Among 417 COVID-19 patients in Shenzhen, demographic characteristics, clinical manifestations and baseline laboratory tests showed significant differences between mild-moderate cohort and severe-critical cohort.Based on these differences, a convenient mathematical model was established to predict the illness severity of COVID-19. The model includes four parameters: age, BMI, CD4+ lymphocytes and IL-6 levels. The AUC of the model is 0.911.The high risk factors for developing to severe COVID-19 are: age ≥ 55 years, BMI > 27 kg / m2, IL-6 ≥ 20 pg / ml, CD4+ T cell ≤ 400 count / μ L.Among 249 discharged COVID-19 patients, those who recovered after 20 days had a lower count of platelet, a higher level of estimated glomerular filtration rate, and higher level of interleukin-6 and myoglobin than those who recovered within 20 days.
Keywords: COVID-19, prediction model, illness severity, CD4+ T lymphocyte
Graphical Abstract
Introduction
COVID-19, caused by SARS-CoV-2, is a highly contagious disease.1 By April 8, 2020, more than 1,350,000 patients were diagnosed with COVID-19 globally, with more than 79,000 deaths worldwide attributable to the disease.2 Recent clinical data reported that mild and critical patients manifested different symptoms. Most of the mild patients with COVID-19 had symptoms such as fever, cough, and mild pneumonia, whereas the critical cases presented dyspnea, respiratory failure, sepsis, organ dysfunction, and even eventual death.3, 4, 5 Therefore, we hypothesize that the clinical and laboratory characteristics may be associated with patients' severity and short-term outcomes. A large cohort study indicated that the total mortality rate of COVID-19 was 2.3%; however, the rate of severe cases was 8.1% while that of critical cases reached 14%.6 In particular, patients with complications, such as septic shock and organ dysfunction, had the highest mortality rate.4,5 The median length of severe patients' aggravation period was 10 days.5 Without specific antiviral agents, an early intervention and supportive treatments are essential for improving clinical outcomes and reducing mortality in severe patients.
The ongoing outbreak of COVID-19 has been overloading medical systems worldwide. An accurate prediction of disease progression is conducive to informative clinical decision making and optimized allocations of hospital resources. Several modeling studies on COVID-19 have been reported to predict the spreading profile using the number of infected patients and transportation,7,8 as well as to predict the illness severity and mortality based on the clinical features.9 In our previous clinical practice and research, we found that the individual immune status plays an important role in the progress and prognosis of the disease.10,11 In this study we aimed to build a model combining clinical characteristics and laboratory findings, including the immune status, to predict the progression and outcomes of COVID-19.
Results
Demographics and Baseline Characteristics
Our study included all 417 confirmed COVID-19 cases in Shenzhen by February 29, 2020, who were all admitted to the Third People's Hospital of Shenzhen. The patients were divided into two groups, comprising 325 (77.9%) mildly to moderately ill patients and 92 (22.1%) severely to critically ill patients. The sample consisted of 198 males (47.5%) and 219 females (52.5%), but the sex ratio was skewed toward males in the severe-critical group (64.1%). No patient had an exposure history (shopping) in the Huanan Seafood Market in Wuhan, although 309 (74.1%) had a traveling history in Hubei province. In addition, 279 (66.9%) patients had a fever before admission and 148 (25.5%) presented with cough. Other symptoms that were not common included expectoration (3.1%), headache (3.4%), myalgia (3.2%), chill (0.7%), nausea or vomiting (0.2%), and diarrhea (2.6%). Most symptom profiles were comparable between the mild-moderate group (n = 325) and the severe-critical group (n = 92) while fever and cough occurred in a significantly higher proportion among severely to critically ill patients (p < 0.001 for fever and p = 0.015 for cough). Hypertension (23 [5.5%]), chronic liver disease (23 [5.5%]), and diabetes (15 [10.1%]) were the most common co-existing conditions. Moreover, 367 (88%) patients had pneumonia first time on admission (Table 1).
Table 1.
Epidemiological and Baseline Clinical Features of 417 Confirmed COVID-19 Patients in the Third People's Hospital of Shenzhen from January 11 to February 18, 2020
| Characteristics | COVID-19 Patients |
|||
|---|---|---|---|---|
| Total (N = 417) | Mild-Moderate (n = 325) | Severe-Critical (n = 92) | p-value | |
| Median age (IQR) | 47 (34–60) | 41 (31–56) | 61 (52–65) | <0.001 |
| Age subgroups | ||||
| 0–17 years | 30 (7.2) | 30 (9.2) | 0 | – |
| 18–60 years | 286 (68.6) | 241 (74.2) | 45 (49) | – |
| >60 years | 101 (24.2) | 54 (16.6) | 47 (51) | – |
| Male (%) | 198 (47.5) | 139 (42.8) | 59 (64.1) | <0.001 |
| BMI (kg/m2) | 23.14 (20.96–25.39) | 22.68 (20.43–24.97) | 24.52 (22.84–26.68) | <0.001 |
| Initial symptoms | ||||
| Fever | 279 (66.9) | 199 (61.2) | 80 (87.0) | <0.001 |
| Cough | 148 (25.5) | 105 (32.3) | 43 (46.7) | 0.015 |
| Expectoration | 13 (3.1) | 9 (2.8) | 4 (4.3) | 0.50 |
| Headache | 14 (3.4) | 12 (3.7) | 2 (2.2) | 0.74 |
| Myalgia | 13 (3.2) | 10 (3.1) | 3 (3.3) | >0.99 |
| Chill | 3 (0.7) | 2 (0.6) | 1 (1.1) | 0.53 |
| Nausea or vomiting | 1 (0.2) | 1 (0.3) | 0 | >0.99 |
| Diarrhea | 11 (2.6) | 7 (2.2) | 4 (4.3) | 0.27 |
| Co-existing chronic medical conditions | ||||
| Chronic heart disease | 9 (2.2) | 2 (0.6) | 7 (7.6) | <0.001 |
| Chronic lung disease | 3 (0.7) | 2 (0.6) | 1 (1.1) | 0.53 |
| Chronic renal disease | 6 (1.4) | 2 (0.6) | 4 (4.3) | 0.023 |
| Chronic liver disease | 23 (5.5) | 15 (4.6) | 8 (8.7) | 0.21 |
| Diabetes | 15 (3.6) | 7 (2.2) | 8 (8.7) | <0.01 |
| Hypertension | 23 (5.5) | 13 (4.0) | 10 (10.9) | 0.022 |
| Cancer | 1 (0.2) | 0 | 1 (1.1) | 0.22 |
| Exposure history | ||||
| Traveling history to Hubei | 309 (74.1) | 236 (72.6) | 73 (79.3) | 0.24 |
| Interval (days), median (IQR) | ||||
| Onset to admission | 3 (1–6) | 3 (1–6) | 4 (2–7) | >0.99 |
| Onset to diagnosis | 2 (0–5) | 1 (0–4) | 2 (1–6) | <0.01 |
| Bilateral pneumonia | 295 (70.7) | 211 (64.9) | 84 (91.3) | <0.001 |
| Unilateral pneumonia | 72 (17.3) | 67 (20.6) | 5 (5.43) | 0.001 |
Values are number (%) except where indicated otherwise. p values indicate differences between mild-moderate and severe-critical patients. p < 0.05 was considered statistically significant.
BMI, body mass index; IQR, interquartile range.
Laboratory Findings in 417 Confirmed COVID-19 Patients
Levels of white blood cells, hemoglobin, triglyceride, low-density lipoprotein, high-density lipoprotein, and cholesterol did not differ between the two groups. These measures were recorded on the day of admission for all patients. They were then divided into groups according to illness severity. There were numerous differences in laboratory findings between the two groups, including the level of lymphocytes (1.44 [1.09–1.96] versus 1.02 [0.83–1.28]), the level of neutrophils (2.55 [1.92–3.42] versus 3.03 [2.28–4.39]), the level of platelets (197 [158–241] versus 151 [129–178]), the cell count of CD4+ T lymphocytes (618 [450–805] versus 334.5 [201–458]), the cell count of CD8+ T lymphocytes (272 [417–571] versus 177.5 [112–289]), the level of D-dimers (0.33 [0.24–0.47] versus 0.53 [0.36–0.83]), the level of alanine aminotransferase (20 [13.45–28] versus 26.65 [20–38]), the level of aspartate aminotransferase (25 [20–33] versus 34[26-48])estimated glomerular filtration rate (108.53 [97.16–118.90] versus 92.50 [76.15–101.76]), the level of chromium (60 [50–74] versus 72 [57–93]), the level of myoglobin (33.02 [26.12–44.03] versus 56.19 [40.63–110.16]), the level of brain natriuretic peptide (1.74 [0.70–3.55] versus 7.55 [4.32–16.32]), the level of troponin I (0.005 [0.005–0.005] versus 0.009 [0.005–0.017]), the level of lactate dehydrogenase (209 [167–372] versus 336 [213–592]), the level of procalcitonin (0.034 [0.023–0.052] versus 0.0645 [0.049–0.088]), the level of C-reactive protein (6.0 [2.2–16.5] versus 27.5 [12.5–52.8]), the level of interleukin-6 (IL-6) (8.51 [3.82–16.03] versus 24.17 [16.26–43.02]), and erythrocyte sedimentation rate (23 [13–41] versus 41 [24–57]) (Table 2).
Table 2.
Laboratory Findings in 417 Confirmed COVID-19 Patients in the Third People's Hospital of Shenzhen from January 11 to February 18, 2020
| Variables | COVID-19 Patients |
p -value | ||
|---|---|---|---|---|
| Total | Mild-Moderate | Severe-Critical | ||
| WBC (×109/L) | 4.67 (3.79–5.83) | 4.64 (3.76–5.72) | 4.82 (3.85–5.95) | >0.99 |
| LYM (×109/L) | 1.31 (0.99–1.82) | 1.44 (1.09–1.96) | 1.02 (0.83–1.28) | <0.001 |
| NEU (×109/L) | 2.68 (2.01–3.59) | 2.55 (1.92–3.42) | 3.03 (2.28–4.39) | <0.01 |
| HGB (g/L) | 137 (127–146) | 137 (126–146) | 138 (127–151) | >0.99 |
| PLT (×109/L) | 186 (149–231) | 197 (158–241) | 151 (129–178) | <0.001 |
| CD4+ cell count | 535 (380–742) | 618 (450–805) | 334.5 (201–458) | <0.001 |
| CD8+ cell count | 350 (202–522) | 272 (417–571) | 177.5 (112–289) | <0.001 |
| CD4+/CD8+ | 1.62 (1.15–2.16) | 1.55 (1.15–1.99) | 1.83 (1.24–2.49) | 0.33 |
| D-DIC (μg/mL) | 0.36 (0.25–0.53) | 0.33 (0.24–0.47) | 0.53 (0.36–0.83) | <0.001 |
| ALT (U/L) | 21 (15–31) | 20 (13.45–28) | 26.65 (20–38) | <0.001 |
| AST (U/L) | 26 (21–35) | 25 (20–33) | 34 (26–48) | <0.001 |
| K (mmol/L) | 3.89 (3.64–4.16) | 3.90 (3.68–4.18) | 3.79 (3.54–4.09) | 0.30 |
| Na (mmol/L) | 138.5 (136.8–139.8) | 138.7 (137.3–140.0) | 136.8 (134.9–139.1) | <0.001 |
| Cr (μmol/L) | 62 (52–76) | 60 (50–74) | 72 (57–93) | <0.001 |
| eGFR (mL/min) | 105.18 (93.55–116.64) | 108.53 (97.16–118.90) | 92.50 (76.15–101.76) | <0.001 |
| CK (U/L) | 67 (51–100) | 66 (49–93) | 83 (56–170) | 0.073 |
| LDH (U/L) | 224 (176–422) | 209 (167–372) | 336 (213–592) | <0.001 |
| MYO (ng/mL) | 37.21 (27.94–53.31) | 33.02 (26.12–44.03) | 56.19 (40.63–110.16) | <0.001 |
| BNP (pg/mL) | 2.91 (0.98–8.23) | 1.74 (0.70–3.55) | 7.55 (4.32–16.32) | <0.001 |
| TnI (μg/L) | 0.005 (0.005–0.005) | 0.005 (0.005–0.005) | 0.009 (0.005–0.017) | <0.001 |
| TG (mmol/L) | 0.95 (0.73–1.31) | 0.91 (0.70–1.37) | 1.02 (0.83–1.24) | >0.99 |
| LDL (mmol/L) | 2.32 (1.91–2.78) | 2.44 (1.98–2.81) | 2.11 (1.83–2.51) | 0.96 |
| HDL (mmol/L) | 1.17 (0.97–1.39) | 1.21 (1.00–1.43) | 1.11 (0.90–1.26) | 0.33 |
| CHOL (mmol/L) | 3.96 (3.43–4.61) | 4.12 (3.57–4.65) | 3.62 (3.28–4.21) | 0.14 |
| PCT (ng/mL) | 0.039 (0.025–0.062) | 0.034 (0.023–0.052) | 0.0645 (0.049–0.088) | <0.001 |
| CRP (mg/L) | 8.7 (2.5–24.8) | 6.0 (2.2–16.5) | 27.5 (12.5–52.8) | <0.001 |
| IL-6 (pg/mL) | 11.88 (4.61–19.63) | 8.51 (3.82–16.03) | 24.17 (16.26–43.02) | <0.001 |
| ESR (mm/h) | 28 (14–45) | 23 (13–41) | 41 (24–57) | <0.001 |
p values indicate differences between mild-moderate and severe-critical patients. p < 0.05 was considered statistically significant.
WBC, white blood cells; LYM, lymphocytes; NEU, neutrophils; HGB, hemoglobin; PLT, platelets; CD4+, CD4+ T lymphocyte; CD8+, CD8+ T lymphocyte; D-DIC, D-dimer; ALT, alanine aminotransferase; AST, aspartate aminotransferase; K, potassium; Na, sodium; Cr, chromium; eGFR, estimated glomerular filtration rate; CK, creatine kinase; MYO, myoglobin; BNP, brain natriuretic peptide; TnI, troponin I; LDH, lactate dehydrogenase; TG, triglyceride; LDL, low-density lipoprotein; HDL, high-density lipoprotein; CHOL, cholesterol; PCT, procalcitonin; CRP, C-reactive protein; IL-6, interleukin-6; ESR, erythrocyte sedimentation rate.
Demographics and Baseline Characteristics of 249 Discharged Patients
The study included 249 patients discharged up until February 24, 2020. In this cohort, the median age was 44 years (interquartile range [IQR], 32–57) and 121 (48.6%) were males. Of these patients, 121 (48.6%) were discharged within 20 days after admission. In addition, 128 (51.4%) recovered after 20 days in hospital, among whom 91 (71.1%) were between 18 and 60 years old. The median duration from onset to hospital admission and from hospitalization to diagnosis was 3 days (IQR, 1–5) and 2 days (IQR, 0–4), respectively. The most common symptoms at the onset were fever (173 [69.5%]) and cough (95 [38.2%]). Less common symptoms included expectoration, headache, diarrhea, nausea, and vomiting. Hypertension (29 [7.6%]), chronic liver disease (21 [8.4%]), and diabetes (12 [4.8%]) were the most common co-existing conditions (Table 3).
Table 3.
Epidemiological and Baseline Clinical Features of 249 Discharged COVID-19 Patients in the Third People's Hospital of Shenzhen until February 24, 2020
| Characteristics | COVID-19 Patients |
p-alue | ||
|---|---|---|---|---|
| Total (N = 249) | ≤20 days (n = 121) | >20 days (n = 128) | ||
| Age (years), median (IQR) | 44 (32–57) | 36 (25–53) | 50 (38–61) | <0.001 |
| Age subgroups | ||||
| 0–17 years | 21 (8.4) | 17 (14.0) | 4 (3.1) | – |
| 18–60 years | 175 (70.3) | 84 (69.4) | 91 (71.1) | – |
| >60 years | 53 (21.3) | 20 (16.5) | 33 (25.8) | – |
| Male (%) | 121 (48.6) | 57 (47.1) | 64 (50.0) | 0.74 |
| BMI (kg/m2) | 23.26 (21.26–25.22) | 23.25 (21.33–25.15) | 23.26 (21.26–25.28) | >0.99 |
| Initial symptoms | ||||
| Fever | 173 (69.5) | 71 (58.7) | 102 (79.7) | <0.001 |
| Cough | 95 (38.2) | 43 (35.5) | 52 (40.6) | 0.49 |
| Expectoration | 3 (1.2) | 2 (1.7) | 1 (0.8) | 0.61 |
| Headache | 9 (3.6) | 3 (2.5) | 6 (4.7) | 0.50 |
| Myalgia | 8 (3.2) | 3 (2.5) | 5 (3.9) | 0.72 |
| Chill | 1 (0.4) | 0 | 1 (0.8) | >0.99 |
| Nausea or vomiting | 1 (0.4) | 0 | 1 (0.8) | >0.99 |
| Diarrhea | 6 (2.4) | 2 (1.7) | 4 (3.1) | 0.68 |
| Co-existing chronic medical conditions | ||||
| Chronic heart disease | 7 (2.8) | 4 (3.3) | 3 (2.3) | 0.72 |
| Chronic lung disease | 3 (1.2) | 1 (0.8) | 2 (1.6) | >0.99 |
| Chronic renal disease | 6 (2.4) | 2 (1.7) | 4 (3.1) | 0.68 |
| Chronic liver disease | 21 (8.4) | 10 (8.3) | 11 (8.6) | >0.99 |
| Diabetes | 12 (4.8) | 4 (3.3) | 8 (6.3) | 0.43 |
| Hypertension | 29 (7.6) | 7 (5.8) | 12 (9.4) | 0.41 |
| Cancer | 1 (0.4) | 0 | 1 (0.8) | >0.99 |
| Exposure history | ||||
| Traveling history to Hubei | 197 (79.1) | 91 (75.2) | 106 (82.8) | 0.19 |
| Interval (days), median (IQR) | ||||
| Onset to admission | 3 (1–5) | 2 (1–4) | 4 (2–7) | <0.001 |
| Onset to diagnosis | 2 (0–4) | 1 (0–3) | 3 (1–6) | <0.001 |
Values are number (%) except where indicated otherwise. p values indicate differences between onset to discharged date ≤20 days and >20 days. p < 0.05 was considered statistically significant.
Compared with patients who recovered within 20 days, those who recovered after 20 days were significantly older (median age, 50 [IQR, 38–61] versus 36 [IQR, 25–53] years; p <.001) and were more likely to have a fever before admission (102 [79.7%] versus 71 [58.7%]) (Table 3).
Laboratory Findings of 249 Discharged Patients
On the day of admission, those of the 249 discharged patients who recovered after 20 days had a lower count of platelets, a higher estimated glomerular filtration rate, and higher level of IL-6 and myoglobin than those who recovered within 20 days; among 249 discharged patients, the median level of platelets, estimated glomerular filtration rate, level of IL-6, and level of myoglobin were 187 × 109/L (IQR, 150–227), 106.58 mL/min (IQR, 94.43–118.12), 10.76 pg/mL (IQR, 4.54–19.07), and 36.85 ng/mL (IQR, 28.59–58.06), respectively (Table 4).
Table 4.
Laboratory Findings in 249 Discharged COVID-19 Patients in the Third People's Hospital of Shenzhen until February 24, 2020
| Variables | COVID-19 Patients |
p-value | ||
|---|---|---|---|---|
| Total (N = 249) | ≤20 days (n = 121) | >20 days (n = 128) | ||
| WBC (×109/L) | 4.63 (3.79–5.72) | 4.71 (4.06–5.83) | 4.54 (3.58–5.71) | >0.99 |
| LYM (×109/L) | 1.33 (1–1.77) | 1.5 (1.12–1.95) | 1.22 (0.98–1.63) | 0.054 |
| NEU (×109/L) | 2.58 (1.98–3.49) | 2.58 (2.03–3.48) | 2.58 (1.98–3.50) | >0.99 |
| HGB (g/L) | 137 (128–148) | 137 (127–147) | 138 (130–148) | >0.99 |
| PLT (×109/L) | 187 (150–227) | 197 (163–244) | 174 (146–204) | 0.012 |
| CD4+ cell count | 550 (414–746) | 618 (453–841) | 519 (377–658) | 0.14 |
| CD8+ cell count | 368 (234–526) | 429 (281–585) | 335 (218–465) | 0.078 |
| CD4+/CD8+ | 1.55 (1.12–2.03) | 1.55 (1.11–1.92) | 1.54 (1.19–2.17) | >0.99 |
| D-DIC (μg/mL) | 0.34 (0.24–0.50) | 0.31 (0.22–0.44) | 0.36 (0.26–0.54) | 0.96 |
| ALT (U/L) | 21.4 (15.1–30.625) | 20 (14.85–32) | 23 (16–30) | >0.99 |
| AST (U/L) | 27 (21–34) | 26 (20–33) | 28 (22–35) | >0.99 |
| K (mmol/L) | 3.90 (3.65–4.16) | 3.94 (3.66–4.17) | 3.89 (3.63–4.15) | >0.99 |
| Na (mmol/L) | 138.5 (136.8–139.7) | 138.7 (137.225–139.7) | 138 (136.1–139.7) | >0.99 |
| Cr (μmol/L) | 64 (52–77) | 62 (49–75) | 64 (54–79) | >0.99 |
| eGFR (mL/min) | 106.58 (94.43–118.12) | 111.11 (99.43–122.05) | 103.56 (92.53–115.10) | <0.01 |
| CK (U/L) | 67 (51–104) | 66 (54–93) | 76 (49–114) | >0.99 |
| LDH (U/L) | 223 (174–406) | 207 (165–384) | 234 (181–429) | >0.99 |
| MYO (ng/mL) | 36.85 (28.59–58.06) | 31.67 (25.30–46.80) | 42.73 (32.29–68.32) | 0.024 |
| BNP (pg/mL) | 1.94 (0.86–8.28) | 1.84 (1.1–3.078) | 4.32 (0.59–12.39) | >0.99 |
| TnI (μg/L) | <0.006 | <0.006 | <0.006 | >0.99 |
| TG (mmol/L) | 0.85 (0.69–1.23) | 0.77 (0.66–1.13) | 1.0 (0.77–1.37) | 0.87 |
| LDL (mmol/L) | 2.27 (1.94–2.82) | 2.23 (1.89–2.77) | 2.32 (1.99–2.86) | >0.99 |
| HDL (mmol/L) | 1.21 (1.00–1.41) | 1.25 (1.00–1.49) | 1.16 (1.01–1.35) | >0.99 |
| CHOL (mmol/L) | 3.96 (3.48–4.64) | 3.87 (3.45–4.58) | 3.96 (3.52–4.72) | >0.99 |
| PCT (ng/mL) | 0.038 (0.025–0.059) | 0.037 (0.025–0.059) | 0.039 (0.026–0.058) | >0.99 |
| CRP (mg/L) | 8.6 (2.5–21.6) | 6.7 (2.5–18.2) | 11.7 (3.2–25.7) | 0.22 |
| IL-6 (pg/mL) | 10.76 (4.54–19.07) | 7.76 (3.44–16.73) | 13.54 (6.90–23.51) | 0.044 |
| ESR (mm/h) | 25 (13–44) | 23 (12–40) | 27 (15–48) | >0.99 |
All laboratory values are in the form of medians(interquartile range). p values indicate differences between mild-moderate and severe-critical patients. p < 0.05 was considered statistically significant.
WBC, white blood cells; LYM, lymphocytes; NEU, neutrophils; HGB, hemoglobin; PLT, platelets; CD4+, CD4+ T lymphocyte; CD8+, CD8+ T lymphocyte; D-DIC, D-dimer; ALT, alanine aminotransferase; AST, aspartate aminotransferase; K, potassium; Na, sodium; Cr, chromium; eGFR, estimated glomerular filtration rate; CK, creatine kinase; MYO, myoglobin; BNP, brain natriuretic peptide; TnI, troponin I; LDH, lactate dehydrogenase; TG, triglyceride; LDL, low-density lipoprotein; HDL, high-density lipoprotein; CHOL, cholesterol; PCT, procalcitonin; CRP, C-reactive protein; IL-6, interleukin-6; ESR, erythrocyte sedimentation rate.
Performance of the Auxiliary Tool for Stratifying Risk of Patients
According to the mentioned methods and analysis, all significant variables were involved in a logistic regression model to predict whether a patient was of a mild-moderate type or a severe-critical type. Insignificant variables during model training were eliminated. This left four interpretable and significant variables, namely IL-6, CD4+ T lymphocyte, body mass index (BMI), and age. Among the 417 patients, 222 possessed complete data of these four parameters, which were thus collected. Therefore, there were four variables (five parameters with intercept) in our model. The sample size is around 44 (222/5) times larger than the free parameters, which is four times larger than the empirical experience. Thus, the sample size is sufficient to train our model. However, since individual differences will influence the modeling results, it is still a limitation that we did not collect completely clinical test results for each patient when hospitalized at the outset.
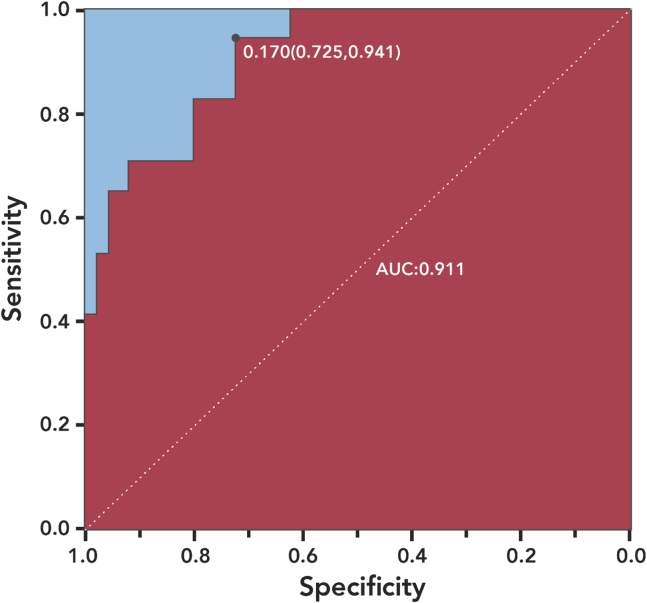
The logistic regression model was interpretable for clinical prediction. Relative weights were assigned according to each variable's regression coefficient. The coefficient values for each variable are 0.0695(IL6), -0.0051(CD4+ T cell), 0.2799(BMI) and 0.0691(Age) respectively. The odds ratios for each variable are 1.0720(IL6), 0.9949(CD4+ T cell), 1.323(BMI) and 1.0716(Age) respectively. All these 4 variables were statistics significant with p<0.05. The area under the curve (AUC) of the training set was 0.921 and that of the testing set was 0.911. Figure 1 shows the sensitivity and specificity of our model.
Figure 1.
ROC Curve from the Testing Set
The sensitivity and specificity of the model are indicated in the text.
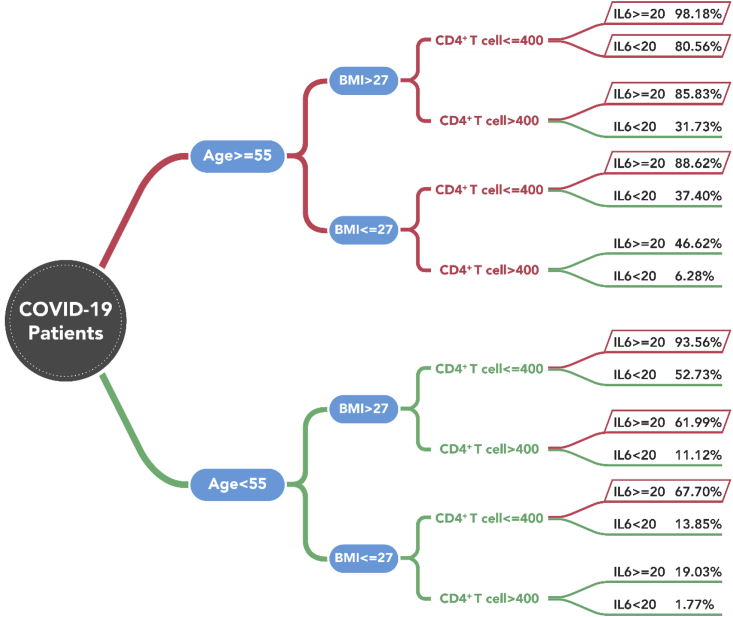
After model training, cut-off values of the four variables were selected based on statistical results and clinical experience. The median probability of progressing to severely to critically ill was significantly higher in patients with a BMI greater than 27 than in those with a BMI of or below 27 (53.18% versus 7.76%). This trend remained for the parameters of CD4+ lymphocytes, IL-6, and age. Patients with a CD4+ T lymphocyte level of ≤400/μL showed a lower median probability of progressing to severe-critical status (62.52%) than those with a level >400/μL. Patients with level of IL-6 ≥20 pg/mL were more likely to develop into a severely to critically ill case (median probability 70.01%) than those whose level was lower than 20 pg/mL (3.80%). Patients aged ≥55 years had a higher probability of developing into a severe-critical case (41.15%) than those younger than 55 (1.73%). The logistic regression model was then retrained with two separate ranges under these four variables. For model validation, we applied a repeated k-fold cross-validation method of 10-fold with 10 repeats. Values of R2, root mean squared error (RMSE), and mean absolute error (MAE) were used for model performance evaluation. Results showed R2 = 0.596, MAE = 0.21, and RMSE = 0.31. In the end, we predicted the probability for each of 16 (24 = 16) types of initial test results, shown in Figure 2. These results represent potential indicators for early initial auxiliary prediction.
Figure 2.
The Probability under 16 Types of Initial Test Results
Older age (≥55 years), high BMI (>27 kg/m2), low level of CD4+ T lymphocytes (≤400/μL), and high level of IL-6 (≥20 pg/mL) have the highest probability of progressing into severe-critical COVID-19. Index units: age (years), BMI (kg/m2), IL-6 (pg/mL), CD4+ T lymphocytes (count/μL).
Discussion
COVID-19 has been spreading worldwide,2 putting a huge amount of pressure on medical systems in a short time. Accurate prediction of disease progression for COVID-19 patients is therefore urgent to facilitate appropriate clinical decision making and optimized medical resource allocations. Risk stratification management will help to alleviate burdens of insufficient medical resources and further reduce mortality.12
Our current research showed that there were significant differences between the mild-moderate group and the severe-critical group concerning several epidemiological and clinical features (age, gender, and fever). Results were consistent with previous reports that age, gender, and underlying diseases might be associated with disease severity.4,9 In terms of laboratory data, 18 out of 27 tested baseline parameters, including levels of blood cells, indicators of myocardial zymogram, liver and renal function, indicators of coagulation function, and levels of infection-related biomarkers were significantly different between the two groups. Hence, we recommended four parameters—age, BMI, and levels of CD4+ T lymphocytes and IL-6—as predictors in future multivariate regression analysis. The elderly (≥55 years), a high level of BMI (>27 kg/m2), a low level of CD4+ T lymphocytes (≤400/μL), and a high level of IL-6 (≥20 pg/mL) are markers associated with the progression to severe and critical illness among COVID-19 patients.
In a large cohort study of COVID-19 in China, the median age of severe cases was 52 years while that of mild cases was 45 years.4 Consistent with our findings, an advanced age was associated with immunity decline characterized by decreases in both humoral and cellular responses according to previous research.13 Although underlying reasons remained unknown, we found a correlation between BMI and disease severity. This finding was in line with results showing BMI as an independent risk factor that could predict the incidence of pneumonia among patients with chronic obstructive pulmonary disease. Lymphocytopenia was present in 83.2% of the hospitalized patients. Severe patients showed more prominent laboratory abnormalities, including lymphocytopenia and leukopenia, than mild ones.6 Among the severe-critical cohort, we also observed lower levels of lymphocytes and CD4+ T lymphocytes. Those with a level of CD4+ T lymphocytes at or below 400/μL suffered a higher risk of progression to severe cases. The cytokine storm caused by a vigorous immune response against the virus was a significant predicting factor for the severity of viral pneumonia. Plasma levels of IL-2, IL-7, IL-10, granulocyte colony-stimulating factor, interferon-γ-inducible protein-10, monocyte chemotattractant protein-1, macrophage inflammatory protein-1α, and tumor necrosis factor α in COVID-19 were higher in intensive care unit (ICU) cases than non-ICU cases.4 Being one of the important cytokines, IL-6 could complement the function of CD8+ T lymphocytes and might aggravate the clinical course.14 Compared with other cytokines, IL-6 is more widely used and more meaningful clinically.
Data for the four clinical test indexes proposed by our model are easy to obtain. Therefore, we recommend that all related characteristics should be tested for patients immediately after admission. Our model reflected the real-world scenario in clinical practice. Receiver-operating characteristic (ROC) analysis suggested that this model had functional capacity in predicting severity among COVID-19 patients (AUC = 0.911). The score may be regarded as a predictor of prognosis to prompt appropriate early intervention. Hence, we present the probability graph of progressing to severe or critical illness, which could be of clinical reference for early diagnosis.
Despite remarkable findings, the study has a few limitations. First, we did not measure broad-spectrum cytokines and viral loads, which may be related to disease progression and severity. Second, the retrospective single-center design led to missing data and unavoidable bias when identifying participants. A larger sample size would be in favor the generalizability of a prediction model. This model would need to be further verified in other cohorts and in clinical practice. Moreover, we did not compare our model performance with alternative machine-learning models, such as the random forest model and the extreme gradient boosting model. It will add value to compare efficacies of different models in the future.
In summary, we proposed a model containing four routine baseline parameters. This allowed accurate prediction of severity among COVID-19 patients. The model accurately stratified these patients into relevant risk categories. It could further help to produce appropriate clinical decisions and optimized allocations of hospital resources when shifting attention to the server during the pandemic.
Material and Methods
Patients
All patients from Shenzhen were admitted to the Third People's Hospital of Shenzhen once their SARS-CoV-2 sample results, collected in upper respiratory tract and tested by real-time PCR, turned out to be positive. All 417 patients included in this retrospective study were confirmed cases who were admitted to this designated infectious disease hospital in Shenzhen from January 11 to February 18, 2020. Ethical approval had been obtained from the Ethics Committee of the Third People's Hospital of Shenzhen.
Procedures
Nasopharyngeal and oropharyngeal specimens for SARS-CoV-2 tests were collected with synthetic fiber swabs under the guidelines introduced by either the World Health Organization (WHO) or the Chinese Center for Disease Control and Prevention (Chinese CDC). Stored at 2°C–8°C, these specimens were packaged with ice and shipped to the Shenzhen Center for Disease Control and Prevention (Shenzhen CDC). Following standard protocols released by Chinese CDC, RNA was extracted and tested by real-time RT-PCR with SARS-CoV-2-specific primers and probes in level-2 biosafety facilities at Shenzhen CDC.1
In addition to treatment outcomes, epidemiological, demographic, clinical, laboratory, and radiological profiles of all patients were followed up. Data were sorted into standardized data collection forms and then reviewed by experienced physicians. We defined the “onset date” as the day when symptoms appeared. For patients without initial symptoms, the onset date was defined as the day of admission. According to the WHO interim guidance,15 all COVID-19 patients were diagnosed and classified into mild to critical pneumonia cases. The National Health Commission of the People's Republic of China issued the seventh version of COVID-19 Treatment Guideline, in which patients were also categorized into mild, moderate (typical), severe, and critically ill cases.16
In the following analysis, we divided patients into two cohorts, namely a mild-moderate cohort and a severe-critical cohort according to the above classification criteria.
Discharge Criteria
The COVID-19 guidelines (seventh version) issued by the National Health Commission of the People’ Republic of China required that all patients must meet certain criteria before being discharged.16
Statistical Analysis
Continuous variables were described using median and interquartile range (IQR) values and were compared using the Mann-Whitney U test. Categorical variables were described as frequency rates, percentages, and proportions and were compared using Chi-square test. When a p value of Chi-square test was around α (0.05), attention was paid to ensure that the approximation of the Chi-square test met with requirements. Under this circumstance, Fisher's test was performed instead when necessary.17 Pairwise comparisons between groups were performed using Bonferroni’s correction. All p values were from two-sided tests, and results were deemed statistically significant at p < 0.05. All statistical analysis was done using R (version 3.6.2).
We selected statistically significant variables to develop a straightforward clinical predicting tool—a logistic regression model—used for early diagnosis. Variables with an insignificant p value were considered as irrelevant and were eliminated during the training process. The area under the ROC curves in the model's sensitivity and specificity results and repeated k-fold cross-validation results were used to assess how well the model performed. Cut-off values of each variable were obtained from statistical results and clinical experience. We then retrained the clinical predicting model to establish an auxiliary diagnosis table showing the probability of developing into a severe-critical case.
Declaration of Interests
None.
Acknowledgments
We thank the patients, nurses, and physicians who provided care for the patients, and the investigators at the Third People's Hospital of Shenzhen. This work is supported by grants from the Science and Technology Innovation Committee of Shenzhen Municipality (202002073000001), National Key Research and Development Program (2020YFC0841700), High-level University Fund (no. G02386301, G02386401), and Guangdong Natural Science Foundation Joint Fund (no. 2019A1515111038).
Published Online: May 21, 2020
Contributor Information
Quanying Liu, Email: liuqy@sustech.edu.cn.
Yingxia Liu, Email: yingxialiu@hotmail.com.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease 2019 (COVID-19) situation report-79. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200408-sitrep-79-covid-19.pdf?sfvrsn=4796b143_4
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan W., Ni Z., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang B., Bragazzi N.L., Li Q., Tang S., Xiao Y., Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect. Dis. Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y., Ma Z., Peppelenbosch M.P., Pan Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob. Health. 2020;8:e480. doi: 10.1016/S2214-109X(20)30068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pawelec G., Barnett Y., Forsey R., Frasca D., Globerson A., McLeod J., Caruso C., Franceschi C., Fülöp T., Gupta S., et al. T cells and aging, January 2002 update. Front. Biosci. 2002;7:d1056–d1183. doi: 10.2741/a831. [DOI] [PubMed] [Google Scholar]
- 14.Nussing S., Sant S., Koutsakos M., Subbarao K., Nguyen T., Kedzierska K. Innate and adaptive T cells in influenza disease. Front. Med. 2018;12:34–47. doi: 10.1007/s11684-017-0606-8. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance. https://www.who.int/internal-publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-oronavirus-(ncov)-infection-is-suspected
- 16.The COVID-19 guidelines of the National Health and Health Commission of China (seventh version) https://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989/files/ce3e6945832a438eaae415350a8ce964.pdf
- 17.Fisher R.A. Oliver and Boyd; 1954. Statistical Methods for Research Workers. [Google Scholar]