Figure 7.
Supracellular Monolayer Alignment Prevents Stress Transmission to the Nucleus and Chromatin
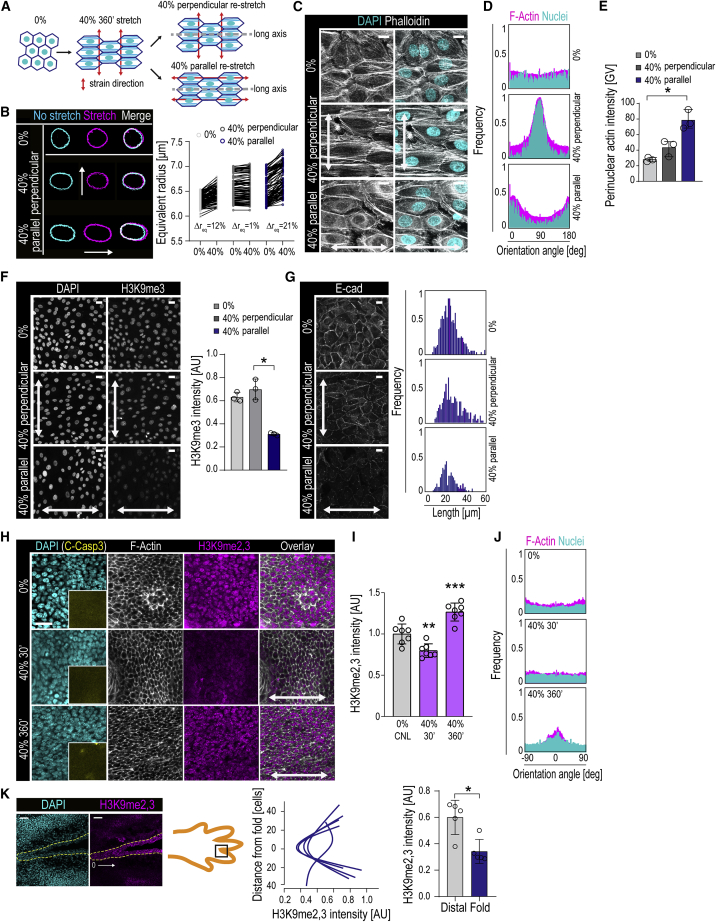
(A) Experimental design to alter stretch direction after monolayer alignment.
(B) Representative nuclear outlines and quantifications of equivalent radius before and during 40% stretch to measure stretch-induced nuclear deformation. is % difference between equivalent radius during and prior to stretch. Note the lack of nuclear deformation in perpendicularly aligned nuclei, whereas nuclei aligned parallel show substantial deformation. (n > 100 cells/condition pooled across three independent experiments).
(C) Representative F-actin (phalloidin) and DAPI images of cells after second 40% stretch regime. Note the alignment of fibers at 40% 360 min before second stretch regime and monolayer disruption and reappearance of perinuclear actin after stretch direction change.
(D) Quantification of F-actin alignment from experiments in (C) (frequency distribution of > 500 cells/condition pooled across 3 independent experiments).
(E) Quantification of perinuclear actin intensity from experiments in (C) (n = 3 independent experiments with > 250 cells/condition/experiment; ∗p = 0.0429, Friedman/Dunn’s).
(F) Representative images and quantification of H3K9me3 intensity before and after stretch direction change (n = 3 independent experiments with > 300 cells/condition/experiment; ∗p = 0.0278, Friedman/Dunn’s).
(G) Representative images and quantification of E-cadherin (E-cad) junctions show loss of junction integrity after stretch direction change (frequency distribution of >300 cells/condition pooled across three independent experiments).
(H–J) Representative images (H) and quantification of H3K9me2,3 intensity (I) and cell alignment (J) in mouse embryonic day 15.5 (E15.5) skin explants exposed to 40% stretch. Note the transient suppression of H3K9me2,3 intensity at 30 min, absence of apoptosis (inset, cleaved caspase-3), and emergence of alignment at 360 min of stretch (n = 7 mice/condition; ∗∗p = 0.0039, ∗∗∗p = 0.0003, ANOVA/Dunnett’s).
(K) Representative images and quantification of H3K9me2,3 intensity in mouse embryo epidermal digit folds. H3K9me2,3 intensity is lower in epidermal stem cells (dotted line) in folds (n = 5 digit folds from 3 mice; 0.0159, Mann-Whitney).
Bar graphs show mean ± SD, Scale bars represent 40 μm in (K) and 10 μm in other panels. White arrows indicate stretch direction. AU, arbitrary units.
See also Figure S7.