Abstract
Leaf senescence is an essential physiological process that is accompanied by the remobilization of nutrients from senescent leaves to young leaves or other developing organs. Although leaf senescence is a genetically programmed process, it can be induced by a wide variety of biotic and abiotic factors. Accumulating studies demonstrate that senescence-associated transcription factors (Sen-TFs) play key regulatory roles in controlling the initiation and progression of leaf senescence process. Interestingly, recent functional studies also reveal that a number of Sen-TFs function as positive or negative regulators of plant immunity. Moreover, the plant hormone salicylic acid (SA) and reactive oxygen species (ROS) have been demonstrated to be key signaling molecules in regulating leaf senescence and plant immunity, suggesting that these two processes share similar or common regulatory networks. However, the interactions between leaf senescence and plant immunity did not attract sufficient attention to plant scientists. Here, we review the regulatory roles of SA and ROS in biotic and abiotic stresses, as well as the cross-talks between SA/ROS and other hormones in leaf senescence and plant immunity, summarize the transcriptional controls of Sen-TFs on SA and ROS signal pathways, and analyze the cross-regulation between senescence and immunity through a broad literature survey. In-depth understandings of the cross-regulatory mechanisms between leaf senescence and plant immunity will facilitate the cultivation of high-yield and disease-resistant crops through a molecular breeding strategy.
Keywords: leaf senescence, plant immunity, hormone, Sen-TF, cross-regulation
1. Introduction
The leaf is the organ that uses the photosynthetic system to convert light into sugars, thus providing energy for plant growth and development. As leaf age increases, the photosynthesis ability gradually decreases and then enters its final stage: leaf senescence [1]. In Arabidopsis, leaf senescence occupies nearly half of the leaf life history from the onset of senescence to completely dead [2]. Although leaf senescence is basically controlled by developmental age, it can be induced by a variety of internal signals, including phytohormones and reproduction, and external factors, such as darkness, UV-B or ozone, nutrient limitation, heat or cold, drought, high salinity, and pathogen attacks [2]. Leaf senescence is critical for plants’ fitness and survival because plants reallocate nutrients from senescent leaves to young leaves and other organs during age- or stress-induced senescence [3,4].
Various molecular, physiological, and biochemical events in cells are genetically regulated in an orderly manner in the entire developmental stage of leaves. Cell cycle, cell wall biogenesis, macromolecule biosynthesis, and photosynthesis are active in the stage before the initiation of senescence. Nevertheless, cell wall disassembly, anthocyanin biosynthesis, and amino acid transport mainly occur during leaf senescence [2]. The metabolic and gene expression profile of cell changes orderly when the leaf senescence is initiated. In addition, the contents of phytohormones, such as ethylene (ET), jasmonates (JA), salicylic acid (SA), and abscisic acid (ABA) increase as leaf ages, whereas gibberellic acid (GA) and cytokinin (CK) decrease, suggesting that they play differential roles in regulating leaf senescence. Reactive oxygen species (ROS) accumulate as leaf ages [5] and function as positive players in leaf senescence [6]. Eventually, leaf senescence displays a programmed cell death (PCD)-like event, which is also termed as developmentally controlled PCD (dPCD) [7].
Plants respond to pathogen attacks using an innate immune system that is broadly divided into pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is initiated by which plants respond to PAMP through cell surface-localized pattern recognition receptors (PRRs) [8]. PTI causes rapid defense responses, such as cell wall enhancement, ion flux, ROS production, mitogen-activated protein kinase (MAPK) cascades, hormone networks, and the expression of defense-related genes [9]. However, successful pathogens can secrete effectors to interfere with PTI signaling pathways. In turn, the hosts use intracellular receptors to recognize effectors, resulting in ETI [8]. ETI usually causes enhanced defense response, such as hypersensitive response (HR), which is a form of PCD.
Plant pathogens are generally divided into biotrophs and necrotrophs according to their lifestyles [10]. Biotrophic pathogens feed nutrients from living host tissues, whereas necrotrophic pathogens take nutrients from dead cells destroyed by their secreted phytotoxins and cell-wall-degrading enzymes. In addition, some pathogens are called hemibiotrophs, because they apply two lifestyles during infection. In plant-pathogen interactions, pathogen-triggered PCD (pPCD) is an effective strategy to limit pathogen spread, especially for biotrophic pathogens. However, pPCD would cause nutrient leakage, which is a benefit for necrotrophic pathogens growth. For pathogens, biotrophic pathogens repress the cell death process, whereas the necrotrophic pathogens promote this case. Thus, plants manipulate their cell death process to cope with pathogens with different lifestyles [11].
PCD is also a typical characteristic of leaf senescence. Although there are many differences between dPCD and pPCD, they are controlled by partial common components [12]. Intriguingly, dPCD-accelerated plants generally show enhanced resistance against biotrophic and hemibiotrophic pathogens, but impaired resistance to necrotrophic pathogens. Nevertheless, dPCD-delayed plants are the opposite (Summarized in Section 3). dPCD-accelerated plants are usually accompanied with higher accumulation of SA and/or ROS. SA and ROS are closely linked to PCD and they are required for plant resistance against biotrophic and hemibiotrophic pathogens. Moreover, they are required for the establishment of both dPCD and pPCD [13]. Thus, SA, ROS, as well as PCD, seem to function as key nodes connecting leaf senescence and plant immunity. In this paper, we review the regulatory roles of SA and ROS on leaf senescence and plant immunity, summarize the transcriptional controls of Sen-TFs on SA and ROS signal pathways, and outline the cross-regulation between leaf senescence and plant immunity by manual curation.
2. The Roles of SA and ROS in Leaf Senescence and Plant Immunity
SA is a well-known hormone for plant disease resistance and is required for systemic acquired resistance (SAR), and plays an important role in the amplification of PTI and ETI [14]. SA-induced and age-dependent gene expression changes exhibit a high degree of overlap in genome-wide transcriptome analysis [2], indicative of the dual roles of SA in leaf senescence and plant immunity. Isochorismate synthase (ICS) mainly synthesizes SA. Arabidopsis genome contains two ICS genes, ICS1 (also known as SID2) and ICS2. Age- and pathogen-induced SA is synthesized through the ICS1 [5,15]. ICS1 is rapidly induced by pathogen attack and it is essential for the establishment of SAR [15,16]. Different from ICS1, ICS2 contributes to the synthesis of basal SA in plants [17]. The mutation in ICS1 leads to more susceptible to Pseudomonas syringae pv. tomato (Pst) pathogens [15]. However, the local resistance against Botrytis cinerea is involved in SA in an ICS1-independent manner [18]. Moreover, the sid2 mutants display delayed leaf senescence phenotypes as compared to Col-0 plants [5]. SA is further perceived by NPR1 (Non-expressor of PR gene 1), which is regarded as a SA receptor demonstrated by recent studies and is responsible for the SA signaling pathway [19,20]. Mutation in NPR1 impairs its function in promoting SA-induced defense gene expression and thereby causes decreased resistance to pathogens [20]. However, the resistance of npr1 mutants to necrotroph Alternaria brassicicola shows a similar level when compared with WT plants [21]. The function of NPR1 in regulating leaf senescence is obvious, as the mutation of NPR1 delays the reaching of chlorotic stage [22]. SAG12, which is a marker gene of leaf senescence, is considerably reduced in npr1 mutants compared to Col-0 plants. In addition, EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1) and PAD4 (PHYTOALEXIN DEFICIENT4) form a heterodimer to regulate plant immunity and are indispensable for the accumulation of SA induced by virulent pathogens [23]. EDS1 and PAD4 positively regulate disease resistance against Pst pathogens [23], but virus-induced gene silencing of EDS1 enhances the tobacco resistance to B. cinerea [24]. Similar to npr1, the pad4 mutant exhibits delayed senescence phenotypes when compared with Col-0 plants [22], and PAD4 positively modulates green peach aphid feeding-induced leaf senescence [25]. The transcriptional level of ICS1 increases during leaf senescence. Consistently, SA content accumulates as leaf aging and has been used as an important inducer of leaf senescence [5]. Thus, the SA signaling pathway plays critical roles in both plant immunity and leaf senescence.
ROS play an integral role in the regulation of numerous biological processes, including leaf senescence and responses to pathogen attacks [13]. ROS are defined as oxygen-containing molecules with higher chemical reactivity than O2. The major forms of ROS in plants include superoxide anion, singlet oxygen, hydroxyl radical, and hydrogen peroxide (H2O2). Among them, H2O2 is the most stable and generally acts as a signal molecule to trigger downstream responses. ROS can be produced in various subcellular compartments, such as plasma membrane, cell wall, mitochondria, chloroplasts, and so on [26]. In general, intracellular ROS are produced in chloroplasts, mitochondria, and peroxisomes, and the apoplastic ROS are produced by plasma membrane-localized NADPH oxidases (RBOHs, respiratory burst oxidase homologs) and cell wall peroxidases, under both normal and stress conditions [26]. ROS act as double-edged swords in cellular processes. Low level of ROS functions as signaling molecules in response to biotic and abiotic stresses, while a high level of ROS causes PCD or even necrosis [27]. In plant-pathogen interaction, an increased production of ROS occurs during PTI and ETI after pathogen recognition and generally contributes to disease resistance. Furthermore, early accumulated ROS could directly strengthen the cell wall and act as signal molecules to activate defense responses [13]. ROS act in combination with SA to regulate the SAR establishment. Changes in the redox status cause the movement of NPR1 to the nucleus to induce defense gene expression [15]. A number of key senescence-associated genes (SAGs), such as ORE1 [28], WRKY53 [29], and WRKY75 [5], are transcriptionally up-regulated in response to H2O2 stimulus. ROS also accumulate during senescence and function as positive players of leaf senescence [5,6].
SA can induce the accumulation of H2O2, and vice versa. SA directly inactivates H2O2 scavengers, such as catalases (CATs) and ascorbates peroxidases (APXs), resulting in the accumulation of H2O2 [30]. Conversely, the exogenous application of H2O2 could induce SA biosynthesis in tobacco [31]. Furthermore, SA and ROS act synergistically to trigger both dPCD and pPCD [12]. PCD plays an essential role in immunity and development. However, dPCD exhibits some distinct characteristics that differ from pPCD. dPCD is an active and long-term process, while pPCD is a passive and rapid process. HR, which is the best-characterized pPCD, is an efficient strategy against pathogen attack. Pathogenesis-related (PR) genes are associated with HR, and HIN1 (hairpin-induced gene 1) is an HR marker gene [32]. Notably, these genes are also induced in an age-dependent manner [33], indicating that some common pathways or cross-talks exist between dPCD and pPCD at the molecular level.
3. The Regulators of SA and ROS
3.1. Transcription Factors
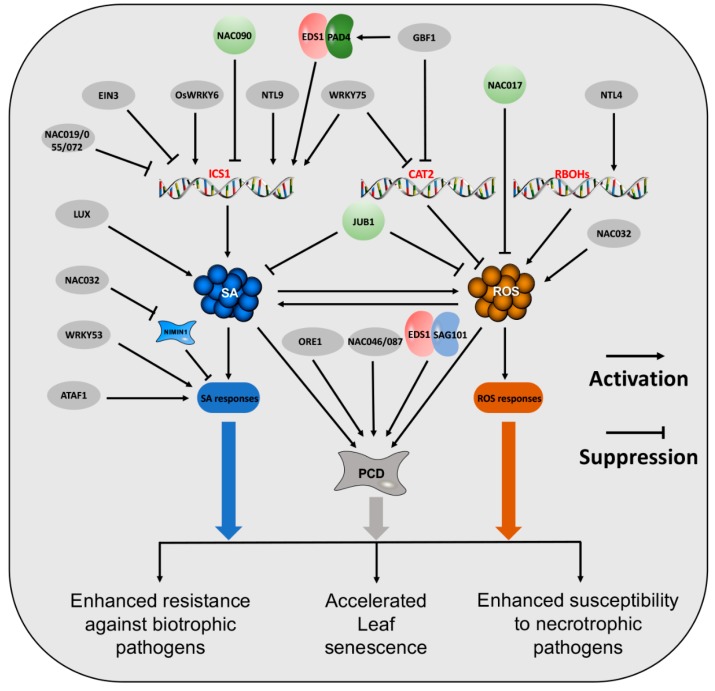
The Arabidopsis developmental transcriptome based on RNA-seq profiling reveals that a large number of genes are upregulated during senescence [34], which are SAGs. Molecular genetic studies demonstrate that numerous SAGs function as key regulators of leaf senescence [35,36]. Intriguingly, many SAGs have also been reported to be implicated in plant immunity. Among them, NAC, WRKY, TCP, MYB, and bZIP family members are associated with leaf senescence and plant resistance, suggesting a pivotal significance of Sen-TFs in the regulation of these two processes [36,37]. As SA and ROS play important roles in the regulation of leaf senescence and plant disease resistance, and ICS1, RBOHs and CAT2 control age- and pathogen-induced SA and ROS biosynthesis and/or metabolism, respectively, they are subjected to complex regulation by Sen-TFs at the transcriptional level (Figure 1).
Figure 1.
Cross-Regulation Network between Leaf Senescence and Plant Immunity Mediated by Salicylic Acid (SA) and Reactive Oxygen Species (ROS). SA, ROS, and programmed cell death (PCD) play a positive role in the regulation of leaf senescence and resistance against biotrophic pathogens but a negative function for plant resistance to necrotrophic pathogens. TFs that positively regulate SA, ROS and/or PCD, including WRKY75, NTL9, GBF1, and ORE1, also positively regulate leaf senescence and plant immunity. Negative regulators, such as JUB1, represses SA and ROS as well as leaf senescence and plant immunity. EIN3 and NAC019/055/072 directly bind to the promoter of ICS1 and suppress its expression, resulting in a decrease of SA production. Abbreviations: LUX, LUX ARRHYTHMO; NAC, NAM/ATAF/CUC; EIN, ETHYLENE INSENSITIVE; EDS, ENHANCED DISEASE SUSCEPTIBILITY; PAD, PHYTOALEXIN DEFICIENT; WRKY; GBF1, G-BOX BINDING FACTOR 1; NTL, NAC with transmembrane motif 1-like; JUB1, JUNGBRUNNEN1; SA, salicylic acid; ROS, reactive oxygen species; NIMIN, NIM1 INTERACTING; ORE, ORESARA; SAG, Senescence-associated Gene; ATAF, ACTIVATING FACTOR; PCD, programmed cell death; CAT2, CATALASE 2; ICS, isochorismate synthase; RBOH, respiratory burst oxidase homolog.
3.1.1. NAC TFs
The plant-specific NAC TF family has more than 100 members in Arabidopsis [38], and many of them are involved in the regulation of leaf senescence and plant immunity (Table 1). Some positive regulators of leaf senescence, such as ORE1 [28], NTL9 [39], NAC032 [40]], and ATAF1 [41], also play important roles in plant immunity. The mutations of these genes delay leaf senescence, while overexpressing plants display precocious senescence phenotypes. Moreover, these mutants support a higher level of hemibiotrophic pathogens, such as Pst DC3000. The overexpression of these genes confers plant resistance against Pst DC3000, but susceptibility to the necrotrophic pathogens, such as B. cinerea [42,43,44,45]. ORE1 has been shown to mediate PCD via directly targeting and activating a senescence-enhanced gene BIFUNCTIONAL NUCLEASE 1 (BFN1), which is associated with PCD [46]. Similarly, NAC087/046 control dPCD by regulating BFN1 in Arabidopsis lateral root cap [47]. Furthermore, BnaNAC87, the homolog of NAC087 in Brassica napus, acts as a positive regulator of ROS metabolism and cell death [48] and it binds the promoter of HIN1 [32,48]. However, the function of NAC087/046 in plant immunity remains unknown. The P. syringae type III effector HopD1 could target NTL9 to suppress ETI and contributes to its virulence [43], indicating that NTL9 is an important component in Arabidopsis immune system against Pst pathogens. Indeed, NTL9 directly binds the promoter of ICS1 in yeast one-hybrid assays and it is required for the induction of ICS1 by flg22 treatment [49]. ANAC032 activates SA signaling by repressing NIMIN1, a key negative regulator of SA-dependent defense, and promotes H2O2 accumulation under stress [40,45]. Additionally, the overexpression of Gossypium hirsutum ATAF1 in cotton also activates SA signaling [44], although the mechanism is unclear.
Table 1.
List of the genes that are involved in leaf senescence and plant immunity.
| Family | Gene | AGI Code | SAG | Leaf Senescence | Resistance a | Resistance b |
|---|---|---|---|---|---|---|
| NAC | ORE1 | AT5G39610 | Y | Positive [28] | Positive [42] | |
| NTL9 | AT4G35580 | Y | Positive [39] | Positive [43] | ||
| NAC032 | AT1G77450 | Y | Positive [40] | Positive [45] | ||
| ATAF1 | AT1G01720 | Y | Positive [41] | Positive [50,51] | Negative [44,52,53] | |
| ANAC046/087 | AT3G04060/AT5G1827 | Y | Positive [54] | |||
| JUB1 | AT2G43000 | Y | Negative [6] | Negative [55] | Positive [56] | |
| NAC017/082/090 | AT1G34190/AT5G09330/AT5G22380 | Y | Negative [57] | |||
| NAC019/055/072 | AT1G52890/AT3G15500/AT4G27410 | Y | Positive [58,59] | Negative [60] | Negative [61] | |
| ORS1 | AT3G29035 | Y | Positive [62] | |||
| NTL4 | AT3G10500 | Y | Positive [63,64] | |||
| WRKY | WRKY53 | AT4G23810 | Y | Positive [29] | Positive [65] | |
| WRKY54/70 | AT2G40750/AT3G56400 | Y | Negative [66] | Positive [67] | Negative [67] | |
| WRKY6 | AT1G62300 | Y | Positive [68] | Positive [68,69] | ||
| WRKY22 | AT4G01250 | N1 | Positive [70] | Positive [71] | ||
| WRKY18/40/60 | AT4G31800/AT1G80840/AT2G25000 | Y | Negative [72] | Negative [73,74] | Positive [74] | |
| WRKY75 | AT5G13080 | Y | Positive [5] | Positive [5] | ||
| bZIP | GBF1 | AT4G36730 | Y | Positive [75] | Positive [76] | |
| MYB | LUX | AT3G46640 | Y | Negative [77] | Positive [78] | |
| TCP | TCP19 | AT5G51910 | N1 | Negative [79] | ||
| TCP20 | AT3G27010 | Y | Negative [79] | |||
| Non-TF | SAG101 | AT5G14930 | Y | Positive [80] | Positive [23] | |
| HLS1 | AT4G37580 | N | Negative [81] | Positive [81] | ||
| JMJ16 | AT1G08620 | Y | Negative [82] | |||
| MPK3 | AT3G45640 | N1 | Positive [83] | Positive [84] | Negative [85] | |
| MPK6 | AT2G43790 | Y | Positive [83] | Positive [84] | Negative [85] | |
| FTSH4 | AT2G26140 | Y | Negative [86] |
a Resistance ability to biotrophic and hemi-biotrophic pathogens. b Resistance ability to necrotrophic pathogens. N1 indicates that the gene is down-regulated during senescence. N represents that the transcriptional level of the gene exhibits no significant changes during senescence. The data of the reference [34] are used for SAGs identification.
JUNGBRUNNEN1 (JUB1, NAC042) is an H2O2-induced NAC TF and functions as a negative regulator of longevity in Arabidopsis [6]. The overexpression of JUB1 causes the accumulation of DELLA proteins and antagonizes SA pathway, resulting in impaired resistance to Pst DC3000, but increases resistance against necrotrophic fungus A. brassicicola [55]. Additionally, higher H2O2 accumulation is observed in jub1 mutants compared to Col-0 plants, whereas overexpressing JUB1 could decrease the cellular H2O2 level, suggesting that JUB1 is involved in the regulation of biosynthesis and/or metabolism of H2O2 [6]. Recently, NAC017, NAC082, and NAC090 have been reported to govern the positive-to-negative regulatory shift in leaf senescence, which is referred to as a “NAC troika”. A single mutant of nac017, nac082, or nac090 accelerates cell death and leaf senescence, while overexpressing lines show the opposite effect, indicative of their negative regulatory roles in leaf senescence [57]. NAC090 exhibits a predominant role in repressing SA accumulation and responses by directly binding the promoters of target genes ICS1 and EDS5. Different from NAC090, ANAC017 predominantly suppresses ROS levels.
NAC019, NAC055, and NAC072 (RD26) positively regulate age-dependent leaf senescence [58]. However, their roles in plant immunity are complicated. The anac019 anac055 anac072 triple mutant shows increased resistance to P. syringae pv. maculicola (Psm) ES4326 [60]. In contrast, anac019 anac055 double mutant exhibits enhanced resistance to necrotrophic fungus B. cinerea, while the overexpression of ANAC019 or ANAC055 has the opposite effects [61]. ANAC019, ANAC055, and ANAC072 function redundantly in the suppression of SA accumulation by repressing the expression of ICS1 [49], suggesting their promotive role of leaf senescence in a SA-independent manner. Indeed, ANAC019 and ANAC055 function as transcription activators to regulate the expression of JA-related defense genes in a COI1- and MYC2-dependent manner [61].
In addition to the above-mentioned transcription factors, several NAC TFs, such as NAC059 (ORS1) [62] and NTL4 [63], have only been implicated in the positive regulation of leaf senescence, which cannot rule out the possibility that they are also involved in plant immunity. Microarray-based expression profiling by using estradiol-inducible ORS1 overexpression lines reveals that several potential targets of ORS1, such as WRKY40 [74], WRKY75 [5], FMO1 [87,88], and ALD1 [89], play critical roles in plant immunity. ChIP assays show that NTL4 induces the accumulation of ROS by directly targeting the promoters of ROS biosynthesis-related genes RBOHs to control PCD during heat- and drought-induced leaf senescence [63,64], which implies that NTL4 is involved in drought/heat stress-triggered disease resistance.
3.1.2. WRKY-TFs
WRKY TFs are one of the largest families of plant-specific TFs, with more than 70 members in Arabidopsis [90]. Although WRKY family TFs are well known for their functions in plant immunity, they are also implicated in the regulation of leaf senescence (Table 1). WRKY53, WRKY75, WRKY22, Oryza sativa WRKY6 (OsWRKY6), and Chimonanthus praecox WRKY71 (CpWRKY71) are positive regulators of leaf senescence [5,29,69,70,91] and/or disease resistance against hemibiotrophic pathogens [5,65,69]. Of them, WRKY53, WRKY75, and OsWRKY6 are involved in the regulation of SA and/or ROS signaling (Figure 1). WRKY53 is partially involved in the SA-signaling pathway [92], and it interacts with the JA-inducible protein EPITHIOSPECIFYING SENESCENCE REGULATOR (ESR/ESP) to antagonistically regulate SA-JA signaling during leaf senescence [93]. WRKY75 promotes SA production by directly binding to the W-box (TTGACT) sequence in the promoter of ICS1 to activate its transcription. Moreover, WRKY75 suppresses the transcription of CAT2 to repress H2O2 scavenging, which results in an accumulation of ROS level. OsWRKY6 directly binds and positively regulates OsICS1 to enhance SA accumulation, leading to constitutive activation of several PR genes [69].
As a negative regulator of leaf senescence, WRKY18 directly binds the W-boxes of WRKY53 promoter and suppresses its expression [72]. WRKY18 acts with its closely related homologs WRKY40 and WRKY60 to redundantly regulate disease resistance. The double mutants wrky18 wrky40 and wrky18 wrky60, as well as the triple mutant wrky18 wrky40 wrky60 display enhanced resistance to P. syringae, but are more susceptible to B. cinerea than Col-0 plants. Moreover, the double mutant wrky18 wrky40 is more resistant to the biotrophic powdery mildew fungus Golovinomyces orontii [73,74]. The triple mutant wrky18 wrky40 wrky60 exhibits a higher level of SA-responsive gene PR1, implying that they negatively regulate SA signaling.
WRKY70 and WRKY54 act redundantly to negatively regulate leaf senescence in Arabidopsis [66], functioning as negative regulators of SA biosynthesis, but positive regulators of SA-mediated gene expression [94]. WRKY70 acts as a common component in both SA- and JA-mediated signal pathways, which is activated by SA and repressed by JA [95]. The elevated expression of WRKY70 results in impaired resistance to necrotroph A. brassicicola, whereas enhances the resistance against biotroph Erysiphe cichoracearum [67], suggesting the dual roles of WRKY54 and WRKY70 in repressing SA biosynthesis and transducing the SA signal.
3.1.3. Other TFs
In addition to NAC and WRKY TF families, other TF family members have also been reported to regulate leaf senescence and plant immunity (Table 1). GBF1, a bZIP protein, controls leaf senescence by directly regulating PAD4 [76] and binding the promoter of CAT2 to suppress its transcription, thus resulting in the decreased H2O2-scavenging activity [75]. Accordingly, the gbf1 mutant shows a decreased H2O2 level and delayed senescence phenotypes [75]. The GBF1 overexpressing plants are more resistant, whereas gbf1 is susceptible, to Pst pathogens when compared to Col-0 [76]. The circadian clock pathway is known to regulate leaf senescence and plant innate immunity [77,96]. One of the core clock components LUX (LUX ARRHYTHMO, also called PHYTOCLOCK1), an MYB family TF, is essential for circadian rhythmicity that functions as a negative regulator of leaf senescence. The leaf of lux-2 shows earlier yellowing as compared with Col-0 [77], and lux mutants display impaired disease resistance against P. syringae and SA- and JA-mediated defense signaling [78]. TCP TFs constitute a small family of plant-specific TFs that play important roles in plant development [97]. The tcp19 or tcp20 single mutant shows slightly early senescence phenotype in the dark, whereas tcp19 tcp20 double mutant displays significantly accelerated senescence, suggesting that TCP19 and TCP20 redundantly regulate leaf senescence [79]. Although the resistance phenotype of tcp19 tcp20 has yet to be studied, the TCP19 was found to bind the promoters of ICS1, PBS3 (avrPphB SUSCEPTIBLE 3), PAD4, and EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1), indicative of their possible involvement in plant immunity [49].
3.2. Non-TFs
In addition to TFs, other types of genes are also involved in the regulation of leaf senescence and disease resistance (Table 1). SAG101, a gene encoding an acyl hydrolase, plays a positive role in leaf senescence [80]. SAG101 has been demonstrated to function as an important component in plant immunity by forming a heterodimer with EDS1, thus mediating TNL-mediated PCD [23]. Arabidopsis HLS1 (HOOKLESS1) encodes a putative histone acetyltransferase and negatively regulates dark-, pathogen-, and ABA-induced leaf senescence [81]. A higher ROS level is observed in ABA-induced senescing leaves in the hls1 mutant. The loss of HLS1 function leads to an increased susceptibility to B. cinerea with larger disease lesions, enhanced necrosis, and chlorosis when compared to controls [81]. However, hls1 mutants also display enhanced disease symptoms after inoculation with Pst DC3000 (AvrRpm1), but they show a level of bacterial growth under Pst DC3000 inoculation, suggesting that HLS1 manipulates defense responses associated with ETI [81]. JMJ16 is a JmjC domain-containing protein and it is a specific H3K4 demethylase in Arabidopsis. The expression of two positive regulators of leaf senescence, WRKY53 and SAG201, is repressed by JMJ16 in an age-dependent manner through reducing H3K4me3 levels at these loci [82]. The jmj16 mutants display an early senescence phenotype, suggesting that JMJ16 is an important epigenetic regulator of leaf senescence via demethylating H3K4 at SAGs in an age-dependent manner. RNA_seq and ChIP_seq reveal that lots of potential target genes of JMJ16, such as WRKY33, CBP60G, EDS5, and PR5, are associated with plant immunity. However, the disease resistance phenotype of jmj16 is not reported so far.
Two Arabidopsis MAPKs, MPK3 and MPK6, play crucial roles in response to biotic and abiotic stresses. MPK3/6 are involved in PTI and ETI [84], and mpk6 mutants show delayed senescence phenotypes [83]. MAPK phosphatase 2 (MKP2) could functionally interact with MPK3/6 and dephosphorylate them, and regulate oxidative stress as well as pathogen defense responses [85]. Contrary to the mpk6 mutants, the MKP2-suppressed line (MPK2_RNAi) displays early senescence phenotypes [35,83]. Another protein phosphatase, phosphatase 2A subunit PP2A-B′γ, was recently reported to negatively regulate leaf senescence and resistance against necrotroph B. cinerea, suggesting that MKP2 and PP2A regulate disease resistance against necrotrophs in different mechanisms [98].
Arabidopsis FtSH4, a mitochondrial AAA-protease, mediates autophagy and senescence [86], and the ftsh4 mutant shows a significantly increased SA level and accelerated leaf senescence and cell death. Although the function of FtSH4 in disease resistance has not been studied, several SA biosynthesis and signaling genes, such as SID2, NDR1, and NPR1, are upregulated in the ftsh4 mutant. Furthermore, the loss of SID2, NDR1, or NPR1 functions in the ftsh4 mutant restores the early senescence phenotypes. In addition, several WRKY genes, including WRKY40, WRKY46, WRKY51, WRKY60, WRKY63, and WRKY75, display elevated expression levels in ftsh4 mutant. Together, the above data imply that FtSH4 plays a negative role in plant immunity.
3.3. The Differences between Age- and Pathogen-Induced SA and ROS
Age- and pathogen-induced changes in SA and ROS can be regulated by a large number of Sen-TFs, as mentioned above. Although Sen-TFs are involved in these two processes, there are still significant differences between them. The spatial and temporal expression of Sen-TFs is strictly regulated, and their transcript levels are gradually upregulated during leaf senescence. Consistently, the contents of SA and ROS gradually increase as the leaf ages. By contrast, pathogen-evoked acute induction of Sen-TFs gene expression as well as the contents of SA/ROS would usually return to basal levels after a certain period of time.
The physiological state of plants, such as age, has complex and delicate regulation in gene expression. For example, the “NAC troika” precisely governs the time-dependent regulatory shift for NAC-TFs in Arabidopsis during leaf senescence. In addition to age, the functional differences of the common regulatory components of SA and ROS are likely dependent on the tissue specificity of gene expression. For instance, NTL9 functions in guard cells for flg22-triggered induction of SA synthesis- related genes and is required for stomatal immunity [49]. However, NTL9 might regulate leaf senescence by regulating the expression of a subset of SAGs in other cell types, such as mesophyll cells. Multi-omics analysis of specific cell types (single-cell sequencing) would contribute to the understanding of the functional differences of Sen-TFs on leaf senescence and plant immunity.
4. Cross-Talks between SA/ROS and Other Hormones during Leaf Senescence and Plant Immunity
Other phytohormones generally regulate senescence and immune responses by antagonistically or synergistically interacting with the SA/ROS signaling pathway because SA and ROS act as central players in leaf senescence and plant immunity [99]. Cross-talks between SA/ROS and other hormones are discussed here, and the key components of plant hormone pathways involved in leaf senescence and/or plant immunity are summarized in Table 2 and shown in Figure 2.
Table 2.
List of phytohormone signal components in leaf senescence and plant immunity.
| Hormone | Component | Function | AGI Code | Role in Leaf Senescence | Role in Resistance a | Role in Resistance b |
|---|---|---|---|---|---|---|
| ET | EIN2 | Involved in ethylene signal transduction | AT5G03280 | Positive [100] | Ambiguous [42,101,102] | Negative [103] |
| EIN3 | Key transcription factor that initiates downstream transcriptional cascades for ethylene responses | AT3G20770 | Positive [100] | Negative [102] | Positive [104] | |
| EIL1 | EIN3-like 1 | AT2G27050 | Positive [100] | Negative [102] | Positive [104] | |
| JA | COI1 | Key component required for JA-induced transcriptional regulation | AT2G39940 | Positive [105,106] | Negative [107] | Positive [21,108] Negative [103,109] |
| JAZ7 | Protein constitutes JA receptor complex with COI1 | AT2G34600 | Negative [110] | Positive [111] | Negative [111] | |
| MYC2 | A MYC-related transcriptional activator interacted with JAZs | AT1G32640 | Positive [112] | Negative [113] | Negative [114,115] | |
| SA | ICS1 | Required for SA biosynthesis | AT1G74710 | Positive [5] | Positive [15] | Non [18] |
| NPR1 | Key regulator of SA-mediated disease resistance | AT1G64280 | Positive [22] | Positive [20] | Non [21] | |
| EDS1 | Component of R gene-mediated disease resistance | AT3G48090 | Positive [22,25] | Positive [23] | Negative [24] | |
| PAD4 | A lipase-like protein that is important for SA signaling.It can interact with EDS1 | AT3G52430 | Positive [22,25] | [23] | ||
| ABA | PYL9 | ABA receptor | AT1G01360 | Positive [116] | ||
| SnRK2.8 | A member of SNF1-related protein kinase involved in ABA signaling | AT1G78290 | Positive [116] | Positive [117] | ||
| ABI5 | A bZIP transcription factor involved in ABA signaling | AT2G36270 | Positive [118] | Non [81] | ||
| GA | DELLAs | Repressor of GA responses and involved in GA-mediated signaling | AT1G14920 | Negative [119] | Negative [120] | Positive [120] |
| CK | AHK3 | Cytokinin receptor | AT1G27320 | Positive [121] | Positive [122] | |
| ARR2 | A transcription factor involved in the cytokinin signaling pathway | AT4G16110 | Non [123] | Positive [122] | ||
| Auxin | TIR1 | Auxin receptor | AT3G62980 | Positive [124] | ||
| ARF2 | An auxin response factor | AT5G62000 | Positive [125,126] | |||
| BR | BRI1 | A plasma membrane localized leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction | AT4G39400 | Positive [11] | Negative [127] |
a Resistance ability to biotrophic and hemi-biotrophic pathogens. b Resistance ability to necrotrophic pathogens.
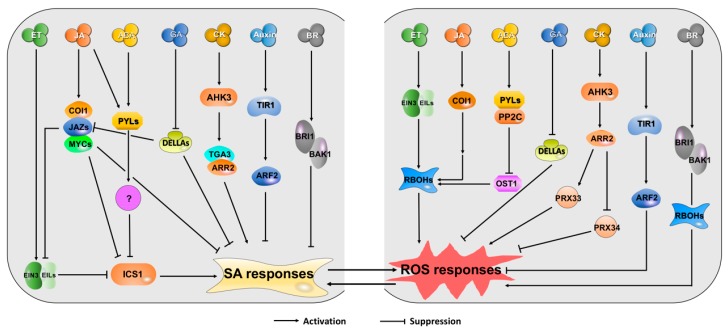
Figure 2.
SA and ROS Are the Hubs that Integrate Regulatory Signals from Plant Hormone Pathways to Control Leaf Senescence and Plant Immunity. Cross-talks between SA/ROS and other hormones are mainly modulated by key components in their signaling pathways. The SA-dependent responses are suppressed by JA, ET, and ABA via repressing SA-related genes such as biosynthesis gene ICS1. Auxin and BRs also antagonize SA signaling pathway. Additionally, GA and CK mainly synergistically interact with SA. Meanwhile, besides Auxin, ET, JA, ABA, GA, CK, and BR can contribute positively to ROS responses. SA can trigger the accumulation of ROS, and vice versa. Abbreviations: ET, ethylene; JA, jasmonic acid; ABA, abscisic acid; GA, gibberellin; CK, Cytokinin; BR, Brassinosteroid.
ET synergistically interacts with JA in plants in response to stresses and generally antagonizes SA signaling [128]. However, the molecular details of the antagonism of JA/ET signaling on the SA pathway are still poorly understood. JA signal is perceived through JA receptor complex that constituted the F-box protein CORONATINE INSENSITIVE 1 (COI1) and JASMONATE ZIM-domain (JAZ) proteins, and activates various downstream TFs, including the IIIe bHLH TFs MYC2, MYC3, and MYC4. Pst pathogen secretes a mimic of JA-Ile virulence factor coronatine (COR) to suppress SA-dependent defenses by activating MYC2 and NAC019/055/072, which repress SA synthesis and/or promote SA metabolism [60]. Consistently, an elevated level of SA is found in the coi1 mutant [107]. JA induces stomatal closure mediated by ROS-dependent signaling pathway. JA increases the ROS level via the NAD(P)H oxidase pathway, but JA-induced ROS accumulation is not observed in coi1 or jar1-1 mutants [129]. ET is perceived by its receptors and then leads to the activation of ETHYLENE-INSENSITIVE 2 (EIN2) [130] and the core TFs EIN3/EIL1 (EIN3-LIKE 1) [131]. EIN3/EIL1 thus activate a large number of downstream genes. For example, EIN3/EIL1 directly target the promoter of SID2 and negatively regulate the accumulation of SA. The ein3 eil1 double mutant thus constitutively accumulates SA, and ROS production is diminished in ethylene-insensitive mutants under Flg22 treatment [132]. In rice, OsEIL1 directly binds the promoters of OsrbohA/OsrbohB to activate their transcription [133]. ET synergistically acts with JA to activate the expression of ERF1 in response to necrotrophic pathogens [99]. JA promotes ABA signaling by inducing the expressions of ABA receptors, such as PYL4 (PYRABACTIN RESISTANCE/PYR1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR (PYR/PYL/RCAR)). Conversely, ABA promotes JA signaling by regulating the expressions of MYC TFs in response to the herbivorous insect [134,135]. As the key components of GA signaling, DELLA proteins interact with JAZ1 to activate MYC-dependent gene expressions in response to stresses [136]. Auxin enhances JA-induced expressions of defense-related genes and JA-Auxin module is involved in resistance to necrotrophic pathogens [137]. However, JA inhibits apical growth of roots and auxin has the opposite effect, suggesting they antagonistically regulate root growth. A possible model is that JA represses the expressions of auxin-responsive TFs, PLETHORAs (PLTs), which are responsible for stem cell niche maintenance [138]. JA antagonistically interacts with CKs to regulate xylem development. CKs negatively regulate xylem differentiation, while JA treatment promotes the formation of extra xylem [139].
Recent studies have greatly expanded our understandings of the cross-talks between ABA and other hormones. ABA antagonizes plant immune responses by repressing SA responses. The ICS1 transcriptional level is down-regulated in ABA receptor mutant pyl. In rice, ABA significantly suppresses the expression levels of OsWRKY45 and OsNPR1 [140]. Meanwhile, increasing evidence indicates that ABA also regulates ROS production through RBOHF. In the presence of ABA, PYLs inhibit PP2Cs, resulting in activation of one of SNF1-related protein kinases (SnRK2s), OPEN STOMATA 1 (OST1), to phosphorylate the N terminus of RBOHF, thus increasing the ROS level [141]. MYC2 is a critical and positive node in the crosstalk between JA and ABA signaling pathways. ABA interacts with JA signaling via PYL6-MYC2 to regulate cotyledon expansion [142] and the tolerance to drought [143]. Moreover, the Plant U-box (PUB) E3 ligase PUB10 directly interacts with MYC2 to destabilize it, negatively regulating both JA and ABA responses [144]. The antagonistic interactions between ABA and ET have been reported in seed dormancy and germination [145]. One of the mechanisms is that ABA inhibits the transcript level and activity of ACO1 [146]. The SnRK2-APC/CTE module is a regulatory hub that is involved in the cross–talks between GA and ABA [147]. Rice Tiller Enhancer (TE), encoding an activator of the APC/CTE E3 ubiquitin ligase complex, interacts with and degrades ABA receptor OsPYLs. SnRK2 phosphorylates TE to interrupt the interaction between TE and OsPYLs. GA reduces SnRK2s levels and inhibits APC/CTE-mediated degradation of OsPYLs. ABI4 positively regulates ARR5 (Arabidopsis RESPONSE REGULATOR 5) expression, a negative regulator of CK signaling, but it suppresses the transcript level of PIN-FORMED 1 (PIN1), indicating that ABI4 mediates the cross-talks among ABA, CK, and auxin [148]. SnRK2.3 interacts with and phosphorylates HAT1, an important transcriptional regulator in BR signaling, to repress its protein stability and binding activity, which increases expressions of ABA-responsive genes and then enhances tolerance to drought, indicating that a relationship between ABA and BR signaling pathways exists [149]. The cross-talks between ABA and other hormones in plant stress responses are discussed in details in several recently published papers [150,151].
GAs, CKs, auxins, and BRs also play important roles in plant development and immune system. GAs regulate these two processes by mediating the degradation of DELLA proteins [152]. DELLAs sequester the JA signaling repressor JAZ family proteins, leading to the activation of MYC2 and JA signaling. Meanwhile, DELLAs also directly antagonize the SA-mediated defense [120]. The accumulation of DELLAs that is induced by stresses causes the enhanced activity of ROS scavenging enzyme and suppresses ROS levels [153]. CKs are reported as modulators of plant immunity by interacting with SA. ARR2, a key TF of CK signaling pathway, interacts with TGA3 and positively enhances the expression of PR1 gene, suggesting that CKs can also function in the SA signaling pathway [154]. CKs receptor HISTIDINE KINASE3 (AHK3) and ARR2 promote PAMP-triggered stomatal closure and ROS accumulation. ARR2 directly activates the expressions of peroxidases PRX33 and PRX34, but not RBOHs, which are required for SA- and PAMP-triggered ROS production [122]. Auxins regulate plant development and plant immunity via repressing the SA signaling pathway [99]. Biotrophic pathogen evolves strategies to produce auxin or manipulate auxin signaling to suppress host SA signaling, thus contributing to its virulence [155]. Auxins limit the H2O2 levels through restraining H2O2 generation to be implicated in the regulation of stomatal opening. BRs are a unique class of plant steroid hormones that mediate growth and development. BRs signals are perceived and transduced by the receptor-like kinase BRASSINOSTEROID INSENSITIVE1 (BRI1) and BRI1-ASSOCIATED KINASE1 (BAK1). Pathogen Pythium graminicola manipulates plant BR pathway to antagonize SA- and GA-mediated defenses to promote its infection [156]. Meanwhile, BRs activate NADPH oxidase activity, which results in an accumulation of H2O2 levels in apoplast [157].
Phytohormones SA, ET, JA, ABA, and BRs play positive roles in regulating leaf senescence, whereas CK, auxins, and GA delay this process [11]. The contents of SA, JA, ET, and ABA increase during leaf senescence and promote this process [1], suggesting that these hormones could synergistically work on leaf senescence process, although many studies describe an antagonistic interaction between the SA and JA/ET/ABA pathways. The application of low concentrations of JA and SA leads to a synergistic effect on the JA- and SA-responsive genes PDF1.2 and PR1, respectively. In addition, a high concentration of JA is required for the induction of H2O2 accumulation, thus JA might promote leaf senescence by partially interacting with ROS [99].
5. Roles of SA/ROS in Biotic and Abiotic Stresses
SA and ROS play important roles in the regulation of plant development as well as biotic and abiotic stress responses (Figure 3).
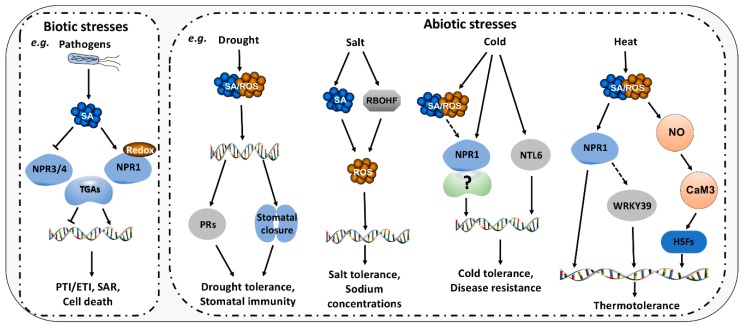
Figure 3.
Roles of SA/ROS in Biotic and Abiotic Stresses. In biotic stresses, SA accumulation occurs after pathogen attack. Subsequently, SA binds to and activates NPR1 that interacts with TGAs to induce defense-related genes. Meanwhile, ROS is required for changes in redox status of NPR1. Besides, SA represses the transcriptional inhibition activities of NPR3/NPR4 to release defense-related genes. The production of SA/ROS is also triggered by various abiotic stresses including drought, salt cold and heat. SA/ROS further regulates NPR1-dependent or NPR1-independent gene expression. Plasma membrane-located NTL6 is relocated into the nuclear to induce a subset of PR genes in response to cold stress. Heat stress-induced ROS cause the accumulation of NO that activates CaM3 and results in DNA targeting of HSFs. TGA, TGACG motif-binding factor; PTI, PAMP-triggered immunity; ETI, Effector-triggered immunity; SAR, systemic acquired resistance; PRs, Pathogenesis-related genes; RBOHF, respiratory burst oxidase homolog; NTL6, NAC with transmembrane motif 1-like; NO, Nitric Oxide; CaM3, Calmodulin 3; HSFs, Heat shock transcription factors.
5.1. Roles of SA/ROS in Biotic Stresses
Biotic stresses (focused on pathogen attacks) cause changes of SA and ROS contents by multiple layers of transcriptional regulation (Figure 1). SA is perceived by its potential receptor NPR1 and NPR3/4 [20]. NPR1 or NPR3/4 interacts with a number of TGA TFs, including TGA2/5/6 to positively or negatively regulate SA-induced PR genes expression and disease resistance, respectively [20,158]. Pathogen-induced SA accumulation promotes the activity of NPR1 to further induce the expression of defense-related genes, while SA represses the transcriptional inhibition activities of NPR3/NPR4 to release defense-related genes. In plant-pathogen interactions, the production of ROS is one of the fastest physiological responses observed in plants after MAMP recognition by PRRs. ROS can induce Ca2+ influx into the cells, activate MAPK cascades, and regulate the redox of TFs, thus activate the expression of defense-related genes [159,160]. Rapid production of ROS leads to HR or HR-like necrosis that contributes to limiting the spread of biotrophs, but it generally benefits the necrotrophs infection [9,10].
5.2. Roles of SA/ROS in Abiotic Stresses
Drought, salt, cold, and heat are the most common adverse abiotic stresses that seriously influence plant growth and development. Water deficit increases the levels of SA/ROS, and drought stress induces SA-responsive PR genes [161], suggesting that SA/ROS might play a key role in drought tolerance. The mutation of a SIZ-type small ubiquitin-related modifier (SUMO) E3 ligase causes stomatal closure and then enhances drought tolerance by elevating SA/ROS accumulation [161]. It should be noted that plants can close stomata via SA/ROS signaling after PAMP recognition, termed as stomatal immunity [49,162], implying that SA/ROS are involved in drought stress and plant immunity.
SA is also involved in salt tolerance. The germination of sid2 seeds is hypersensitive to salt stress. The endogenous H2O2 is elevated in wild type and sid2 seeds under high salinity. However, the level of H2O2 is reduced after treatment with SA, suggesting that SA plays an essential role in seed germination under salt stress by balancing ROS [163]. The loss-of-function of RBOHF decreases salinity-induced ROS and increases sodium concentrations in root vasculature, indicating that RBOHF functions in salinity-induced vasculature-specific ROS accumulation and salt tolerance [164].
Cold stress also increases the levels of SA/ROS [165], indicative of a close link between cold stress and immune responses. In Arabidopsis, NTL6 has been demonstrated to be an important regulator in linking cold signals with pathogenesis [166]. Cold could enhance disease resistance by activating NTL6, which upregulates several PR genes by directly binding their promoters, such as PR1, PR2, and PR5. However, NTL6-mediated PR genes expression is independent of SA [166]. Recently, NPR1 was found to mediate a novel regulatory pathway in cold acclimation independently of SA and TGA [167]. However, the exogenous application of SA improves the cold tolerance in maize, cucumber, and rice [168].
Benzothiadiazole (BTH), an SA analogue, can induce thermosensitive genes, indicating that SA is involved in heat stress [169]. Application of BTH induces bacterial resistance in a NPR1-dependent manner [169]. cpr5 (constitutive expressor of PR genes), a SA constitutive accumulation mutant, displays a high thermotolerance, while SA-deficient NahG transgenic plants exhibit an opposite phenotype. The overexpression of WRKY39 enhances thermotolerance by upregulating SA-related genes [170]. ROS accumulation is required for heat stress responses in plants. Heat stress-induced ROS are mainly produced in chloroplasts by RBOHB and RBOHD. ROS causes the accumulation of nitric oxide (NO), which subsequently activates Calmodulin 3 (CaM3), and then enhances the binding activity of HSFs (heat shock transcription factors) [171].
6. Concluding Remarks
Numerous signaling components are involved in both leaf senescence and plant immunity, which suggests that these two events are not independent biological processes and share partial regulatory networks. It is apparent that SA and ROS play important and overlapping roles in leaf senescence and disease resistance. The co-players promote leaf senescence and immune response by converging on the SA and ROS pathways, which explains the phenomenon that the early-senescing mutants with increased SA and/or ROS levels are more resistant to biotrophic pathogens, but more susceptible to necrotrophic pathogens. The relationships between these two processes have not been systematically studied, despite outstanding progresses have been made over the past few decades in the individual field of leaf senescence and plant immunity. Based on our above-mentioned gene network analysis, it is reasonable to speculate that a large number of SAGs are also involved in regulating plant immunity, and vice versa. Thus, in-depth understandings of the cross-regulatory mechanisms between leaf senescence and plant immunity will facilitate the cultivation of high-yield and disease-resistant crops through molecular breeding strategy.
Acknowledgments
We sincerely apologize to those authors whose work is not included in this review due to space limitations.
Author Contributions
Y.Z., Z.L. and H.G. conceived and wrote the manuscript. Y.Z. and H.-L.W. collected the data and organized the tables and the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from the National Natural Science Foundation of China [31570286 to H.G.; 31970196 to Z.L.; 31900173 to H.W.], Chinese Postdoctoral Science Foundation [2019M650514 to Y. Z. and 2019M650516 to H.W.], the startup funding for plant aging research from “Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Forestry University”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Woo H.R., Kim H.J., Lim P.O., Nam H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019;70:347–376. doi: 10.1146/annurev-arplant-050718-095859. [DOI] [PubMed] [Google Scholar]
- 2.Lim P.O., Kim H.J., Gil Nam H. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 3.Masclaux C., Valadier M.H., Brugière N., Morot-Gaudry J.F., Hirel B. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta. 2000;211:510–518. doi: 10.1007/s004250000310. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe M., Balazadeh S., Tohge T., Erban A., Giavalisco P., Kopka J., Mueller-Roeber B., Fernie A.R., Hoefgen R. Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol. 2013;162:1290–1310. doi: 10.1104/pp.113.217380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo P., Li Z., Huang P., Li B., Fang S., Chu J., Guo H. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell. 2017;29:2854–2870. doi: 10.1105/tpc.17.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu A., Allu A.D., Garapati P., Siddiqui H., Dortay H., Zanor M.I., Asensi-Fabado M.A., Munné-Bosch S., Antonio C., Tohge T., et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24:482–506. doi: 10.1105/tpc.111.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daneva A., Gao Z., Van Durme M., Nowack M.K. Functions and regulation of programmed cell death in plant development. Annu. Rev. Cell Dev. Biol. 2016;32:441–468. doi: 10.1146/annurev-cellbio-111315-124915. [DOI] [PubMed] [Google Scholar]
- 8.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 9.Yu X., Feng B., He P., Shan L. From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 2017;55:109–137. doi: 10.1146/annurev-phyto-080516-035649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glazebrook J. Contrasting Mechanisms of Defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 11.Häffner E., Konietzki S., Diederichsen E. Keeping control: The role of senescence and development in plant pathogenesis and defense. Plants. 2015;4:449–488. doi: 10.3390/plants4030449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huysmans M., Lema A S., Coll N.S., Nowack M.K. Dying two deaths - programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 2017;35:37–44. doi: 10.1016/j.pbi.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Torres M.Á., Jones J.D.G., Dangl J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Li X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019;50:29–36. doi: 10.1016/j.pbi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Cao H., Bowling S.A., Gordon A.S., Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.2307/3869945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Z.Q., Dong X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 17.Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J.-P. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008;147:1279–1287. doi: 10.1104/pp.108.119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari S., Plotnikova J.M., De Lorenzo G., Ausubel F.M. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003;35:193–205. doi: 10.1046/j.1365-313X.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y., Zhang D., Chu J.Y., Boyle P., Wang Y., Brindle I.D., De Luca V., Després C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1:639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y., Sun T., Ao K., Peng Y., Zhang Y., Li X., Zhang Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 2018;173:1454–1467.e15. doi: 10.1016/j.cell.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 21.Thomma B.P., Eggermont K., Penninckx I.A., Mauch-Mani B., Vogelsang R., Cammue B.P., Broekaert W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris K., MacKerness S.A., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- 23.Lapin D., Kovacova V., Sun X., Dongus J.A., Bhandari D.D., von Born P., Bautor J., Guarneri N., Rzemieniewski J., Stuttmann J., et al. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell. 2019;31:2430–2455. doi: 10.1105/tpc.19.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Oirdi M., Bouarab K. Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol. 2007;175:131–139. doi: 10.1111/j.1469-8137.2007.02086.x. [DOI] [PubMed] [Google Scholar]
- 25.Pegadaraju V. Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol. 2005;139:1927–1934. doi: 10.1104/pp.105.070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waszczak C., Carmody M., Kangasjärvi J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018;69:209–236. doi: 10.1146/annurev-arplant-042817-040322. [DOI] [PubMed] [Google Scholar]
- 27.Van Breusegem F., Dat J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006;141:384–390. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.I., Köhler B., Mueller-Roeber B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010;62:250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]
- 29.Miao Y., Laun T., Zimmermann P., Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004;55:853–867. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 30.Rao M.V., Paliyath G., Ormrod D.P., Murr D.P., Watkins C.B. Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 1997;115:137–149. doi: 10.1104/pp.115.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leon J., Lawton M.A., Raskin I. Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol. 1995;108:1673–1678. doi: 10.1104/pp.108.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frías M., González C., Brito N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011;192:483–495. doi: 10.1111/j.1469-8137.2011.03802.x. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Zhang Y., Zou D., Zhao Y., Wang H.L., Zhang Y., Xia X., Luo J., Guo H., Zhang Z. LSD 3.0: A comprehensive resource for the leaf senescence research community. Nucleic Acids Res. 2020;48:D1069–D1075. doi: 10.1093/nar/gkz898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D., Penin A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- 35.Li Z., Peng J., Wen X., Guo H. Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 2012;54:526–539. doi: 10.1111/j.1744-7909.2012.01136.x. [DOI] [PubMed] [Google Scholar]
- 36.Li Z., Zhao Y., Liu X., Peng J., Guo H., Luo J. LSD 2.0: An update of the leaf senescence database. Nucleic Acids Res. 2014;42:D1200–D1205. doi: 10.1093/nar/gkt1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.-F., Wu S.-H., Swidzinski J., Ishizaki K., et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 38.Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Yoon H.K., Kim S.G., Kim S.Y., Park C.M. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells. 2008;25:438–445. [PubMed] [Google Scholar]
- 40.Mahmood K., El-Kereamy A., Kim S.-H., Nambara E., Rothstein S.J. ANAC032 positively regulates age-dependent and stress-induced senescence in Arabidopsis thaliana. Plant Cell Physiol. 2016;57:2029–2046. doi: 10.1093/pcp/pcw120. [DOI] [PubMed] [Google Scholar]
- 41.Garapati P., Xue G.P., Munné-Bosch S., Balazadeh S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015;168:1122–1139. doi: 10.1104/pp.15.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Daoud F., Cameron R.K. ANAC055 and ANAC092 contribute non-redundantly in an EIN2-dependent manner to age-related resistance in Arabidopsis. Physiol. Mol. Plant Pathol. 2011;76:212–222. doi: 10.1016/j.pmpp.2011.09.003. [DOI] [Google Scholar]
- 43.Block A., Toruño T.Y., Elowsky C.G., Zhang C., Steinbrenner J., Beynon J., Alfano J.R. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014;201:1358–1370. doi: 10.1111/nph.12626. [DOI] [PubMed] [Google Scholar]
- 44.He X., Zhu L., Xu L., Guo W., Zhang X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 2016;35:2167–2179. doi: 10.1007/s00299-016-2027-6. [DOI] [PubMed] [Google Scholar]
- 45.Allu A.D., Brotman Y., Xue G.-P., Balazadeh S. Transcription factor ANAC032 modulates JA/SA signalling in response to Pseudomonas syringae infection. EMBO Rep. 2016;17:1578–1589. doi: 10.15252/embr.201642197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matallana-Ramirez L.P., Rauf M., Farage-Barhom S., Dortay H., Xue G.P., Dröge-Laser W., Lers A., Balazadeh S., Mueller-Roeber B. NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant. 2013;6:1438–1452. doi: 10.1093/mp/sst012. [DOI] [PubMed] [Google Scholar]
- 47.Huysmans M., Buono R.A., Skorzinski N., Radio M.C., De Winter F., Parizot B., Mertens J., Karimi M., Fendrych M., Nowack M.K. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell. 2018;30:2197–2213. doi: 10.1105/tpc.18.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan J., Tong T., Li X., Chen Q., Dai M., Niu F., Yang M., Deyholos M.K., Yang B., Jiang Y.-Q. A novel NAC-type transcription factor, NAC87, from oilseed rape modulates reactive oxygen species accumulation and cell death. Plant Cell Physiol. 2018;59:290–303. doi: 10.1093/pcp/pcx184. [DOI] [PubMed] [Google Scholar]
- 49.Zheng X.Y., Zhou M., Yoo H., Pruneda-Paz J.L., Spivey N.W., Kay S.A., Dong X. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. USA. 2015;112:9166–9173. doi: 10.1073/pnas.1511182112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen M.K., Rung J.H., Gregersen P.L., Gjetting T., Fuglsang A.T., Hansen M., Joehnk N., Lyngkjaer M.F., Collinge D.B. The HvNAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007;65:137–150. doi: 10.1007/s11103-007-9204-5. [DOI] [PubMed] [Google Scholar]
- 51.Nakashima K., Tran L.-S.P., Van Nguyen D., Fujita M., Maruyama K., Todaka D., Ito Y., Hayashi N., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B., Zhao Q., Zhang L., Li Y., Yang C., et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19:1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- 53.Wang X., Basnayake B.M.V.S., Zhang H., Li G., Li W., Virk N., Mengiste T., Song F. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. MPMI. 2009;22:1227–1238. doi: 10.1094/MPMI-22-10-1227. [DOI] [PubMed] [Google Scholar]
- 54.Oda-Yamamizo C., Mitsuda N., Sakamoto S., Ogawa D., Ohme-Takagi M., Ohmiya A. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 2016;6:23609. doi: 10.1038/srep23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shahnejat-Bushehri S., Nobmann B., Devi Allu A., Balazadeh S. JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav. 2016;11:e1181245. doi: 10.1080/15592324.2016.1181245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saga H., Ogawa T., Kai K., Suzuki H., Ogata Y., Sakurai N., Shibata D., Ohta D. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. MPMI. 2012;25:684–696. doi: 10.1094/MPMI-09-11-0244. [DOI] [PubMed] [Google Scholar]
- 57.Kim H.J., Park J.-H., Kim J., Kim J.J., Hong S., Kim J., Kim J.H., Woo H.R., Hyeon C., Lim P.O., et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2018;115:E4930–E4939. doi: 10.1073/pnas.1721523115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hickman R., Hill C., Penfold C.A., Breeze E., Bowden L., Moore J.D., Zhang P., Jackson A., Cooke E., Bewicke-Copley F., et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013;75:26–39. doi: 10.1111/tpj.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S., Gao J., Yao L., Ren G., Zhu X., Gao S., Qiu K., Zhou X., Kuai B. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 2016;35:1–14. doi: 10.1007/s00299-016-1991-1. [DOI] [PubMed] [Google Scholar]
- 60.Zheng X.Y., Spivey N.W., Zeng W., Liu P.P., Fu Z.Q., Klessig D.F., He S.Y., Dong X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11:587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bu Q., Jiang H., Li C.B., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Li C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008;18:756–767. doi: 10.1038/cr.2008.53. [DOI] [PubMed] [Google Scholar]
- 62.Balazadeh S., Kwasniewski M., Caldana C., Mehrnia M., Zanor M.I., Xue G.P., Mueller-Roeber B. ORS1, an H2O2-Responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant. 2011;4:346–360. doi: 10.1093/mp/ssq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee S., Seo P.J., Lee H.J., Park C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012;70:831–844. doi: 10.1111/j.1365-313X.2012.04932.x. [DOI] [PubMed] [Google Scholar]
- 64.Lee S., Lee H.J., Huh S.U., Paek K.H., Ha J.H., Park C.M. The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 2014;227:76–83. doi: 10.1016/j.plantsci.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Murray S.L., Ingle R.A., Petersen L.N., Denby K.J. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. MPMI. 2007;20:1431–1438. doi: 10.1094/MPMI-20-11-1431. [DOI] [PubMed] [Google Scholar]
- 66.Besseau S., Li J., Palva E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J., Brader G., Kariola T., Palva E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006;46:477–491. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- 68.Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi C., Hwang S.H., Fang I.R., Kwon S.I., Park S.R., Ahn I., Kim J.B., Hwang D.J. Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015;208:846–859. doi: 10.1111/nph.13516. [DOI] [PubMed] [Google Scholar]
- 70.Zhou X., Jiang Y., Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 2011;31:303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abbruscato P., Nepusz T., Mizzi L., del Corvo M., Morandini P., Fumasoni I., Michel C., Paccanaro A., Guiderdoni E., Schaffrath U., et al. OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol. Plant Pathol. 2012;13:828–841. doi: 10.1111/j.1364-3703.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Potschin M., Schlienger S., Bieker S., Zentgraf U. Senescence Networking: WRKY18 is an Upstream Regulator, a Downstream Target Gene, and a Protein Interaction Partner of WRKY53. J. Plant Growth Regul. 2013;33:106–118. doi: 10.1007/s00344-013-9380-2. [DOI] [Google Scholar]
- 73.Schön M., Töller A., Diezel C., Roth C., Westphal L., Wiermer M., Somssich I.E. Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. MPMI. 2013;26:758–767. doi: 10.1094/MPMI-11-12-0265-R. [DOI] [PubMed] [Google Scholar]
- 74.Xu X., Chen C., Fan B., Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smykowski A., Zimmermann P., Zentgraf U. G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol. 2010;153:1321–1331. doi: 10.1104/pp.110.157180. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Giri M.K., Singh N., Banday Z.Z., Singh V., Ram H., Singh D., Chattopadhyay S., Nandi A.K. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J. 2017;91:802–815. doi: 10.1111/tpj.13608. [DOI] [PubMed] [Google Scholar]
- 77.Kim H., Kim H.J., Vu Q.T., Jung S., McClung C.R., Hong S., Nam H.G. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2018;115:8448–8453. doi: 10.1073/pnas.1722407115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang C., Gao M., Seitz N.C., Angel W., Hallworth A., Wiratan L., Darwish O., Alkharouf N., Dawit T., Lin D., et al. LUX ARRHYTHMO mediates cross-talk between the circadian clock and defense in Arabidopsis. Nat. Commun. 2019;10:2543. doi: 10.1038/s41467-019-10485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danisman S., van Dijk A.D.J., Bimbo A., van der Wal F., Hennig L., de Folter S., Angenent G.C., Immink R.G.H. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013;64:5673–5685. doi: 10.1093/jxb/ert337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He Y., Gan S. A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell. 2002;14:805–815. doi: 10.1105/tpc.010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao C.-J., Lai Z., Lee S., Yun D.-J., Mengiste T. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell. 2016;28:1662–1681. doi: 10.1105/tpc.16.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu P., Zhang S., Zhou B., Luo X., Zhou X.F., Cai B., Jin Y.H., Niu D., Lin J., Cao X., et al. The histone H3K4 demethylase JMJ16 represses leaf senescence in Arabidopsis. Plant Cell. 2019;31:430–443. doi: 10.1105/tpc.18.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou C., Cai Z., Guo Y., Gan S. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009;150:167–177. doi: 10.1104/pp.108.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodriguez M.C.S., Petersen M., Mundy J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 85.Lumbreras V., Vilela B., Irar S., Solé M., Capellades M., Valls M., Coca M., Pagès M. MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 2010;63:1017–1030. doi: 10.1111/j.1365-313X.2010.04297.x. [DOI] [PubMed] [Google Scholar]
- 86.Zhang S., Li C., Wang R., Chen Y., Shu S., Huang R., Zhang D., Li J., Xiao S., Yao N., et al. The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 2017;173:2294–2307. doi: 10.1104/pp.16.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hartmann M., Zeier T., Bernsdorff F., Reichel-Deland V., Kim D., Hohmann M., Scholten N., Schuck S., Bräutigam A., Hölzel T., et al. Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell. 2018;173:456–469.e16. doi: 10.1016/j.cell.2018.02.049. [DOI] [PubMed] [Google Scholar]
- 88.Chen Y.C., Holmes E.C., Rajniak J., Kim J.G., Tang S., Fischer C.R., Mudgett M.B., Sattely E.S. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2018;115:E4920–E4929. doi: 10.1073/pnas.1805291115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song J.T., Lu H., McDowell J.M., Greenberg J.T. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- 90.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 91.Huang R., Liu D., Huang M., Ma J., Li Z., Li M., Sui S. CpWRKY71, a WRKY transcription factor gene of Wintersweet (Chimonanthus praecox), promotes flowering and leaf senescence in Arabidopsis. Int. J. Mol. Sci. 2019;20:5325. doi: 10.3390/ijms20215325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu Y., Dong Q., Yu D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012;185–186:288–297. doi: 10.1016/j.plantsci.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 93.Miao Y., Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 2007;19:819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang D., Amornsiripanitch N., Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J. The WRKY70 Transcription Factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu H., McClung C.R., Zhang C. Tick tock: Circadian regulation of plant innate immunity. Annu. Rev. Phytopathol. 2017;55:287–311. doi: 10.1146/annurev-phyto-080516-035451. [DOI] [PubMed] [Google Scholar]
- 97.Martín-Trillo M., Cubas P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 98.Durian G., Jeschke V., Rahikainen M., Vuorinen K., Gollan P.J., Brosché M., Salojärvi J., Glawischnig E., Winter Z., Li S., et al. PROTEIN PHOSPHATASE 2A-B’γ controls Botrytis cinerea resistance and developmental leaf senescence. Plant Physiol. 2019;182:1161–1181. doi: 10.1104/pp.19.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pieterse C.M.J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 100.Li Z., Peng J., Wen X., Guo H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell. 2013;25:3311–3328. doi: 10.1105/tpc.113.113340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zipfel C. Combined roles of ethylene and endogenous peptides in regulating plant immunity and growth. Proc. Natl. Acad. Sci. USA. 2013;110:5748–5749. doi: 10.1073/pnas.1302659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen H., Xue L., Chintamanani S., Germain H., Lin H., Cui H., Cai R., Zuo J., Tang X., Li X., et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009;21:2527–2540. doi: 10.1105/tpc.108.065193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trusov Y., Sewelam N., Rookes J.E., Kunkel M., Nowak E., Schenk P.M., Botella J.R. Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J. 2008;58:69–81. doi: 10.1111/j.1365-313X.2008.03755.x. [DOI] [PubMed] [Google Scholar]
- 104.Berrocal-Lobo M., Molina A., Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- 105.He Y., Fukushige H., Hildebrand D.F., Gan S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002;128:876–884. doi: 10.1104/pp.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee S.-H., Sakuraba Y., Lee T., Kim K.-W., An G., Lee H.Y., Paek N.-C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015;57:562–576. doi: 10.1111/jipb.12276. [DOI] [PubMed] [Google Scholar]
- 107.Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001;26:509–522. doi: 10.1046/j.1365-313x.2001.01050.x. [DOI] [PubMed] [Google Scholar]
- 108.Wang C., Ding Y., Yao J., Zhang Y., Sun Y., Colee J., Mou Z. Arabidopsis elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 2015;83:1019–1033. doi: 10.1111/tpj.12946. [DOI] [PubMed] [Google Scholar]
- 109.Thatcher L.F., Manners J.M., Kazan K. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 2009;58:927–939. doi: 10.1111/j.1365-313X.2009.03831.x. [DOI] [PubMed] [Google Scholar]
- 110.Yu J., Zhang Y., Yuan J.S., Xu W., Su Z. JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 2016;67:751–762. doi: 10.1093/jxb/erv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thatcher L.F., Cevik V., Grant M., Zhai B., Jones J.D.G., Manners J.M., Kazan K. Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J. Exp. Bot. 2016;67:2367–2386. doi: 10.1093/jxb/erw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qi T., Wang J., Huang H., Liu B., Gao H., Liu Y., Song S., Xie D. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell. 2015;27:1634–1649. doi: 10.1105/tpc.15.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cui H., Qiu J., Zhou Y., Bhandari D.D., Zhao C., Bautor J., Parker J.E. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant. 2018;11:1053–1066. doi: 10.1016/j.molp.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 114.Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kazan K., Manners J.M. MYC2: The master in action. Mol. Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 116.Zhao Y., Chan Z., Gao J., Xing L., Cao M., Yu C., Hu Y., You J., Shi H., Zhu Y., et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA. 2016;113:1949–1954. doi: 10.1073/pnas.1522840113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee H.J., Park Y.J., Seo P.J., Kim J.H., Sim H.J., Kim S.G., Park C.M. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell. 2015;27:3425–3438. doi: 10.1105/tpc.15.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Su M., Huang G., Zhang Q., Wang X., Li C., Tao Y., Zhang S., Lai J., Yang C., Wang Y. The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci. 2016;247:93–103. doi: 10.1016/j.plantsci.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 119.Chen M., Maodzeka A., Zhou L., Ali E., Wang Z., Jiang L. Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci. 2014;219–220:26–34. doi: 10.1016/j.plantsci.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 120.Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D.G. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 121.Kim J., Park S.J., Lee I.H., Chu H., Penfold C.A., Kim J.H., Buchanan-Wollaston V., Nam H.G., Woo H.R., Lim P.O. Comparative transcriptome analysis in Arabidopsis ein2/ore3 and ahk3/ore12 mutants during dark-induced leaf senescence. J. Exp. Bot. 2018;69:3023–3036. doi: 10.1093/jxb/ery137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arnaud D., Lee S., Takebayashi Y., Choi D., Choi J., Sakakibara H., Hwang I. Cytokinin-mediated regulation of reactive oxygen species homeostasis modulates stomatal immunity in Arabidopsis. Plant Cell. 2017;29:543–559. doi: 10.1105/tpc.16.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim H.J., Ryu H., Hong S.H., Woo H.R., Lim P.O., Lee I.C., Sheen J., Nam H.G., Hwang I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim J.I., Murphy A.S., Baek D., Lee S.-W., Yun D.-J., Bressan R.A., Narasimhan M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011;62:3981–3992. doi: 10.1093/jxb/err094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lim P.O., Lee I.C., Kim J., Kim H.J., Ryu J.S., Woo H.R., Nam H.G. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 2010;61:1419–1430. doi: 10.1093/jxb/erq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellis C.M., Nagpal P., Young J.C., Hagen G., Guilfoyle T.J., Reed J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 127.Goddard R., Peraldi A., Ridout C., Nicholson P. Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare) MPMI. 2014;27:1095–1106. doi: 10.1094/MPMI-03-14-0069-R. [DOI] [PubMed] [Google Scholar]
- 128.Kunkel B.N., Brooks D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002;5:325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 129.Munemasa S., Oda K., Watanabe-Sugimoto M., Nakamura Y., Shimoishi Y., Murata Y. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 2007;143:1398–1407. doi: 10.1104/pp.106.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ju C., Yoon G.M., Shemansky J.M., Lin D.Y., Ying Z.I., Chang J., Garrett W.M., Kessenbrock M., Groth G., Tucker M.L., et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mersmann S., Bourdais G., Rietz S., Robatzek S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010;154:391–400. doi: 10.1104/pp.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang C., Li W., Cao J., Meng F., Yu Y., Huang J., Jiang L., Liu M., Zhang Z., Chen X., et al. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 2017;89:338–353. doi: 10.1111/tpj.13388. [DOI] [PubMed] [Google Scholar]
- 134.Lackman P., González-Guzmán M., Tilleman S., Carqueijeiro I., Pérez A.C., Moses T., Seo M., Kanno Y., Häkkinen S.T., Van Montagu M.C.E., et al. Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. USA. 2011;108:5891–5896. doi: 10.1073/pnas.1103010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fernández-Calvo P., Chini A., Fernández-Barbero G., Chico J.-M., Gimenez-Ibanez S., Geerinck J., Eeckhout D., Schweizer F., Godoy M., Franco-Zorrilla J.M., et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hou X., Xia K., Yan Y. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 137.Qi L., Yan J., Li Y., Jiang H., Sun J., Chen Q., Li H., Chu J., Yan C., Sun X., et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012;195:872–882. doi: 10.1111/j.1469-8137.2012.04208.x. [DOI] [PubMed] [Google Scholar]
- 138.Chen Q., Sun J., Zhai Q., Zhou W., Qi L., Xu L., Wang B., Chen R., Jiang H., Qi J., et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell. 2011;23:3335–3352. doi: 10.1105/tpc.111.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jang G., Yoon Y., Choi Y.D. Crosstalk with Jasmonic Acid Integrates Multiple Responses in Plant Development. Int. J. Mol. Sci. 2020;21:305. doi: 10.3390/ijms21010305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang C.-J., Shimono M., Sugano S., Kojima M., Yazawa K., Yoshida R., Inoue H., Hayashi N., Sakakibara H., Takatsuji H. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in Rice– Magnaporthe grisea interaction. MPMI. 2010;23:791–798. doi: 10.1094/MPMI-23-6-0791. [DOI] [PubMed] [Google Scholar]
- 141.Qi J., Wang J., Gong Z., Zhou J.-M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017;38:92–100. doi: 10.1016/j.pbi.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 142.Aleman F., Yazaki J., Lee M., Takahashi Y., Kim A.Y., Li Z., Kinoshita T., Ecker J.R., Schroeder J.I. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci. Rep. 2016;6:28941. doi: 10.1038/srep28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ollas C., Dodd I.C. Physiological impacts of ABA–JA interactions under water-limitation. Plant Mol. Biol. 2016;91:641–650. doi: 10.1007/s11103-016-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Seo J.S., Zhao P., Jung C., Chua N.H. PLANT U-BOX PROTEIN 10 negatively regulates abscisic acid response in Arabidopsis. Appl. Biol. Chem. 2019;62:39. doi: 10.1186/s13765-019-0446-0. [DOI] [Google Scholar]
- 145.Arc E., Sechet J., Corbineau F., Rajjou L., Marion-Poll A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013;4:1–20. doi: 10.3389/fpls.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Linkies A., Müller K., Morris K., Turečková V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E., et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009;21:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin Q., Wu F., Sheng P., Zhang Z., Zhang X., Guo X., Wang J., Cheng Z., Wang J., Wang H., et al. The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nature Commun. 2015;6:7981. doi: 10.1038/ncomms8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.O’Brien J.A., Benková E. Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 2013;4:451. doi: 10.3389/fpls.2013.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tan W., Zhang D., Zhou H., Zheng T., Yin Y., Lin H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018;14:e1007336. doi: 10.1371/journal.pgen.1007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ku Y.-S., Sintaha M., Cheung M.-Y., Lam H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018;19:3206. doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kumar M., Kesawat M.S., Ali A., Lee S.-C., Gill S.S., Kim A.H.U. Integration of abscisic acid signaling with other signaling pathways in plant stress responses and development. Plants. 2019;8:592. doi: 10.3390/plants8120592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sun T.-P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 2011;21:R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 153.Achard P., Renou J.-P., Berthomé R., Harberd N.P., Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 2008;18:656–660. doi: 10.1016/j.cub.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 154.Choi J., Huh S.U., Kojima M., Sakakibara H., Paek K.-H., Hwang I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell. 2010;19:284–295. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 155.Chen Z., Agnew J.L., Cohen J.D., He P., Shan L., Sheen J., Kunkel B.N. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I.-R., Vera-Cruz C., Kikuchi S., Höfte M. Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 2012;158:1833–1846. doi: 10.1104/pp.112.193672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xia X.-J., Wang Y.-J., Zhou Y.-H., Tao Y., Mao W.-H., Shi K., Asami T., Chen Z., Yu J.-Q. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009;150:801–814. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y., Tessaro M.J., Lassner M., Li X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell. 2003;15:2647–2653. doi: 10.1105/tpc.014894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Torres M.Á. ROS in biotic interactions. Physiol. Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 160.Wrzaczek M., Brosché M., Kangasjärvi J. ROS signaling loops—Production, perception, regulation. Curr. Opin. Plant Biol. 2013;16:575–582. doi: 10.1016/j.pbi.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 161.Miura K., Okamoto H., Okuma E., Shiba H., Kamada H., Hasegawa P.M., Murata Y. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 2013;73:91–104. doi: 10.1111/tpj.12014. [DOI] [PubMed] [Google Scholar]
- 162.Melotto M., Zhang L., Oblessuc P.R., He S.Y. Stomatal Defense a Decade Later. Plant Physiol. 2017;174:561–571. doi: 10.1104/pp.16.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee S., Kim S.-G., Park C.-M. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010;188:626–637. doi: 10.1111/j.1469-8137.2010.03378.x. [DOI] [PubMed] [Google Scholar]
- 164.Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R., Smith J.A.C., Harberd N.P. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012;31:4359–4370. doi: 10.1038/emboj.2012.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Baier M., Bittner A., Prescher A., van Buer J. Preparing plants for improved cold tolerance by priming. Plant Cell Environ. 2018;42:782–800. doi: 10.1111/pce.13394. [DOI] [PubMed] [Google Scholar]
- 166.Seo P.J., Kim M.J., Park J.-Y., Kim S.-Y., Jeon J., Lee Y.-H., Kim J., Park C.-M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- 167.Olate E., Jiménez-Gómez J.M., Holuigue L., Salinas J. NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat. Plants. 2018;4:811–823. doi: 10.1038/s41477-018-0254-2. [DOI] [PubMed] [Google Scholar]
- 168.Kang H.-M., Saltveit M.E. Chilling tolerance of maize, cucumber and rice seedling leaves and roots are differentially affected by salicylic acid. Physiol. Plant. 2002;115:571–576. doi: 10.1034/j.1399-3054.2002.1150411.x. [DOI] [PubMed] [Google Scholar]
- 169.Huot B., Castroverde C.D.M., Velásquez A.C., Hubbard E., Pulman J.A., Yao J., Childs K.L., Tsuda K., Montgomery B.L., He S.Y. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat. Commun. 2017;8:1808–1812. doi: 10.1038/s41467-017-01674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li S., Zhou X., Chen L., Huang W., Yu D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells. 2010;29:475–483. doi: 10.1007/s10059-010-0059-2. [DOI] [PubMed] [Google Scholar]
- 171.Ohama N., Sato H., Shinozaki K., Yamaguchi-Shinozaki K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]