Abstract
Background & Aims
Although severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects gastrointestinal tissues, little is known about the roles of gut commensal microbes in susceptibility to and severity of infection. We investigated changes in fecal microbiomes of patients with SARS-CoV-2 infection during hospitalization and associations with severity and fecal shedding of virus.
Methods
We performed shotgun metagenomic sequencing analyses of fecal samples from 15 patients with Coronavirus Disease 2019 (COVID-19) in Hong Kong, from February 5 through March 17, 2020. Fecal samples were collected 2 or 3 times per week from time of hospitalization until discharge; disease was categorized as mild (no radiographic evidence of pneumonia), moderate (pneumonia was present), severe (respiratory rate ≥30/min, or oxygen saturation ≤93% when breathing ambient air), or critical (respiratory failure requiring mechanical ventilation, shock, or organ failure requiring intensive care). We compared microbiome data with those from 6 subjects with community-acquired pneumonia and 15 healthy individuals (controls). We assessed gut microbiome profiles in association with disease severity and changes in fecal shedding of SARS-CoV-2.
Results
Patients with COVID-19 had significant alterations in fecal microbiomes compared with controls, characterized by enrichment of opportunistic pathogens and depletion of beneficial commensals, at time of hospitalization and at all timepoints during hospitalization. Depleted symbionts and gut dysbiosis persisted even after clearance of SARS-CoV-2 (determined from throat swabs) and resolution of respiratory symptoms. The baseline abundance of Coprobacillus, Clostridium ramosum, and Clostridium hathewayi correlated with COVID-19 severity; there was an inverse correlation between abundance of Faecalibacterium prausnitzii (an anti-inflammatory bacterium) and disease severity. Over the course of hospitalization, Bacteroides dorei, Bacteroides thetaiotaomicron, Bacteroides massiliensis, and Bacteroides ovatus, which downregulate expression of angiotensin-converting enzyme 2 (ACE2) in murine gut, correlated inversely with SARS-CoV-2 load in fecal samples from patients.
Conclusions
In a pilot study of 15 patients with COVID-19, we found persistent alterations in the fecal microbiome during the time of hospitalization, compared with controls. Fecal microbiota alterations were associated with fecal levels of SARS-CoV-2 and COVID-19 severity. Strategies to alter the intestinal microbiota might reduce disease severity.
Keywords: Coronavirus, Bacteria, Gut Microbiome, Fecal Nucleic Acid
Abbreviations used in this paper: ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019; GI, gastrointestinal; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Graphical abstract
What You Need to Know.
Backgroud and Context
SARS-CoV-2 infects gastrointestinal tissues. The authors investigated changes in fecal microbiomes of patients with SARS-CoV-2 infection during hospitalization and associations with severity and fecal shedding of virus.
New Findings
Fecal microbiomes from patients with COVID-19 had depletion of symbionts and enrichment of opportunistic pathogens, which persisted after clearance of SARS-CoV-2. Baseline microbiome composition associated with COVID-19 severity. Multiple species from the Bacteroidetes phylum correlated inversely with fecal shedding of SARS-CoV-2.
Limitations
This was a pilot exploratory study of 15 patients with COVID-19; further studies are needed of alterations in intestinal microbiomes of these patients over time.
Impact
These findings indicate the prolonged effect of SARS-CoV-2 infection on the gut microbiomes of patients with COVID-19. Strategies to alter the gut microbiome might be developed to manage gastrointestinal effects of the virus in these patients.
Coronavirus Disease 2019 (COVID-19) is a respiratory illness caused by a novel coronavirus (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]) and more than 3.3 million people worldwide have been infected as of May 1, 2020. Although most cases of COVID-19 are mild, disease can be severe, resulting in hospitalization, respiratory failure, or death.1 Early reports from Wuhan showed that 2% to 10% of patients with COVID-19 had gastrointestinal (GI) symptoms, including diarrhea, but a recent meta-analysis reported that up to 20% had GI symptoms.2, 3, 4, 5 Studies have detected SARS-CoV-2 virus in anal swabs and stool samples in almost 50% of patients with COVID-19, suggesting that the digestive tract might be an extrapulmonary site for virus replication and activity.6 , 7 Moreover, fecal calprotectin was found to be elevated in patients with COVID-19 with diarrhea,8 an indicator of inflammatory responses in the gut. SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) receptor to enter the host, and this receptor is highly expressed in both the respiratory and GI tracts.9, 10, 11 ACE2 is important in controlling intestinal inflammation and gut microbial ecology.12 With trillions of diverse bacteria dwelling in our gut, the gut microbiome has a myriad of effects on gene regulation of immune response and metabolism. The commensal microbiota ecosystem in the gut is dynamic and can be regulated by invading viruses to facilitate a stimulatory or suppressive response.13 Studies have shown that respiratory viral infections may be associated with altered gut microbiome, which predispose patients to secondary bacterial infections.14 , 15 Recent meta-transcriptome sequencing of bronchoalveolar lavage fluid showed that the microbiota in SARS-CoV-2–infected patients was dominated by pathogens or oral and upper respiratory commensal bacteria.16 In addition, comorbidities commonly associated with severe COVID-19 are known to be associated with alterations in bacteria taxa from the phyla Bacteroidetes and Firmicutes,17, 18, 19, 20 which were reported to regulate ACE2 expression in rodents.21 There is an urgent need to understand host microbial perturbations that underlie SARS-CoV-2 infection, which may affect response to infection and efficacy of various future immune interventions, such as vaccines.22
In this pilot study, we hypothesize that the intestinal microbiota is altered in SARS-CoV-2 infection and is associated with susceptibility to severe disease. We prospectively included 15 hospitalized patients with COVID-19 admitted between February 16, 2020, and March 2, 2020, in Hong Kong, China, followed from hospital admission until discharge. Through the application of deep shotgun metagenomics, we investigated longitudinal changes of the gut microbiome in COVID-19.
Methods
Study Subject and Design
This prospective study involved 15 patients with COVID-19 hospitalized with laboratory-confirmed SARS-CoV-2 infection, 6 patients hospitalized with community-acquired pneumonia (pneumonia controls), and 15 healthy individuals (healthy controls) (Table 1 , Supplementary Table 1, Figure 1 ). SARS-CoV-2 infection was confirmed by 2 consecutive reverse-transcriptase polymerase chain reaction (RT-PCR) tests targeting different regions of the RdRp gene performed by the local hospital and Public Health Laboratory Service. Pneumonia controls were patients admitted with community-acquired pneumonia tested negative for SARS-CoV-2 PCR on 2 respiratory samples. Patients with COVID-19 and pneumonia controls were admitted to the Prince of Wales Hospital or the United Christian Hospital, Hong Kong. Healthy controls were individuals with no past medical history or history of antibiotic intake in the past 3 months recruited via advertisement from the general population and tested negative for SARS-CoV-2. All subjects were recruited between February 5 and March 17, 2020. Severity of COVID-19 infection was categorized as (1) mild, if there was no radiographic evidence of pneumonia; (2) moderate, if pneumonia was present along with fever and respiratory tract symptoms; (3) severe, if respiratory rate ≥30/min, oxygen saturation ≤93% when breathing ambient air, or PaO2 / FiO2 ≤300 mm Hg (1 mm Hg = 0.133 kPa); or (4) critical, if there was respiratory failure requiring mechanical ventilation, shock, or organ failure requiring intensive care.23 This study was approved by the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committees (2020.076). All patients provided informed consent to participate in this study. Data including demographics, laboratory results, imaging results, and medical therapy were extracted from the electronic medical records in the Hong Kong Hospital Authority clinical management system. Fecal samples from patients with COVID-19 were collected serially 2 to 3 times per week until discharge. This study was conducted in accordance with the Declaration of Helsinki.
Table 1.
Subject Characteristics
| Variables | COVID-19 cases | Pneumonia controls | Healthy controls |
|---|---|---|---|
| Number | 15 | 6 | 15 |
| Male | 7 (47) | 4 (67) | 9 (60) |
| Median age, y (IQR) | 55 (44, 67.5) | 50 (44, 65) | 48 (45, 48) |
| Comorbidities, n (%) | 6 (40) | 6 (100) | 0 (0) |
| Recent exposure history, n (%) | |||
| Travel to cities of Hubei province | 1 (7) | 0 (0) | 0 (0) |
| Contact with person with COVID-19 | 5 (33) | 0 (0) | 0 (0) |
| Have family cluster outbreak | 4 (27) | 0 (0) | 0 (0) |
| Symptoms at admission, n (%) | |||
| Fever | 9 (60) | 4 (67) | |
| Gastrointestinal symptoms | |||
| Diarrhea | 1 (7) | 2(33) | |
| Respiratory symptoms | |||
| Cough | 11 (73) | 4 (67) | |
| Sputum | 5 (33) | 3 (50) | |
| Rhinorrhea | 3 (20) | 1 (17) | |
| Shortness of breath | 4 (27) | 3 (50) | |
| Blood result | |||
| Lymphocyte counts (x 109/L, normal range 1.1–2.9) | 0.9 (0.7, 1.1) | 1.1 (0.9, 1.2) | |
| Antibiotic therapy at presentation, n (%) | 7 (47) | 6 (100) | |
| Amoxycillin Clavulanate | 4 (27) | 3 (50) | |
| Cephalosporin | 5 (33) | 6 (100) | |
| Tetracycline | 4 (27) | 0 (0) | |
| Antiviral therapy, n (%) | 13 (87) | 0 (0) | |
| Lopinavir-Ritonavir | 13 (87) | 0 (0) | |
| Ribavirin | 7 (47) | 0 (0) | |
| Interferon beta-1b | 1 (7) | 0 (0) | |
| Death, n (%) | 0 (0) | 0 (0) |
NOTE. Values are expressed in number (percentage) and median (interquartile range).
Figure 1.
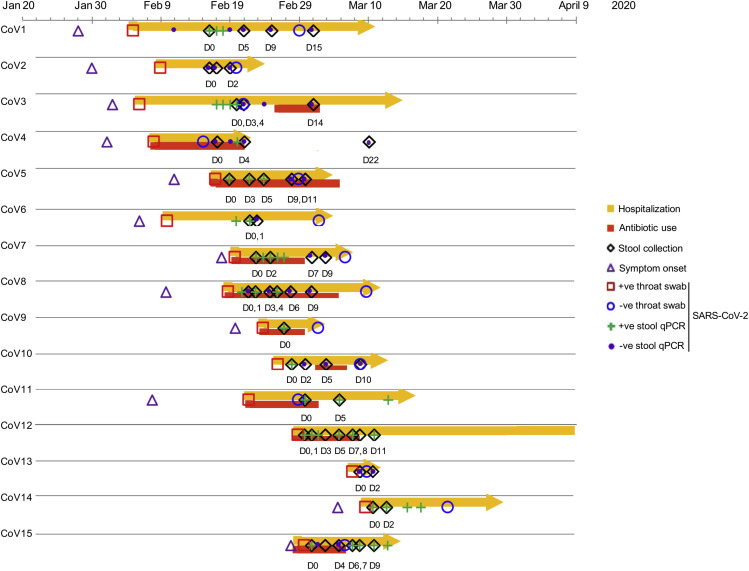
Schematic diagram of stool sample collection, SARS-CoV-2 PCR test results and hospitalization duration in patients with COVID-19 (n = 15). “CoV” denotes patient with COVID-19. Stool specimens were serially collected for shotgun metagenomics sequencing and quantitative RT-PCR test for SARS-CoV-2 virus; “D0” denotes baseline date when the first stool was collected after hospitalization; the following timepoints starting with “D” represent days since baseline stool collection. “+ve throat swab”: the first positive result for SARS-CoV-2 virus in nasopharyngeal/throat/pooled swabs; “-ve throat swab”: the first negative result for SARS-CoV-2 virus in 2 consecutive negative nasopharyngeal/throat/pooled swab tests, on which patient was then discharged.
Detection of Fecal SARS-CoV-2 Viral Load
SARS-CoV-2 viral loads in stool were measured using real-time RT-PCR assay. Viral RNA from stool samples was extracted using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany); 0.1g of stool was suspended in 1 mL of viral transport medium (in 1:10 dilution) and centrifuged for 20 minutes at 4000g. A 140-μL aliquot of the filtrate was used as starting material following the manufacturer’s protocol. SARS-CoV-2 RNA was quantified using real-time RT-PCR. The primer-probe set N1 (2019-nCoV_N1-F: 5ʹ-GAC CCC AAA ATC AGC GAA AT-3ʹ, 2019-nCoV_N1-R: 5ʹ-TCT GGT TAC TGC CAG TTG AAT CTG-3ʹ, and 2019-nCoV_N1-P: 5ʹ-FAM-ACC CCG CAT TAC GTT TGG ACC-BHQ1-3ʹ) designed by US Centers for Disease Control and Prevention were purchased from Integrated DNA Technologies (Coralville, IA). The 1-step real-time RT-PCR reaction contained 10 μL of the extracted preparation, 4 μL TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems, Foster City, CA) in a final reaction volume of 20 μL. The primer and probe concentration were 0.5 μM and 0.125 μM, respectively. The cycling conditions, 25°C for 2 minutes, 50°C for 15 minutes, 95°C for 2 minutes, followed by 45 cycles of 95°C for 15 seconds, and 55°C for 30 seconds, were performed with the StepOnePlus Real-Time PCR System (Applied Biosystems). The cycle threshold values of real-time RT-PCR were converted into viral RNA copies based on a standard curve prepared from 10-fold serial dilutions of known copies of plasmid containing the full N gene (2019-nCoV_N_Positive Control; Integrated DNA Technologies). Samples were considered as negative if the cycle threshold values exceeded 39.9 cycles. The detection limit of real-time RT-PCR was 347 copies/mL.
Microbial Profiling of Fecal Samples With Metagenomic Sequencing
An approximately 0.1 g fecal sample was prewashed with 1 mL double-distilled H2O and pelleted by centrifugation at 13,000g for 1 minute. The fecal DNA was subsequently extracted from the pellet using Maxwell RSC PureFood GMO and Authentication Kit (Promega, Madison, WI) following the manufacturer’s instructions. Briefly, the fecal pellet was added to 1 mL of CTAB buffer and vortexed for 30 seconds, then the sample was heated at 95°C for 5 minutes. After that, the samples were vortexed thoroughly with beads at maximum speed for 15 minutes. Then, 40 μL of proteinase K and 20 μL of RNase A was added to the sample and the mixture was incubated at 70°C for 10 minutes. The supernatant was then obtained by centrifuging at 13,000g for 5 minutes and was added into the Maxwell RSC machine for DNA extraction. Extracted DNA was subject to DNA libraries construction, completed through the processes of end repairing, adding A to tails, purification and PCR amplification, using Nextera DNA Flex Library Preparation kit (Illumina, San Diego, CA). Libraries were subsequently sequenced on our in-house sequencer Illumina NextSeq 550 (150 base pairs paired-end) at the Center for Microbiota Research, The Chinese University of Hong Kong. Raw sequence reads were filtered and quality-trimmed using Trimmomatic v0.3624 as follows: (1) trimming low-quality base (quality score <20); (2) removing reads shorter than 50 base pairs; (3) removing sequencing adapters. Contaminating human reads were filtering using Kneaddata (Reference database: GRCh38 p12) with default parameters. Profiling of bacterial communities was performed using MetaPhlAn2 (V2.9) by mapping reads to clade-specific markers.25
Statistical Analysis
Relative abundance data from MetaPhlAn2 were imported into R v3.5.1. Nonmetric multidimensional scaling analyses were performed on all baseline fecal microbiomes between groups, and serial fecal microbiomes in each COVID-19 case during the disease course, based on Bray-Curtis dissimilarities using vegan package (v2.5–3). Differential bacterial taxa between patients with COVID-19 (with or without antibiotics treatment at inclusion), patients with community-acquired pneumonia, and healthy controls were identified using Multivariate Association with Linear Models (MaAsLin).26 Spearman correlation analyses were conducted to associate baseline microbiome profiles of 7 antibiotics-naïve patients at baseline with COVID-19 severity, and to associate longitudinal fecal SARS-CoV-2 loads with timepoint-matched bacterial profiles across all 15 patients with COVID-19, while adjusting for confounding factors.
Data Availability
Metagenomics Sequencing dataset was deposited to the National Center for Biotechnology Information Sequence Read Archive under BioProject accession number PRJNA624223.
Results
Fecal Microbiome Alterations in COVID-19
Among the 15 patients with COVID-19, 7 were antibiotics-naïve (COVID-19[abx−]) and 8 received empirical antibiotics (COVID-19[abx+]) at baseline (defined as date of the first stool collection after hospitalization). The median ages of patients with COVID-19, pneumonia controls, and healthy controls were 55, 50, and 48 years, respectively; 40% and 100% of patients with COVID-19 and pneumonia controls, respectively, had underlying comorbidities (Table 1, Supplementary Table 1). All patients with COVID-19 presented with respiratory symptoms but only 1 had diarrhea at presentation. None of the patients developed GI symptoms during hospitalization. Median duration of hospitalization was 21 ± 2.4 days (mean ± SE) in COVID-19 and pneumonia cases.
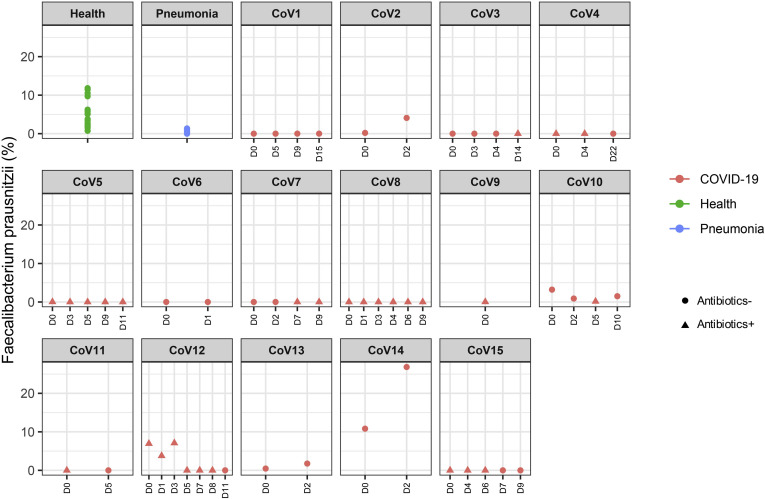
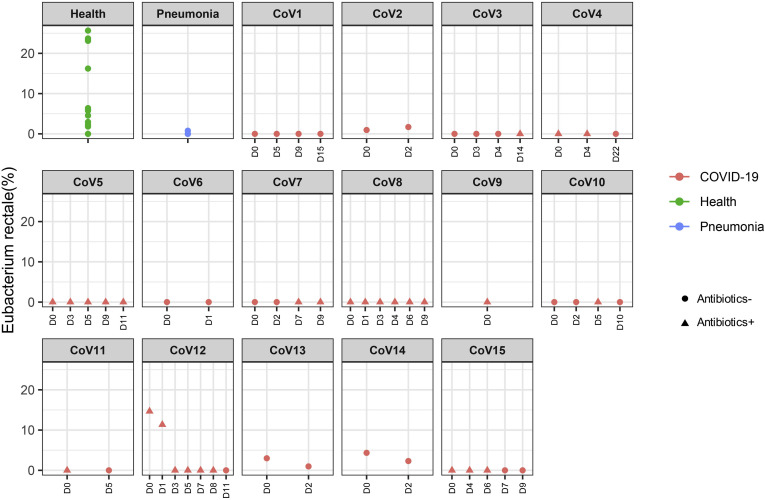
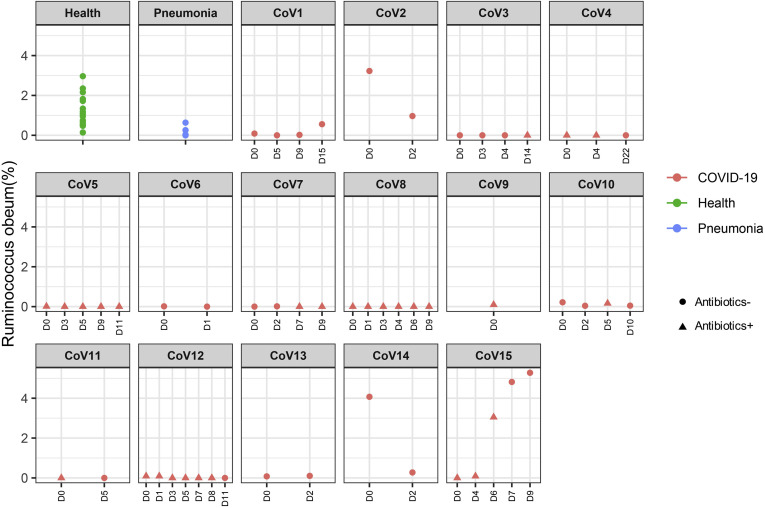
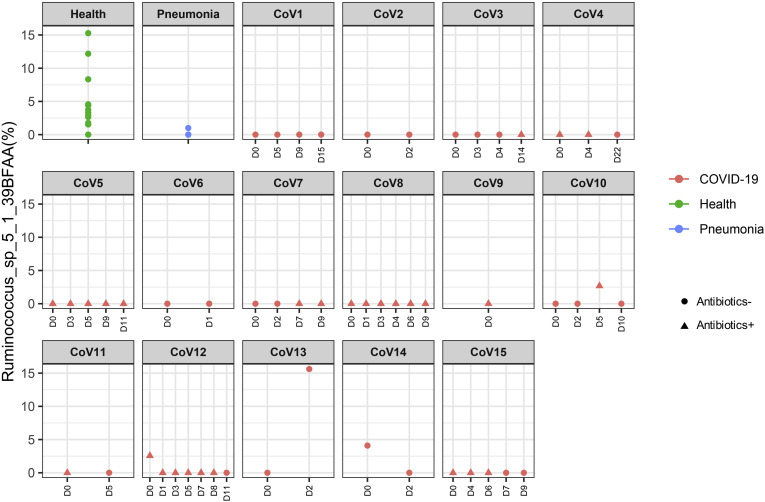
To understand alterations of the gut microbiome that underlie SARS-CoV-2 infection, we compared baseline fecal microbiome of patients with COVID-19 with healthy controls and pneumonia controls adjusting for age, gender, antibiotic use, and comorbidities. Antibiotic-naïve patients with COVID-19 were enriched in opportunistic pathogens known to cause bacteremia,27 , 28 including Clostridium hathewayi, Actinomyces viscosus, and Bacteroides nordii compared with controls (Table 2 ). COVID-19(abx+) patients demonstrated a further depletion of multiple bacterial species, which are symbionts beneficial to host immunity including Faecalibacterium prausnitzii, Lachnospiraceae bacterium 5_1_63FAA, Eubacterium rectale, Ruminococcus obeum, and Dorea formicigenerans compared with COVID-19(abx−) patients (Table 2). Regardless of antibiotic use, underrepresented bacterial species in patients with COVID-19 were consistently absent or present at very low abundance during the disease course, even when SARS-CoV-2 virus was cleared from the nasopharyngeal swab and stool, and respiratory symptoms had resolved (Supplementary Figures 1–6).
Table 2.
Gut Microbiome Features in Patients With COVID-19
| Gut microbiome feature | Taxon | Group | Coefficient | P value | Q value |
|---|---|---|---|---|---|
| COVID-19 (antibiotics naïve: abx−) | |||||
| Specifically enriched in COVID-19(abx−) | p_Actinobacteria|c_Actinobacteria|o_Actinomycetales|f_Actinomycetaceae|g_Actinomyces|s_Actinomyces_viscosus | COVID-19 (Abx−) | 0.243 | 8.2E-08 | 6.4E-05 |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Clostridiaceae|g_Clostridium|s_Clostridium_hathewayi | COVID-19 (Abx−) | 1.130 | 4.8E-06 | 2.5E-03 | |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Bacteroidaceae|g_Bacteroides|s_Bacteroides_nordii | COVID-19 (Abx−) | 0.164 | 2.3E-05 | 8.5E-03 | |
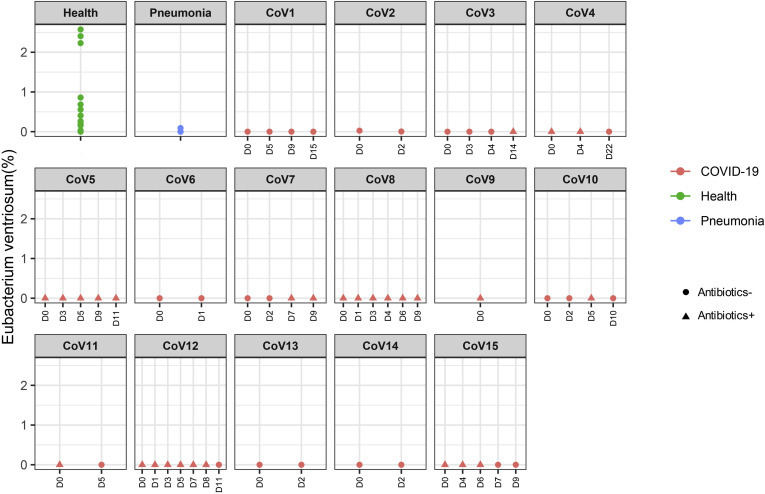
| Underrepresented in both COVID-19 and pneumonia | p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae|g_Eubacterium|s_Eubacterium_ventriosum | COVID-19 (Abx−) | -0.280 | 1.2E-04 | 2.5E-02 |
| COVID-19 (antibiotics exposed: abx+) | |||||
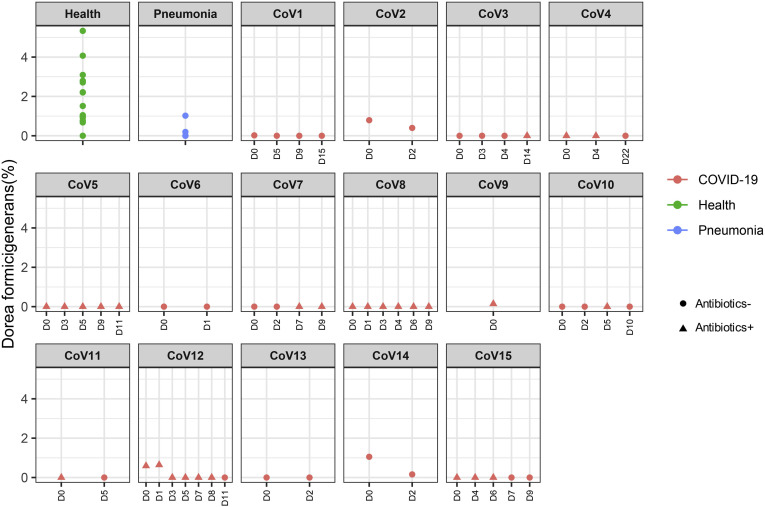
| Underrepresented in COVID-19(abx+) | p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Dorea|s_Dorea_formicigenerans | COVID-19 (Abx+) | -0.812 | 2.6E-05 | 0.00853 |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Blautia | COVID-19 (Abx+) | -0.441 | 6.1E-05 | 0.01491 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Ruminococcaceae|g_Faecalibacterium | COVID-19 (Abx+) | -0.537 | 3.2E-05 | 0.00924 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Ruminococcaceae|g_Faecalibacterium|s_Faecalibacterium_prausnitzii | COVID-19 (Abx+) | -0.537 | 3.2E-05 | 0.00924 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae | COVID-19 (Abx+) | -0.289 | 4.6E-05 | 0.01236 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae|g_Eubacterium | COVID-19 (Abx+) | -0.289 | 4.6E-05 | 0.01236 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae|g_Eubacterium|s_Eubacterium_rectale | COVID-19 (Abx+) | -0.903 | 1.7E-04 | 0.03316 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Ruminococcaceae | COVID-19 (Abx+) | -0.300 | 2.0E-04 | 0.03709 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Roseburia | COVID-19 (Abx+) | -0.598 | 2.3E-04 | 0.04018 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Coprococcus | COVID-19 (Abx+) | -0.447 | 1.6E-04 | 0.03239 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Blautia|s_Ruminococcus_obeum | COVID-19 (Abx+) | -0.623 | 4.3E-06 | 0.00243 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Lachnospiraceae_noname|s_Lachnospiraceae_bacterium_5_1_63FAA | COVID-19 (Abx+) | -0.341 | 2.5E-05 | 0.00853 | |
| Underrepresented in both COVID-19 and pneumonia | p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae|g_Eubacterium|s_Eubacterium_ventriosum | COVID-19 (Abx+) | -0.307 | 8.6E-06 | 0.00376 |
| Pneumonia patients | |||||
| Underrepresented in pneumonia | p_Firmicutes|c_Bacilli|o_Lactobacillales|f_Enterococcaceae|g_Enterococcus|s_Enterococcus_faecium | Pneumonia | 0.228 | 5.0E-05 | 0.01261 |
| p_Firmicutes|c_Erysipelotrichia|o_Erysipelotrichales|f_Erysipelotrichaceae|g_Erysipelotrichaceae_noname|s_Clostridium_ramosum | Pneumonia | 0.195 | 3.1E-06 | 0.00190 | |
| p_Firmicutes|c_Erysipelotrichia|o_Erysipelotrichales|f_Erysipelotrichaceae|g_Coprobacillus | Pneumonia | 0.402 | 5.2E-06 | 0.00257 | |
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Lachnospiraceae_noname|s_Lachnospiraceae_bacterium_5_1_63FAA | Pneumonia | -0.348 | 7.6E-05 | 0.01752 | |
| Underrepresented in both COVID-19 and pneumonia | p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae|g_Eubacterium|s_Eubacterium_ventriosum | Pneumonia | -0.256 | 3.5E-04 | 0.03539 |
Supplementary Figure 1.
Longitudinal changes of fecal abundance of Eubacterium ventriosum in patients with COVID-19 over the disease course. Bacterial species abundance is expressed as fractional abundance (%). “D0” denotes baseline date when the first stool was collected after hospitalization; the following timepoints starting with “D” represent days since baseline stool collection.
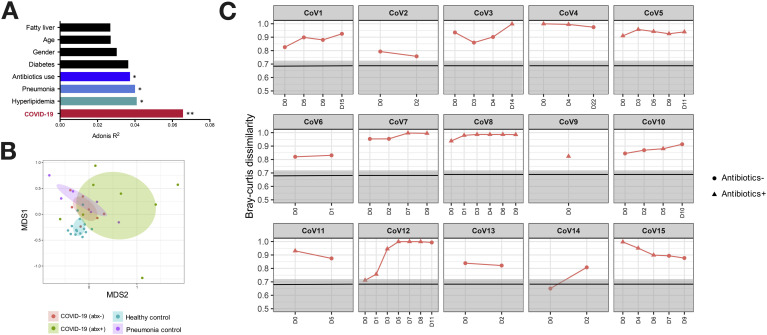
Among all host factors, COVID-19 infection showed the largest effect size in affecting the gut microbiome (PERMANOVA test, R 2 = 0.066, P = .002, Figure 2 A), followed by hyperlipidemia, pneumonia, and antibiotics, whereas age and gender showed no significant effects on gut microbiome alterations (Figure 2 A). At the whole microbiome community level, healthy subjects’ fecal microbiome clustered together, whereas that of COVID-19(abx−) patients clustered separately (PERMANOVA test, P = .001) and were more heterogeneous (Figure 2 B). Antibiotic treatment in patients with COVID-19 was associated with a more heterogeneous microbiome configuration and accompanied by further shift of the gut microbiome away from a healthy microbiome (Figure 2 B).
Figure 2.
Gut microbiome alterations in patients with COVID-19 and longitudinal changes over the disease course. (A) The effect size of subject metadata in gut microbiome composition, as determined by PERMANOVA test. ∗∗P < .01; ∗P < .05. (B) Microbiome community alterations in COVID-19, viewed by NMDS (nonmetric multidimensional scaling) plot based upon Bray-Curtis dissimilarities. The microbiomes were compared among healthy controls (n = 15), COVID-19 (abx−, n = 7), COVID-19 (abx+, n = 8), and pneumonia controls (n = 6). (C) Dissimilarity of the gut microbiome of patients with COVID-19 to that of healthy controls during the disease course. The microbiome dissimilarity was calculated as Bray-Curtis dissimilarity. The gray area denotes the range of Bray-Curtis dissimilarities among gut microbiomes of healthy controls, and the solid black line indicates the median dissimilarity among healthy individuals. “CoV” denotes patient with COVID-19. “D0” denotes baseline date when the first stool was collected after hospitalization; the following timepoints starting with “D” represent days since baseline stool collection.
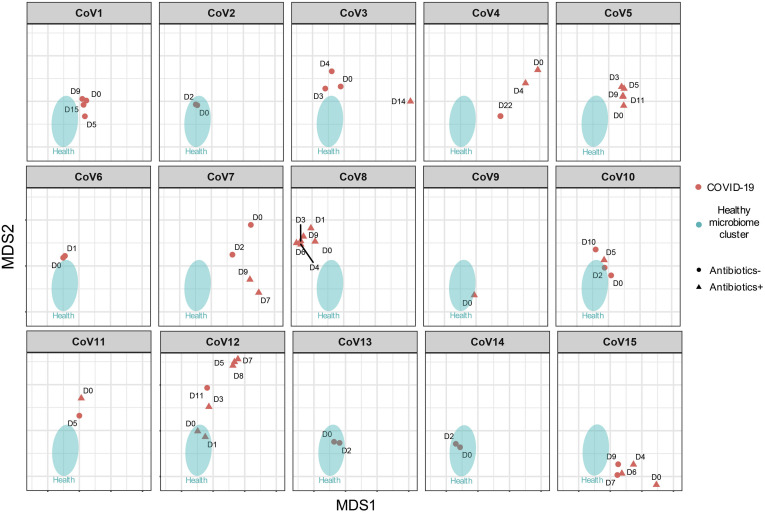
We next explored whether recovery from SAR-CoV-2 infection was associated with restoration of gut microbiome to a community level similar to that of healthy individuals. Overall, the gut microbiome of all patients with COVID-19 remained stable but were markedly disparate from that of healthy controls, both during the disease course and after clearance of SARS-CoV-2 (Figure 2 C). Although the microbiome of 5 patients with COVID-19 (CoV1, 4, 7, 11, 15) showed closer proximity to healthy microbiomes over time, patients CoV3, 5, 8, 10, and 12 became more disparate from healthy microbiomes over time (Supplementary Figure 7). At the last follow-up, the gut microbiome of these 10 patients remained substantially different from that of healthy controls, despite clearance of SARS-CoV-2 infection as defined by negative SARS-CoV-2 tests on nasopharyngeal swab or deep throat saliva (Figure 2 C, Supplementary Figure7). Of note, patient CoV4 was discharged on day 5 but his gut microbiome on day 22 was persistently different from that of healthy individuals.
Supplementary Figure 7.
Longitudinal changes of the fecal microbiome in patients with COVID-19, at the community level, during the disease course. “CoV” denotes patient with COVID-19. “D0” denotes baseline date when the first stool was collected after hospitalization; the following timepoints starting with “D” represent days since baseline stool collection.
Baseline Gut Microbiome and Disease Severity of COVID-19
To understand whether baseline gut microbiome affects the severity of COVID-19, we assessed association between baseline fecal microbiome and COVID-19 severity (mild, moderate, severe, or critical) in 7 antibiotic-naïve COVID-19 cases. A total of 23 bacterial taxa were found to be significantly associated with COVID-19 disease severity, most of which (15 of 23) were from the Firmicutes phylum (Table 3 ). Among them, 8 and 7 Firmicutes members, respectively, showed positive and negative correlation with disease severity. These data are in line with a report showing that different Firmicutes bacteria have diverse roles in upregulating or downregulating ACE2 expression in the murine gut.21 Our finding of the association of gut Firmicutes bacteria with COVID-19 severity highlights the potential importance of bacterial membership in modulating human response to SARS-CoV-2 infection.
Table 3.
Correlation of Gut Bacteria With COVID-19 Severity
| Correlation | Bacteria taxa | Correlation coefficient Rho | P value |
|---|---|---|---|
| Positive correlation with COVID-19 severity | p__Firmicutes|c__Erysipelotrichia|o__Erysipelotrichales|f__Erysipelotrichaceae|g__Coprobacillus | 0.92 | .003 |
| p__Firmicutes|c__Erysipelotrichia|o__Erysipelotrichales|f__Erysipelotrichaceae|g__Erysipelotrichaceae_noname|s__Clostridium_ramosum | 0.92 | .003 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Clostridiaceae|g__Clostridium|s__Clostridium_hathewayi | 0.90 | .005 | |
| p__Firmicutes|c__Erysipelotrichia | 0.90 | .006 | |
| p__Firmicutes|c__Erysipelotrichia|o__Erysipelotrichales | 0.90 | .006 | |
| p__Firmicutes|c__Erysipelotrichia|o__Erysipelotrichales|f__Erysipelotrichaceae | 0.90 | .006 | |
| p__Firmicutes|c__Erysipelotrichia|o__Erysipelotrichales|f__Erysipelotrichaceae|g__Erysipelotrichaceae_noname | 0.90 | .006 | |
| p__Actinobacteria|c__Actinobacteria|o__Actinomycetales|f__Actinomycetaceae|g__Actinomyces|s__Actinomyces_odontolyticus | 0.87 | .011 | |
| p__Firmicutes|c__Erysipelotrichia|o__Erysipelotrichales|f__Erysipelotrichaceae|g__Erysipelotrichaceae_noname|s__Erysipelotrichaceae_bacterium_6_1_45 | 0.87 | .011 | |
| p__Proteobacteria|c__Gammaproteobacteria|o__Enterobacteriales|f__Enterobacteriaceae|g__Enterobacter | 0.87 | .011 | |
| p__Proteobacteria|c__Gammaproteobacteria|o__Enterobacteriales|f__Enterobacteriaceae|g__Enterobacter|s__Enterobacter_cloacae | 0.87 | .011 | |
| p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Porphyromonadaceae|g__Parabacteroides|s__Parabacteroides_unclassified | 0.81 | .029 | |
| p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Rikenellaceae|g__Alistipes|s__Alistipes_indistinctus | 0.81 | .029 | |
| Negative correlation with COVID-19 severity | p__Actinobacteria|c__Actinobacteria|o__Bifidobacteriales|f__Bifidobacteriaceae|g__Bifidobacterium|s__Bifidobacterium_pseudocatenulatum | −0.81 | .026 |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Dorea | −0.81 | .026 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Dorea|s__Dorea_longicatena | −0.81 | .026 | |
| p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Bacteroidaceae|g__Bacteroides|s__Bacteroides_ovatus | −0.84 | .019 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Anaerostipes|s__Anaerostipes_hadrus | −0.87 | .011 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Lachnospiraceae_noname|s__Lachnospiraceae_bacterium_5_1_63FAA | −0.87 | .011 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Lachnospiraceae|g__Roseburia | −0.87 | .011 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Faecalibacterium | −0.87 | .011 | |
| p__Firmicutes|c__Clostridia|o__Clostridiales|f__Ruminococcaceae|g__Faecalibacterium|s__Faecalibacterium_prausnitzii | −0.87 | .011 | |
| p__Bacteroidetes|c__Bacteroidia|o__Bacteroidales|f__Rikenellaceae|g__Alistipes|s__Alistipes_onderdonkii | −0.90 | .005 |
Three bacterial members from the Firmicutes phylum, the genus Coprobacillus, the species Clostridium ramosum and C. hathewayi, were the top bacteria positively associated with COVID-19 disease severity (Spearman correlation coefficient Rho >0.9, P < .01, Table 3). Both C. ramosum and C. hathewayi have been associated with human infection and bacteremia.27 , 29 Importantly, Coprobacillus bacterium has been shown to strongly upregulate colonic expression of ACE2 in the murine gut.21 In contrast, 2 beneficial species, Alistipes onderdonkii and Faecalibacterium prausnitzii, were top bacterial species to show a negative correlation with COVID-19 severity (Table 3). Alistipes species are indole positive, involved in the serotonin precursor tryptophan metabolism and in maintaining gut immune homeostasis,30 , 31 whereas F. prausnitzii has anti-inflammatory properties.32 Although we cannot assign a causative or preventive role of these bacteria in disease pathogenesis or severity, our data underscore a potential role for bacteria in determining response to SARS-CoV-2 infection and intensity of the infection in the host.
Fecal SARS-CoV-2 Virus Load and Gut Bacterial Abundance
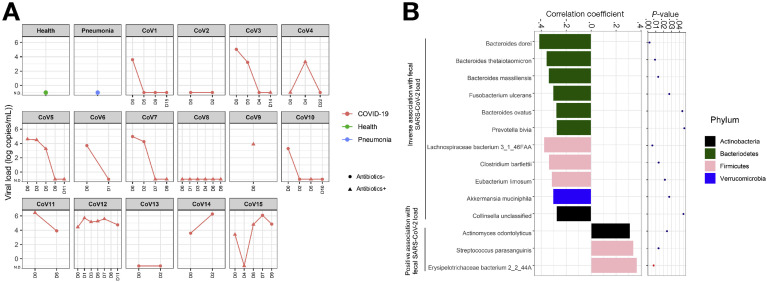
Eleven of the 15 patients had SARS-CoV-2 nucleic acid detected in feces at hospitalization (median 3.86 × 103 copies per mL inoculum, as determined by RT-PCR) and 5 of them cleared the SARS-CoV-2 virus over time (Figure 3 A). We investigated whether gut bacteria were associated with fecal SARS-CoV-2 load over the course of hospitalization. A total of 14 bacterial species were identified to be significantly associated with fecal viral load of SARS-CoV-2 across all fecal samples (Figure 3 B). Among them, 6 species were from the Bacteroidetes phylum. Four Bacteroides species, including Bacteroides dorei, Bacteroides thetaiotaomicron, Bacteroides massiliensis, and Bacteroides ovatus, showed significant inverse correlation with fecal SARS-CoV-2 load (all Spearman correlation coefficient Rho <−0.2, P < .05, Figure 3 B). Interestingly, all these 4 species were associated with downregulation of ACE2 expression in the murine colon.21 Taken together, these data suggest that Bacteroides species may have a potential protective role in combating SARS-CoV-2 infection by hampering host entry through ACE2. In contrast, Erysipelotrichaceae bacterium 2_2_44A, a Firmicutes species, showed the strongest positive correlation with fecal SARS-CoV-2 load (Spearman correlation coefficient Rho = 0.89, P = .006, Figure 3 B). Erysipelotrichaceae has been implicated in inflammation-related disorders of the GI tract.33 Considering the strong association of baseline abundance of Erysipelotrichaceae with COVID-19 severity (Spearman correlation Rho = 0.89, P = .006, Table 3), gut Erysipelotrichaceae may play a role in augmenting SARS-CoV-2 infection in the host gut.
Figure 3.
Correlation between gut bacteria and fecal SARS-CoV-2 shedding in patients with COVID-19 over the disease course. (A) Longitudinal changes in fecal viral loads of patients with COVID-19. (B) Bacteria significantly associated with fecal viral load during disease course, as determined by Spearman correlation test.
Discussion
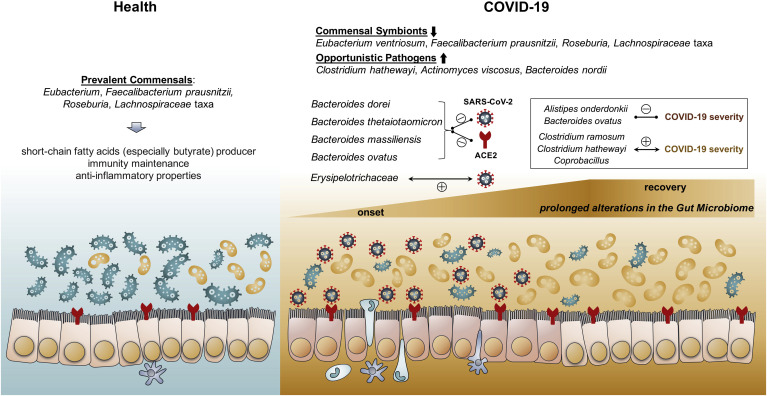
We showed for the first time that the gut microbiome was disturbed in patients with COVID-19. The alterations, observed even in patients with COVID-19 naïve to antibiotic therapy, were characterized by enrichment of opportunistic pathogens and depletion of beneficial commensals (Figure 4 ). Loss of salutary species in COVID-19 persisted in most patients despite clearance of SARS-CoV-2 virus, suggesting that exposure to SARS-CoV-2 infection and/or hospitalization may be associated with a more long-lasting detrimental effect to the gut microbiome.
Figure 4.
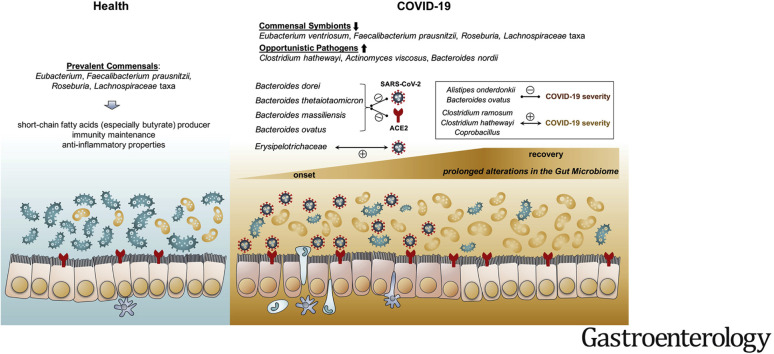
Schematic summary of the gut microbiome alterations in COVID-19. In healthy individuals, Eubacterium, Faecalibacterium prausnitzii, Roseburia, and Lachnospiraceae taxa are prevalent in their gut microbiome. However, the gut microbiome of patients with COVID-19 is characterized by enrichment of opportunistic pathogens and depletion of commensals in the gut. Such gut dysbiosis persists during the COVID-19 disease course, even after clearance/recovery of SARS-CoV-2 infection. Baseline fecal abundance of the bacteria Coprobacillus, Clostridium ramosum, and Clostridium hathewayi showed significant correlation with COVID-19 severity, whereas an anti-inflammatory bacterium Faecalibacterium prausnitzii showed an inverse correlation. Four Bacteroidetes members, including Bacteroides dorei, Bacteroides thetaiotaomicron, Bacteroides massiliensis, and Bacteroides ovatus, known to downregulate ACE2 expression in the murine gut, showed significant inverse correlation with fecal SARS-CoV-2 viral load in patients with COVID-19.
Studies have shown that respiratory viral infections can alter the gut microbiome, such as pulmonary infections by influenza and respiratory syncytial virus.15 , 34, 35, 36 Viral infections predispose patients to secondary bacterial infections, which often have a more severe clinical course.14 , 37 We found that a number of pathogens and opportunistic pathogens were enriched in the gut microbiome of patients with COVID-19, including C. hathewayi, B. nordii, A. viscosus, and a higher baseline abundance of C. hathewayi correlated with more severe COVID-19. Most of these bacteria are bacteremia-associated bacteria, indicating susceptibility for severe disease course due to potential secondary bacterial infection. We also identified an opportunistic pathogen of the oral cavity and upper respiratory tract, A. viscosus, in the gut of patients with COVID-19.38 Its presence suggests the passage or transmission of extra-intestinal microbes into the gut.
Recently, a study provided direct evidence that SARS-CoV-2 can bind to human ACE2 as host entry point.9 ACE2 is highly expressed in the intestine especially in colonocytes of healthy subjects and in patients with inflammatory bowel disease,10 and can regulate amino acid transport, microbial ecology, and inflammation in the gut.12 Interestingly, Bacteroidetes species have been shown to downregulate ACE2 expression in the murine colon, whereas Firmicutes species showed variable effects in modulating ACE2 expression.21 We found that baseline abundance of Bacteroidetes species, A. onderdonkii and B. ovatus, negatively correlated with COVID-19 severity, and 4 species from the genus Bacteroides of the phylum Bacteroidetes (B. dorei, B. thetaiotaomicron, B. massiliensis, and B. ovatus) showed inverse correlation with fecal viral load of SARS-CoV-2 (Figure 4). Among them, B. dorei has been reported to suppress colonic ACE2 expression21 and to calibrate host immune response.39 , 40 The highest SARS-CoV-2 mortality and morbidity have been reported in older patients and in those with underlying chronic diseases that are associated with inflammation, such as hypertension, obesity, diabetes mellitus, and coronary artery disease.41, 42, 43 Interestingly, these subjects were also reported to have a lower abundance of Bacteroides species than healthy individuals.17, 18, 19, 20 These findings altogether suggest that an individual’s gut microbiome configuration may affect the subject’s susceptibility and response to SARS-CoV-2 infection.
Cytokine profile associated with hyperinflammation state in severe COVID-19 has been characterized by increased interferon-γ inducible protein and other cytokines. Given limited proven treatment for COVID-19, understanding host cytokine pathways and microbiota interactions with cytokine responses in SARS-CoV-2 infection is essential in developing new treatment approaches.44
One major limitation of this exploratory study is the modest sample size. Although assigning a causative relationship between COVID-19 and gut dysbiosis requires larger validation studies, this pilot study presents the first data to examine the influence of SARS-CoV2 infection on gut microbiome composition and dynamics. We attempted to adjust for factors, such as age, gender, therapy, and comorbidities, which may explain the observed variance in the data. As we included only hospitalized patients with moderate/severe disease, such findings may not be generalizable to all COVID-19 cases, including those with mild or asymptomatic COVID-19. Stool collected after hospitalization for microbiome analysis does not represent the bona fide baseline microbiome at COVID-19 onset, nor the baseline microbiome before disease onset. Further studies should prospectively include asymptomatic subjects, and if infected with SARS-CoV-2, followed up at disease onset, during disease course, and long term after discovery to delineate the role of microbiome changes in SARS-CoV-2 infection and postinfection recovery.
The use of empirical antibiotics (which was common in the initial outbreak of SARS-CoV-2 when secondary bacterial infection was a concern) led to further loss of salutary symbionts and exacerbation of gut dysbiosis in patients with COVID-19, and our data support avoidance of unnecessary antibiotics use in the treatment of viral pneumonitis, as antibiotics can eliminate beneficial bacteria and weaken the gut barrier.45 In addition, antibiotics-driven gut microbiome perturbation can alter immunity to vaccines in humans.46 Improving efficacy of future immune interventions such as vaccines, through modulating the gut microbiome, in combating COVID-19 should be considered. One approach for promoting a healthy microbiome may include measures to enhance intestinal butyrate production through the promotion of microbial interactions by dietary changes, and reduction of proinflammatory states.
In conclusion, our study provides evidence of prolonged gut microbiome dysbiosis in COVID-19 and its association with fecal SARS-CoV-2 virus shedding and disease severity. These data highlight a new concept that novel and targeted approach of modulation of the gut microbiota may represent a therapeutic avenue for COVID-19 and its comorbidities.
Acknowledgments
We thank all health care workers working in isolation wards of Prince of Wales Hospital, Hong Kong, China. We thank Apple C.M. Yeung, Wendy C.S. Ho, Miu L. Chin, Rity Wong, and Vickie Li for their technical contributions in this study. We thank Whitney Tang for her assistance with the graphical abstract.
CRediT Authorship Contributions
Tao Zuo, PhD (Formal analysis: Lead; Investigation: Equal; Methodology: Lead; Writing – original draft: Lead; Writing – review & editing: Lead). Fen Zhang, PhD (Formal analysis: Equal; Methodology: Equal). Grace C.Y. Lui, Dr (Conceptualization: Equal; Resources: Lead; Writing – review & editing: Equal). Yun Kit Yeoh, PhD (Writing – review & editing: Lead). Amy Y.L. Li, Bachelor (Data curation: Lead). Hui Zhan, PhD (Investigation: Supporting). Yating Wan, PhD (Methodology: Supporting). Arthur Chung, PhD (Methodology: Supporting). Chun Peng Cheung, Bachelor (Project administration: Lead). Nan Chen, Master (Investigation: Supporting). Christopher K.C. Lai, PhD (Writing – review & editing: Supporting). Zigui Chen, PhD (Writing – review & editing: Equal). Eugene Y.K. Tso, MD (Resources: Equal). Kitty S.C. Fung, MD (Resources: Equal). Veronica Chan, MD (Resources: Equal). Lowell Ling, MD (Resources: Equal). Gavin Joynt, PhD (Supervision: Supporting). David S.C. Hui, MD (Supervision: Supporting). Francis K.L. Chan, MD (Supervision: Lead). Paul K.S. Chan, PhD (Project administration: Lead). Siew C. Ng, PhD (Conceptualization: Lead; Funding acquisition: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead).
Footnotes
Conflict of interest The authors disclose no conflicts.
Funding This study was funded by D. H. Chen Foundation and Health and Medical Research Fund, Hong Kong.
Author names in bold designate shared co-first authorship.
Supplementary Table 1.
| Case | Sex | Age | Comorbidities | Recent exposure history | Symptoms at admission |
Admitted to ICU | Chest radiograph findings | COVID-19 severity | |
|---|---|---|---|---|---|---|---|---|---|
| Fever and respiratory | GI | ||||||||
| CoV1 | F | 65 | Hypertension, Chronic hepatitis B carrier | No | Fever, cough, sputum | Nil | Yes | Bilateral LZ haziness | Critical |
| CoV2 | F | 55 | None | Contact with person with COVID-19 | Fever, runny nose | Nil | No | Bilateral LZ haziness | Moderate |
| CoV3 | M | 42 | None | Travel to Hubei province | Fever, cough | Nil | Yes | RLZ haziness, RLL collapse |
Critical |
| CoV4 | M | 70 | Hyperlipidemia, duodenal ulcer | No | Cough, shortness of breath | Nil | No | Bilateral lung haziness | Severe |
| CoV5 | M | 58 | None | No | Fever, cough | Diarrhea | No | Slight RLZ haziness | Moderate |
| CoV6 | M | 71 | None | No | Fever, cough, shortness of breath | Nil | No | Bilateral lung infiltration | Severe |
| CoV7 | M | 48 | Diabetes mellitus, hypertension, hyperlipidemia | No | Fever, cough | Nil | No | LLZ haziness | Moderate |
| CoV8 | F | 38 | None | No | Fever, cough, sputum, runny nose | Nil | No | Bilateral LZ infiltrates | Moderate |
| CoV9 | M | 33 | None | Contact with person with COVID-19 | Fever, cough | Nil | No | Bilateral LZ haziness | Moderate |
| CoV10 | F | 70 | Obesity, hypertension | No | Cough | Nil | No | Bilateral LZ haziness | Moderate |
| CoV11 | M | 62 | Diabetes, hyperlipidemia, left subclavian artery occlusion | No | Fever, cough, sputum, shortness of breath | Nil | No | Bilateral lung infiltrates | Severe |
| CoV12 | F | 71 | Hypertension, renal impairment, hyperlipidemia | Contact with person with COVID-19 | Cough | Nil | No | Bilateral lung infiltrates | Moderate |
| CoV13 | F | 47 | None | Contact with person with COVID-19 | Cough | Nil | No | Bilateral lung infiltrates | Moderate |
| CoV14 | F | 22 | None | Contact with person with COVID-19 | Fever, runny nose | Nil | No | Bilateral lung infiltrates | Moderate |
| CoV15 | F | 46 | None | Contact with person with COVID-19 | Cough, shortness of breath | Nil | No | Clear | Mild |
| P1 | F | 69 | Hypertension, diabetes mellitus, Tricuspid regurgitation | No | Fever | Nil | No | LMZ pneumonia | N/A |
| P2 | M | 43 | Nonalcoholic Fatty liver disease | No | Cough | Nil | No | Right sided infiltrates | N/A |
| P3 | F | 92 | Diabetes, Hypertension, pulmonary fibrosis, Paroxysmal atrial fibrillation, Acute coronary syndrome | No | Cough, sputum, shortness of breath | Nil | No | Bilateral lung infiltrate | N/A |
| P4 | M | 47 | Diabetes mellitus | No | Fever, sputum | Nil | No | Left effusion, LMZ infiltrates | N/A |
| P5 | M | 36 | Ischemic priapism | No | Fever, cough, sputum, shortness of breath | Diarrhea | No | N/A | N/A |
| P6 | M | 52 | Epilepsy, Hepatitis | No | Fever, cough, runny nose, shortness of breath | Diarrhea | No | N/A | N/A |
References
- 1.Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy [published online ahead of print March 23, 2020]. JAMA https://doi.org/10.1001/jama.2020.4683. [DOI] [PubMed]
- 2.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang W., Feng Z., Rao S. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 5.Cheung K.S., Hung I.F.N., Chan P.P.Y. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wölfel R., Corman V.M., Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y., Li X., Zhu B. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nature Medicine. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Effenberger M., Grabherr F., Mayr L. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J., Ye G., Shi K. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Zhao S, Liu M, et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism [published online ahead of print February 5, 2020]. medRxiv doi: 10.1101/2020.02.05.20020545. [DOI]
- 11.Xiao F., Tang M., Zheng X. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto T., Perlot T., Rehman A. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma W.-T., Pang M., Fan Q.-L. The commensal microbiota and viral infection: a comprehensive review. Front Immunol. 2019;10:1551. doi: 10.3389/fimmu.2019.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanada S., Pirzadeh M., Carver K.Y. Respiratory viral infection-induced Microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz S., Mazel-Sanchez B., Kandasamy M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. 2018;6:9. doi: 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Z., Xiao Y., Kang L. Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin Infect Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh P.J., Ley R.E., Mahowald M.A. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 18.Emoto T., Yamashita T., Sasaki N. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. Journal of atherosclerosis and thrombosis. 2016;23:908–921. doi: 10.5551/jat.32672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang T., Santisteban M.M., Rodriguez V. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65:1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley R.E., Turnbaugh P.J., Klein S. Human gut microbes associated with obesity. nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 21.Geva-Zatorsky N., Sefik E., Kua L. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943.e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalantar-Zadeh K., Ward S.A., Kalantar-Zadeh K. Considering the effects of microbiome and diet on SARS-CoV-2 infection: nanotechnology roles. ACS Nano. 2020;14:5179–5182. doi: 10.1021/acsnano.0c03402. [DOI] [PubMed] [Google Scholar]
- 23.Wu J., Liu J., Zhao X. Clinical characteristics of imported cases of COVID-19 in Jiangsu province: a multicenter descriptive study. Clin Infect Dis. 2020;71:706–712. doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N., Waldron L., Ballarini A. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan X.C., Tickle T.L., Sokol H. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsayed S., Zhang K. Human infection caused by Clostridium hathewayi. Emerg Infect Dis. 2004;10:1950. doi: 10.3201/eid1011.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dakshinamoorthy M., Venkatesh A., Arumugam K. A literature review on dental caries vaccine-a prevention strategy. Indian J Public Health. 2019;10:3041–3043. [Google Scholar]
- 29.Forrester J.D., Spain D.A. Clostridium ramosum bacteremia: case report and literature review. Surg Infect. 2014;15:343–346. doi: 10.1089/sur.2012.240. [DOI] [PubMed] [Google Scholar]
- 30.Gao J., Xu K., Liu H. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdu E.F., Hayes C.L., O’Mahony S.M. Elsevier; London: 2016. Importance of the microbiota in early life and influence on future health. In: The Gut-Brain Axis; pp. 159–184. [Google Scholar]
- 32.Miquel S., Martin R., Rossi O. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Kaakoush N.O. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Li F., Wei H. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell–dependent inflammation. J Exp Med. 2014;211:2397–2410. doi: 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deriu E., Boxx G.M., He X. Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groves H.T., Cuthbertson L., James P. Respiratory disease following viral lung infection alters the murine gut microbiota. Front Immunol. 2018;9:182. doi: 10.3389/fimmu.2018.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brundage J.F. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis. 2006;6:303–312. doi: 10.1016/S1473-3099(06)70466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habib S., Siddiqui A.H., Azam M. Actinomyces viscosus causing disseminated disease in a patient on methotrexate. Respir Med Case Rep. 2018;25:158–160. doi: 10.1016/j.rmcr.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vatanen T., Kostic A.D., d’Hennezel E. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida N., Emoto T., Yamashita T. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;138:2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]
- 41.Qingxian C., Fengjuan C., Wang T. Obesity and COVID-19 severity in a designated hospital in Shenzhen, China. Diabetes Care. 2020;43:1392–1398. doi: 10.2337/dc20-0576. [DOI] [PubMed] [Google Scholar]
- 42.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill M.A., Mantzoros C., Sowers J.R. Commentary: COVID-19 in patients with diabetes. Metabolism. 2020;107:154217. doi: 10.1016/j.metabol.2020.154217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendes V., Galvao I., Vieira A.T. Mechanisms by which the gut microbiota influences cytokine production and modulates host inflammatory responses. J Interferon Cytokine Res. 2019;39:393–409. doi: 10.1089/jir.2019.0011. [DOI] [PubMed] [Google Scholar]
- 45.Tulstrup M.V.-L., Christensen E.G., Carvalho V. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagan T., Cortese M., Rouphael N. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Metagenomics Sequencing dataset was deposited to the National Center for Biotechnology Information Sequence Read Archive under BioProject accession number PRJNA624223.