Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has had devastating global impacts and will continue to have dramatic effects on public health for years to come. A better understanding of the immune response to SARS-CoV-2 will be critical for the application and development of therapeutics. The degree to which the innate immune response confers protection or induces pathogenesis through a dysregulated immune response remains unclear. In this review, we discuss what is known about the role of the innate immune system during SARS-CoV-2 infection, suggest directions for future studies, and evaluate proposed COVID-19 immunomodulating therapeutics.
Keywords: COVID-19, SARS-CoV-2, innate immune response, cytokines, myeloid cells, natural killer cells, complement
In this Minireview, Julia McKechnie and Catherine Blish discuss recent studies on the role of the innate immune response during SARS-Cov2 infection. They provide insights into the potential of immunomodulatory drugs as therapies and identify gaps for furthering our understanding of COVID-19.
Main Text
Introduction
The recent pandemic of coronavirus disease 2019 (COVID-19) began in December 2019 and is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). As of May 12, 2020, COVID-19 has resulted in more than four million cases and 291,000 deaths globally. Approximately 80% of COVID-19 patients experience mild symptoms, such as cough and fever, which do not require hospitalization (Huang et al., 2020). Of the 20% that do require hospitalization, approximately 50% develop severe respiratory failure, currently considered a form of acute respiratory distress syndrome (ARDS) (Guan et al., 2020). Some reports suggest that the severe respiratory failure observed in COVID-19 is atypical of ARDS (Gattinoni et al., 2020, Tang et al., 2020). This, combined with the fact that some patients’ symptoms suddenly worsen around one week after symptom onset, suggests that severe COVID-19 pathogenesis could be mediated by a uniquely dysregulated immune response.
The components of the innate immune system act as first responders for the detection and clearance of viral infections. Innate immune cells secrete proinflammatory cytokines that inhibit viral replication, stimulate the adaptive immune response, and recruit other immune cells to the site of infection. Granulocytes degranulate in response to extracellular pathogens, releasing enzymes and toxic proteins. Monocytes traffic to tissues and differentiate into monocyte-derived macrophages and dendritic cells (mDCs). Macrophages and neutrophils phagocytose and destroy pathogens as well as infected cells. Activated DCs present pathogen-derived antigens to naive helper T cells to initiate the adaptive immune response. Natural killer (NK) cells kill virally infected cells via degranulation, receptor-mediated apoptosis, and antibody-dependent cell-mediated cytotoxicity. Finally, the complement system plays a role in immune cell recruitment, activation, and destruction of pathogens. In spite of these critical antiviral functions, an overactive innate immune response can contribute to disease pathogenesis. This Minireview will cover what is currently known about the innate immune response to SARS-CoV-2 infection, how this relates to the pathogenesis of COVID-19, and the implications for development of therapeutics. Although lung epithelial cells also express innate immune receptors and produce inflammatory cytokines in response to SARS-CoV-2 infection (Blanco-Melo et al., 2020), this antiviral response has been discussed previously (Vareille et al., 2011) and will not be the focus of this Minireview.
The Cytokine Response to SARS-CoV-2 Infection
Several studies have reported an association between progression to severe COVID-19 and dysregulated secretion of proinflammatory cytokines. Both intensive care unit (ICU) and non-ICU COVID-19 patients in Wuhan, China had increased plasma concentrations of IL-1β, IL-1Ra, IL-7, IL-8, IL-9, IL-10, basic FGF, GCSF, GM-CSF, IFN-ɣ, CXCL10, CCL2, CCL3, CCL4, PDGF, TNFɑ, and VEGF in comparison with levels seem in healthy controls (Huang et al., 2020). Further, in comparison with non-ICU patients, ICU patients had higher concentrations of IL-2, IL-7, IL-10, GCSF, CXCL10, CCL2, CCL3, and TNFɑ. Similar results were reported in a USA cohort where increases in circulating IL-6, IL-1Ra, CCL2, CCL8, CXCL2, CXCL8, CXCL9, and CXCL16 were observed in SARS-CoV-2+ patients in comparison with patients with non-COVID-19-related respiratory issues (Blanco-Melo et al., 2020). Neither study reported significant increases in type I or type III interferons (IFNs), which could be due to the short-lived nature of these cytokines in vivo. Importantly, Blanco-Melo et al. reported a moderate IFN response to SARS-CoV-2 infection in primary cells and found that IFN can efficiently restrict SARS-CoV-2 replication in vitro. These data suggest that unlike SARS-CoV, SARS-CoV-2 triggers an IFN response which could limit viral spread.
Further analysis of the COVID-19 cytokine response was performed by classifying critically ill patients according to criteria used in sepsis, which are categorized as (1) macrophage-activation syndrome (MAS), (2) immune dysregulation characterized by low human leukocyte antigen DR (HLA-DR) expression on CD14+ monocytes without elevated ferritin, and (3) an intermediate state lacking obvious dysregulation (Giamarellos-Bourboulis et al., 2020). COVID-19 patients falling into the dysregulation and MAS categories had significantly increased IL-6 concentrations in comparison with the intermediate group. Upon stimulation with lipopolysaccharide (LPS), peripheral blood mononuclear cells (PBMCs) from intermediate, dysregulated, and MAS patients produced the same amounts of TNFɑ and IL-6, but IL-1β production was significantly decreased in dysregulated and MAS PBMCs in comparison with intermediate PBMCs. This suggests some level of immunoparalysis; however, no comparison was made to stimulated healthy control PBMCs.
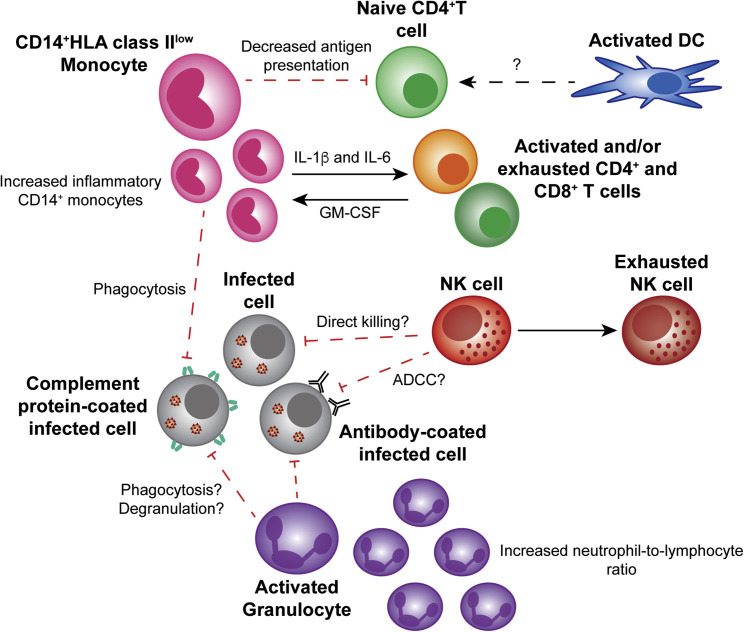
Interestingly, the proinflammatory cytokine IL-1β and/or its agonist IL-1Ra have been implicated in all the aforementioned studies. Longitudinal transcriptional analysis of whole blood from one severe COVID-19 patient suggests that only elevated IL-1A and IL-1B expression preceded the lowest point in respiratory function, with other cytokines, including IL-6, peaking after respiratory improvement. These data suggest that IL-1 could play a unique role in driving pathogenesis (Ong et al., 2020). However, it is difficult to draw conclusions from one patient, and it is important to note that many other proinflammatory cytokines were also elevated prior to decompensation, even if they had not yet peaked (Ong et al., 2020). A unique aspect of this study was the ability to compare this severe case to two mild COVID-19 cases, in which proinflammatory cytokines were not markedly elevated. A second study also demonstrated that proinflammatory cytokines were not elevated in mild COVID-19 (Thevarajan et al., 2020). Together, these studies indicate that a heightened proinflammatory response is characteristic of severe COVID-19. The potential factors driving this proinflammatory state in the peripheral blood are summarized in Figure 1 .
Figure 1.
The Peripheral Innate Immune Response to Severe SARS-CoV-2 Infection
Some peripheral CD14+ monocytes have an inflammatory phenotype and secrete T cell-activating cytokines, whereas others have decreased HLA class II expression, which could result in decreased antigen presentation to naive T cells. Monocytes and activated granulocytes, such as neutrophils, might phagocytose or degranulate in response to opsonized infected cells. Prior to exhaustion, NK cells might kill infected cells via direct killing or ADCC. Although a decrease in the abundance of DCs is reported, the behavior of DCs is currently unknown. Solid lines represent interactions that have been reported. Dashed lines represent interactions that have not been reported and warrant future studies. Abbreviations: DC, dendritic cell; NK cell, natural killer cell; ADCC, antibody-dependent cellular cytotoxicity.
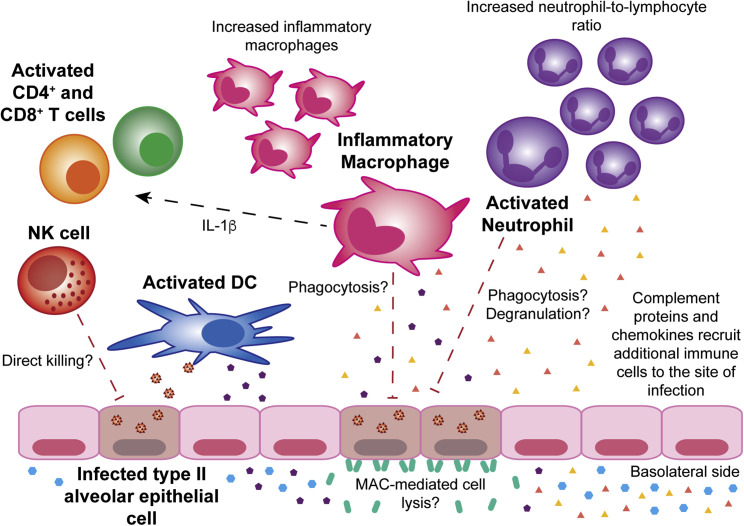
To date, two studies have investigated the local cytokine response to SARS-CoV-2 infection in bronchoalveolar lavage fluid (BALF). Global functional analyses of differentially expressed genes by Zhou et al. revealed an upregulation in inflammatory pathways, such as “chemokine signaling” and “chemokine signaling pathway” (Zhou et al., 2020b). Conversely, Xiong et al. found that upregulated genes were related to viral infection with the most enriched biological processes being “co-translational protein targeting to membrane” and “protein targeting to the ER [endoplasmic reticulum]” (Xiong et al., 2020). Unlike Xiong et al., Zhuo et al. found significant upregulation of CXCL17, a chemoattractant for immature DCs and monocytes, as well as IL1RN and IL1β. Although the differences between these two studies could be due to the lower sample size used by Xiong et al., both studies found significant upregulation of chemokines likely responsible for neutrophil (CXCL1, CXCL2, CXCL6) and monocyte (CCL2 and CCL8) recruitment to the lungs (Xiong et al., 2020, Zhou et al., 2020b). Upregulation of CCL2 and CCL8 was also observed in postmortem lung samples from two COVID-19 patients (Blanco-Melo et al., 2020). Zhou et al. also investigated expression of interferon-stimulated genes (ISGs) and found 83 to be significantly upregulated, including those with direct antiviral activity, such as IFITMs. Upregulation of IFIT2 and IFITM3 were confirmed by Blanco-Melo et al. (Blanco-Melo et al., 2020). With one exception, patients who were sampled later from the date of symptom onset had lower levels of cytokine-related genes and ISGs (Zhou et al., 2020b). The potential factors driving this proinflammatory state in the lung are summarized in Figure 2 .
Figure 2.
The Innate Immune Response to Severe SARS-CoV-2 Infection of the Lung
There are increased levels of both inflammatory macrophages and activated neutrophils in the lung. Inflammatory macrophages secrete IL-1β, activating T cells. Activated DCs are also present and likely take up viral antigens to present to naive T cells. NK cells, inflammatory macrophages, and activated neutrophils could kill infected type II alveolar epithelial cells by a variety of mechanisms. Additionally, formation of the MAC might also result in lysis of infected cells. Complement proteins and chemokines produced by lung epithelial cells and other cell types at the site of infection recruit additional immune cells. Solid lines represent interactions that have been reported. Dashed lines represent interactions that have not been reported and warrant future studies. Abbreviations: DC, dendritic cell; NK cell, natural killer cell; MAC, complement membrane attack complex.
Together, these data present several common themes. The first is that COVID-19 consistently results in the upregulation of chemokines, particularly those that act as chemoattractants for neutrophils and monocytes. This suggests that influx of these cell types into infected tissues could contribute to tissue damage and increased cytokine production. The second is that high levels of proinflammatory cytokines such as IL-1β, IL-2, and IL-6 could be a hallmark of more severe disease. The third is that there seems to be a robust ISG signature in the lungs, which supports the idea that SARS-CoV-2 stimulates the IFN response to some degree. Future studies with larger sample sizes and longitudinal sampling are required. It will be critical to determine how the course of the proinflammatory response relates to symptoms and patient outcomes. It would also be important to determine to what extent the potentially moderate IFN response is suppressing viral replication versus contributing to immunopathogenesis.
Myeloid Cells in the Context of COVID-19
A study of 61 COVID-19 patients found that the blood neutrophil count and neutrophil-to-lymphocyte ratio (NLR) was significantly higher in severe cases (Liu et al., 2020) (Figure 1). This study went on to find that the NLR was the most accurate predictor of progression to severe COVID-19. A significantly higher proportion of activated mast cells and neutrophils as well as a higher NLR were also observed in the BALF of COVID-19 patients (Zhou et al., 2020b) (Figure 2). Similarly, Wilk et al. observed that COVID-19 patients requiring ventilation had a significant increase in peripheral activated granulocytes in comparison with unventilated COVID-19 patients and healthy controls (Wilk et al., 2020). Wilk et al.’s data, along with the fact that chemoattractants for neutrophils are upregulated during COVID-19 (Blanco-Melo et al., 2020, Huang et al., 2020, Xiong et al., 2020, Zhou et al., 2020b), suggest that granulocytes could significantly contribute to pathogenesis.
Significant dysregulation of monocytes and macrophages seems to be a feature of severe COVID-19. In severe cases, there is a significant decrease in CD16+ monocytes and a shift toward CD14+ monocytes (Wilk et al., 2020) (Figure 1). However, this increase in CD14+ monocytes appears to resolve during the late recovery stage of infection (Wen et al., 2020). Several groups have reported decreased CD14+ monocyte expression of HLA class II genes (Giamarellos-Bourboulis et al., 2020, Ong et al., 2020, Wilk et al., 2020). This decrease suggests that severe COVID-19 patients might be unable to mount a robust T cell response, which could contribute to the observed decrease in T cell levels (Diao et al., 2020, Giamarellos-Bourboulis et al., 2020, Ong et al., 2020, Zheng et al., 2020). Interestingly, a negative correlation between serum IL-6 levels and the absolute number of HLA-DR molecules on CD14+ monocytes was reported (Giamarellos-Bourboulis et al., 2020), suggesting IL-6 is associated with the downregulation of HLA-DR.
The role of peripheral monocytes in the proinflammatory response is unclear. Guo et al. identified a unique subpopulation of monocytes present in the severe phase of COVID-19 (Guo et al., 2020). This severe phase-specific subpopulation expressed high levels of inflammatory genes potentially regulated by the transcription factors ETS2, NFIL3, and PHLDA2. Zhou et al. also found an increase in inflammatory monocytes secreting IL-6 and GM-CSF in COVID-19 patients, particularly in patients experiencing severe COVID-19 (Zhou et al., 2020a) (Figure 1). Even COVID-19 patients in recovery stages, early or late, were reported to have more CD14+IL-1β+ and IFN-activated monocytes than did healthy controls (Wen et al., 2020). In contrast, Wilk et al. found that peripheral monocytes did not significantly upregulate genes for proinflammatory cytokines (Wilk et al., 2020). There was also significant heterogeneity in ISG upregulation in monocytes, with only some patients upregulating ISGs, but no clear association with disease severity (Wilk et al., 2020). The inconsistencies between the three single-cell RNA-seq (scRNA-seq) studies could be due to the differences in study design. Guo et al. evaluated only two subjects, and Wen et al. used recovery stage patients, whereas Wilk et al. used seven acute stage patients but only one with serial samples. It is also possible that the increase in IL-6 and GM-CSF protein observed by Zhou et al. is not reflected in the transcriptome.
In the lung, Zhou et al. did not observe a significant increase in M0, M1, or M2 macrophages in COVID-19 patients in comparison with healthy controls (Zhou et al., 2020b). A different study reported the opposite, finding that severe COVID-19 cases had more macrophages than did mild cases (Liao et al., 2020) (Figure 2). Liao et al. also found that the composition of lung macrophages was dramatically different in severe cases in comparison with mild cases and healthy controls. Severe cases had an increase in inflammatory monocyte-derived macrophages and SPP1 + macrophages with almost no alveolar macrophages. In contrast, the composition of lung macrophages in mild cases and healthy controls was dominated by alveolar macrophages. Interestingly, in a mouse model of SARS-CoV infection, alveolar macrophages were identified as acting in an immunosuppressive capacity (Zhao et al., 2009). Overall, Liao et al.’s study suggests that trafficking of peripheral monocytes to the lung and their differentiation into macrophages could enhance the proinflammatory response and the recruitment of other innate immune cells.
Thus far, very little has been published on the role of DCs in SARS-CoV-2 infection. A significant decrease in conventional DCs and plasmacytoid DCs has been reported in COVID-19 patients in comparison with healthy controls (Wilk et al., 2020) (Figure 1). Another study found a decrease in the abundance of resting DCs and an increase in the abundance of activated DCs in the lungs of COVID-19 patients in comparison with healthy controls (Zhou et al., 2020b) (Figure 2). Clearly, more research is needed to determine whether or not DCs, like monocytes and macrophages, are dysregulated in the setting of SARS-CoV-2 infection. Given that DCs are the canonical antigen-presenting cells, this will have important implications for the antigen-specific T cell response.
Currently, there is no evidence to suggest that innate immune cells are targets of SARS-CoV-2 infection (Wilk et al., 2020).
The NK Cell Response to SARS-CoV-2
Studies have shown that severe COVID-19 patients have depleted peripheral NK cell counts in comparison with counts in mild cases and healthy controls (Wen et al., 2020, Wilk et al., 2020, Zheng et al., 2020). This is consistent with previous findings in SARS (Xia et al., 2004). Zheng et al. found that the percentage of peripheral NK cells expressing the inhibitory marker NKG2A was higher in COVID-19 patients than in healthy controls whereas the percentage of NK cells expressing the activation markers CD107a, IFN-ɣ, IL-2, and TNFɑ was lower (Zheng et al., 2020) (Figure 1). They also showed that the number of NK cells increased upon convalescence with a decrease in the percentage of more naive NKG2A+ NK cells. scRNA-seq analysis of PBMCs from COVID-19 patients revealed upregulation of transcripts for exhaustion markers, LAG3 and HAVCR2, in comparison with healthy controls (Wilk et al., 2020). Taken together, these results suggest functional exhaustion of peripheral NK cells during SARS-CoV-2 infection.
Despite the fact that a decrease in peripheral NK cells is consistently observed in severe COVID-19 cases (Giamarellos-Bourboulis et al., 2020, Wilk et al., 2020, Zheng et al., 2020), it is unclear whether this decrease is due to NK cell trafficking to infected tissues or cell death. One study found that several upregulated genes in PBMCs from COVID-19 patients are involved in the apoptosis and P53 signaling pathways, suggesting lymphopenia could be due to SARS-CoV-2-mediated apoptosis (Xiong et al., 2020). To date, there have been two studies investigating NK cells in BALF (Figure 2). Zhou et al. performed bulk RNA-seq on BALF from eight COVID-19 cases and found a significant decrease in the estimated abundance of resting NK cells in COVID-19 patients in comparison with healthy controls but found no change in the abundance of activated NK cells (Zhou et al., 2020b). Conversely, scRNA-seq analysis of BALF samples from three severe patients, three mild patients, and eight healthy controls found higher proportions of NK cells in COVID-19 patients, suggesting NK cell trafficking to the lungs (Liao et al., 2020). The discrepancies between these two studies could be explained by the fact that Zhou et al.’s cohort was sampled closer to the day of symptom onset than the cohort used by Liao et al., suggesting that the influx of NK cells into the lungs might only occur during the later stages of infection.
Overall, these data suggest that upon well-established SARS-CoV-2 infection, NK cells exit the periphery and traffic to the lung. NK cells remaining in the periphery might not contribute to proinflammatory cytokine production but instead have an exhausted phenotype that could facilitate virus dissemination to other sites of the body. Given the observed differences in NK cell count between mild and severe COVID-19 cases (Wilk et al., 2020, Zheng et al., 2020), it is important that we stratify COVID-19 cases on the basis of severity in future analyses. To clarify the role of NK cells during SARS-CoV-2 infection, the interaction between SARS-CoV-2-infected cells and primary NK cells needs to be studied. Do infected cells upregulate ligands for activating NK cell receptors, facilitating targeting by NK cells? Conversely, do infected cells downregulate activating ligands or upregulate inhibitory ligands as a mechanism of NK cell evasion? Addressing these questions will also require HLA and killer immunoglobulin-like receptor (KIR) typing of COVID-19 patients to determine whether certain haplotypes correlate with disease severity.
COVID-19 and the Complement System
There is a potential role for complement during SARS-CoV-2 infection. Similar to what has been reported in SARS (Huang et al., 2005), serum levels of complement proteins are increased in severe COVID-19 patients in comparison with mild cases and healthy controls (Gao et al., 2020) (Figure 1). Additionally, gene functional enrichment analysis of differentially regulated genes in the PBMCs of COVID-19 patients showed an enrichment in genes associated with complement activation and the classical pathway (Xiong et al., 2020).
Mannose-binding lectin (MBL) is a pattern-recognition protein present in the serum that, together with MBL-associated serine protease 2 (MASP-2), mediates activation of the lectin pathway by binding to sugars expressed by a variety of pathogens. Interestingly, Gao et al. found that SARS-CoV-2 nucleocapsid (N) protein interacts with MASP-2, inducing MASP-2 auto-activation and cleavage of complement protein, C4 (Gao et al., 2020). Moreover, lung tissue from severe COVID-19 patients revealed significant deposits of MBL, MASP-2, C3, C4a, C4d, and C5b-9 (components of the membrane-attack complex), suggesting that complement contributes to lung injury (Figure 2). Contrary to these results, studies in SARS have suggested that the lectin pathway, mediated by MBL, plays a role in clearance of SARS-CoV (Ip et al., 2005, Zhou et al., 2010). One of these studies found that genotypes associated with low MBL levels were more prevalent in SARS patients (Ip et al., 2005). However, these results are controversial as another group found no significant difference in MBL genotypes between SARS patients and healthy controls (Yuan et al., 2005).
Based on the aforementioned studies, complement, while perhaps unessential for viral clearance, might contribute to SARS-CoV-2 immunopathogenesis. Clinical studies need to evaluate the presence of complement proteins, MBL, and MASP-2 in the serum of COVID-19 patients. If a robust correlation exists between increased disease severity and high concentrations of these proteins, complement-targeted therapies should be considered to mitigate pathogenesis. Additionally, determining whether genotypes associated with low MBL levels are more prevalent in severe COVID-19 patients would be important for identifying at-risk populations.
COVID-19 Therapeutics
Based on the evidence that a dysregulated immune response plays a role in SARS-CoV-2 pathogenesis, there has been significant excitement about the potential to improve COVID-19 outcomes by using immunomodulators. As of May 12, 2020, Clinicaltrials.gov had 1,409 COVID-related clinical trials. Many immunomodulatory agents are being explored alone and in combination with other drugs. A summary of these agents is provided in Table 1 . At the forefront of this debate are the IL-6 blocking agents Tocilizumab and Sarilumab, with 52 studies of these agents alone. There have been several anecdotal reports of benefit with Tocilizumab, including a study profiling the immune response by RNA-seq (Guo et al., 2020). However, understanding the usefulness of this and other drugs will require randomized, placebo-controlled clinical trials because such anecdotal reports can often be misleading. An important consideration in such trials is the timing of administration, because premature administration could hinder the ability to fight the virus. Current clinical guidelines from the Infectious Diseases Society of America highlight these knowledge gaps and recommend that any unproven immune modulating agent only be used in the contexts of a clinical trial (Bhimraj et al., 2020).
Table 1.
A Summary of Drugs That Are Being Tested for Treatment of COVID-19 or That Have Been Suggested for Use in this Setting
| Drug Class | Drug Name | Mechanism of Action | References |
|---|---|---|---|
| Monoclonal antibodies | Gimsilumab | Anti-GM-CSF antibody. GM-CSF promotes the proinflammatory response. GM-CSF expression was shown to increase in TH1 cells and monocytes in COVID-19 patients, particularly ICU patients. | Zhou et al., 2020a |
| Sarilumab and Tocilizumab | Anti-IL-6 antibodies. Higher blood concentrations of IL-6 were reported to be predictive of fatal outcome in COVID-19 patients. | Ruan et al., 2020 | |
| IL-1 receptor agonist | Anakinra | Competitively inhibits IL-1 binding to the IL-1 type I receptor. Increased concentrations of IL-1 have been reported in COVID-19 patients. IL-1⍺ and IL-1β have been implicated in playing a role in severe COVID-19. | Giamarellos-Bourboulis et al., 2020, Huang et al., 2020, Ong et al., 2020 |
| Tyrosine kinase inhibitors | Ruxolitinib | JAK1 and JAK2 inhibitor. Inhibits NK cell activity and the production of proinflammatory cytokines. It also impacts DC differentiation, migration, and function, which could suppress antigen-specific T cell responses. | Elli et al., 2019 |
| Baricitinib | JAK1 and JAK2 inhibitor. Identified as a numb-associated kinase (NAK) inhibitor, with high affinity for AAK1. AAK1 is a regulator of clathrin-mediated endocytosis, the pathway utilized by SARS-CoV-2 to enter cells. Could prevent viral entry into cells in addition to its anti-inflammatory activity. | Stebbing et al., 2020 | |
| Fedratinib | JAK2-specific inhibitor. Many of the cytokines found to be elevated in the serum of COVID-19 patients either promote TH17 responses or are produced by TH17 cells. IL-6 and IL-23 activate STAT3, the transcription factor responsible for TH17 differentiation and function, through JAK2. Inhibition of JAK2 could limit the proinflammatory activity of TH17 cells. | Wu and Yang, 2020 | |
| Quinoline | Chloroquine and Hydroxychloroquine | Inhibit replication of other viruses by interfering with virion binding to cellular receptors and increasing endosomal pH during viral entry. These drugs can also inhibit antigen processing and presentation by APCs, prevent TLR signaling, and reduce production of proinflammatory cytokines. | Devaux et al., 2020, Schrezenmeier and Dörner, 2020 |
| Interferon | IFN-β | Binds to the IFNAR complex (IFNAR1/IFNAR2) which is expressed by most cells. Stimulates transcription of ISGs via the JAK/STAT/IRF9 pathway. Interferes with viral replication and dissemination. | Hemann et al., 2017, Sallard et al., 2020 |
| IFN-λ | Binds to the IFNL complex (IFNLR1/IL10R2). Expression of the IFNL complex is limited to epithelial cells and some immune cell subsets, such as neutrophils. Stimulates transcription of ISGs via the JAK/STAT/IRF9 pathway. Interferes with viral replication and dissemination. | Hemann et al., 2017, Prokunina-Olsson et al., 2020 | |
| Vaccine | BCG | An attenuated strain of Mycobacterium bovis. Thought to provide broad protection against respiratory infections through its similarity to viral antigens, antigen-independent activation of adaptive immune cells, and/or long-term activation and reprogramming of innate immune cells. | Redelman-Sidi, 2020 |
| Corticosteroid | Methylprednisolone | Acts on the transcriptional level to inhibit the production and function of proinflammatory mediators. | Zha et al., 2020 |
Similar to our experience with ARDS, there is unlikely to be a one-size-fits-all therapy to treat COVID-19-associated severe respiratory failure. Instead, different treatments will likely be effective in different subsets of patients. Although we are all anxious for new treatments, it is critical that we carefully evaluate the benefit of such interventions in a controlled fashion through randomized trials.
Concluding Remarks
This Minireview has detailed several components of what is considered to be a uniquely dysregulated innate immune response to SARS-CoV-2 infection. But a critical question we must ask ourselves is, what is a “regulated” response to SARS-CoV-2? The answer could lie with a patient population that, up until now, has been relatively understudied. The vast majority of COVID-19 studies have focused on patients requiring hospitalization. A critical part of furthering our understanding of SARS-CoV-2 infection, and potentially identifying a regulated immune response, will be to study COVID-19 patients not requiring hospitalization. What is different about their reaction to SARS-CoV-2 infection that allows them to resolve the infection without medical intervention? A longitudinal study comparing the immune responses of hospitalized versus non-hospitalized patients would help provide this important insight. Such studies will provide a necessary guide as to how we need to modulate the immune response for maximum benefit and improved patient outcomes.
The COVID-19 pandemic has become unlike any other we have seen in modern history. It has impacted the global economy, health care systems, the way we interact, and, indeed, our way of life. The world is looking to scientific researchers for answers and although we have made significant progress in a short amount of time, there is still plenty of work to be done.
Acknowledgments
The authors would like to thank everyone working on the front lines of the COVID-19 pandemic. J.L.M. is supported by the National Science Foundation Graduate Research Fellowship DGE-1656518 and NIH training grant T32-AI007290 (PI Olivia Martinez). C.A.B. is supported by NIH/NIDA DP1 DA04608902, a 2019 Sentinel Pilot Project from the Bill & Melinda Gates Foundation, and Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases #1016687. C.A.B. is the Tashia and John Morgridge Faculty Scholar in Pediatric Translational Medicine from the Stanford Maternal Child Health Research Institute and an Investigator of the Chan Zuckerberg Biohub.
References
- Bhimraj A., Morgan R.L., Shumaker A.H., Lavergne V., Baden L., Cheng V.C.-C., Edwards K.M., Gandhi R., Muller W.J., O’Horo J.C. Infectious Diseases Society of America Guidelines on the Treatment and Management of Patients with COVID-19. Clin. Infect. Dis. 2020:ciaa478. doi: 10.1093/cid/ciaa478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.04.026. S0092-8674(20)30489-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020 doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elli E.M., Baratè C., Mendicino F., Palandri F., Palumbo G.A. Mechanisms Underlying the Anti-inflammatory and Immunosuppressive Activity of Ruxolitinib. Front. Oncol. 2019;9:1186. doi: 10.3389/fonc.2019.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T., Hu M., Zhang X., Li H., Zhu L., Liu H., Dong Q., Zhang Z., Wang Z., Hu Y. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv. 2020 [Google Scholar]
- Gattinoni L., Coppola S., Cressoni M., Busana M., Rossi S., Chiumello D. COVID-19 Does Not Lead to a “Typical” Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2020;201:1299–1300. doi: 10.1164/rccm.202003-0817LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. S1931-3128(20)30236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Li B., Ma H., Wang X., Cai P., Yu Q., Zhu L., Jin L., Jiang C., Fang J. Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. bioRxiv. 2020 [Google Scholar]
- Hemann E.A., Gale M., Jr., Savan R. Interferon Lambda Genetics and Biology in Regulation of Viral Control. Front. Immunol. 2017;8:1707. doi: 10.3389/fimmu.2017.01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-L., Huang J., Duan Z.-H., Wei J., Min J., Luo X.-H., Li J.-G., Tan W.-P., Wu L.-Z., Liu R.-Y. Th2 predominance and CD8+ memory T cell depletion in patients with severe acute respiratory syndrome. Microbes Infect. 2005;7:427–436. doi: 10.1016/j.micinf.2004.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip W.K.E., Chan K.H., Law H.K.W., Tso G.H.W., Kong E.K.P., Wong W.H.S., To Y.F., Yung R.W.H., Chow E.Y., Au K.L. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191:1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Chen L., Li J., Wang X., Wang F. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 [Google Scholar]
- Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R. Neutrophil-to-lymphocyte ratio predicts severe illness patients with 2019 novel coronavirus in the early stage. MedRxiv. 2020 doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Haja Mohideen S.M., Chan K.S., Tan A.T., Bertoletti A., Ooi E.E., Low J.G.H. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.03.021. S1931-3128(20)30185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Alphonse N., Dickenson R.E., Durbin J.E., Glenn J.S., Hartmann R., Kotenko S.V., Lazear H.M., O’Brien T.R., Odendall C. COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 2020;217:e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelman-Sidi G. Could BCG be used to protect against COVID-19? Nat. Rev. Urol. 2020 doi: 10.1038/s41585-020-0325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical Predictors of Mortality Due to COVID-19 Based on an Analysis of Data of 150 Patients From Wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallard E., Lescure F.X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeier E., Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Du R., Wang R., Cao T., Guan L., Yang C., Zhu Q., Hu M., Li X., Li Y. Comparison of Hospitalized Patients With ARDS Caused by COVID-19 and H1N1. Chest. 2020 doi: 10.1016/j.chest.2020.03.032. S0012-3692(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vareille M., Kieninger E., Edwards M.R., Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 2011;24:210–229. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0944-y. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.005. S1684-1182(20)30065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C., Xu L., Wang Z., Qin Z., Tong Z., Huang K., Xiao B., Qi M., Jiang B., Wang C. The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am. J. Clin. Pathol. 2004;121:507–511. doi: 10.1309/WPK7Y2XKNF4CBF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F.F., Tanner J., Chan P.K.S., Biffin S., Dyer W.B., Geczy A.F., Tang J.W., Hui D.S.C., Sung J.J.Y., Sullivan J.S. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens. 2005;66:291–296. doi: 10.1111/j.1399-0039.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L., Li S., Pan L., Tefsen B., Li Y., French N., Chen L., Yang G., Villanueva E.V. Corticosteroid treatment of patients with coronavirus disease 2019 (COVID-19) Med. J. Aust. 2020;212:416–420. doi: 10.5694/mja2.50577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Van Rooijen N., Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lu K., Pfefferle S., Bertram S., Glowacka I., Drosten C., Pöhlmann S., Simmons G. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010;84:8753–8764. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.017. S1931-3128(20)30244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]