Abstract
Background
Zengye decoction (ZYD) has been widely used in the treatment of type 2 diabetes mellitus (T2DM). Exploring the fate of various components of ZYD in vivo is of considerable significance for pharmacological research and molecular mechanism elaboration. However, the systematic analysis on the metabolic behavior of chemical components of ZYD in T2DM rats has not been reported.
Methods
To screen and characterize the complex chemical compositions of ZYD, and metabolism fate in plasma, urine, bile, and feces of T2DM rats, the model of T2DM rats was prepared. A rapid procedure using high-performance liquid chromatography coupled with electrospray ionization quadrupole time of flight tandem mass spectrometry (HPLC-ESI-Q-TOF–MS/MS) was established. Data were acquired and analyzed by Agilent MassHunter Workstation Qualitative Analysis software version B.07.00 and PCDL manager B.07.00.
Results
A total of 80 compounds were identified or tentatively characterized in ZYD, 31 more than previously detected. Besides, 36 prototype components and 49 metabolites of ZYD were found and characterized in T2DM rats, and the proposed fragmentation pathways and possible metabolic behaviors of the main types of compounds were described.
Conclusions
This study developed the understanding of the composition of ZYD as well as the cleavage rules and metabolic pathways of the prototype compounds. Besides, this study provided abundant data for further research and for study of the metabolism of traditional Chinese medicine prescriptions.
Keywords: Zengye decoction, Type 2 diabetes mellitus, HPLC-ESI-Q-TOF–MS/MS, Qualitative analysis, Metabolite identification
Background
Type 2 diabetes mellitus (T2DM) is the most common form of diabetes, accounting for 90%–95% of all diabetic patients, which is primarily due to the relative lack of insulin secretion or reduced sensitivity to insulin [1, 2]. According to the latest report, the worldwide prevalence of adult diabetes has reached 9.3%, equivalent to 463 million adults worldwide with diabetes [3]. T2DM has become a serious global public health problem. Therefore, it is of practical significance to develop new drugs for the treatment of T2DM.
Traditional Chinese medicine (TCM) has been widely used in health care in many Asian countries for thousands of years. With the release of the detailed description of TCM by the 11th version of the International Statistical Classification of Diseases and Related Health Problems (ICD), TCM is more widely and gradually accepted around the world. Zengye decoction (ZYD) is a well-known TCM prescription used to treat ‘wasting thirst syndrome’, which would probably be diagnosed as T2DM according to the nationwide unified western medicine diagnostic criteria [4, 5]. ZYD was initially recorded in wen bing tiao bian written by Wu tang in Qing Dynasty of Chinese history (1936 AD-1912 AD) and is composed of Scrophulariae Radix, Rehmanniae Radix and Ophiopogonis Radix. Modern pharmacological studies show that ZYD exhibits hypoglycemic effect [6, 7]. However, the mechanism corresponding to its hypoglycemic effect is still unclear, due to the sophisticated features of multi-components and biological multi-effect of TCM [8]. Therefore, it is necessary to evaluate the therapeutic substances of hypoglycemic effect using modern scientific research methods, not only the herbal phytochemical compositions but also the absorption and metabolism of active ingredients in vivo of T2DM.
There have been some researches on the chemical constituents and herbal ingredients of ZYD in previous studies [9, 10]. A few reports have studied the absorption of a few compounds of ZYD [11]. We have demonstrated that ZYD improves insulin resistance in T2DM rats [12]. However, the metabolism of ZYD in experimental diabetes models has not reported. Actually, in the pathological state of diabetes, the absorption, distribution, metabolism, and excretion of ZYD may be different from those in the natural and healthy state [13, 14]. This ambiguity presents the greatest obstacle to deeper pharmacological mechanism investigation and scientific connotation interpretation. Therefore, a comprehensive, systematic analysis of the absorption and metabolic components of ZYD in vivo under the diabetic state is urgent.
In this paper, a rapid procedure using high-performance liquid chromatography coupled with electrospray ionization quadrupole time of flight tandem mass spectrometry (HPLC-ESI-Q-TOF–MS/MS) was established to characterize complex chemical compounds and metabolic components. In actual, HPLC-ESI-Q-TOF–MS/MS has been substantially applied to qualitative analysis of multiple components and metabolites in the complex mixture especially for the TCM prescriptions owing to its extraordinary performance, high resolution, accurate mass measurement, and rapid scan speed [15]. In the present study, a rat model of T2DM was established, and the absorbed components and metabolic ingredients in plasma, bile, urine, feces were screened after oral administration of ZYD. At the same time, the proposed fragmentation pathways and possible metabolic behaviors of the composition of ZYD in vivo were described in detail.
Methods
Chemicals and reagents
HPLC-grade acetonitrile was purchased from Tedia (Fairfield, OH, USA). HPLC-grade methanol was purchased from CINC High Purity Solvents Co. Ltd (Shanghai, China). The purified water was obtained using a Milli-Q water purification system (Millipore, Bedford, MA, USA). Formic acid of HPLC-grade was purchased from Aladdin Bio-Chem Technology Co. Ltd (Shanghai, China). n-Butanol was purchased from Sinopharm Chemical Reagent Co. Ltd (Nanjing, China). Catalpol, leonuride, acteoside, isoacteoside, harpagide, harpagoside, were obtained from Sichuan Weikeqi biological technology Co., Ltd (purity ≥ 98%). Cinnamic acid, p-coumaric acid, ferulic acid, were obtained from Chengdu Biopurify Phytochemicals Ltd (purity ≥ 98%). Streptozotocin (STZ) was purchased from Sigma (St. Louis, MO, USA). All other chemicals and solvents were of analytical grade.
Preparation of Zengye decoction extract
The crude drugs of Scrophulariae Radix, dried Rehmanniae Radix, Ophiopogonis Radix were purchased from Nanjing Traditional Chinese Medicine Clinics (Nanjing, China). The three herbs were authenticated by professor Jin Qi (Jiangsu Key Laboratory of TCM Evaluation and Translational Research). The crude drugs (130 g. Scrophulariae Radix: dried Rehmanniae Radix: Ophiopogonis Radix, 5:4:4, w/w/w) were soaked in double-distilled water and extracted three times by boiling in distilled water (1300 mL, 1040 mL, 780 mL) under reflux for 1 h. And then, the collected filtrates were combined, concentrated, and freeze-dried to obtain a lyophilized powder.
Preparation model of T2DM rats and drug administration
Male Wistar rats weighing 140–160 g were purchased from Comparative Medicine Center of Yangzhou University (Yangzhou, China). The rats were raised in an air-conditioned room at 23 ± 1 °C and a 12 h light/dark cycle. The animals were adaptively fed for 1 week prior to use. All the operations were allowed by the Animal Ethics Committee of China Pharmaceutical University, China Pharmaceutical University, Nanjing, Jiangsu, China.
The model of T2DM rats was induced by high-fat diet combined with low-dose STZ (35 mg/kg) [16]. The method was improved according to the previous reports and laboratory studies [10, 17]. Briefly, the animals were fed a high-fat diet for 3 weeks followed by intraperitoneal injection of STZ (35 mg/kg) which dissolved in cold citrate buffer (pH 4.3, 0.05 M). The fasting blood glucose (FBG) of rats was measured 3 days after injection, the level was higher than 11.1 mmol/L for subsequent experiments.
The T2DM rats were randomly divided into two groups. One group received ZYD (13 g/kg body weight, twice a day) via oral administration for 7 days. The other group received water, as the model control group. In addition, a normal healthy rats group received water, as the normal control group. The rats were fasted but with free access to water for 12 h before experiment.
Samples collection and pretreatment
All samples were obtained after drug administration. The blood samples (n = 4) were collected from T2DM rats in the heparinized centrifugal tube at 0.5 h, 1 h, 2 h, 4 h and 8 h by retro-orbital venipuncture and immediately centrifuged at 1200×g for 15 min to obtain plasma. The urine and feces (n = 4) were collected at 0–24 h in independent metabolic cages. The feces samples were naturally dried in the fume hood and then crushed into powder. The bile (n = 4) was collected at 0–8 h by bile duct intubation and drainage under general anesthesia induced by 1% pentobarbital sodium, 55 mg/kg. All biological samples of the same type in the same group at each time point were equally combined into one sample, and stored at − 80 °C before pretreatment and analysis.
An aliquot of 1 mL plasma sample was added into triple volume of acetonitrile and vigorously vortexed for 1 min. Then the mixture was centrifuged at 1500 xg for 15 min. The supernatant was transferred to another centrifuge tube and evaporated to dryness under a gentle stream of nitrogen at 37 °C. The residual was reconstituted in 200 μL methanol: water mixture (7:3, v/v) and then centrifuged at 13,700×g for 15 min. The supernatant was filtered through 0.22 μm nylon microporous filter membrane. The filtrates were analyzed by HPLC-ESI-Q-TOF–MS/MS.
Likewise, an aliquot of 1 mL urine sample was added into 3 mL methanol and vortexed for 1 min. A weight of 0.8 g feces was extracted within 8 mL methanol for 30 min under ultrasonic. Afterwards, the mixture was centrifuged at 13,700×g for 15 min. The supernatant was transferred to another centrifuge tube and evaporated to dryness under a gentle stream of nitrogen at 37 °C. The residual was re-dissolved in 200 μL reconstituted solvent (methanol: water, 7:3, v/v) and centrifuged at 13,700×g for 15 min, and the solution was filtered through 0.22 μm nylon microporous filter membrane. The bile was pretreatment in the same way as urine.
HPLC-ESI-Q-TOF–MS/MS condition
Chromatographic separation was performed on an Agilent 1260 Infinity HPLC system (Agilent Technologies, Santa Clara, CA, USA) using a Diamonsil C18 column (4.6 × 250 mm, 5 μm) and a precolumn with the same packing at 30 °C. The mobile phase A was water containing 0.01% formic acid and phase B was acetonitrile containing 0.01% formic acid. The gradient elution conditions were as follows: 2% B at 0–10 min; 2–7% B at 10–20 min; 7–16% B at 20–45 min; 16–22% B at 45–60 min; 22–34% B at 60–85 min; 34–80% B at 85–115 min; 80–100% B at 115–125 min. The total flow rate was 1 mL/min, and an aliquot of 20 μL injected for analysis.
Mass spectrometry data were obtained using an Agilent 6530 Q-TOF MS/MS system (Agilent Technologies, Santa Clara, CA, USA) equipped with an ESI interface. The optimum parameters were set as follows: ESI source in the negative mode; drying gas (N2) flow rate 10.0 L/min; drying gas temperature 320 °C; nebulizer pressure 35 psi; capillary voltage 3500 V; skimmer 65 V; fragmentor voltage 120 V; MS/MS collision energy 25 V; scan range 100–2000 Da.
The HPLC-ESI-Q-TOF–MS/MS data were acquired and analyzed by an Agilent MassHunter Workstation Qualitative Analysis software version B.07.00 and PCDL manager B.07.00 (Agilent Technologies, Santa Clara, CA, USA).
Results
Identification of chemical profile of Zengye Decoction
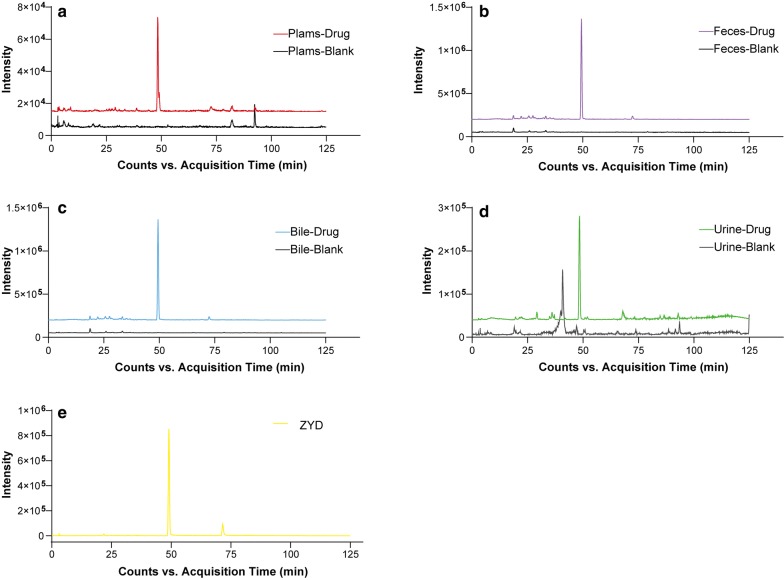
In order to more accurately characterize the intracorporal process of ZYD in the T2DM rats, the chemical constituents of ZYD were initially identified by HPLC-ESI-Q-TOF–MS/MS. In the present study, 80 compounds from ZYD were tentatively identified by comparing with the reference standards, the retention time, and reviewing literature [9, 10, 19–25]. The total ion chromatogram (TIC) of ZYD in negative mode was showed in Fig. 1a, and the detailed compounds information was summarized in Additional file 1: Table S1. Thirty-one previously undetected compounds were found compared with previous reports [9, 10]. In addition, the structural types of all compounds in ZYD were mainly iridoid glycosides, phenethylalchohol glycosides and phenylpropanoid glycosides, aromatic acid, homoisoflavonoids, steroidal saponins.
Fig. 1.
a Total ion chromatogram (TIC) of Zengye decoction (ZYD) in the negative ion mode and b–e extracted ion chromatograms (EICs) of ZYD in biological and blank samples in the negative ion mode. b-1 ZYD plasma sample and b-2 blank plasma sample; c-1 ZYD bile sample and c-2 blank bile sample; d-1 ZYD urine sample and d-2 blank urine sample; and e-1 ZYD feces sample and e-2 blank feces sample
Characterization of ZYD prototype components absorbed in T2DM rats
In this study, the biological samples, including plasma, urine, feces, and bile of T2DM rats treated with ZYD, were analyzed by HPLC-ESI-Q-TOF–MS/MS under constant conditions. Peaks which displayed at the same position on the chromatograms of both drug-containing biological samples and ZYD, but not in controlled blank biological samples were considered as absorbed and metabolic components of ZYD. Both TIC and EIC (extracted ion chromatograms) profiles were used to screen the absorbed prototype components. Finally, 11 compounds were observed in plasma, 17 were in urine, 5 were in bile, 35 were in feces in T2DM rats treated with ZYD. The EICs were showed in Fig. 1b1–e2 and the prototype compounds were listed in Table 1. Furthermore, rehmapicrogenin was chosen as a representative absorbed component to demonstrate EICs further in Fig. 2. In contrast to the controlled blank biological samples, rehmapicrogenin showed remarkable peaks in drug-containing groups.
Table 1.
Characterization of ZYD prototype components in vivo by HPLC-ESI-Q-TOF-MS/MS
| Peak No. | tR(min) | Precursor ions (m/z) | Formula | Error (mDa) | Fragment ions | Identification | P | U | F | B | Refs. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimental | Theoretical | |||||||||||
| P1 | 10.332 | 407.1282* | 407.1195 | C17H22O10 | − 8.70 | 361.1038, 199.0607, 181.0507, 169.0505 | Catalpol | + | + | + | − | [10] |
| P2 | 20.658 | 391.1284* | 391.1246 | C15H22O19 | − 3.81 | 211.1011, 183.0662, 165.0543, 139.0410 | Aucubin | + | + | + | − | [10] |
| P4 | 23.285 | 731.2325* | 731.2251 | C27H42O20 | − 7.35 | 685.2276, 505.1599, 341.1118, 179.0561 | Rehmannioside D | − | + | + | − | [10] |
| P5 | 25.065 | 409.1384* | 409.1351 | C15H24O10 | − 3.05 | 201.0771, 183.0663, 165.0556, 157.0504 | Harpagide | + | + | + | + | [10] |
| P6 | 26.149 | 393.1443* | 393.1402 | C15H24O9 | − 4.06 | 185.0560, 179.0526, 167.0716, 113.0238 | Leonuride | + | + | + | + | [10] |
| P8 | 29.733 | 373.1216 | 373.1140 | C16H22O10 | − 7.58 | 331.0989, 221.0893, 167.0426, 149.0622 | Geniposidic acid | − | + | + | − | [18] |
| P9 | 30.113 | 421.1332* | 421.1351 | C16H24O10 | 1.95 | 213.0732, 195.0662, 183.0657, 169.0437 | 6-O-methylcatalpol | + | + | + | + | [19] |
| P10 | 30.783 | 461.1698 | 461.1664 | C20H30O12 | − 3.35 | 461.1698 | Decaffeoylacteoside | − | − | + | − | [10] |
| P11 | 31.790 | 375.1317 | 375.1297 | C16H24O10 | 2.03 | 375.1317 | 8-epilogonic acid | − | + | + | − | [9] |
| P12 | 36.172 | 487.1527 | 487.1457 | C21H28O13 | − 6.99 | 487.1527 | Cistanoside F | − | − | + | − | [10] |
| P14 | 41.765 | 313.0938 | 313.0929 | C14H18O8 | − 0.91 | 313.0938, 229.1749, 137.0605, 123.0439 | Rhamnopyranosyl vanilloyl | − | + | − | − | [20] |
| P15 | 42.066 | 475.1845 | 475.1821 | C21H32O12 | − 2.4 | 475.1845 | Darendoside B | − | − | + | − | [10] |
| P16 | 43.203 | 607.2290 | 607.2244 | C26H40O16 | − 4.64 | 607.2298, 475.1859, 461.1697, 443.1586, 149.0457, 131.0351 | β-(3-hydroxy-4-methoxyhenyl) ethyl-O-α-L-arabinopyranosyl-(1 → 6)-O-[6-α-L-rhamnpyranosyl-(1 → 3)-β-D-glucopyranoside | − | + | + | − | [10] |
| P20 | 49.444 | 183.1028 | 183.1027 | C10H16O3 | − 0.13 | 183.1036, 139.1123, 123.0792 | Rehmapicrogenin | + | + | + | + | [10] |
| P23 | 54.309 | 435.2269* | 435.2236 | C19H34O8 | − 3.33 | 389.2370, 179.0539, 161.0451, 119.0348 | Rehmaionoside A/B | − | + | + | − | [10] |
| P26 | 56.018 | 799.2778 | 799.2666 | C36H48O20 | − 11.18 | 799.2778 | Jionoside A1/A2 | − | + | + | − | [21] |
| P24 | 56.295 | 163.0399 | 163.0401 | C9H8O3 | 0.17 | 145.0270, 119.0499 | p-coumaric acid | + | + | + | − | [10] |
| P29 | 60.152 | 193.0511 | 193.0500 | C10H10O4 | − 0.47 | 178.0269, 149.0598, 134.0369, 121,0282 | Ferulic acid | − | + | + | − | [10] |
| P31 | 61.894 | 623.2049 | 623.1981 | C29H36O15 | − 6.76 | 461.1676, 315.1084, 161.0241, 135.0445 | Acteoside | − | − | + | − | [10] |
| P34 | 63.221 | 769.2634 | 769.2561 | C35H46O19 | − 7.35 | 769.2634 | Scrophuloside B1/B2 | − | − | + | − | [22] |
| P36 | 63.473 | 623.2021 | 623.1981 | C29H36O15 | − 3.96 | 623.2021 | Isoacteoside or Forsythoside A | − | − | + | − | [10] |
| P37 | 65.538 | 525.1665 | 525.1614 | C24H30O13 | − 5.14 | 525.1665 | 8-O-caffeoyl harpagide | − | − | + | − | [19] |
| P39 | 67.938 | 429.2171 | 429.2130 | C21H34O9 | − 4.09 | 249.1512, 231.1370, 187.1472 | Jiocarotenoside A1/A2 | − | + | + | − | [23] |
| P41 | 69.475 | 783.2798 | 783.2717 | C36H48O19 | − 8.10 | 607.2294, 589.2154, 461.1666, 193.0500 | Angoroside C | + | − | + | − | [10] |
| P42 | 70.273 | 783.2840 | 783.2717 | C36H48O19 | − 12.30 | 783.2840, 829.4135 | Isoangoroside C | + | − | + | − | [10] |
| P47 | 70.428 | 637.2199 | 637.2138 | C30H38O15 | − 6.11 | 461.1706, 193.0512, 149.0577, 134.0372 | Leucosceptoside A | − | − | + | − | [21] |
| P52 | 75.561 | 651.2397 | 651.2294 | C31H40O15 | − 10.26 | 651.2397 | Cistanoside D | − | − | + | − | [24] |
| P55 | 79.343 | 539.1823* | 539.1770 | C24H30O11 | − 5.29 | 493.1715, 345.1210, 183.0662, 165.0556 | Harpagoside | + | − | + | + | [10] |
| P56 | 81.455 | 147.0450 | 147.0452 | C9H8O2 | 0.15 | 147.0450 | Cinnamic acid | + | − | + | − | [10] |
| P62 | 98.429 | 345.1014 | 345.0980 | C18H18O7 | − 3.42 | 345.1014 | 5,7,2′,4′-tetradihydroxy-8-methoyl-6-methyl-homoisoflavanone | − | − | + | − | [10] |
| P74 | 104.020 | 359.1161 | 359.1136 | C19H20O7 | − 2.47 | 359.1161 | Ophiopogonanone E | − | + | + | − | [10] |
| P75 | 105.884 | 343.1215 | 343.1187 | C19H20O6 | − 2.79 | 343.1215 | 5-7-4′-trihydroxy-5′-methoxy-6,8-dimethyl hamoisoflavanone | − | − | + | − | [25] |
| P77 | 107.848 | 327.0905 | 327.0874 | C18H16O6 | − 3.09 | 327.0905 | Ophiopogonone A | − | − | + | − | [10] |
| P78 | 110.065 | 339.0898 | 339.0874 | C19H16O6 | − 2.39 | 339.0898 | Methylophiopogone A | − | + | + | − | [10] |
| P79 | 110.853 | 341.1059 | 341.1031 | C19H18O6 | − 2.84 | 206.0587, 178.0258 | Methylophiopogonanone A | − | − | + | − | [10] |
| P80 | 111.626 | 327.1260 | 327.1238 | C19H20O5 | − 2.20 | 327.1260 | Methylophiopogone B | − | − | + | − | [10] |
a), tR, retention time; *, [M+HCOOH−H]−, other, [M−H]−; P, plasma; U, urine; F, feces; B, bile. +, containing; −, not
Fig. 2.
EICs of rehmapicrogenin at m/z 183.1027, [M−H]− in negative ion mode; a ZYD-containing and blank plasma sample; b ZYD-containing and blank feces sample; c ZYD-containing and blank bile sample; d ZYD-containing and blank urine sample; e ZYD
Identification of phenethylalchohol glycosides and phenylpropanoid glycosides
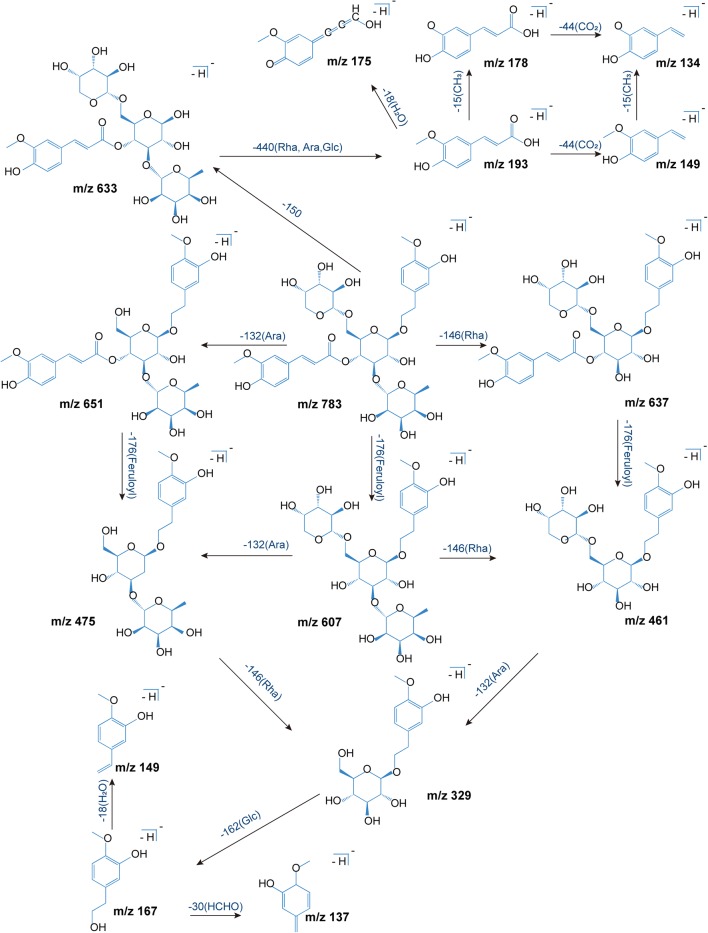
Totally 11 phenethylalchohol glycosides and phenylpropanoid glycosides were detected in vivo, and their structures usually included phenylethanol, phenylpropanoid, and glycoside units. Phenylethanol group often involved loss of neutral molecules like H2O, HCHO, and CH3OH. Phenylpropionic acid easily loses H2O due to the presence of hydroxyl and carboxyl groups. In another way, different phenylpropionic acid shows different typical fragments, such as ferulic acid (193, 178, 175, 149, 134), p-coumaric acid (163, 145, 119), cinnamic acid (147, 129, 103). In the case of angoroside C, the fragmentation pathway diagram was shown in Fig. 3. The quasi-molecular ion peak of angoroside C (783.2798 [M−H]−) could lose a series of residuals to generate ion peaks including 651.2173 [M-Ara(132 Da)–H]−, 633.2078 [M-Ara-H2O–H]−, m/z 607.2299 [M-Feruloyl(176 Da)-H]−, m/z 589.2192 [M-Feruloyl-H2O–H]−, m/z 637.2163 [M-Rha(146 Da)–H]−, m/z 475.1792 [M-Ara-Feruloyl-H]–, m/z 461.1707 [M-Rha-Feruloyl-H]−, m/z 443.1553 [M-Rha-Feruloyl-H2O–H]–, m/z 329.1226 [M-Ara-Rha-Feruloyl-H]−, m/z 311.1474 [M-Rha-Ara-H2O–H]–, and typical fragments where the residues further to break, such as feruloyl (175,134), 3-methylhydroxytyrosol (167), etc. Likewise, other compounds could produce similar fragments.
Fig. 3.
Proposed fragmentation pathway diagram of angoroside C in the negative ion mode. Ara, arabinosyl; Rha, rhamnosyl; Glc, glucosyl; Feruloyl, Ferulic acid dehydration
Identification of iridoid glycosides
Iridoid glycosides are the main components of Scrophulariae Radix and dried Rehmanniae Radix, which have similar mother nuclei and mass spectral fragmentation pattern during cracking. These compounds generally produce precursor ions like [M−H]−, [M+HCOOH–H]−, [M+Cl]−, and [2M−H]− in the negative ion mode. In the secondary ion mode, the glucosyl group (Glc, C6H10O5, 162 Da) were usually preferentially lost to expose iridoid mother nucleus. Subsequent loss of a series of H2O (18 Da) molecules occurred because of the presence of many hydroxyl groups (OH, 17 Da) in the mother nucleus. Fracture of the iridoid at enol-ether bonds was accompanied by the loss of acetaldehyde (CH3CHO, 44 Da), formaldehyde (HCHO, 30 Da), and H2O. Typical fragmentation pathway provided reliable information for the identification of iridoid glycosides. For instance, leonuride was the main iridoid glycosides in Rehmanniae Radix. The precursor ions were m/z 393.1503 [M+HCOOH–H]−, m/z 383.1210 [M+Cl]−, m/z 347.1439 [M−H]−, which produced m/z 185.0860 [M-Glc-H]−, m/z 167.0741 [M-Glc-H2O–H]−, m/z 149.0606 [M-Glc-2H2O–H]−, m/z 137.0203 [M-Glc-HCHO–H]−, m/z 123.0444 [M-Glc-CH3CHO–H]−. At the same time, the glucosyl residue also produced typical fragments such as m/z 161.0462, m/z 113.0227. In order to more intuitively display the fragmentation pathway of this type components, the proposed fragmentation pathways of leonuride in negative ion mode was exhibited in Fig. 4a.
Fig. 4.
Proposed fragmentation pathways of a leonuride and b methylophiopogonanone A in the negative ion mode
Identification of homoisoflavones
Seven homoisoflavones were detected in T2DM rats. Homoisoflavones are a particular class of flavonoids, which are connected by methylene group (CH2) between B and C rings. Compound P79 was confirmed as methyl-ophiopogonanone A, the MS/MS spectrum and proposed fragmentation diagram were shown in Fig. 4b. P79 gave [M−H]− at m/z 341.1051, in which fragmentation at m/z 206.0580 that originated from the initial loss of B-ring and CH2 to generate [M-B-ring-CH2–H]−. The ion at m/z 178.0629 was attributed to a loss of CO from m/z 206.0580. Besides, [M−H]− ion just eliminated B-ring to produce the [M-B-ring-H]− at m/z 219.1341. Analogously, the remaining six compounds were identified, respectively.
Identification of other compounds
Based on their exact molecular mass and MS/MS spectra, P14, P15, P20, P23, and P39 were temporarily identified as rhamnopyranosyl vanilloyl, darendoside B, rehmapicrogenin, rehmaionoside A/B, jiocarotenoside A1/A2, respectively.
Tentative characterization of the ZYD metabolites in T2DM rats
Potential metabolic pathways of the ZYD components were determined by comparing data from public databases and relevant publications with the cleavage results of ZYD component mother nuclei. As a result, a total of 49 presumptive metabolites in plasma, feces, urine, and bile were preliminary illuminated. All metabolic components were listed in Table 2.
Table 2.
Identification of ZYD metabolic components in T2DM rats
| Peak No. | tR(min) | Precursor ions (m/z) | Error (mDa) | Formula | Fragment ions(m/z) | Identification | Metabolic type | P | U | F | B | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exp | The | |||||||||||
| M1 | 12.985 | 201.0776 | 201.0768 | − 0.75 | C9H14O5 | 201.0762, 157.0529, 139.0751 | Harpagenin | Hydrolyzation | + | − | + | − |
| M2 | 24.773 | 377.1460 | 377.1453 | − 0.68 | C16H26O10 | 377.1460, 217.0938, 169.0841, 161.0465 | Dihydrogen methylcatalpol | Methylation, hydrogenation | + | + | − | − |
| M3 | 26.057 | 263.0240 | 263.0231 | − 0.90 | C9H12O7S | 263.0240, 183.0455, 165.0588, 121.0291 | Dehydrated harpagenin sulfate | Dehydration, sulfation | − | + | − | − |
| M4 | 26.304 | 153.0555 | 153.0557 | 0.22 | C8H10O3 | 153.0576, 123.0448, 135.0412, 121.0281 | Hydroxytyrosol | Hydrolyzation | + | − | + | − |
| M5 | 26.862 | 583.1938 | 583.2032 | 9.43 | C27H36O14 | 583.2279, 195.0671, 151.0671, 179.0566, 161.0460, 149.0501 | Acetyl 6-O- dihydro-feruloyl harpagide | Hydrogenation, acetylation | − | − | + | − |
| M6 | 29.227 | 185.0819 | 185.0819 | 0.13 | C9H14O4 | 185.0819, 141.0891 | Deglucosylated leonuride | Deglucosylation | + | + | − | + |
| M7 | 36.029 | 225.0774 | 225.0768 | − 0.55 | C11H14O5 | 225.0801, 210.0793, 165.0556 | Methyl hydrated ferulic acid | Methylation, hydration | − | − | + | − |
| M8 | 31.271 | 181.0506 | 181.0506 | 0.03 | C9H10O4 | 181.0513, 163.0398, 137.0514, 119.0498 | Hydrated p-coumaric acid | Hydration | + | + | + | + |
| M9 | 35.171 | 451.2234* | 451.2185 | − 4.91 | C19H34O9 | 451.2234, 225.0505, 179.0553, 161.0482 | Hydroxy rehmaionoside A | Hydroxylation | − | − | + | − |
| M10 | 36.81 | 363.1109 | 363.1085 | − 2.36 | C18H20O8 | 363.1136, 345.1053, 183.0626, 179.0445, 165.0555, 139.0667 | Deglucosylated 8-O-caffeoyl-harpagide | Deglucosylation | − | − | + | − |
| M11 | 37.114 | 137.0602 | 137.0608 | 0.6 | C8H10O2 | 137.0602, 122.0397, 111.0424, 107.0455 | Deoxyhydroxytyrosol | Deoxidation | − | − | + | − |
| M12 | 37.366 | 489.1641 | 489.1614 | − 2.74 | C21H30O13 | 489.1641, 179.0363, 165.0542, 113.0249 | Didehydrated 8-O-caffeoyl harpagide | Dehydration | − | − | − | + |
| M13 | 37.895 | 339.0709 | 339.0722 | 1.26 | C15H16O9 | 339.0709 | p-coumaric acid glucuronide | Glucuronidation | − | − | − | + |
| M14 | 37.895 | 181.0506 | 181.0506 | 0.03 | C9H10O4 | 181.0506, 163.0773, 137.0601, | Dihydro-caffeic acid | Hydrogenation | − | + | + | − |
| M15 | 39.276 | 151.0405 | 151.0405 | − 0.43 | C8H8O3 | 151.0405, 123.0439, 107.0501 | Dehydrogen hydroxytyrosol | Dehydrogenation | − | + | + | − |
| M16 | 39.401 | 371.0993 | 371.0984 | − 0.93 | C16H20O10 | 371.0993, 195.0657, 177.0549, 193.0334, 175.0255, 113.0242 | Dihydro-ferulic acid glucuronide | Hydrogenation, glucuronidation | + | + | − | − |
| M17 | 41.295 | 369.0842 | 369.0827 | − 1.48 | C16H18O10 | 369.0842, 193.0504, 178.0272, 149.0599, 134.0369, 113.0232 | Ferulic acid glucuronide | Glucuronidation | + | + | − | + |
| M18 | 41.618 | 233.0131 | 233.0125 | − 0.57 | C8H10O6S | 233.0131, 153.0554, 135.0444, 123.0449, 121.0299, 109.0274 | Hydroxytyrosol sulfate | Sulfation | − | + | + | − |
| M19 | 42.554 | 341.0890 | 341.0878 | − 1.19 | C15H18O9 | 341.0890, 165.0556, 121.0657, 175.0233, 149.0598, 113.0236 | Dihydro-p-coumaric acid glucuronide | Hydrogenation, glucuronidation | − | + | − | − |
| M20 | 42.703 | 357.0818 | 357.0827 | 0.92 | C15H18O10 | 357.0822, 339.0646, 175.0585, 131.0406 | Hydrated p-coumaric acid glucuronide | Glucuronidation, hydration | − | − | + | − |
| M21 | 45.675 | 181.0870 | 181.0870 | 0.02 | C10H14O3 | 181.0376, 137.0971, 122.0663, 121.0653 | Dehydro-rehmapicrogenin | Dehydrogenation | + | − | + | − |
| M22 | 45.826 | 179.0350 | 179.0350 | − 0.02 | C9H8O4 | 179.0613, 135.0450, 161.4731, | Caffeic acid | Hydrolyzation | − | + | + | − |
| M23 | 47.591 | 359.1361 | 359.1348 | − 1.34 | C16H24O9 | 359.1361, 183.1025, 139.1122, 113.0232 | Rehmapicrogenin glucuronide | Glucuronidation | − | + | − | + |
| M24 | 47.813 | 151.0398 | 151.0401 | 0.27 | C8H8O3 | 151.0398, 107.0500 | Dehydrogen hydroxytyrosol | Dehydrogenation | − | + | − | − |
| M25 | 48.821 | 247.0288 | 247.0282 | − 0.62 | C9H12O6S | 247.0288, 167.0711, 152.0474, 149.0285 | Methyl hydroxytyrosol sulfate | Methylation, sulfation | − | + | − | − |
| M26 | 49.757 | 385.1154 | 385.1140 | − 1.38 | C17H22O10 | 385.1154, 209.0821, 191.0689, 175.0120, 113.0244 | Dihydro-methyl ferulic acid glucuronide | Hydrogenation, methylation, glucuronidation | − | + | − | + |
| M27 | 50.395 | 369.0827 | 369.0827 | 0.02 | C16H18O10 | 369.0842, 193.0504, 178.0272, 149.0599, 134.0369, 113.0232 | Ferulic acid glucuronide | Glucuronidation | − | − | − | + |
| M28 | 54.181 | 275.0239 | 275.0231 | − 0.8 | C10H12O7S | 275.0239, 195.0662, 177.0558, 151.0761, 136.0524, 121.0293 | Dihydro-ferulic acid sulfate | Hydrogenation, sulfation | − | + | − | − |
| M29 | 54.078 | 195.0667 | 195.0663 | − 0.42 | C10H12O4 | 195.0676, 177.0570, 136.0524 | Dihydro-ferulic acid | Hydrogenation | + | + | + | − |
| M30 | 55.035 | 165.0556 | 165.0557 | 0.12 | C9H10O3 | 165.0559, 147.0427, 129.0326, 121.0655 | Hydrated cinnamic acid | Hydration | − | − | + | + |
| M31 | 55.744 | 233.0139 | 233.0125 | − 1.37 | C8H10O6S | 233.0131, 153.0554, 135.0444, 123.0449 | Hydroxytyrosol sulfate | Sulfation | − | − | + | − |
| M32 | 55.195 | 331.1208 | 331.1187 | − 2.09 | C18H20O6 | 331.1222, 313.1108, 287.0830, 165.0553, 147.0440, 103.0543 | Deglucosylated harpagoside | Deglucosylation | − | − | + | − |
| M33 | 56.101 | 521.1820 | 521.1664 | − 12.05 | C25H30O12 | 521.1785, 503.1661, 183.0665, 157.0509, 193.0404, 113.0247 | Dehydrated harpa-gide glucuronide | Dehydration, glucuronidation | − | − | + | − |
| M34 | 56.427 | 245.0133 | 245.0125 | − 0.77 | C9H10O6S | 245.0133, 165.0554, 147.0432, 121.0657 | Dihydro-p-coumaric acid sulfate | Hydrogenation, sulfation | − | + | − | − |
| M35 | 58.309 | 209.0822 | 209.0819 | − 0.27 | C11H14O4 | 209.0813, 191.0713, 165.0926, 149.0616 | Dihydro-methyl ferulic acid | Hydrogenation, methylation | + | − | + | − |
| M36 | 58.362 | 273.0083 | 273.0074 | − 0.85 | C10H10O7S | 273.0083, 193.0503, 178.0268, 134.0369 | Ferulic acid sulfate | Sulfation | − | + | − | − |
| M37 | 59.756 | 625.2194 | 625.2194 | − 5.61 | C29H38O15 | 625.2194, 461.1695, 315.1104, 181.0510, 163.0407, 153.0538 | Dihydro-acteoside | Hydrogenation | − | − | + | − |
| M38 | 60.258 | 217.1088 | 217.1081 | − 0.65 | C10H18O5 | 217.1087, 199.0936, 186.2198, 171.1025, 155.1062, 153.0895 | Dihydro-methyl harpagenin | Hydrogenation, methylation | − | − | + | − |
| M39 | 61.965 | 135.0449 | 135.0452 | 0.25 | C8H8O2 | 135.0449, 123.0065, 107.0468, 100.9257 | Dehydrated hydroxytyrosol | Dehydration | + | − | + | − |
| M40 | 63.176 | 361.1518 | 361.1504 | − 1.39 | C16H26O9 | 361.2317, 185.1180, 141.1279, 113.0242 | Dihydrogen rehmapicrogenin glucuronide | Hydrogenation, glucuronidation | − | + | − | − |
| M41 | 64.035 | 247.0294 | 247.0282 | − 1.22 | C9H12O6S | 247.0537, 167.0706, 152.0476 | Methyl hydroxytyrosol sulfate | Methylation, sulfation | − | − | + | − |
| M42 | 66.026 | 377.1471 | 377.1453 | − 1.78 | C16H26O10 | 377.1471, 201.1129, 183.1002, 165.0567, 175.0242, 113.0242 | Harpagenin glucuronide | Glucuronidation | − | + | − | + |
| M43 | 70.428 | 637.2199 | 637.2138 | − 6.11 | C30H38O15 | 637.2155, 461.1706, 193.0512, 135.0407 | Methyl acteoside | Methylation | − | − | + | − |
| M44 | 72.673 | 275.0246 | 275.0231 | − 1.50 | C10H12O7S | 275.0240, 195.0665, 177.0575, 151.0772 | Hydrated ferulic acid sulfate | Hydration, sulfation | + | − | + | − |
| M45 | 72.935 | 245.0135 | 245.0125 | − 0.97 | C9H10O6S | 245.0135, 165.0666, 147.0549, 121.0355 | Dihydro-p-coumaric acid sulfate | Hydrogenation, sulfation | − | − | + | − |
| M46 | 78.821 | 273.0081 | 273.0074 | − 0.65 | C10H10O7S | 273.0090, 193.0506, 178.0271, 134.0372 | Ferulic acid sulfate | Sulfation | + | − | − | + |
| M47 | 78.166 | 149.0609 | 149.0608 | − 0.10 | C9H10O2 | 149.0609, 107.0480, 105.0703 | Dihydro-cinnamic acid | Hydrogenation | − | − | + | − |
| M48 | 79.350 | 583.2092 | 583.2032 | − 5.97 | C27H36O14 | 583.2100, 193.0525, 149.0439, 201.1145, 183.0656, 165.0568 | Acetyl-6-O-dihydro-feruloyl harpagide | Hydrogenation, acetylation | − | − | + | − |
| M49 | 74.738 | 315.1270 | 315.1297 | 2.67 | C14H20O8 | 315.1278, 297.1136, 161.0582, 135.0442 | Hydroxytyrosol glucosylate | Hydrolyzation | − | − | + | − |
a): P, plasma; U, urine; F, feces; B, bile. +, containing; −, not
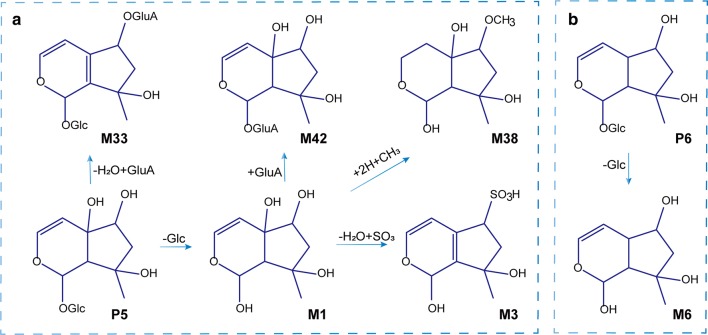
Identification of iridoid glycoside-related metabolites
Totally 12 metabolites of the iridoids were initially identified, most of which were derived from the metabolism of harpagide and its derivatives. Harpagide and its derivatives easily lose a glycosyl group and are converted to harpagenin by hydrolases. M1 displayed [M−H]− at m/z 201.1129, which typical product ions were basically consistent with the above description of harpagide. M42 showed [M−H]− at m/z 377.1471, and the fragment ion at m/z 201.1129 was yielded obviously by the loss of a GluA group (176 Da). Simultaneously, the deglucose products of some other iridoid glycosides were also found. M6 displayed [M−H]− at m/z 185.0819, a neutral loss of 162 Da (Glc) comparing with leonuride, indicating it was deglucosylated leonuride. M32 exhibited [M−H]− at m/z 331.1208. Comparing to harpagoside, the metabolite underwent a loss of glucosyl group. Possible metabolic pathways of harpagide and leonuride were shown in Fig. 5a, b.
Fig. 5.
Possible metabolic pathways of major iridoid glycosides in T2DM rats. a Harpagide; b Leonuride
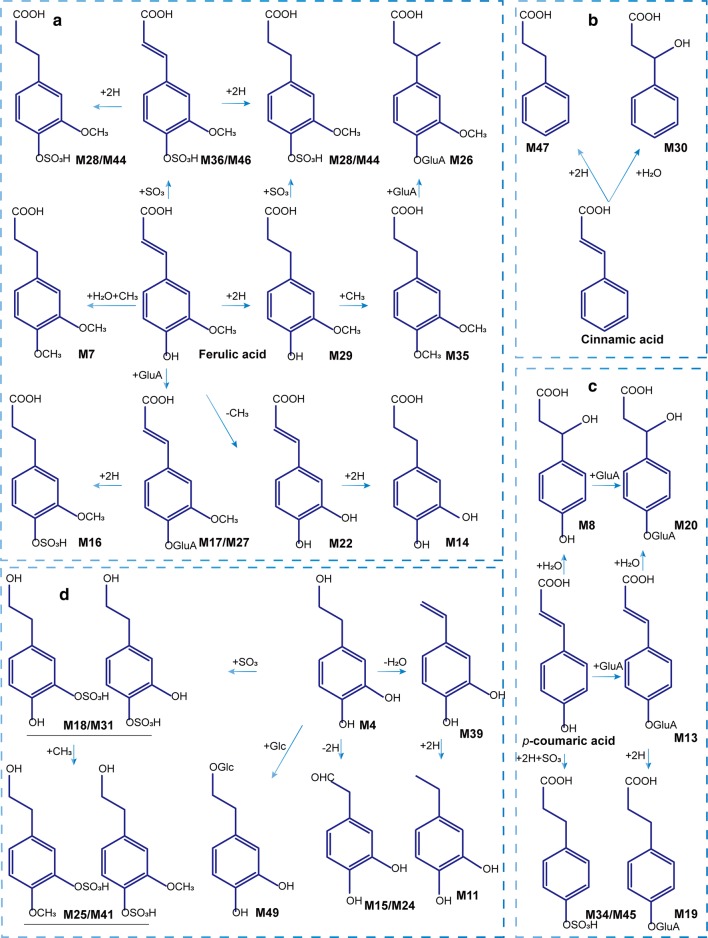
Identification of phenylpropanoid-related metabolites
A total of 23 constituents were initially identified as generating from the metabolism of phenylpropanoid-related compounds. The metabolic pathways were shown in Fig. 6a–c. [M−H]– ions, M19 at m/z 341.0890 and M34 at m/z 245.0133, had formed by the addition of 176 Da (GluA, C6H8O7) and 80 Da (SO3), respectively, to m/z 165.0554. Both had neutral losses of H2O and CO2 (44 Da). The metabolic profile was consistent with p-coumaric acid, indicating that the two metabolites were the glucuronidation and sulfonation products of dihydro p-coumaric acid. At the same time, the demethylation and hydrogenation products of ferulic acid were also observed. The demethylated product M22, called caffeic acid, neutral losing CO2 to produce abundant fragments of m/z 135.0450. The neutral loss of hydrogenated product M29 was the same as that of ferulic acid, including the loss of H2O to produce m/z 177.0570, and the loss of CO2 and methyl group (CH3, 15 Da) to produce typical debris m/z 136.0524, etc. The other 19 metabolites were identified in a similar manner.
Fig. 6.
Possible metabolic pathways for major ZYD compounds in T2DM rats. a ferulic acid; b cinnamic acid; cp-coumaric acid; and d Hydroxytyrosol
Identification of hydroxytyrosol-related metabolites
Hydroxytyrosol (HT) was mainly derived from phenylethanoid glycosides, which was produced by hydrolysis and formed different metabolites through various metabolic pathways in vivo. In this study, ten HT-related metabolites were identified in vivo, and the specific metabolic pathways were shown in Fig. 6d. M15 showed [M−H]– at m/z 151.0405, the loss of 2 Da (2H) from HT, suggesting that the metabolite was generated from the dehydrogenation of HT. Moreover, M18 and M31 both yielded [M−H]− at m/z 233.0139, linkage of an SO3 group separately, suggesting it to be hydroxytyrosol sulfated. Furthermore, both M25 and M41 showed [M−H]− at m/z 247.0288, the addition of methyl to M18 and M31, respectively.
Identification of metabolic components of other compounds
Analysis of the fragmentation pattern of rehmaionoside A found that M9 showed [M+HCOOH-H]– at m/z 451.2234, an addition of 16 Da to [rehmaionoside A-H]−, suggesting that M9 was tentatively identified as hydroxyrehmaionoside A. The product ion m/z 243.1105, and 225.0782 were losses of glucosyl group and water from M9. The product ions of M23 included m/z 183.1025, m/z 139.1122, m/z 175.0294, and m/z 113.0232. m/z 183.1025, and m/z 139.1122 were product ions of P20 and m/z 175.0294, m/z 113.0232 were the characteristic fragments of glucuronic acid. M23 was thus initially identified as a glucuronic acid-binding product of rehmapicrogenin. Also, the dehydrogenation product M21 (m/z 181.087) of rehmapicrogenin, and the hydrogenation product M40 (m/z 361.1518) of M23 were observed. The above fragmentation regularity was consistent with rehmapicrogenin.
Distribution of ZYD metabolites in T2DM rats
According to the distribution of ZYD metabolites in T2DM rats, Fourteen metabolites were found in plasma samples, 23 in urine, 33 in feces, and 11 in bile, which all have been identified and the details were listed in Table 2. Harpagoside, an iridoid glycoside, was the primary bioactive constituent of ZYD, which was detected in plasma, bile and feces samples of T2DM rats and its possible metabolites could be found in all bio-samples. For example, deglucosylation of harpagoside was observed in feces due to the transformation of harpagoside by glycoside hydrolase in the gut. Besides, glucuronic conjugates of p-coumaric acid, harpagenin, ferulic acid, and rehmapicrogenin were detected in bile, which was consistent with the glucuronic conjugates as the primarily metabolites in bile excretion. Meanwhile, p-coumaric acid was found in plasma, urine, and feces, but not in bile. It was likely that p-coumaric acid was metabolized to a glucuronic conjugate in the liver and then excreted into the duodenum via the bile duct and regenerated the prototype by a glucuronidase, and some of them were reabsorbed into the liver through the enterohepatic circulation.
Discussion
According to the results of the pre-experiment, there was no significant difference between the previously established method and the current method in the HPLC optimization process [10]. Hence, the previous performance was used for the next experiment. Besides, the negative ion mode was chosen for further analysis because most of the compounds in ZYD contain functional groups such as hydroxyl, carboxyl, etc., and most components can be detected in the negative ion mode.
Sufficient detection level was the premise of instrument analysis [26]. The low concentrations of TCM components in vivo and possibly substantial interference by the matrix effect posed significant challenges for the analysis in vivo [27]. Samples handling has become the essential part of biological sample analysis, so it was of considerable significance to choose the appropriate pretreatment method. In the current research, dialysis, protein precipitation (PPT), solid phase extraction (SPE), immunoaffinity extraction, and other methods were widely used for the biological samples pretreatment. Among them, the PPT shows the advantages of simplicity, rapidity, and convenience, and it has been widely used in the qualitative analysis of TCM in vivo [28]. So PPT was the preferred method for this experiment.
The effectiveness of TCM for disease prevention and treatment depends on the active ingredients contained in TCM [29]. The absorption of a drug is a prerequisite for its pharmacological activity within the body. Therefore, it was assumed that the absorbable component might be an active ingredient, and that the disease condition might have some impacts on the absorption process [30]. Diabetes may reduce the expression and function of P-glycoprotein (P-gp) in the intestine [31]. When the intestinal P-gp activity is inhibited, the absorption of some drugs will be enhanced in the intestine [32]. A previous study found that the plasma concentrations of catalpol and harpagide in diabetes-model rats were increased compared with normal rats, and that the clearance rate was slower [11]. Diabetes also changes the experesion of cytochrome P450 enzymes, including CYP3A4, CYP2E1, CYP2C9, and CYP2D4 participate in phase I drug metabolism [33–35]. Catalpol has been shown to effect the activity of CYP3A4, CYP2E1 and CYP2C9 that resulted in pharmacokinetic interactions of coadministered drugs [36]. It is possible that differences in the pharmacokinetics of ZYD compounds in diabetic and normal rats may be caused by disease-associated changes in some functional enzymes.
Some of the prototype and metabolic components identified in ZYD have had pharmacological effects in the treatment of T2DM in animal models and in cell lines. Harpagoside in ZYD could activate the PPAR-γ pathway in 3T3-L1 adipocytes to regulate lipid and glucose metabolism similar to the hypoglycemic effects of thiazolidinedione [37, 38]. p-Coumaric acid was shown to promote glucose uptake and utilization by activating the AMPK pathway and upregulating GLUT2 expression [39, 40]. Ferulic acid was reported to promote glucose uptake by activating PI3K-Akt pathway and upregulating the expression of GLUT4. It has also been found to promote glycogen synthesis and inhibit gluconeogenesis by downregulating the expression of PEPCK, G6PC and upregulating glucokinase expression [41–44]. In addition, Ferulic acid was also found to increase intracellular Ca2+ to promote insulin secretion [44, 45]. Among metabolites, caffeic acid was shown to increase insulin sensitivity in HepG2 cells, reduce hepatic glucose output, enhance glucose uptake, promote insulin secretion, and increase antioxidant activity in adipocytes [45–48]. Our previous study showed that ZYD had hypoglycemic activity, improved dyslipidemia, and promoted pancreatic islet-cell function in T2DM model rats [12]. Knowing which of the chemical constituents of ZYD presented in vivo is essential for further investigation of the material basis and mechanism of ZYD in the treatment of T2DM.
Conclusions
ZYD is increasingly used for the treatment of T2DM. However there have been few reports on the component analysis of ZYD in vivo and even fewer in the pathological state of T2DM. This study identified and tentatively characterized previously undetected ZYD ingredients and was the first to systematically analyze the metabolism of ZYD ingredients in T2DM rats. As a consequence, thirty-six prototype components and 49 metabolites were presumed and characterized in vivo, and the proposed fragmentation pathways and possible metabolic behaviors of the main types of compounds were analyzed. In summary, this study added to the understanding of the chemical profile of ZYD and its metabolism information in T2DM rats. It provided essential data for the detailed pharmacokinetic study and pharmacodynamic material basis of ZYD in T2DM. Meanwhile, the study methods are applicable to the study of the metabolism of other TCM prescriptions.
Supplementary information
Additional file 1: Table S1. Compounds information of ZYD by HPLC-ESI-Q-TOF MS/MS.
Acknowledgements
Not applicable.
Abbreviations
- Ara
Arabinosyl group
- FBG
Fasting blood glucose
- Glc
Glucosyl group
- GluA
Glucuronic acid group
- HT
Hydroxytyrosol
- PPT
Protein precipitation
- Rha
Rhamnosyl group
- T2DM
Type 2 diabetes mellitus
- TCM
Traditional Chinese medicine
- STZ
Streptozotocin
- ZYD
Zengye decoction
Authors’ contributions
JQ and ZQ conceived and designed all the experiments; SC, MW and YT carried out the experiments; SC performed the data analyses and wrote the manuscript, JQ and ZQ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No: 81673555) and the “Double First-Class” University project (CPU2018GF06 and CPU2018GY32).
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article.
Ethics approval and consent to participate
The study protocol and experiments were approved by the Animal Ethics Committee of China Pharmaceutical University, China Pharmaceutical University, Nanjing, Jiangsu, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shanquan Chang, Email: csq0821@hotmail.com.
Mei Wang, Email: meiwang777@163.com.
Yushan Tian, Email: yushan126163@126.com.
Jin Qi, Email: qijin2006@163.com.
Zhixia Qiu, Email: qiuzhixia_cpu@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13020-020-00331-z.
References
- 1.Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2019. Diabetes Care. 2019; 42: S13–28. 10.2337/dc19-s002. [DOI] [PubMed]
- 2.World Health Organization. Global Report on Diabetes. Book. 2016;88.
- 3.Forouzanfar Mohammad H, Afshin Ashkan, Alexander Lily T, Anderson H Ross, Bhutta Zulfiqar A, Biryukov Stan, Brauer Michael, Burnett Richard, Cercy Kelly, Charlson Fiona J, Cohen Aaron J, Dandona Lalit, Estep Kara, Ferrari Alize J, Frostad Joseph J, Fullman Nancy, Gething Peter W, Godwin William W, Griswold Max, Hay Simon I, Kinfu Yohannes, Kyu Hmwe H, Larson Heidi J, Liang Xiaofeng, Lim Stephen S, Liu Patrick Y, Lopez Alan D, Lozano Rafael, Marczak Laurie, Mensah George A, Mokdad Ali H, Moradi-Lakeh Maziar, Naghavi Mohsen, Neal Bruce, Reitsma Marissa B, Roth Gregory A, Salomon Joshua A, Sur Patrick J, Vos Theo, Wagner Joseph A, Wang Haidong, Zhao Yi, Zhou Maigeng, Aasvang Gunn Marit, Abajobir Amanuel Alemu, Abate Kalkidan Hassen, Abbafati Cristiana, Abbas Kaja M, Abd-Allah Foad, Abdulle Abdishakur M, Abera Semaw Ferede, Abraham Biju, Abu-Raddad Laith J, Abyu Gebre Yitayih, Adebiyi Akindele Olupelumi, Adedeji Isaac Akinkunmi, Ademi Zanfina, Adou Arsène Kouablan, Adsuar José C, Agardh Emilie Elisabet, Agarwal Arnav, Agrawal Anurag, Kiadaliri Aliasghar Ahmad, Ajala Oluremi N, Akinyemiju Tomi F, Al-Aly Ziyad, Alam Khurshid, Alam Noore K M, Aldhahri Saleh Fahed, Aldridge Robert William, Alemu Zewdie Aderaw, Ali Raghib, Alkerwi Ala'a, Alla François, Allebeck Peter, Alsharif Ubai, Altirkawi Khalid A, Martin Elena Alvarez, Alvis-Guzman Nelson, Amare Azmeraw T, Amberbir Alemayehu, Amegah Adeladza Kofi, Amini Heresh, Ammar Walid, Amrock Stephen Marc, Andersen Hjalte H, Anderson Benjamin O, Antonio Carl Abelardo T, Anwari Palwasha, Ärnlöv Johan, Artaman Al, Asayesh Hamid, Asghar Rana Jawad, Assadi Reza, Atique Suleman, Avokpaho Euripide Frinel G Arthur, Awasthi Ashish, Quintanilla Beatriz Paulina Ayala, Azzopardi Peter, Bacha Umar, Badawi Alaa, Bahit Maria C, Balakrishnan Kalpana, Barac Aleksandra, Barber Ryan M, Barker-Collo Suzanne L, Bärnighausen Till, Barquera Simon, Barregard Lars, Barrero Lope H, Basu Sanjay, Batis Carolina, Bazargan-Hejazi Shahrzad, Beardsley Justin, Bedi Neeraj, Beghi Ettore, Bell Brent, Bell Michelle L, Bello Aminu K, Bennett Derrick A, Bensenor Isabela M, Berhane Adugnaw, Bernabé Eduardo, Betsu Balem Demtsu, Beyene Addisu Shunu, Bhala Neeraj, Bhansali Anil, Bhatt Samir, Biadgilign Sibhatu, Bikbov Boris, Bisanzio Donal, Bjertness Espen, Blore Jed D, Borschmann Rohan, Boufous Soufiane, Bourne Rupert R A, Brainin Michael, Brazinova Alexandra, Breitborde Nicholas J K, Brenner Hermann, Broday David M, Brugha Traolach S, Brunekreef Bert, Butt Zahid A, Cahill Leah E, Calabria Bianca, Campos-Nonato Ismael Ricardo, Cárdenas Rosario, Carpenter David O, Carrero Juan Jesus, Casey Daniel C, Castañeda-Orjuela Carlos A, Rivas Jacqueline Castillo, Castro Ruben Estanislao, Catalá-López Ferrán, Chang Jung-Chen, Chiang Peggy Pei-Chia, Chibalabala Mirriam, Chimed-Ochir Odgerel, Chisumpa Vesper Hichilombwe, Chitheer Abdulaal A, Choi Jee-Young Jasmine, Christensen Hanne, Christopher Devasahayam Jesudas, Ciobanu Liliana G, Coates Matthew M, Colquhoun Samantha M, Manzano Alejandra G Contreras, Cooper Leslie Trumbull, Cooperrider Kimberly, Cornaby Leslie, Cortinovis Monica, Crump John A, Cuevas-Nasu Lucia, Damasceno Albertino, Dandona Rakhi, Darby Sarah C, Dargan Paul I, das Neves José, Davis Adrian C, Davletov Kairat, de Castro E Filipa, De la Cruz-Góngora Vanessa, De Leo Diego, Degenhardt Louisa, Del Gobbo Liana C, del Pozo-Cruz Borja, Dellavalle Robert P, Deribew Amare, Jarlais Don C Des, Dharmaratne Samath D, Dhillon Preet K, Diaz-Torné Cesar, Dicker Daniel, Ding Eric L, Dorsey E Ray, Doyle Kerrie E, Driscoll Tim R, Duan Leilei, Dubey Manisha, Duncan Bruce Bartholow, Elyazar Iqbal, Endries Aman Yesuf, Ermakov Sergey Petrovich, Erskine Holly E, Eshrati Babak, Esteghamati Alireza, Fahimi Saman, Faraon Emerito Jose Aquino, Farid Talha A, Farinha Carla Sofia e Sa, Faro André, Farvid Maryam S, Farzadfar Farshad, Feigin Valery L, Fereshtehnejad Seyed-Mohammad, Fernandes Jefferson G, Fischer Florian, Fitchett Joseph R A, Fleming Tom, Foigt Nataliya, Foreman Kyle, Fowkes F Gerry R, Franklin Richard C, Fürst Thomas, Futran Neal D, Gakidou Emmanuela, Garcia-Basteiro Alberto L, Gebrehiwot Tsegaye Tewelde, Gebremedhin Amanuel Tesfay, Geleijnse Johanna M, Gessner Bradford D, Giref Ababi Zergaw, Giroud Maurice, Gishu Melkamu Dedefo, Giussani Giorgia, Goenka Shifalika, Gomez-Cabrera Mari Carmen, Gomez-Dantes Hector, Gona Philimon, Goodridge Amador, Gopalani Sameer Vali, Gotay Carolyn C, Goto Atsushi, Gouda Hebe N, Gugnani Harish Chander, Guillemin Francis, Guo Yuming, Gupta Rahul, Gupta Rajeev, Gutiérrez Reyna A, Haagsma Juanita A, Hafezi-Nejad Nima, Haile Demewoz, Hailu Gessessew Bugssa, Halasa Yara A, Hamadeh Randah Ribhi, Hamidi Samer, Handal Alexis J, Hankey Graeme J, Hao Yuantao, Harb Hilda L, Harikrishnan Sivadasanpillai, Haro Josep Maria, Hassanvand Mohammad Sadegh, Hassen Tahir Ahmed, Havmoeller Rasmus, Heredia-Pi Ileana Beatriz, Hernández-Llanes Norberto Francisco, Heydarpour Pouria, Hoek Hans W, Hoffman Howard J, Horino Masako, Horita Nobuyuki, Hosgood H Dean, Hoy Damian G, Hsairi Mohamed, Htet Aung Soe, Hu Guoqing, Huang John J, Husseini Abdullatif, Hutchings Sally J, Huybrechts Inge, Iburg Kim Moesgaard, Idrisov Bulat T, Ileanu Bogdan Vasile, Inoue Manami, Jacobs Troy A, Jacobsen Kathryn H, Jahanmehr Nader, Jakovljevic Mihajlo B, Jansen Henrica A F M, Jassal Simerjot K, Javanbakht Mehdi, Jayaraman Sudha P, Jayatilleke Achala Upendra, Jee Sun Ha, Jeemon Panniyammakal, Jha Vivekanand, Jiang Ying, Jibat Tariku, Jin Ye, Johnson Catherine O, Jonas Jost B, Kabir Zubair, Kalkonde Yogeshwar, Kamal Ritul, Kan Haidong, Karch André, Karema Corine Kakizi, Karimkhani Chante, Kasaeian Amir, Kaul Anil, Kawakami Norito, Kazi Dhruv S, Keiyoro Peter Njenga, Kemmer Laura, Kemp Andrew Haddon, Kengne Andre Pascal, Keren Andre, Kesavachandran Chandrasekharan Nair, Khader Yousef Saleh, Khan Abdur Rahman, Khan Ejaz Ahmad, Khan Gulfaraz, Khang Young-Ho, Khatibzadeh Shahab, Khera Sahil, Khoja Tawfik Ahmed Muthafer, Khubchandani Jagdish, Kieling Christian, Kim Cho-il, Kim Daniel, Kimokoti Ruth W, Kissoon Niranjan, Kivipelto Miia, Knibbs Luke D, Kokubo Yoshihiro, Kopec Jacek A, Koul Parvaiz A, Koyanagi Ai, Kravchenko Michael, Kromhout Hans, Krueger Hans, Ku Tiffany, Defo Barthelemy Kuate, Kuchenbecker Ricardo S, Bicer Burcu Kucuk, Kuipers Ernst J, Kumar G Anil, Kwan Gene F, Lal Dharmesh Kumar, Lalloo Ratilal, Lallukka Tea, Lan Qing, Larsson Anders, Latif Asma Abdul, Lawrynowicz Alicia Elena Beatriz, Leasher Janet L, Leigh James, Leung Janni, Levi Miriam, Li Xiaohong, Li Yichong, Liang Juan, Liu Shiwei, Lloyd Belinda K, Logroscino Giancarlo, Lotufo Paulo A, Lunevicius Raimundas, MacIntyre Michael, Mahdavi Mahdi, Majdan Marek, Majeed Azeem, Malekzadeh Reza, Malta Deborah Carvalho, Manamo Wondimu Ayele Ayele, Mapoma Chabila C, Marcenes Wagner, Martin Randall V, Martinez-Raga Jose, Masiye Felix, Matsushita Kunihiro, Matzopoulos Richard, Mayosi Bongani M, McGrath John J, McKee Martin, Meaney Peter A, Medina Catalina, Mehari Alem, Mejia-Rodriguez Fabiola, Mekonnen Alemayehu B, Melaku Yohannes Adama, Memish Ziad A, Mendoza Walter, Mensink Gert B M, Meretoja Atte, Meretoja Tuomo J, Mesfin Yonatan Moges, Mhimbira Francis Apolinary, Millear Anoushka, Miller Ted R, Mills Edward J, Mirarefin Mojde, Misganaw Awoke, Mock Charles N, Mohammadi Alireza, Mohammed Shafiu, Mola Glen Liddell D, Monasta Lorenzo, Hernandez Julio Cesar Montañez, Montico Marcella, Morawska Lidia, Mori Rintaro, Mozaffarian Dariush, Mueller Ulrich O, Mullany Erin, Mumford John Everett, Murthy Gudlavalleti Venkata Satyanarayana, Nachega Jean B, Naheed Aliya, Nangia Vinay, Nassiri Nariman, Newton John N, Ng Marie, Nguyen Quyen Le, Nisar Muhammad Imran, Pete Patrick Martial Nkamedjie, Norheim Ole F, Norman Rosana E, Norrving Bo, Nyakarahuka Luke, Obermeyer Carla Makhlouf, Ogbo Felix Akpojene, Oh In-Hwan, Oladimeji Olanrewaju, Olivares Pedro R, Olsen Helen, Olusanya Bolajoko Olubukunola, Olusanya Jacob Olusegun, Opio John Nelson, Oren Eyal, Orozco Ricardo, Ortiz Alberto, Ota Erika, PA Mahesh, Pana Adrian, Park Eun-Kee, Parry Charles D, Parsaeian Mahboubeh, Patel Tejas, Caicedo Angel J Paternina, Patil Snehal T, Patten Scott B, Patton George C, Pearce Neil, Pereira David M, Perico Norberto, Pesudovs Konrad, Petzold Max, Phillips Michael Robert, Piel Frédéric B, Pillay Julian David, Plass Dietrich, Polinder Suzanne, Pond Constance D, Pope C Arden, Pope Daniel, Popova Svetlana, Poulton Richie G, Pourmalek Farshad, Prasad Noela M, Qorbani Mostafa, Rabiee Rynaz H S, Radfar Amir, Rafay Anwar, Rahimi-Movaghar Vafa, Rahman Mahfuzar, Rahman Mohammad Hifz Ur, Rahman Sajjad Ur, Rai Rajesh Kumar, Rajsic Sasa, Raju Murugesan, Ram Usha, Rana Saleem M, Ranganathan Kavitha, Rao Puja, García Christian Aspacia Razo, Refaat Amany H, Rehm Colin D, Rehm Jürgen, Reinig Nikolas, Remuzzi Giuseppe, Resnikoff Serge, Ribeiro Antonio L, Rivera Juan A, Roba Hirbo Shore, Rodriguez Alina, Rodriguez-Ramirez Sonia, Rojas-Rueda David, Roman Yesenia, Ronfani Luca, Roshandel Gholamreza, Rothenbacher Dietrich, Roy Ambuj, Saleh Muhammad Muhammad, Sanabria Juan R, Sanchez-Riera Lidia, Sanchez-Niño Maria Dolores, Sánchez-Pimienta Tania G, Sandar Logan, Santomauro Damian F, Santos Itamar S, Sarmiento-Suarez Rodrigo, Sartorius Benn, Satpathy Maheswar, Savic Miloje, Sawhney Monika, Schmidhuber Josef, Schmidt Maria Inês, Schneider Ione J C, Schöttker Ben, Schutte Aletta E, Schwebel David C, Scott James G, Seedat Soraya, Sepanlou Sadaf G, Servan-Mori Edson E, Shaddick Gavin, Shaheen Amira, Shahraz Saeid, Shaikh Masood Ali, Levy Teresa Shamah, Sharma Rajesh, She Jun, Sheikhbahaei Sara, Shen Jiabin, Sheth Kevin N, Shi Peilin, Shibuya Kenji, Shigematsu Mika, Shin Min-Jeong, Shiri Rahman, Shishani Kawkab, Shiue Ivy, Shrime Mark G, Sigfusdottir Inga Dora, Silva Diego Augusto Santos, Silveira Dayane Gabriele Alves, Silverberg Jonathan I, Simard Edgar P, Sindi Shireen, Singh Abhishek, Singh Jasvinder A, Singh Prashant Kumar, Slepak Erica Leigh, Soljak Michael, Soneji Samir, Sorensen Reed J D, Sposato Luciano A, Sreeramareddy Chandrashekhar T, Stathopoulou Vasiliki, Steckling Nadine, Steel Nicholas, Stein Dan J, Stein Murray B, Stöckl Heidi, Stranges Saverio, Stroumpoulis Konstantinos, Sunguya Bruno F, Swaminathan Soumya, Sykes Bryan L, Szoeke Cassandra E I, Tabarés-Seisdedos Rafael, Takahashi Ken, Talongwa Roberto Tchio, Tandon Nikhil, Tanne David, Tavakkoli Mohammad, Taye Belaynew Wasie, Taylor Hugh R, Tedla Bemnet Amare, Tefera Worku Mekonnen, Tegegne Teketo Kassaw, Tekle Dejen Yemane, Terkawi Abdullah Sulieman, Thakur J S, Thomas Bernadette A, Thomas Matthew Lloyd, Thomson Alan J, Thorne-Lyman Andrew L, Thrift Amanda G, Thurston George D, Tillmann Taavi, Tobe-Gai Ruoyan, Tobollik Myriam, Topor-Madry Roman, Topouzis Fotis, Towbin Jeffrey Allen, Tran Bach Xuan, Dimbuene Zacharie Tsala, Tsilimparis Nikolaos, Tura Abera Kenay, Tuzcu Emin Murat, Tyrovolas Stefanos, Ukwaja Kingsley N, Undurraga Eduardo A, Uneke Chigozie Jesse, Uthman Olalekan A, van Donkelaar Aaron, van Os Jim, Varakin Yuri Y, Vasankari Tommi, Veerman J Lennert, Venketasubramanian Narayanaswamy, Violante Francesco S, Vollset Stein Emil, Wagner Gregory R, Waller Stephen G, Wang Jian Li, Wang Linhong, Wang Yanping, Weichenthal Scott, Weiderpass Elisabete, Weintraub Robert G, Werdecker Andrea, Westerman Ronny, Whiteford Harvey A, Wijeratne Tissa, Wiysonge Charles Shey, Wolfe Charles D A, Won Sungho, Woolf Anthony D, Wubshet Mamo, Xavier Denis, Xu Gelin, Yadav Ajit Kumar, Yakob Bereket, Yalew Ayalnesh Zemene, Yano Yuichiro, Yaseri Mehdi, Ye Pengpeng, Yip Paul, Yonemoto Naohiro, Yoon Seok-Jun, Younis Mustafa Z, Yu Chuanhua, Zaidi Zoubida, Zaki Maysaa El Sayed, Zhu Jun, Zipkin Ben, Zodpey Sanjay, Zuhlke Liesl Joanna, Murray Christopher J L. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet. 2016;388(10053):1659–1724. doi: 10.1016/S0140-6736(16)31679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cyranoski D. Why Chinese medicine is heading for clinics around the world. Nature. 2018 doi: 10.1038/d41586-018-06782-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen W. Clinical observation on treating 45 cases of diabetes. Clin J Chin Med. 2013;5:12–13. doi: 10.1186/1749-8546-5-12. [DOI] [Google Scholar]
- 6.Yang F, Qi J, Zhu D. Experimental study on hypoglycemic effects of zengyetang. Chin J Exp Trad Med Formul. 2010;16:98–102. [Google Scholar]
- 7.Duan W, Zhang Z, Kong Y, Chen Z, Li M. Comparison hypoglycemic effects of Zengye Decoction with different processed product of rehmanniae in diabetes rats. China J Trad Chin Med Pharm. 2014;29:266–268. [Google Scholar]
- 8.Zhao M, Chen Y, Wang C, Xiao W, Chen S, Zhang S, et al. Systems pharmacology dissection of multi-scale mechanisms of action of Huo-Xiang-Zheng-Qi formula for the treatment of gastrointestinal diseases. Front Pharmacol. 2019 doi: 10.3389/fphar.2018.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Kuang W, Xu X, Li D, Zhu W, Lan Z, et al. Putative identification of components in Zengye Decoction and their effects on glucose consumption and lipogenesis in insulin-induced insulin-resistant. J Chromatogr. 2018;1073:145–153. doi: 10.1016/j.jchromb.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Tian YS, Du ZY, Xiao Y, Yu B, Qi J. Screening and identification of potential hypoglycemic components in Zeng Ye Tang by high-performance liquid chromatography coupled with tandem quadrupole time-of-flight mass spectrometry. J Sep Sci. 2017;40:4709–4717. doi: 10.1002/jssc.201700507. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Liu Z, Peng Y, Zhang L, Ju P, Bi K, et al. Validated LC-MS method for simultaneous quantitation of catalpol and harpagide in rat plasma: application to a comparative pharmacokinetic study in normal and diabetic rats after oral administration of Zeng-Ye-Decoction. Biomed Chromatogr. 2013;27:1503–1510. doi: 10.1002/bmc.2949. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Chang S, Tian Y, Zhang G, Qi J. Zengye Decoction ameliorates insulin resistance by promoting glucose uptake. Rejuvenation Res. 2020 doi: 10.1089/rej.2019.2228. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Li J, Sun Z, Wu C, Ma J, Wang J, et al. Comparative pharmacokinetics of six coumarins in normal and breast cancer bone-metastatic mice after oral administration of Wenshen Zhuanggu Formula. J Ethnopharmacol. 2018;224:36–44. doi: 10.1016/j.jep.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 14.Deng Y, Shi Q, Chen B, Zhang X, Liu S, Qiu X. Comparative pharmacokinetics of baicalin in normal and the type 2 diabetic rats after oral administration of the Radix scutellariae extract. Fitoterapia. 2012;83:1435–1442. doi: 10.1016/J.FITOTE.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Cheng Y, Liu Z, Ding L, Qiu T, Chai L, et al. Systematic screening and characterization of multiple constituents in Guizhi Fuling capsule and metabolic profiling of bioactive components in rats using ultra-high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. J Chromatogr B. 2017;1061–1062:474–486. doi: 10.1016/j.jchromb.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Lv X, Li J, Xu Z, Chen L. The characterization of high-fat diet and multiple low-dose streptozotocin induced type 2 diabetes rat model. Exp Diab Res. 2008 doi: 10.1155/2008/704045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia Y, Xu B, Xu J. Effects of type 2 diabetes mellitus on the pharmacokinetics of berberine in rats. Pharm Biol. 2016;55:510–515. doi: 10.1080/13880209.2016.1255649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Zhu P, Liu B, Wei L, Xu Y. Simultaneous determination of fourteen compounds of Hedyotis diffusa Willd extract in rats by UHPLC–MS/MS method: application to pharmacokinetics and tissue distribution study. J Pharm Biomed Anal. 2018;159:490–512. doi: 10.1016/J.JPBA.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Yuan Q, Liu E, Qi L, Bi Z, Li P. Fragmentation study of iridoid glycosides and phenylpropanoid glycosides in Radix Scrophulariae by rapid resolution liquid chromatography with diode-array detection and electrospray ionization time-of-flight mass spectrometry. Biomed Chromatogr. 2010;24:808–819. doi: 10.1002/bmc.1368. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Liang D, Luo H, Hao Z, Wang Y, Zhang C, et al. Chemical constituents from root tubers of Rehmannia glutinosa. Chin Trad Herbal Drugs. 2014;45:16–22. doi: 10.7501/j.issn.0253-2670.2014.01.003. [DOI] [Google Scholar]
- 21.Gong P, Tian Y, Guo Y, Gu L, Li J, Qi J, et al. Comparisons of antithrombosis, hematopoietic effects and chemical profiles of dried and rice wine-processed Rehmanniae Radix extracts. J Ethnopharmacol. 2019;231:394–402. doi: 10.1016/J.JEP.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Xu F, Liu Z, Ma L, Shang M, Liu G, et al. Identification of chemical constituents in Scrophulariae Radix by HPLC-IT-TOF-MS. Chin J Nat Med. 2016;41:1257–1268. doi: 10.4268/cjcmm20160717. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki H, Nishimura H, Morota T, Katsuhara T, Chin M, Mitsuhashi H. Norcarotenoid glycosides of Rehmannia glutinosa var Purpurea. Phytochemistry. 1991;30:1639–1644. doi: 10.1016/0031-9422(91)84224-g. [DOI] [Google Scholar]
- 24.Hiroko K, Hiromi K, Nobuo T, et al. Studies on the constituents of Cistanchis Herba. Yakugaku Zasshi. 1986;106:562–566. doi: 10.1248/yakushi1947.106.7_562. [DOI] [Google Scholar]
- 25.Lanzetta R, Laonigro G, Mangoni L. Three 3-Benzyl-Cchromanones from muscari comosum. Phytochemistry. 1985;24:624–626. doi: 10.1016/s0031-9422(00)80790-6. [DOI] [Google Scholar]
- 26.Li S, Lin H, Qu C, Tang Y, Shen J, Li W, et al. Urine and plasma metabonomics coupled with UHPLC-QTOF/MS and multivariate data analysis on potential biomarkers in anemia and hematinic effects of herb pair Gui-Hong. J Ethnopharmacol. 2015;170:175–183. doi: 10.1016/J.JEP.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Yan G, Huang Y, Wang X. Identification technique for in vivo ingredients of traditional Chinese medicines based on LC-MS analysis. China J Chin Materia Med. 2012 doi: 10.4268/cjcmm20121216. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Yang L, He S, Qin X, Li A. Metabolomics coupled with integrative pharmacology reveal the protective effect of FangjiHuangqi Decoction against adriamycin-induced rat nephropathy model. J Pharm Biomed Anal. 2019;174:525–533. doi: 10.1016/J.JPBA.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Lai W. Research on material basis of traditional chinese medicine compound based on correlation analysis of metabolism and efficacy. Chin J Exp Trad Med Formul. 2011;17:10–12. [Google Scholar]
- 30.Li P, Qi L, Wen X, Sheng L. Methods for the elucidation of bioactive components and quality control of traditional chinese medicines. Chin J Nat Med. 2007;5:66–77. [Google Scholar]
- 31.Zhang L, Lu L, Jin S, Jing X, Yao D, Hu N, et al. Tissue-specific alterations in expression and function of P-glycoprotein in streptozotocin-induced diabetic rats. Acta Pharmacol Sin. 2011;32:956–966. doi: 10.1038/aps.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chula S, Hang L, Yinying B, Jianning S, Shi R. The effects of notoginsenoside R1 on the intestinal absorption of geniposide by the everted rat gut sac model. J Ethnopharmacol. 2012;142:136–143. doi: 10.1016/J.JEP.2012.04.027. [DOI] [PubMed] [Google Scholar]
- 33.Shimojo N. Cytochrome P450 changes in rats with streptozocin-induced diabetes. Int J Biochem. 1994;26:1261–1268. doi: 10.1016/0020-711x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 34.Shimojo N, Ishizaki T, Imaoka S, Funae Y, Fujii S, Okuda K. Changes in amounts of cytochrome P450 isozymes and levels of catalytic activities in hepatic and renal microsomes of rats with streptozocin-induced diabetes. Biochem Pharmacol. 1993;46:621–627. doi: 10.1016/0006-2952(93)90547-a. [DOI] [PubMed] [Google Scholar]
- 35.Taheri A, Lavasani H, Kasirzadeh S, Sheikholeslami B, Ardakani YH, Rouini M-R. Changes in CYP2D enzyme activity following induction of type 2 diabetes, and administration of cinnamon and metformin: an experimental animal study. Xenobiotica. 2018;48:984–989. doi: 10.1080/00498254.2017.1390626. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Cao X, Li T, Li X. Effects of catalpol on the activity of human liver cytochrome P450 enzymes. Xenobiotica. 2019;49:1289–1295. doi: 10.1080/00498254.2018.1558309. [DOI] [PubMed] [Google Scholar]
- 37.Kim TK, Park KS. Inhibitory effects of harpagoside on TNF-α-induced pro-inflammatory adipokine expression through PPAR-γ activation in 3T3-L1 adipocytes. Cytokine. 2015 doi: 10.1016/j.cyto.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Fajas L, Auboeuf D, Raspé E, Schoonjans K, Lefebvre AM, Saladin R, et al. The organization, promoter analysis, and expression of the human PPARγ gene. J Biol Chem. 1997 doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 39.Yoon SA, Kang S, Shin HS, Kang SW, Kim JH, Ko HC, et al. P-Coumaric acid modulates glucose and lipid metabolism via AMP-activated protein kinase in L6 skeletal muscle cells. Biochem Biophys Res Commun. 2013 doi: 10.1016/j.bbrc.2013.02.067. [DOI] [PubMed] [Google Scholar]
- 40.Amalan V, Vijayakumar N, Indumathi D, Ramakrishnan A. Antidiabetic and antihyperlipidemic activity of p-coumaric acid in diabetic rats, role of pancreatic GLUT 2: in vivo approach. Biomed Pharmacother. 2016 doi: 10.1016/j.biopha.2016.09.039. [DOI] [PubMed] [Google Scholar]
- 41.Kang BB, Chiang BH. Amelioration of insulin resistance using the additive effect of ferulic acid and resveratrol on vesicle trafficking for skeletal muscle glucose metabolism. Phytother Res. 2019 doi: 10.1002/ptr.6561. [DOI] [PubMed] [Google Scholar]
- 42.Nankar R, Prabhakar PK, Doble M. Hybrid drug combination: combination of ferulic acid and metformin as anti-diabetic therapy. Phytomedicine. 2017 doi: 10.1016/j.phymed.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 43.Prabhakar PK, Doble M. Synergistic effect of phytochemicals in combination with hypoglycemic drugs on glucose uptake in myotubes. Phytomedicine. 2009 doi: 10.1016/j.phymed.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Son MJ, Rico CW, Nam SH, Kang MY. Effect of oryzanol and ferulic acid on the glucose metabolism of mice fed with a high-fat diet. J Food Sci. 2011 doi: 10.1111/j.1750-3841.2010.01907.x. [DOI] [PubMed] [Google Scholar]
- 45.Azay-Milhau J, Ferrare K, Leroy J, Aubaterre J, Tournier M, Lajoix AD, et al. Antihyperglycemic effect of a natural chicoric acid extract of chicory (Cichorium intybus L.): a comparative in vitro study with the effects of caffeic and ferulic acids. J Ethnopharmacol. 2013 doi: 10.1016/j.jep.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 46.Chen L, Teng H, Cao H. Chlorogenic acid and caffeic acid from Sonchus oleraceus Linn synergistically attenuate insulin resistance and modulate glucose uptake in HepG2 cells. Food Chem Toxicol. 2019 doi: 10.1016/j.fct.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 47.Alicic RZ, Neumiller JJ, Johnson EJ, Dieter B, Tuttle KR. Sodium–glucose cotransporter 2 inhibition and diabetic kidney disease. Diabetes. 2019;68:248–257. doi: 10.2337/dbi18-0007. [DOI] [PubMed] [Google Scholar]
- 48.Un JJ, Lee MK, Yong BP, Jeon SM, Choi MS. Antihyperglycemic and antioxidant properties of caffeic acid in db/db mice. J Pharmacol Exp Ther. 2006 doi: 10.1124/jpet.106.105163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Compounds information of ZYD by HPLC-ESI-Q-TOF MS/MS.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.