Abstract
We aimed to investigate the occurrence of acquired AmpC β-lactamases (qAmpC), and characterize qAmpC-producing Enterobacteriaceae from different non-clinical environments in Portugal. We analysed 880 Enterobacteriaceae resistant to third-generation cephalosporins recovered from 632 non-clinical samples [healthy human and healthy animal (swine, chickens) faeces; uncooked chicken carcasses; aquatic and trout aquaculture samples]. Bacterial and qAmpC identification, antibiotic susceptibility, clonal (PFGE, MLST) and plasmid (S1-/I-CeuI-PFGE, replicon typing, hybridization) analysis were performed using standard methods. The occurrence of qAmpC among Enterobacteriaceae from non-clinical origins was low (0.6%; n = 4/628 samples), corresponding to CMY-2-producing Escherichia coli from three healthy humans (HH) and one uncooked chicken carcass (UCC). We highlight a slight increase in CMY-2 human faecal carriage in the two periods sampled [1.0% in 2013–2014 versus 0% in 2001–2004], which is in accordance with the trend observed in other European countries. CMY-2-producing E. coli belonged to B22-ST4953 (n = 2, HH), A0-ST665 (n = 1, HH) or A1-ST48 (n = 1, UCC) clones. blaCMY-2 was identified in non-typeable and IncA/C2 plasmids. This study is one of the few providing an integrated evaluation of the qAmpC-producing Enterobacteriaceae occurrence, which was low, from a very large collection of different non-clinical origins. Further surveillance in contemporary collections can provide an integrated epidemiological information of potential shifts in reservoirs, transmission routes and mechanisms of dissemination of blaqAmpC in non-clinical settings.
Keywords: CMY-2, Escherichia coli, ST48, ST665, plasmids
Enterobacteriaceae resistant to third-generation cephalosporins are endemic in many parts of the world, mainly by the production of extended-spectrum β-lactamases (ESBLs) or acquired AmpC β-lactamases (qAmpC) [1]. While the occurrence and characterization of ESBL-producers across different niches has been characterized in detail, far less data concerning qAmpC-producing Enterobacteriaceae are available. In fact, existing studies have been focused particularly on clinical niches, and/or specific bacterial species (Escherichia coli, non-typhoidal Salmonella) and/or qAmpC-types (mostly CMY-2) [2,3], while the occurrence of qAmpC-producing Enterobacteriaceae across different non-clinical settings is barely known.
In Portugal, DHA-1 and CMY-2 are the most commonly found qAmpC-types among Enterobacteriaceae associated with hospital- or community-acquired human infections, although a recent increase in CMY-2 was observed in recent years (44%, 2010–2013 versus 6%, 2002–2008) [1]. Since blaCMY-2 is the most common qAmpC-type among non-clinical niches [3], we aimed to investigate the occurrence of qAmpC producers among a very large collection of Enterobacteriaceae isolates resistant to third-generation cephalosporins, recovered from a wide range of non-clinical samples, spanning the periods described above [1].
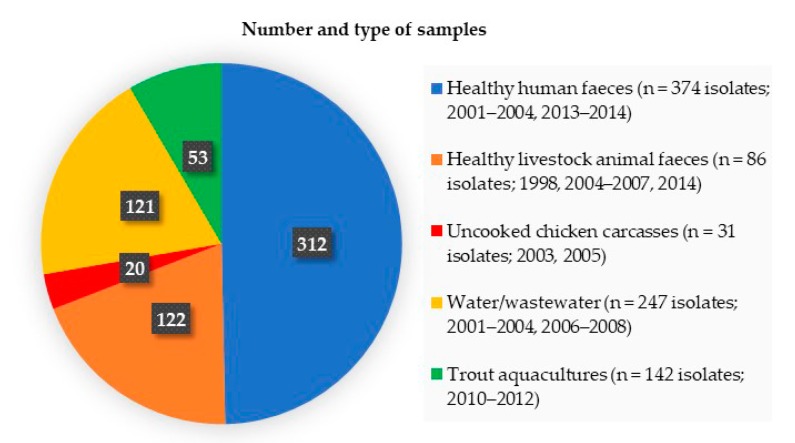
We analysed 880 Enterobacteriaceae isolates resistant to third-generation cephalosporins recovered from different non-clinical samples (n = 628): (i) healthy human faeces; (ii) healthy livestock animal faeces (78 swine, 44 chickens), (iii) uncooked chicken carcasses for human consumption, (iv) water/wastewater samples of aquatic environments, (v) and water/sediment/feed samples of trout aquaculture environments (Figure 1). Samples were collected from different regions of Portugal, and processed as described [4,5,6,7]. Briefly, the aliquots of uncooked chicken carcasses (0.2 mL) and aquaculture samples (0.1 mL) pre-enriched in buffered peptone water, and aliquots of the remaining samples (0.2 mL of human or animal faeces, 0.1 mL of wastewaters, filters from filtered water of aquatic environments) were seeded on CHROMagarTM Orientation/MacConkey agar plates supplemented with ceftazidime (1 mg/L) or cefotaxime (1 mg/L) [4,5,6,7]. Representative Enterobacteriaceae isolates (oxidase negative, approximately one to five unique morphotypes per plate) were recovered from the agar plates supplemented with antibiotics. qAmpC producers were preliminary identified by phenotypic criteria and/or polymerase chain reaction and the sequencing of genes coding for qAmpC enzymes (CMY, MOX, FOX, LAT, ACT, MIR, DHA, ACC) [1]. Bacterial identification, antibiotic susceptibility testing, and blaqAmpC genetic context were performed in qAmpC positive isolates as previously described [1]. Clonal relatedness was investigated by XbaI-PFGE and MLST (http://enterobase.warwick.ac.uk/species/index/ecoli), and E. coli phylogenetic groups were identified [1]. Plasmid analysis included replicon typing, S1-/I-CeuI-PFGE and hybridization [1].
Figure 1.
Number and type of samples (number of Enterobacteriaceae isolates resistant to third-generation cephalosporins; year of collection) analysed in this study.
We identified four CMY-2-producing E. coli recovered from three healthy humans [1 female (aged 65) and 2 males (aged 25 and 67)], and one uncooked chicken carcass (Table 1). This represents a very low occurrence (0.6%; n = 4/628 samples) of qAmpC producers in the samples analysed. It is of note that, although the human faecal carriage rate of qAmpC-producers was low, a slight increase between 2001–2004 (0%) and 2013–2014 (1.0%; n = 3/312) was identified, possibly suggesting a higher colonization density that should be monitored. The current rate (1.0%) is comparable to those reported in a few other European countries [8].
Table 1.
Features of CMY-2-producing E. coli recovered from non-clinical origins in Portugal.
| Origin (Sample) a | Year of Collection | PhG b | ST/CC/PFGE-Type c | Genetic Environment of blaCMY-2 | Plasmid Replicon Content [Inc Family (Size in kb)] | Resistance to Non-β-Lactams f | |
|---|---|---|---|---|---|---|---|
| Associated with blaCMY-2 |
Other | ||||||
| HH (33) | 2014 | B22 | ST4953/EC2 | ΔISEcp1-blaCMY-2-blc-sugE | ND d | FII + I1 | GEN, NET, TOB, STR, TET, CHL, NAL, CIP, SUL |
| HH (34) | 2014 | B22 | ST4953/EC2 | ΔISEcp1-blaCMY-2-blc-sugE | ND d | FII | GEN, NET, TOB, STR, TET, CHL, NAL, CIP, SUL |
| HH (97) | 2014 | A0 | ST665/EC3 | ISEcp1-blaCMY-2-blc-sugE | NT e (75) | K + B/O | STR, TET, NAL, SUL |
| UCC (6) | 2003 | A1 | ST48/CC10/EC4 | ISEcp1-blaCMY-2-blc-sugE | A/C2 (150) | - | KAN, GEN, TOB, STR, TET, CHL, SUL |
a HH, healthy humans; UCC, uncooked chicken carcass; b PhG, E. coli phylogenetic group; c ST, Sequence Type, CC, clonal complex; d ND, not determined due to multiple plug degradations; e NT, non-typeable; f CIP, ciprofloxacin; CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; NET, netilmicin; STR, streptomycin; SUL, sulphonamides; TET, tetracycline; TOB, tobramycin.
The absence of qAmpC producers among livestock animals (swine, chickens) is surprising since livestock animals are known as reservoirs of blaCMY-2 [3], which might be explained by the low number of analysed samples from each period. However, the detection of CMY-2-producing E. coli in an uncooked chicken carcass (n = 1/20; 5.0%), is of note since it can have either animal origin or cross-contamination of meat by humans during processing or at retail. In any case, this rate can be underestimated considering the low number of samples tested, and highlights the need of further monitorization in animals or at retail settings. To the best of our knowledge, this study represents the first analysing such a diverse and representative sample from aquatic environments, with the absence of qAmpC producers suggesting a particularly low burden in the Portuguese setting.
CMY-2-producing E. coli belonged to phylogroups B22 (n = 2, HH), A0 (n = 1, HH) or A1 (n = 1, UCC) (Table 1). The indistinguishable PFGE pattern found among B2-ST4953 E. coli isolates recovered from two-family related individuals (husband and wife), indicates a common source for CMY-2 faecal carriage or human-to-human transmission. The other two A-E. coli isolates were assigned to ST665 (A0) and ST48 (A1), the latter belonging to the clonal complex 10. These clones have been sporadically identified as CMY-2 producers from clinical (ST665 in South Africa) or non-clinical (ST48 in Tunisia and Poland) settings [9,10,11], or associated with other mechanisms of antibiotic resistance (e.g., ESBL/carbapenemase production or plasmid-mediated colistin resistance) in different reservoirs (hospitals, animals or food products) [11,12], further supporting circulation of these clones along the food chain. All the CMY-2-producing E. coli isolates were multidrug-resistant (non-susceptible to at least one agent in three or more antimicrobial categories tested). CMY-2 was identified in frequently reported genetic contexts (ΔISEcp1/ISEcp1-blaCMY-2-blc-sugE) (1) (Table 1). When characterizable, plasmids carrying blaCMY-2 were identified as IncA/C2 (150 kb) (Table 1), a plasmid type previously associated with blaCMY-2 among clinical isolates in our country [1].
To the best of our knowledge, this is one of the few studies that determined the occurrence of qAmpC-producing Enterobacteriaceae among environmental, animal and human niches. Our data confirm the presence of qAmpC producers outside the clinical setting in our country and highlight the need for further surveillance in contemporary collections, that will provide integrated epidemiological information on potential shifts in reservoirs, trends of qAmpC-producers and blaqAmpC, and transmission routes among different reservoirs.
Author Contributions
Conceptualization, E.M. and Â.N.; methodology, T.G.R.; formal analysis, T.G.R.; investigation, T.G.R.; writing—original draft preparation, T.G.R.; writing—review and editing, E.M., Â.N., and L.P.; supervision, E.M. and Â.N.; funding acquisition, L.P. All authors have read and agree to the published version of the manuscript.
Funding
This research was funded by European Union (FEDER funds) through Programa Operacional Fatores de Competitividade–COMPETE, and from National Funds [Fundação para a Ciência e a Tecnologia (FCT)] through project UIDB/04378/2020. Teresa Gonçalves Ribeiro was supported by a Ph.D. fellowship from Fundação para a Ciência e a Tecnologia (Grant SFRH/BD/75752/2011), and Ângela Novais was supported by national funds through FCT, I.P., in the context of the transitional norm (DL57/2016/CP1346/CT0032).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Ribeiro T.G., Novais Â., Rodrigues C., Nascimento R., Freitas F., Machado E., Peixe L. Dynamics of clonal and plasmid backgrounds of Enterobacteriaceae producing acquired AmpC in Portuguese clinical settings over time. Int. J. Antimicrob. Agents. 2019;53:650–656. doi: 10.1016/j.ijantimicag.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Liebana E., Carattoli A., Coque T.M., Hasman H., Magiorakos A.-P., Mevius D., Peixe L., Poirel L., Schuepbach-Regula G., Torneke K., et al. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 2013;56:1030–1037. doi: 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 3.Carattoli A., Coque T., Hasman H., Mevius D., Peixe L., Poirel L., Schupbach G., Threlfall J., Torneke K., Torres C. Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 2011;9:2322 [Google Scholar]
- 4.Machado E., Coque T.M., Cantón R., Sousa J.C., Peixe L. Antibiotic resistance integrons and extended-spectrum {beta}-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 2008;62:296–302. doi: 10.1093/jac/dkn179. [DOI] [PubMed] [Google Scholar]
- 5.Machado E., Coque T.M., Cantón R., Sousa J.C., Silva D., Ramos M., Rocha J., Ferreira H., Peixe L. Leakage into Portuguese aquatic environments of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2009;63:616–618. doi: 10.1093/jac/dkn510. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues C., Machado E., Fernandes S., Peixe L., Novais Â. An update on faecal carriage of ESBL-producing Enterobacteriaceae by Portuguese healthy humans: detection of the H30 subclone of B2-ST131 Escherichia coli producing CTX-M-27. J. Antimicrob. Chemother. 2016;71:1120–1122. doi: 10.1093/jac/dkv443. [DOI] [PubMed] [Google Scholar]
- 7.Antunes P., Campos J., Mourão J., Pereira J., Novais C., Peixe L. Inflow water is a major source of trout farming contamination with Salmonella and multidrug resistant bacteria. Sci. Total Environ. 2018;642:1163–1171. doi: 10.1016/j.scitotenv.2018.06.143. [DOI] [PubMed] [Google Scholar]
- 8.Ruh E., Zakka J., Hoti K., Fekrat A., Guler E., Gazi U., Erdogmus Z., Suer K. Extended-spectrum β-lactamase, plasmid-mediated AmpC β-lactamase, fluoroquinolone resistance, and decreased susceptibility to carbapenems in Enterobacteriaceae: fecal carriage rates and associated risk factors in the community of Northern Cyprus. Antimicrob. Resist. Infect. Control. 2019;8:98. doi: 10.1186/s13756-019-0548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mbelle N.M., Feldman C., Osei Sekyere J., Maningi N.E., Modipane L., Essack S.Y. The Resistome, Mobilome, Virulome and Phylogenomics of Multidrug-Resistant Escherichia coli Clinical Isolates from Pretoria, South Africa. Sci. Rep. 2019;9:16457. doi: 10.1038/s41598-019-52859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben Said L., Jouini A., Alonso C.A., Klibi N., Dziri R., Boudabous A., Ben Slama K., Torres C. Characteristics of extended-spectrum β-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total Environ. 2016;550:1103–1109. doi: 10.1016/j.scitotenv.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 11.Zając M., Sztromwasser P., Bortolaia V., Leekitcharoenphon P., Cavaco L.M., Ziȩtek-Barszcz A., Hendriksen R.S., Wasyl D. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019;10:1753. doi: 10.3389/fmicb.2019.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myrenås M., Slettemeås J.S., Thorsteinsdottir T.R., Bengtsson B., Börjesson S., Nilsson O., Landén A., Sunde M. Clonal spread of Escherichia coli resistant to cephalosporins and quinolones in the Nordic broiler production. Vet. Microbiol. 2018;213:123–128. doi: 10.1016/j.vetmic.2017.11.015. [DOI] [PubMed] [Google Scholar]