Abstract
The molecular interactions between a pest and its host plant are the consequence of an evolutionary arms race based on the perception of the phytophagous arthropod by the plant and the different strategies adopted by the pest to overcome plant triggered defenses. The complexity and the different levels of these interactions make it difficult to get a wide knowledge of the whole process. Extensive research in model species is an accurate way to progressively move forward in this direction. The two-spotted spider mite, Tetranychus urticae Koch has become a model species for phytophagous mites due to the development of a great number of genetic tools and a high-quality genome sequence. This review is an update of the current state of the art in the molecular interactions between the generalist pest T. urticae and its host plants. The knowledge of the physical and chemical constitutive defenses of the plant and the mechanisms involved in the induction of plant defenses are summarized. The molecular events produced from plant perception to the synthesis of defense compounds are detailed, with a special focus on the key steps that are little or totally uncovered by previous research.
Keywords: Tetranychus urticae, plant defense, constitutive and inducible defenses, signalling events, mite counter-defenses
1. Introduction
Plants are constantly subjected to a combination of abiotic and biotic stresses which provoke a strong selection pressure leading to plant adaptation to growth–defense trade-off. In the context of the plant interaction with pathogens and pests, plants have developed complex strategies to survive and preserve their fitness by combining constitutive and inducible defenses [1,2,3]. However, defenses against phytophagous arthropods require different strategies than those towards pathogens due to their mobility and precise feeding modes, which produce specific plant injuries. Plant responses to arthropods are based on inherited traits and are generally divided into three categories: antixenosis or deterrence, antibiosis or resistance and tolerance [4,5]. Antixenosis represents a clear nonpreference of an arthropod for a plant and is denoted by the presence of deterrent factors (colours, odors, textures) that alter arthropod behaviour and select an alternative host. Antibiosis is the plant–pest antagonistic association in which chemical and morphological defensive plant factors adversely affect the biology of the arthropod. Tolerance is the plant’s ability to interact with the arthropod and to withstand, repair or recover from the potential damage produced by the pest. These three strategies can be exclusive or can overlap mechanistically and functionally since their defensive purpose is plant specific and dependent on arthropod species and its feeding mode.
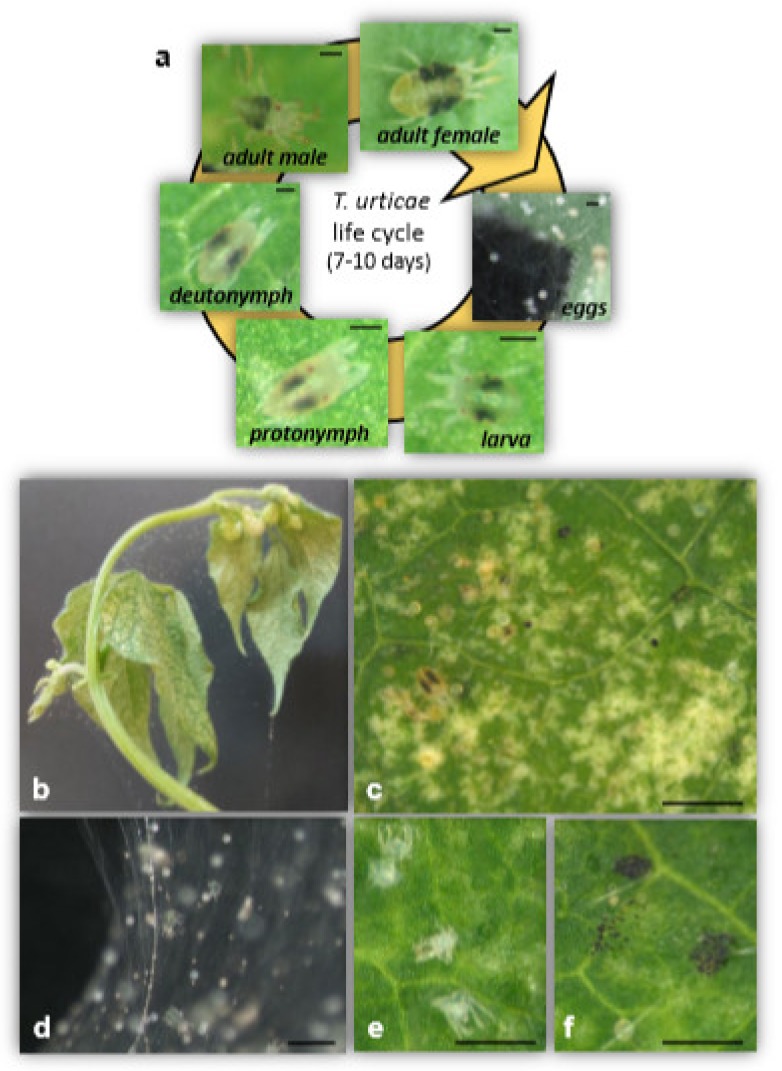
Most studies on plant–arthropod interactions have been focused in insects but mites (Chelicerata, Arachnida, Acari) also use plants as a food source, causing significant damage and yield losses. Among phytophagous acari, spider mites (Acari, Tetranychidae) are the most important family with nearly 1,300 described species in about 77 genera, of which approximately 10% are plant feeders [6]. In particular, the two-spotted spider mite Tetranychus urticae Koch is considered a serious threat for agriculture because it is an extremely polyphagous species with a short life cycle, high offspring production and a remarkable ability to develop pesticide resistance [7,8,9]. T. urticae females may produce over 100 eggs of different size depending on the embryo sex since this species reproduces by arrhenotoky, a form of parthenogenesis in which males develop from unfertilized eggs while diploid females derived from fertilized eggs [10]. Egg laying is followed by larvae hatching and the subsequent development of protonymph, deutonymph and adult stages, to complete a life cycle that takes place in less than 10 days under optimal conditions (Figure 1a).
Figure 1.
Phenotypic features of the plant–spider mite interaction. T. urticae life cycle (a), symptoms of mite-infested bean plants (b,c), and spider mite silk web (d), molts (e) and feces (f). Bar scales are indicated: 100 µM (a), 500 µM (c), 200 µM (d) and 250 µM (e,f).
T. urticae is a cosmopolitan pest that feeds on more than 1,100 documented plant species, of which 150 are important agronomic crops [11]. Besides, under the current climate change scenario associated with dry and hot conditions, T. urticae shortens its life cycle, produces more generations per year and broadens the host range [12]. Spider mites feed, mainly but not only, on the leaves by piercing individual mesophyll cells. They introduce a retractable stylet between epidermal cells or through the open stomata without inferring any cell damage, inject saliva to predigest the mesophyll cell content and suck its content. Consequently, the chlorophyll is lost, the photosynthetic rate is reduced and typical chlorotic lesions appear (Figure 1b,c). Under severe infestations, leaf defoliation and crop losses are produced [13,14]. When a plant host is overexploited and food resources become limited, T. urticae uses its ability to synthesize silk as a strategy to migrate and colonize new plants. They generate silk balls that may give refuge to hundreds of mites and are used as aerial dispersal elements spread either by wind or animal transport [15]. Besides, silk webbing over leaf surfaces protects the mite colony from external aggressions, is used for mite locomotion and acts as a pheromone substrate [16,17] (Figure 1d).
The sequence of the genome of T. urticae identified specific expansions of gene families involved in digestion, detoxification and transport of xenobiotic compounds to cope with defense molecules of different host plants [18]. These features support the mite polyphagous character and demonstrate its capability to counteract the effects of a variety of active components either synthesized by the plant as defenses or as components of the acaricides used for pest control. In addition, the presence of endosymbionts, mainly Wolbachia spp, in spider mites may alter their interaction with the host, manipulate the plant defenses and modify mite performance [19,20]. Biological control of spider mites through natural predators is a useful practice in closed greenhouses but not in open fields. Unfortunately, up to now, there are no plant cultivars with resistance to spider mites [21,22].
Working knowledge of the spider mite biology together with the development of the wide range of genomic tools and protocols established for T. urticae [23,24,25,26,27] make this species a model within the phytophagous chelicerate to address different aspects of the plant–spider mite interactions. To understand the underlying molecular mechanisms of plant defense against spider mites has been a major challenge in the last years [22,28]. However, plant defense responses to T. urticae encompass a complex and highly regulated process involving many factors, signaling molecules and pathways. In this puzzle, where all pieces are essential, there are still many knowledge gaps to understand the whole defensive landscape. This review compiles all the current knowledge of plant–spider mite T. urticae interplay identifying host defense mechanisms to be unveiled.
2. Plant Defenses Against T. urticae: Step by Step
2.1. Physical and Chemical Constitutive Barriers
The defensive response of the plant to T. urticae challenge includes a combination of pre-existing constitutive defenses evolutionary developed to avoid pest damage, and inducible defenses specifically generated upon spider mite infestation. There are several examples of the protective role of constitutive barriers, either physical or chemical, to combat this phytophagous acari. For instance, an increased leaf trichome density in raspberry (Rubus idaeus L.) correlated negatively with spider mite presence and demonstrated that this antixenosis trait hampered the spider mite movement and limited the mite egg deposition [29]. Gomez-Sanchez et al. [30] have recently shown that the thicker leaf cuticles detected in barley (Hordeum vulgare) HvPap-1 knockdown plants protected leaves from T. urticae feeding probably by hindering stylet penetration into the leaf mesophyll.
Regarding the chemical barriers, a clear association between glandular trichomes in wild tomato genotypes and spider mites has been described and identified as a source of resistance (Table 1). Solanum habrochaites Kanpp and Spooner accession LA 407 accumulates methyl ketones that had deterrence activity against spider mites [31]. Solanum pennelli Correll LA-716, Solanum pimpinellifolium TO-937 and Solanum galapagense Darwin and Perelata VI057400, VI045262, VI037869 and VI037239 accessions, were resistant to spider mites since the acylsucrose exudated by type IV trichomes caused mortality and reduced the oviposition rates of spider mite females [32,33,34]. Tomato genotypes rich in zingiberene, a sesquiterpene exuded by type IV and VI trichomes, presented antibiosis-type resistance by increasing nymph mortality and decreasing fecundity [35]. These defense traits make wild-tomato genotypes also resistant to other phytophagous species different than acari, and even to phytopathogens. Thus, they should be considered a source of general antibiosis-type resistant traits that could be introduced in cultivated species [36]. Bleekler et al. [37] studied an atypical terpene pathway producing high concentrations of 7-epizingiberene in wild tomato. The introduction of two genes involved in this pathway into a cultivated greenhouse variety produced the accumulation of this sesquiterpene in the transgenic tomato trichomes. The mortality of mites that fed on these tomatoes increased up to 40%; fecundity was also reduced in comparison to non-transformed cultivated tomatoes [37]. Likewise, the accumulation of other allochemicals such as flavonoids in citrus, phenolic compounds in chrysanthemum or terpenoids in cucumber and citrus have also been associated with resistance to T. urticae [38,39,40]. Taken together, these data indicate that constitutive barriers, whether physical (anatomical) or chemical against T. urticae infestation are dependent on the specific features of the host species, accession or cultivar rather than a specific response to the feeder species.
Table 1.
Plant-derived molecules with defense properties directly targeted to T. urticae.
| Molecules | Effects on Spider Mites | Plants | Reference |
|---|---|---|---|
| Indole-glucosinolates | Toxicity | Thale cress | Zhurov et al. 2014 [41] Santamaria et al. 2019 [42] |
| Flavonoids | Palatability/Toxicity | Chrysanthemum | Kielkiewicz & van de Vrie 1990 [38] |
| Citrus | Agut et al. 2014, 2015 [40,43] | ||
| Tomato | Martel et al. 2015 [44] | ||
| Common bean | Hoseinzadeh et al. 2020 [45] | ||
| pepper | Zhang et al. 2020 [46] | ||
| Phenyl propanoids | Toxicity | Thale cress | Zhurov et al. 2014 [41] |
| Tomato | Martel et al. 2015 [44] | ||
| Terpenoids | Repellence/Toxicity | Citrus | Agut et a. 2015 [43] |
| Tomato | Kant et al. 2004 [47] | ||
| Bleeker et al. 2012 [37] | |||
| Martel et al. 2015 [44] | |||
| Oliveira et al., 2018 [35] | |||
| Cucumber | Balkema-Boomstra et al. 2003 [39] | ||
| Pepper | Zhang et al. 2020 [46] | ||
| Alkaloids | Deterrence | Citrus | Agut et al. 2014, 2015 [40,43] |
| Pepper | Zhang et al. 2020 [46] | ||
| Phenolics | Tomato | Kant eta al. 2004 [47] | |
| Martel et al. 2015 [44] | |||
| Acyl-sugars | Repellence | Tomato | Alba eta l. 2009 [32] |
| Lucini et al. 2015 [33] | |||
| Rakha et al. 2017 [34] | |||
| Methyl ketones | Deterrence | Tomato | Antonious & Snyder 2015 [31] |
| Phenolics | Toxicity | Star anise | Koeduka et al. 2014 [48] |
| Protease inhibitors | Atinutritive | Tomato | Li et al. 2002 [49] Kant et al. 2004 [47] |
| Martel et al. 2015 [44] | |||
| Thale cress | Santamaria et al. 2018 [50] |
2.2. Inducible Defenses: Plant Perception
Most of the plant defenses against pests, including spider mites, are induced by the herbivore, first in the tissue where the infestation takes place (local response), and then in distal undamaged tissues of the same plant (systemic response). Both local and systemic responses involve structural and chemical modifications, particularly developed against each precise feeder. Induced defenses result in both energy saving and the avoidance of self-intoxication. However, these differences entail risks due to their delay on being operational [4]. The induction of plant defenses is initiated when specific receptors (pattern recognition receptors, PRRs) detect the presence of herbivore-associated molecular patterns (HAMPs), microbe-associated molecular patterns (MAMPs) derived from herbivore and associated microbes, or damage-associated molecular patterns (DAMPs) consequence of herbivore injury. Rapidly, this set of elicitors/effectors triggers a signal transduction cascade modulated by hormones, mainly jasmonic acid (JA), salicylic acid (SA) and ethylene (ET). Subsequently, the activated pathway leads to a reprogramming of gene expression and the activation of transcription factors that regulate the synthesis of molecules with defensive properties against pests, which are recognized as direct defenses. In addition, indirect defenses, mainly volatiles, are also produced to attract natural enemies of the phytophagous arthropod or to send messages to neighboring plants [22,51,52,53].
Plant perception begins with the phytophagous arthropod contact. Initially, arthropod touching, plant cell disruption and deposition of chemicals from the herbivore induce a rapid and precise defensive response [54]. Thus, plant damage, pest-associated cues like insect/acari behavior, feeding vibrations, oral secretions and oviposition fluids that release specific compounds, may elicit/suppress plant defenses [2,55,56]. In the T. urticae case, Villarroel et al. [57] identified a battery of putative HAMPs, salivary-secreted peptides, with potential elicitor/effector function to interfere with the plant defenses. Two of these peptides, Tu28 and Tu84, suppressed mite-induced SA defenses when these were transiently expressed in Nicotiana benthamiana Domin plants that promoted the reproductive performance of the acari. However, the molecular role of these effector molecules is yet to be determined. Recently, two novel proteins termed tetranin 1 and 2 (Tet1 and Tet2), which are located in the salivary glands of spider mites and have been shown to induce defenses in the host plants, have been characterized [58]. The infiltration of recombinant Tet1 and Tet2 proteins in bean leaves elicited early cellular responses such as cytosolic Ca2+ influx, membrane depolarization and reactive oxygen species (ROS) production. Moreover, spider mite feeding assays in bean and eggplant leaves resulted in improved T. urticae performance due to reduced mortality and presence of fewer predatory mites. These findings showed that plants integrate specific signals and stimuli from T. urticae to induce defenses, but how plants modulate the simultaneous recognition of different salivary proteins/peptides requires more investigation. Nevertheless, salivary glands or oral secretions are not the only source of spider mite signals to be potentially recognized by the plant as a threat to induce defense responses. Faeces, eggs, silk and acari molts (Figure 1d–f) can be an alternative source of HAMPs to alert the plant about the spider mite attack [59]. How plant responses are induced or suppressed by these HAMPs constitutes a set of unknown events or gaps to be explored.
Despite being one of the key players in the initial steps of plant defense signalling, the identification and characterization of PRRs in the plant–herbivore interaction is largely missing. A few receptor-like-kinases with an ecto-domain involved in ligand binding, a transmembrane region and an intracellular kinase domain have been functionally characterized. They act as active participants in the recognition of HAMPs derived from phytophagous insects to trigger defense pathways. Some PRRs have been identified in plants interacting with certain lepidopteran and aphid species [60,61,62,63,64]. In contrast, the information on PRRs associated with the recognition of other insect species is very limited.
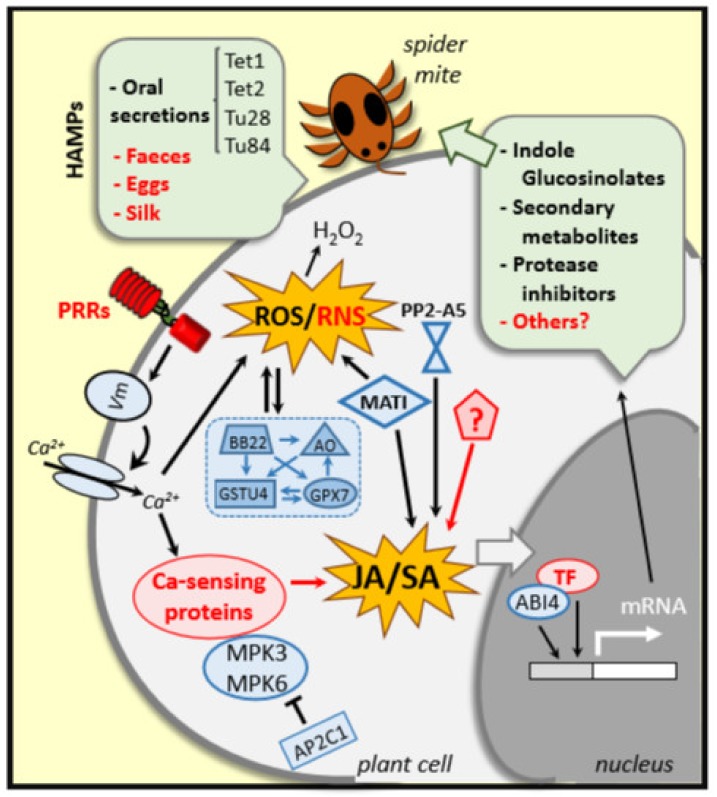
Comparative genome-wide transcriptome analyses of plant responses to T. urticae have revealed the upregulation of several receptor-encoding genes either across plant species or in a single plant host [44,65]. Alysm-containing receptor-like kinase (LYK4) is involved in chitin-triggered signalling induced by mite feeding in Arabidopsis thaliana (thale cress), grapevine and tomato. However, its ligands/binding pairs and downstream defense signalling events have to be deciphered. Recently, Santamaria et al. [42] have characterized a spider-mite-induced thale cress gene, PP2-A5. This gene encodes a protein with two domains, a Toll/Interleukin-1 receptor (TIR) and a phloem protein 2 with lectin activity (PP2), located at the N- and C-terminal regions of the protein, respectively. In PP2-A5 overexpressing or knock-out plants transcriptional reprogramming takes place leading to alter the plant defense responses against T. urticae that are mediated by a hormonal crosstalk. However, whether the receptor domain participates in the spider mite recognition by binding to derived-HAMPs has yet to be determined. Further investigation is required to have a clear understanding of the function of each single domain. Likewise, a recent functional characterization of a thale cress bidirectional promoter shared by two divergent genes has demonstrated the simultaneous induction of the two genes by T. urticae feeding. Interestingly, one of these two divergent genes encoded an LRR-receptor-kinase that could be involved in the perception of the phytophagous mites to initiate the defense transduction cascade [66]. Further investigations are needed to demonstrate its role. In conclusion, the arthropod ligand-PRR binding in the plant–spider mite interaction is one of the weakest links and presents an important knowledge gap to our understanding of the plant-spider mite interaction (Figure 2).
Figure 2.
Plant event in responses to T. urticae infestation. Specific plant receptors (PRRs) recognize elicitors/effectors (HAMPs) derived from either the plant or the spider mite that induce alterations in the membrane potential (Vm), cytosolic Ca2+ influxes and ROS/RNS burst. Ca2+-sensing proteins, MPKs and phosphatases (APC21) participate in the defense transduction pathway. H2O2 content is highly regulated by ROS-related enzymes (BB22, AO, GSTU and CPX7). Besides, genes such as MATI, PP2A5 and others still unknown participate in the tight regulation of the hormonal crosstalk, mainly in the Jasmonic Acid/Salicylic Acid balance. All together plus some transcription factors (ABI4 and other unknown TFs) regulate the induction of the synthesis of a battery of defense molecules. Unknown genes, molecules, pathways and responses are indicated in red.
2.3. Inducible Defenses: Early Signalling Events
The recognition of phytophagous signals, particularly those that bind to specific PRRs at the plasma membrane, produces variations in the electrochemical gradient between intra- and extra-cellular sites. Concomitant to a depolarization of the membrane potential (Vm), the cytosolic Ca2+6influx and the ion channel activity increased, whereas a ROS and reactive nitrogen species (RNS) burst occurs in response to arthropod feeding. These changes take place rapidly after infestation and activate a defense-signalling cascade. Calcium sensors such as calmodulin, calcineurin, Ca-dependent protein kinases and Ca-ATPases play a crucial role to regulate downstream targets in the signal transduction cascade that eventually result in a generation of battery of defenses [2,52,67]. So far, most of the information on the early defense events comes from studies on plant–insect interaction. Also, some reports have demonstrated that T. urticae infestation produces cell damage that leads to alterations in cytosolic Ca2+ levels and Vm variations accompanied by ROS production [58]. Accumulation of ROS and phenolic compounds at the wounding sites caused by T. urticae in barrelclover (Medicago truncatula) and thale cress plants have been reported [2,68,69,70]. ROS molecules, particularly H2O2, are essential in defense signalling and in oxidative pathways to regulate cell proliferation and promote cellular processes [71]. Excess of H2O2 causes oxidative stress and induces programmed cell death while moderate abundance of H2O2 functions as a defense-signaling molecule. Thus, ROS are essential to keep levels below damaging concentrations but triggering signals. Santamaria et al. [50] demonstrated the role of thale cress ROS homeostasis related genes that are involved in the synthesis of H2O2 and or degradation of H2O2 and ascorbate, in response to T. urticae infestation. Silencing of the three genes involved in ROS degradation resulted in higher leaf damage and better mite performance in comparison to wild-type plants. It has also been reported that the effect of the mite attack triggered immunity (MATI) gene involved in the maintenance of the thale cress antioxidant status was correlated with the spider mite performance. MATI-overexpressing plants led to a moderate H2O2 accumulation and increased thale cress resistance to spider mite attack as reflected by lesser leaf damage and the reduced mite fecundity observed in comparison to the symptoms and oviposition rates quantified in control and mutant plants [70].
There is some evidence demonstrating that ROS production depends on the activity of plant oxidases such NADPH oxidases, also known as respiratory burst oxidase homologues (RBOHs). They have the ability to integrate Ca2+ signalling, protein phosphorylation and ROS production as key mediators of rapid local and systemic signalling in plants in response to pathogens and pests [72,73]. Nicotiana attenuata Torr. ex S. Watson ROBH-silencing plants significantly reduced ROS levels and were more susceptible to the generalist lepidopteran Spodoptera littoralis Boisduval feeding than control plants [74]. Likewise, chemical inhibition of RBOH activity in wheat plants reduced H2O2 accumulation and altered the expression of downstream defense enzymes such as β-1,3-glucanases and peroxidases involved in wheat resistance to the aphid Diuraphis noxia Mordvilko ex Kurdiumov [75]. There is no known information on the role of RNS, particularly nitrogen oxide (NO), which is considered a signalling molecule in plant defense against a plethora to of pest and pathogens [76,77].
Another early event in response to herbivores is the activation of kinases and phospholipase C leading to the formation of phosphatidic acid, a secondary messenger involved in oxidative stress [47]. In addition, mitogen-activated protein kinases (MAPKs) are also key players of the transduction pathway. Schweighofer et al. [78] showed that the thale cress Ser/Thr phosphatase type 2C, AP2C1, acted as negative regulator of the stress-responsive MPK4 and MPK6 activities, which had a defense role not only against pathogens but also in the thale cress–spider mite interaction. Mutant ap2c1 plants, where both kinases were highly activated, resulted more resistant to T. urticae than wild-type and AP2C1 overexpressing plants. Moreover, the complementation of the ap2c1 mutation restored the normal fecundity of spider mites [78].
Among the early plant responses to T. urticae infestation, is very important to mention the alteration of the hormone levels. Generally, JA is the main regulator of induced defenses triggered by spider mites. As Rioja et al. [28] reported, elevated JA correlates with the degree of plant resistance to T. urticae infestation since plants with constitutive JA-mediated responses were more resistant to spider mite populations while mutants in JA synthesis were more susceptible [44,79]. However, not only the JA pathway is activated after spider mite infestation. The induction of both JA and SA pathways has been reported in spider mite infested thale cress [42,64,70], tomato [44,47], citrus [40,43,80] and pepper [46], among other plant species. Moreover, He et al. [81] also reported the accumulation of SA and conjugated-SA forms induced by spider mite attack in bean (Phaseolus vulgaris) leaves but the JA content was not analyzed in this study. Although JA and SA are considered antagonist hormones, SA does not seem to antagonize JA responses in the plant–spider mite context since SA content also increases after spider mite attack. Most likely, the potential reciprocal crosstalk between JA and SA signalling pathways allows a fine-tune modulation to sense the synthesis of specific direct and indirect defensive molecules against the acari. The JA/SA dosage, their temporal dynamics and the level of activation vary among plant hosts and plant developmental stage. There are many other factors, which may have different effects on the plant defense and on spider mite behavior [82].
Besides JA and SA, other phytohormones participate in the signalling and regulation of plant responses against T. urticae. It has been demonstrated the emission of ET and other volatiles in tomato and lima bean, which are triggered by spider mite attack [44,47,83]. In addition, the abscisic acid (ABA) insensitive 4 transcription factor (ABI4) has been identified as a crucial component of chloroplast retrograde signaling that regulates mite-associated thale cress defense controlled by ABA [84]. More recently, differences have also been found in spider mite-regulated auxin levels in thale cress [42]. In addition, metabolic and hormonal pathways are shared with other environmental biotic and abiotic stimuli, and such mutual interference was reported for light stress-mites and aphid-mite interaction [84,85].
Taken together, these results suggest that the modulation of plant resistance/susceptibility to spider mites is dependent on the hormonal crosstalk but it remains to be clarified how such complex signalling cascades affect plant defenses (Figure 2). These phytohormone-mediated mechanisms are not only specific to spider mite infestation but are considered the hallmark for plant defense signalling against both biotic and abiotic stressors [86,87].
2.4. Inducible Defenses: Late Defense Events
Early signaling triggers a series of later events that culminate to the production of compounds functioning as toxins, repellents and/or antifeedants that are involved in direct and indirect responses. Examples of plant-derived metabolites with defense properties that directly or indirectly target spider mites are presented in Table 1.
Transcriptomic and metabolomic approaches performed on different plants species following spider mite infestation have allowed the identification of a plethora of secondary metabolites involved in defense, mainly flavonoid and terpenoid products, alkaloids derived from the shikimate pathway, and some compounds resulting of the aminobenzoate degradation. Among them, glucosinolates exclusively synthesized by Brassicaceae, were highlighted in the thale cress–spider mite relationship. In particular, indole-glucosinolates (IG), compounds derived from the tryptophan pathway, whose expression is dependent of JA, constituted the central defenses against spider mites in this model species. Zhurov et al. [41] reported that some IG metabolites were induced by mite feeding and acted as deterrents and antifeedant agents. Accumulation of IG in thale cress dramatically increased spider mite mortality. IG-deficient thale cress mutants with reduced levels of a set of IG metabolites were highly susceptible to spider mite feeding. Furthermore, the overexpression of the ATR1 transcription factor, a positive regulator of IG gene expression made plants more resistant to spider mite infestation and increased larval mortality [41]. Although the mechanism of action of IG as antiacaricidal molecules has not been fully validated, it is well-known that thiocyanates, isothiocyanates, nitriles and other toxic compounds are derived from the glucosinolate hydrolysis mediated by myrosinases [87]. This hydrolytic process takes place in damaged tissues and the toxic products directly target the phytophagous arthropod physiology either reacting with biological nucleophiles or modifying nucleic acids and proteins [52].
Likewise, transcriptomic assays performed at different time points of mite-infested tomato plants revealed the upregulation of genes involved in the synthesis of secondary metabolites with known functions in plant defense [44,47]. Besides, the emission of volatile organic compounds, including phenolics and several classes of terpenes, was detected over a period of the first five days after infestation. The increase in the volatile production coincided with the greater olfactory preference of predatory mites for infested plants. Agut et al. [40] found flavonoid compounds such as naringenin, hesperitin and p-coumaric acid accumulated in a mite-resistant citrus genotype, and demonstrated that these metabolites were over-accumulated after mite infestation. Since some flavonoids, such as p-coumaric acid, participate in the biosynthesis of lignin polymers, their defense role was related to the formation of physical barriers to reduce the palatability of the plant. The upregulation of genes encoding flavonoids have been recently found in a comparative transcriptomic study performed in resistant common bean cultivar to spider mites [49]. In addition, macarpine, a benzophenanthridine alkaloid derived from shikimate, was also identified in mite-infested citrus [40,43]. In the same citrus genotype was detected the production of the terpene volatiles α-ocimene, α-farnesene, pinene and D-limonene, and the green leaf volatile 4-hydroxy-4-methyl-2-pentanone, all of them with a marked repellent effect on spider mites. Interestingly, volatiles released from the resistant genotype after spider mite infestation promoted mite-induced resistance in a neighbouring susceptible citrus genotype, thereby reducing oviposition rates [43]. Very recently, the combination of transcriptome and metabolome analyses in spider-mite infested pepper (Capsicum annuum) has allowed us to appreciate the role of terpenoids, including di- and tri-terpene glucosides, some mono-terpenes, sesquiterpenes and homoterpenes, during T. urticae infestation as well as alkaloids and aromatic compounds [46]. Other secondary metabolites such as O-dimethylallyleugenol, a renylated phenylpropene identified in the Japanese star anise, Illicium anisatum L., exhibited deterrent activity to spider mites [45]. In addition, as previously described, secondary compounds such as methyl-ketones and acyl-sugars provide chemical defense against T. urticae infestation [31,32,33,34,35]. These results indicate that each plant species, and even each genotype, specifically respond to spider mite infestation generating different molecules with distinct defense properties.
Besides the induction of secondary metabolites, primary defensive metabolites including proteins and peptides are also accumulated in plants after T. urticae infestation. The well-known example is the rapid increase of proteinase inhibitors (PIs), generally encoded by JA-dependent genes, and mainly but not exclusively found in Solanaceae plants. These proteins exert direct effects on phytophagous arthropods by interfering with their physiology, either inhibiting the gut digestive protease activities or the function of other proteases involved in growth and development [23,48]. In tomato, two serine PI genes, PIN-I and PIN-II, were shown to be consistently induced by spider mite attack [47,88,89]. Moreover, measurements of the corresponding PI activities showed about twofold increase in mite-infested leaves compared to control ones. Martel et al. [44] corroborated these results and indicated that tomato PI genes were among the most highly induced in the microarray analyses performed in the spider mite-tomato system, suggesting that they represent one of the major tomato defense response outputs upon spider mite herbivory. Likewise, the induction of two genes, AtKTI4 and AtKTI5, encoding Kunitz trypsin inhibitors (KTIs) in spider mite-infested thale cress, and the fact that their corresponding silenced lines conferred higher susceptibility to T. urticae than wild-type plants, strongly support the potential defense role of KTIs [90]. In addition, barley cystatins, inhibitors of cysteine-proteases, have also been used as defense transgenes. When they were expressed alone or in combination with serine-PIs in maize and thale cress, an enhanced resistance was observed against phytophagous mites by altering mite cysteine-proteases [23,91]. In conclusion, although the list of molecules potentially involved in the plant defense against T. urticae is high, for most of them the mode of action and the targets in the mite remain to be remains yet to be identified.
The mentioned early- and late-term defenses induced by the phytophagous mites can be altered by prior experiences. The earlier exposure of plants to specific stimuli may prepare it for upcoming stresses. This phenomenon termed priming, is a physiological event of immune adaptation that prepares the plant to react faster, to generate a more effective defense response with no trade-offs in the absence of challenge. A clear example of priming in plant defense to T. urticae has been reported by Agut et al. [43]. They demonstrated the effect of volatiles emitted from spider mite-infested sour orange and Cleopatra mandarin plants on ono-infested plants. Notably, macarpine was identified as one of the defensive compounds accumulated in response to priming treatment [43]. If the function of this compound is to induce resistance has to be proven. If so, volatiles might be used as new inducers of plant resistance to spider mites under field conditions. This hypothesis is supported by a recent publication [92] that reported how spider mites’ biological parameters, mainly oviposition on strawberry leaves, are affected by aromatic compounds derived from intercropping. Priming was also observed in the case of sequential and different herbivore infestation leading to mutual pest suppression [85]. Priming is another gap to be explored to reduce costs of plant defense.
3. Spider Mite Counter-Defenses
Many phytophagous organisms, and particularly the generalist T. urticae, have acquired traits to overcome plant defenses through three main strategies: avoidance, metabolic resistance and suppression [4,93]. The avoidance of induced plant defenses entails a behavioral feature while the other two strategies make the herbivore cope with ingested plant metabolites. Metabolic resistance against toxic molecules can arise from target-site insensitivity or detoxification mechanisms and may imply metabolite modification, degradation and/or secretion. Defense suppression is achieved via sabotage of the host plant’s molecular machinery. Thus, phytophagous mites have evolved specialized molecules secreted into or onto their host to interfere in different manners with the host’s ability to defend itself [4]. In the T. urticae case, the most striking example of polyphagy with a high ability to adapt to novel plants and developing rapid resistance to pesticides, the combination of the three mentioned strategies is used to maximize its performance [94]. Its own feeding mechanism may explain how spider mites try to avoid the induction of plant defenses. The stylet usually penetrates through stomata or epidermal pavement cells to reach single mesophyll cells, avoiding epidermal damage, which minimizes the detection of the attack, therefore delaying the plant response [14]. In addition, T. urticae genome revealed strong signatures of polyphagy linked to the expansion of gene families involved in digestion, detoxification and transport of xenobiotics. These gene families include cytochrome P450, carboxyl/cholinesterases (CCEs), glutathione S-transferases (GSTs) and ATP-binding cassette transporters (ABC). Besides, the integration of new detoxifying genes acquired by lateral gene transfer from bacteria enhances the spider mite protection to host plant toxic compounds [18,95]. However, the suppression of defenses is probably the main and most interesting strategy developed by spider mites to cope with toxicity and survive [91,94,95]. T. urticae and the Solanaceae-specialist T. evansi may suppress SA- and JA-dependent defenses in tomato plants by acting downstream of the phytohormone pathway, but each mite strain affects the expression of tomato defense genes in a different way [96]. The mite T. evansi can suppress JA-dependent responses by stimulating SA pathway to activate the negative crosstalk between their signalling pathways [97]. As previously commented, T. urticae also secretes effectors via its saliva into plant tissues to interfere with host immune response and to promote its reproductive performance [56,57]. However, the ecological costs and benefits of defense suppression are still unclear since these observations come from laboratory assays lacking the natural ecological context. In nature, mites coexist with other herbivorous and predatory mites, which may also be favored by the mite-suppression of plant defenses. Furthermore, in the case of T. urticae the puzzle becomes much more complicated because of the extraordinary number of genes associated with metabolic resistance, which, a priori, seems unnecessary to spend mite resources in the suppression of plant defenses. In fact, Blaazer et al. [93] predicted that a generalist herbivore such as this mite confined to a host for a long period of time will replace the suppression trait by resistance traits based on they are ecologically safer and promote mite performance more strongly.
4. Conclusions and Future Prospects
To fully understand the complex molecular interaction between a pest and its host plant is currently a major challenge for plant–pest researchers. The present compilation of the known information, regarding the molecular mechanisms involved in the arms race between the mite T. urticae and its host plants, highlights the great deal of key knowledge that remains unknown. In the near future, efforts should be focused on unveiling how plants perceive and signal the mite attack. To that end, it is necessary to determine the receptors of the plant that play a role in the interaction as well as the molecules of the mite that bind and form complexes with plant molecules. Besides, it will be essential to establish the consequences of these interactions, if the mite molecules act as elicitors and trigger plant immune response, or as effectors by hampering plant defense. Broad transcriptome, proteome and metabolome analysis of the responses to the mite in different plant species, as well as similar experimental approaches related to the mites feeding on them, would provide a wide core of molecular data to establish working hypotheses. Then, genetic modifications of plants and mites could be done to check these hypotheses and to fill in the existing gaps in the molecular mechanisms that control the plant–mite interplay. An advanced understanding of the mite–plant interaction will be a strong tool to enhance crop performance by improving specific plant defenses against T. urticae attack.
Author Contributions
I.D. conceived the idea and wrote the original draft of the manuscript. M.M. and M.E.S. made substantial contributions to enhance a final version of the manuscript. A.A., I.R.-D. and P.G.-M. prepared samples and photos. All authors, including G.R.-H., D.A.O.-M., A.G., and E.C. participated in the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
Research was supported by the Ministerio de Economia, Industria y Competitividad (MEIC, grants BIO2017-83472-R and RyC17MESFB), the “Severo Ochoa Program for Centres of Excellence in R&D” from the Agencia Estatal de Investigación of Spain (grant SEV-2016-0672 (2017–2021) to the CBGP and the Programa de Apoyo a la realización de Proyectos I+D para jóvenes investigadores de la Universidad Politécnica de Madrid (UPM). RyC grant (RyC2017-21814) from the MEIC and Plan Propio from the UPM financed M.E.S. Studenships from: MEIC to I.R-D (PRE2018-083375), Comunidad de Madrid to G.R-H (PEJD-2019-PRE/BIO-15882), La Caixa Foundation to D.O-M (LCF/BQ/IN18/11660014), UPM to A.G., and the postdoctoral contract associated to the Severo Ochoa Program to E.C. (EoI-TSP2-06), are gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest
References
- 1.Wu J., Baldwin I.T. New insights into plant responses to the attack from insect herbivore. Annu. Rev. Genet. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- 2.Santamaria M.E., Arnaiz A., Gonzalez-Melendi P., Martinez M., Diaz I. Plant perception and short-term responses to phytophagous insects and mites. Int. J. Mol. Sci. 2018;19:1356. doi: 10.3390/ijms19051356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson S.W., Magerøy M.H., Lopez Sanchez A., Smith L.M., Furci L., Cotton T.E.A., Krokene P., Ton J. Surviving in a hostile world: Plant strategies to resist pests and diseases. Annu. Rev. Phytopathol. 2019;57:505–529. doi: 10.1146/annurev-phyto-082718-095959. [DOI] [PubMed] [Google Scholar]
- 4.Kant M.R., Jonckheere W., Knegt B., Lemos F., Liu J., Schimmel B.C.J., Villarroel C.A., Ataide L.M.S., Dermauw W., Glas J.J., et al. Mechanisms and ecological consequences of plant defense induction and suppression in herbivore communities. Ann. Bot. 2015;115:1015–1051. doi: 10.1093/aob/mcv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoorooei M., Hoseinzadeh A.H., Maali-Amiri R., Allahyari H., Torkzadeh-Mahanti M. Antisenosis and antibiosis response of common bean (Phaseolus vulgaris) to two-spotted spider mite (Tetranychus uriticae) Exp. Appl. Acarol. 2018;74:365–381. doi: 10.1007/s10493-018-0240-4. [DOI] [PubMed] [Google Scholar]
- 6.Vacante V. The Handbook of Mites of Economic Plants: Identification, Bio-Ecology and Control. CABI International; Wallingford, UK: 2016. [Google Scholar]
- 7.Agrawal A.A. Host-range evolution: Adaptation and trade-off in fitness of mites on alternative hosts. Ecology. 2000;81:500–508. doi: 10.1890/0012-9658(2000)081[0500:HREAAT]2.0.CO;2. [DOI] [Google Scholar]
- 8.Van Leeuwen T., Tirry L., Yamamoto A., Nauen R. The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pestic. Biochem. Phsyiol. 2015;121:12–21. doi: 10.1016/j.pestbp.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Rincon R., Rodriguez D., Coy-Barrera E. Botanicals against Tetranychus urticae Koch under laboratory conditions: A survey of alternatives for controlling pest mites. Plants. 2019;8:272. doi: 10.3390/plants8080272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macke E., Magalhaes S., Khan H.D.T., Luciano A., Frantz A., Facon B., Olivieri I. Sex allocation in haplodiploids is mediated by egg size: Evidence in the spider mite Tetranychus urticae Koch. Proc. Biol. Sci. 2011;278:1054–1063. doi: 10.1098/rspb.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migeon A., Dorkeld F. Institute for Agronomy Research, Center for Biology and Management of Populations; Montpellier, France: [(accessed on 2 March 2020)]. Spider Mites Web: A Comprehensive Database for the Tetranychidae. 2006–2020. Available online: ww1.montpellier.inra.fr/CBGP/spmweb. [Google Scholar]
- 12.Ximenez-Embun M.G., Castañera P., Ortego F. Drought stress in tomato increases the performance of adapted and non-adapted strains of Tetranychus urticae. J. Insect Physiol. 2017;96:73–81. doi: 10.1016/j.jinsphys.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Park Y.L., Lee J.H. Leaf cell and tissue damage of cucumber caused by two spotted spider mite (Acari: Tetranychidae) J. Econ. Entomol. 2002;95:952–957. doi: 10.1093/jee/95.5.952. [DOI] [PubMed] [Google Scholar]
- 14.Bensoussan N., Santamaria M.E., Zhurov V., Diaz I., Grbic M., Grbic V. Plant-herbivore interaction: Dissection of the cellular pattern of Tetranychus urticae feeding on the host plant. Front. Plant Sci. 2016;7:1105. doi: 10.3389/fpls.2016.01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clotuche G., Mailleux A.C., Fernandez A.A., Deneubourg J.L., Detrain C., Hance T. The formation of collective silk balls in the spider mite Tetranychus urticae Koch. PLoS ONE. 2011;6:e18854. doi: 10.1371/journal.pone.0018854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clotuche G., Mailleux A.C., Deneubourg J.L., Detrain C., Hance T. The silk road of Tetranychus urticae: Is it a single or a double lane? Exp. Appl. Acarol. 2012;56:345–354. doi: 10.1007/s10493-012-9520-6. [DOI] [PubMed] [Google Scholar]
- 17.Dogan S., Caglar M., Caglar B., Cirak C., Zeytun E. Structural characterization of the silk of two-spotted spider mite Tetranychus urticae Koch (Acari: Tetranychidae) Syst. Appl. Acarol. 2017;22:572–583. [Google Scholar]
- 18.Grbic M., van Leeuwen T., Clark R.M., Rombauts S., Rouze P., Grbic V., Osborne E.J., Dermauw W., Thi Ngoc P.C., Ortego F., et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staudacher H., Schimmel B.C.J., Lamers M.M., Wybouw N., Groot A.T., Kant M.R. Independent effects of a herbivore’s bacterial symbionts on its performance and induced plant defenses. Int. J. Mol. Sci. 2017;18:182. doi: 10.3390/ijms18010182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schausberger P. Herbivore-associated bacteria as potential mediators and modifiers of induced plant defense against spider mites and thrips. Front. Plant Sci. 2018;9:1107. doi: 10.3389/fpls.2018.01107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attia G., Grissa K.L., Lognay G., Butume E., Hance T., Mailleux A.C. A review of the major biological approaches to control the worldwide pest Tetranychus urticae (Acari: Tetranuychidae) with special reference to natural pesticides. J. Pest Sci. 2013;86:361–386. doi: 10.1007/s10340-013-0503-0. [DOI] [Google Scholar]
- 22.Agut B., Pastor V., Jaques J.A., Flors V. Can plant defense mechanisms provide new approaches for the sustainable control of the two-spotted spider mite Tetranychus urticae? Int. J. Mol Sci. 2018;19:614. doi: 10.3390/ijms19020614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santamaria M.E., Hernández-Crespo P., Ortego F., Grbic V., Grbic M., Diaz I., Martinez M. Cysteine peptidases and their inhibitors in Tetranychus urticae: A comparative genomic approach. BMC Genom. 2012;13:307. doi: 10.1186/1471-2164-13-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazaux M., Navarro M., Bruinsma K.A., Zhurov V., Negrave T., van Leeuwen T., Grbic V., Grbic M. Application of two-spotted spider mite Tetranychus urticae for plant-pest interaction studies. J. Vis. Exp. 2014;89:e51738. doi: 10.3791/51738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T., España M.U., Nunes M.A., Zhurov V., Dermauw W., Osakabe M., Van Leeuwen T., Grbic M., Grbic V. Protocols for the delivery of small molecules to the two-spotted spider mite, Tetranychus urticae. PLoS ONE. 2017;12:e0180658. doi: 10.1371/journal.pone.0180658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki T., Nunes M.A., España M.U., Namin H.H., Jin P., Bensoussan N., Zhurov V., Rahman T., De Clercq R., Hilson P., et al. RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: A comparative analysis of five methods for gene silencing. PLoS ONE. 2017;12:e0180654. doi: 10.1371/journal.pone.0180654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abouelmaaty H.G., Fukushi M., Abouelmaaty A.G., Ghazy N.A., Suzuki T. Leaf disc-mediated oral delivery of small molecules in the absence of surfactant to the two-spotted spider mite, Tetranychus urticae. Exp. Appl. Acarol. 2019;77:1–10. doi: 10.1007/s10493-018-0335-y. [DOI] [PubMed] [Google Scholar]
- 28.Rioja C., Zhurov V., Bruinsma K., Grbic M., Grbic V. Plant-herbivore interactions: A case of an extreme generalist, the two-spotted spide rmite Tetranychus urticae. Mol. Plant Microbe Interact. 2017;30:935–945. doi: 10.1094/MPMI-07-17-0168-CR. [DOI] [PubMed] [Google Scholar]
- 29.Karley A.J., Mitchell C., Brookes C., McNicol J., O’Neill O.T., Roberts H., Graham J., Johnson N.S. Exploiting physical defense traits for crop protection: Leaf trichomes of Rubus idaeus have deterrent effects on spider mites but not aphids. Ann. Appl. Biol. 2015;168:159–172. doi: 10.1111/aab.12252. [DOI] [Google Scholar]
- 30.Gomez-Sanchez A., Gonzalez-Melendi P., Santamaria M.E., Arbona V., Lopez-Gonzalvez A., Garcia A., Hensel G., Kumlehn J., Martinez M., Diaz I. Repression of drought-induced cysteine-protease genes alters barley leaf structure and responses to abiotic and biotic stresses. J. Exp. Bot. 2019;70:2143–2155. doi: 10.1093/jxb/ery410. [DOI] [PubMed] [Google Scholar]
- 31.Antonious G.F., Snyder J.C. Repellency and oviposition deterrence of wild tomato leaf extracts to spider mites, Tetranychus urticae Koch. J. Environ. Sci. Health B. 2015;50:667–673. doi: 10.1080/03601234.2015.1038960. [DOI] [PubMed] [Google Scholar]
- 32.Alba J.M., Montserrat M., Fernandez-Muñoz R. Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucrose of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant imbred line population. Exp. Appl. Acarol. 2009;47:35–47. doi: 10.1007/s10493-008-9192-4. [DOI] [PubMed] [Google Scholar]
- 33.Lucini T., Faria M.V., Rohde C., Resende J.T.V., Oliveira J.R.F. Acylsugar and the role of trichomes in tomato genotypes resistance to Tetranychus urticae. Arthropod Plant Interact. 2015;9:45–53. doi: 10.1007/s11829-014-9347-7. [DOI] [Google Scholar]
- 34.Rakha M., Bouba N., Ramasamy S., Regnard J.L., Hanson P. Evaluation of wild tomato accessions (Solanum spp.) for resistance to two-spotted spider mite (Tetranychus urticae Koch) based on trichome type and acylsugar content. Genet. Resour. Crop Evol. 2017;64:1011–1022. doi: 10.1007/s10722-016-0421-0. [DOI] [Google Scholar]
- 35.Oliveira J.R.F., de Resende J.T.V., Maluf W.R., Lucini T., de Lima Filho R.B., de Lima I., Nardi C. Trichomes and allelochemicals in tomato genotypes have antagonistic effects upon behavior and biology of Tetranychus urticae. Front. Plant Sci. 2018;9:1132. doi: 10.3389/fpls.2018.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baier J.E., Resende J.T.V., Faria M.V., Schwarz K., Meert L. Indirect selection of industrial tomato genotypes that are resistant to spider mites (Tetranychus urticae) Genet. Mol. Res. 2015;14:244–252. doi: 10.4238/2015.January.16.8. [DOI] [PubMed] [Google Scholar]
- 37.Bleeker P.M., Mirabella R., Diergaarde P.J., VanDoorn A., Tissier A., Kant M.R., Prins M., Vos M., Haring M.A., Schuurink R.C. Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proc. Natl. Acad. Sci. USA. 2012;109:20124–20129. doi: 10.1073/pnas.1208756109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kielkiewicz M., van de Vrie M. Within-leaf differences in nutritive value and defense mechanism in chrysanthemum to the two spotted spider mite (Tetranychus urticae) Exp. Appl. Acarol. 1990;10:33–43. doi: 10.1007/BF01193971. [DOI] [Google Scholar]
- 39.Balkema-Boomstra A.G., Zijlstra S., Verstappen F.W., Inggamer H., Mercke P.E., Jongsma M.A., Bouwmeester H.J. Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.) J. Chem. Ecol. 2003;29:225–235. doi: 10.1023/A:1021945101308. [DOI] [PubMed] [Google Scholar]
- 40.Agut B., Gamir J., Jacas J.A., Hurtado M., Flors V. Different metabolic and genetic responses in citrus may explain relative susceptibility to Tetranychus urticae. Pest Manag. Sci. 2014;70:1728–1741. doi: 10.1002/ps.3718. [DOI] [PubMed] [Google Scholar]
- 41.Zhurov V., Navarro M., Bruinsma K.A., Arbona V., Santamaria M.E., Cazaux M., Wybouw N., Osborne E.J., Ens C., Rioja C., et al. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 2014;164:384–399. doi: 10.1104/pp.113.231555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santamaria M.E., Martinez M., Arnaiz A., Rioja C., Burow M., Grbic V., Diaz I. An Arabidopsis TIR-lectin two-domain protein confers defense properties against Tetranychus urticae. Plant Physiol. 2019;179:1298–1314. doi: 10.1104/pp.18.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agut B., Gamir J., Jaques J.A., Flors V. Tetranychus urticae-triggered responses promote genotype dependent conspecific repellence or attractiveness in citrus. New Phytol. 2015;207:790–804. doi: 10.1111/nph.13357. [DOI] [PubMed] [Google Scholar]
- 44.Martel C., Zhurov V., Martinez M., Cazaux M., Auger P., Migeon A., Santamaria M.E., Wybouw N., Diaz I., Van Leeuwen T., et al. Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Mol. Plant Microbe Interact. 2015;3:343–361. doi: 10.1094/MPMI-09-14-0291-FI. [DOI] [PubMed] [Google Scholar]
- 45.Koeduka T., Sugimoto K., Watanabe B., Someya N., Kawanishi D., Gotoh T., Ozawa R., Takabayashi J., Matsui K., Hiratake J. Bioactivity of natural O-prenylated phenylpropenes from Illicium anisatum leaves and their derivatives against spider mites and fungal pathogens. Plant Biol. 2014;16:451–456. doi: 10.1111/plb.12054. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Bouwmeester H.J., Kappers I.F. Combined transcriptome and metabolome analysis identifies defense responses in spider mite-infested pepper (Capsicum annuum) J. Exp. Bot. 2020;71:330–343. doi: 10.1093/jxb/erz422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kant M.R., Ament K., Sabelis M.W., Haring M.A., Schuurink R.C. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol. 2004;135:483–495. doi: 10.1104/pp.103.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez M., Santamaria M.E., Diaz-Mendoza M., Arnaiz A., Carrillo L., Ortego F., Diaz I. Phytocystatins: Defense proteins against phytophagous insects and acari. Int. J. Mol. Sci. 2016;17:1747. doi: 10.3390/ijms17101747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoseinzadeh A.H., Soorni A., Shoorooei M., Torkzadeh M., Amiri R.M., Allahyari H., Mohammadi R. Comparative transcriptome provides molecular insights into defense-associated mechanisms against spider mites in resistant and susceptible common bean cultivars. PLoS ONE. 2020 doi: 10.1371/journal.pone.0228680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santamaria M.E., Arnaiz A., Velasco-Arroyo B., Grbic V., Diaz I., Martinez M. Arabidopsis response to the spider mite Tetranychus urticae depends on the regulation of reactive oxygen species homeostasis. Sci. Rep. 2018;8:9432. doi: 10.1038/s41598-018-27904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santamaria M.E., Martinez M., Cambra I., Grbic V., Diaz I. Understanding plant defense responses against herbivore attacks: An essential first step towards the development of sustainable resistance against pests. Transgenic Res. 2013;22:697–708. doi: 10.1007/s11248-013-9725-4. [DOI] [PubMed] [Google Scholar]
- 52.Stahl E., Hilfiker O., Reymond P. Plant-arthropod interactions: Who is the winner? Plant J. 2018;93:703–728. doi: 10.1111/tpj.13773. [DOI] [PubMed] [Google Scholar]
- 53.Erb M., Reymond P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019;70:527–557. doi: 10.1146/annurev-arplant-050718-095910. [DOI] [PubMed] [Google Scholar]
- 54.Bonaventure G. Perception of insect feeding by plants. Plant Biol. 2012;14:872–880. doi: 10.1111/j.1438-8677.2012.00650.x. [DOI] [PubMed] [Google Scholar]
- 55.Body M.A.J., Neer W.C., Vore C., Lin H.J., Vu D.C., Schultz J.C., Cocroft R.B., Appel H.M. Caterpillar chewing vibrations cause changes in plant hormones and volatile emissions in Arabidopsis thaliana. Front. Plant Sci. 2019;10:810. doi: 10.3389/fpls.2019.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reymond P. Perception, signaling and molecular basis of oviposition-mediated plant responses. Planta. 2013;238:247–258. doi: 10.1007/s00425-013-1908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villarroel C.A., Jonckheere W., Alba J.M., Glas J.J., Dermauw W., Haring M.A., Van Leeuwen T., Schuurink R.C., Kant M.R. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 2016;86:119–131. doi: 10.1111/tpj.13152. [DOI] [PubMed] [Google Scholar]
- 58.Iida J., Desaki Y., Hata K., Uemura T., Yasuno A., Islam M., Maffei M.E., Ozawa R., Nakajima T., Galis I., et al. Tetranins: New putative spider mite elicitors of host plant defense. New Phytol. 2019;224:875–886. doi: 10.1111/nph.15813. [DOI] [PubMed] [Google Scholar]
- 59.Santamaria M.E., Gonzalez-Cabrera J., Martinez M., Grbic V., Castañera P., Diaz I., Ortego F. Digestive proteases in bodies and faeces of the two-spotted spider mite, Tetranychus urticae. J. Insect Physiol. 2015;78:69–77. doi: 10.1016/j.jinsphys.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Gilardoni P.A., Hettenhausen C., Baldwin I.T., Bonaventure G. Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell. 2011;23:3512–3532. doi: 10.1105/tpc.111.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mantelin S., Peng H.C., Li B., Atamian H.S., Takken F.L., Kaloshian I. The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. Plant J. 2011;67:459–471. doi: 10.1111/j.1365-313X.2011.04609.x. [DOI] [PubMed] [Google Scholar]
- 62.Peng H.C., Mantelin S., Hicks G.R., Takken F.L., Kaloshian I. The conformation of a plasma membrane-localized somatic embryogenesis receptor kinase complex is altered by a potato aphid-derived effector. Plant Phsyiol. 2016;171:2211–2222. doi: 10.1104/pp.16.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu L., Ye M., Kuai P., Ye M., Erb M., Lou Y. OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 2018;219:1097–1111. doi: 10.1111/nph.15247. [DOI] [PubMed] [Google Scholar]
- 64.Gouhier-Darimont C., Stahl E., Glausen G., Reymond P. The Arabidopsis lectin receptor kinase LecRK-I.8 is involved in insect egg perception. Front. Plant Sci. 2019;10:623. doi: 10.3389/fpls.2019.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diaz-Riquelme J., Zhurov V., Rioja C., Perez-Moreno I., Torres-Perez R., Grimplet J., Carbonell-Bejerano P., Bajda S., Van Leeuwen T., Martinez-Zapater J., et al. Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranyhus urticae. BMC Genom. 2016;17:74. doi: 10.1186/s12864-016-2401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arnaiz A., Martinez M., Gonzalez-Melendi P., Grbic P., Diaz I., Santamaria M.E. Plant defenses against pests driven by a bidirectional promoter. Front. Plant Sci. 2019;10:930. doi: 10.3389/fpls.2019.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zebelo S.A., Maffei M.E. Role of early signalling events in plant-insect interactions. J. Exp. Bot. 2015;66:435–448. doi: 10.1093/jxb/eru480. [DOI] [PubMed] [Google Scholar]
- 68.Leitner M., Boland W., Mithöfer A. Direct and indirect defenses induced by piercing-sucking and chewing herbivores in Medicago truncatula. New Phytol. 2005;167:597–606. doi: 10.1111/j.1469-8137.2005.01426.x. [DOI] [PubMed] [Google Scholar]
- 69.Santamaria M.E., Cambra I., Martinez M., Pozancos C., Gonzalez-Melendi P., Grbic V., Castañera P., Ortego F., Diaz I. Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae. PLoS ONE. 2012;8:e43011. doi: 10.1371/journal.pone.0043011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santamaria M.E., Martinez M., Arnaiz A., Ortego F., Grbic V., Diaz I. MATI, a novel protein involved in the regulation of herbivore-associated signaling pathways. Front. Plant Sci. 2017;8:975. doi: 10.3389/fpls.2017.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mitler R. ROS are good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 72.Suzuki N., Miller G., Morales J., Shulaev V., Torres M.A., Mittler R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011;14:691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 73.Gilroy S., Bialasek M., Suzuki N., Gorecka M., Devireddy A.R., Karpinski S., Mittler R. ROS, Calcium, and electric signals: Key mediators of rapid systemic signalling in plants. Plant Physiol. 2016;171:1606–11615. doi: 10.1104/pp.16.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu J., Wang L., Wuensche H., Baldwin I.T. Narboh D, a respisratory burst oxidase homolog in Nicotiana attenuata, is required for late defense resposnes after herbivore attack. J. Integr. Plant Biol. 2013;55:187–198. doi: 10.1111/j.1744-7909.2012.01182.x. [DOI] [PubMed] [Google Scholar]
- 75.Moloi M.J., van der Westhuizen A.J. The reactive oxygen species are involved in resistance responses of wheat to the Russian wheat aphid. J. Plant Physiol. 2006;163:1118–1125. doi: 10.1016/j.jplph.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 76.Arimura G., Ozawa R., Maffei M.E. Recent advances in plant early signalling in response to herbivory. Int. J. Mol. Sci. 2011;12:3623–3739. doi: 10.3390/ijms12063723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., He J., Jiang L., Wu H., Xiao Y., Liu Y., Du Y., Liu C., Wan J. Nitric oxide production is associated with response to Brown planthopper infestation in rice. J. Plant Physiol. 2011;168:739–745. doi: 10.1016/j.jplph.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 78.Schweighofer A., Kazanaviciute V., Scheikl E., Teige M., Doczi R., Hirt H., Schwanninger M., Kant M., Schuurink R., Mauch F., et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell. 2007;19:2213–2224. doi: 10.1105/tpc.106.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ament K., Kant M.R., Sables M.W., Haring M.A., Schuurink R.C. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salycilate emission in tomato. Plant Physiol. 2004;135:2025–2037. doi: 10.1104/pp.104.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maserti B.E., del Carratore R., Della Croce C.M. Comparative analysis of proteome changes induced by the two-spotted spider mite Tetranychus urticae and methyl-jamonate in citrus leaves. J. Plant Physiol. 2011;168:392–402. doi: 10.1016/j.jplph.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 81.He W., Li H., Li X., Li M., Chen Y. Tetranychus urticae Koch induced accumulation of salicylic acid in frijole leaves. Pestic. Biochem. Physiol. 2007;88:78–81. doi: 10.1016/j.pestbp.2006.09.002. [DOI] [Google Scholar]
- 82.Wei J., van Loon J.J.A., Gols R., Menzel T.R., Li N., Kang L., Dicke M. Reciprocal crosstalk between jasmonate and salicylate defense-signalling pathways modulates plant volatile emission and herbivore host-selection behaviour. J. Exp. Bot. 2014;65:3289–3298. doi: 10.1093/jxb/eru181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arimura G., Ozawa R., Nishioka T., Boland W., Koch T., Kühnemann F., Takabayashi J. Herbivore-induced volatiles induce the emission of ethylene in neighboring lima bean plant. Plant J. 2002;29:87–98. doi: 10.1046/j.1365-313x.2002.01198.x. [DOI] [PubMed] [Google Scholar]
- 84.Barczak-Brzyzek A., Kiełkiewicz M., Gorecka M., Kot K., Karpinska B., Filipecki M. Abscisic acid insensitive 4 transcription factor is an important player in the response of Arabidopsis thaliana to two-spotted spider mite (Tetranychus urticae) feeding. Exp. Appl. Acarol. 2017;73:317–326. doi: 10.1007/s10493-017-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kielkiewicz M., Barczak-Brzyzek A., Karpinska B., Filipecki M. Unravelling the complexity of plant defense induced by a simultaneous and sequential mite and aphud insfestation. Int. J. Mol. Sci. 2019;20:806. doi: 10.3390/ijms20040806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verma V., Ravindran P., Kumar P. Plant hormone-medaited regulation od stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ku Y.S., Sintaha M., Cheung M.Y., Lam H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018;19:3206. doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li C.Y., Williams M.M., Loh Y.T., Lee G.I., Howe G.A. Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 2002;130:494–503. doi: 10.1104/pp.005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kant M.R., Sabelis M.W., Haring M.A., Schuurink R.C. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defenses. Proc. Biol. Sci. 2008;275:443–452. doi: 10.1098/rspb.2007.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arnaiz A., Talavera-Mateo L., Gonzalez-Melendi P., Martinez M., Diaz I., Santamaria M.E. Kunitz trypsin inhibitors in defense against spider mites. Front. Plant Sci. 2018;9:986. doi: 10.3389/fpls.2018.00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carrillo L., Martinez M., Ramessar K., Cambra I., Castañera P., Ortego F., Diaz I. Expression of a barley cystatin gene in maize enhances resistance against phytophagous mites by altering their cysteine-proteases. Plant Cell Rep. 2011;30:101–112. doi: 10.1007/s00299-010-0948-z. [DOI] [PubMed] [Google Scholar]
- 92.Hata F.T., Bega V.L., Ventura M.U., dos Santos F., Poloni da Silva J.E., Machado R.R., Sousa V. Plant acceptance for oviposition of Tetranychus urticae on strawberry leaves is influenced by aromatic plants in laboratory and greenhouse intercropping experiments. Agronomy. 2020;10:193. doi: 10.3390/agronomy10020193. [DOI] [Google Scholar]
- 93.Blaazer C.J.H., Villacis-Perez E., Chafi R., Van Leeuwen T., Kant M.R., Schimmel B.C.J. Why do herbivorous mites suppress plant defenses? Front. Plant Sci. 2018;9:1057. doi: 10.3389/fpls.2018.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Snoeck S., Wybouw N., van Leeuwen T., Dermauw W. Transcriptomic plasticity in the arthropod generalist Tetranychus urtciae upon long-term acclimation to different host plants. Genes Genomes Genet. 2018;8:3865–3879. doi: 10.1534/g3.118.200585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wybouw N.T., van Leeuwen T., Dermauw W. A massive incorporation of microbial genes into the genome of Tetranycus urticae, a polyphagous arthropod herbivore. Insect Mol. Biol. 2018;27:333–351. doi: 10.1111/imb.12374. [DOI] [PubMed] [Google Scholar]
- 96.Alba J.M., Schimmel B.C.J., Glas J.J., Ataide L.M., Pappas M.L., Villarroel C.A., Schuurink R.C., Sabelis M.W., Kant M.R. Spider mites suppress tomato defenses downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytol. 2015;205:828–840. doi: 10.1111/nph.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sarmento R.A., Lemos F., Bleeker P.M., Schuurink R.C., Pallini A., Oliveira M.G.A., Lima E.R., Kant M.R., Sabelis M.W., Janssen A. A herbivore that manipulates plant defense. Ecol. Lett. 2011;14:229–236. doi: 10.1111/j.1461-0248.2010.01575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]