Abstract
Cardiac allograft vasculopathy (CAV) is associated with intragraft B cell infiltrates. Here, we studied the clonal composition of B cell infiltrates using 4 graft specimens with CAV. Using deep sequencing, we analyzed the immunoglobulin heavy chain variable region repertoire in both graft and blood. Results showed robust B cell clonal expansion in the graft but not in the blood for all cases. Several expanded B cell clones, characterized by their uniquely rearranged complementarity-determining region 3, were detected in different locations in the graft. Sequences from intragraft B cells also showed elevated levels of mutated rearrangements in the graft compared to blood B cells. The number of somatic mutations per rearrangement was also higher in the graft than in the blood, suggesting that B cells continued maturing in situ. Overall, our studies demonstrated B cell clonal expansion in human cardiac allografts with CAV. This local B cell response may contribute to the pathophysiology of CAV through a mechanism that needs to be identified.
Keywords: B cell biology, basic (laboratory) research/science, heart (allograft) function/dysfunction, heart transplantation/cardiology, immunobiology, rejection: chronic
1 |. INTRODUCTION
Cardiac allograft vasculopathy (CAV) is a serious complication responsible for the majority of long-term graft failures following heart transplantation.1,2 Although the pathophysiological mechanisms of CAV are still poorly understood, B cells are now thought to be central to its development.3 In humans, B cells and plasma cells are frequently observed as diffuse or nodular infiltrates in the adventitia of the allograft coronary arteries in the context of CAV.4–7 The function of these cells, however, is still unclear. A closer examination is needed to understand how they participate in the local immune response associated with the development of CAV. In recent studies, we used chronically rejected cardiac allografts removed at time of retransplantation to characterize B cell infiltrates at the single cell level. Specifically, we derived 102 B cell clones from infiltrates around the coronary arteries of three different allografts and interrogated their reactivity. None of these cells was specific to donor HLA. In contrast, a majority of the clones displayed a polyreactive profile, characteristic of “innate” B cells.7 These findings revealed the nature of B cells present in transplanted human hearts during CAV. Here, we used next-generation sequencing to further examine the clonal composition of B cell infiltrates and compare their repertoire and diversity to that of circulating B cell populations found in the transplant recipients’ blood.
2 |. METHODS
2.1 |. Patients and biological specimens
Explanted cardiac tissue specimens and blood samples used in this study were collected from 4 patients with cardiac allograft vasculopathy during a second transplant procedure at Massachusetts General Hospital and Columbia University Medical Center. The study was approved by the Massachusetts General Hospital/Partners and Columbia University Medical Center Institutional Review Boards. Fresh, unfixed tissue samples were obtained in the operating room and collected from different areas of the allografts, including the right coronary artery (RCA), left anterior descending coronary artery (LAD), circumflex coronary artery (CIR), and the ventricular endomyocardium (Endo).
2.2 |. Immunohistochemistry and immunofluorescence
Fresh graft tissue samples were fixed in 10% phosphate buffered formalin (Thermo Fisher Scientific, Waltham, MA) and embedded in paraffin blocks. Five-micron sections were deparaffinized, and antigen retrieval was performed using a decloaker as previously described.7 The slides were rinsed and cooled in phosphate buffered saline. Endogenous peroxidase activity was blocked by incubation for 5 minutes with Dual Endogenous Enzyme-Blocking Reagent (Agilent, Santa Clara, CA, USA). Successive sections were then stained to assess for the expression of CD20 (Clone L26; Agilent). For immunofluorescence, the following antibodies were used: CD27 (clone EPR8569; Abcam, Cambridge, MA) and CD20 (Clone L26; Agilent). Donkey antirabbit (AF488; Life Technologies, Carlsbad, CA) and donkey anti-mouse (NL557; R & D Systems, Minneapolis, MN) were used as secondary antibodies.
2.3 |. Immunoglobulin heavy chain variable region (IGHV) repertoire analysis
Fragments from the explant specimens that were approximately equal in size and that corresponded to tissue directly adjacent to the large vessels (right coronary artery, left descending CA, and circumflex CA) were cut and used for the IGHV analysis. Genomic DNA extracted from explanted graft tissue and peripheral blood mononuclear cells (PBMC) using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and was shipped to Adaptive Biotechnology for sequencing. IGHV sequencing was carried out for all specimens using the next generation sequencing-based ImmunoSEQ platform. Briefly, the IGH complementarity-determining region 3 (CDR3) region was amplified and sequenced from 400 ng of total genomic DNA using a multiplex polymerase chain reaction (PCR) system. A 130 base-pair fragment spanning each unique CDR3 and sufficient to identify the VDJ region was obtained. Additionally, primers are included so B cell precursor DJ rearrangements are also amplified. Amplicons were sequenced using the Illumina HiSeq platform. IGH V, D, and J gene definitions were identified using the IMGT database (www.imgt.org) and binned using a modified nearest-neighbor algorithm to merge closely related sequences and remove both PCR and sequencing errors. Data were analyzed using the ImmunoSEQ analyzer toolset and the R statistical language.
2.4 |. IGHV mutation analysis
Nucleic acid sequence data for each sample were downloaded from ImmunoSEQ and processed with IgBLAST8 to annotate and align the sequences to the IMGT database (www.imgt.org) of known V, D, and J gene segments. Only productive sequences with a V segment alignment score >70 and annotated as containing a CDR3 were considered. For each read, the mutation level was calculated as the fraction of V segment nucleotides that differed from the best matching germline V segment. The fraction of reads with >0% mutation level (Figure 3A) and the mean mutation level over all reads in a sample (Figure 3B) where calculated in R using the plyr package.
FIGURE 3.
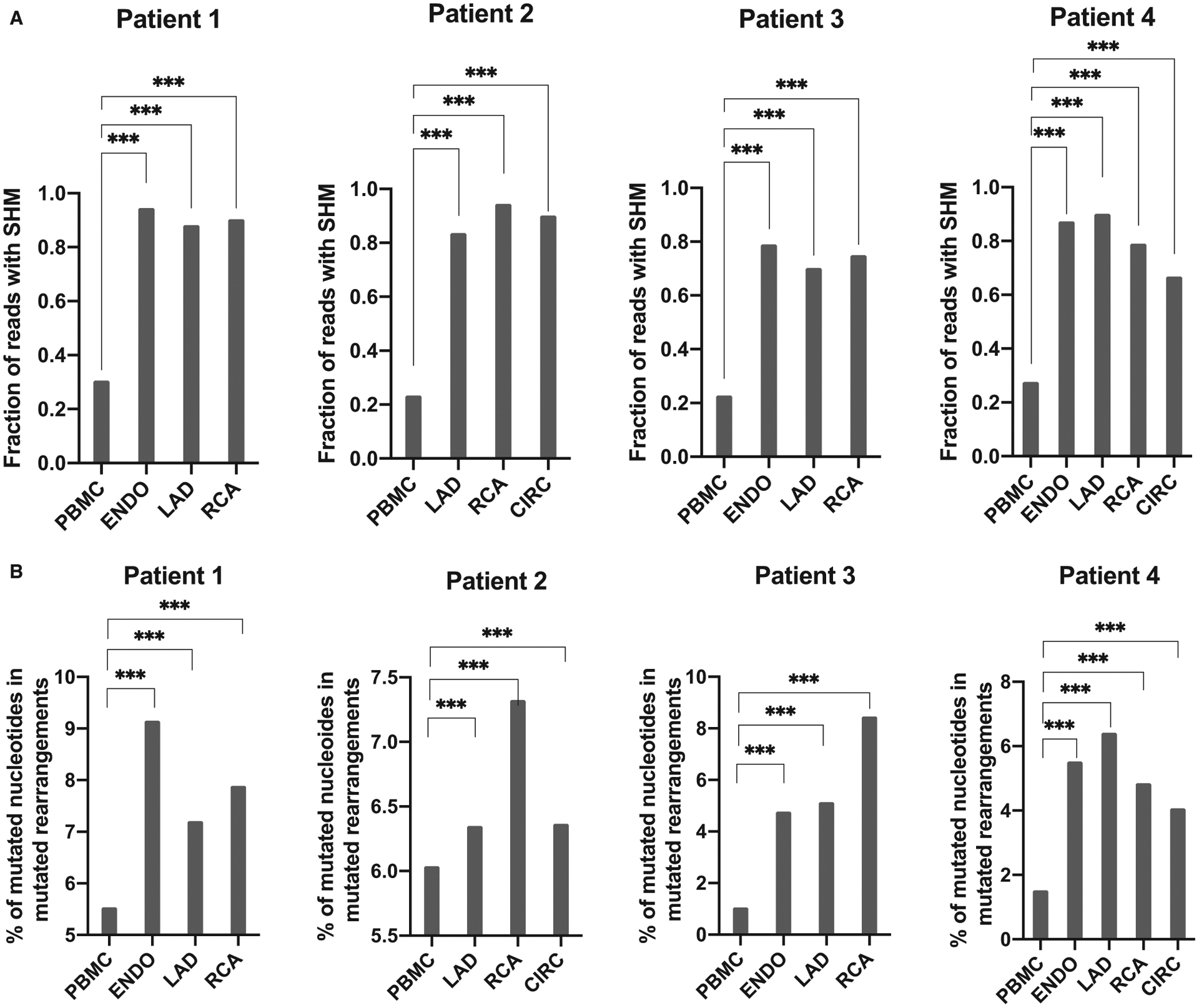
Blood and intragraft B cell IGHV mutation rates. A, Overall percentage of productive rearrangements with SHM for all graft locations and blood for the 4 patients. B, Average percentage of mutated nucleotides among productive mutated rearrangements. Differences between locations were assessed using a two-tailed Mann-Whitney test (***P < .001). IGHV, immunoglobulin heavy chain variable; SHM, somatic hypermutations
2.5 |. Statistical methods
Summary statistics were obtained from the ImmunoSEQ website. Statistical analysis was performed using the R statistical programming language (version 2.15.2 [2012–10-26]) within the RStudio wrapper (version 0.97.443), additional packages used include the plyr package (version 1.8). Graphs were made in R. A P < .05 was regarded as significant. To compare the proportion of CDR3 overlap between cardiac compartments and PBMC samples, we calculated proportion shared as (#CDR3 sequencing reads from clones found in both samples)/(# of total CDR3 sequencing reads from both samples). Significance was determined by the nonparametric Wilcoxon Mann-Whitney U test. Box-whisker plots were done in R using the ggplot2 package and show median (horizontal line), interquartile range (box), and 1.5 times the interquartile range (whiskers).
2.6 |. Clonality analysis
To compare repertoire clonality between samples, we calculated the Shannon diversity index (also known as Shannon entropy) of the observed distribution of IGH clones. The Shannon diversity index is commonly used in ecology to measure species diversity. Here we use it to measure clonality, the inverse of diversity, with IGH clones in place of species. The maximum value of the Shannon index depends on sampling depth. To compare samples sequenced at varying depths, we normalized the index for each location by dividing the Shannon index value S by the maximum possible value (Smax) to obtain a value between zero and one (S/Smax = normalized Shannon index). The inverse of the normalized Shannon index of productive IgH sequences is show in Figure 1A. To compare clonality between groups, we used the nonparametric two-tailed Wilcoxon signed-rank test.
FIGURE 1.
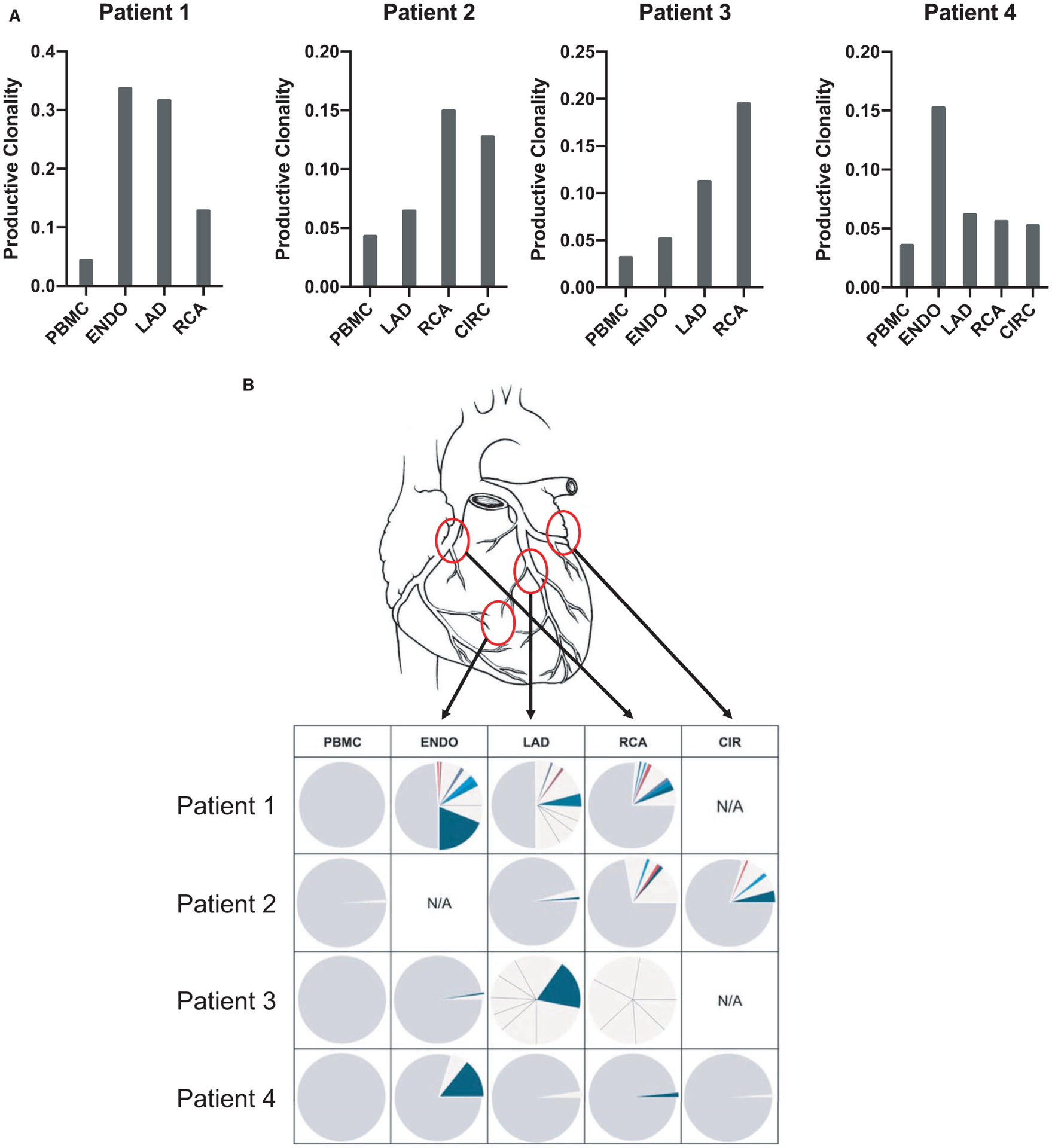
B cell repertoire analysis in graft infiltrates and peripheral blood. A, IGH clonality of graft locations and blood for 4 transplant recipients with active CAV, calculated from productive rearrangements by taking the inverse of the normalized (for sequencing depth) Shannon diversity index. B, Pie chart representation of the frequency of individual CDR3 sequences observed in 4 distinct allograft sites and in the blood for 4 explants. Each pie section corresponds to a unique CDR3 sequence with a frequency above 1%. Sections corresponding to sequences present at less than 1% are merged and depicted as gray. The noncolored sections correspond to clones that were only detected in the indicated location. Colored pie chart sections (blue, red…) correspond to clones that were detected in more than one locale within the graft. CAV, cardiac allograft vasculopathy; ENDO, endocardium; LAD, left anterior descending coronary artery; RCA, right coronary artery; CIR, circumflex coronary artery
3 |. RESULTS
Four heart transplant recipients were included in this study. All experienced cardiac allograft failure due to CAV or recurrent restrictive cardiomyopathy resulting in congestive heart failure (patient 4) 5–15 years after the original transplantation and underwent re-transplantation at Massachusetts General Hospital or Columbia University Medical Center (Table 1). Patient 4 had CAV International Society for Heart and Lung Transplantation (ISHLT) score 1 at the time of re-transplantation. Three out of four (patients 1, 2, and 4) also had evidence of acute cellular rejection. None of the patients had circulating DSA at the time of re-transplantation. Tissue samples from the original cardiac allografts explanted during the procedure were collected for all four patients and used to study B cell infiltrates. The specimens included tissue surrounding the RCA, left coronary artery (LCA), LAD, circumflex coronary artery as well as tissue sampled from the endomyocardium (ENDO). Immunohistochemical staining revealed B cell infiltration throughout the graft for the 4 patients. B cells were located around the vessels or in areas surrounding the vessels (Figure S1A). Both nodular and diffuse infiltrates were observed for all four patients. These infiltrates were often adjacent to adipose tissue regardless of their pattern. Immunofluorescence staining further determined that a majority of CD20+ B cells also expressed the memory marker CD27 (Figure S1B).
TABLE 1.
Patient characteristics
| Patient | Agea (y) | Sex | Original disease | Time to re-Tx (y) | ISHLT CAV scorea | Immunosuppressiona | DSAa | Rejectiona |
|---|---|---|---|---|---|---|---|---|
| 1 | 31 | M | Endocardial cushion defect | 15 | 3 | CsA, MMF, Prednisone | Neg. | ACR |
| 2 | 57 | M | Nonischemic cardiomyopathy | 15 | 2 | CsA, Aza, Prednisone | Neg. | ACR |
| 3 | 5 | F | Idiopathic cardiomyopathy | 5 | 2 | FK506, MMF, Rapamycin | Neg. | - |
| 4 | 28 | F | Restrictive cardiomyopathy | 6 | 1b | CsA, Aza, Prednisone | Neg. | ACR |
ACR, acute cellular rejection; CAV, cardiac allograft vasculopathy; CsA, cyclosporine; DSA, donor-specific antibodies; ISHLT, International Society for Heart and Lung Transplantation; MMF, mycophenolate mofetil; AZA, azathioprine.
At time of retransplantation.
Patient 4 had congestive heart failure due to recurrent restrictive cardiomyopathy.
3.1 |. B cell clonal expansion in cardiac allografts
We used a next generation sequencing (NGS) approach to analyze the IGHV repertoire in the different locales of the 4 explanted allografts. Genomic DNA was extracted from the collected specimens (RCA, LAD, CIRC, ENDO) as well as from the patients’ PBMC and used as template. Deep sequencing was performed by Adaptive Biotechnologies using their repertoire analysis platform (Immunoseq) and generated between 82 935 and 1 093 855 sequences for a total of 13 graft and 4 PBMC specimens (average 452 694 sequences/sample; Table S1). Tissue fragments from a minimum of three different locations were used for each explanted graft. Two small graft fragments generated a low number of IGHV sequences (patient 3 LAD and RCA). Even though the sample size was limited for these two specimens, we included them in our report as the sequences proved informative in terms of clonal expansion. On the whole, B cell populations were significantly less diverse in graft infiltrates compared to the blood as illustrated by higher Shannon diversity indexes (Figure 1A). However, differences were noticeable between cases. The diversity observed for some locales was only slightly elevated when compared to the blood (eg, patient 4), indicating that clonal expansion was not as pronounced in these areas. All sequences were clustered by CDR3 identity and the frequency of each unique rearrangement was calculated. Figure 1B reports the frequency of clonotypic CDR3 sequences in the blood and graft compartments using a pie chart representation for the four cases examined. Each section of the pie corresponds to a unique CDR3 sequence. The size of the sections corresponds to the frequency of the specific CDR3 sequence among all sequences. As shown in this figure, several unique CDR3 sequences were found at high frequency in the graft. As an example, a unique sequence represented >20% of all sequences in the Patient 1 endocardium (dark blue section). Such high frequency indicated that the corresponding B cell clones had undergone robust expansion in the graft. In contrast, sequences obtained from the patient blood did not reveal any clonal predominance.
3.2 |. B cell repertoire overlap between graft locales (Figure 2)
FIGURE 2.
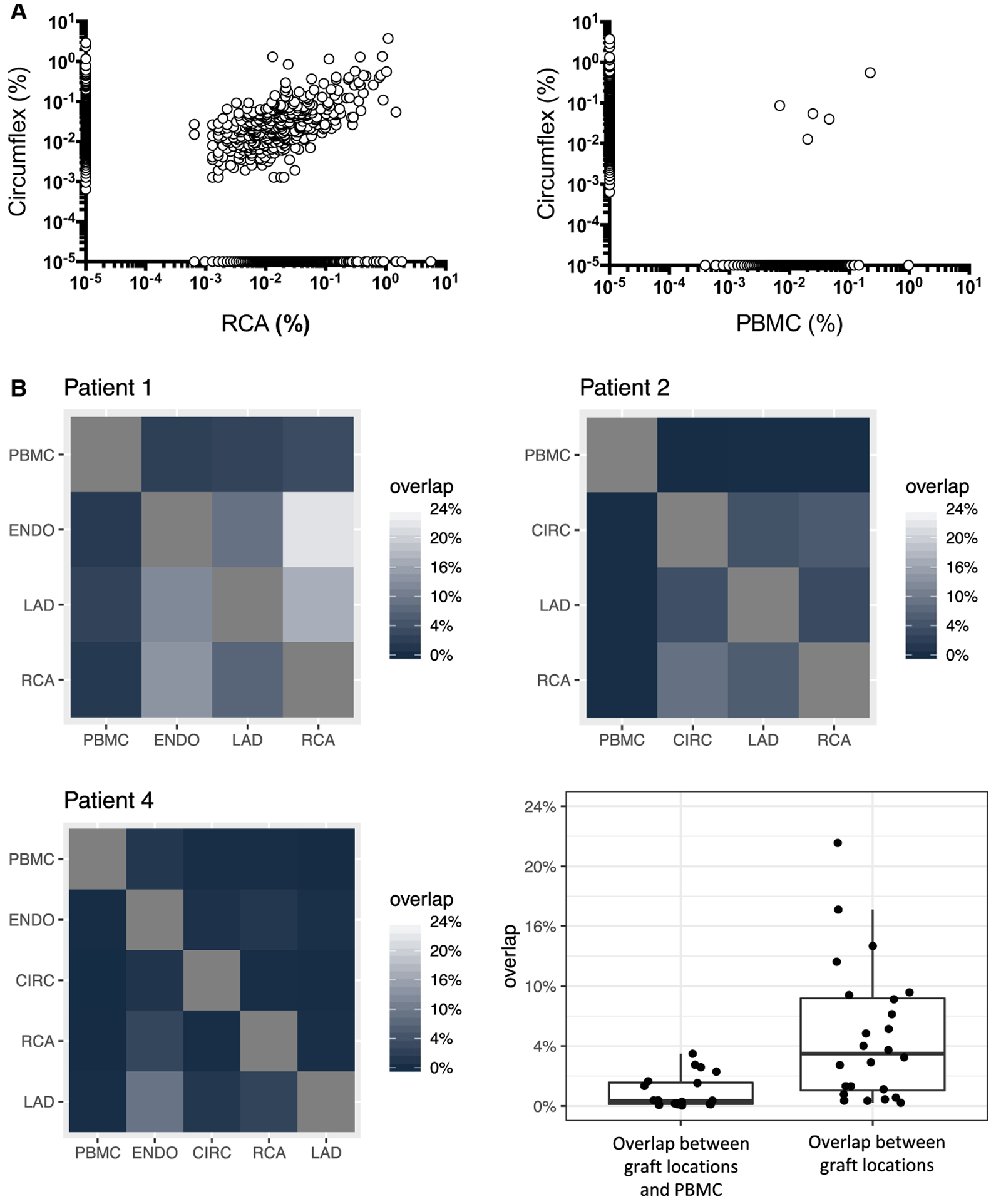
Overlap between B cell repertoires. A, Example of IGHV sequence repertoire overlap between 2 distinct locations within the same graft (RCA and CIR, left panel) and between the graft and PBMC (right panel) for patient 2. Each dot represents a unique sequence. The frequency of each sequence is reported on the x and y axes. Sequences along the x and y axis were detected in only one compartment. B, Summary of the CDR3 productive rearrangements repertoire overlap between the different graft locations and blood for 3 patients. The percentage corresponds to CDR3 sequences from the locations shown on the y-axis that are also found at the location shows on the x-axis. Note that for patient 3, the number of sequences obtained for the coronary arteries was not sufficient to calculate the repertoire overlap. The box-whisker plot shows the distribution of overlaps between graft locations and PBMC (left) or between different graft locations (right). CIR, circumflex coronary artery; IGHV, immunoglobulin heavy chain variable; PBMC, peripheral blood mononuclear cells; RCA, right coronary artery
Clones detected at different locations and sharing the same CDR3 were considered as being clonally related. Remarkably, we detected several unique CDR3 sequences at multiple sites (Figure 1B, pie chart sections filled with the same colors for each patient) revealing that the same clones were present at different locales in the graft. In general, we observed a significant overlap between B cell infiltrates at different locations in the graft specimens, whereas the overlap between graft infiltrates and PBMC was minimal. An example of such overlap between RCA and CIRC infiltrates for patient 2 is depicted in Figure 2A. In contrast, the overlap was virtually nonexistent between CIRC and PBMC. Figure 2B depicts the CDR3 overlap between graft locales for all cases with the exception of patient 3 for whom the number of sequences was too low. The heatmap shows the percentage of CDR3 sequences from a given locale (y-axis) that overlap with another locale (x-axis). Percent overlap with PBMCs was significantly lower than overlap with non-PBMC tissues (box-whisker plot, P < .0005, two-tailed Wilcoxon rank sum test with continuity correction).
3.3 |. Enrichment for somatically mutated B cells in graft infiltrates
An analysis of the VH region sequenced obtained through the Immunoseq platform revealed the presence of somatic hypermutations (SHM) in a number of rearrangements. The percentage of mutated sequences was significantly higher in the graft than in the blood for all four patients (Figure 3A, P < .001). The level of SHM for all mutated rearrangements was also significantly higher for graft IGHV sequences compared to PBMC sequences, suggesting that infiltrating B cells underwent mutation in situ (Figure 3B, P < .001).
4 |. DISCUSSION
B cells infiltrates are common in heart and kidney allografts during rejection. Paradoxically, intragraft B cells are not associated with circulating DSA, a sign of systemic humoral alloimmunity or even antibody-mediated rejection.6,7,9 In kidney transplant recipients, B cells are often observed in the context of acute cellular rejection (ACR).10–12 Following heart transplantation, they are detected in endocardial biopsies as immune components of Quilty lesions13–15 or around coronary arteries in the context of active CAV.3,4,7,16 Several questions remain unanswered regarding B cell infiltrates. In particular, their clonal composition and role in the local immune reactions are poorly understood.
Repertoire analysis offers a window into the dynamics of a given immune response by tracking individual clones in different locations.17 The development of NGS greatly expanded the capacity of repertoire analysis. Remarkably, our study is the first to examine the clonal composition of B cell subsets infiltrating heart transplants in the context of CAV. Furthermore, we compared B cell infiltrates in different locations of the allografts as well as in the peripheral blood. Our results revealed sequences represented at very high frequency in the graft compared to the blood, indicating that the corresponding B cell clones had undergone robust expansion. All graft specimens examined showed the presence of B cell clones expanded to a frequency above 1%. This is in marked contrast with peripheral blood populations, which were far more diverse as evidenced by a lower clonality index. This observation suggests the activation in situ of specific clones presumably through the recognition of antigens exposed in the graft. Another possibility is that B cells were previously activated in secondary lymphoid organs before being recruited in the graft. While all samples used for this IGHV repertoire analysis showed B cell infiltrates by immunohistochemistry, we could not find any correlation between the level of clonal expansion and the pattern of B cell infiltrates (diffuse, nodular).
Another significant observation is the presence of the individual B cell clones identified by the same uniquely rearranged CDR3 at different locations within the graft. In some instances, massively expanded clones were found in the endocardium and around two main arteries. The overlap between locales reached more than 20% for some explants, while it was virtually negligible between the graft and the blood. The fact that identical clones are stimulated and retained in different locales, most probably through the recognition of antigenic determinants present throughout the graft, further supports an important role for infiltrating B cells in the pathophysiology of CAV.
Furthermore, we observed an increased level of SHM in IGHV sequences in all graft locations when compared to blood B cell sequences. These mutation data can be explained in different ways. B cells with higher levels of SHM may have been preferentially recruited and expanded at the graft site. Alternatively, B cell clones could have undergone somatic mutation upon activation in situ. The detection of activation-induced cytidine deaminase (AID) in cardiac graft-infiltrating B cells by Huibers and colleagues favors the latter hypothesis.18 A combination of these two scenarios is also plausible in accordance with the memory phenotype of infiltrating B cells expressing CD27.
Clonal expansion, spatial distribution, and differentiation through the accumulation of SHM are important elements to appreciate the dynamics of local B cell responses. Yet, the antigen specificity of the B cells at the center of the local immunity is crucial to understand how they interact with donor cells and contribute to the pathophysiology of CAV. A report published by Huibers et al18 in 2017 suggests that some plasma cells found in structures reminiscent of germinal centers in the graft could produce donor HLA specific antibodies. This notion was based on the reactivity of immunoglobulins eluted directly from explant tissue. In previous studies, we characterized the reactivity of over 100 B cell clones derived from 3 different cardiac grafts explanted because of CAV. In contrast to the observations made by Huibers et al, we could not detect any donor-specific reactivity among these cells. However, a majority of the clones displayed a polyreactive profile typical of “innate B cells,” also known as B1 B cells in mice.7,19,20 These cells react to apoptotic cell determinants, oxidation-related antigens, DNA, cardiolipin, or a combination of these.21 Such autoantigens are usually cryptic. Upon inflammation and tissue injury, they become accessible to immune cells. It is therefore not surprising that individual B cell clones reactive to these structures can be detected in different parts of the rejected allografts. Their presence and expansion in situ reported here is likely an indication of the local destruction.
Our study was restricted by the limited number of cases examined. There were also inherent limitations due to the type, size, and quality of surgical explant specimens available for our repertoire analysis. Nevertheless, our results are particularly significant as they provide additional elements to appreciate the immune response elicited amid CAV. In particular, our repertoire analysis reported here revealed a vigorous clonal expansion throughout the graft as well as accumulation of SHM in situ. In a previous study, we observed that a majority of B cells infiltrating the graft in the context of CAV were polyreactive and not donor specific.7 The latter observation is inviting us to revisit common assumptions about immune mechanisms of graft rejection. The allospecific component may not be as prominent as commonly accepted, at least with respect to B cells in chronically rejected heart transplants. Whether broadly reactive innate B cells are recruited secondarily to donor-specific clones is still unknown. It is noteworthy that a majority of CAV patients in our cohort had not developed donor-specific antibodies, arguing against the initial presence of a specific response. An alternate hypothesis is that graft-infiltrating B1-like B cells are active element of an ongoing innate response to graft wounding. This scenario would fit well with a growing literature involving non-HLA antibody responses in host responses to solid organ grafts in general and cardiac transplants in particular. Autoantigenic targets include membrane proteins such as angiotensin II type 1 receptor, apoptosis-related proteins such as perlecan as well as other immunogenic determinants.22–26 Even if not truly donor specific, graft-infiltrating B cells could nevertheless contribute to late allograft failure through fibrosis as suggested by Jansen et al.27 Still, the exact role of graft-infiltrating B cells is difficult to ascertain. In nonhuman primates, preemptive B cell depletion appeared to lower the incidence of CAV.28 In humans, a comparable treatment led to opposite results as reported by Starling et al29 for the Clinical Trials in Organ Transplantation-11 (CTOT-11) study investigators. In this trial, patients receiving rituximab on days 0 and 12 posttransplant showed higher increase in percentage of atheroma volume at 1 year posttransplant compared to patients who received the placebo. These findings, suggesting a protective role of B cells against CAV, have to be interpreted with caution though. Rituximab deletes both regulatory B cells and effector B cells. Depending on the timing, the treatment can have opposite effects. Clatworthy et al30 observed a comparable increase in acute cellular rejection following kidney transplantation after rituximab used as induction therapy. Yet, the same drug has shown success as a treatment of rejection when administered posttransplant.31–33 In the context of advanced CAV, rituximab could be envisaged to primarily target infiltrating B cells, hypothesizing that these cells play an important part in the pathophysiological process.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01-AI116814.
Funding information
National Institute of Allergy and Infectious Diseases, Grant/Award Number: AI116814
Abbreviations:
- CAV
cardiac allograft vasculopathy
- CDR3
complementarity-determining region 3
- Ig
immunoglobulin
- IGHV
immunoglobulin heavy chain variable region
- RCA
right coronary artery
- LAD
left anterior descending coronary artery
- CIR
circumflex coronary artery
- Endo
ventricular endomyocardium
- PCR
polymerase chain reaction
- PBMC
peripheral blood mononuclear cells
Footnotes
DISCLOSURES
The authors of this manuscript have no conflicts to disclose as described by the American Journal of Transplantation.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Mehra MR. Contemporary concepts in prevention and treatment of cardiac allograft vasculopathy. Am J Transplant. 2006;6(6):1248–1256. [DOI] [PubMed] [Google Scholar]
- 2.Sipahi I, Starling RC. Cardiac allograft vasculopathy: an update. Heart Fail Clin. 2007;3(1):87–95. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin WM 3rd, Halushka MK, Valujskikh A, Fairchild RL. B cells in cardiac transplants: from clinical questions to experimental models. Semin Immunol. 2012;24(2):122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gareau A, Hirsch GM, Lee TD, Nashan B. Contribution of B cells and antibody to cardiac allograft vasculopathy. Transplantation. 2009;88(4):470–477. [DOI] [PubMed] [Google Scholar]
- 5.Wehner JR, Fox-Talbot K, Halushka MK, Ellis C, Zachary AA, Baldwin WM 3rd. B cells and plasma cells in coronaries of chronically rejected cardiac transplants. Transplantation. 2010;89(9):1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huibers MMH, Gareau AJ, Vink A, et al. The composition of ectopic lymphoid structures suggests involvement of a local immune response in cardiac allograft vasculopathy. J Heart Lung Transplant. 2015;34(5):734–745. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee D, Moore C, Gao B, et al. Prevalence of polyreactive innate clones among graft-infiltrating B cells in human cardiac allograft vasculopathy. J Heart Lung Transplant. 2018;37(3):385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41(Web Server issue):W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wehner JR, Baldwin WM 3rd. Cardiac allograft vasculopathy: do adipocytes bridge alloimmune and metabolic risk factors? Curr Opin Organ Transplant. 2010;15(5):639–644. [DOI] [PubMed] [Google Scholar]
- 10.Sarwal M, Chua M-S, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. New Engl J Med. 2003;349(2):125–138. [DOI] [PubMed] [Google Scholar]
- 11.Zarkhin V, Kambham N, Li LI, et al. Characterization of intra-graft B cells during renal allograft rejection. Kidney Int. 2008;74(5): 664–673. [DOI] [PubMed] [Google Scholar]
- 12.Zarkhin V, Li L, Sarwal M. “To B or not to B?” B-cells and graft rejection. Transplantation. 2008;85(12):1705–1714. [DOI] [PubMed] [Google Scholar]
- 13.Chu KE, Ho EK, de la Torre L, Vasilescu ER, Marboe CC. The relationship of nodular endocardial infiltrates (Quilty lesions) to survival, patient age, anti-HLA antibodies, and coronary artery disease following heart transplantation. Cardiovasc Pathol. 2005;14(4): 219–224. [DOI] [PubMed] [Google Scholar]
- 14.Marboe CC, Billingham M, Eisen H, et al. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant. 2005;24(7 Suppl):S219–S226. [DOI] [PubMed] [Google Scholar]
- 15.Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24(11): 1710–1720. [DOI] [PubMed] [Google Scholar]
- 16.Wehner JR, Fox-Talbot K, Halushka MK, Ellis C, Zachary AA, Baldwin WM 3rd. B cells and plasma cells in coronaries of chronically rejected cardiac transplants. Transplantation. 2010;89(9): 1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babel N, Stervbo U, Reinke P, Volk HD. The identity card of T cells-clinical utility of T-cell receptor repertoire analysis in transplantation. Transplantation. 2019;103(8):1544–1555. [DOI] [PubMed] [Google Scholar]
- 18.Huibers MMH, Gareau AJ, Beerthuijzen JMT, et al. Donor-specific antibodies are produced locally in ectopic lymphoid structures in cardiac allografts. Am J Transplant. 2017;17(1):246–254. [DOI] [PubMed] [Google Scholar]
- 19.Zorn E, See SB. Is there a role for natural antibodies in rejection following transplantation? Transplantation. 2019;103(8):1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zorn E New insights on innate B-cell immunity in transplantation. Xenotransplantation. 2018;25(3):e12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zorn E, See SB. Polyreactive natural antibodies in transplantation. Curr Opin Organ Transplant. 2017;22(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardinal H, Dieude M, Hebert MJ. The emerging importance of non-HLA autoantibodies in kidney transplant complications. J Am Soc Nephrol. 2017;28(2):400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delville M, Charreau B, Rabant M, Legendre C, Anglicheau D. Pathogenesis of non-HLA antibodies in solid organ transplantation: where do we stand? Hum Immunol. 2016;77(11):1055–1062. [DOI] [PubMed] [Google Scholar]
- 24.Dieude M, West LJ, Muruve DA, et al. New answers to old conundrums: what antibodies, exosomes and inflammasomes bring to the conversation. Canadian National Transplant Research Program International Summit Report. Transplantation. 2018;102(2):209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobashigawa J, Colvin M, Potena L, et al. The management of antibodies in heart transplantation: An ISHLT consensus document. J Heart Lung Transplant. 2018;37(5):537–547. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Reed EF. The importance of non-HLA antibodies in transplantation. Nat Rev Nephrol. 2016;12(8):484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen MA, Otten HG, de Weger RA, Huibers MM. Immunological and fibrotic mechanisms in cardiac allograft vasculopathy. Transplantation. 2015;99(12):2467–2475. [DOI] [PubMed] [Google Scholar]
- 28.Kelishadi SS, Azimzadeh AM, Zhang T, et al. Preemptive CD20 + B cell depletion attenuates cardiac allograft vasculopathy in cyclosporine-treated monkeys. J Clin Invest. 2010;120(4):1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starling RC, Armstrong B, Bridges ND, et al. Accelerated allograft vasculopathy with rituximab after cardiac transplantation. J Am Coll Cardiol. 2019;74(1):36–51. [DOI] [PubMed] [Google Scholar]
- 30.Clatworthy MR, Watson CJE, Plotnek G, et al. B-cell-depleting induction therapy and acute cellular rejection. New Engl J Med. 2009;360(25):2683–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clatworthy MR. Targeting B cells and antibody in transplantation. Am J Transplant. 2011;11(7):1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zarkhin V, Li L, Kambham N, Sigdel T, Salvatierra O, Sarwal MM. A randomized, prospective trial of rituximab for acute rejection in pediatric renal transplantation. Am J Transplant. 2008;8(12):2607–2617. [DOI] [PubMed] [Google Scholar]
- 33.Becker YT, Becker BN, Pirsch JD, Sollinger HW. Rituximab as treatment for refractory kidney transplant rejection. Am J Transplant. 2004;4(6):996–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


