absence of H3K9me3 recovery at 360 min stretch in siα-catenin cells (n = 3 independent experiments with >300 cells/condition/experiment; ∗p = 0.0269, ∗∗p = 0.0099, repeated measures-ANOVA).
(E) Representative heatmaps and images of Ca2+- sensor intensity during 10 cycles of 40% stretch (n > 100 cells from a representative of three independent experiments).
(F) Representative images and quantification of H3K9me3 in cells treated with GdCl3. Note lack of stretch-induced H3K9me3 reduction in GdCl3-treated cells (n = 3 independent experiments with n > 200 cells/condition/experiment; ∗p = 0.0164, Friedman/Dunn’s).
(G) Representative F-actin images and quantification indicating lack of perinuclear actin rings in GdCl3-treated cells (n > 300 cells pooled across three independent experiments, ∗p = 0.0338, Friedman/Dunn’s).
(H) Representative images and quantification show an increase in γH2AX-positive cells in GdCl3-treated cells upon stretch (n = 3 independent experiments with n > 300 cells/condition/experiment; ∗p = 0.0113, Friedman/Dunn’s).
(I) Quantification of H3K9me3 in Piezo1-depleted cells (siPiezo1) shows absence of H3K9me3 reduction by stretch (n = 3 independent experiments with >350 cells/condition/experiment; ∗p = 0.0164, Friedman/Dunn’s).
(J) Representative staining and quantification showing an increase in γH2Ax-positive cells in stretched siPiezo1 cells (n = 3 independent experiments with >300 cells/condition/experiment; ∗p = 0.0274, Friedman/Dunn’s).
(K) Representative nuclear outlines and quantifications of equivalent nuclear radius imaged before and during 40% stretch. is % difference between equivalent radius during and prior to stretch in control (GFP) and SUV-IRES-GFP expressing cells (n > 40 cells pooled across three independent experiments).
(L) Representative heatmaps of Ca2+- sensor dye Cal-590AM intensity prior and during 10 cycles of 40% stretch with or without thapsigargin pretreatment. Thapsigargin application triggers initial Ca2+ flash, after which the cells do not respond to stretch (n > 100 cells/condition from a representative of three independent experiments).
(M) Representative immunofluorescence images and quantification of H3K9me3 in cells treated with BAPTA-AM to chelate intracellular calcium and subjected to stretch. Note lack of stretch-induced reduction in H3K9me3 in BAPTA-AM-treated cells (n = 3 independent experiments with n > 200 cells/condition/experiment; ∗p = 0.0133, Friedman/Dunn’s).
Bar graphs show mean ± SD, scale bars represent 20 μm, and white arrows indicate stretch direction. AU, arbitrary units.
See also Figure S5.
Figure S5.
Intracellular Ca2+ Regulates Heterochromatin, Related to Figure 5
(A) Representative immunofluorescence images and quantification of α-catenin-depleted EPCs (siα-catenin) and scrambled siRNA controls (siCNL) exposed to stretch. Note efficient depletion of α-catenin.
(B) Quantification of adherens junction length (E-cad) show loss of junctions in α-catenin-depleted cells (n = 1000 cells/condition pooled across 3 independent experiments; ∗p = 0.0009, Friedman/Dunn’s).
(C) Motif-enrichment analysis of significantly altered phosphopeptide sequences using Phosida posttranslational modification database indicates over-representation of CDK1-5, CAMK, and AKT kinase consensus motifs.
(D) Quantification of F-actin and nuclei of EPC monolayers treated with GdCl3 and exposed to stretch at indicated amplitudes and times. No effect of time-dependent reorientation of F-actin and nuclear axes perpendicular are seen with GdCl3.
(E) Quantitative RT-PCR analyses of Piezo1 mRNA expression, normalized to B2M (upper panel) in EPC monolayers subjected to siRNA-mediated silencing of Piezo1 (n = 3 independent experiments). Ct values for Piezo2 mRNA in EPC monolayers are in the same range as no-template controls (NTC), indicating lack of expression and compensation upon Piezo1 silencing (lower panel).
(F) Representative immunofluorescence image of siPiezo1 cells transfected with Piezo1-FLAG and stained with ER marker PDI and FLAG antibodies to detect Piezo1 localization.
(G) Representative images of siPiezo1 cells show loss of stretch effect on H3K9me3 intensity (n = 3 independent experiments with > 300 cells/condition/experiment.
(H) Representative immunofluorescence images and quantification of γH2AX in cells treated with BAPTA-AM (3 independent experiments with n > 200 cells/condition/experiment; ∗∗∗p < 0.0342, Friedman/Dunn’s).
(I) Representative immunofluorescence images and quantification of H3K9me3 in the absence of extracellular Ca2+ (n > 350 cells/condition pooled across 3 independent experiments; ∗∗∗p < 0.0228, Friedman/Dunn’s).
(J) Representative immunofluorescence images of E-cadherin (E-cad) at adherens junctions of cells cultured in the presence of 1.8mM Ca2+ but not in the absence of calcium (representative of 3 independent experiments).
(K) Frequency distribution of quantification of F-actin (phalloidin; magenta) and nuclear (dapi; cyan) orientation in EPC monolayers exposed to 40% stretch from in the absence of calcium (n > 300 cells/condition pooled across 3 independent experiments). Bar graphs show mean ± SD, boxplots show 95% confidence interval, scale bars represent 5 μm in (F) and 10 μm in other panels, white arrows indicate stretch direction, AU = arbitrary units.
As motif enrichment analyses of the phosphoproteomics data revealed enrichment of calcium (Ca2+)/calmodulin-dependent protein kinase II consensus sites (Figure S5C), we asked if Ca2+ signaling could play a role in nuclear mechanotransduction. Indeed, live imaging of intracellular Ca2+ revealed that stretch triggered elevated cytoplasmic Ca2+ (Figure 5E; Video S4). Strikingly, blocking stretch-induced calcium channels with gadolinium trichloride (GdCl3) (Adding et al., 2001) prevented the H3K9me3 decrease but had no effect on supracellular alignment (Figures 5F and S5D), further confirming that the chromatin and supracellular mechanoresponses occur independently of each other. GdCl3 also blocked perinuclear actin ring formation (Figure 5G), consistent with previous observations demonstrating that perinuclear actin polymerization is Ca2+-dependent (Wales et al., 2016). Further, the absence of the chromatin mechanoresponse in GdCl3-treated cells led to increased stretch-induced DNA damage, as predicted (Figure 5H).
Time-lapse epifluorescence video (in pseudocolor) of an EPC monolayer stained with Cal-590 calcium dye shows induction of intracellular calcium upon cyclic stretch. Images of were acquired continuously for 5 mins prior and during 10 cycles of 40% stretch application 0.3 s/frame.
To identify the specific stretch-induced ion channel, we investigated Piezo channels that are present both at the ER and plasma membrane and play important roles in mechanotransduction (Coste et al., 2010, Gudipaty et al., 2017, McHugh et al., 2010). Piezo1 was highly expressed in EPCs, whereas Piezo2 was not detected, even upon Piezo1 knockdown (Figure S5E). FLAG-tagged Piezo1 expression in Piezo1-depleted cells confirmed previous reports that Piezo1 is present at the ER and to some extent at the plasma membrane (Coste et al., 2010) (Figure S5F). Importantly, depletion of Piezo1 blocked the chromatin mechanoresponse (Figures 5I and S5G) and substantially enhanced stretch-induced DNA damage (Figure 5J), indicating that Piezo1 is the responsible stretch-induced ion channel.
We next sought to identify the trigger for the Ca2+ release and hypothesized that cell stretching could induce direct deformation of the NE and the associated ER, thus prompting Ca2+ release. Live imaging of cells under 40% stretch indeed revealed immediate deformation of the nucleus in the direction of stretch (Figure 5K). This nuclear deformation/elongation was well within the range (<3 μm) where heterochromatin has been described to be the dominant component of nuclear stiffness (Stephens et al., 2017). Consistently, expression of Suv39H1 to increase H3K9me3 and nuclear stiffness reduced stretch-induced nuclear deformation (Figure 5K). Interestingly, this nuclear deformation was accompanied by Ca2+ release, which, despite abundant extracellular calcium present in the culture medium, could be prevented using thapsigargin pretreatment to block the sarco/ER Ca2+ ATPase to empty ER calcium stores prior to stretch (Gerasimenko et al., 2003) (Figure 5L). Consistently, chelating intracellular calcium using BAPTA-AM (Tang et al., 2007, Tsien, 1981) also prevented stretch-induced heterochromatin remodeling (Figure 5M), resulting in DNA damage (Figure S5H).
In contrast to manipulation of ER Ca2+ stores, removing extracellular Ca2+ from the extracellular space did not prevent the chromatin mechanoresponse (Figure S5I), confirming the outer nuclear membrane/ER as the source for the Ca2+. However, removal of extracellular Ca2+, as expected, prevented formation of E-cadherin-based junctions (Meng and Takeichi, 2009), thus preventing supracellular alignment and recovery of H3K9me3 levels (Figures S5J and S5K). This was consistent with the observation that adherens junctions are dispensable for the chromatin mechanoresponse but required for monolayer reorientation and concomitant restoration of heterochromatin.
Minimum Nuclear Stiffness Is Required for Ca2+ Release and Subsequent Heterochromatin Response
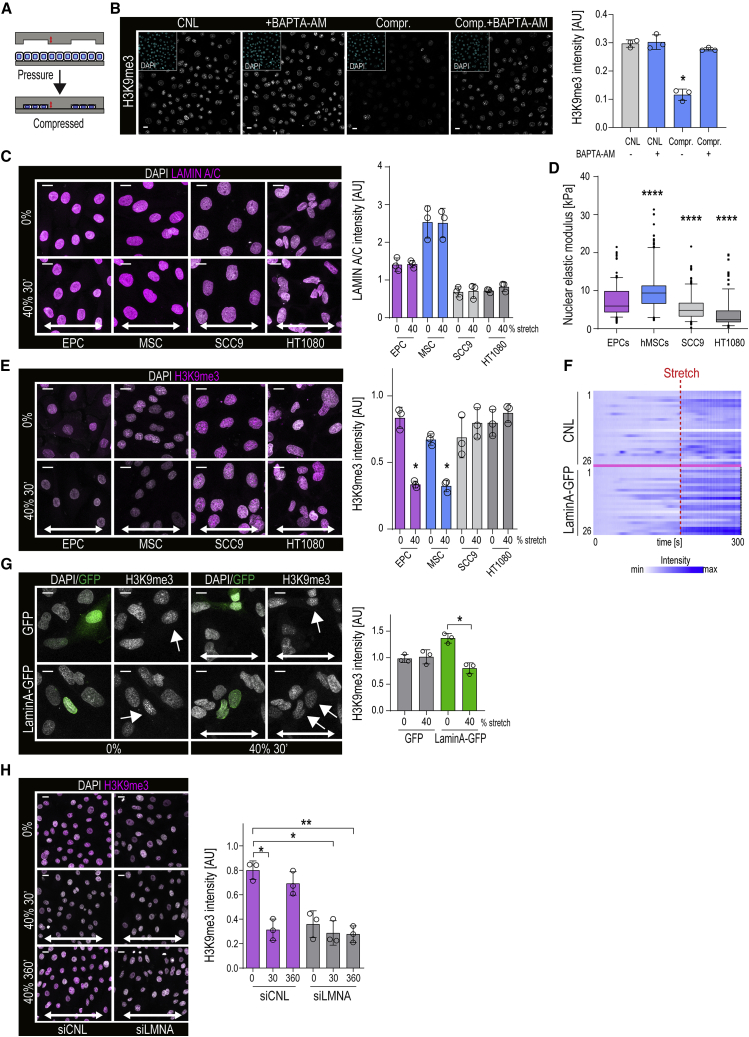
To understand if direct cell/nuclear deformation is capable of triggering ER Ca2+ release and subsequent nuclear mechanotransduction, we compressed cells using pressure-controlled pillars whose height was calculated to induce nuclear deformation (Figure 6A). As predicted, elevated cytosolic Ca2+ was triggered by both compression and its release (Figure S6A). Intriguingly, compression was also sufficient to reduce H3K9me3 (Figure 6B). As for stretch, chelating intracellular calcium using BAPTA-AM prevented the compression-induced reduction in H3K9me3 (Figure 6B).
Figure 6.
Minimum Nuclear Stiffness Is Required for Ca2+ Release and Subsequent Heterochromatin Response
(A) Schematic representation of the compression system where compression depth depends on pillar height (red arrows) designed to compress nuclei.
(B) Representative H3K9me3 images and quantification of cells compressed with and without 10 μm BAPTA-AM. Insets show DAPI stain. Compression triggers reduction in H3K9me3, prevented by BAPTA-AM (n = 3 independent experiments with >300 cells/condition/experiment; ∗p = 0.0052, Friedman/Dunn’s).
(C) Representative Lamin A/C images and quantification of EPCs, MSCs, and SCC9 and HT1080 cells exposed to 40% stretch for 30 min. MSCs contain highest Lamin A/C levels, whereas SCC9 and HT1080 cells have low Lamin A/C. Stretch has no effect on Lamin A/C (n = 3 independent experiments with n > 120 cells/condition/experiment).
(D) Force indentation experiments indicating reduced nuclear stiffness in SCC9 and HT1080 cells (n > 60 cells/condition pooled across three independent experiments; ∗∗∗∗p = 0.0015, Kruskal-Wallis/Dunn’s).
(E) Representative H3K9me3 images and quantification of EPCs, MSCs, and SCC9 and HT1080 cells exposed to 40% stretch for 30 min. Stretch reduces H3K9me3 in EPCs and MSCs but not in SCC9 and HT1080 cells (n = 3 independent experiments with n > 120 cells/condition/experiment ∗p = 0.0303, Friedman/Dunn’s).
(F) Representative heatmaps of Ca2+-sensor dye Cal-590AM intensity prior and during 10 cycles of 40% stretch in HT1080 cells with or without Lamin A overexpression. Note stronger Ca2+ response in Lamin-A-expressing cells (n = 26 cells/condition from a representative of three independent experiments).
(G) Representative H3K9me3 immunofluorescence images and quantification of HT1080 cells with or without Lamin A-GFP overexpression exposed to 40% stretch for 30 min. Lamin A overexpression increases H3K9me3 and allows H3K9me3 decrease upon stretch (n = 3 independent experiments with n > 20 cells/condition/experiment; ∗p = 0.0266, Friedman/Dunn’s).
(H) Representative H3K9me3 immunofluorescence images and quantification of EPCs depleted of Lamin A (siLMNA) or scrambled control (siCNL) exposed to 40% stretch at for 30 min. siLMNA cells have lower baseline H3K9me3 levels which does not further decrease upon stretch (n = 3 independent experiments with n > 200 cells/condition/experiment; ∗p = 0,0495, ∗∗p = 0.0023, Friedman/Dunn’s).
Bar graphs show mean ± SD, boxplots show 95% confidence interval, scale bars represent 10 μm, and white arrows indicate stretch direction. AU, arbitrary units.
See also Figure S6.
Figure S6.
Analyses of Intracellular Ca2+ during Compression and Nuclear Stiffness upon Lamin A Depletion, Related to Figure 6
(A) Representative heatmap of calcium sensor dye Cal-590 intensity during compression and decompression of EPC monolayers show calcium release upon application of compression and upon compression release (red dotted lines; n = 3 independent experiments with > 40 cells /experiment).
(B) AFM force indentation experiments of Lamin A-depleted cells (siLMNA) exposed to 30 min 40% stretch show decreased nuclear elastic modulus and lack of further effect by stretch in siLMNA cells (Boxplots with 95% confidence interval, n = 3 independent experiments with > 50 nuclei/condition/experiment; ∗∗∗∗p < 0.001, Kruskal-Wallis/Dunn’s).
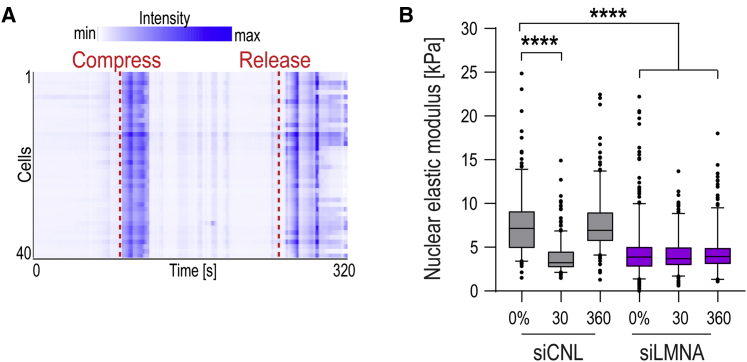
Piezo1 activity can be tuned by plasma membrane tension (Lewis and Grandl, 2015). We thus asked if nuclear membrane tension could similarly threshold the effect of stretch/ER Ca2+-triggered reduction in H3K9me3. As nuclear membrane tension has previously been shown to scale with Lamin A levels (Buxboim et al., 2017), we first asked if the effect of stretch on H3K9me3 scales with Lamin A. To this end, we compared two stem cell types, EPCs and mesenchymal stem cells (MSCs), with two cancer cell lines, the epithelial cancer SCC9 and fibrosarcoma HT1080. Both stem cell types had high Lamin A and high nuclear elastic moduli, and they responded to 40% stretch by reducing H3K9me3 (Figures 6C–6E). In contrast, both cancer cell lines had low Lamin A levels and low nuclear elastic moduli, and they showed a slight increase in H3K9me3 levels upon stretch (Figures 6C–6E), consistent with previous reports (Stephens et al., 2019). Intriguingly, increasing Lamin A expression in HT-1080 cells rendered them sensitive to stretch-induced intracellular Ca2+ release (Figure 6F), and these cells now also responded to stretch by decreasing H3K9me3 levels (Figure 6G). Conversely, reducing Lamin A in EPCs using small interfering RNA (siRNA) strongly reduced the nuclear elastic modulus and prevented stretch from further decreasing H3K9me3 and nuclear stiffness (Figures 6H and S6B).
Collectively, these results suggest that the nucleus and the associated ER membranes sense deformation, the sensitivity of which is defined by steady-state nuclear stiffness/membrane tension. Cell/nuclear deformation of cells with taut nuclei and high membrane tension triggers Ca2+ release from the ER, inducing perinuclear actin polymerization and heterochromatin changes to promote chromatin mobility and decrease apparent NE tension. These changes are likely to facilitate dissipation of mechanical energy to prevent DNA damage.
Supracellular Monolayer Alignment Prevents Stress Transmission to the Nucleus and Chromatin
We next set out to understand how cells experiencing long-term stretch are able to switch off nuclear mechanotransduction to restore steady-state chromatin and nuclear architecture. We hypothesized that anisotropic positioning of the long axes of cells, nuclei, and F-actin stress fibers perpendicular to stretch direction represented the phenomenon of strain avoidance (De et al., 2007, Obbink-Huizer et al., 2014) to minimize nuclear strain. To test this, EPC monolayers were pre-stretched for 360 min at 40% to allow full alignment (see Figures 1B and 1C). Live imaging of these fully aligned monolayers during exposure to a second cycle of 40% stretch in the same stretch direction showed absence of nuclear deformation (Figures 7A and 7B). In contrast, substantial deformation was detected in monolayers that were re-exposed to a second cycle of 40% stretch but now in perpendicular direction relative to the first stretch cycle (Figures 7A and 7B).
Figure 7.
Supracellular Monolayer Alignment Prevents Stress Transmission to the Nucleus and Chromatin
(A) Experimental design to alter stretch direction after monolayer alignment.
(B) Representative nuclear outlines and quantifications of equivalent radius before and during 40% stretch to measure stretch-induced nuclear deformation. is % difference between equivalent radius during and prior to stretch. Note the lack of nuclear deformation in perpendicularly aligned nuclei, whereas nuclei aligned parallel show substantial deformation. (n > 100 cells/condition pooled across three independent experiments).
(C) Representative F-actin (phalloidin) and DAPI images of cells after second 40% stretch regime. Note the alignment of fibers at 40% 360 min before second stretch regime and monolayer disruption and reappearance of perinuclear actin after stretch direction change.
(D) Quantification of F-actin alignment from experiments in (C) (frequency distribution of > 500 cells/condition pooled across 3 independent experiments).
(E) Quantification of perinuclear actin intensity from experiments in (C) (n = 3 independent experiments with > 250 cells/condition/experiment; ∗p = 0.0429, Friedman/Dunn’s).
(F) Representative images and quantification of H3K9me3 intensity before and after stretch direction change (n = 3 independent experiments with > 300 cells/condition/experiment; ∗p = 0.0278, Friedman/Dunn’s).
(G) Representative images and quantification of E-cadherin (E-cad) junctions show loss of junction integrity after stretch direction change (frequency distribution of >300 cells/condition pooled across three independent experiments).
(H–J) Representative images (H) and quantification of H3K9me2,3 intensity (I) and cell alignment (J) in mouse embryonic day 15.5 (E15.5) skin explants exposed to 40% stretch. Note the transient suppression of H3K9me2,3 intensity at 30 min, absence of apoptosis (inset, cleaved caspase-3), and emergence of alignment at 360 min of stretch (n = 7 mice/condition; ∗∗p = 0.0039, ∗∗∗p = 0.0003, ANOVA/Dunnett’s).
(K) Representative images and quantification of H3K9me2,3 intensity in mouse embryo epidermal digit folds. H3K9me2,3 intensity is lower in epidermal stem cells (dotted line) in folds (n = 5 digit folds from 3 mice; 0.0159, Mann-Whitney).
Bar graphs show mean ± SD, Scale bars represent 40 μm in (K) and 10 μm in other panels. White arrows indicate stretch direction. AU, arbitrary units.
See also Figure S7.
Next, we directly tested the causative relationship between alignment and nuclear mechanotransduction using the setup described above. Strikingly, forcing parallel orientation of the pre-aligned monolayers to stretch resulted in re-formation of perinuclear actin rings, strong suppression of H3K9me3, and disruption of adherens junctions (Figures 7C–7G). Consistent with the importance of alignment in allowing H3K9me3 restoration, applying 20% biaxial stretch where alignment does not occur led to long-term suppression of H3K9me3 (Figures S7A–S7C). Thus, we conclude that supracellular monolayer alignment blocks nuclear mechanotransduction to allow restoration of the chromatin state and to minimize stress-induced load on adhesion structures, which together ensure global mechanoprotection of the EPC monolayers. This notion was consistent with the observations that depletion of α-catenin or removal of extracellular Ca2+ not only attenuated alignment but also prevented restoration of heterochromatin during long-term 40% stretch (see Figures 5C, 5D and S5I).
Figure S7.
Stretch Responses in Intact Skin, Related to Figure 7
(A) Representative F-actin (phalloidin; magenta), nuclear (dapi; cyan), and H3K9me3 (gray) immunofluorescence images of EPC monolayers exposed to biaxial cyclic stretch at 20% amplitude for 360 min.
(B) Quantification of immunofluorescence images in (A) shows lack of F-actin and nuclear major axes reorientation (frequency distribution of > 500 cells/condition pooled across 3 independent experiments).
(C) Quantification of H3K9me3 intensity from images in (A) shows a decrease in H3K9me3 in EPC monolayers subjected to 20% biaxial stretch (n = 3 independent experiments with > 200 cells/condition/experiment; ∗p = 0.0113, paired t test).
(D) Schematic illustration and images of ex vivo stretch experiments with E15.5 embryonic whole skin explants.
(E) Quantification of H3K9me2,3 intensity from the first suprabasal layer of ex vivo stretched epidermis shows a decrease in H3K9me2,3 in EPC monolayers subjected to 40% uniaxial stretch (n = 3 independent experiments with > 200 cells/condition/experiment; ∗p = 0.0435, ANOVA/Dunnet’s).
(F–H) Schematic illustration, representative images (F), and quantification of α-catenin tension sensitive epitope antibody (α-18, gray) and phospho-myosin light chain 2 (pMLC2, magenta) immunofluorescence images in digit folds of E15.5 embryos (n = 8 digit folds from 3 mice; ∗p = 0.0148, ∗∗p = 0.0070, Mann Whitney). (G) shows α-18 and pMLC2 intensity as a function of distance, (H) shows α-18 and pMLC2 mean intensity in the digit fold and distal from the fold. Bar graphs show mean ± SD, scale bars represent 20 μm, white arrows indicate stretch direction, AU = arbitrary units.
Finally, we asked if the observed nuclear mechanotransduction and altered heterochromatin states observed in vitro are also relevant in intact tissue. To this end, we established an ex vivo system to stretch intact skin from mouse embryos (Figure S7D). Strikingly, exposing embryonic stage 15.5 skin to 40% uniaxial, cyclic stretch induced a transient reduction in heterochromatin both in the basal stem cell and suprabasal layers, after which H3K9me3 was restored to levels even slightly higher than at stretch onset (Figures 7H, 7I, and S7E). This reduced H3K9me3 was subsequently overridden by supracellular alignment (Figures 7H and 7J). As expected, the alignment in intact skin occurred in the direction of stretch, resulting from differences in 2D versus 3D tissue ability of cell intercalation, F-actin remodeling, and/or differences in the ratios of tissue compaction strains to applied cyclic strains (Blanchard et al., 2009, Chen et al., 2018, Obbink-Huizer et al., 2014). Importantly, no signs of tissue damage in response to stretch was observed, as also indicated by the lack of apoptosis (Figure 7H).
To assess whether physiological stresses generated by tissue deformation also trigger heterochromatin changes, we analyzed epithelial folds during digit morphogenesis where high lateral tension is present (Sui et al., 2018), also demonstrated by increased mechanical unfolding of α-catenin and myosin activity (Figures S7F–S7H). Importantly, reduction of H3K9me3 in stem cells showed strong spatial correlation with the folding curvature of the tissue at these sites (Figure 7K). Collectively these data demonstrate that alterations in chromatin states are associated with dynamic changes in tissue tension in vivo.
Discussion
The functional demands and longevity of tissue-specific stem cells require particularly efficient mechanisms to counteract damage (Mandal et al., 2011). DNA damage compromises stem cell function and tissue integrity, and much is known about how DNA damage is mitigated in stem cells (Blanpain et al., 2011). In contrast, little is known about the mechanisms that prevent unwanted chromatin rearrangements and DNA damage upon nuclear deformation that occurs when cells experience mechanical forces. We propose that chromatin mechanoprotection employs two distinct mechanisms: (1) a rapid, sensitive chromatin response that relies on ER Ca2+ release to reduce lamina-associated heterochromatin, thereby increasing chromatin mobility and lowering nuclear membrane tension; and (2) a slower, less sensitive supracellular response that relies on cell-cell contacts to reorient cell and nuclear long axes, adherens junctions, and F-actin to prevent nuclear strain. We further propose that the nuclear mechanoprotection, via the induction of changes in chromatin state to alter its mechanics, provides an unspecific “emergency break” mechanism for tissues to endure sudden and dynamic deformations without damaging their genomes. The observed chromatin “fluidification” provides an effective means to dissipate mechanical energy (Bonakdar et al., 2016), thus reducing direct force propagation to the DNA, preventing torsion or even breaks. This finding is consistent with the observed fluidization of cytoplasmic material in response to mechanical stress (Blanchard et al., 2009, Harris et al., 2012, Kollmannsberger and Fabry, 2011, Trepat et al., 2008).
The observed chromatin mechanoresponse is triggered by nuclear deformation, which results in Ca2+ release from the ER in a Piezo1-dependent manner. The sensitivity of the ER calcium release is determined by Lamin A levels and nuclear stiffness, where stiffer nuclei are more likely to respond. One possible explanation is that stiffer nuclei deform less upon stretch, and the associated ER membranes are subjected to more stretch and increased membrane tension. Also, higher levels of Lamin A may allow stronger mechanointegration with the cytoskeleton via the linker of nucleoskeleton and cytoskeleton (LINC) complex (Cho et al., 2017). Parallel to H3K9me3 remodeling, Ca2+ release catalyzes perinuclear actin ring formation. Previous work shows that biaxial stretch-induced actin polymerization reduces free nuclear G-actin to attenuate transcription in an H3K27me3-dependent manner (Le et al., 2016, Wales et al., 2016). Interestingly, we now recapitulate these findings also in the context of low-amplitude uniaxial stretch, where similarly to biaxial stretch, the monolayers do not undergo supracellular alignment. The Ca2+ dependency of both H3K9me3 and H3K27me3-mediated chromatin remodeling suggests that nuclear deformation is upstream of both effects.
Since precise control of epigenetic chromatin states and the ability to efficiently silence specific genomic loci are essential for proper control of genetic programs and cell identity (Bickmore and van Steensel, 2013), the chromatin-driven mechanoresponse might not be a sustainable, long-term means to bear loads. Thus, a more effective form of long-term, mechanoprotection, such as the observed supracellular rearrangement of the epidermis is required. In situations where supracellular alignment is not induced, due to insufficient stretch amplitude or lack of unidirectionality of stretch, sustained chromatin fluidification leads to transcriptional repression and decreased expression of cell identity and differentiation genes. Whether this represents a detrimental effect of mechanical stress or a mechanism of long-term physiological adaptation remains open for future studies. Importantly, the molecular mechanisms described here are generic and functional in a broad range of cell types and model systems. Thus, the nuclear mechanoadaptation mechanisms that reduce nuclear membrane tension, increase chromatin mobility, and reorient monolayers are likely to have broad significance in vivo.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-H3K9me2,3 | Cell Signaling | Cat# 5327; RRID:_AB10695295 |
| Rabbit monoclonal anti-H3K9me3 | Cell Signaling | Cat# 13969; RRID:AB_2798355 |
| Rabbit monoclonal anti-H3K27me3 | Cell Signaling | Cat# 9733; RRID:AB_2616029 |
| Rabbit polyclonal anti-H3K9me2 | Thermo Fisher | Cat# PA5-16195; RRID:AB_10986203 |
| Rabbit polyclonal anti-phospho-Histone H2A.X (Ser139) | Cell Signaling | Cat# 9718; RRID:AB_2118009 |
| Mouse monoclonal anti-Lamin A/C | Cell Signaling | Cat# 4777; RRID:AB_10545756 |
| Rabbit monoclonal anti-Lamin A/C | Abcam | Cat# ab133256; RRID:AB_2813767 |
| Rabbit polyclonal anti-Lamin B1 | Abcam | Cat# ab16048; RRID:AB_10107828 |
| Rabbit polyclonal anti-RNA-Pol II (s2P) | Abcam | Cat# ab5095; RRID:AB_304749 |
| Rabbit polyclonal anti-SUV39H1 | Thermo Fisher | Cat# PA5-29470; RRID:AB_2546946 |
| Rabbit polyclonal anti-phospho-Myosin LC2 (Thr18/Ser19) | Cell Signaling | Cat# 3674; RRID:AB_2147464 |
| Mouse monoclonal anti-E-Cadherin | BD Biosciences | Cat# 610181; RRID:AB_397580 |
| Rabbit monoclonal anti-Cleaved caspase3 | Cell Signaling | Cat# 9664; RRID:AB_2070042 |
| Rabbit polyclonal anti-α-Catenin | Sigma-Aldrich | Cat# C2081; RRID:AB_476830 |
| Mouse monoclonal anti-α18 | Yonemura et al., 2010 | N/A |
| Rabbit monoclonal anti-GAPDH | Cell Signaling | Cat# 2118; RRID:AB_561053 |
| Mouse monoclonal anti-total Histone 3 | Cell Signaling | Cat# 3638; RRID:AB_1642229 |
| Rabbit polyclonal anti-Lamin B1 | Cell Signaling | Cat# 9087; RRID:AB_10896336 |
| Rabbit polyclonal anti-Lamin B2 | Cell Signaling | Cat# 9622; RRID:AB_10950813 |
| Rabbit polyclonal anti-Setdb1 | Abcam | Cat# ab12317; RRID:AB_299005 |
| Rabbit polyclonal anti-pLamin A (S22) | Invitrogen | Cat# PA5-17113; RRID:AB_10989809 |
| Rabbit polyclonal anti-pPaxillin (Tyr31) | Thermo Fisher | Cat# 44-720G; RRID:AB_2533732 |
| Rabbit polyclonal anti-pCDK1-3 (T14) | Cell Signaling | Cat# 9477; RRID:AB_2714143 |
| Rabbit polyclonal anti-pCofilin (S3) | Cell Signaling | Cat# 3313; RRID:AB_2080597 |
| Rabbit polyclonal anti-Hsp90 | Cell Signaling | Cat# 4874; RRID:AB_2121214 |
| Rabbit monoclonal anti-PDI | Cell Signaling | Cat# 3501; RRID:AB_2156433 |
| Mouse anti-DYKDDDDK Tag (9A3) Flag | Cell Signaling | Cat# 8146; RRID:AB_10950495 |
| Rabbit IgG | Cell Signaling | Cat# 2729; RRID:AB_1031062 |
| Rabbit polyoclonal anti-H3K9me3 (ChIP grade) | Abcam | Cat# ab8898; RRID:AB_306848 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BAPTA-AM | Abcam | Cat# ab120503 |
| JIB4 | Tocris | Cat# 4972 |
| gadolinium (III) chloride hexahydrate | Sigma-Aldrich | Cat# 203289-5G |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# 472301 |
| Thapsigargin | Abcam | Cat# ab120286 |
| Cal-590 AM calcium dye | AAT Bioquest | Cat# 20510 |
| FLIPPER-TR | Spirochrome | Cat# SC020 |
| Critical Commercial Assays | ||
| Titansphere Phos-TiO kit | GL Sciences | Cat# 5010-21309 |
| NucleoSpin RNA Plus kit | Macherey & Nagel | Cat# 740955 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| DyNAmo ColorFlash SYBR Green Mix | Thermo Fisher | Cat# F416 |
| TruSeq Stranded Total RNA kit | Illumina | Cat# RS-122-2301 |
| TrueSeq ChIP library preparation kit | Illumina | Cat# IP-202 |
| Qubit DS high-sensitivity kit | Invitrogen | Cat# Q32851 |
| Deposited Data | ||
| Raw and analyzed sequencing data | This paper | GEO: GSE143519 |
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Experimental Models: Cell Lines | ||
| Human: Juvenile Epidermal Keratinocyte Progenitors, Pooled | CellNTec | HPEKp |
| Human: Mesenchymal Stem Cells | Lonza | Cat# PT-2501 |
| Human: HT1080 fibrosarcoma cells | ATCC | ATCC CCL-121; RRID:CVCL_0317 |
| Human: SCC9 tongue squamous carcinoma cells | ATCC | ATCC CRL-1629; RRID:CVCL_1685 |
| Experimental Models: Organisms/Strains | ||
| Mouse: NMRI E15.5 embryos | Charles River | Strain 605 |
| Oligonucleotides | ||
| Primers for qPCR, see table in STAR Methods | This paper | N/A |
| siRNA hPiezo1 | Ambion (Silencer Select) | Cat# s18892 and s18893 |
| siRNA hα-catenin | Ambion (Silencer Select) | Cat# s3717 s3718 |
| siRNA hLamin A | Ambion (Silencer Select) | Cat# s8221 s8222 |
| siRNA hSuv39H1 | Ambion (Silencer Select) | Cat# s13658 |
| Recombinant DNA | ||
| Suv39H1-IRES-EGFP | Sino Biological | Cat# 28113 |
| cGAS-EGFP | Raab et al., 2016 | Addgene ID: 86675 |
| hPiezo1 3xFlag | GeneCopoeia | ID: Z6777 |
| pHAGE-TO-dCas9 | Ma et al., 2016 | Addgene ID: 64107 |
| pLH-sgRNA1-2xPP7 | Ma et al., 2016 | Addgene ID: 75390 |
| pHAGE-EFS-PCP-3xGFPnls | Ma et al., 2016 | Addgene ID: 75386 |
| Lamin A-GFP | Kochin et al., 2014 | N/A |
| Lamin A S22A - GFP | Kochin et al., 2014 | N/A |
| Lamin A S22A/S329A - GFP | Kochin et al., 2014 | N/A |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji |
| MaxQuant | Cox and Mann | https://www.maxquant.org/; RRID:SCR_014485 |
| Kallisto version 0.45.0 0 | Bray et al., 2016 | http://pachterlab.github.io/kallisto/download; RRID:SCR_016582 |
| FeatureCounts version v1.6.2. | Liao et al., 2014 | https://bioconductor.org/; RRID:SCR_012919 |
| DEseq2 version 1.22.2 | Love et al., 2014 | https://bioconductor.org/; RRID:SCR_015687 |
| Bowtie2 version 2.3.4.1 | Langmead and Salzberg 2012 | https://bioconductor.org/ |
| Macs2 | Ma et al., 2016 | https://bioconductor.org/ |
| DiffBind version 2.10.0 | Ross-Innes et al., 2012 | https://bioconductor.org/; RRID:SCR_012918 |
| ChIPseeker version 1.5.1 | Yu et al., 2015 | https://bioconductor.org/ |
| Bedtools intersect version 2.26.0 | Quinlan and Hall 2010 | https://bioconductor.org/; RRID:SCR_006646 |
| R Studio | RStudio Inc | https://rstudio.com/ |
| GraphPad Prism 8 | GraphPad | N/A |
| SoftWoRx 7.0 | GE Healthcare | N/A |
| MATLAB | MathWorks | N/A |
| SerialEM | Mastronarde, 1997 | https://bio3d.colorado.edu/SerialEM/; RRID:SCR_017293 |
| Amira software (version 5.3.2) | Thermo Fisher | N/A |
| SymPhoTime 64 software | PicoQuant | N/A |
| IMOD software package (versions 4.9.0, 4.10.18 and 4.10.32) | Mastronarde, 1997 | https://bio3d.colorado.edu/imod/; RRID:SCR_003297 |
| Microscopy Image Browser (MIB) | Belevich et al., 2016 | http://mib.helsinki.fi/; RRID:SCR_016560 |
| Thunder STORM Fiji plugin | Ovesný et al., 2014 | RRID:SCR_016897 |
| 3D Objects Counter | Bolte and Cordelières, 2006 | RRID:SCR_017066 |
| PVCamTest software version 3.4.183 | Photometrics | N/A |
| Leica Application Suite software (LAS X version 2.0.0.14332), | Leica | N/A |
| FLIMfit 5.1.1. software | Imperial College London | https://flimfit.org/; RRID:SCR_016298 |
| JPK SPM Control Software v.5 | Bruker Nano | N/A |
| JPK Data Processing Software | Bruker Nano | N/A |
| Other | ||
| Sequence data analyses | This paper | https://github.com/mpg-age-bioinformatics |
Lead Contact and Materials Availability
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sara A. Wickström (sara.wickstrom@helsinki.fi).
Experimental Model and Subject Details
Primary cells and cell lines
Human epidermal stem/progenitor cells (EPCs) isolated from pooled human male foreskin biopsies were purchased from CellnTec and grown in epithelial culture medium (CnT-Prime; CellnTec). Cells were used between passages 1-3. Human mesenchymal stem cells were purchased from Lonza and were cultured in Mesenchymal Stem Cell Growth Medium (PT-3001, Lonza). SCC9 human tongue squamous cell carcinoma (CRL-1629) and HT1080 human fibrosarcoma cells (CCL-121) were from ATCC and were cultured in Dulbecco’s MEM with 10% fetal calf serum (GIBCO). All cells were cultured in 5% CO2 at 37°C.
Mice
Embryonic day 15.5 NMRI embryos of both genders were used according to the guidelines of local authorities (Southern Finland Regional State Administrative Agency, ESAVI) and University of Helsinki Center for Animal Experiments (KEK, permit 18-020).
Method Details
Cell culture
Human epidermal stem/progenitor cells (EPCs) isolated from pooled neonatal human foreskin biopsies were purchased from CellnTec (HPEKp, lots ES1110044 and ES1204665) and grown in supplier's cell culture medium (CnT-Prime; CellnTec). Cells were used between passages 2-4. Human mesenchymal stem cells were purchased from Lonza and were cultured in suppliers Mesenchymal Stem Cell Growth Medium (PT-3001, Lonza). SCC9 human tongue squamous cell carcinoma (CRL-1629) and HT1080 human fibrosarcoma cells (CCL-121) were from ATCC and were cultured in Dulbecco’s MEM with 10% fetal calf serum (GIBCO).
Mechanical stretching
The custom-built uniaxial cell stretcher, the molds for elastic silicone cell-culture chambers and their calibration has been described in detail previously (Faust et al., 2011, Noethel et al., 2018). Polydimethylsiloxane (PDMS) elastomers were prepared from a two-component formulation (Sylgard 184, Dow Corning) by mixing base and crosslinker in a ratio of 40 to 1 (weight/weight) to obtain substrates with 50 kPa stiffness. The solutions were mixed vigorously for 5 minutes after which the mix was degassed using a vacuum pump. After pouring solution on the molds the PDMS was cured by incubation at 60°C for 16 h. After removing the silicon chambers from the molds, chambers were UV sterilized for 1h and finally coated with fibronectin (20 μg/ml) in phosphate-buffered saline (PBS) for 1 h at 37°C prior to cell seeding. 300 000 cells per elastomer (4 cm2) were seeded 16 h before experiment start. 3 h before initiation of cyclic stretch, culture medium was replaced by medium containing 1.8 mM Ca2+ to promote cell-cell contact formation, unless indicated otherwise. Chambers were clamped into the stretching device where a linear stage for uniaxial stretch was driven by a direct current motor with integrated gearbox (RB35, Conrad Electronic SE). Cells were then exposed to 5, 20, 40% stretch at 0.1 Hz frequency. Stretch amplitudes were preselected with an accuracy of 20 μm and they were stable during the experiment.
For biaxial stretch, culture plates with a silicon elastomere membrane (Bioflex; FlexCell International Corporation) were coated with fibronectin (20 μg/ml) in PBS for 1 h at 37°C prior to cell seeding. 600 000 cells per elastomer were seeded 16 h prior to experiment start, and 3 h before initiation of stretch culture medium was replaced by medium containing 1.8 mM Ca2+. Cells were then exposed to cyclic mechanical strain using the Flexcell Tension System (FX4000T; FlexCell International Corporation) at 20% elongation, 0.1Hz frequency.
Chemical treatments
Where indicated, cells were treated with gadolinium (III) chloride hexahydrate (25 μM, Sigma-Aldrich), BAPTA-AM (10 μM, Abcam) or JIB4 (10 μM, Tocris), which were added to the culture 15 min prior to mechanical stretch/compression. Thapsigargin (2 μM, Abcam) was added at the start of live imaging of stretch. The vehicle dimethyl sulfoxide (DMSO; Sigma-Aldrich) was used as a control for all treatments.
Transfections, plasmids and RNAi
siRNAs targeting hPiezo1 (ID: s18892 and s18893), hα-catenin (ID: s3717 and s3718), Lamin A (ID: s8221 and s8222), Suv39H1 (ID: s13658), and negative control siRNA (AM4635) were from Ambion (Silencer Select). Suv39H1-IRES-EGFP was from Sino Biological (ID: 28113), cGAS-EGFP was from Addgene (ID: 86675; Raab et al., 2016) and hPiezo1 3xFlag from GeneCopoeia (ID: Z6777). Telomeres were tagged using co-transfection of a 3-vector CRISPR Rainbow system as described in Ma et al. (2016) using pHAGE-TO-dCas9/pHAGE-EFS-PCP-3xGFPnls/ pLH-sgRNA1-2xPP7 vectors (Addgene ID: 64107, 75390, 75386). Wild-type GFP-tagged Lamin A, S22A and S22A/S329A phosphomutants were a gift from John Eriksson (University of Turku, Finland) and have been described earlier (Kochin et al., 2014). Transfections were performed using Lipofectamine RNAiMax or Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. After 72 h (siRNA), or 24 h (protein expression) of transfection cells were subjected to specific experimental analyses.
Immunofluorescence and confocal microscopy
Cells were fixed in 4% paraformaldehyde (PFA) or ice-cold methanol, permeabilized with 0.3% Triton X-100 in PBS, and blocked in 5% bovine serum albumin (BSA). Samples were subsequently incubated overnight in primary antibody in 1% BSA/0.3% Triton X-100/PBS, followed by washing in PBS and incubation in secondary antibody in 1% BSA/0.3% Triton X-100/PBS. Finally, samples were mounted in Elvanol. The following antibodies were used: H3K9me2,3 (Cell Signaling 5327; 1:1000), H3K9me3 (Cell Signaling 13969; 1:1600) H3K27me3 (Cell Signaling 9733; 1:1000), H3K9me2 (Thermo Fisher PA5-16195; 1:1000) Phospho-Histone H2A.X (Ser139) (Cell Signaling 9718; 1:500), Lamin A/C (Cell Signaling 4777; 1:200), Lamin A/C (Abcam; ab133256; 1:1000), Lamin B1 (Abcam; ab16048; 1:1000), RNA-Pol II (s2P) (Abcam; ab5095; 1:1000), SUV39H1 (Thermo Fisher PA5-29470; 1:1000). Phospho-Myosin LC2 (Thr18/Ser19) (Cell Signaling; 3674; 1:200), E-Cadherin (BD Biosciences; 610181; 1:300), Cleaved caspase3 (Cell Signaling; 9664; 1:1000) α-Catenin (Sigma-Aldrich; C2081 1:500), α-18 (Yonemura et al., 2010), DYKDDDDK Tag (9A3) Flag (Cell Signaling; 8146; 1:1600), PDI (Cell Signaling; 3501; 1:1000). Alexa Fluor 488, 568 and 647 conjugated secondary antibodies (1:300, all from Invitrogen). Actin was labeled with Alexa Fluor 488, 568, or 647-conjugated phalloidin (Invitrogen; 1:600).
All fluorescence images were collected by laser scanning confocal microscopy (SP8X; Leica) with Leica Application Suite software (LAS X version 2.0.0.14332), using 40x, 63x or 100x immersion objectives. Images where acquired at room temperature using sequential scanning of frames of 1 μm thick confocal planes (pinhole 1) after which 10 planes encompassing complete cell nuclei were projected as a maximum intensity confocal stack. Images were collected with the same settings for all samples within an experiment.
Image analyses
Images were analyzed using Fiji (Schindelin et al., 2012). For nuclear imaging of histone modifications, fields were randomly selected based exclusively on the presence of nuclei, as assessed by DAPI staining. Areas of interest were generated using automated thresholding of the DAPI staining, after which mean fluorescence intensities of nuclear stainings were quantified within the areas of interest from maximum projection images obtained as described above. The measured mean nuclear intensities were normalized to DAPI intensity of the corresponding individual nuclei to account for possible unevenness in sample topology.
To measure the distribution of F-actin fiber direction, maximum projections of phalloidin fluorescence were analyzed using the Directionality plug-in in Fiji (Liu, 1991), which is based on Fourier spectrum analysis. E-cadherin-positive adherens junctions were traced to measure the orientation (range −90 to +90°) with respect to a line parallel to the y axis of the image. Two-dimensional nuclear features were quantified from maximum projections of DAPI fluorescence. Nuclear aspect ratio (calculated as major (a) to minor axis (b) ratio and orientation angle (the angle between the long axis and a line parallel to the x axis of the image) were measured by using the “Analyze particles” function in Fiji. Three-dimensional nuclear features were measured from confocal stacks by running 3D Objects Counter and 3D Ellipsoid Fitting plug-ins (Bolte and Cordelières, 2006, Orré et al., 2019).
To quantify perinuclear actin ring intensity, nuclear areas of interest were generated by automatic thresholding on maximum projections of DAPI fluorescence. These areas of interests were then duplicated and dilated by a factor of 0.2 to obtain 0.2 μm-thick bands. The resulting 0.2um-thick bands were redirected on midplanes of phalloidin-stained confocal stacks to quantify perinuclear F-actin intensity.
Live nuclear deformation was quantified by generating areas of interest by automatic thresholding on maximum projections of nuclear dye fluorescence directly before and during stretching the elastomere substrate. We measured nuclear areas , and the equivalent radius was calculated as . was defined as the percentage difference between the equivalent radius during and prior to stretch.
To quantify nuclear rupture, cells were transfected with cGAS-GFP as described above. Total cGAS-GFP intensity was measured by automatic thresholding on maximum projections of cGAS-GFP fluorescence, and in parallel nuclear areas of interest were generated by automatic thresholding on maximum projections of DAPI fluorescence. Nuclear areas of interests were then duplicated and dilated by a factor of 0.4 to obtain 0.4μm-thick bands. The resulting 0.4μm-thick bands were redirected on maximum projections of cGAS-GFP images quantify perinuclear cGAS-GFP intensity. Perinuclear cGAS-GFP intensity values were then normalized to the corresponding cGAS-GFP total intensity.
Mass spectrometry
All mass spectrometry samples, performed in biological triplicates, were lysed in 6 M guanidinium chloride buffer supplemented with 5 mM Tris(2-carboxyethyl)phosphine, 10 mM chloroacetamide in 100mM Tris-HCl. Following lysis samples were heated at 95°C for 10 min, sonicated at high performance for 10 cycles (30 s on/off) using Bioruptor Plus Ultrasonicator (Diagenode), and span down for 20 minutes at room temperature at 20000 g. Supernatants were then Trypsin-Gold digested (Promega Corp., V5280) overnight at 37°C. Following digestion samples were acidified to block trypsin activity and peptides were cleaned with custom-packed C18-SD. Stage Tips. Eluted peptides were vacuum dried at 30°C and either subjected to LC/MS or further processed to enrich for phosphopeptides. Phosphopeptides were enriched using the 3 mg/200 μL Titansphere Phos-TiO kit (GL Sciences) according to manufacturer instructions. Briefly, spin tips were assembled with centrifugal adaptors and connected to waste fluid tubes; 3000 g spins for 2 min at room temperature were used for all steps. Spin tips were first conditioned with 0.4% trifluoroacetic acid/8 0% acetonitrile solution and then equilibrated with 25% lactic acid in equilibration buffer. Peptides were then adsorbed to the spin tips, rinsed twice, and eluted with 5% ammonium hydroxide. After elution phosphopeptides were vacuum dehydrated for 2 hours at 30°C, cleaned with custom-packed C18-SD. Stage Tips. Both phosphopeptides and total peptides were separated on a 25 cm, 75 μm internal diameter PicoFrit analytical column (New Objective) packed with 1.9 μm ReproSil-Pur 120 C18-AQ media (Dr. Maisch) using an EASY-nLC 1200 (Thermo Fisher Scientific). The column was maintained at 50°C. Buffer A and B were 0.1% formic acid in water and 0.1% formic acid in 80% acetonitrile. Peptides were separated on a segmented gradient from 6% to 31% buffer B for 120 min and then from 31% to 50% for 10 min followed by washing and re-equilibration. Eluted peptides from cells were analyzed with an Orbitrap QExactive HF mass spectrometer (Thermo Fisher Scientific). Peptide precursor m/z measurements were carried out at resolution of 60000 in the range of 300 to 1800 m/z. The ten most intense precursors with charge state from 2 to 7 were selected for HCD fragmentation using 25% normalized collision energy. The m/z values of the peptide fragments were measured at a resolution of 30000 using an AGC target of 2e5 and 55 ms maximum injection time. For phophopeptides an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) was used for analyzing. Peptide precursor m/z measurements were carried out at resolution of 60000 in the range of 300 to 1500 m/z. Top speed mode was used to select precursors with charge state from 2 to 7 for HCD fragmentation using 27% normalized collision energy. The m/z values of the peptide fragments were measured at a resolution of 60000 using an AGC target of 2e5 and 120 ms maximum injection time. The raw data were analyzed with MaxQuant version 1.6.1.0 using the integrated Andromeda search engine. Peptide fragmentation spectra were searched against the canonical and isoform sequences of the human reference proteome (proteome ID UP000005640, downloaded September 2018 from UniProt). Methionine oxidation and protein N-terminal acetylation were set as variable modifications; cysteine carbamidomethylation was set as fixed modification. The digestion parameters were set to “specific” and “Trypsin/P,” The minimum number of peptides and razor peptides for protein identification was 1 and the minimum number of unique peptides was 0. Protein identification was performed at a peptide spectrum matches and protein false discovery rate of 0.01. The “second peptide” option was on. Successful identifications were transferred between the different raw files using the “Match between runs” option. Label-free quantification (LFQ) was performed using an LFQ minimum ratio count of 2. LFQ intensities were filtered for a number of valid values that was equal to the minimum number of replicates in an experimental group minus one. Missing values were imputed from a normal distribution with a width of 0.3 and down shift of 1.8. Differential abundance analysis was performed with the Perseus statistical framework (http://maxquant.net/perseus/) version 1.5.2.4. After removing the contaminants and reverse identifications, the intensities were transformed to log2. One-way ANOVA was performed to identify the significantly up- and downregulated proteins. Benjamini-Hochberg FDR 0.05 was used for truncation.
RNA sequencing and analysis
Total RNA was isolated using the NucleoSpin RNA Plus kit (Macherey&Nagel). After quantification and quality control using Agilent 2200 TapeStation, total RNA amounts were adjusted and libraries were prepared using the TruSeq Stranded Total RNA kit with Ribo-zero gold rRNA depletion (Illumina). Paired-end RNA sequencing was carried out on Illumina HiSeq2000 machines by using the 2_100-bp protocol and V3 chemistry from three biological replicates per condition. After quality control, adaptor sequences were removed with flexbar63. Reads mapping to rRNA-related genes were filtered out by using a custom ribosomal RNA-only reference. After preprocessing, reads were mapped using Kallisto version 0.45.0 0 (Bray et al., 2016) creating pseudo-alignments to the human reference genome build hg38. Gene expression was subsequently estimated using featureCounts version v1.6.2. (Liao et al., 2014). FeatureCounts results were aggregated over all samples and pairwise differential gene expression was calculated using the R package DEseq2 version 1.22.2 (Love et al., 2014). Gene ontology term analyses were performed using Panther.
Chromatin immunoprecipitation
400 000 cells per elastomer were seeded a day prior to the experiment. Cells from 6 elastomers were pooled to generate a single biological replicate. Cells were exposed to 40% stretch for 30 min after which both stretched and control cells plated on elastomers were fixed in 1% methanol-free paraformaldehyde (Pierce) in CnT medium for 5 min at room temperature. Fixation was terminated by removing fixative and adding 0.125M Glycine in PBS for 5 min in room temperature. Cells were subsequently rinsed twice with PBS after which cells were scraped in ice-cold PBS containing protease inhibitors, collected, and pelleted by centrifugation at 5000 RPM for 10 minutes at 4°C. Pellets were snap frozen in liquid nitrogen and stored at −80°C. After all biological replicates had been collected, all samples were further processed together. Cell pellets were lysed in lysis buffer (25 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.1% NP-40) after which nuclei were passed through 10 strokes of Dounce homogenization to aid nuclear release and collected by centrifugation at 4600xg for 10 min at 4°C. Pellets were resuspended in 130 μl of sonication buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS + protease inhibitors) and sonicated using a Covaris M20 sonicator (4-9°C temperature range; 5% duty factor 75% peak power) for 4.5 min. After sonication, samples were subjected to centrifugation at 21000xg at 4°C and the supernatant was collected as the chromatin fraction. DNA concentration was measured using Qubit DS High-sensitivity kit (Invitrogen) and sample concentrations were adjusted to be equal. 10% of chromatin was collected as input and 2μl was analyzed using using Agilent 2200 TapeStation to ensure optimal shearing (300-600 bp fragments) and the rest was subjected to immunoprecipitation. For this, 380 μl of sonication buffer and 3 μg of antibody (H3K9me3, Abcam ab8898; H3K27me3, Cell Signaling #9733; or IgG control, Cell Signaling #2729) was added and samples were incubated in an end-over mixer for 16 h at 4°C. 30 μl protein G Dynabeads was subsequently added to each sample, which were then further rotated in an end-over mixer for 4 h at 4°C. Beads were collected using a magnet and washed once with sonication buffer, followed by washes in wash buffer 1 (50 mM HEPES pH 7.9, 500 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS), wash buffer 2 (20 mM Tris pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate), and finally with 20 mM Tris pH 8,0, 1 mM EDTA. Samples were subsequently eluted in 50 mM Tris pH 8.0, 0,1 mM EDTA, 1% SDS at 65°C for 30 min. To reverse crosslinks, 8 μl 1M NaCl and 5 μl of 20mg/mL Proteinase K were added to the samples, which were then incubated first 3h at 42°C and then 65°C for 16 h. Subsequently, 10mg/ml RNase A was added for 2 h 37°C. DNA was purified by phenol:chloroform extraction, followed by ethanol precipitation, after which DNA content was quantified using Qubit. Samples were analyzed by qPCR or next generation sequencing.
ChIP sequencing and analysis
Libraries were prepared from 20 ng ChIP or input DNA using TruSeq ChIP library preparation kit (Illumina) and sequenced using Illumina NovaSeq6000 machines using the 2_100-bp protocol from three biological replicates. After quality control and adaptor removal as described above for RNaseq, reads were mapped to the reference genome hg38 using bowtie2 version 2.3.4.1 (Langmead and Salzberg, 2012). For each sample, broad ChIP peaks were detected using macs2 (Ma et al., 2016) with merged ChIP input files. Consensus peaks for H3K9me3 binding and the subsequent analysis of differential H3K9me3 binding sites were performed using the R package DiffBind version 2.10.0 (Ross-Innes et al., 2012). Consensus peaks were annotated with biotypes using the R package ChIPseeker version 1.5.1 (Yu et al., 2015) and the Bioconductor annotation databases TxDb.Hsapiens.UCSC.hg38.knownGene and org.Hs.eg.db.
Chromosome occupancy profiles were generated using a custom R script with the following steps: Genomic location for summit of consensus peaks was calculated, followed by calculation of relative position of summit by dividing by the respective chromosome size which was rounded to 2 decimal points. The number of peaks with the same relative position were counted, essentially binning the peaks into 1% chromosomal intervals. All chromosomes were compiled by calculating the average number of down/upregulated peaks for each bin after which data was smoothed by averaging over 5 bins.
To quantify overlap between RNaseq and ChIPseq datasets, genomic location of genes was expanded to include the region 5000 bp upstream of the transcription start site. ChIP peaks were intersected with expanded genomic locations of genes expressed in the RNaseq using bedtools intersect version 2.26.0 (Quinlan and Hall, 2010).
Western blotting
Cells were rinsed in phosphate buffered saline (PBS), suspended in lysis buffer (50 mM Tris-HCl buffer (pH 8.0), containing 150 mM NaCl, 1% Triton X-100, 0.05% sodium deoxycholate, 10 mM EDTA, protease and phosphatase inhibitors) and cleared by centrifugation. The lysates were then reduced in Laemmli sample buffer at 95°C, separated by polyacrylamide gel electrophoresis in the presence of SDS and transferred onto PVDF membranes. Membranes were blocked with 5% milk powder in Tris-buffered saline containing 0.05% Tween (TBS-Tween) for 1 h at room temperature, after which primary antibodies were added in 5% BSA, TBS-Tween and incubated over night at +4°C. The membranes were subsequently washed in TBS-Tween after which secondary horseradish peroxidase conjugated antibodies (Bio-Rad) were added in 5% milk powder in TBS-Tween and incubated for 30 min room temperature. After extensive washing in TBS-Tween, antibody binding was detected by chemiluminescence (Immobilon Western, Millipore) using the Bio-Rad ChemiDoc Imaging System. The following antibodies were used: GAPDH (Cell Signaling 2118; 1:5000), total H3 (Cell Signaling 3638; 1:5000), H3K9me2,3 (Cell Signaling 5327; 1:5000), Lamin A/C (Cell Signaling 4777; 1:2500), Lamin B1 (Cell Signaling 9087; 1:1000), Lamin B2 (Cell Signaling 9622; 1:1000), Setdb1 (Abcam ab12317; 1:2000), Suv39h1 (Thermo PA-29470; 1:2000), pLamin A (S22; Invitrogen PA5-17113; 1:2500), pPaxillin (Tyr31; Thermo 44-720G; 1:2500), pCDK1-3 (T14; Cell Signaling 9477; 1:2500), pCofilin (S3; Cell Signaling 3313; 1:2500), Hsp90 (Cell Signaling 4874; 1:2500).
Atomic force microscopy (AFM)
AFM measurements were performed on cell monolayers plated on silicon elastomers using JPK NanoWizard 2 (Bruker Nano) atomic force microscope mounted on an Olympus IX73 inverted fluorescent microscope (Olympus) and operated via JPK SPM Control Software v.5. Elastomers were mounted on the AFM directly after cyclic stretch and measurements were performed within 15 minutes. Triangular non-conductive Silicon Nitride cantilevers (MLCT, Bruker Daltonics) with a nominal spring constant of 0.01 Nm−1 were used for the nanoindentation experiments of the apical surface of cells and the nucleus. For all indentation experiments, forces of up to 3 nN were applied, and the velocities of cantilever approach and retraction were kept constant at 2 μm s−1 ensuring an indentation depth of 500nm. All analyses were performed with JPK Data Processing Software (Bruker Nano). Prior to fitting the Hertz model corrected by the tip geometry to obtain Young’s Modulus (Poisson’s ratio of 0.5), the offset was removed from the baseline, contact point was identified, and cantilever bending was subtracted from all force curves.
Chromatin mobility/rheology
For chromatin rheology studies, confluent EPC monolayers were transfected as described above with pHAGE-TO-dCas9/pHAGE-EFS-PCP-3xGFPnls/ pLH-sgRNA1-2xPP7 vectors to label telomeres, as well as either pEGFP-N1 or Suv91H1-IRES-EGFP. 24 h post transfection, confluent monolayers were subjected to stretch for indicated times and subsequent immediate live imaging at a frame rate of 1000 ms for 5 min using a 60X/1.42 Plan Apo N oil objective and a CoolSnap HQ2 camera mounted on an inverted Olympus IX73 epifluorescence microscope and visualized with PVCamTest software version 3.4.183.
One-dimensional tracking of the fluorophores was performed using custom Laptrack71 programs designed in MATLAB (Natick) as previously published (Booth-Gauthier et al., 2012). Briefly, images were cropped and aligned to remove artifacts (imaging drift, nuclear translation and rotation), only persistent tracks of particles were used for analysis. The ensemble-averaged mean squared displacement (MSD) was calculated versus lag time, τ with error bars of SEM. For tracks that were linear MSD versus τ, the effective diffusivity (Deff) was calculated as MSD = Deffτ on this timescale of measurement. The 95% confidence intervals for Deff values were used for statistical comparison. If 95% confidence intervals did not overlap, values of Deff were considered significantly different. Error bars comparing the Deff represent the 95% confidence interval.
Electron microscopy
The following electron microscopy grade reagents were used - formaldehyde (Electron Microscopy Sciences, EMS-15710), Glutaraldehyde (EM-grade, Sigma, G7651), Osmium tetroxide (Electron Microscopy Sciences, RT 19130), 0.4M sodium cacodylate buffer (Electron Microscopy Sciences, 11654).
Samples for transmission electron microscopy analyses were fixed and prepared exactly as described previously (Ou et al., 2017). Briefly, samples were fixed with 2.5% electron microscopy grade glutaraldehyde in 5 mM CaCl2, 0.1 M sodium cacodylate buffer, pH 7.4 at room temperature for 5 min, following 1 h fixation on ice. Following fixation all the steps were performed on ice. Samples were first washed 5 times with 0.1 M sodium cacodylate buffer and blocked for 15 min in 10 mM glycine/10 mM potassium cyanide in 0.1 M sodium cacodylate buffer (blocking buffer). Following blocking samples were rinsed 1X with 0.1 M sodium cacodylate buffer and then stained with 10 μM DRAQ5 in 0.1% saponin in 0.1 M sodium cacodylate buffer for 10 min, followed by washing 3x for 5 min with above-described blocking buffer. Samples were then incubated with 2.5 mM diaminobezidine tetrahydrochloride in 0.1 M sodium cacodylate buffer and placed onto a glass-bottom dish, monolayer side down, then mounted onto Zeiss confocal microscope surrounded by ice-packs to maintain a near 4°C temperature and imaged using a 63X oil immersion objective lens and 633 nm filter set to continuously illuminate the membranes for 10 min. Samples were then rinsed 3x for 5 min with 0.1 M sodium cacodylate buffer, following a 1 h stain with 1% osmium tetroxide, 2 mM CaCl2, 1.5% potassium ferrocyanide in 0.15 M sodium cacodylate buffer. Post staining, samples were washed twice for 2 min with double-distilled water, dehydrated in ethanol series prior to gradual infiltration into Epon (TAAB 812), then cut into 80 nm sections and mounted on copper slot grids for imaging on a conventional 80 kV Jeol JEM-1400 (Jeol Ltd).
Electron tomography
For 3D electron tomography samples were prepared as described above. After polymerization in Epon overnight at 60°C, a pyramid was made using a razor blade on the area of cells of darker, photo-oxidized nucleus. 230-nm-thick sections were cut with a 35° diamond knife (Diatome) on Leica EM Ultracut UC7 ultramicrotome (Leica) and collected on Pioloform-coated single slot copper grids.
Dual axis tilt series were recorded from a semi-thick section using SerialEM (Mastronarde, 1997) software running on a Tecnai FEG 20 transmission electron microscope (Thermo Fisher Scientific) operated at 200 kV, high tilt specimen holder (model 2020; E.A. Fischione Instruments) and a 4k by 4k Ultrascan CCD camera (Gatan Inc). The tilt series were acquired at one-degree intervals between ± 62° at nominal magnification of 14,500x and 10-nm gold particles placed on both grid faces served as fiducial markers for alignment. Prior alignment and reconstruction with IMOD software package (versions 4.9.0 and 4.10.18) (Kremer et al., 1996) the images were binned by two providing a pixel size of 1.5 nm. The tomograms were reconstructed with a simultaneous iterative reconstruction technique using 14 iterations.
For visualization of the condensed chromatin the tomograms were binned by 2, scaled to a common mean and standard deviation using IMOD software package (version 4.10.32). The condensed DNA was then selected within the nucleus (nuclei segmented using MIB, version 2.501 (Belevich et al., 2016)) by gray level thresholding (center value 127), removing separate pixels by smoothing and excluding objects smaller than 800, fraction 50 using Amira software (version 5.3.2).
For quantification of lamina-associated condensed chromatin, the surface area of condensed chromatin with direct contact to the lamina was measured by manual tracking. The surface area was subsequently normalized to the nuclear perimeter.
Membrane tension measurements
Flipper-TR fluorescent tension probe (Spirochrome, SC020) was utilized to quantify membrane tension (Colom et al., 2018). To stain the outer nuclear membranes, 1 μM Flipper-TR was applied 16 h prior to initiating cell stretching and subsequent FLIM measurements. Elastomers were mounted on the microscope directly after stretching and imaged for maximum 10 min. FLIM imaging was performed using a Zeiss LSM 880 confocal microscope equipped with a time correlated single photon counting module from PicoQuant. Zen Black and SymPhoTime 64 software were used to record the data in photon-counting mode using a 40x Plan-Apochromat 1.40 NA oil immersion objective. Excitation was performed using a pulsed 488 nm laser operating at 20 MHz with emission collected through bandpass 565/610 nm filter gated with a GaAsP detector and a TimeHarp 260 PICO board. FLIMfit 5.1.1. software (Imperial College London) was used to fit fluorescence decay data from regions of interest to a dual exponential model after deconvolution for the instrument response function/IRF, measured using backscattered emission light of a saturated Erythrosin B with KI. The longest lifetime with the higher fit amplitude was used to quantify membrane tension.
Super resolution microscopy
For Stochastic Optical Reconstruction Microscopy (STORM) cells were fixed in 3% paraformaldehyde/0.1% glutaraldehyde for 10 min at room temperature then washed with 0.1% sodium borohydride for 5 min then twice with PBS. Samples were then blocked and stained as described above for immunofluorescence and confocal microscopy (using exclusively Alexa-647 as a secondary antibody) following a post-stain with 3% paraformaldehyde/0.1% glutaraldehyde and subsequent three washes in PBS. Samples were then stored at 4°C until imaging. Imaging was performed in β-mercaptoethanol/glucose oxidase/catalase buffer on a GE Deltavision OMX SR using a 63X Apo N 60x/1.49 NA oil TIRF objective. Image reconstruction was performed using Thunder STORM Fiji plugin (Ovesný et al., 2014).
For 3D structured illumination microscopy (SIM) cells were fixed with 4% paraformaldehyde for 20 min at room temperature, washed 3 times with PBS, and subjected to immunofluorescence as described above using Alexa 488 and 561 secondary antibodies. Samples were mounted in hardening Prolong Glass mounting media (Thermo P36982) on high performance coverslips (Zeiss) and allowed to cure for 2 days at room temperature prior to imaging in order to draw silicone substrates close to the coverslips. Imaging was performed on GE Deltavision OMX SR using 1.520 oil with PlanApo N 60X/1.42 Oil. Image reconstruction was performed using SoftWoRx 7.0 software.
Calcium and nuclear deformation imaging
For live imaging of intracellular Ca2+, Cal-590 AM calcium dye (2 μM; AAT Bioquest 20510) was applied on cells for 10 min at 37°C, then washed twice with PBS and imaged in fresh medium 20 min post staining. For real-time nuclear deformation measurements, cells were stained with 1μM live cell nuclear dye (Vybrant; Thermo Fisher V35004). Cells were imaged using a 20X/0.45 PlanFL N air objective and a CoolSnap HQ2 camera mounted on an inverted Olympus IX73 epifluorescence microscope, with the uniaxial stretch device described above mounted on the microscope stage. Images were acquired with PVCamTest software version 3.4.183. Images were acquired continuously under basal conditions and during stretch, after which the effect of bleaching was corrected by normalizing all fluorescence signals to corresponding background fluorescence signals.
qPCR
RNA was isolated using the Nucleospin RNA Plus kit (Macherey&Nagel), after which cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed on the StepOne Plus Real Time PCR System (Applied Biosystems) using the DyNAmo ColorFlash SYBR Green Mix (Thermo Fisher). Gene expression changes were calculated following normalization to B2M using the comparative Ct (cycle threshold) method. The primers used are provided in Table S4.
Nuclear compression
Compression was performed with a previously published cell confiner system (Le Berre et al., 2012). Briefly, suction cup-bound coverslips with polydimethylsiloxane (PDMS) pillars of either 4.2 μm (about 20% compression) or 20 μm (uncompressed controls) height, based on nuclear height measured from live cells, were driven on top of EPC monolayers using a controlled pressure pump (Elveflow). Compression was held on the nuclei for 15 minutes in the presence/absence of chemical treatments, as indicated, and fixation in 4% PFA was performed directly at the end of the compression period.
Mice, ex vivo stretch and immunofluorescence of tissue
Embryonic day 15.5 NMRI embryos were isolated and their full-thickness back skin was excised. The skin biopsies were placed in keratinocyte growth medium (Chacón-Martínez et al., 2017), attached to silicon elastomers using small pins and subjected to 40% uniaxial cyclic stretch parallel to the medial-lateral axis. Control skins were treated in identical manner without mechanical stretch. Cell viability and tissue integrity after stretch was confirmed by histology and cleaved caspase-3 staining.
Tissue biopsies were subsequently fixed in 4% paraformaldehyde for 1h at room temperature after which whole mount stainings were performed as described above for immunofluorescence staining, with the exception of stronger permeabilization (0.6% Triton X-100 in PBS) and overnight primary antibody incubation being carried out at 37C°.
Limbs from E15.5 NMRI embryos were fixed with 4% paraformaldehyde for 2 h at room temperature and embedded in paraffin. 8 μm sections were cut with a microtome and deparaffinized using standard protocols. After antigen retrieval in citrate buffer using a pressure cooker, sections were blocked with 5% goat serum/3%BSA and subjected to immunofluorescence staining and analysis as described above.
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (GraphPad, version 8). Statistical significance was determined by the specific tests indicated in the corresponding figure legends. In all cases where a test for normally distributed data was used, normal distribution was confirmed with the Kolmogorov–Smirnov test (α = 0.05). All experiments presented in the manuscript were repeated at least in 3 independent experiments/biological replicates. No datapoints were excluded from the analyses.
Data and Code Availability
The accession number for the ChIP and RNA sequencing data reported in this paper is GEO: GSE143519. All custom analysis scripts and data that support the conclusions are available from the authors on request.
Acknowledgments
We thank Matthieu Piel (Institute Curie) for support with compression experiments and manuscript comments, Samuli Ollila (University of Helsinki) for advice on membrane tension, Anu Luoto and Mervi Lindman for technical assistance, Karlo Skube for help with protein analyses, John Eriksson (University of Turku) for the Lamin A plasmids, the FACS & Imaging Facility of MPI for Biology of Ageing, and Marko Crivaro and Jussi Kenkkilä (the Biomedicum Helsinki Imaging Unit) for imaging and image analysis support. We thank Janine Altmüller and Christian Becker (Cologne Center for Genomics) for sequencing. This work was supported by the Helsinki Institute of Life Science, the Wihuri Research Institute, the Academy of Finland, the Max Planck Society, the Max Planck Förderstiftung, and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement 770877 - STEMpop) (all to S.A.W.); the Deutsche Forschungsgemeinschaft (DFG; project number 73111208 - SFB 829 to S.A.W. and C.M.N.); Germany’s Excellence Strategy (CECAD; EXC 2030 – 390661388 to S.A.W. and C.M.N.; SPP 1782 to C.M.N., B.H., and R.M.; and FOR 2743 to B.H. and R.M.); the NSF (CMMI 1300476 to K.N.D.); and the NIH (EB003392 to D.B.W.). M.M.N. is supported by the ERC under the European Union’s Horizon 2020 research and innovation program (Marie Sklodowska-Curie Actions, grant agreement 748004 - MiMEtiC). Y.A.M. is the recipient of EMBO Long-Term fellowship ALTF 728-2017 and Human Frontier Science Program fellowship LT000861/2018.
Author Contributions
M.M.N. and Y.A.M. designed, performed, and analyzed most experiments. L.C.B. performed the in vivo experiments. D.B.W. and K.N.D. performed chromatin rheology measurements. F.M. and J.B. analyzed sequencing data. J.M.G.A. designed compression experiments. B.H. and R.M. designed and produced the stretch device. H.V. and E.J. performed electron microscopy. X.L. performed the proteomics. C.M.N. provided conceptual advice, designed experiments, and analyzed data. S.A.W. conceived and supervised the study and wrote the paper. All authors edited the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: April 16, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.03.052.
Supplemental Information
Differential distance-based cluster analysis of phosphosites from EPCs subjected to either 0% or 40% uniaxial stretch for 30 and 360 min.
Differential peak analyses of H3K9me3 chromatin immunoprecipitation sequencing of cells exposed to either 0% or 40% uniaxial stretch for 30 min.
Differential gene expression analyses of cells exposed to either 0% or 40% uniaxial stretch for 30 or 360 min.
References
- Adding L.C., Bannenberg G.L., Gustafsson L.E. Basic experimental studies and clinical aspects of gadolinium salts and chelates. Cardiovasc. Drug Rev. 2001;19:41–56. doi: 10.1111/j.1527-3466.2001.tb00182.x. [DOI] [PubMed] [Google Scholar]
- Anlaş A.A., Nelson C.M. Tissue mechanics regulates form, function, and dysfunction. Curr. Opin. Cell Biol. 2018;54:98–105. doi: 10.1016/j.ceb.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.S., Nicetto D., Zaret K.S. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 2016;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belevich I., Joensuu M., Kumar D., Vihinen H., Jokitalo E. Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLoS Biol. 2016;14:e1002340. doi: 10.1371/journal.pbio.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q., Khanna N., Alvikas J., Belmont A.S. β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol. 2013;203:767–783. doi: 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore W.A., van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Blanchard G.B., Kabla A.J., Schultz N.L., Butler L.C., Sanson B., Gorfinkiel N., Mahadevan L., Adams R.J. Tissue tectonics: morphogenetic strain rates, cell shape change and intercalation. Nat. Methods. 2009;6:458–464. doi: 10.1038/nmeth.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Mohrin M., Sotiropoulou P.A., Passegué E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8:16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Bonakdar N., Gerum R., Kuhn M., Spörrer M., Lippert A., Schneider W., Aifantis K.E., Fabry B. Mechanical plasticity of cells. Nat. Mater. 2016;15:1090–1094. doi: 10.1038/nmat4689. [DOI] [PubMed] [Google Scholar]
- Booth-Gauthier E.A., Alcoser T.A., Yang G., Dahl K.N. Force-induced changes in subnuclear movement and rheology. Biophys. J. 2012;103:2423–2431. doi: 10.1016/j.bpj.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Buchwalter A., Kaneshiro J.M., Hetzer M.W. Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 2019;20:39–50. doi: 10.1038/s41576-018-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A., Irianto J., Swift J., Athirasala A., Shin J.-W., Rehfeldt F., Discher D.E. Coordinated increase of nuclear tension and lamin-A with matrix stiffness outcompetes lamin-B receptor that favors soft tissue phenotypes. Mol. Biol. Cell. 2017;28:3333–3348. doi: 10.1091/mbc.E17-06-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares L., Vincent R., Zalvidea D., Campillo N., Navajas D., Arroyo M., Trepat X. Hydraulic fracture during epithelial stretching. Nat. Mater. 2015;14:343–351. doi: 10.1038/nmat4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-Martínez C.A., Klose M., Niemann C., Glauche I., Wickström S.A. Hair follicle stem cell cultures reveal self-organizing plasticity of stem cells and their progeny. EMBO J. 2017;36:151–164. doi: 10.15252/embj.201694902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalut K.J., Höpfler M., Lautenschläger F., Boyde L., Chan C.J., Ekpenyong A., Martinez-Arias A., Guck J. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys. J. 2012;103:2060–2070. doi: 10.1016/j.bpj.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Vigliotti A., Bacca M., McMeeking R.M., Deshpande V.S., Holmes J.W. Role of boundary conditions in determining cell alignment in response to stretch. Proc. Natl. Acad. Sci. USA. 2018;115:986–991. doi: 10.1073/pnas.1715059115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Irianto J., Discher D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017;216:305–315. doi: 10.1083/jcb.201610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom A., Derivery E., Soleimanpour S., Tomba C., Molin M.D., Sakai N., González-Gaitán M., Matile S., Roux A. A fluorescent membrane tension probe. Nat. Chem. 2018;10:1118–1125. doi: 10.1038/s41557-018-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B., Mathur J., Schmidt M., Earley T.J., Ranade S., Petrus M.J., Dubin A.E., Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubiles M.D., Barroso S., Vaquero-Sedas M.I., Enguix A., Aguilera A., Vega-Palas M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018;46:2347–2355. doi: 10.1093/nar/gky006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De R., Zemel A., Safran S.A. Dynamics of cell orientation. Nat. Phys. 2007;3:655–659. [Google Scholar]
- De R., Zemel A., Safran S.A. Do cells sense stress or strain? Measurement of cellular orientation can provide a clue. Biophys. J. 2008;94:L29–L31. doi: 10.1529/biophysj.107.126060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi B., Niethammer P. Nuclear membrane stretch and its role in mechanotransduction. Nucleus. 2017;8:156–161. doi: 10.1080/19491034.2016.1263411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust U., Hampe N., Rubner W., Kirchgessner N., Safran S., Hoffmann B., Merkel R. Cyclic stress at mHz frequencies aligns fibroblasts in direction of zero strain. PLoS ONE. 2011;6:e28963. doi: 10.1371/journal.pone.0028963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimenko J., Maruyama Y., Tepikin A., Petersen O.H., Gerasimenko O. Calcium signalling in and around the nuclear envelope. Biochem. Soc. Trans. 2003;31:76–78. doi: 10.1042/bst0310076. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- Gudipaty S.A., Lindblom J., Loftus P.D., Redd M.J., Edes K., Davey C.F., Krishnegowda V., Rosenblatt J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature. 2017;543:118–121. doi: 10.1038/nature21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger S., Laurell E., Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Harris A.R., Peter L., Bellis J., Baum B., Kabla A.J., Charras G.T. Characterizing the mechanics of cultured cell monolayers. Proc. Natl. Acad. Sci. USA. 2012;109:16449–16454. doi: 10.1073/pnas.1213301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jor J.W.Y., Parker M.D., Taberner A.J., Nash M.P., Nielsen P.M.F. Computational and experimental characterization of skin mechanics: identifying current challenges and future directions. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013;5:539–556. doi: 10.1002/wsbm.1228. [DOI] [PubMed] [Google Scholar]
- Kochin V., Shimi T., Torvaldson E., Adam S.A., Goldman A., Pack C.-G., Melo-Cardenas J., Imanishi S.Y., Goldman R.D., Eriksson J.E. Interphase phosphorylation of lamin A. J. Cell Sci. 2014;127:2683–2696. doi: 10.1242/jcs.141820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmannsberger P., Fabry B. Linear and nonlinear rheology of living cells. Annu. Rev. Mater. Res. 2011;41:75–97. [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E., Kale S., Casares L., Gómez-González M., Uroz M., Valon L., Nair R.V., Garreta E., Montserrat N., Del Campo A. Active superelasticity in three-dimensional epithelia of controlled shape. Nature. 2018;563:203–208. doi: 10.1038/s41586-018-0671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H.Q., Ghatak S., Yeung C.Y.C., Tellkamp F., Günschmann C., Dieterich C., Yeroslaviz A., Habermann B., Pombo A., Niessen C.M., Wickström S.A. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 2016;18:864–875. doi: 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- Le Berre M., Aubertin J., Piel M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr. Biol. 2012;4:1406–1414. doi: 10.1039/c2ib20056b. [DOI] [PubMed] [Google Scholar]
- Lewis A.H., Grandl J. Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. eLife. 2015;4:e12088. doi: 10.7554/eLife.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu Z.-Q. Scale space approach to directional analysis of images. Appl. Opt. 1991;30:1369–1373. doi: 10.1364/AO.30.001369. [DOI] [PubMed] [Google Scholar]
- Liu X., Maiti R., Lu Z.H., Carré M.J., Matcher S.J., Lewis R. New non-invasive techniques to quantify skin surface strain and sub-surface layer deformation of finger-pad during sliding. Biotribology (Oxf.) 2017;12:52–58. [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., Pederson T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016;34:528–530. doi: 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti R., Gerhardt L.C., Lee Z.S., Byers R.A., Woods D., Sanz-Herrera J.A., Franklin S.E., Lewis R., Matcher S.J., Carré M.J. In vivo measurement of skin surface strain and sub-surface layer deformation induced by natural tissue stretching. J. Mech. Behav. Biomed. Mater. 2016;62:556–569. doi: 10.1016/j.jmbbm.2016.05.035. [DOI] [PubMed] [Google Scholar]
- Mandal P.K., Blanpain C., Rossi D.J. DNA damage response in adult stem cells: pathways and consequences. Nat. Rev. Mol. Cell Biol. 2011;12:198–202. doi: 10.1038/nrm3060. [DOI] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- McHugh B.J., Buttery R., Lad Y., Banks S., Haslett C., Sethi T. Integrin activation by Fam38A uses a novel mechanism of R-Ras targeting to the endoplasmic reticulum. J. Cell Sci. 2010;123:51–61. doi: 10.1242/jcs.056424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W., Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb. Perspect. Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnikova Y.A., Nava M.M., Wickström S.A. Emerging roles of mechanical forces in chromatin regulation. J. Cell Sci. 2017;130:2243–2250. doi: 10.1242/jcs.202192. [DOI] [PubMed] [Google Scholar]
- Nicetto D., Donahue G., Jain T., Peng T., Sidoli S., Sheng L., Montavon T., Becker J.S., Grindheim J.M., Blahnik K. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science. 2019;363:294–297. doi: 10.1126/science.aau0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noethel B., Ramms L., Dreissen G., Hoffmann M., Springer R., Rübsam M., Ziegler W.H., Niessen C.M., Merkel R., Hoffmann B. Transition of responsive mechanosensitive elements from focal adhesions to adherens junctions on epithelial differentiation. Mol. Biol. Cell. 2018;29:2317–2325. doi: 10.1091/mbc.E17-06-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbink-Huizer C., Oomens C.W.J., Loerakker S., Foolen J., Bouten C.V.C., Baaijens F.P.T. Computational model predicts cell orientation in response to a range of mechanical stimuli. Biomech. Model. Mechanobiol. 2014;13:227–236. doi: 10.1007/s10237-013-0501-4. [DOI] [PubMed] [Google Scholar]
- Obropta E.W., Newman D.J. Skin strain fields at the shoulder joint for mechanical counter pressure space suit development. Proceedings of the 2016 IEEE Aerospace Conference. 2016:1–9. [Google Scholar]
- Orré T., Rossier O., Giannone G. The inner life of integrin adhesion sites: From single molecules to functional macromolecular complexes. Exp. Cell Res. 2019;379:235–244. doi: 10.1016/j.yexcr.2019.03.036. [DOI] [PubMed] [Google Scholar]
- Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., O’Shea C.C. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný M., Křížek P., Borkovec J., Švindrych Z., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.-R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.-M., Manel N., Piel M. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K., Faix J., Bogdan S., Linder S., Kerkhoff E. Actin assembly mechanisms at a glance. J. Cell Sci. 2017;130:3427–3435. doi: 10.1242/jcs.206433. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnol S.T., Dahl K.N. Active cytoskeletal force and chromatin condensation independently modulate intranuclear network fluctuations. Integr. Biol. 2014;6:523–531. doi: 10.1039/c3ib40226f. [DOI] [PubMed] [Google Scholar]
- Spagnol S.T., Dahl K.N. Spatially resolved quantification of chromatin condensation through differential local rheology in cell nuclei fluorescence lifetime imaging. PLoS ONE. 2016;11:1–19. doi: 10.1371/journal.pone.0146244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Banigan E.J., Adam S.A., Goldman R.D., Marko J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 2017;28:1984–1996. doi: 10.1091/mbc.E16-09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Liu P.Z., Banigan E.J., Almassalha L.M., Backman V., Adam S.A., Goldman R.D., Marko J.F. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell. 2018;29:220–233. doi: 10.1091/mbc.E17-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Liu P.Z., Kandula V., Chen H., Almassalha L.M., Herman C., Backman V., O’Halloran T., Adam S.A., Goldman R.D. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell. 2019;30:2320–2330. doi: 10.1091/mbc.E19-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui L., Alt S., Weigert M., Dye N., Eaton S., Jug F., Myers E.W., Jülicher F., Salbreux G., Dahmann C. Differential lateral and basal tension drive folding of Drosophila wing discs through two distinct mechanisms. Nat. Commun. 2018;9:4620. doi: 10.1038/s41467-018-06497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Jin M.W., Xiang J.Z., Dong M.Q., Sun H.Y., Lau C.P., Li G.R. The membrane permeable calcium chelator BAPTA-AM directly blocks human ether a-go-go-related gene potassium channels stably expressed in HEK 293 cells. Biochem. Pharmacol. 2007;74:1596–1607. doi: 10.1016/j.bcp.2007.07.042. [DOI] [PubMed] [Google Scholar]
- Trepat X., Lenormand G., Fredberg J.J. Universality in cell mechanics. Soft Matter. 2008;4:1750–1759. [Google Scholar]
- Tsien R.Y. A non-disruptive technique for loading calcium buffers and indicators into cells. Nature. 1981;290:527–528. doi: 10.1038/290527a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Belmont A.S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales P., Schuberth C.E., Aufschnaiter R., Fels J., García-Aguilar I., Janning A., Dlugos C.P., Schäfer-Herte M., Klingner C., Wälte M. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife. 2016;5:e19850. doi: 10.7554/eLife.19850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walko G., Castañón M.J., Wiche G. Molecular architecture and function of the hemidesmosome. Cell Tissue Res. 2015;360:363–378. doi: 10.1007/s00441-014-2061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H.-C., Goldschmidt-Clermont P., Wille J., Yin F.C.-P. Specificity of endothelial cell reorientation in response to cyclic mechanical stretching. J. Biomech. 2001;34:1563–1572. doi: 10.1016/s0021-9290(01)00150-6. [DOI] [PubMed] [Google Scholar]
- Wang L., Chang J., Varghese D., Dellinger M., Kumar S., Best A.M., Ruiz J., Bruick R., Peña-Llopis S., Xu J. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessendorf A.M., Newman D.J. Dynamic understanding of human-skin movement and strain-field analysis. IEEE Trans. Biomed. Eng. 2012;59:3432–3438. doi: 10.1109/TBME.2012.2215859. [DOI] [PubMed] [Google Scholar]
- Xia Y., Ivanovska I.L., Zhu K., Smith L., Irianto J., Pfeifer C.R., Alvey C.M., Ji J., Liu D., Cho S. Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol. 2018;217:3796–3808. doi: 10.1083/jcb.201711161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T., Poduch K., Ryba T., Lu J., Hiratani I., Tachibana M., Shinkai Y., Gilbert D.M. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc. Natl. Acad. Sci. USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EPC monolayers were subjected to 0% (control) stretch, fixed and processed for electron tomography. Movie shows a model of condensed chromatin (purple) and nuclear envelope (yellow). Tomograms were scaled to a common mean and segmented applying thresholding, smoothing, and size exclusion. High-density compacted chromatin is seen at the nuclear lamina. See also Videos S2 and S3.
EPC monolayers were subjected to 40% stretch for 30 min, fixed, and processed for electron tomography. Movie shows a model of condensed chromatin (purple) and nuclear envelope (yellow). Tomograms were scaled to a common mean and segmented applying thresholding, smoothing, and size exclusion. High-density compacted chromatin at the nuclear lamina is decreased with 30 min of stretch. See also Videos S1 and S3.
EPC monolayers were subjected to 40% stretch for 360 min, fixed, and processed for electron tomography. Movie shows a model of condensed chromatin (purple) and nuclear envelope (yellow). Tomograms were scaled to a common mean and segmented similarly applying thresholding, smoothing, and size exclusion. High-density compacted chromatin is restored to baseline levels at 360 min of stretch. See also Videos S1 and S2.
Time-lapse epifluorescence video (in pseudocolor) of an EPC monolayer stained with Cal-590 calcium dye shows induction of intracellular calcium upon cyclic stretch. Images of were acquired continuously for 5 mins prior and during 10 cycles of 40% stretch application 0.3 s/frame.
Differential distance-based cluster analysis of phosphosites from EPCs subjected to either 0% or 40% uniaxial stretch for 30 and 360 min.
Differential peak analyses of H3K9me3 chromatin immunoprecipitation sequencing of cells exposed to either 0% or 40% uniaxial stretch for 30 min.
Differential gene expression analyses of cells exposed to either 0% or 40% uniaxial stretch for 30 or 360 min.
Data Availability Statement
The accession number for the ChIP and RNA sequencing data reported in this paper is GEO: GSE143519. All custom analysis scripts and data that support the conclusions are available from the authors on request.