Abstract
Recombinase polymerase amplification (RPA) assays are valuable molecular diagnostic tools that can detect and identify plant pathogens in the field without time-consuming DNA extractions. Historically, RPA assay reagents were commercially available as a lyophilized pellet in microfuge strip tubes, but have become available in liquid form more recently—both require the addition of primers and probes prior to use, which can be challenging to handle in a field setting. Lyophilization of primers and probes, along with RPA reagents, contained within a single tube limits the risk of contamination, eliminates the need for refrigeration, as the lyophilized reagents are stable at ambient temperatures, and simplifies field use of the assays. This study investigates the potential effect of preformulation on assay performance using a previously validated Phytophthora genus-specific RPA assay, lyophilized with primers and probes included with the RPA reagents. The preformulated lyophilized Phytophthora RPA assay was compared with a quantitative polymerase chain reaction (qPCR) assay and commercially available RPA kits using three qPCR platforms (BioRad CFX96, QuantStudio 6 and Applied Biosystems ViiA7) and one isothermal platform (Axxin T16-ISO RPA), with experiments run in four separate labs. The assay was tested for sensitivity (ranging from 500 to 0.33 pg of DNA) and specificity using purified oomycete DNA, as well as crude extracts of Phytophthora-infected and non-infected plants. The limit of detection (LOD) using purified DNA was 33 pg in the CFX96 and ViiA7 qPCR platforms using the preformulated kits, while the Axxin T16-ISO RPA chamber and the QuantStudio 6 platform could detect down to 3.3 pg with or without added plant extract. The LOD using a crude plant extract for the BioRad CFX96 was 330 pg, whereas the LOD for the ViiA7 system was 33 pg. These trials demonstrate the consistency and uniformity of pathogen detection with preformulated RPA kits for Phytophthora detection when conducted by different labs using different instruments for measuring results.
Keywords: diagnostics, isothermal amplification, oomycetes, point of care diagnostics
1. Introduction
Oomycetes within the genus Phytophthora constitute a large group of destructive plant pathogens. Phytophthora species cause root, crown, stem, foliar and fruit diseases on agriculturally and ecologically important species of plants [1,2,3,4]. These diseases can be difficult or impossible to distinguish by symptoms alone and in-lab diagnostic testing is required for accurate pathogen identification. Identification of ambiguous Phytophthora species has traditionally relied on techniques such as baiting, isolation onto a Phytophthora semi-selective medium, DNA extraction and polymerase chain reaction (PCR) or antibodies (i.e., immunostrips) to identify the genus or species present [5,6,7,8]. However, this is time consuming and some species, such as the causal agent of sudden oak death, Phytophthora ramorum [9], are of regulatory importance, requiring a rapid and accurate identification. Likewise, the generic antibody used in commercial immunostrips to detect Phytophthora species cross reacts with some Pythium or Phytopythium species, making this detection method fast but not specific [10].
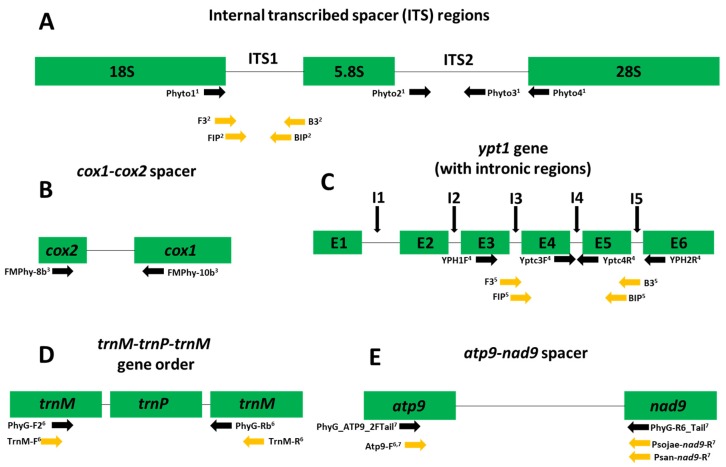
Non-isothermal molecular-based assays (e.g., polymerase chain reaction (PCR) and quantitative PCR (qPCR)) have been developed to identify Phytophthora species using several nuclear and mitochondrial loci (e.g., ypt1 gene and atp9–nad9 spacer region) [1,11,12,13,14,15,16,17,18,19,20] (Figure 1). Figure 1 details the loci commonly used for Phytophthora species identification in both isothermal and non-isothermal assays; citations for the primers used are included in the figure legend (Figure 1). While non-isothermal molecular assays can identify Phytophthora species, depending on the loci amplified, molecular assays have the potential to cross react with non-target DNA [6,11]. Polymerase chain reaction-based assays require a significant amount of setup and run time, and some can cross react with certain Pythium or Phytopythium species, making them not suitable for the fast turnaround times and accuracy needed when detecting and identifying plant pathogens [1]. Likewise, PCR assays require gel electrophoresis of the DNA product and cannot be performed in the field for onsite detection. qPCR assays that do not need gel electrophoresis for results, instead using a probe containing a fluorophore to detect amplification on a fluorometer with the ability to quantify the target DNA, are available. However, significant time input is still needed to perform DNA extractions and run the qPCR assay itself.
Figure 1.
Visualization of intergenic regions used for diagnostic assays of oomycetes with particular emphasis on Phytophthora species. (A) Internal transcribed spacer region, (B) cox1-cox2 spacer region, (C) ras-related ypt1 gene with intronic regions, (D) trnM-trnP-trnM gene order and (E) atp9–nad9 spacer region. Also denoted are the primers used in various reported isothermal (yellow arrows) and non-isothermal (black arrows) assays for Phytophthora species. 1 Phytophthora ramorum-specific primers from Garbelotto et al., 2002. Phyto2 and Phyto3 are nested primers to be used after a preliminary amplification with Phyto1 and Phyto4. 2 Phytophthora ramorum-specific primers from Tomlinson et al., 2010. 3 Phytophthora genus-specific primers from Martin et al., 2004. 4 Phytophthora kernoviae-specific primers from Schena et al., 2006. Yptc3F and Yptc4R are nested primers to be used after a preliminary amplification with YPH1F and YPH2R. 5 Phytophthora infestans-specific primers from Khan et al., 2017. 6 Universal primers for Phytophthora species from Miles et al., 2015. Isothermal assay primers TrnM-F and TrnM-R are used for genus-specific detection. 7 Phytophthora genus-specific (PhyG_ATP9_2FTail and PhyG-R6_Tail, Atp9-F) and species-specific (Psojae-nad9-R for Phytophthora sojae and Psan-nad9-R for Phytophthora sansomeana) primers from Rojas et al., 2017.
Isothermal assays used in plant diagnostics are predominately loop-mediated isothermal amplifications (LAMPs) or recombinase polymerase amplifications (RPA) [19,20,21,22,23,24,25,26,27,28,29,30,31,32]. These assays achieve amplification at stable temperatures (65 °C for LAMP and 39–42 °C for RPA assays) and thus do not need thermocyclers to amplify target DNA. As with any molecular-based assay, there can be issues with cross contamination in a laboratory setting. However, isothermal assays such as LAMP and RPA have the potential to be a fast and reliable test to determine the presence or absence of an organism within a sample [31].
The majority of the currently available isothermal diagnostic assays for Phytophthora are LAMP assays. Currently, LAMP assays have been reported for Phytophthora kernoviae [21], Phytophthora infestans [22], Phytophthora cinnamomi [23], Phytophthora melonis [24], Phytophthora nicotianae [25], Phytophthora sojae, and Phytophthora ramorum [21,26]. LAMP assays utilize four primers designed to anneal to different regions of the target DNA, as well as a unique DNA polymerase, with strand displacement activity enabling target amplification at a constant temperature (65 °C) [33]. Monitoring to determine a successful amplification can be performed visually, as a magnesium pyrophosphate precipitate is produced as the assay runs or fluorescent dyes, such as SYBR Green, can be incorporated so that the assay can be detected on a fluorometer [34]. The primary disadvantage is that the chemistry is quite different from PCR and so it may take significant optimization to achieve successful and specific amplification [35]. Likewise, the large number of amplicons produced in LAMP reactions make them difficult to use in the lab without amplicon contamination occurring. Limited information is available about multi-plexing LAMP reactions or whether it is possible to use this technology for detection of specific SNPs.
RPA isothermal assays have been developed for many plant pathogens [19,20,27,28,29,30,31,32]. For Phytophthora species specifically, a genus-specific assay (targeting the trnM-trnP-trnM gene order) and four species-specific assays targeting the atp9–nad9 spacer region have been validated as specific [19,20]. RPA assays are more specific than using antibody based immunostrips and enzyme-linked immunosorbent assays (ELISA), as they rely on conserved DNA sequences instead of a generic antigen for detection [10,36]. RPA assays, like LAMP, are isothermal, with amplification typically occurring between 39 and 42 °C, and thus do not need a thermocycler to amplify the target region of DNA. Due to the simplicity in hardware and reagents required, these RPA assays are suitable for field deployment and can be used to provide real-time identification of targeted microbes within a single sample in under 30 min with minimal equipment inputs [19,20], making them a suitable replacement for current serological techniques [37]. Unlike LAMP assays, commercially available RPA exo kits (TwistAmp® exo, TwistDX Ltd., Cambridge, UK) have a DNase to digest the amplified template, thus reducing the probability of amplicon contamination.
Commercially available RPA kits with either lyophilized or liquid reagents are available through limited sources (TwistDX Ltd., Cambridge, UK and Agdia, Elkhart, IN, USA). The lyophilized formulation only requires a rehydration buffer, magnesium acetate, user-supplied primers, probes and DNA template to start the amplification. However, there are currently no preformulated kits with primers and probes incorporated into the lyophilized pellet commercially available for oomycete pathogen detection. Having preformulated kits containing primers and probes from previously validated Phytophthora genus detection assays [20] would facilitate use by regulatory agencies and diagnostic laboratories working with Phytophthora species, as it would simplify their use under field conditions. In this manuscript, a previously developed assay that was validated against more than 136 Phytophthora, 21 Pythium and 1 Phytopythium species to assure its specificity to only amplify Phytophthora sp. was used to determine whether lyophilization negatively affects the sensitivity and specificity of the assay. The goal of this study was to (1) evaluate whether lyophilization of Phytophthora genus-specific primers and probes within RPA assay tubes affects the sensitivity and specificity of the assay and (2) evaluate whether the preformulated lyophilized Phytophthora genus-specific RPA assay is transferable between multiple qPCR platforms and labs using a single-blind DNA sample testing method.
2. Results
2.1. Comparable Results Were Observed between Preformulated and Commercially Available RPA Reactions
To test the preformulation of the RPA assay, the authors collaborated with TwistDx to manufacture a preliminary batch of the preformulated assay using the TwistDx TwistAmp exo kits. These kits contained primers and probes lyophilized with nearly all other reagents, requiring relatively few reagent additions and minimal equipment (Figure 2 and Figure 3). This preliminary batch of the preformulated assay was used to identify whether there was an effect of preformulation on assay sensitivity or specificity. Initially, the preformulated assay was validated using the Axxin T16 to identify whether the limit of detection (LOD) of the assay would be comparable to the commercially available assay. Overall, the preformulated assay performed very similar to the commercially available assay, with an average onset of amplification time difference of 43.8 s between the preformulated and commercially available kits, a negligible difference in assays that run for 30 min (Table 1). The preformulated assay was able to identify P. ramorum in crude infected samples, as well as purified P. cinnamomi DNA. The plant internal control was detected in all samples containing plant tissue but was not detected when using purified DNA extractions from cultured oomycetes only (Table 1).
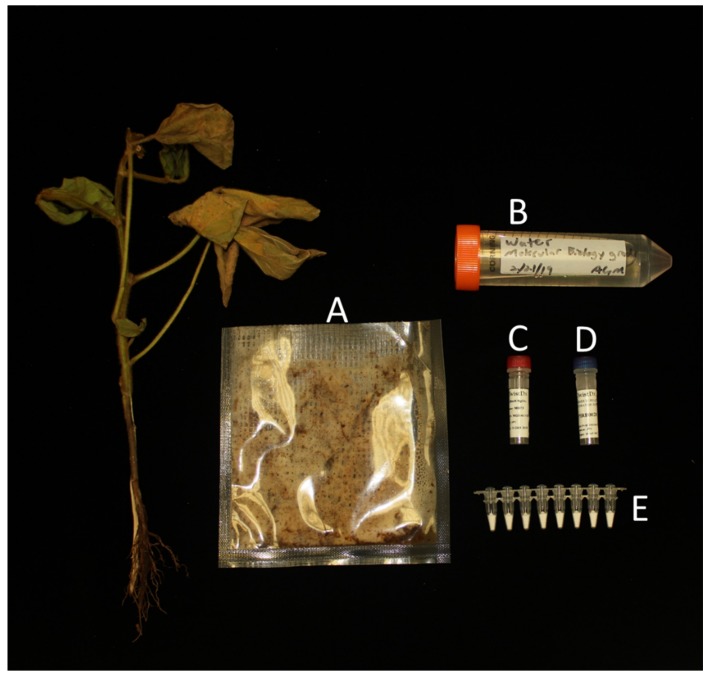
Figure 2.
Visualization of the minimal space and equipment needed to perform the preformulated recombinase polymerase amplification (RPA) assay outside of the laboratory. AxxinT16-ISO used for visualization.
Figure 3.
All necessary reagents to run the lyophilized RPA assay. (A) Crude extract produced with diseased plant sample and GEB2 buffer within an Agdia mesh bag. (B) Sterile water. (C) Magnesium acetate to initiate the reaction. (D) Rehydration buffer. (E) Preformulated lyophilized RPA reagents in pre-loaded tubes (TwistDX, Cambridge, UK).
Table 1.
Comparison of recombinase polymerase amplification (RPA) assays using commercially available kits requiring addition of primers and probes and preformulated kits with the primers and probes lyophilized with the reaction mixture. Recombinase polymerase amplification using primers and probes for Phytophthora genus-specific detection and a plant internal control reported by Miles et al. [20]. Data collected with an Axxin T16-ISO platform.
| Sample | Average Time Onset of Amplification with the Commercially Available Reaction a | Average Time Onset of Amplification with the Preformulated Lyophilized Reaction a |
|---|---|---|
| Crude plant extract Phytophthora rubi-infected raspberry cane | 17.4 b | 17.6 |
| 8.0 b | 8.0 | |
| Crude plant extract infected Phytophthora ramorum-leaf | 15.7 | 15.2 |
| 8.1 | 8.1 | |
| Crude plant extract infected P. ramorum-leaf | 14.8 | 12.3 |
| 8.0 | 8.0 | |
| Purified Pythium splendens DNA (1 ng) | - c | - |
| - | - | |
| Purified Phytophthora cinnamomi DNA (3500 pg) | 6.5 | 5.85 |
| Purified P. cinnamomi DNA (350 pg) | 9.0 | 8.5 |
| Purified P. cinnamomi DNA (35 pg) | 11.4 | 11.0 |
| Purified P. cinnamomi DNA (3.5 pg) | 16.5 | 16.1 |
| Purified P. cinnamomi DNA (0.35 pg) | - | - |
a Values are expressed as the average (n = 2) onset of amplification in minutes, including the pre-agitation step. b Bold (top) values are Phytophthora (FAM). Italic (bottom) values are plant internal control (ROX). c (-) indicates a reaction no different than the water control.
2.2. Multiple Platforms Were Effective at Detecting Phytophthora Species by RPA
The Axxin T16-ISO isothermal (Martin lab), BioRad CFX96 (Chilvers lab), QuantStudio 6 (Blomquist lab) and Applied Biosystems ViiA7 (Bilodeau lab) platforms were used to further evaluate the transferability between labs of the preformulated Phytophthora RPA assay using Phytophthora ramorum-purified DNA, tested at various concentrations ranging from 0.33 to 0.33 pg/µL, with and without a crude plant extract (Table 2). Crude plant extracts were added to the purified DNA to verify assay specificity and to simulate reactions with actual plant samples at various pathogen DNA concentrations. A positive control (500 pg/µL purified P. ramorum DNA) and negative control (500 pg/µL purified Pythium splendens DNA) were also tested in each experiment. The limit of detection (LOD) for purified DNA without plant extract was 33 pg/µL for the CFX96 and ViiA7 qPCR platforms (Table 2). The addition of a crude plant extract to the purified DNA had no effect on LOD for the ViiA7 qPCR platform, while the CFX96 could only detect down to 0.33 ng/µL in the presence of a crude plant extract (Table 2). Py. splendens DNA did not amplify in all four platforms. The Axxin T16-ISO platform had a single instance of a false-positive reading for the plant internal control (Table 2). The Axxin T16-ISO and QuantStudio 6 were slightly more sensitive and able to detect P. ramorum consistently at 3.3 pg of DNA, with and without plant extracts added.
Table 2.
Initial validation of preformulated lyophilized RPA assay on crude plant extracts and purified Phytophthora ramorum or Pythium splendens DNA.
| Sample | Platform | Average Time Onset of Amplification without Plant Extract a | Average Time Onset of Amplification with Plant Extract | ||
|---|---|---|---|---|---|
|
Phytophthora Genus (FAM) b |
Plant Internal Control (ROX) c |
Phytophthora Genus (FAM) |
Plant Internal Control (ROX) |
||
|
P. ramorum (500 pg/µL) (Positive control) |
Axxin T16-ISO | 7.66 | NR d | - e | - |
| Bio-Rad CFX96 | 12.38 | NR | - | - | |
| ViiA7 RT-PCR | 13.01 | NR | - | - | |
| QuantStudio 6 | 6.69 | NR | - | - | |
| Phytophthora-infected citrus | Axxin T16-ISO | - | - | 7.24 | 8.21 |
| Bio-Rad CFX96 | - | - | 9.08 | 10.46 | |
| ViiA7 RT-PCR | - | - | 12.68 | 14.35 | |
| QuantStudio 6 | - | - | 5.55 | 5.34 | |
|
Pythium splendens (500 pg/µL) (Negative Control) |
Axxin T16-ISO | NR | NR | - | - |
| Bio-Rad CFX96 | NR | NR | - | - | |
| ViiA7 RT-PCR | NR | NR | - | - | |
| QuantStudio 6 | NR | NR | - | - | |
| P. ramorum (0.33 ng/µL) | Axxin T16-ISO | 10.54 | 12.67 | 12.11 | 10.04 |
| Bio-Rad CFX96 | 14.97 | NR | 17.36 | 29.81 | |
| ViiA7 RT-PCR | 12.6 | NR | 13.99 | 8.95 | |
| QuantStudio 6 | 7.53 | NR | 8.11 | 23.21 | |
| P. ramorum (33 pg/µL) | Axxin T16-ISO | 18.90 | NR | 19.71 | 14.22 |
| Bio-Rad CFX96 | 22.66 | NR | NR | 20.45 | |
| ViiA7 RT-PCR | 16.51 | NR | 16.98 | 9.68 | |
| QuantStudio 6 | 9.31 | NR | 11.83 | 22.77 | |
| P. ramorum (3.3 pg/µL) | Axxin T16-ISO | 27.32 | NR | 31.12 | 13.77 |
| Bio-Rad CFX96 | NR | NR | NR | 10.57 | |
| ViiA7 RT-PCR | NR | NR | NR | 9.57 | |
| QuantStudio 6 | 12.99 | NR | 16.01 | 23.09 | |
| P. ramorum (0.33 pg/uL) | Axxin T16-ISO | NR | NR | NR | 10.06 |
| Bio-Rad CFX96 | NR | NR | NR | 15.46 | |
| ViiA7 RT-PCR | NR | NR | NR | 9.28 | |
| QuantStudio 6 | NR | NR | NR | 22.97 | |
a Values are expressed as the average (n = 2) onset of amplification in minutes. b Phytophthora genus-specific probe. c Plant internal control probe. d (NR) refers to reactions that were run but did not have any observed amplification. e (-) refers to reactions that could not be run with or without a plant extract, respectively.
2.3. Accurate Identification of Phytophthora-Infected Material Was Possible in a Single Blind Sample Evaluation of the Preformulated Assay
Samples collected during a 2015 Phytophthora survey of California nurseries [20] were used as a single-blind validation panel for further assay validation. The causal agent of the diseased tissues was identified with high confidence using isolation, qPCR and commercial RPA methods previously [20]. A blind validation panel was constructed by taking a subset of these samples and removing the identities before being sent to all labs participating in the evaluation. Results from Miles et al. [20] (Axxin T16-ISO) were then treated as a standard for pathogen identification when testing the preformulated Phytophthora genus-specific RPA assay. The BioRad CFX96 platform was able to accurately replicate the Axxin T16’s Phytophthora genus qPCR and commercial RPA results on both Phytophthora-infected and non-infected plant tissues (Table 3). The Applied Biosystems ViiA7 had a single technical rep with a false-negative reading with the plant internal control. However, all Phytophthora-infected samples tested with the ViiA7 platform had a positive amplification, while all Phytophthora free samples did not amplify (Table 3). The QuantStudio 6 platform had a single instance of false-positive readings for the Phytophthora genus-specific assay: Asparagus officianalis infected with Pythium sp. Likewise, this platform also had three instances of false-negative readings for amplification with the Phytophthora genus-specific assay. There was only one instance of a false-negative reading for the plant internal control (Table 3).
Table 3.
Blind validation panel used to evaluate the preformulated lyophilized recombinant polymerase amplification (RPA) reaction and commercially available RPA reactions compared to the results of qPCR assays as reported in Miles et al. [20].
| Axxin T16 a | Bio-Rad CFX96 RPA | ViiA7 System RPA | QuantStudio 6 RPA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Pathogen Identified |
Phytophthora Genus RPA (FAM) b |
Phytophthora Genus qPCR (FAM) |
Phytophthora Genus (FAM) |
Plant Internal Control (ROX) c |
Phytophthora Genus (FAM) |
Plant Internal Control (ROX) |
Phytophthora Genus (FAM) |
Plant Internal Contro
(ROX) |
|
| Rhamnus californica | Phytophthora cactorum | + d | + | + | + | + | + | + | + | |
| Prunus avium | Pythium sp. | − e | − | − | + | − | + | − | − | |
| Gardenia jasminoides ‘Radicans’ | P. nicotianae | + | + | + | + | + | + | − | + | |
| Gardenia jasminoides ‘Mystery’ | P. nicotianae | + | + | + | + | + | − | + | + | |
| Aucuba japonica ‘Mr. Goldstrike’ | P. citricola | + | + | + | + | + | + | + | + | |
| Asparagus officinalis | Pythium sp. | − | − | − | + | − | + | + | + | |
| Rhus integrifolia | P. nicotianae | + | + | + | + | + | + | + | + | |
| Fragaria x ananassa | P. cactorum | + | + | + | + | + | + | − | + | |
| Rubus sp. | Pythium sp. | − | − | − | + | − | + | − | + | |
| Rubus sp. | P. rubi | + | + | + | + | + | + | + | + | |
| Citrus sp. 1 | P. citrophthora | + | + | + | + | + | + | + | + | |
| Citrus sp. 2 | P. citrophthora | + | + | + | + | + | + | + | + | |
| Citrus sp. 3 | P. citrophthora | + | + | + | + | + | + | + | + | |
| Citrus sp. 4 | P. citrophthora | + | + | + | + | + | + | + | + | |
| Citrus sp. 5 | P. citrophthora | + | + | + | + | + | + | + | + | |
| Myrtus sp. | P. nicotianae | + | + | + | + | + | + | + | + | |
| Pseudotsuga menziesii | P. cambivora | + | + | + | + | + | + | + | + | |
| Hedera sp. | P. tropicalis | + | + | + | + | + | + | + | + | |
| Rhododendron sp. | P. ramorum | + | + | + | + | + | + | + | + | |
| Rhododendron sp. | P. ramorum | + | + | + | + | + | + | − | + | |
| Umbellularia californica | P. ramorum | + | + | + | + | + | + | + | + | |
| Viburnum sp. | P. ramorum | + | + | + | + | + | + | + | + | |
| Rhododendron sp. | P. ramorum | + | + | + | + | + | + | + | + | |
| Water | N/A | − | − | − | − | − | − | − | − | |
a Data collected by Miles et al. 2015. b Phytophthora genus-specific probe. c Plant internal control probe. d (+) indicates a positive amplification. e (−) indicates a negative amplification, no different than the water control.
2.4. Phytophthora-Infected Fresh Plant Material Can Be Identified in under 15 Min
Leaves of a Rhododendron plant were inoculated individually with five Phytophthora species and a mock-inoculation control. All Phytophthora-infected leaves exhibited symptoms 10 dpi and were used to make crude plant extracts. Mock-inoculated leaves did not develop symptoms and were also harvested 10 dpi. The lyophilized assay was able to accurately identify all Phytophthora-infected leaves when using a crude plant extract, with no false-positive readings for the mock-inoculated or water-only controls (Table 4). In one amplification for the sample inoculated with Phytophthora boehmeriae, the plant internal control amplification was no different than the water only control (Table 4).
Table 4.
Horticulture (Rhododendron sp.) and agriculture (soybean) samples tested with the preformulated lyophilized recombinase polymerase amplification (RPA) assay.
| Phytophthora Species a | Host Plant | Platform | Phytophthora Genus (FAM) | Plant Internal Control (ROX) |
|---|---|---|---|---|
| Mean OT b | Mean OT | |||
| P. boehmeriae | Rhododendron sp. | ViiA7 RT-PCR | 6.78 * | NR |
| P. pseudosyringae | Rhododendron sp. | ViiA7 RT-PCR | 10.63 | 5.92 * |
| P. cryptogea | Rhododendron sp. | ViiA7 RT-PCR | 9.52 | 6.96 |
| P. ramorum | Rhododendron sp. | ViiA7 RT-PCR | 9.7 | 7.41 |
| P. infestans | Rhododendron sp. | ViiA7 RT-PCR | 6.20 | 10.74 * |
| P. infestans | Rhododendron sp. | ViiA7 RT-PCR | 8.01 | 9.84 * |
| Leaf only | Rhododendron sp. | ViiA7 RT-PCR | NR c | 8.15 * |
| Water only | Rhododendron sp. | ViiA7 RT-PCR | NR * | NR * |
| P. sojae 1 | Glycine max | Bio-Rad CFX96 | 8.82 | 11.76 |
| P. sojae 2 | Glycine max | Bio-Rad CFX96 | 6.88 | 8.04 |
| P. sojae 3 | Glycine max | Bio-Rad CFX96 | 5.36 | 7.46 |
| P. sojae 4 | Glycine max | Bio-Rad CFX96 | 4.35 | 7.92 |
| P. sojae 5 | Glycine max | Bio-Rad CFX96 | 3.97 | 10.8 |
| P. sojae 6 | Glycine max | Bio-Rad CFX96 | 10.58 | 13.45 |
| P. sojae 7 | Glycine max | Bio-Rad CFX96 | 9.54 | 15.85 |
| P. sojae 8 | Glycine max | Bio-Rad CFX96 | 9.25 | 12.99 |
| P. sojae 9 | Glycine max | Bio-Rad CFX96 | 4.36 | 27.18 |
| P. sojae 10 | Glycine max | Bio-Rad CFX96 | 5.25 | 22.84 |
a Species of Phytophthora inoculated (ViiA7) or isolated (CFX96) from host. b Values are expressed as the average (n = 2) onset of amplification (OT) in minutes. c (NR) indicates a negative reaction, no different than the water control. * n = 1.
Soybeans used in an oomycete soil baiting assay resulted in soybean seedlings displaying symptoms of Phytophthora sojae infection. Diseased seedlings were cut in half vertically, where half was used to culture the causal agent and half was used to produce crude plant extracts. P. sojae was successfully isolated from all seedlings, and 100% of these seedlings also resulted in a positive Phytophthora amplification (Table 4).
Environmental samples suspected of having a Phytophthora ramorum infection were submitted to the California Department of Food and Agriculture Plant and Pest Diagnostics (CDFA-PPD) lab for further testing. Causal organisms of disease were identified as described below. All Phytophthora ramorum-infected samples resulted in a positive amplification. One sample, “Phytophthora chlamydospora” had no amplification on either the Phytophthora genus-specific or the plant internal control. Likewise, there was one more instance of a false-negative reading with the plant internal control (Table 5). Samples were tested on the QuantStudio 6 platform.
Table 5.
Environmental samples tested with the preformulated RPA assay on the QuantStudio 6 platform.
| Sample a | Host Name | Phytophthora Genus (FAM) | Plant Internal Control (ROX) |
|---|---|---|---|
| Mean OT b | Mean OT b | ||
| Phytophthora ramorum 1 | Rhododendron sp. ‘Cunningham’ | 3.80 | 6.23 |
| P. ramorum 2 | Rhododendron sp. ‘Cunningham’ | 7.83 | 26.9 |
| P. ramorum 3 | Rhododendron sp. ‘Cunningham’ | 5.64 | 5.55 |
| P. ramorum 4 | Rhododendron sp. ‘Cunningham’ | 5.90 | 5.99 |
| P. ramorum 5 | Rhododendron sp. ‘Cunningham’ | 7.07 | 6.32 |
| P. ramorum 6 | Umbellularia californica | 8.39 | 26.9 |
| P. ramorum 7 | Rhododendron sp. ‘Grace Seabrook’ | 10.28 | 5.19 |
| P. ramorum 8 | Rhododendron sp. ‘Grace Seabrook’ | 8.09 | 5.56 |
| P. ramorum 9 | Rhododendron sp. ‘Grace Seabrook’ | 11.14 | 5.12 |
| P. ramorum 10 | Rhododendron sp. ‘Grace Seabrook’ | 12.78 | 6.12 |
| P. ramorum 11 | Rhododendron sp. ‘Grace Seabrook’ | 10.32 | NR c |
| P. ramorum 12 | Rhododendron sp. ‘Taurus’ | 9.05 | 5.57 |
| P. ramorum 13 | Arctostaphylos refugioensis | 11.19 | 6.93 |
| P. ramorum 14 | Rhododendron sp. ‘Rangoon’ | 7.75 | 5.27 |
| Spumella-like flagellate | Fragaria sp. | NR | 27.11 |
| Pythium debaryanum | Fragaria sp. | NR | 24.24 |
| Plasmopara viticola | Vitis vinifera | NR | 18.57 |
| P. multivora | Camellia sinensis | 4.80 | 22.06 |
| P. chlamydospora | Rosa sp. | NR | NR |
| P. brassicae | Brassica oleracea | 5.85 | 1.38 |
| P. syringae 1 | Rhododendron sp. ‘President Roosevelt’ | 5.22 | 6.07 |
| P. syringae 2 | Rhododendron sp. | 4.76 | 5.38 |
| P. syringae 3 | Rhododendron sp. | 5.55 | 6.31 |
| P. syringae 4 | Rhododendron sp. | 5.85 | 5.69 |
| P. syringae 5 | Rhododendron sp. | 5.54 | 6.04 |
| P. syringae 6 | Arctostaphylos densiflora | 7.41 | 25.90 |
| Unknown Isolate 1 | Chiosya ternate | NR | 26.0 |
| Unknown Isolate 2 | Mahonia repens | NR | 9.91 |
| Unknown Isolate 3 | Camellia sasanqua | NR | 24.24 |
| Water control | NA | NR | NR |
a Species identified within the sample via ELISA and ITS sequencing. b Values are expressed as the average (n = 2) onset of amplification (OT) in minutes. c (NR) indicates a negative reaction, no different than the water control.
3. Discussion
During evaluations of the preformulated RPA kits, to determine the effect of lyophilization on primers and probes, nearly no difference between the preformulated and commercially available kits was found. An average onset of amplification time difference of 43.8 s when using the Axxin T16-ISO RPA chamber was observed between the preformulated and commercially available kits (Table 1). Nearly indistinguishable results indicated that preformulation via lyophilization of primers and probes did not significantly affect the sensitivity and specificity of the Phytophthora genus-specific assay. This assay was found to be specific to Phytophthora, with the ability to consistently detect and differentiate between Phytophthora-infected and non-infected tissue. Likewise, the limit of detection (LOD) was identified using four separate platforms. The LOD using purified DNA was 33 pg with the BioRad CFX96 and ViiA7 platforms using the preformulated kits, while the Axxin T16-ISO RPA chamber and QuantStudio 6 could detect down to 3.3 pg with and without a crude plant extract. The LOD using a crude plant extract for the BioRad CFX96 was 330 pg, whereas the LOD for the ViiA7 system was 33 pg.
There was an observed difference in sensitivity of the assay when using the Axxin T16-ISO compared to the qPCR platforms, with the Axxin being 10–100-fold more sensitive. This is likely due to the Axxin FAM fluorophore channel being scaled to a high level of sensitivity prior to testing. The Axxin T16-ISO platform had a single instance of a false-positive reading with the plant internal control. Similarly, when testing fresh samples of soybean, Rhododendron and environmental samples, differences in detection of Phytophthora and the plant internal control were identified. The BioRad CFX96 had consistent amplification of both the plant internal control as well as positive Phytophthora amplification in Phytophthora sojae-infected soybeans. However, the ViiA7 RT-PCR system and QuantStudio 6 had instances of inconsistent amplification of the plant internal control and Phytophthora-infected plants, where one of the two technical reps did not amplify when both plant material and Phytophthora were present, or in the case of the QuantStudio 6, no amplification was detected. This could be due to amplification starting in some reactions before the samples were placed into the respective platforms. Inadequate mixing of the crude plant extract and reagents within tubes could cause variable amplification efficiency between technical replications. Varying levels of plant DNA within a plant extract could also lead to variable amplification times of the plant internal control. In general, reaction times under 30 min would be considered positive for both the Phytophthora genus-specific marker and the plant internal control. Despite the observed instances of variability in amplification, approximately 90% of reactions were identical across the three tested qPCR platforms as compared to the commercially available RPA kit as well as the Phytophthora genus-specific qPCR assay run on the Axxin T16-ISO (Table 3). These results reinforce our hypothesis that preformulation within a single pellet will not negatively affect assay performance.
When used in a plant inspection setting, the preformulated RPA was able to accurately identify all Phytophthora ramorum-infected samples (Table 5). Likewise, closely related genera like Plasmopara and Pythium were not amplified—another example of the specificity of this assay. Interestingly, one Phytophthora sample, Phytophthora chlamydospora, did not amplify. This could be due to high levels of inhibitors being present in the lysate, as the plant internal control also did not amplify. Nevertheless, in a diagnostic setting, when interpreting a molecular diagnostic test, it is imperative to take into account the history of the sample to accurately identify the causal organism. Samples displaying symptoms typical of a Phytophthora disease (i.e., water soaked, necrotic stem vascular tissue in soybeans indicative of P. sojae, necrosis on the margins of Rhododendron leaves indicative of P. ramorum) which have ambiguous results from molecular tests should be more thoroughly investigated via microscopy, culturing techniques or DNA sequencing to identify the causal organism.
The ability to test and diagnose quarantined plant pathogens quickly is imperative to stop the spread of these destructive plant diseases. Furthermore, the ability to conduct in field testing for these pathogens could reduce the time between diseased plant observation and causal agent identification. Isothermal assays, such as RPA, are useful in this regard, as they can be field deployable when using an isothermal detection chamber, such as the Axxin T16-ISO (Figure 2). Likewise, the assay validated in this manuscript requires minimal reagents to be added, as the primers and probes are preformulated into the lyophilized pellet (Figure 3). While the Axxin T16-ISO was used in this study, there are similar fluorimeters available from various manufacturers that could also be used: BioRanger (Diagenetix Inc., Honolulu, HI, USA), Amplifire (Agdia Inc., Elkhart, IN, USA), Genie® II or Genie® III (OptiGene Ltd., West Sussex, UK).
Commercial availability of an accurate, field-deployable, isothermal assay preformulated with necessary primers and probes that does not require expensive lab equipment, DNA extraction or constant refrigeration of reagents would benefit in-field diagnostics. Wide availability of such an assay could decrease the time between observation of disease and identification of the causal agent of disease, something that would be of particular importance for regulated Phytophthora species like Phytophthora ramorum and P. kernoviae. Having an assay that can detect regulated and non-regulated pathogens increases its potential use from diagnostics labs working with regulated Phytophthora species to labs that receive potentially infected plant samples from the horticultural and agricultural industries for identification, as well as labs conducting surveys on Phytophthora species. The lyophilized isothermal RPA assay developed and validated in this manuscript could be used for any such aforementioned study on Phytophthora species.
The isothermal RPA assay described in this manuscript uses mitochondrial gene order to provide sensitivity and specificity to Phytophthora species. The mitochondrial gene order, trnM-trnP-trnM, is conserved among Phytophthora species, making it highly specific [19,20], unlike other regions previously used for diagnostic assays [1,11,12,13,14,15,16,17,18,19,20,38]. Interestingly, this region is not conserved in the closely related genera Pythium, making it an ideal marker for Phytophthora-specific diagnostics. This gene region has been previously developed and validated into other TaqMan and RPA Phytophthora genus- and species-specific assays [19,20,39,40]. Miles et al. [20] developed a Phytophthora genus-specific isothermal assay with a detection limit between 200 and 300 femtograms (0.2–0.3 pg). The TaqMan assay for Phytophthora genus developed by Rojas et al. [19] had a limit of detection of 100 fg (0.1 pg) when using purified DNA, while the RPA assay developed had a limit of detection of 10 pg [19]. The preformulated lyophilized assay evaluated herein had nearly identical results to that of the standard commercially prepared kit; it was able to detect purified Phytophthora species DNA down to 3.3–33 pg depending on which platform was used to read amplification (Table 1 and Table 2) [20].
In addition to the trnM-trnP-trnM gene order, another gene order (atp9–nad9) was also found to be useful for development of species-specific detection capabilities. Bilodeau et al. [40] and Miles et al. [39] also developed fifty species-specific Phytophthora TaqMan probes based on the atp9–nad9 spacer region for regulated and non-regulated Phytophthora species, as well as one-hundred and twenty-four unique TaqMan probes for various Phytophthora species. RPA assays targeting this locus were developed for detection of Phytophthora of regulatory importance (such as Phytophthora ramorum and P. kernoviae) or of agronomic importance (Phytophthora sojae, P. cactorum, P. citrophthora) and could be used with the trnM-trnP-trnM assay to provide further, species-specific, diagnostic capabilities [20,39,40].
The potential for false-negative readings and false-positive readings has been a long-standing issue in plant pathogen serological and molecular diagnostics [38,41]. In order to control for false-positive readings, samples should always be run in two to three technical reps, so that false-positive readings for amplification can be identified by consensus. Likewise, including a water control reaction can identify potential contamination of reagents, which is of concern with molecular assays and can lead to false-positive readings for amplification. Unlike LAMP assays that generate DNA end products that could contaminate pipettes or the workspace leading to future false-positive readings, the RPA exo formulation used in this experimentation has an exonuclease component to the amplification mixture that digests the DNA during amplification, thereby reducing the potential for cross contamination. To control for false-negative readings due to the presence of amplification inhibitors, our assay utilizes a plant mitochondrial internal control primer and probe set which targets the plant cytochrome oxidase (cox) gene [20]. This was used to determine whether inhibitors in the plant extract prevented amplification, which provides a control amplification for quality of the extract. Providing an internal control for each reaction allows users to quickly identify whether reactions needs to be repeated. Assays performed with purified DNA from Phytophthora species could enable a similar internal control by ‘spiking’ their reactions with pathogen-free plant DNA.
One interesting aspect of the isothermal RPA assay is the potential use of a reverse-transcriptase (RT) to target RNA without the need to construct cDNA before use [42]. Previous studies have shown that this can be done with viral plant pathogens such as plum pox virus of Prunus sp. [30], little cherry virus 2 [29], as well as Potato virus Y and Wheat dwarf virus [42]. Using RT in an RPA assay could provide an in-field diagnostic test to identify living Phytophthora pathogens within samples by targeting mRNA for a constitutively expressed gene [43,44]. Real-time identification of a living pathogen-causing disease would be of great benefit to plant pathology as a whole, as the main criticism of current molecular diagnostic procedures is that there is no way to identify whether the DNA that was amplified was from viable or dead cells. Having the ability to amplify RNA templates by RPA would solve this problem, as RNA is not persistent in the environment like DNA is, thus ensuring that the amplification is from a viable cell. Other procedures to ensure that only DNA from living cells is amplified have been developed using propidium monoazide, a membrane-impermeant dye that intercalates into DNA in the absence of an intact membrane, inhibiting amplification [45]. However, more testing needs to be done in this area before it can be widely adopted for practical use in molecular diagnostics.
In conclusion, given the consistency of results obtained by different labs using different equipment, the preformulated assay described here would be a useful tool to plant diagnostic labs and points of inspection if commercially available. Specifically, this assay could be used during routine inspections at nurseries and ports of entry to screen materials for regulated Phytophthora species, such as Phytophthora ramorum and P. kernoviae. Samples with Phytophthora-positive amplification could be identified in under 30 min without the need to refrigerate the reagents or use a non-portable thermocycler, giving inspectors the ability to perform these reactions on site and in real time. This assay is sensitive (LOD of 3.3–33 pg DNA) and specific to Phytophthora species, does not cross react with Pythium or closely related organisms [20], and does not require extensive DNA preparations before use. Based on the ease of use, minimal equipment needed, sensitivity and specificity, the assay described in this manuscript would be of benefit and use to plant diagnostic labs and inspectors at ports of entry to monitor regulated or non-regulated Phytophthora species.
4. Materials and Methods
4.1. Reagents and Assay Conditions
Preformulated lyophilized RPA assay tubes in the TwistAmp exo formulation containing (1) the Phytophthora genus-specific primers and probes for the trnM-trnP-trnM locus and (2) the primers and probes for the plant internal control, targeting the plant COX1 gene, of Miles et al. (2015) were prepared by TwistDX (Cambridge, UK) (Table 6). Concentrations were as previously reported. The pellet was rehydrated and completely dissolved with 37.5 μL TwistAmp exo rehydration buffer and 7 μL DNase-free water prior to adding 3 μL of crude plant extract or purified DNA obtained as described below. This mixture was homogenized via pipette and 2.5 μL of 280 mM magnesium acetate (MgAc) was pipetted into the cap of each reaction tube and closed gently so that the MgAc remained in the cap. The samples were centrifuged for 10 s to initiate a uniform amplification starting time, mixed, and incubated at room temperature for 4 min, mixed again, centrifuged again, and placed into the Axiin T16 and qPCR platforms (BioRad CFX96, Bio-Rad Laboratories, Hercules, CA, USA, QuantStudio 6, Thermo Fisher Scientific Inc., Waltham, MA, USA and the ViiA7 Real-Time PCR system, Thermo Fisher Scientific Inc.). FAM (Phytophthora genus probe) and ROX (plant internal control probe) channels were used on each qPCR platform to measure fluorescence over a 30 min period at 39 °C. The fluorescence cycle threshold (CT) baseline for FAM and ROX was set just above the water control for each platform, so that samples with equal or less fluorescence than the water control were identified as negative. Data was exported into Microsoft® Excel (2007) to produce tables.
Table 6.
Primers and probes used in this study for Phytophthora genus-specific detection and plant internal control.
| Primers, Probes a | Sequence (5′–3′) | Target |
|---|---|---|
| Primers | ||
| Phytophthora genus specific | ||
| TrnM-F | ATGTAGTTTAATGGTAGAGCGTGGGAATC | tRNA-M |
| TrnM-R | GAACCTACATCTTCAGATTATGAGCCTGATAAG | tRNA-M |
| Plant internal control | ||
| Cox1-IPC-F | CATGCGTGGACCTGGAATGACTATGCATAGA | COX1 |
| Cox1-IPC-R | GGTTGTATTAAAGTTTCGATCGGTTAATAACA | COX1 |
| Probes | ||
| Phytophthora genus specific | ||
| TrnM-P | TAGAGCGTGGGAATCATAATCCTAATGTTG [FAM-dT] A [THF] G [BHQ1-dT] TCAAATCCTACCATCAT [3′-C3SPACER] | tRNA-M |
| Plant internal control | ||
| Cox1-IPC-P | GGTCCGTTCTAGTGACAGCATTCCYACTTTTATTA [ROX- dT] C [THF] C [BHQ2-dT] YCCGGTACTGGC [3′-C3SPACER] | COX1 |
a Primers and probes from Miles et al. 2015 [20].
4.2. Production of Pure DNA and Crude Plant Extracts
Pure DNA samples used in this study were obtained via a modified phenol-chloroform extraction protocol [46]. DNA samples were quantified using the Quant-iT™ dsDNA High Sensitivity assay on the Invitrogen™ Qubit™ 2 system (Thermo Fisher Scientific). Serial dilutions of Phytophthora ramorum and P. cinamommi were made to determine the LOD of the assay, as explained below.
Crude plant extracts were made from healthy Umbellularia californica leaves. Briefly, 0.5 g of leaf tissue was added to 5 mL to GEB2 buffer within a netted bag and ground. A volume of 1 µL of this crude plant extract was added to purified DNA to determine the LOD of the assay with and without a crude plant extract, as described below.
4.3. Initial Evaluation of Preformulated lyophilized Kits Reaction
A preliminary experiment was conducted to identify the effect, if any, that primer/probe lyophilization had on the RPA assay. Purified Phytophthora cinnamomi DNA [20], ranging from 0.35 to 3500 pg, was used to compare the reaction containing lyophilized primers/probe to the commercially available formulation. Crude plant Phytophthora ramorum-infected extracts were used to validate that the assay still worked when lyophilized, and purified Pythium splendens DNA was used as a negative control (Table 1). The Axxin T16 platform was used for data collection.
4.4. Limit of Detection Determination for Preformulated Lyophilized Kits
Once the assay with lyophilized primers/probe was observed to work as expected, the BioRad CFX96, Axxin T16, QuantStudio 6 and the ViiA7 Real-Time PCR system were used for further evaluation of the preformulated reactions. Purified DNA from Phytophthora ramorum was used as a positive control (500 pg/µL) and purified Pythium splendens DNA was used as a negative control (500 pg/µL) in both systems [20,39]. Purified DNA from P. ramorum was further used to determine the LOD of the platforms using serial dilutions ranging from 0.33 to 0.33 pg/µL. The assay LOD was determined with and without plant extract for each qPCR platform (Table 2).
4.5. Single Blind Multi-Lab Evaluation of Preformulated Lyophilized Kits Reaction
Plant samples used for the single blind lyophilized RPA evaluations were obtained during a 2015 survey of California nurseries for Phytophthora diseases [20]. Putative Phytophthora-infected samples were diagnosed and identified via culturing, qPCR, and RPA to identify the causal organism of infection. A subset of samples from this survey were randomly selected to be used in the preformulated RPA evaluation. Single blind testing was performed on Phytophthora- and Pythium-infected plant samples and compared to commercial RPA and qPCR reactions using the Axxin T16, as well as direct isolations from the infected material for identification (Table 3). The BioRad CFX96, QuantStudio 6 and ViiA7 Real-Time PCR platforms were used for data collection.
4.6. Fresh Sample Testing
Fresh plant samples from soybean seedlings and Rhododendron leaves were used to further test the assay on the BioRad CFX96 and ViiA7 platforms, respectively (Table 4). Soybean seedlings were used from a soil baiting assay for Phytophthora sojae [47]. Diseased seedlings were identified and carefully removed from soil. Seedlings were then washed of any debris and surface sterilized in a 70% ethanol solution before being cut in half vertically, with half of a single seedling used as a crude extract for RPA assays and the other half used for isolation and pathogen identification. Isolations were conducted on corn meal agar medium (CMA-PARP) amended with, pentachloronitrobenzene (PCNB) (50 mg/L), ampicillin (250 mg/L), rifampicin (10 mg/L and pimaricin (5 mg/L), selecting for oomycetes [48]. Phytophthora sojae was isolated from all soybean seedlings and identified based on host, culture morphology on CMA-PARP media as well as its inability to grow on full-strength PDA [47]. Crude soybean extracts were prepared by adding 2.5 g plant tissue into 5 mL GEB2 buffer (product number: ACC 00130; Agdia) in plastic extraction bags containing netting (product number ACC 00930; Agdia).
Detached Rhododendron leaves were inoculated individually with Phytophthora boehmeriae, P. pseudosyringae, P. cryptogea, P. ramorum, P. infestans, or a mock-inoculated leaf and then tested with the lyophilized assay on the ViiA7 qPCR platform (Table 4). Leaves were surface sterilized with a 10% bleach solution for 1 min and subsequently washed twice with sterile water before inoculation. Inoculations were performed by injuring the leaf using a needle and placing a colonized agar plug of the pathogen onto the injury site, or a non-colonized agar plug for the mock inoculation. All leaves inoculated with Phytophthora species exhibited symptoms, while the mock-inoculated leaf did not develop symptoms. Inoculated leaves were harvested 10 days post inoculation (DPI) and a crude plant extract was made as noted above.
Environmental samples submitted to the California USDA-ARS for Sudden Oak Death (Phytophthora ramorum) testing were used to validate this assay in a plant inspection setting. Samples submitted were ground in Lysing Matrix A (product number: SKU 6910100, MP Biomedicals) following the Agdia Phytophthora ELISA Protocol (product number: PSA 92601; Agdia). The samples were then centrifuged to pellet the solid organic material and the lysate was used for RPA testing on the QuantStudio 6 platform. Causal organisms of disease were verified via ELISA and ITS sequencing (Table 5).
Acknowledgments
The authors would like to acknowledge Twistdx Ltd. (Cambridge, UK) for their willingness to produce these preformulated lyophilized pellets for us to test. Matthew Forrest (Twistdx Ltd.) has also been extremely helpful for the project and aided us with various technical molecular aspects of the project. The authors would like to thank Debbie Shearlaw from the Canadian Food Inspection Agency (CFIA) for offering support with sample processing and running the experiment at CFIA. The authors would also like to thank Mitchell Roth and Janette Jacobs for their thorough and insightful edits to this manuscript.
Author Contributions
Conceptualization, F.N.M. and T.D.M.; methodology, T.D.M.; validation, A.G.M., G.J.B., P.W. and T.D.M.; formal analysis, A.G.M.; writing—original draft preparation, A.G.M.; writing—review and editing, A.G.M., G.J.B., T.D.M., F.N.M., M.I.C., P.W. and C.B. All authors have read and agreed to the published version of the manuscript.
Funding
We would like to acknowledge funding from Michigan State University’s Project GREEEN, The North Central Soybean Research Promotion, Michigan Soybean Promotion Committee, USDA-SCBGP with CDFA.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Martin F., Abad Z.G., Balci Y., Ivors K. Identification and detection of Phytophthora: Reviewing our progress, identifying our needs. Plant Dis. 2012;96:1080–1103. doi: 10.1094/PDIS-12-11-1036-FE. [DOI] [PubMed] [Google Scholar]
- 2.Barber P., Paap T., Burgess T., Dunstan W., Hardy G. A diverse range of Phytophthora species are associated with dying urban trees. Urban For. Urban Green. 2013;12:569–575. doi: 10.1016/j.ufug.2013.07.009. [DOI] [Google Scholar]
- 3.Erwin D.C., Ribeiro O.K. Phytophthora Diseases Worldwide. Vol. 90 American Phytopathological Society; St. Paul, MN, USA: 1996. [Google Scholar]
- 4.Ali-Shtayeh M.S., MacDonald J.D., Kabashima J. A method for using commercial ELISA tests to detect zoospores of Phytophthora and Pythium species in irrigation water. Plant Dis. 1991;75:305–311. doi: 10.1094/PD-75-0305. [DOI] [Google Scholar]
- 5.Bulluck R., Shiel P., Berger P., Kaplan D., Parra G., Li W., Palm M. A Comparative analysis of detection techniques used in US regulatory programs to determine Presence of Phytophthora ramorum in Camellia japonica ‘Nucio’s Gem’ in an infested nursery in southern California. Plant Health Prog. 2006;7:9. doi: 10.1094/PHP-2006-1016-01-RS. [DOI] [Google Scholar]
- 6.Kox L.F.F., Van Brouwershaven I.R., Van De Vossenberg B.T.L.H., Van Den Beld H.E., Bonants P.J.M., De Gruyter J. Diagnostic values and utility of immunological, morphological, and molecular methods for in planta detection of Phytophthora ramorum. Phytopathology. 2007;97:1119–1129. doi: 10.1094/PHYTO-97-9-1119. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien P., Williams N., Hardy G. Detecting Phytophthora. Crit. Rev. Microbiol. 2009;35:169–181. doi: 10.1080/10408410902831518. [DOI] [PubMed] [Google Scholar]
- 8.Timmer L.W., Menge J.A., Zitko S.E., Pond E., Miller S.A., Johnson E.L. Comparison of ELISA techniques and standard isolation methods for Phytophthora detection in citrus orchards in Florida and California. Plant Dis. 1993;77:791–796. doi: 10.1094/PD-77-0791. [DOI] [Google Scholar]
- 9.Goheen E.M., Hansen E., Kanaskie A., Osterbauer N., Parke J., Pscheidt J., Chastagner G. Sudden Oak Death and Phytophthora Ramorum: A Guide for Forest Managers, Christmas Tree Growers, and Forest-Tree Nursery Operators in Oregon and Washington. Oregon State University Extension Services; Corvallis, OR, USA: 2006. [Google Scholar]
- 10.MacDonald J.D., Stites J., Kabashima J. Comparison of serological and culture plate methods for detecting species of Phytophthora, Pythium, and Rhizoctonia in ornamental plants. Plant Dis. 1990;74:655–659. doi: 10.1094/PD-74-0655. [DOI] [Google Scholar]
- 11.Bilodeau G., Pelletier G., Pelletier F., Lévesque C.A., Hamelin R.C. Multiplex real-time polymerase chain reaction (PCR) for detection of Phytophthora ramorum, the causal agent of sudden oak death. Can. J. Plant Pathol. 2009;31:195–210. doi: 10.1080/07060660909507593. [DOI] [Google Scholar]
- 12.Bilodeau G.J., Lévesque C.A., De Cock A.W.A.M., Duchaine C., Brière S., Uribe P., Hamelin R.C. Molecular detection of Phytophthora ramorum by real-time polymerase chain reaction using TaqMan, SYBR Green, and molecular beacons. Phytopathology. 2007;97:632–642. doi: 10.1094/PHYTO-97-5-0632. [DOI] [PubMed] [Google Scholar]
- 13.Garbelotto M., Rizzo D.M., Hayden K., Meija-Chang M., Davidson J.M., Tjosvold S. In: Phytophthora Ramorum and Sudden Oak Death in California: III. Preliminary Studies in Pathogen Genetics. US Dep. Agric. For. Serv. Gen. Tech. PSW-GTR-184, 5th Symp. Calif. Oak Woodlands. Standiford R., McCreary D., editors. Pacific Southwest Research Station, Forest Service, U.S. Department of Agriculture; Albany, CA, USA: 2002. pp. 765–774. [Google Scholar]
- 14.Hussain S., Lees A.K., Duncan J.M., Cooke D.E.L. Development of a species-specific and sensitive detection assay for Phytophthora infestans and its application for monitoring of inoculum in tubers and soil. Plant Pathol. 2005;54:373–382. doi: 10.1111/j.1365-3059.2005.01175.x. [DOI] [Google Scholar]
- 15.Martin F.N., Tooley P.W., Blomquist C. Molecular detection of Phytophthora ramorum, the causal agent of sudden oak death in California, and two additional species commonly recovered from diseased plant material. Phytopathology. 2004;94:621–631. doi: 10.1094/PHYTO.2004.94.6.621. [DOI] [PubMed] [Google Scholar]
- 16.Schena L., Hughes K.J.D., Cooke D.E.L. Detection and quantification of Phytophthora ramorum, P. kernoviae, P. citricola and P. quercina in symptomatic leaves by multiplex real-time PCR. Mol. Plant Pathol. 2006;7:365–379. doi: 10.1111/j.1364-3703.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- 17.Schena L., Duncan J.M., Cooke D.E.L. Development and application of a PCR-based “molecular tool box” for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathol. 2008;57:64–75. doi: 10.1111/j.1365-3059.2007.01689.x. [DOI] [Google Scholar]
- 18.Winton L.M., Hansen E.M. Molecular diagnosis of Phytophthora lateralis in trees, water, and foliage baits using multiplex polymerase chain reaction. For. Pathol. 2001;31:275–283. doi: 10.1046/j.1439-0329.2001.00251.x. [DOI] [Google Scholar]
- 19.Rojas J.A., Miles T.D., Coffey M.D., Martin F.N., Chilvers M.I. Development and application of qPCR and RPA genus- and species-specific detection of Phytophthora sojae and P. sansomeana root rot pathogens of soybean. Plant Dis. 2017;101:1171–1181. doi: 10.1094/PDIS-09-16-1225-RE. [DOI] [PubMed] [Google Scholar]
- 20.Miles T.D., Martin F.N., Coffey M.D. Development of rapid isothermal amplification assays for detection of Phytophthora spp. in plant tissue. Phytopathology. 2015;105:265–278. doi: 10.1094/PHYTO-05-14-0134-R. [DOI] [PubMed] [Google Scholar]
- 21.Tomlinson J., Dickinson M., Boonham N. Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology. 2010;100:143–149. doi: 10.1094/PHYTO-100-2-0143. [DOI] [PubMed] [Google Scholar]
- 22.Khan M., Li B., Yue J., Weng Q., Chen Q. Evaluation of different PCR-Based assays and LAMP method for rapid detection of Phytophthora infestans by targeting the Ypt1 gene. Gene Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai T.-T., Yang X., Hu T., Li Z., Xu Y., Lu C. A novel LAMP assay for the detection of Phytophthora cinnamomi utilizing a new target gene identified from genome sequences. Plant Dis. 2019 doi: 10.1094/PDIS-04-19-0781-RE. first look. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q., Li B., Liu P., Lan C., Zhan Z., Weng Q. Development and evaluation of specific PCR and LAMP assays for the rapid detection of Phytophthora melonis. Eur. J. Plant Pathol. 2013;137:597–607. doi: 10.1007/s10658-013-0273-9. [DOI] [Google Scholar]
- 25.Li B., Liu P., Xie S., Yin R., Weng Q., Chen Q. Specific and sensitive detection of Phytophthora nicotianae by nested PCR and loop-mediated isothermal amplification assays. J. Phytopathol. 2014;163:185–193. doi: 10.1111/jph.12305. [DOI] [Google Scholar]
- 26.Dai T.-T., Lu C.-C., Lu J., Dong S., Ye W., Wang Y., Zheng X. Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol. Lett. 2012;334:27–34. doi: 10.1111/j.1574-6968.2012.02619.x. [DOI] [PubMed] [Google Scholar]
- 27.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:1–7. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlinson J.A., Barker I., Boonham N. Faster, simpler, more-specific methods for improved molecular detection of Phytophthora ramorum in the field. Appl. Environ. Microbiol. 2007;73:4040–4047. doi: 10.1128/AEM.00161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point of-care diagnostics: A critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 30.Strayer-Scherer A., Jones J.B., Paret M.L. Recombinase polymerase amplification assay for field detection of tomato bacterial spot pathogens. Phytopathology. 2019;109:690–700. doi: 10.1094/PHYTO-03-18-0101-R. [DOI] [PubMed] [Google Scholar]
- 31.Londoño M.A., Harmon C.L., Polston J.E. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol. J. 2016;13:48. doi: 10.1186/s12985-016-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mekuria T.A., Zhang S., Eastwell K.C. Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification. J. Virol. Methods. 2014;205:24–30. doi: 10.1016/j.jviromet.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S., Ravelonandro M., Russell P., McOwen N., Briard P., Bohannon S., Vrient A. Rapid diagnostic detection of plum pox virus in Prunus plants by isothermal AmplifyRP® using reverse transcription-recombinase polymerase amplification. J. Virol. Methods. 2014;207:114–120. doi: 10.1016/j.jviromet.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 34.Burkhardt A., Koike S.T., Henry P., Gordon T., Martin F.N. Detection of Fusarium oxysporum f. species fragariae from infected strawberry plants. Plant Dis. 2018;103:1006–1013. doi: 10.1094/PDIS-08-18-1315-RE. [DOI] [PubMed] [Google Scholar]
- 35.Burkhardt A., Ramon M.L., Smith B., Koike S.T., Martin F.N. Development of molecular methods to detect Macrophomina phaseolina from strawberry plants and soil. Phytopathology. 2018;108:1386–1394. doi: 10.1094/PHYTO-03-18-0071-R. [DOI] [PubMed] [Google Scholar]
- 36.Lobato I.M., O’Sullivan C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane C.R., Hobden E., Walker L., Barton V.C., Inman A.J., Hughes K.J.D., Barker I. Evaluation of a rapid diagnostic field test kit for identification of Phytophthora species, including P. ramorum and P. kernoviae at the point of inspection. Plant Pathol. 2007;56:828–835. doi: 10.1111/j.1365-3059.2007.01615.x. [DOI] [Google Scholar]
- 38.Maniatis T., Fritsch E.F., Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1982. [Google Scholar]
- 39.Miles T., Martin F., Robideau G., Bilodeau G., Coffey M. Systematic development of Phytophthora species-specific mitochondrial diagnostic markers for economically important members of the genus. Plant Dis. 2017;101:1162–1170. doi: 10.1094/PDIS-09-16-1224-RE. [DOI] [PubMed] [Google Scholar]
- 40.Dorrance A.E., Berry S.A., Anderson T.R., Meharg C. Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant Health Prog. 2008;10:1094. doi: 10.1094/PHP-2008-0118-01-DG. [DOI] [Google Scholar]
- 41.Jeffers S.N., Martin S.B. Comparison of two media selective for Phytophthora and Pythium species. Plant Dis. 1986;70:1038–1043. doi: 10.1094/PD-70-1038. [DOI] [Google Scholar]
- 42.Si Ammour M., Bilodeau G.J., Tremblay D.M., Van der Heyden H., Yaseen T., Varvaro L., Carisse O. Development of real-time isothermal amplification assays for on-site detection of Phytophthora infestans in potato leaves. Plant Dis. 2017;101:1269–1277. doi: 10.1094/PDIS-12-16-1780-RE. [DOI] [PubMed] [Google Scholar]
- 43.Bilodeau G.J., Martin F., Coffey M., Blomquist C. Development of a multiplex assay for genus and species-specific detection of Phytophthora based on differences in mitochondrial gene order. Phytopathology. 2014;104:733–748. doi: 10.1094/PHYTO-09-13-0263-R. [DOI] [PubMed] [Google Scholar]
- 44.Martin R., James D., Levesque C.A. Impacts of molecular diagnostic technologies on plant disease management. Annu. Rev. Phytopathol. 2000;38:207–239. doi: 10.1146/annurev.phyto.38.1.207. [DOI] [PubMed] [Google Scholar]
- 45.Glais L., Jacquot E. Detection and characterization of viral species/subspecies using isothermal recombinase polymerase amplification (RPA) assays. Methods Mol. Biol. 2015;1302:207–225. doi: 10.1007/978-1-4939-2620-6_16. [DOI] [PubMed] [Google Scholar]
- 46.Chimento A., Cacciola S.O., Garbelotto M. Detection of mRNA by reverse-transcription PCR as an indicator of viability in Phytophthora ramorum. For. Pathol. 2012;42:14–21. doi: 10.1111/j.1439-0329.2011.00717.x. [DOI] [Google Scholar]
- 47.Vettraino A.M., Tomassini A., Vannini A. Detection and quantification of mRNA by reverse transcription real time PCR as indicator of viability of Phytophthora cambivora in soil. Acta Hortic. 2009;844:361–366. doi: 10.17660/ActaHortic.2009.844.50. [DOI] [Google Scholar]
- 48.Nocker A., Sossa-Fernandez P., Burr M., Camper A. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 2007;73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]